Abstract
The mature HIV-1 nucleocapsid protein NCp7 (NC) plays a key role in reverse transcription facilitating the two obligatory strand transfers. Several properties contribute to its efficient chaperon activity: preferential binding to single-stranded regions, nucleic acid aggregation, helix destabilization, and rapid dissociation from nucleic acids. However, little is known about the relationships between these different properties, which are complicated by the ability of the protein to recognize particular HIV-1 stem–loops, such as SL1, SL2, and SL3, with high affinity and without destabilizing them. These latter properties are important in the context of genome packaging, during which NC is part of the Gag precursor. We used NMR to investigate destabilization of the full-length TAR (trans activating response element) RNA by NC, which is involved in the first strand transfer step of reverse transcription. NC was used at a low protein:nucleotide (nt) ratio of 1:59 in these experiments. NMR data for the imino protons of TAR identified most of the base pairs destabilized by NC. These base pairs were adjacent to the loops in the upper part of the TAR hairpin rather than randomly distributed. Gel retardation assays showed that conversion from the initial TAR–cTAR complex to the fully annealed form occurred much more slowly at the 1:59 ratio than at the higher ratios classically used. Nevertheless, NC significantly accelerated the formation of the initial complex at a ratio of 1:59.
Keywords: HIV-1 nucleocapsid protein, nucleic acid dynamics, imino protons, NMR spectroscopy, kinetic model
INTRODUCTION
The mature HIV-1 nucleocapsid protein NCp7 (NC), which is generated by proteolytic cleavage of the Gag precursor, is a small retroviral protein consisting of a poorly folded highly basic N-terminal region and two highly conserved CCHC zinc finger motifs connected by a basic linker displaying some flexibility. Several forms of this protein are observed during the course of the replication cycle. NC exists as a domain of the Gag precursor protein and the partially matured products NCp15 and NCp9, which are generated by the retroviral protease (Mirambeau et al. 2010). NC is involved in many steps in the HIV-1 replication cycle, including reverse transcription, selective genome packaging, genome dimerization, genome protection, and integration (Levin et al. 2005, 2010; Darlix et al. 2007, 2011, 2014; Sleiman et al. 2012). During reverse transcription, the nucleic acid chaperone activity of NC facilitates the rearrangement of nucleic acids into their most thermodynamically stable structures. This activity results from three different properties of NC: (i) nucleic acid aggregation; (ii) duplex destabilization; (iii) rapid on-off kinetics (Cruceanu et al. 2006a,b; Stewart-Maynard et al. 2008; Godet and Mély 2010; Levin et al. 2010). Nucleic acid aggregation is thought to be dependent on the N-terminal basic part of NC, whereas duplex destabilization is associated with the zinc fingers (Beltz et al. 2005; Levin et al. 2005, 2010; Vo et al. 2009a, b; Godet and Mély 2010). However, some studies have suggested a role of zinc fingers in nucleic acid aggregation (Krishnamoorthy et al. 2003; Levin et al. 2005; Mirambeau et al. 2006; Mitra et al. 2013) and others have implicated basic residues in nucleic acid destabilization (Wu et al. 2014).
The first-strand transfer occurring during reverse transcription of the HIV-1 genome has been widely studied (Basu et al. 2008; Godet and Mély 2010; Levin et al. 2010). After the initiation of reverse transcription at the PBS (primer binding site) with tRNAlys (Levin et al. 2010; Sleiman et al. 2012, 2013), the minus-strand strong-stop DNA [(−)ssDNA] is synthesized from the 5′ terminal part of the RNA genome. Continuation of the reverse transcription process requires the translocation of (−)ssDNA from the 5′-end to the 3′-end of the RNA genome. This translocation requires the presence of the R (Repeat) sequence at both ends of the genome. The R sequence consists of the TAR (trans activating response element) and poly(A) elements, both of which can self-fold into stem–loop structures in their RNA and DNA forms (Berkhout et al. 2001). The upper part of the TAR stem–loop is required for efficient transcription of the integrated HIV-1 genome. The lower part of the TAR stem–loop also plays an important role in HIV-1 replication (Klaver and Berkhout 1994). NC promotes the annealing of the TAR RNA hairpin with the cTAR DNA hairpin. This is an essential step in the first strand transfer process and has thus been the subject of many studies (Godet et al. 2006; Liu et al. 2007; Vo et al. 2009a,b; Godet and Mély 2010). The NC-mediated destabilization of the secondary structures of TAR and cTAR stem–loops is one of the initial steps in the annealing process. NC destabilizes the cTAR DNA to a much greater extent than the more stable TAR RNA (Bernacchi et al. 2002). More generally, the destabilizing activity of NC has been shown to be inversely correlated with oligonucleotide stability (Bernacchi et al. 2002; Beltz et al. 2003, 2005). It has been shown, by time-resolved fluorescence spectroscopy, that there are preferentially destabilized segments in the lower part of the cTAR DNA molecule (Godet et al. 2013). The TAR RNA hairpin appears to be only weakly destabilized by NC, but precise data are lacking. A precise investigation of the destabilization process is thus required to improve our understanding of the destabilizing activity of NC and the initial steps of the NC-promoted annealing of cTAR and TAR.
The ability of NC to destabilize secondary structures in nucleic acids is a key component of its chaperone activity. It has been shown that this protein significantly accelerates the annealing of complementary sequences only if these sequences display a significant level of secondary structures (You and McHenry 1994; Golinelli and Hughes 2003; Heilman-Miller et al. 2004). However, despite some recent advances in the field (Bourbigot et al. 2008), the details of the destabilization mechanism and its structural and/or dynamic basis remain unknown. Globally, NC appears to be a weak duplex destabilizer able to destabilize only segments of <8 or 11 base pairs (bp) in length, depending on the study considered (Beltz et al. 2004; Levin et al. 2005). NC thus destabilizes low-stability stem–loop structures, such as the cTAR hairpin. Numerous bulges and internal loops in the TAR and cTAR hairpins are required to lower the stability of the neighboring double-stranded segments and permit their destabilization by NC (Beltz et al. 2004). Phylogenetic comparisons of TAR RNA structures in different HIV-1 isolates have demonstrated a conservation of bulges and loops (Berkhout 1992; Zarudyana et al. 2014). It appears likely that the stabilities of the TAR and cTAR stem–loops coevolved with the destabilizing activity of NC (Beltz et al. 2004).
The relationship between nucleic acid binding affinity and nucleic acid secondary structure destabilization is complex. One previous study suggested that the strong NC-interaction domains do not correspond to the domains of the HIV-1 genome destabilized by NC (Wilkinson et al. 2008). Consistent with this observation, structural studies have shown that NC binding to the SL2 and SL3 loops does not significantly destabilize the adjacent stems (De Guzman et al. 1998; Amarasinghe et al. 2000; Johnson et al. 2000). The high affinity of binding may prevent the rapid on–off binding kinetics of NC required for its chaperone activity (Cruceanu et al. 2006a; Levin et al. 2010). Consistent with this hypothesis, Gag and NCp15 proteins have higher affinities than NC for nucleic acids but their chaperone activity is weaker (Cruceanu et al. 2006b; Wu et al. 2010; Wang et al. 2014).
In this context, we used NMR spectroscopy to investigate the NC-induced destabilization of the full-length TAR RNA stem–loop (59 nt) (Fig. 1A) to improve our understanding of the mode of action of NC in first-strand transfer. The TAR RNA has been the subject of many NMR studies, but these previous investigations focused on the upper part of the hairpin and the dynamic aspects (Puglisi et al. 1992; Aboul-ela et al. 1995, 1996; Al-Hashimi et al. 2002; Pitt et al. 2004; Musselman et al. 2007; Dethoff et al. 2008; Bailor et al. 2010; Dethoff and Al-Hashimi 2011). We analyzed the NC-mediated destabilization process by monitoring imino proton resonance shifts and line broadening after the addition of NC. We followed the destabilization events as a function of time using very fast two-dimensional NMR methods (Schanda and Brutscher 2005; Schanda et al. 2005; Farjon et al. 2009).
FIGURE 1.
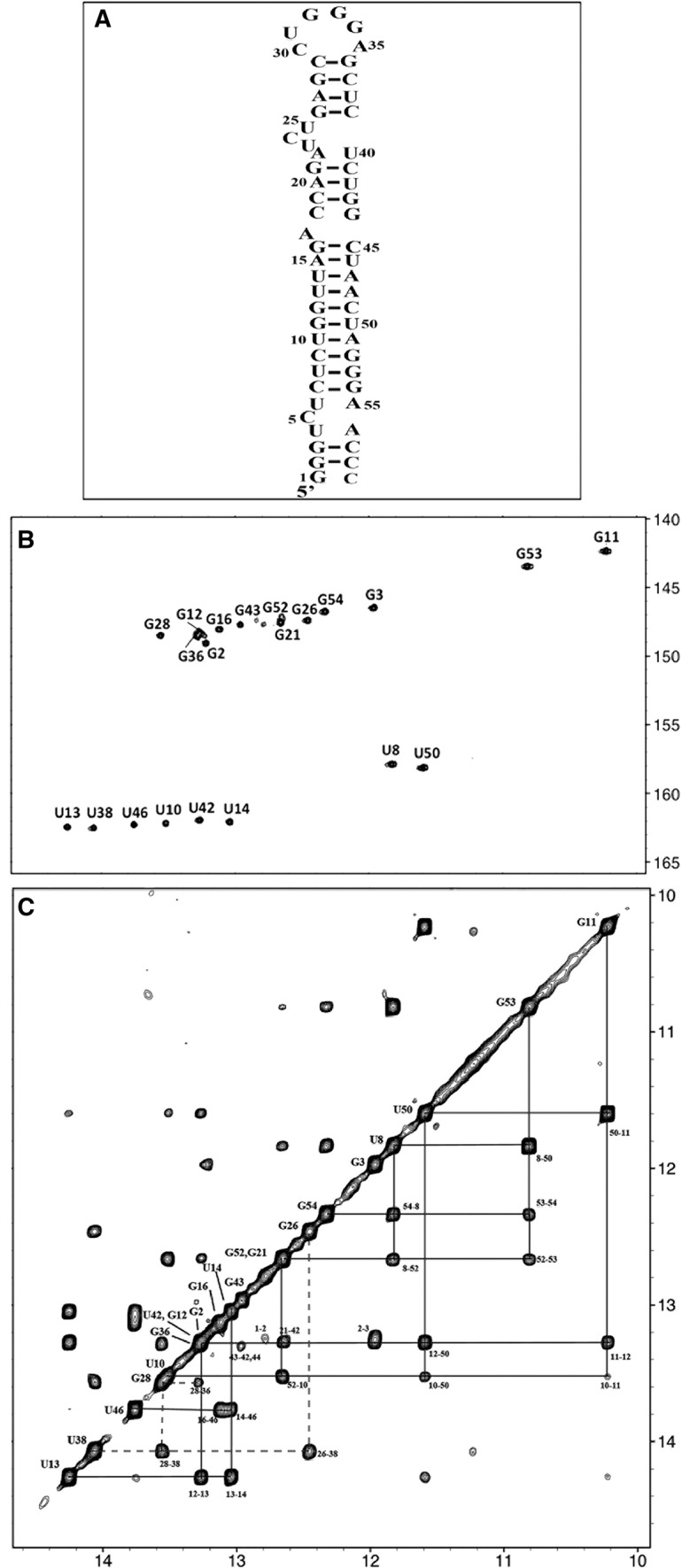
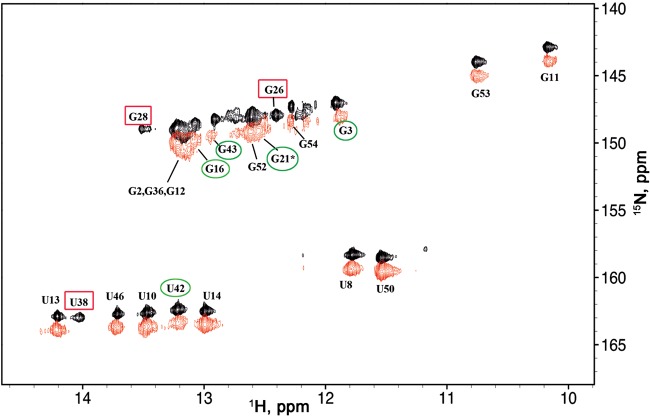
Schematic diagram of TAR with the detected imino protons indicated, together with the associated NMR spectra. (A) Diagram of the TAR molecule with black marks indicating the base pairs for which imino protons were successfully detected and assigned in NMR experiments. The observation of G44 was ambiguous, and therefore no black mark was assigned to this base pair. (B) 1H–15N TROSY spectrum of the imino groups of labeled 15N,13C-labeled TAR at 10°C and 950 MHz. The peaks are labeled with the name of the residue bearing the imino proton. (C) The imino group part of the 2D NOESY spectrum of 15N,13C-labeled TAR at 10°C and 950 MHz. Imino protons are labeled with the name of the residue bearing this proton. The NOE connectivities involving the different parts of TAR are indicated with lines of different types: solid lines for NOE connectivities in the long stem (base pairs C7–G54 to G16–C45); dotted lines for the upper stem (base pairs G26–C39 to C29–G36). The connectivities of the intermediate stem (base pairs C18–G44 to G21–C39) are not indicated by lines, because some ambiguity remains for the detection of G44 (with U42) near 13.25 ppm (spectral overlap). A corresponding cross-peak labeled “43–42,44” indicates the possible cross-peak within this stem. Note that the distance corresponding to the separation of G23 and U42 imino protons would be expected to exceed 5 Å in a standard A-RNA helix.
RESULTS
NMR analysis of the TAR RNA (59 nt)
The imino region of the 1H-15N TROSY spectrum of the TAR RNA obtained at 10°C is shown in Figure 1B. Imino protons were identified with a combination of 1H–15N TROSY and NOESY spectra (Fig. 1B,C). Twenty-two imino protons were assigned, whereas 26 signals were expected on the basis of secondary structure predictions (taking into account the expected two signals for each G:U base pair) (Fig. 1A). No NMR signals were observed for the G1–C59, U4–A56, U6–A55, and A22–U40 imino groups corresponding to base pairs that are located at the ends of stems. Some weaker and broader additional resonances were observed close to 12.7–12.8 ppm in the proton dimension, but could not be assigned due to the absence or weakness of associated correlations on 2D NOESY (Fig. 1). With the exception of the terminal base pair G1–C59, most of the missing imino resonances corresponded to A–U base pairs located near a bulge (Fig. 1A; the status of the C18–G44 imino proton is difficult to assess due to overlap). Thus, the imino protons were not observed for any of the A–U base pairs adjacent to a bulge, i.e., U4–A56, U6–A55, and A22–U40 (Fig. 1B,C).
TAR-cTAR annealing at a low NC:nt ratio
We then added NC to the TAR RNA at an NC:RNA ratio of 1:1 (1 NC:59 nucleotides [nt]). We chose to use this low ratio to prevent aggregation effects and excessive broadening of resonances leading to the loss of large amounts of information. It was important to check that the TAR–cTAR annealing was accelerated by NC at this low NC:nt ratio. These conditions are very different from those observed in vivo (Darlix et al. 2011), but they make it possible to investigate the role of NC chaperone activity in the various steps of the annealing process. Gel retardation assays displayed a striking pattern of biexponential behavior, with fast and slow components (Fig. 2). We analyzed the results obtained with a kinetic model initially developed for a similar experimental system. This model (described in the “Materials and Methods” section) involves the rapid formation of a transient duplex that is gradually converted to the fully annealed TAR–cTAR duplex (Vo et al. 2006, 2009b). The first step of the reaction was analyzed in the framework of a bimolecular and reversible reaction, because it was not possible to use pseudo-first order simplification here (see “Materials and Methods” section). Indeed, the determination of rate constants at a low NC:nt ratio requires the use of high DNA and RNA concentrations. The rate constants are reported in Table 1. We can compare the values obtained at the 1:14 ratio common to our study and that of Vo and colleagues (Fig. 9 in Vo et al. [2009b]), the values for k1, k−1, and k2 were very similar (k1 = 4.4 × 103 versus 2.5 × 103 M−1 sec−1; k−1 = 4 × 10−3 versus 5 × 10−3 sec−1 and k2 = 3.0 × 10−4 versus 5 × 10−4 sec−1).
FIGURE 2.
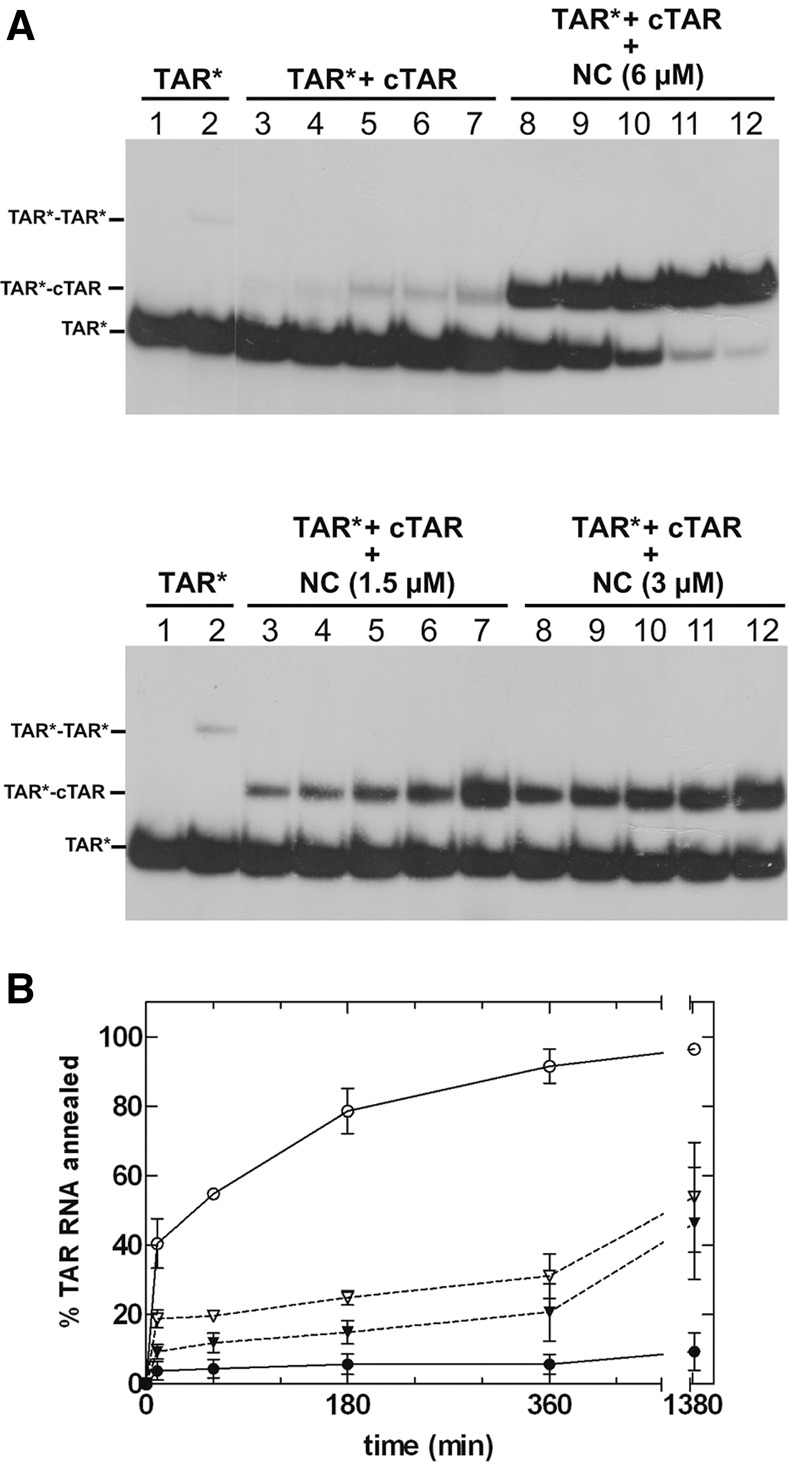
(A) Time course of TAR RNA annealing with cTAR DNA in the presence or absence of various concentrations of NC. Lane 1, heat-denatured TAR 32P-RNA. TAR 32P-RNA alone (lanes 2) or mixed with cTAR DNA (lanes 3–12) was incubated at 20°C for 10 min (lanes 2,3,8), 1 h (lanes 4,9), 3 h (lanes 5,10), 6 h (lanes 6,11), or 24 h (lanes 7,12). The monomeric form of TAR is indicated by TAR*. The TAR RNA-cTAR DNA duplex is indicated by TAR*–cTAR. In the absence of cTAR and NC (lanes 2), the band (TAR*–TAR*) above the monomer probably corresponds to the dimeric form of TAR described elsewhere (Andersen et al. 2004). (B) The graph was derived from the experiments shown in A. Filled circles, in the absence of NC; filled triangle, NC (1.5 µM); open triangles, NC (3 µM); open circles, NC (6 µM).
TABLE 1.
Kinetic parameters for the annealing of TAR-cTAR hairpins in the presence and absence of NC at various protein:nucleotide ratios
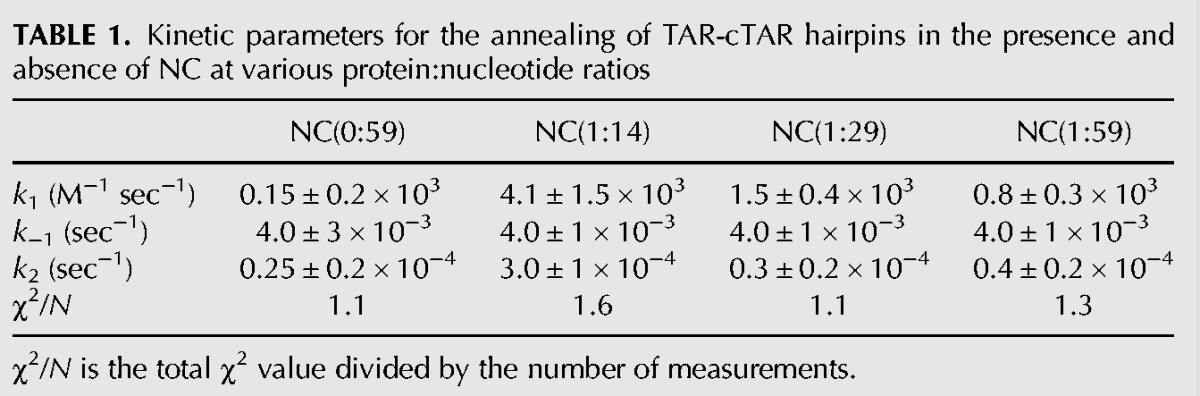
When the NC:nt ratio decreases from 1:14 to 1:29, 1:59, and 0:59 (no NC), k1 decreases by factors of 2, 5, and 27, respectively, whereas the k−1 remains constant. Moreover, the k2 elementary constant decreases by more than an order of magnitude from 1:14 to 0:29. Our data thus indicate that the transition from the initial RD* duplex (R and D stand for RNA and DNA, respectively, and RD* is the intermediate RNA:DNA complex, see “Materials and Methods” section for more information) to the extended duplex is extremely slow at NC:nt ratios below 1:14. Interestingly, the decrease in k1 value as the ratio decreases from 1:59 to 0:59 was significant (factor of 5). Thus, even at this low ratio (1:59), NC significantly increases the rate of formation of the initial RD* duplex. Therefore, from the results of this kinetic analysis, we would expect an NMR study of the interaction of NC with TAR at a ratio of 1:59 to identify the TAR destabilization events important for formation of the initial TAR-cTAR (RD*) duplex.
NMR analysis of NC-induced TAR secondary structure destabilization
The addition of NC to TAR RNA at a 1:1 ratio (NC:nt ratio of 1:59) induced considerable line broadening for all the imino proton resonances (Fig. 3). However, some resonances were affected to a larger degree than others and some even completely disappeared. Assignments of resonances were mostly deduced from analysis of 2D 1H-15N TROSY and SOFAST-HMQC experiments (Figs. 3, 4). We classified the imino proton resonances into three categories: (i) those broadened to such an extent that they were not observable (G26, G28, and U38) (Figs. 3, 4); (ii) those broadened but still observable (G16, G21, U42, and G43), although there was some ambiguity in the assignment for G21 and G52, which partly overlap, and (iii) those displaying little broadening (G3, U10, G11, U13, U14, U46, G52, G53, and G54) (Figs. 3, 4). The imino protons not mentioned above were difficult to assess due to overlapping. In addition, the resonances of the second group (G21, U42, and G43) displayed slight chemical shift variations, whereas no such variations were observed for the resonances of the third group (except G3).
FIGURE 3.
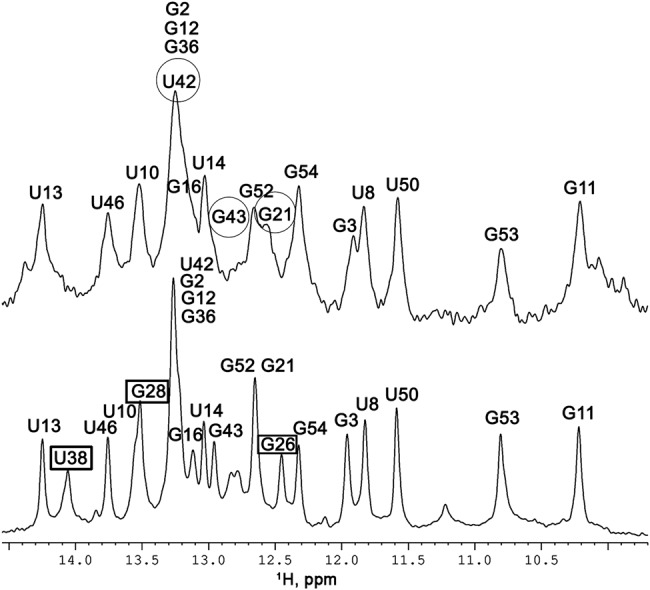
Imino proton region of 15N labeled TAR at 10°C. (Bottom) Free TAR, (top) TAR with NC in a 1:1 ratio (1:59 NC:nt). The resonances that disappear totally are indicated by rectangles in spectrum at the bottom; the resonances showing significant broadening and/or chemical shifts are indicated by circles in the top spectrum.
FIGURE 4.
2D 1H–15N SOFAST-HMQC spectra at 10°C and 950 MHz of free TAR (black) and TAR-NC in a 1:1 ratio (1:59 NC:nt) (red) at 10°C and 950 MHz. The imino groups are labeled with the name of the residue. The resonances that disappear totally are indicated by red rectangles and those that display significant broadening and/or chemical shifts are indicated by green circles. The two spectra have been shifted to facilitate visualization of the sets of resonances.
The three different classes of imino protons described above are located in different parts of the molecule (Fig. 5A): the most affected resonances (first group) involved the 4-bp stem G26–C39 to C29–G36 located between the apical loop and the three-pyrimidine bulge (Fig. 5A). Three imino protons in this stem were not observable in the presence of NC, and the signal of the fourth guanine (G36) may be overlapped so it was therefore difficult to take it into account. The imino protons of the second group belong to the short stem located in the middle of the hairpin (base pairs C18–G44 to G21–C41). Those of the third group belong to the large central stem extending from base pairs C7–G54 to G16–C45 and to a 2 bp stem at the extreme end of the molecule. Nine imino protons (of 13 in total) from the long central stem were still clearly observable. They displayed a weak overlap and no shift relative to the other resonances of the TAR molecule.
FIGURE 5.
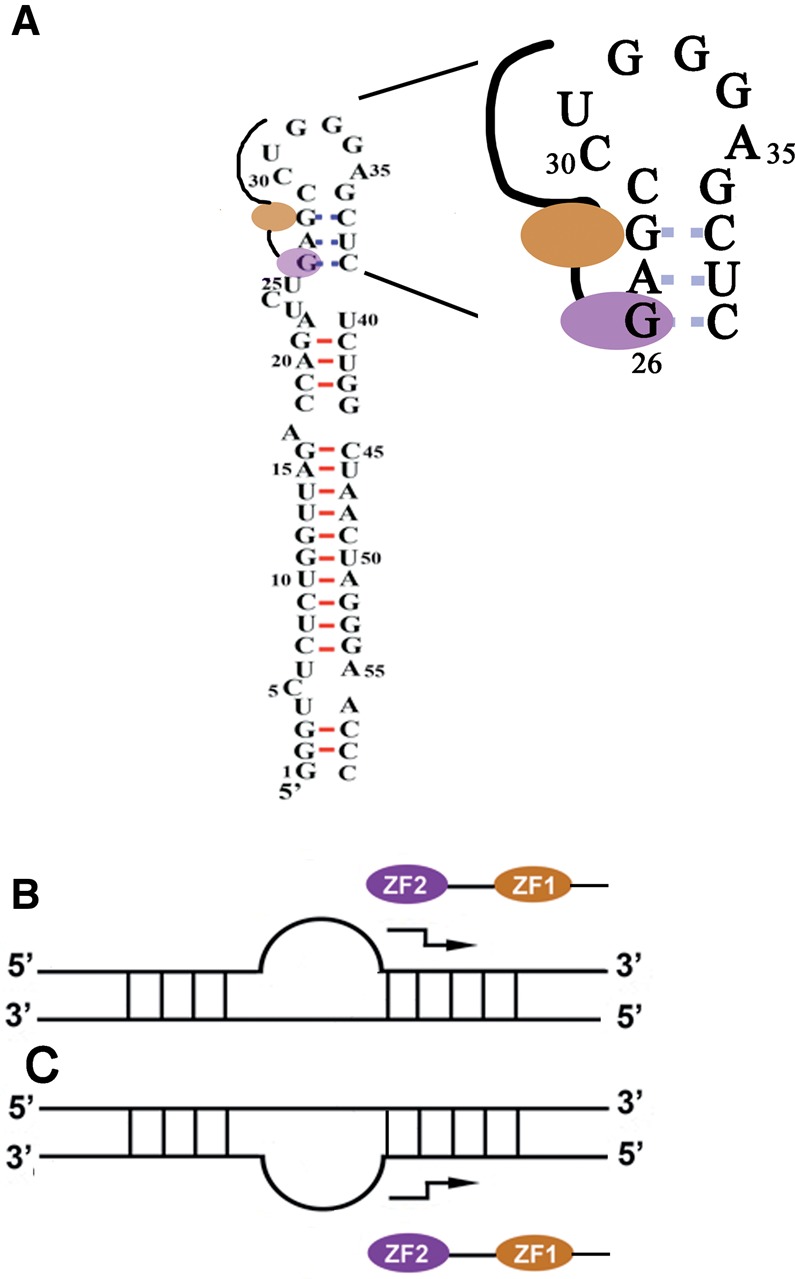
Hypothetical diagrams of RNA-DNA binding and destabilization by NC. (A) View of the TAR hairpin with a report of the observed imino protons (red) observed in the NC–TAR complex and those observable in the free RNA but not in the NC–RNA complex (dashed blue lines). NC is depicted with its two zinc fingers (ZF1: orange ellipse; ZF2: violet ellipse), the linker between the two zinc fingers and the N-terminal basic region are indicated by solid lines. We show also an expanded view of the destabilized region highlighting the hypothetical binding site at the G26 base for the ZF2 domain, whereas the ZF1 domain is more involved in the destabilization process. This image is consistent with both the data presented here and the work of McCauley et al. (2015). (B) Diagram of the destabilization of RNA secondary structure mediated by NC. The example shows a double-stranded RNA interrupted by a bulge. The binding polarity of the zinc fingers along the 5′-3′ nucleic acid chain is as observed in the three solved structures of RNA–NC complexes, with the N- to C-terminal polarity of the peptide chain antiparallel to the 5′-3′ direction of the nucleic acid chain (Bazzi et al. 2011). The NC is thought to bind preferentially to the strand containing single-stranded regions, and a staggered arrow indicates the preferentially destabilized region. This orientation of the destabilization process results from the ordering of the two domains of NC along the nucleic acid chain, with ZF1 responsible for stem destabilization (see “Discussion” section). (C) Diagram of the destabilization of DNA secondary structure mediated by NC. This example is similar to that shown in B, but with an opposite binding polarity deduced from that of known DNA–NC complexes (Bazzi et al. 2011). The position of the bulge is reversed relative to B as it is the case for the cTAR hairpin. The mechanisms involved in RNA and DNA destabilization converge to destabilize the same region of the two molecules, to the right of the bulge.
The specific broadening observed for particular imino protons is consistent with a preferential destabilization of these base pairs, corresponding to an initial phase of TAR destabilization by NC. We therefore suggest that NC preferentially destabilizes the apical stem of the TAR hairpin, having a much lesser effect on the long stem in the center of this molecule. The short stem at the 5′/3′ end was only moderately affected by NC, because the G3 imino proton remained clearly observable. Overall, the NMR data indicated a selective destabilization of the apical stem adjacent to the UCU bulge.
DISCUSSION
We provide here the first description of NMR studies on the full-length TAR hairpin in the absence and presence of NC in a 1:1 ratio. TAR is probably the RNA molecule most widely studied by NMR, but previous studies focused exclusively on the upper part of the TAR hairpin. Moreover, most previous studies on the upper part of TAR focused on the relative motion of the two upper helices. These helices, separated by the UCU bulge, which acts as a hinge, display independent and collective motions at the nanosecond timescale (Zhang et al. 2006, 2007; Salmon et al. 2013). The bottom part of the molecule has also been shown to have an important biological role (Klaver and Berkhout 1994). However, destabilization of the full-length TAR stem–loop by NC has never been investigated by NMR spectroscopy.
We assigned the imino protons of the TAR RNA and, therefore, determined base-pairing status for the whole molecule. The assignments of the imino protons of the upper part of the TAR hairpin were carefully compared with those obtained in previous studies dealing with this portion of the molecule. The assignments were similar, except for the G43 residue, which has a different sequence context due to the truncation of the TAR sequence in the previous studies (Hennig and Williamson 2000; Dethoff et al. 2008). The assigned resonances and the inter-base pair NOE networks were consistent with the self-folding of TAR proposed in Figure 1A. Interestingly, the imino protons of 3 bp were not observable. All these base pairs were U.A base pairs adjacent to a bulge (in Fig. 1A, no imino protons are indicated for C18–G44 due to overlap, resulting in ambiguity, and G1-C59 in the terminal base pair). The A22-U40 imino proton which was not observed here, was also unobservable in previous studies dealing with the upper half of the TAR hairpin (Puglisi et al. 1992; Aboul-ela et al. 1996; Zhang et al. 2006).
Overall, our experimental data are consistent with the structure predicted by mfold (Zuker 2003), but the imino protons of several A–U base pairs adjacent to a bulge were not observed, revealing significant fraying at these locations (Fig. 1A). Two of these A–U base pairs are located in the U4-U6 region, resulting in an unpaired U4C5U6 motif similar to the U23C24U25 bulge. These findings are consistent with chemical probing data showing some reactivity for the residues of the U4C5U6 motif, but less than for the U23C24U25 bulge and the apical loop (Kanevsky et al. 2005). Significant motions, like those already described for the U23C24U25 bulge (Zhang et al. 2006, 2007) probably occur for the U4C5U6 motif, increasing thus the potential complexity and flexibility of the TAR hairpin.
NC has nucleic acid destabilizing activity associated with chaperone properties. It is thought to exert this activity by binding preferentially to single-stranded nucleic acid regions and destabilizing the adjacent double-stranded regions (Levin et al. 2005). This activity would be expected to result in line broadening for imino protons, revealing changes to the dynamics of the base pair. Other factors may also contribute to NMR line broadening: (i) the higher correlation time associated with complex formation; (ii) fast- to intermediate-regime exchange associated with free and bound forms of the oligonucleotide, as previously described (Barraud et al. 2008). Many biophysical studies on DNA and RNA oligonucleotides have demonstrated destabilization of the nucleic acid secondary structure by NC (Beltz et al. 2005; Levin et al. 2005; Godet and Mély 2010; Godet et al. 2013) and some NMR studies described the direct impact on base pair dynamics (Johnson et al. 2000; Tisné et al. 2001). Line broadening preferentially affected the short stem surrounded by two single-stranded regions (apical loop and bulge U23C24U25). There are probably broadening effects linked to NC binding (chemical exchange), but we suggest that most of these broadening effects are due to the direct modification of base pair dynamics in the double-stranded region following the binding of NC to the adjacent single-stranded region (Fig. 5A). In contrast, the long central stem (nucleotides 6–16/45–55) was only slightly affected. The imino protons of the G16–C45 to G21–C41 base pairs displayed intermediate behavior, with significantly more broadening than those of the long central stem but less than for the apical stem. The short stem at the TAR end was only mildly affected by NC, because the G3 imino proton was detected in the NC:TAR complex.
Several studies have reported data for NC binding sites within the TAR stem–loop. Isothermal calorimetry titrations showed that TAR contained one high affinity site (110 nM), possibly with a second site of intermediate affinity (Heng et al. 2012). Kanevsky et al. (2005) showed that the apical loop G32 and G33 residue constitute a binding site for NC, this is probably not the case for G34, which has been shown to be involved in a transient cross-loop base pair (Huthoff et al. 2004; Lee et al. 2014). This finding was supported by the results of biophysical studies on the dynamic properties of the TAR apical loop demonstrating that G32 was highly dynamic (Dethoff et al. 2008), constituting a favorable binding site for NC. Indeed several studies have shown that NC binds preferentially to dynamic, accessible guanine residues (Sun et al. 2007; Bazzi et al. 2012). These data converge to suggest that there is a moderate- or high-affinity site in the apical loop. In this study, it was not possible to check for the presence of this site with 1H–15N NMR data due to the absence of observable imino protons in the apical loop.
However, the presence of a putative strong binding site in the apical loop was not necessarily correlated with destabilization of the adjacent stem. Indeed, one key property of the chaperone activity of NC is the rapid on/off kinetics of nucleic acid binding (Cruceanu et al. 2006a). This property suggests that NC does not need to bind to high-affinity sites to destabilize DNA/RNA structures. Indeed, its ability to recognize specific high-affinity stem–loops is important for the selective recognition of the viral RNA genome by the NC domain of the Gag protein during the packaging process, but not for reverse transcription. Several structural studies on NC–nucleic acid complexes (De Guzman et al. 1998; Amarasinghe et al. 2000; Johnson et al. 2000; Bourbigot et al. 2008) have provided support for this assertion, by showing that the strong binding of NC to the apical loops of the SL2 and SL3 elements does not destabilize the adjacent stems. In contrast, NC binding to stem–loops with lower affinities than SL2 and SL3 induces destabilization of the adjacent stems (Johnson et al. 2000; Bourbigot et al. 2008). Consistent with these findings, the sequential processing of Gag by protease enhances the nucleic acid chaperone activity of the resulting polypeptides (NCp15, NCp9, NCp7), and this improvement is associated with a lower affinity and faster binding/dissociation kinetics with nucleic acids (Cruceanu et al. 2006b; Rein 2010; Wu et al. 2013; Wang et al. 2014).
The destabilized stems are probably adjacent to binding sites of low or moderate affinity for NC. There should be a balance between the preferential binding of NC to exposed guanines residues and the need for rapid on/off kinetics. Consistent with this, a SHAPE (selective 2′-hydroxyl acylation analyzed by primer extension) study of the HIV-1 NL4-3 RNA genome suggested that the strong NC interaction domains and the domains destabilized by NC are not located in the same regions (Wilkinson et al. 2008). Indeed, Wilkinson and colleagues showed that the strong NC interaction domains in the SL1, SL2, and SL3 stem–loops were characterized by single-stranded regions containing unpaired G residues, whereas the destabilized domains contained single-stranded A/U-rich motifs adjacent to a duplex in which the first base pair included a guanosine nucleotide. The UCU bulge followed by the G26–C39 pair of the TAR hairpin is one of the six destabilized sites identified in the HIV-1 genome (Wilkinson et al. 2008). SHAPE analysis detected no strong NC binding sites within the TAR element.
Previous studies have shown that the destabilization of the TAR RNA hairpin by NC(12–55) is weaker than that of the cTAR DNA hairpin by the same peptide (Bernacchi et al. 2002; Azoulay et al. 2003; Beltz et al. 2005). Bernacchi et al. (2002) showed that destabilization of the TAR RNA hairpin by NC(12–55) involved only 1 bp at a peptide to nucleotide molar ratio of 1:7.5. TAR destabilization in our conditions was limited because NC destabilizes only three consecutive base pairs with high efficiency. However, the NC:nt ratio was significantly different in the two studies (1:7.5 versus 1:59) (Bernacchi et al. 2002) and NC(1–55) has a greater destabilization capacity that NC(12–55) (Levin et al. 2005; Vo et al. 2009b; Mitra et al. 2013). Finally, the extensive broadening of imino proton resonance does not necessarily indicate a loss of the corresponding base pair. It may, instead, indicate an increase in the proton exchange rate of the observed base pair.
One interesting finding of this study was the difference in behavior of the two stems adjacent to the internal UCU bulge, the upper stem (nucleotides 26–29/36–39) being significantly more destabilized by NC than the lower stem (nucleotides 18–22, 40–44). The SHAPE data (Wilkinson et al. 2008) indicated that the UCU bulge was less reactive after the removal of NC, suggesting a partial destabilization of one or both adjacent stems by the protein. We suggest that there may be an initial binding event at the G26 position adjacent to the internal loop, as the structure of this site is close to the consensus site already identified by SHAPE with a guanine located in a double-stranded segment (Fig. 11 in Wilkinson et al. 2008). The breathing of the G26–C39 base pair adjacent to a bulge might facilitate an interaction of NC with G26, thereby inducing destabilization of the four base pairs of this stem from G26 to C29 (Fig. 5A). This mechanism is very similar to that described for RNA helicases, in which a first binding event allows the adjacent base pairs to be destabilized, thereby increasing their fraying (Woodson 2010; Jankowsky 2011; Mustoe et al. 2014). We suggest that the UCU bulge and the apical loop constitute binding sites of moderate or low affinity for NC, allowing fast dissociation kinetics, with the protein sliding toward the double-stranded segments. According to this hypothesis, the upper stem would be preferentially destabilized because it is located between two single-stranded regions, whereas the lower stem is close to only one single-stranded region (bulge UCU). Alternatively, the upper stem may be preferentially destabilized because the NC functions with a definite polarity. The ability of NC to bind nucleic acids with a defined polarity has been suggested in a number of studies (Fig. 5B,C; Vuilleumier et al. 1999; Bourbigot et al. 2008; Bazzi et al. 2011; Darlix et al. 2011). In the three RNA–NC complexes for which high-resolution structures have been obtained (De Guzman et al. 1998; Amarasinghe et al. 2000; Spriggs et al. 2008), the protein is oriented anti-parallel to the 5′-3′ direction of the nucleic acid chain. As a result, the N-terminal zinc finger (ZF1) binds a residue 3′ to the residue binding to the C-terminal zinc finger (ZF2) (Fig. 5B). This binding polarity may be responsible for the polarity of destabilization observed for the adjacent stems (Fig. 5B). This hypothesis is consistent with the SHAPE data for the 5′ UTR region of the HIV-1 genome, suggesting that the destabilized stems are located on the 3′-side of single-stranded regions (Wilkinson et al. 2008). The relationship between binding polarity and the polarized destabilization of stem–loops may result from the lack of equivalence of the two zinc fingers, ZF1 and ZF2, with ZF1 more involved in the destabilization than ZF2. In contrast, ZF2 is involved in the specific recognition of guanine residues (Guo et al. 2002; Heath et al. 2003; Beltz et al. 2005; Narayanan et al. 2006; Hergott et al. 2013; Mitra et al. 2013). Binding polarity may therefore be responsible for the localization of ZF1 close to a double-stranded region. The binding of NC to G26 may position the ZF1 toward the upper stem (Fig. 5A) triggering the destabilization of this stem (Fig. 5B).
Our results are very similar to those of a recent single-molecule stretching paper investigating TAR destabilization by NC (McCauley et al. 2015). This recent study, like ours, identified specific targeted sites corresponding to guanines adjacent to unstable stem regions, such as mismatches, loops, and bulges. G26 was identified as a key site for the initiation of destabilization in both studies. The two studies differed in the NC:nt ratio used, with a ratio of 1:4 in the single-molecule study and 1:59 here. We used a low ratio to detect the initial destabilizing event preceding the binding of NC to other sites as its concentration increases (Darlix et al. 2011; McCauley et al. 2015).
At the low NC:nt ratio (1:59) used in our NMR study, destabilization was limited to the upper part of TAR. Thus, the long stem involving nucleotides 6–16/45–55 and the short terminal stem (nucleotides 1–4/56–59) displayed no significant destabilization. Additional NC molecules are probably required to destabilize these stems. Our results provide an opportunity to compare rate constants from the different parts of the reactional scheme (see Equation 2). The formation of the initial TAR-cTAR binary complex appears to be the first event after the addition of NC, because the associated bimolecular rate k1 displays considerable variation, by a factor of almost 30 when the NC:nt ratio is increased from 0 to 1:14 (Table 1). In contrast, under the same conditions, the dissociation rate k−1 displayed no variation. The strand-exchange rate k2 increased by a factor of 10 when the NC:nt ratio is increased from 0 to 1:14. Finally, our data are consistent with the destabilizing activity of NC being important only for the steps of the annealing reaction requiring duplex disruption. Indeed, the formation of the initial binary complex, involving the melting of the upper 4-bp stem, is highly dependent on the destabilizing activity of NC, whereas the reciprocal reaction does not require any structure melting. The final conversion into the extended duplex is also highly dependent on NC, but to a lesser extent than binary complex formation, and this step requires higher NC concentrations. In conclusion, our results are consistent with the formation of an extended kissing loop complex TAR-cTAR as suggested, in previous studies performed at low NC:nt ratios. However, they do not exclude the possibility that the “zipper” pathway is preferred at high NC:nt ratios (Godet et al. 2006; Liu et al. 2007; Vo et al. 2009b).
MATERIALS AND METHODS
Samples
The 15N/13C labeled TAR RNA sequence was obtained from Silantes GmbH (Germany). The TAR sequence used was that of the NL4-3 isolate. The NC(1–55) sequence (NL4-3 isolate) was overexpressed and the product was purified, as previously described (Barraud et al. 2007).
Samples were dissolved in 20 mM sodium phosphate buffer, pH 6.5, supplemented 50 mM NaCl and 0.1 mM EDTA in a 300 µL Shigemi tube. The RNA strand concentration in the tube was 340 µM, TAR RNA, and NC(1–55) were mixed in a 1:1 molar ratio (NC:nt 1:59).
NMR
NMR spectra were recorded on a Bruker Avance 950 MHz (TGIR, Gif sur Yvette, France) equipped with a triple-resonance cryoprobe with z-axis field gradient. All experiments were performed in 90% H2O and 10% 2H2O, at 283 K. For NOESY experiments, two mixing times of 150 and 200 msec were used, and the spectral widths were 20 833 Hz in both dimensions. 512 t1 experiments were recorded in the States-TPPI mode, each with 4029 data points per experiment. 1H–15N TROSY sensitivity enhanced gradient (Pervushin et al. 1997; Nietlispach 2005) experiments were carried out with 256 points in t1 and 1024 data points per experiment. 1H–15N 2D SOFAST-HMQC experiments were recorded with a PC9 and REBURP pulses for the band selective excitation and refocusing of imino protons (Schanda and Brutscher 2005; Farjon et al. 2009) and for recording the changes in the complexes over time. The relaxation delay was set to 200 msec and 200 experiments were recorded in the 15N dimension; the total experiment time per 2D was 295 sec.
Gel retardation assays
A heat-denatured TAR (7.5 pmol of TAR 32P-RNA at 3 × 103 cpm/pmol in 10 µL of double-distilled water) was used as a control. It was prepared by heating at 90°C for 2 min and chilling for 2 min on ice, and mixing with 4 µL of loading buffer (50% w/v glycerol, 0.05% w/v bromophenol blue, 0.05% w/v xylene cyanol). The annealing assays were carried out in a final volume of 10 µL. TAR 32P-RNA (7.5 pmol at 3 × 103 cpm/pmol) in 3.6 µL of water was heated at 90°C for 2 min and chilled for 2 min on ice. We then added 0.9 µL of annealing buffer (final concentrations: 30 mM NaCl, 0.2 mM MgCl2, and 25 mM Tris–HCl pH 7.5) and the sample was incubated at 20°C for 30 min. Unlabeled DNA (7.5 pmol) was subjected to the same renaturation treatment and was then added to refolded TAR 32P-RNA. The reaction mixture was incubated at 20°C in the absence or presence of NC (1.5, 3, or 6 µM) for various periods of time. Reactions were stopped by adding SDS (at a final concentration of 0.5%) and by proteinase K digestion (2 µg) at 20°C for 30 min. At the end of the incubation period, we added 4 µL of loading buffer to the reaction mixture. The samples were analyzed by electrophoresis on a 12% polyacrylamide gel (37.5:1 [w/w], acrylamide/bisacrylamide) at 25°C in 1× TBE buffer (90 mM Tris–borate [pH 8.3], 2 mM EDTA). After electrophoresis, the gel was fixed, dried and autoradiographed. Unannealed and annealed TAR RNAs were quantified with a Typhoon TRIO (GE Healthcare) and ImageQuant software. The percent annealed TAR RNA was determined as 100 × [annealed/(annealed + unannealed)].
Kinetic analysis of the gel-retardation assays
We performed gel retardation assays to follow the extended duplex formation over time. Extended duplex formation at various NC:nt ratios appeared to follow biexponential kinetics with two processes occurring at very different rates. This behavior could be described by Equation 1, where P(t) is the percentage of annealed molecules (Vo et al. 2009b). The kinetic phases could therefore be described by two components with rate constants kf for the fast component and ks for the slow component, where f is the fraction of the fast component and corresponds to the probability of intermediate complex RD* formation:
| (1) |
This analysis is similar to those used in previous studies (Vo et al. 2006, 2009b). We used this approach because it was developed for the same experimental system (analysis by gel-retardation assays of TAR-cTAR annealing in the presence of NC). The TAR and cTAR hairpins were assumed to anneal via a two-step pathway:
| (2) |
This reaction scheme describes a first rapid step in which a productive and transient complex is formed. This complex is slowly converted to the extended duplex corresponding to the fully annealed state. The RD* and RD complexes (R and D indicate RNA and DNA strands, respectively) cannot be distinguished on the basis of gel electrophoresis data.
Unlike Vo et al. (2009b), we were unable to make use of an approximation to pseudo-first-order conditions because [D]0 and [R]0 concentrations were identical. The reasons for the choice of these concentrations are explained below. The first step of the reaction (2) corresponds to a second-order bimolecular reversible reaction. Some simplifications can be used to describe this reaction: [D] = [R] and [RD*] = [D]0–[D] due to our initial conditions, [D]0 = [R]0 and the stoichiometry of the reaction. In these conditions
| (3) |
and
| (4) |
After integration [of the form dx/(a + bx + cx2)], the time dependence of the concentration of free cTAR was obtained as follows (Baker and Weng 1992):
| (5) |
where
and
Using the expression of [D] and [RD*] = [D]0–[D] as a function of the time, we can then deduce the fractions of the fast (f) component throughout the reaction, this fraction corresponding to the probability of intermediate formation:
| (6) |
Equation 1 can be therefore written for all values of time, using kf = kobs. According to which the fast component assigned to the first step of the reaction is related to the rate constant kobs described in Equation 5 and the slow component rate constant corresponding to the second and slower component of Equation 2 is proportional to the fraction of the preannealed intermediate RD* such that ks = f.k2 (Vo et al. 2009b). We simulated the values of Equation 1 using Equations 5 and 6 and numerical values for k1, k−1, and k2 (k−2 was not taken into account as its value was very low) and retained the values giving the lowest χ2 value. Statistical procedures were used to estimate the uncertainty on the various rate constants. Using the uncertainty of the various experimental measurements, we generated files of synthetic experimental data, using a Gaussian distribution and the Box–Muller method (Bevington and Robinson 2003). The global χ2 calculations were used to identify the best values for the rate constants. The criterion for selecting uncertainty was a doubling of the χ2 values.
Choice of nucleic acid concentrations
In the gel-retardation experiments described above, the kinetic analysis was carried out with a second-order bimolecular and reversible mechanism. The associated equations are much more complex than those used when pseudo-first order conditions can be assumed (i.e., one of the components [DNA] is in large excess over the other [RNA]). We chose to work with equal DNA and RNA concentrations and with an NC:RNA ratio (1:59) similar to those of the NMR studies. These conditions are probably also more similar to those in vivo with similar DNA and RNA concentrations. These conditions necessitated the use of an analysis method appropriate for a bimolecular association mechanism. In addition to the choice concerning stoichiometry, we also worked at higher DNA concentrations than in previous studies. This point was discussed by Vo et al. (2006, 2009b) when they described the limitations of the experimental conditions associated with pseudo-first order conditions and the concentrations used. Indeed, the elementary rates k1 and k−1 could be determined only for NC:nt > 1:16, because these rates can be determined only if there is a large fast component (f). However, at the concentrations used by these authors ([D]0 = 150 nM), the f fraction, the fraction corresponding to the fast component, could be deduced from pseudo-first order conditions as follows:
| (7) |
From Equation 7 it is clear that in the conditions used, k1D ≪ k−1. The f fraction is small, at ∼5%. However, using the same NC:nt ratio and therefore the same k1 and k−1 rate values but with a larger DNA concentration (a larger D term), k1D is larger and the relationship k1D ≪ k−1 no longer holds; this implies the existence of a significant f fraction, as calculated below. A second limitation is that, at a high NC:nt ratio (>1:7), the fast component is dominant and the elementary rate constant associated with the second step (k2) cannot be determined. Indeed, under the conditions used by Vo et al. (2006, 2009b), k2 could be only determined at ratios of 1:16 to 1:8, a rather limited range.
These limitations can be overcome and rate constants determined correctly even at low NC:nt ratios in the case of a high DNA concentration ([D]0 = 750 nM) is used, indeed in these conditions, k1D became close from k−1. In such conditions, the fraction f is 30%, 18%, 9%, and 2% at the 1:14, 1:29, 1:59 and 0:59 ratios, respectively. The fast component is therefore clearly observable (except at the 0:59 ratio) as shown by the experimental data (Fig. 4). Furthermore, as the fast component is never present in a large excess, it was also possible to determine k2.
ACKNOWLEDGMENTS
We gratefully acknowledge the financial support received from the TGIR-RMN-THC Fr3050 CNRS grant. We thank ANRS (Agence Nationale Française de Recherche contre le SIDA et les Hépatites) for financial support for our research.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.054445.115.
REFERENCES
- Aboul-ela F, Karn J, Varani G. 1995. The structure of the human immunodeficiency virus type-1 TAR RNA reveals principles of RNA recognition by Tat protein. J Mol Biol 253: 313–332. [DOI] [PubMed] [Google Scholar]
- Aboul-ela F, Karn J, Varani G. 1996. Structure of HIV-1 TAR RNA in the absence of ligands reveals a novel conformation of the trinucleotide bulge. Nucleic Acids Res 24: 3974–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hashimi HM, Gosser Y, Gorin A, Hu W, Majumdar A, Patel DJ. 2002. Concerted motions in HIV-1 TAR RNA may allow access to bound state conformations: RNA dynamics from NMR residual dipolar couplings. J Mol Biol 315: 95–102. [DOI] [PubMed] [Google Scholar]
- Amarasinghe GK, De Guzman RN, Turner RB, Chancellor KJ, Wu ZR, Summers MF. 2000. NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the Ψ-RNA packaging signal. Implications for genome recognition. J Mol Biol 301: 491–511. [DOI] [PubMed] [Google Scholar]
- Andersen ES, Contera SA, Knudsen B, Damgaard CK, Bsenbacher F, Kjems J. 2004. Role of the trans-activation response element in dimerization of HIV-1 RNA. J Biol Chem 279: 22243–22249. [DOI] [PubMed] [Google Scholar]
- Azoulay J, Clamme JP, Darlix JL, Roques BP, Mély Y. 2003. Destabilization of the HIV-1 complementary sequence of TAR by the nucleocapsid protein through activation of conformational fluctuations. J Mol Biol 326: 691–700. [DOI] [PubMed] [Google Scholar]
- Bailor MH, Sun XY, Al-Hashimi HM. 2010. Topology links RNA secondary structure with global conformation, dynamics, and adaptation. Science 327: 202–206. [DOI] [PubMed] [Google Scholar]
- Baker GM, Weng LC. 1992. Apparent first-order behavior under second-order kinetic conditions: a general concept illustrated by the reversible binding of hydrogen peroxide to cytochrome C oxidase. J Theor Biol 158: 221–229. [DOI] [PubMed] [Google Scholar]
- Barraud P, Gaudin C, Dardel F, Tisné C. 2007. New insights into the formation of HIV-1 reverse transcription initiation complex. Biochimie 89: 1204–1210. [DOI] [PubMed] [Google Scholar]
- Barraud P, Dardel F, Tisne C. 2008. Optimizing HSQC experiment for the observation of exchange broadened signals in RNA–protein complexes. Cr Chim 11: 474–479. [Google Scholar]
- Basu VP, Song M, Gao L, Rigby ST, Hanson MN, Bambara RA. 2008. Strand transfer events during HIV-1 reverse transcription. Virus Res 134: 19–38. [DOI] [PubMed] [Google Scholar]
- Bazzi A, Zargarian L, Chaminade F, Boudier C, De Rocquigny H, René B, Mély Y, Fossé P, Mauffret O. 2011. Structural insights into the cTAR DNA recognition by the HIV-1 nucleocapsid protein: role of sugar deoxyriboses in the binding polarity of NC. Nucleic Acids Res 39: 3903–3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzi A, Zargarian L, Chaminade F, De Rocquigny H, Rene B, Mély Y, Fossé P, Mauffret O. 2012. Intrinsic nucleic acid dynamics modulates HIV-1 nucleocapsid protein binding to its targets. PLoS One 7: e38905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltz H, Azoulay J, Bernacchi S, Clamme JP, Ficheux D, Roques B, Darlix JL, Mély Y. 2003. Impact of the terminal bulges of HIV-1 cTAR DNA on its stability and the destabilizing activity of the nucleocapsid protein NCp7. J Mol Biol 328: 95–108. [DOI] [PubMed] [Google Scholar]
- Beltz H, Piémont E, Schaub E, Ficheux D, Roques B, Darlix JL, Mély Y. 2004. Role of the structure of the top half of HIV-1 cTAR DNA on the nucleic acid destabilizing activity of the nucleocapsid protein NCp7. J Mol Biol 338: 711–723. [DOI] [PubMed] [Google Scholar]
- Beltz H, Clauss C, Piémont E, Ficheux D, Gorelick RJ, Roques B, Gabus C, Darlix JL, de Rocquigny H, Mély Y. 2005. Structural determinants of HIV-1 nucleocapsid protein for cTAR DNA binding and destabilization, and correlation with inhibition of self-primed DNA synthesis. J Mol Biol 348: 1113–1126. [DOI] [PubMed] [Google Scholar]
- Berkhout B. 1992. Structural features in TAR RNA of human and simian immunodeficiency viruses: a phylogenetic analysis. Nucleic Acids Res 20: 27–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkhout B, Vastenhouw NL, Klasens BI, Huthoff H. 2001. Structural features in the HIV-1 repeat region facilitate strand transfer during reverse transcription. RNA 7: 1097–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernacchi S, Stoylov S, Piémont E, Ficheux D, Roques BP, Darlix JL, Mély Y. 2002. HIV-1 nucleocapsid protein activates transient melting of least stable parts of the secondary structure of TAR and its complementary sequence. J Mol Biol 317: 385–399. [DOI] [PubMed] [Google Scholar]
- Bevington PR, Robinson DK. 2003. Data reduction and error analysis for the physical sciences, 3rd ed. McGraw-Hill, NY. [Google Scholar]
- Bourbigot S, Ramalanjaona N, Boudier C, Salgado GF, Roques BP, Mély Y, Bouaziz S, Morellet N. 2008. How the HIV-1 nucleocapsid protein binds and destabilises the (−)primer binding site during reverse transcription. J Mol Biol 383: 1112–1128. [DOI] [PubMed] [Google Scholar]
- Cruceanu M, Gorelick RJ, Musier-Forsyth K, Rouzina I, Williams MC. 2006a. Rapid kinetics of protein-nucleic acid interaction is a major component of HIV-1 nucleocapsid protein's nucleic acid chaperone function. J Mol Biol 363: 867–877. [DOI] [PubMed] [Google Scholar]
- Cruceanu M, Urbaneja MA, Hixson CV, Johnson DG, Datta SA, Fivash MJ, Stephen AG, Fisher RJ, Gorelick RJ, Casas-Finet JR, et al. 2006b. Nucleic acid binding and chaperone properties of HIV-1 Gag and nucleocapsid proteins. Nucleic Acids Res 34: 593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlix JL, Garrido JL, Morellet N, Mély Y, de Rocquigny H. 2007. Properties, functions, and drug targeting of the multifunctional nucleocapsid protein of the human immunodeficiency virus. Adv Pharmacol 55: 299–346. [DOI] [PubMed] [Google Scholar]
- Darlix JL, Godet J, Ivanyi-Nagy R, Fossé P, Mauffret O, Mély Y. 2011. Flexible nature and specific functions of the HIV-1 nucleocapsid protein. J Mol Biol 410: 565–581. [DOI] [PubMed] [Google Scholar]
- Darlix JL, de Rocquigny H, Mauffret O, Mély Y. 2014. Retrospective on the all-in-one retroviral nucleocapsid protein. Virus Res 193: 2–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Guzman RN, Wu ZR, Stalling CC, Pappalardo L, Borer PN, Summers MF. 1998. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science 279: 384–388. [DOI] [PubMed] [Google Scholar]
- Dethoff EA, Al-Hashimi HM. 2011. Characterization of ps-Ms dynamics in the TAR apical loop by NMR. Biophys J 100: 238a. [Google Scholar]
- Dethoff EA, Hansen AL, Musselman C, Watt ED, Andricioaei I, Al-Hashimi HM. 2008. Characterizing complex dynamics in the transactivation response element apical loop and motional correlations with the bulge by NMR, molecular dynamics, and mutagenesis. Biophys J 95: 3906–3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farjon J, Boisbouvier J, Schanda P, Pardi A, Simorre JP, Brutscher B. 2009. Longitudinal-relaxation-enhanced NMR experiments for the study of nucleic acids in solution. J Am Chem Soc 131: 8571–8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godet J, Mély Y. 2010. Biophysical studies of the nucleic acid chaperone properties of the HIV-1 nucleocapsid protein. RNA Biol 7: 687–699. [DOI] [PubMed] [Google Scholar]
- Godet J, de Rocquigny H, Raja C, Glasser N, Ficheux D, Darlix JL, Mély Y. 2006. During the early phase of HIV-1 DNA synthesis, nucleocapsid protein directs hybridization of the TAR complementary sequences via the ends of their double-stranded stem. J Mol Biol 356: 1180–1192. [DOI] [PubMed] [Google Scholar]
- Godet J, Kenfack C, Przybilla F, Richert L, Duportail G, Mély Y. 2013. Site-selective probing of cTAR destabilization highlights the necessary plasticity of the HIV-1 nucleocapsid protein to chaperone the first strand transfer. Nucleic Acids Res 41: 5036–5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golinelli MP, Hughes SH. 2003. Secondary structure in the nucleic acid affects the rate of HIV-1 nucleocapsid-mediated strand annealing. Biochemistry 42: 8153–8162. [DOI] [PubMed] [Google Scholar]
- Guo J, Wu T, Kane BF, Johnson DG, Henderson LE, Gorelick RJ, Levin JG. 2002. Subtle alterations of the native zinc finger structures have dramatic effects on the nucleic acid chaperone activity of human immunodeficiency virus type 1 nucleocapsid protein. J Virol 76: 4370–4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath MJ, Derebail SS, Gorelick RJ, DeStefano JJ. 2003. Differing roles of the N- and C-terminal zinc fingers in human immunodeficiency virus nucleocapsid protein-enhanced nucleic acid annealing. J Biol Chem 278: 30755–30763. [DOI] [PubMed] [Google Scholar]
- Heilman-Miller SL, Wu T, Levin JG. 2004. Alteration of nucleic acid structure and stability modulates the efficiency of minus-strand transfer mediated by the HIV-1 nucleocapsid protein. J Biol Chem 279: 44154–44165. [DOI] [PubMed] [Google Scholar]
- Heng X, Kharytonchyk S, Garcia EL, Lu K, Divakaruni SS, LaCotti C, Edme K, Telesnitsky A, Summers MF. 2012. Identification of a minimal region of the HIV-1 5′-leader required for RNA dimerization, NC binding, and packaging. J Mol Biol 417: 224–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig M, Williamson JR. 2000. Detection of N-H···N hydrogen bonding in RNA via scalar couplings in the absence of observable imino proton resonances. Nucleic Acids Res 28: 1585–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hergott CB, Mitra M, Guo J, Wu T, Miller JT, Iwatani Y, Gorelick RJ, Levin JG. 2013. Zinc finger function of HIV-1 nucleocapsid protein is required for removal of 5′-terminal genomic RNA fragments: a paradigm for RNA removal reactions in HIV-1 reverse transcription. Virus Res 171: 346–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huthoff H, Girard F, Wijmenga SS, Berkhout B. 2004. Evidence for a base triple in the free HIV-1 TAR RNA. RNA 10: 412–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowsky E. 2011. RNA helicases at work: binding and rearranging. Trends Biochem Sci 36: 19–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson PE, Turner RB, Wu ZR, Hairston L, Guo J, Levin JG, Summers MF. 2000. A mechanism for plus-strand transfer enhancement by the HIV-1 nucleocapsid protein during reverse transcription. Biochemistry 39: 9084–9091. [DOI] [PubMed] [Google Scholar]
- Kanevsky I, Chaminade F, Ficheux D, Moumen A, Gorelick R, Negroni M, Darlix JL, Fossé P. 2005. Specific interactions between HIV-1 nucleocapsid protein and the TAR element. J Mol Biol 348: 1059–1077. [DOI] [PubMed] [Google Scholar]
- Klaver B, Berkhout B. 1994. Evolution of a disrupted TAR RNA hairpin structure in the HIV-1 virus. EMBO J 13: 2650–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamoorthy G, Roques B, Darlix JL, Mély Y. 2003. DNA condensation by the nucleocapsid protein of HIV-1: a mechanism ensuring DNA protection. Nucleic Acids Res 31: 5425–5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Dethoff EA, Al-Hashimi HM. 2014. Invisible RNA state dynamically couples distant motifs. Proc Natl Acad Sci 111: 9485–9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin JG, Guo J, Rouzina I, Musier-Forsyth K. 2005. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Prog Nucleic Acid Res Mol Biol 80: 217–286. [DOI] [PubMed] [Google Scholar]
- Levin JG, Mitra M, Mascarenhas A, Musier-Forsyth K. 2010. Role of HIV-1 nucleocapsid protein in HIV-1 reverse transcription. RNA Biol 7: 754–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HW, Zeng Y, Landes CF, Kim YJ, Zhu Y, Ma X, Vo MN, Musier-Forsyth K, Barbara PF. 2007. Insights on the role of nucleic acid/protein interactions in chaperoned nucleic acid rearrangements of HIV-1 reverse transcription. Proc Natl Acad Sci 104: 5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCauley MJ, Rouzina I, Manthei KA, Gorelick RJ, Musier-Forsyth K, Williams MC. 2015. Targeted binding of nucleocapsid protein transforms the folding landscape of HIV-1 TAR RNA. Proc Natl Acad Sci 112: 13555–13560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirambeau G, Lyonnais S, Coulaud D, Hameau L, Lafosse S, Jeusset J, Justome A, Delain E, Gorelick RJ, Le Cam E. 2006. Transmission electron microscopy reveals an optimal HIV-1 nucleocapsid aggregation with single-stranded nucleic acids and the mature HIV-1 nucleocapsid protein. J Mol Biol 364: 496–511. [DOI] [PubMed] [Google Scholar]
- Mirambeau G, Lyonnais S, Gorelick RJ. 2010. Features, processing states, and heterologous protein interactions in the modulation of the retroviral nucleocapsid protein function. RNA Biol 7: 724–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra M, Wang W, Vo MN, Rouzina I, Barany G, Musier-Forsyth K. 2013. The N-terminal zinc finger and flanking basic domains represent the minimal region of the human immunodeficiency virus type-1 nucleocapsid protein for targeting chaperone function. Biochemistry 52: 8226–8236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musselman C, Al-Hashimi HM, Andricioaei I. 2007. IRED analysis of TAR RNA reveals motional coupling, long-range correlations, and a dynamical hinge. Biophys J 93: 411–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustoe AM, Brooks CL, Al-Hashimi HM. 2014. Hierarchy of RNA functional dynamics. Annu Rev Biochem 83: 441–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan N, Gorelick RJ, DeStefano JJ. 2006. Structure/function mapping of amino acids in the N-terminal zinc finger of the human immunodeficiency virus type 1 nucleocapsid protein: residues responsible for nucleic acid helix destabilizing activity. Biochemistry 45: 12617–12628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nietlispach D. 2005. Suppression of anti-TROSY lines in a sensitivity enhanced gradient selection TROSY scheme. J Biomol NMR 31: 161–166. [DOI] [PubMed] [Google Scholar]
- Pervushin K, Riek R, Wider G, Wuthrich K. 1997. Attenuated T2 relaxation by mutual cancellation of dipole–dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc Natl Acad Sci 94: 12366–12371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitt SW, Majumdar A, Serganov A, Patel DJ, Al-Hashimi HM. 2004. Argininamide binding arrests global motions in HIV-1 TAR RNA: comparison with Mg2+-induced conformational stabilization. J Mol Biol 338: 7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puglisi JD, Tan R, Calnan BJ, Frankel AD, Williamson JR. 1992. Conformation of the TAR RNA-arginine complex by NMR spectroscopy. Science 257: 76–80. [DOI] [PubMed] [Google Scholar]
- Rein A. 2010. Nucleic acid chaperone activity of retroviral Gag proteins. RNA Biol 7: 700–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon L, Bascom G, Andricioaei I, Al-Hashimi HM. 2013. A general method for constructing atomic-resolution RNA ensembles using NMR residual dipolar couplings: the basis for inter-helical motions revealed. J Am Chem Soc 135: 5457–5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schanda P, Brutscher B. 2005. Very fast two-dimensional NMR spectroscopy for real-time investigation of dynamic events in proteins on the time scale of seconds. J Am Chem Soc 127: 8014–8015. [DOI] [PubMed] [Google Scholar]
- Schanda P, Kupce E, Brutscher B. 2005. SOFAST-HMQC experiments for recording two-dimensional heteronuclear correlation spectra of proteins within a few seconds. J Biomol NMR 33: 199–211. [DOI] [PubMed] [Google Scholar]
- Sleiman D, Goldschmidt V, Barraud P, Marquet R, Paillart JC, Tisné C. 2012. Initiation of HIV-1 reverse transcription and functional role of nucleocapsid-mediated tRNA/viral genome interactions. Virus Res 169: 324–339. [DOI] [PubMed] [Google Scholar]
- Sleiman D, Barraud P, Brachet F, Tisne C. 2013. The interaction between tRNALys3 and the primer activation signal deciphered by NMR spectroscopy. PLoS One 8: e64700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spriggs S, Garyu L, Connor R, Summers MF. 2008. Potential intra- and intermolecular interactions involving the unique-5′ region of the HIV-1 5′-UTR. Biochemistry 47: 13064–13073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart-Maynard KM, Cruceanu M, Wang F, Vo MN, Gorelick RJ, Williams MC, Rouzina I, Musier-Forsyth K. 2008. Retroviral nucleocapsid proteins display nonequivalent levels of nucleic acid chaperone activity. J Virol 82: 10129–10142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XY, Zhang Q, Al-Hashimi HM. 2007. Resolving fast and slow motions in the internal loop containing stem-loop 1 of HIV-1 that are modulated by Mg2+ binding: role in the kissing-duplex structural transition. Nucleic Acids Res 35: 1698–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisné C, Roques BP, Dardel F. 2001. Heteronuclear NMR studies of the interaction of tRNALys3 with HIV-1 nucleocapsid protein. J Mol Biol 306: 443–454. [DOI] [PubMed] [Google Scholar]
- Vo MN, Barany G, Rouzina I, Musier-Forsyth K. 2006. Mechanistic studies of mini-TAR RNA/DNA annealing in the absence and presence of HIV-1 nucleocapsid protein. J Mol Biol 363: 244–261. [DOI] [PubMed] [Google Scholar]
- Vo MN, Barany G, Rouzina I, Musier-Forsyth K. 2009a. Effect of Mg2+ and Na+ on the nucleic acid chaperone activity of HIV-1 nucleocapsid protein: implications for reverse transcription. J Mol Biol 386: 773–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo MN, Barany G, Rouzina I, Musier-Forsyth K. 2009b. HIV-1 nucleocapsid protein switches the pathway of transactivation response element RNA/DNA annealing from loop-loop “kissing” to “zipper”. J Mol Biol 386: 789–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuilleumier C, Bombarda E, Morellet N, Gérard D, Roques BP, Mély Y. 1999. Nucleic acid sequence discrimination by the HIV-1 nucleocapsid protein NCp7: a fluorescence study. Biochemistry 38: 16816–16825. [DOI] [PubMed] [Google Scholar]
- Wang W, Naiyer N, Mitra M, Li J, Williams MC, Rouzina I, Gorelick RJ, Wu Z, Musier-Forsyth K. 2014. Distinct nucleic acid interaction properties of HIV-1 nucleocapsid protein precursor NCp15 explain reduced viral infectivity. Nucleic Acids Res 42: 7145–7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson KA, Gorelick RJ, Vasa SM, Guex N, Rein A, Mathews DH, Giddings MC, Weeks KM. 2008. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol 6: e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson SA. 2010. Taming free energy landscapes with RNA chaperones. RNA Biol 7: 677–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Datta SA, Mitra M, Gorelick RJ, Rein A, Levin JG. 2010. Fundamental differences between the nucleic acid chaperone activities of HIV-1 nucleocapsid protein and Gag or Gag-derived proteins: biological implications. Virology 405: 556–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Mitra M, McCauley MJ, Thomas JA, Rouzina I, Musier-Forsyth K, Williams MC, Gorelick RJ. 2013. Aromatic residue mutations reveal direct correlation between HIV-1 nucleocapsid protein's nucleic acid chaperone activity and retroviral replication. Virus Res 42: 2525–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Mitra M, Naufer MN, McCauley MJ, Gorelick RJ, Rouzina I, Musier-Forsyth K, Williams MC. 2014. Differential contribution of basic residues to HIV-1 nucleocapsid protein's nucleic acid chaperone function and retroviral replication. Nucleic Acids Res 42: 2525–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You JC, McHenry CS. 1994. Human immunodeficiency virus nucleocapsid protein accelerates strand transfer of the terminally redundant sequences involved in reverse transcription. J Biol Chem 269: 31491–31495. [PubMed] [Google Scholar]
- Zarudyana MI, Potyahaylo AL, Kolomiets IM, Hovorun DM. 2014. Phylogenetic study on structural elements of HIV-1 poly(A) region. 2. USE domain and TAR hairpin. Biopolym Cell 30: 29–36. [Google Scholar]
- Zhang Q, Sun X, Watt ED, Al-Hashimi HM. 2006. Resolving the motional modes that code for RNA adaptation. Science 311: 653–656. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Stelzer AC, Fisher CK, Al-Hashimi HM. 2007. Visualizing spatially correlated dynamics that directs RNA conformational transitions. Nature 450: 1263–1267. [DOI] [PubMed] [Google Scholar]
- Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]