Abstract
Mitochondria provide the main source of energy to eukaryotic cells, oxidizing fats and sugars to generate ATP. Mitochondrial fatty acid β-oxidation (FAO) and oxidative phosphorylation (OXPHOS) are two metabolic pathways which are central to this process. Defects in these pathways can result in diseases of the brain, skeletal muscle, heart and liver, affecting approximately 1 in 5000 live births. There are no effective therapies for these disorders, with quality of life severely reduced for most patients. The pathology underlying many aspects of these diseases is not well understood; for example, it is not clear why some patients with primary FAO deficiencies exhibit secondary OXPHOS defects. However, recent findings suggest that physical interactions exist between FAO and OXPHOS proteins, and that these interactions are critical for both FAO and OXPHOS function. Here, we review our current understanding of the interactions between FAO and OXPHOS proteins and how defects in these two metabolic pathways contribute to mitochondrial disease pathogenesis.
Keywords: disease, mitochondria, protein complex assembly, protein interactions, supercomplex
MITOCHONDRIAL METABOLISM
Mitochondria occupy almost all human cell types and are involved in essential metabolic and cellular processes, including calcium and iron homoeostasis, apoptosis, innate immunity and haeme biosynthesis [1]. However, the primary function of mitochondria is the production of energy in the form of ATP [2–4]. ATP is the body's energy currency, playing vital roles in cell differentiation, growth and reproduction, thermogenesis and powering the contraction of muscles for movement [1]. In humans, ATP is produced by two different processes; through the breakdown of glucose or other sugars in the absence of oxygen in the cytoplasm (glycolysis), or by the metabolism of fats, sugars and proteins in the mitochondria in the presence of oxygen. Although both processes produce ATP, oxidative metabolism accounts for 95% of ATP produced and yields 20 times the amount of ATP as its anaerobic counterpart.
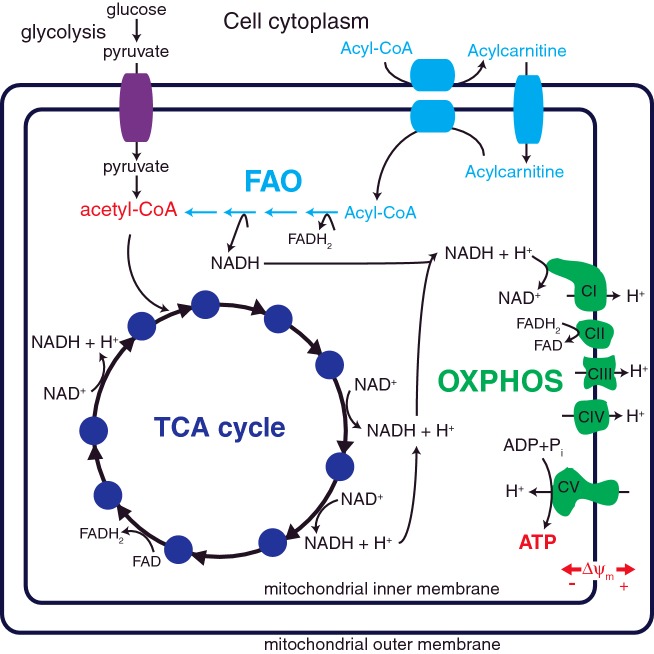
Mitochondria utilize three main enzymatic pathways to generate ATP; the tricarboxylic acid (TCA) cycle, oxidative phosphorylation (OXPHOS) and fatty acid β-oxidation (FAO). The TCA cycle oxidizes acetyl-CoA, derived from sugars, fats and amino acids, to generate NADH and flavin adenine dinucleotide (FADH2), which can be used by the OXPHOS system to generate ATP (Figure 1).
Figure 1. Mitochondrial metabolism.
Glucose breakdown through glycolysis and the TCA cycle (dark blue) generates reduced NADH and FADH2. Fatty acid β-oxidation (FAO, light blue) of fatty acyl-CoA esters is performed in four enzymatic reactions that also generates NADH and FADH2, as well as acetyl-CoA. Electrons derived from NADH and FADH2 are utilized by the five OXPHOS complexes (green) to generate ATP. Complex I (CI, NADH: ubiquinone oxidoreductase), complex III (CIII, ubiquinol: ferricytochrome c oxidoreductase) and complex IV (CIV, cytochrome c oxidase) pump electrons out of the mitochondrial matrix to generate a membrane potential (Δψm) that drives the synthesis of ATP by complex V (CV, FoF1-ATP synthetase). CII, complex II (succinate: ubiquinone oxidoreductase).
OXIDATIVE PHOSPHORYLATION (OXPHOS)
OXPHOS involves a series of oxidation–reduction reactions which results in the phosphorylation of ADP to produce ATP. This process is performed by five protein complexes which reside in the inner mitochondrial membrane: complex I (CI), NADH: ubiquinone oxidoreductase, EC 1.6.5.3; complex II (CII), succinate: ubiquinone oxidoreductase, EC 1.3.5.1; complex III (CIII) ubiquinol: ferricytochrome c oxidoreductase, EC 1.10.2.2; complex IV (CIV) cytochrome c oxidase, EC 1.9.3.1 and complex V (CV), FoF1-ATP synthetase, EC; 3.6.3.14.
Complex I (CI) accepts electrons from NADH, whereas CII accepts electrons from FADH2, both of which are derived from the TCA cycle and FAO. CI and CII then reduce ubiquinone, the substrate of CIII. CIII then transfers electrons from reduced ubiquinone to cytochrome c. Next, CIV passes the electrons from cytochrome c to O2, reducing it to form H2O. As the electrons are transferred between the OXPHOS complexes, protons are pumped across the inner mitochondrial membrane by CI, CIII and CIV to create an electrochemical potential (∆ψm). ∆ψm is used by CV to drive the phosphorylation of ADP to produce ATP (Figure 1).
OXPHOS SUPERCOMPLEXES
Since the identification of the five OXPHOS complexes, their orientation in situ has been debated. Two main theories have been postulated; the fluid model and the solid-state model [5]. The fluid model suggests that the OXPHOS complexes localize individually and diffuse laterally in the mitochondrial inner membrane. In this model, electron transfer is dependent on random collisions between CI, CII, CIII and CIV [6]. Conversely, the solid-state theory proposes that the constituents of OXPHOS combine to form stable structures, termed ‘supercomplexes’, that contain two or more of the OXPHOS complexes.
Initial findings identified possible physical interactions between CI and CIII [7,8], as well as CII and CIII [9]. Further research resulted in the isolation of a supercomplex containing CIII/CIV2 in several strains of bacteria [10–12] and CI/CIII2, CIII2, CI2/CIV and CI/CIII2/CIV with varying stoichiometry in potato mitochondria [13,14]. In yeast, a CV dimer (CV2) [15] and CIII2/CIV1–2 supercomplex have also been detected [16].
In mammalian mitochondria, CI/CIII2 and CI/CIII2/CIV1-3 supercomplexes have been identified. In particular, the CI/CIII2/CIV1–3 supercomplex has been described as the ‘respirasome’, representing a single, functional respiratory unit [16]. Although the existence of OXPHOS supercomplexes within the inner mitochondrial membrane is now widely accepted, the function of these structures is still debated. A large body of research has revealed that the supercomplexes are integral for OXPHOS complex stability. Experimental evidence showed that a mutation in MT-CYB, which encodes the cytochrome b subunit of CIII, results in the disruption of CIII assembly in both mice and humans. In addition, destabilization of CI was also observed. This suggests that the presence of CIII is essential for CI assembly and/or stability via their interaction in a supercomplex [17].
Similarly, knockout of one of the CIV assembly factor genes, COX10, results in the loss of CIV activity and steady-state levels, with an associated reduction in mitochondrial respiratory capacity. Perturbed assembly of CI was also observed in COX10 knockout mouse mitochondria, suggesting that stable CIV is required to maintain CI stability [18]. Overall, these findings highlight the interdependence of the OXPHOS complexes for their stability, via their association in the OXPHOS supercomplex.
In addition, the phospholipid cardiolipin is required for OXPHOS supercomplex assembly and stability [19]. The majority of cardiolipin is found in the inner mitochondrial membrane where it is essential for mitochondrial function. Nuclear magnetic resonance imaging of bovine heart mitochondria has identified cardiolipin attached to CV [20]. It has also been shown that cardiolipin is required to maintain CI, CIII and CIV structure and function [21].
In humans, mutations to TAZ, which encodes the cardiolipin acyltransferase Tafazzin, result in Barth syndrome, a disease characterized by dilated cardiomyopathy, skeletal myopathy and neutropenia [22]. Barth syndrome patients exhibit increased accumulation of monolysocardiolipin precursors and reduced mature tetralinoleoylcardiolipin production, with associated CIII and CIV deficiencies [23]. It has also been shown that Tafazzin defects result in destabilized OXPHOS supercomplexes, which in turn results in reduced steady-state levels of CI. These findings suggest that cardiolipin is essential for OXPHOS supercomplex stability, and that loss of supercomplex stability contributes to Barth syndrome pathogenesis [19].
Apart from stabilizing the OXPHOS complexes, the supercomplex structure may also play a role in substrate channelling. Flux experiments have shown that the formation of the respirasome allows for substrate channelling by decreasing the distance in which electrons travel between mobile electron carriers and the OXPHOS complexes. This finding is supported by in-gel enzymatic assays that demonstrate respirasome catalytic activity in a range of eukaryotes [16,24,25]. In addition, the formation of the respirasome has also been proposed to limit oxidative stress [26]. By forcing closer interactions between CI and CIII, the leakage of electrons to form superoxide is less likely. Indeed, oxidative stress is a common attribute in diseases where supercomplex assembly is disturbed (reviewed in [27,28]).
However, recent findings suggest that the main function of supercomplexes is not to channel substrates and stabilize the OXPHOS complexes, but may instead be a protein packaging and space saving phenomenon. Recent flux control analyses have discounted electron channelling in supercomplexes [29], while electron microscopy of the supercomplex structure has revealed that the distances between CIII and CIV may be too large for efficient substrate channelling [30]. In addition, it has been proposed that the interdependence of complex stability is most likely due to downstream effects of increased oxidative stress and not due to their presence in the same supercomplex [29].
In summary, although the existence of OXPHOS supercomplexes is now largely established, there is still debate regarding their functional significance. Irrespective of their purpose, the OXPHOS supercomplexes play an important role in mitochondrial respiration and their disruption contributes to mitochondrial disease pathogenesis, possibly in ways we are yet to fully understand.
MITOCHONDRIAL FATTY ACID β-OXIDATION (FAO)
Fatty acids are vital constituents of enzymes, hormones and cell membranes. In addition, they are a major source of energy. Fatty acids are metabolized in mitochondria by FAO, a critical pathway of energy production in a variety of cell types, including the heart [31,32]. In fact, under normal physiological conditions, FAO provides the majority of ATP (60–70%) required for proper heart contraction [33]. FAO yields a high amount of energy for the cell. For example, the complete oxidation of a 16-carbon palmitic acid will yield a total of 112 molecules of ATP.
FAO is crucial for homoeostatic regulation, specifically in times of fasting or endurance exercise that requires high energy resources. During this high energy demand, fat stores are broken down for metabolism by tissues in need [34]. In particular, the liver metabolizes fatty acids to produce ketone bodies for consumption by the brain when glucose is unavailable [35,36].
At least 20 separate transport proteins and enzymes are required for activation and breakdown of fatty acids via FAO (Table 2) [37]. Fatty acids are transported through the blood as non-esterified fatty acids bound to lipoproteins or serum albumin. Upon reaching their target cell, short and medium chain fatty acids (C4–C12) traverse the cell membrane by passive diffusion. However, saturated and unsaturated long chain fatty acids cross the cell membrane by sodium dependent fatty acid transporters [38]. These include fatty acid transport proteins (FATPs), plasma membrane fatty acid binding proteins and the fatty acid translocase protein CD36.
Table 2. Proteins involved in mitochondrial fatty acid β-oxidation (FAO).
| Protein | Gene | Pathogenic mutation | Clinical presentations |
|---|---|---|---|
| Carnitine transport cycle and transport | |||
| Carnitine O-palmitoyltransferase 1A (CPT1A) | CPT1A | Yes | Reye-like syndrome, hypoketosis, coma, hyperammonaemia, hypertriglyceridemia, renal tubular acidosis, hypoglycaemia, hepatomegaly, lethargy, hypotonia, hyperemesis, diarrhoea, hyperbilirubinemia, acute fatty liver of pregnancy, hyperemesis |
| Carnitine O-palmitoyltransferase 1B (CPT1B) | CPT1B | No | |
| Carnitine O-palmitoyltransferase 1C (CPT1C) | CPT1C | Yes | Spastic paraplegia |
| Carnitine O-palmitoyltransferase 2 (CPT2) | CPT2 | Yes | Hypothermia, lethargy, seizures, hypotonia, cardiomegaly, hyperreflexia, cardiac arrhythmias, lipid accumulation in liver, heart and kidney, polymicrogyria in brain, microcephaly |
| Carnitine acylcarnitine translocase (CACT) | SLC25A20 | Yes | Cardiomyopathy, liver dysfunction, apnoea, seizures, tachycardia, hypotension, coma, hypoglycaemia, dicarboxylic aciduria, hypocarnitinemia, < tex − math/ > hepatomegaly, sudden infant death |
| Organic cation/carnitine transporter 2 | SLC22A5 | Yes | Systemic carnitine deficiency, hypoketotic hypoglycaemia, skeletal myopathy, cardiomyopathy |
| Fatty acid β-oxidation cycle | |||
| Very-long chain acyl-CoA dehydrogenase (VLCAD) | ACADVL | Yes | Rhabdomyolysis, hypoglycaemia, myopathy, myoglobinuria, hepatomegaly, cardiomegaly, cardiac arrest, hypotonia, lipid accumulation in various tissues |
| Long-chain acyl-CoA dehydrogenase (LCAD) | ACADL | No | |
| Medium-chain acyl-CoA dehydrogenase (MCAD) | ACADM | Yes | Sudden Infant Death, hypoglycaemia, lethargy, coma, fatty deposits in liver, Reye-like syndrome, hyperammonaemia, cardiomyopathy |
| Short chain acyl-CoA dehydrogenase (SCAD) | ACADS | Yes | Acidosis, neurological impairment, myopathy, muscle weakness, emesis, failure to thrive, developmental delay, hypertonia, hyperactivity, reduced consciousness |
| Short/branched chain specific acyl-CoA dehydrogenase, mitochondrial (SBCAD) | ACADSB | Yes | 2-Methylbutyryl glycinuria |
| Mitochondrial trifunctional protein (MTP) | |||
| Long chain enoyl-CoA hydratase (LCEH) | HADHA | Yes | Cardiomyopathy, Reye-like Syndrome, liver dysfunction, myopathy, rhabdomyolysis, metabolic acidosis, neuropathy, maternal HELLP syndrome, preeclampsia, acute liver failure of pregnancy, developmental delay, myoglobinuria, hypoparathyroidism |
| Long-chain 3-hydroxyacyl-CoA dehydrogenase (LCHAD) | HADHA | Yes | |
| Long-chain 3-ketoacyl-CoA thiolase (LCKAT) | HADHB | Yes | |
| 3-Ketoacyl-CoA thiolase (KAT) | ACAA2 | No | |
| Hydroxyacyl-CoA dehydrogenase (HADH) | HADH | Yes | Familial hyperinsulinaemic hypoglycaemia |
| Others | |||
| Acyl-CoA dehydrogenase 9 (ACAD9) | ACAD9 | Yes | Leigh Syndrome, complex I deficiency, cardiomyopathy, muscle weakness, metabolic acidosis |
| Acyl-CoA dehydrogenase 10 (ACAD10) | ACAD10 | No | |
| Acyl-CoA dehydrogenase 11 (ACAD11) | ACAD11 | No | |
| Electron transfer flavoprotein (ETF) | ETFA, ETFB | Yes | Glutaric aciduria 2A and 2B, multiple Acyl-CoA dehydrogenase deficiency, isolated myopathy |
| Electron transfer flavoprotein: ubiquinone oxidoreductase (ETF-QO) | ETFDH | Yes | Glutaric aciduria 2C, multiple acyl-CoA dehydrogenase deficiency |
| Enoyl-CoA hydratase, short chain 1 (ECHS1) | ECHS1 | Yes | Development delay, cardiomyopathy, apnoea, Leigh syndrome |
| Enoyl-CoA delta isomerase, 1 (ECI1) | ECI1 | No | |
| Enoyl-CoA delta isomerase, 2 (ECI2) | ECI2 | No | |
| 2,4-Dienoyl-CoA reductase (DECR1) | DECR1 | No | |
| Delta(3,5)-delta(2,4)-dienoyl-CoA isomerase, mitochondrial | ECH1 | No | |
| propionyl-CoA carboxylase (PCC) | PCCA, PCCB | Yes | Propionic academia type I and II, episodic vomiting, lethargy, ketosis, neutropenia, thrombocytopenia, hyperglycinuria, hyperglycinaemia, hypogammaglobulinemia, developmental delay, protein intolerance |
| Methylmalonyl-CoA epimerase (MCEE) | MCCE | Yes | Methylmalonic aciduria, retarded motor development, spasticity, dystonia, failure to thrive, gastroesophageal reflux, metabolic acidosis, dehydration, tachypnea, ketonuria, hydrocephalus and macrocephaly |
| Methylmalonyl-CoA mutase (MCM) | MUT | Yes | Methylmalonic aciduria type mut, poor feeding, dehydration, metabolic acidosis, valine intolerance, lethargy, ketoacidosis, multi-organ failure, developmental delay, interstitial nephritis, seizures, basal ganglia infarct |
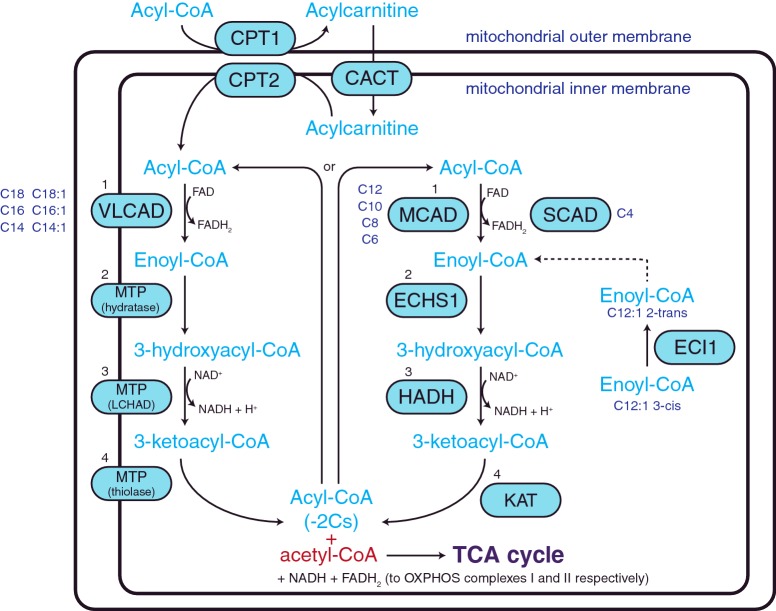
Once inside the cell, acyl-CoA synthetases activate the fatty acid by converting it from its non-esterified form to a fatty acyl-CoA ester. These esters can form the preliminary substrates for cholesterol, phospholipid and triacylglycerol synthesis, or enter the mitochondria via the carnitine shuttle system for FAO (Figure 2). The carnitine transport of fatty acyl-CoAs involves three steps. Firstly, the fatty acyl-CoA is bound to carnitine by carnitine O-palmitoyltransferase 1 (CPT1) to form a fatty acylcarnitine. Fatty acylcarnitines are then transported across the mitochondrial inner membrane by the carnitine acylcarnitine translocase (CACT). Once inside the mitochondrial matrix, carnitine O-palmitoyltransferase 2 (CPT2) converts the fatty acylcarnitine back to a fatty acyl-CoA ester [4].
Figure 2. Mitochondrial fatty acid β-oxidation (FAO) spiral.
Fatty acyl-CoA esters are converted to fatty acylcarnitines by CPT1 for transport into the mitochondria by CACT. Acylcarnitines are subsequently converted back to fatty acyl-CoA esters once inside the mitochondria by CPT2 for metabolism by the fatty acid β-oxidation (FAO) spiral. FAO consists of four reactions (numbered 1–4 in black) which are performed by enzymes that are fatty acid chain length specific (chain lengths shown in dark blue). (1) Dehydrogenation of the fatty acyl-CoA by very long chain (VLCAD), medium chain (MCAD) or short chain (SCAD) acyl-CoA dehydrogenases to create enoyl-CoA, (2) hydration by the enoyl-CoA hydratase activity of the MTP or ECHS1 to add water to enoyl-CoA to form 3-hydroxyacyl-CoA, (3) a second dehydrogenation by MTP or HADH to generate 3-ketoacyl-CoA and (4) thiolysis by the thiolase activity of the MTP or KAT to produce a shortened fatty acyl-CoA and acetyl-CoA. Oxidation of unsaturated fatty acids requires the action of ECI1.
Metabolism of fatty acyl-CoAs by FAO requires four enzymatic reactions; dehydrogenation, hydration, a second dehydrogenation and thiolysis. The end products of these reactions are one acetyl-CoA molecule, two electrons (which enter OXPHOS) and a fatty acyl-CoA ester that has been shortened by two carbon atoms. As such, the FAO pathway is often referred to as a spiral pathway, as the resulting shortened fatty acyl-CoA ester returns to the beginning of the pathway and is re-oxidized until only two acetyl-CoA molecules remain (Figure 2).
In the first step of FAO, fatty acyl-CoAs undergo dehydrogenation, removing two hydrogen atoms to create enoyl-CoA. Dehydrogenation is performed by a family of acyl-CoA dehydrogenases with particular fatty acid chain length specificity, ranging from 4 to 24 carbons. This family of enzymes includes: very long chain acyl-CoA dehydrogenase (VLCAD, C12–C24), long chain acyl-CoA dehydrogenase (LCAD, C14–C18) (low expression in humans), medium chain acyl-CoA dehydrogenase (MCAD, C6–C12) and short chain acyl-CoA dehydrogenase (SCAD, C4–C6). Dehydrogenation is flavin adenine dinucleotide (FAD) dependent, with FAD reduced to FADH2. The liberated electrons are passed from FADH2 to the electron transfer flavoprotein (ETF), a heterodimer consisting of an alpha (ETFA) and beta (ETFB) subunit. The electrons are then transferred to ubiquinone by the ETF: ubiquinone oxidoreductase (ETF-QO). Finally, ubiquinone enters the OXPHOS pathway by being oxidized by CIII.
For long chain fatty acids (C14–C18), the remaining three steps are catalysed by the mitochondrial trifunctional protein (MTP). Encoded by two genes, MTP is a hetero-octamer consisting of four α-subunits (encoded by HADHA) and four β-subunits (encoded by HADHB). The α-subunit comprises long chain enoyl-CoA hydratase (LCEH) and the NAD-dependent enzyme long chain 3-hydroxyacyl-CoA dehydrogenase (LCHAD), whereas long chain 3-ketoacyl-CoA thiolase (LKAT) is found in the β-subunit.
For medium (C6–C12) and short chain fatty acyl-CoAs (C4–C6), the last three steps of FAO are performed by different enzymes that are located in the mitochondrial matrix. These include: step 2, hydration by short chain enoyl-CoA hydratase 1 (ECHS1 or crotonase), step 3, dehydrogenation by hydroxyacyl-CoA dehydrogenase (HADH) and step 4, thiolysis by 3-ketoacyl-CoA thiolase (KAT) (Figure 2).
Fatty acids that have an uneven number of carbon atoms result in a three-carbon propanoyl-CoA at the last spiral, which is subsequently converted to succinyl-CoA in three steps. Propanoyl-CoA is carboxylated to (S)-methylmalonyl-CoA by propionyl-CoA carboxylase (PCC), a dodecameric enzyme comprised of six alpha (PCCA) and six beta (PCCB) subunits. (S)-Methylmalonyl-CoA is then isomerized by methylmalonyl-CoA epimerase (MCEE) to form (R)-methylmalonyl-CoA. The final step is performed by methylmalonyl-CoA mutase (MCM) and requires co-factor Vitamin B12 to produce succinyl-CoA, which then enters the TCA cycle.
MITOCHONDRIAL DISEASE
Mitochondrial disease comprises a heterogeneous group of disorders, owing to the structural and functional complexity of the mitochondrion itself. Mitochondrial biogenesis and function requires the coordinated effort of over 1100 proteins that are encoded by both the nuclear DNA (nDNA) and the maternally inherited mtDNA. Mitochondrial disorders can be caused by pathogenic mutations in mtDNA or nDNA, resulting in a variety of inheritance patterns, including maternal, autosomal dominant, autosomal recessive and X-linked [39].
Mitochondrial disease symptoms are wide ranging, from mild to severe, early onset to late onset, and can affect one organ or multiple systems. In this regard, mitochondrial disorders do not always show a genotype–phenotype correlation. Two patients may possess the same pathogenic mutation but present differently, or have different mutations that result in the same clinical phenotype. For example, mutations in MT-ND1 and MT-ND4 can cause Leber hereditary optic neuropathy (LHON), a disease characterized by bilateral, painless central vision loss in early to late adulthood resulting from the specific degeneration of retinal ganglion cells in the optic nerve [40]. However, these same mutations can also be associated with more severe symptoms, including dystonia and short stature [41–43].
The most common mitochondrial disorders are attributed to dysfunction of the OXPHOS system and occur at an estimated frequency of 1 in 5000 live births. OXPHOS disorders are characterized by deficiencies in OXPHOS complex activity and/or reductions to the steady-state levels of the OXPHOS complexes, with subsequent diminished ATP production. These may be isolated complex disorders or a combination thereof. Defects can be caused by mutations in genes that encode protein subunits of the OXPHOS complexes, proteins required for OXPHOS complex biogenesis, as well as proteins that are vital for the replication, transcription and translation of mtDNA (Table 1) [39].
Table 1. Genes with pathogenic mutations resulting in OXPHOS disorders.
| Complex subunits | Proteins for import/processing or assembly | mtDNA expression or replication | Nucleotide transport or synthesis | Membrane composition | |
|---|---|---|---|---|---|
| Complex I | MT-ND1, MT-ND2, MT-ND3, MT-ND4, MT-ND4L, MT-ND5, MT-ND6, NDUFA1, NDUFA2, NDUFA4, NDUFA8, NDUFA9, NDUFA10, NDUFA11, NDUFA12, NDUFB3, NDUFB8, NDUFB9, NDUFS1, NDUFS2, NDUFS3, NDUFS4, NDUFS6, NDUFS7, NDUFS8, NDUFV1, NDUFV2, NDUFV3 | ACAD9, FOXRED1, NDUFAF1, NDUFAF2, NDUFAF3, NDUFAF4, NDUFAF5, NDUFAF6, NUBPL | AARS2, AGK, C10orf2, C12orf65, DARS2, EARS2, FARS2, GFM1, GFM2, LRPPRC, MPV17, MRPL3, MRPS16, MRPS22, MT01, MTFMT, MTPAP, MTTL1, MTTW, POLG, POLG2, RARS2, RMND1, SARS2, TACO1, TRMU, TSFM, TUFM, YARS2 | DGUOK, POS1, RRM2B, SLC25A4, SLC25A3, SUCLA2, SUCLG1, TK2, TYMP | CABC1, COQ2, COQ6, COQ9, CYCS, DNM1L, MPC1 MFN2, NMT, OPA1, PDSS1, PDSS2, SERAC1, TAZ |
| Complex II | SDHA, SDHB, SDHC, SDHD | SDHAF1, SDHAF2 | |||
| Complex III | CYC1,MT-CYB, UQCRB, UQCRQ, UQCRC2 | BCS1L, HCCS, LYRM7, TTC19, UQCC2, UQCC3 | |||
| Complex IV | COX4I2, COX6B1, COX7B, MT-CO1, MT-CO2, MT-CO3 | APOPT1, COA5, COA6, COX10, COX14, COX15, COX20, ETHE1, FASTKD2, PET100, SCO1, SCO2, SURF1 | |||
| Complex V | ATP5E, MT-ATP6, MT-ATP8 | ATPAF2, TMEM70 | |||
| Multiple complexes | ABCB7, AFG3L2, BOLA3, DNAJC19, FXN, GFER, GLRX5, HSPD1, ISCU, LYRM4, NFU1, SPG7, TIMM8A |
OXPHOS complex I
Isolated CI deficiencies (OMIM #252010) are the most common causes of OXPHOS disorders, accounting for approximately 30% of affected patients. CI diseases comprise a range of clinically heterogeneous conditions. Complex I consists of 44 different structural subunits, one of which is found twice, for a total of 45 subunits. Pathogenic mutations have been identified in 21 CI subunit genes, including all seven of the mtDNA-encoded subunits [44,45] (Table 1).
In recent years, significant advances have also been made in our understanding of the machinery involved in CI biogenesis. This process requires additional proteins, termed ‘assembly factors’, which aid complex I assembly [46]. The first pathogenic human mutation in a CI assembly factor was found in NDUFAF2 [47]. Since this initial discovery, several other pathogenic mutations have been identified in CI assembly factor genes, including NDUFAF1 [48], NDUFAF3 [49], NDUFAF4 [50], NDUFAF5 [51], NDUFAF6 [52], FOXRED1 [53], ACAD9 [54] and NUBPL [53] (Table 1). The identification of these mutations, and the examination of how they disrupt CI assembly, have greatly aided our understanding of CI biogenesis.
The clinical presentation of CI disorders can range from the neonatal period to adult onset, with symptoms including cardiomyopathy, liver disease and neurological disorders [55–58]. The most common clinical presentations include Leigh Syndrome, a multiple organ disorder with degeneration of the muscular, peripheral and central nervous systems [55,59–61], fatal infantile lactic acidosis and other infancy/early childhood onset neuropathological disorders [44].
In addition, CI defects have been linked to Parkinson's disease (PD), a neurodegenerative condition which results from the progressive loss of dopaminergic neurons in the substantia nigra. Isolated CI respiratory deficiencies are a common feature, specifically in the substantia nigra of PD patients [62,63]. The underlying pathogenic mechanisms are not well understood, although a growing body of research suggests mitochondrial respiratory dysfunction and oxidative stress contribute to disease pathogenesis [62–65]. In some Parkinson's disease patients, mtDNA mutations have also been detected, specifically in the D-loop region and the MT-ND5 gene [65].
OXPHOS complex II
Complex II (CII) is composed of four highly conserved nuclear encoded subunits and participates in both OXPHOS and the TCA cycle. In OXPHOS, CII transfers electrons to reduce ubiquinone, whereas in the TCA cycle it metabolizes fumarate to succinate.
Although CII deficiencies (OMIM # 252011) are rare, they still demonstrate the typical clinical heterogeneity associated with mitochondrial diseases. Many CII disorders present in childhood as retinopathies [66] or as encephalopathies [67,68], namely Leigh Syndrome [69], with accompanying cardiomyopathy [70]. Conversely, adult onset CII disorders have also been reported [71]. Mutations in the CII subunit genes SDHA [72], SDHB [73], SDHC [74] and SDHD [75] have also been associated with paragangliomas and pheochromocytoma (Table 1).
Pathogenic mutations in CII assembly factors have also been identified, including SDHAF1 [67,76], which can result in infantile leukoencephalopathy [76], and SDHAF2 [77] (Table 1). SDHAF2 mutations may present similarly to mutations in SDHA [72], SDHB [73], SDHC [74] and SDHD [75] as paraganglioma and pheochromocytoma [77,78].
OXPHOS complex III
Complex III (CIII) subunits are largely encoded by the nDNA with only one being mtDNA encoded, cytochrome b [79]. Conditions attributed to CIII deficiency (OMIM #124000) are uncommon, with wide ranging clinical variability. Clinical presentations can include lactic acidosis, sensorineural loss, liver failure, LHON, developmental delay, cardiomyopathies and encephalopathy [80–83]. In addition, mutations in genes encoding CIII assembly factors have also been identified [84,85]. For example, mutations in BCS1L, which encodes the mitochondrial chaperone BCS1, disrupt CIII assembly with increased reactive oxygen species production, resulting in a severe multiple systems condition [84] (Table 1).
OXPHOS complex IV
Complex IV (CIV) is the terminal enzyme of the respiratory chain and possesses 13 subunits, three of which are encoded by mtDNA. These three mtDNA-encoded subunits form the catalytic site of CIV [60]. Interestingly, mutations to these three subunits are rare. First identified in 1977 [86], CIV deficiencies (OMIM # 220110) can present as myopathies, facial dimorphism and lactic acidosis [86,87]. The majority of known pathogenic mutations are in nDNA genes that encode CIV structural subunits or assembly factors [60], including COX6B1 [88], and PET100 [89] (Table 1).
OXPHOS complex V
Complex V (CV) is made up of two functional units that span the inner mitochondrial membrane (Fo) and the mitochondrial matrix (F1). Complex V phosphorylates ADP to produce ATP by utilizing the electrochemical gradient produced during CI to CIV electron transfer [90].
Human pathologies arising from isolated CV deficiencies are the rarest of the OXPHOS disorders [91]. Pathogenic mutations to MTATP6 [92] can result in maternally inherited Leigh syndrome (MILS) [93] and neuropathy, ataxia, retinal pigmentosa (NARP) [90,92]. Additionally, mutations to CV assembly factors, TMEM70 [94] and ATPAF2 [95] have been identified and present with cardiomyopathy, hypotonia, intrauterine growth restriction and oligohydramnios [94,95] (Table 1).
FAO DISEASE
FAO disorders present as a variety of clinical phenotypes and almost always demonstrate an autosomal recessive inheritance pattern. Of note, there is one reported case of a patient with an autosomal dominant CPT-2 mutation [96] (Table 2). The incidence of FAO disease is believed to be approximately 1:10000, with clinical phenotypes attributed to energy deficiency in tissues that rely heavily on the FAO pathway, such as the heart, liver and skeletal muscle. In infants, where stored glycogen levels are low and metabolic rates are high, FAO is the primary pathway for generating ATP [97,98]. As such, disruption to FAO can result in severe disease during the early stages of life, and can present as intrauterine growth restriction, prematurity, cardiomyopathies, neuropathies, liver failure, rhabdomyolysis, lactacidemia, Reye-like Syndrome (a condition that mimics the metabolic disease Reye Syndrome with hypoglycaemia and hypoketonemia) and neonatal death. In late-onset FAO disease, symptoms can include myopathy and exercise intolerance, cardiac arrhythmias and neuropathy [4]. Of note, women who are heterozygous for an FAO gene mutation in HADHA may experience pregnancy induced disease which commonly manifests as preeclampsia and acute liver failure [99,100].
In addition to the primary FAO enzyme deficiency, disruption of FAO can result in excess metabolic intermediates in affected tissues, including the heart, liver, brain and eyes. This is considered to contribute to organ dysfunction due to the toxicity of these intermediates. Excess intermediates are often found in the blood, and can also be expelled in the urine of affected individuals (and as such are used as biomarkers of FAO disease [4,101]).
The first recognized case of human FAO disease was described in 1973 as a CPT2 deficiency in muscle cells [102]. Since then, mutations in at least 19 transport proteins and enzymes involved in FAO have been identified (Table 2). FAO disease presentation is not always persistent and can appear in bouts. These bouts are triggered by stimuli that require fatty acid breakdown, including fasting, endurance exercise, cold exposure and increased dietary fat consumption. Disorders of FAO have also been attributed to sudden infant death syndrome (SIDS) fatalities [103], resulting in the employment of newborn screening for many FAO deficiencies. Since its implementation, many asymptomatic newborn babies have been diagnosed with an inborn error in FAO metabolism, with an increase in MCAD deficiency diagnoses [104]. Since many patients die during their first metabolic stress induced episode, quick diagnosis, allowing for immediate treatment, has been praised for reductions in FAO mortalities in recent years [104]. Still, proper management of many FAO disorders is hindered by the inability to recognize clear biochemical abnormalities in asymptomatic patients.
At present, no cures exist for FAO disease, with treatment strategies focusing primarily on reducing fat intake. Fasting is also avoided, particularly by increasing carbohydrate consumption at night before sleep or during illness [4]. For carnitine transport disorders, carnitine supplementation has been tested, although there is some disagreement as to its effectiveness [105,106]. Supplementation with medium chain fatty acids may also be a way to bypass deficiencies in long chain fatty acid metabolism, such as VLCAD deficiencies, but this is yet to be verified [4].
DISORDERS OF FAO ENZYMES
MCAD deficiency (OMIM #201450) is the most well studied FAO disease [107], with the majority of symptomatic MCAD deficiencies thought to result from a common point mutation (985A>G) in ACADM [108]. Clinical presentations include hypotonia, Reye-like Syndrome, seizures, apnoea, hepatomegaly, fever, vomiting, diarrhoea and coma [109]. In the Caucasian population of several western countries, the number of ACADM mutation carriers is estimated to be less than 1:110; specifically, England (1:68), Australia (1:71), Denmark (1:100) and the United States (1:107) [110].
VLCAD deficiency (OMIM #201475) can affect multiple tissues, including heart and muscle, as well as the liver, and as such can result in a severe clinical phenotype. VLCAD deficiency can be classified into three main subgroups based on age of onset, each of which correlates closely with clinical severity and prognosis. The first subgroup, with neonatal presentation, is characterized by cardiomyopathy and is frequently fatal in early life. The second subgroup presents in infancy, with hypoketosis and hypoglycaemia, and frequently mimics Reye-Syndrome [111,112]. The third subgroup has a milder phenotype with adolescent to adult onset, with clinical phenotypes including exercise intolerance and myopathies [111].
Deficiencies in the octameric MTP (OMIM #609015) result in mitochondrial disease which is primarily neuropathological. The build-up of FAO intermediates, which result in tissue toxicity, has been proposed as a primary pathogenic factor in these diseases. MTP deficiencies are generally characterized by reductions in the enzymatic activities of all three of its constituent enzymes. However, isolated deficiencies have been identified in LCHAD that are caused by HADHA mutations [113,114]. In addition, women who are heterozygous for mutations in HADHA commonly present with acute fatty liver failure and haemolysis, elevated liver enzymes and low platelet counts (HELLP) during pregnancy [113]. Similar to VLCAD deficiency, MTP deficiencies also follow a correlation between age and severity. Age of onset varies substantially and reported cases range from mild to severe, affecting newborns to adults [115].
SCAD deficiency (OMIM #20170) displays phenotypic variability, although neurological impairment appears to be a common theme. This suggests that there may be other factors, such as environmental or epigenetic, which can trigger the disease symptoms, resulting in the wide range of clinical variability associated with SCAD deficiency [116,117]. Similar to other FAO diseases, SCAD disease can be divided into subgroups based on age of presentation, which can either be in infancy/early childhood or in late adulthood. In infants, developmental delay, hypotonia and myopathy are common [118]. In addition, ethylmalonic aciduria is a common feature of SCAD deficiency, and as such is a commonly used biomarker of the disease [116].
Pathogenic mutations in ACADS have been identified in patients with SCAD deficiency, whereas others carry two ACADS polymorphisms (G625A and C511T) that are considered susceptibility variants [116,117]. Newborn screening of SCAD has identified these variants in patients with ethylmalonic aciduria, however, they have also been detected in many asymptomatic individuals. As such, the clinical relevance of SCAD deficiencies has now come into question [119].
ECHS1 encodes a mitochondrial hydratase which catalyses the second step of FAO. In addition, ECHS1 is also involved in the isoleucine and valine pathways, converting methacrylyl-CoA to (S)-3-hydroxyisobutyryl-CoA and acryloyl-CoA to 3-hydroxypropionyl-CoA [120].
ECHS1 deficiency (OMIM #616277) is phenotypically variable, with relatively early onset, ranging from neonatal presentation to early childhood. Almost all of the patients identified to date exhibit bilateral lesions to the brain consistent with the primary OXPHOS disorder, Leigh Syndrome. In addition, cardiomyopathy, developmental delay and metabolic acidosis are common, with death mostly under the age of 1 year [120–124] (Table 3). Given the involvement of ECHS1 in both the amino acid and FAO pathways, build-up of intermediates from both pathways are a common feature [120–125]. Interestingly, ECHS1 has been demonstrated to be most active in the valine pathway and may be expendable in FAO [122].
Table 3. Pathogenic ECHS1 mutations and their associated clinical and biochemical features.
Biochemical and clinical characteristics of ECHS1 patients identified to date. A. B.–at birth, R. C.–respiratory chain, n. d.–not determined, CS, centrum semiovale; Pu, putamen; GP, globus pallidus; NC, nuclear caudatus; BG, basal ganglia; SN, substantia nigra and PV, periventricular; do, days old; mo, months old; yo, years old.
| Patient information | Clinical | Biochemistry | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Author | ID | M/F | Age of onset | Age now | Symptoms | Neuroimaging (MRI and MRS) | ECHS1 protein levels | Respiratory chain activity and metabolic enzyme analysis | BN-PAGE |
| Haack et al., 2015 | #MRB166 c.(161G>A); (394G>A) p.(Arg54His); (Ala132Thr) | F | 1 yo | Alive 8 yo | Hearing loss, development delay, increased lactate, hypotonia, ataxia | n. d. | n. d. | n. d. | n. d. |
| #346 c.(176A>G); (476A>G) p.(Asn59Ser); (Gln159Arg) | F | A. B. | Died 4 mo | Hearing loss, epilepsy, cardiomyopathy, increased lactate | Brain atrophy and white matter abnormalities | Reduced | Reduced CI | n. d. | |
| #376 c.(98T<C); (176A>G) p.(Phe33Ser); (Asn59Ser) | F | A. B. | Alive 3 yo | Hearing loss, developmental delay, epilepsy, cardiomyopathy, increased lactate | Symmetrical bilateral abnormalities BG | Reduced | Reduced CIV | n. d. | |
| #42031 c.(197T<C); (449A>G) p.(Ile66Thr); (Asp150Gly) | M | A. B. | Died 11 mo | Hearing loss, optic atrophy, developmental delay, epilepsy, dystonia, cardiomyopathy, excessive 2-methyl-1,3,dihydroxybutyrate | Symmetrical punctiform hyper-sensitivities in CS | Reduced | R. C. normal, reduced pyruvate | n. d. | |
| #52236 c.(229G>C); (476A>G) p.(Glu77Gln); (Gln159Arg) | F | 11 mo | Alive 31 yo | Hearing loss, optic atrophy, wheelchair bound by 9yo, spastic tetra paresis, developmental delay, epilepsy, dystonia, increased lactate | Signal hyper-sensitivities in NC and Pu | Reduced | R. C. normal | n. d. | |
| #57277 C(161G>A); (431dup) p.(Arg54His); (Leu145Alafs*6) | F | A. B. | Alive 16 yo | Hearing loss, optic atrophy, communicates through voice computer, developmental delay, dystonia, increased lactate | Increased T2-signal in Pu and GP until 2yo | Reduced | R. C. normal | n. d. | |
| #68552 c.(476A>G); (476A>G) p.(Gln159Arg); (Gln159Arg) | F | A. B. | Died 2.3 yo | Developmental delay, epilepsy, dystonia, increased lactate | Symmetrical white matter abnormalities | n. d. | Reduced CI | n. d. | |
| #68761 c.(161G>A); (817A>G) p.(Arg54His); (Lys273Glu) | M | A. B. | Died 7.5 yo | Developmental delay, epilepsy, dystonia | Brain atrophy | n. d. | Decreased ATP production | n. d. | |
| #73663 c.(673T>C); (673T>C) p.(Cys225Arg); (Cys225Arg) | F | A. B. | Alive 2 yo | Developmental delay, epilepsy, cardiomyopathy, increased lactate, increased 2-methyl-1,3,dihydroxybutyrate | Delayed myelination, white matter lesions | Reduced | R. C. normal | n. d. | |
| #76656 c.(268G<A); (583G>A) p.(Gly90Arg); (Gly195Ser) | F | 2 yo | Alive 5 yo | Hearing loss, developmental delay, dystonia, increased 2-methyl-1,3,dihydroxybutyrate | Signal hyper-sensitivities in Pu, GP, NC and PV white matter | n. d. | R. C. normal | n. d. | |
| Yamada et al., 2015 | III-2 c.(176A>G); (413C>T) p.(Asn59Ser); (Ala138Val) | F | 10 mo | Alive 7 yo | Developmental delay, dystonia, intellectual disability, increased lactate and N-acetyl-S-(2-carboxypropyl) cysteine | Bilateral hyper-sensitivities to Pu, GP, NC and SN | Reduced | R. C. normal | n. d. |
| III-3 c.(176A>G); (413C>T) p.(Asn59Ser); (Ala138Val) | M | 7 mo | Died 5 yo | Developmental delay, dystonia, intellectual disability, increased lactate, N-acetyl-S-(2-carboxypropyl) cysteine and 2-methyl-1,3, dihydroxybutyrate | Bilateral hypersensitivities to Pu, GP, NC and SN | Reduced | R. C. normal | n. d. | |
| Tetrault et al., 2015 | P1 c.(583A>G); (583G>A) p.(Thr180Ala); (Gly195Ser) | F | 2.5 mo | Died 10 mo | Failure to thrive, developmental delay, nystagmus, reduced pyruvate dehydrogenase activity | Bilateral T2 hyper intensity of BG | n. d. | Pyruvate dehydrogenase reduced. R. C. normal in fibroblasts | Reductions to Complexes I and III in muscle |
| P2 c.(583A>G); (713C>T) p.(Thr180Ala); (Ala238Val) | M | 2.9 yo | Alive 18 yo | Failure to thrive, developmental delay, dystonia, nystagmus, hearing loss, truncal ataxia, microcephaly, increased lactate | Bilateral hyper sensitivity of BG | n. d. | R. C. normal in fibroblasts | n. d. | |
| P3 c.(583A>G); (713C>T) p.(Thr180Ala); (Ala238Val) | M | 10 mo | Alive 13 yo | Failure to thrive, developmental delay, optic atrophy, hearing loss, nystagmus, truncal ataxia, increased lactate | T2 hyperintensities of the BG | n. d. | R. C. normal in muscle | n. d. | |
| P4 c.(583A>G); (476A>G) p.(Thr180Ala); (Gln159Arg) | F | 6 mo | Alive 12 yo | Failure to thrive, hypotonia, dystonia, optic atrophy, nystagmus, hearing loss, microcephaly, hyperketosis and encephalopathy | Hypersensitivity of BG | n. d. | R. C. normal in muscle | Reductions to Complex III and IV | |
| Ferdinandusse et al., 2015 | Patient 1 c.(817A > G); (817A > G) p.(Lys273Glu); (Lys273Glu) | F | A. B. | Died 1 do | Depressed respiration, increased lactate, hyperammonaemia, cardiomyopathy, hepatomegaly, degeneration of white matter in brain, spongy myelinopathy, Alzheimer's type II metabolic gliosis, muscularization in intralobular arterioles (lungs) | Multiple cystic lesions | n. d. | n. d. | n. d. |
| Patient 2 c.(817A > G); (817A > G) p.(Lys273Glu); (Lys273Glu) | F | A. B. | Died 2 do | Apnoea, increased lactate, encephalopathy, increased short medium and long chain acylcarnitine, increased triacylglycerols, hypoxic respiratory failure, increased alanine and proline, liver steatosis | Multiple cystic lesions | Reduced | n. d. | n. d. | |
| Patient 3 c.(433C > T); (476A > G) p.(Leu145Phe); (Gln159Arg) | F | 4 mo | Alive 7 yo | Hypotonia, developmental delay, microcephaly, hearing loss, dysphagia, apnoea, cardiomyopathy, oedema, increased lactate, 2-methyl-2,3, dihydroxybutyrate and cysteine and reduced E3 lipoamide dehydrogenase | Symmetrical atrophy of cerebellum and atrophy of grey matter | Reduced | R. C. normal in muscle | n. d. | |
| Patient 4 c.(673 T > C); (674G > C) p.(Cys225Arg); (Cys225Ser) | M | A. B. | Alive 3 yo | Bilateral glaucoma, psychomotor, failure to thrive, retardation, lower limb hypotonia, upper limb dystonia, Kussmaul breathing, increased lactate, metabolic acidosis, hyperketosis, increased acylcarnitines, increased 2-methyl-2,3, dihydroxybutyrate, increased cysteine | T2 hyperintensities detected in GP, Pu and bilateral symmetrical lesions in CP. Atrophy of the midbrain nuclei | Reduced | Normal pyruvate dehydrogenase and R. C. complex activity in muscle | n. d. | |
| Sakai et al., 2014 | Patient 1 c.(2T>G); (5C>T) p.(Met1Arg); (Ala2Val) | M | 2 mo | Alive (assumed) | Hearing loss, developmental delay, hypotonia, nystagmus, spasticity, increased lactate | Bilateral T2 hyper intensity of the Pu | Reduced | Patient cells–reduced CI, III and IV. Immortalized myoblasts, reduced CI, IV and V | No differences |
| Peters et al., 2014 | Patient 1 c.(473C>A); (414+3G>C) p.(Ala158Asp) | F | A. B. | Died 4 mo | Apnoea, cardiomyopathy, increased lactate, increased cysteine | Atrophy of the brain and symmetrical T2 hypersensitivity in Pu. Large lactate peak | Reduced | Reduced pyruvate dehydrogenase in patient fibroblasts. R. C. was normal | n. d. |
| Patient 2 c.(473C>A); (414+3G>C) p.(Ala158Asp) | M | A. B. | Died 8 mo | Apnoea, hypotonia, developmental delay, nystagmus, cardiomyopathy, increased cysteine | Reduced bilateral myelination of GP and Pu | Reduced | Reduced pyruvate dehydrogenase | n. d. | |
Aside from enzymes directly involved in FAO catalysis, pathogenic mutations have also been identified in other FAO genes including ACAD9, ACADSB, ETFA, ETFB, ETFDH, PCCA, PCCB, MCEE, MUT, SLC25A20, HADH, CPT1A, CPT1C and CPT2 [4] (Table 2).
FAO–OXPHOS PROTEIN INTERACTIONS
Reduced NAD and FADH2 produced during FAO pass their electrons to the OXPHOS complexes. Therefore, these pathways share substrates and are linked biochemically. Interestingly, primary disorders of one of these pathways have been shown to inhibit or disturb the other.
These secondary defects are thought to arise from the build-up of toxic intermediates [54,121,123,126–131]. However, there is growing evidence that physical links between FAO and OXPHOS proteins exist, raising the possibility that loss of these interactions may cause the secondary defects observed and therefore contribute to disease pathology [113,128–130,132,133]
The first descriptions of FAO–OXPHOS protein interactions were the identification of the FAO proteins, MTP and thiolase, bound to OXPHOS CI [132]. It was hypothesized that these physical associations were necessary for NADH oxidation/reduction coupling and provided an efficient mechanism of substrate channelling [132].
Other findings also suggest that physical interactions exist between OXPHOS and FAO proteins that are crucial for their combined function and stability [54,133,134]. ETF was found in purified complexes from porcine liver mitochondria containing CIII [135]. In rat liver mitochondria, the fatty acid proteins VLCAD, ETF, TFP, LCHAD and MCAD were shown to co-migrate with monomeric CI and the CI/III2/IV1–3 OXPHOS supercomplex by blue native polyacrylamide gel electrophoresis (BN-PAGE) and by sucrose density gradients [134]. Conversely, isovaleryl-CoA dehydrogenase (IVD), a dehydrogenase involved in amino acid metabolism and not FAO, did not co-migrate with OXPHOS complexes or supercomplexes [134]. High levels of ACAD activity were detected in purified OXPHOS supercomplexes by the addition of straight chain acyl-CoA substrates for VLCAD, LCAD, MCAD and SCAD. Furthermore, when palmitoyl-CoA was added to the sucrose gradient fractions that contained OXPHOS supercomplexes (in conjunction with ETF, NAD and ATP), palmitoyl-CoA was completely metabolized without the accumulation of any FAO intermediates [134]. These findings suggest that FAO–OXPHOS supercomplexes are present in mitochondria and that they are metabolically active structures which can oxidize fatty acids.
COMBINED LCHAD AND OXPHOS DEFECTS
Given the direct physical interactions between FAO and OXPHOS proteins, it is possible that FAO protein defects will affect OXPHOS CI and/or OXPHOS supercomplex respiratory activity. Indeed, patients with LCHAD deficiency frequently exhibit secondary OXPHOS CI deficiencies [127,136]. This is associated with enlarged mitochondria and increased mitochondrial proliferation, which may occur as part of a compensatory mechanism [126,136,137]. LCHAD deficient patients usually present with elevated FAO intermediates and excretion of organic acids in the urine [126], with a combination of FAO/CI clinical phenotypes, including severe hypotonia, developmental delay, seizures and hepatic dysfunction.
As LCHAD interacts with OXPHOS CI and the OXPHOS supercomplex directly [134], it is possible that a primary LCHAD deficiency will disrupt the stability and/or function of CI, resulting in secondary CI defect. Another possible mechanism by which LCHAD deficiency may alter OXPHOS CI stability is via the interaction with cardiolipin.
Cardiolipin has been shown to be vital for OXPHOS complex and supercomplex formation and stability [19–21]. As described previously, mutations in TAZ, which encodes the mitochondrial cardiolipin acyl-transferase, Tafazzin, result in reduced levels of mature tetralinoleoylcardiolipin in the inner mitochondrial membrane, with the subsequent destabilization of OXPHOS CI/CIII2/CIV1–3 supercomplex and monomeric CI [19].
LCHAD also exhibits acyl-CoA acyltransferase activity, and can generate mature tetralinoleoylcardiolipin from its precursor, monolysocardiolipin, in the presence of lineolyl-CoA, oleoyl-CoA and palmitoyl-CoA. Interestingly, the overexpression of LCHAD in Barth Syndrome patient lymphoblasts increases mature cardiolipin production, with an associated increase in the steady-state level of OXPHOS complex subunits [133]. Conversely, knockdown of LCHAD in Barth Syndrome patient lymphoblasts results in the accumulation of monolysocardiolipin [133]. Therefore, LCHAD may be essential for OXPHOS CI/CIII2/CIV1–3 supercomplex and CI monomer stability and assembly via its ability to generate mature cardiolipin [19,133].
COMBINED ECHS1 DEFICIENCY AND OXPHOS COMPLEX DEFECTS
Of the 23 patients described thus far with reported ECHS1 deficiency, 19 have undergone OXPHOS respiratory analysis, with six patients exhibiting detectable OXPHOS enzyme defects. Notably, these OXPHOS deficiencies are not consistent, and vary from isolated CI or CIV deficiencies to combined CI, CIII, CIV or CI, CIV, CV deficiencies (Table 3).
BN-PAGE analysis has been performed using cells from three different ECHS1 patients, again with varied results. Reduced steady-state levels were observed in CI and CIII [123], in CIII and CIV [123], or not at all [121]. Of note, the patients with reduced OXPHOS complex steady-state levels did not exhibit OXPHOS enzymatic defects [123]. However, patients with detectable OXPHOS enzymatic defects typically have a more severe form of Leigh Syndrome than patients with no OXPHOS deficits (Table 3).
In addition to the OXPHOS defects, pyruvate dehydrogenase activity was reduced in all but one of the ECHS1 deficiencies that was fatal [120–125] (Table 3). Interestingly, the pyruvate dehydrogenase complex has been shown to bind to CI in vitro. It has been suggested that this binding may act to couple NADH oxidation/reduction and increase respiratory efficiency [132]. As such, defects in ECHS1 may disrupt pyruvate dehydrogenase activity and/or stability, resulting in a secondary disruption of OXPHOS CI activity/stability. This may explain in part the CI defects observed in some ECHS1 patients. Alternatively, the accumulation of the toxic intermediates methacylyl-CoA and acryloyl-CoA in ECHS1 deficient patients may be responsible for the pathology observed. These molecules can spontaneously react with sulfhydryl groups, potentially disrupting OXPHOS enzyme complex and pyruvate dehydrogenase activities [120].
FAO PROTEINS AND OXPHOS COMPLEX I ASSEMBLY
OXPHOS CI is the largest of the respiratory chain complexes. It is a multimeric complex that is arranged into an L-shape structure. This L-shaped structure is composed of a hydrophilic arm and a hydrophobic arm and is highly conserved from bacteria to eukaryotes [138–140]. The proper assembly of mature CI involves the coordinated assembly of 45 structural subunits, the majority of which are encoded by the nDNA and must be transported into the mitochondria via membrane bound transport systems [55,141,142]. Furthermore, a group of proteins termed ‘assembly factors’ are required to form the final, mature holocomplex [143].
One FAO protein that has been identified as a bona fide CI assembly factor is acyl-CoA dehydrogenase 9 (ACAD9) [54,128]. ACAD9 was initially identified as a homologue of the FAO protein VLCAD, and can compensate in the absence of VLCAD by producing C12 and C14:1 carnitines [128]. However, Nouws et al. [54] showed that ACAD9 also interacts with two CI assembly factors, NDUFAF1 and Ecsit, and that knockdown of ACAD9 results in altered CI biogenesis and isolated CI deficiencies. Complementation with catalytically inactive ACAD9 was able to rescue CI biogenesis and CI activity in two patients with ACAD9 deficiency (although it was less effective than its wild type counterpart in one patient) [54,128], suggesting that ACAD9 activity is not required for complex I assembly [128]. However, studies have shown that mutations in ACAD9 affect both FAO and OXPHOS activities simultaneously, suggesting that ACAD9 plays two important roles as both a fatty acid dehydrogenase and a complex I assembly factor.
In addition to ACAD9, the FAO proteins 3-hydroxyacyl-CoA dehydrogenase (HADH) and enoyl-CoA delta isomerase 1 (ECI1) are predicted by phylogenetic profiling to be involved in CI biogenesis [144]. A list of genes that have co-evolved with genes encoding CI subunits (complex I phylogenetic profile (COPP) gene list) contains both HADH and ECI1, suggesting putative roles for their FAO protein products in CI biogenesis. Interestingly, immunoprecipitation experiments have shown that HADH interacts with the CI subunits NDUFV2 and NDUFS2, further suggesting a role for HADH in CI biogenesis [145]. The COPP gene list also contains genes that have been confirmed experimentally as bona fide CI assembly factors, including FOXRED1 [53], NDUFAF6 [52] and NDUFAF5 [51,144], substantiating the validity of the COPP list in predicting the involvement of HADH and ECI1 in CI biogenesis.
PRIMARY OXPHOS DEFICIENCIES ASSOCIATED WITH SECONDARY FAO DEFECTS
Primary OXPHOS CII deficiencies can result in metabolic disorders associated with secondary defects in FAO, presenting with cardiomyopathy, short stature, lactic acidosis, craniofacial dysmorphic features, hypertrichosis and myopathy [129,130]. Deficiencies in CII have been shown to inhibit FAO, resulting in toxic levels of butyrylcarnitine [129]. It has been suggested that this may be due to disruption of the FAD pool; defects in CII activity will result in FADH2 remaining reduced, resulting in a lack of oxidized FAD to accept electrons from the fatty acyl-CoA dehydrogenases involved in the first step of FAO. However, as FAD does not frequently shuttle between the ACADs and CII, this mechanism may only partially explain the combined OXPHOS CII and FAO defects observed in some patients [129]. Alternatively, physical interactions between CII and enzymes involved in FAO may exist, however this is yet to be shown experimentally. Whichever the case, treatment of patients with combined OXPHOS CII/FAO defects has proved problematic, as supplementation with essential fatty acids results in metabolic crises [129].
CONCLUDING REMARKS
Secondary defects in OXPHOS function, due to primary FAO deficiencies, were initially attributed to reduced co-factor sharing and the build-up of toxic fatty acid intermediates, resulting in the inhibition of OXPHOS complex activities [113,121,123,127,136,146]. Although this may be the case in some FAO disorders, the build-up of toxic intermediates does not sufficiently explain the reduced steady-state levels of OXPHOS complexes observed in patients with mutations in FAO genes [54,123].
However, it has now been shown that FAO proteins interact physically with the OXPHOS complexes [134], and that specific FAO protein defects can result in respiratory chain defects [54,121,127,136,146]. Conversely, primary OXPHOS deficiencies can result in secondary FAO disease [129,130], highlighting the importance of the interactions between the FAO and OXPHOS pathways. Given the large range of metabolic diseases caused by OXPHOS and FAO deficiencies, a detailed understanding of the protein interactions between both pathways will be crucial for our comprehension of the pathogenesis involved and for the development of new therapies for the treatment of mitochondrial disease that will target both the FAO and OXPHOS pathways simultaneously.
Abbreviations
- ACAD9
acyl-CoA dehydrogenase 9
- BN-PAGE
blue native polyacrylamide gel electrophoresis
- CACT
carnitine acylcarnitine translocase
- CI
oxidative phosphorylation complex I (NADH: ubiquinone oxidoreductase, EC 1.6.5.3)
- CII
oxidative phosphorylation complex II (succinate: ubiquinone oxidoreductase, EC 1.3.5.1)
- CIII
oxidative phosphorylation complex III (ubiquinol: ferricytochrome c oxidoreductase, EC 1.10.2.2)
- CIV
oxidative phosphorylation complex IV (cytochrome c oxidase, EC 1.9.3.1)
- COPP
complex one phylogenetic profile
- CPT1
carnitine O-palmitoyltransferase 1
- CPT2
carnitine O-palmitoyltransferase 2
- CV
OXPHOS complex V (FoF1-ATP synthetase, EC; 3.6.3.14)
- ECHS1
enoyl-CoA hydratase, short chain 1, mitochondrial
- ECI1
enoyl-CoA delta isomerase, 1
- ETF
electron transfer flavoprotein
- ETFA
electron transfer flavoprotein alpha subunit
- ETFB
electron transfer flavoprotein beta subunit
- ETF-QO
electron transfer flavoprotein: ubiquinone oxidoreductase
- FAD
flavin adenine dinucleotide
- FADH2
reduced flavin adenine dinucleotide
- FAO
fatty acid β-oxidation
- H2O
water
- HADH
hydroxyacyl-CoA dehydrogenase
- KAT
3-ketoacyl-CoA thiolase, mitochondrial
- LCAD
long chain acyl-CoA dehydrogenase
- LCEH
long chain enoyl-CoA hydratase
- LCHAD
long chain 3-hydroxyacyl-CoA dehydrogenase
- LHON
Leber hereditary optic neuropathy
- LKAT
long chain 3-ketoacyl-CoA thiolase
- MCAD
medium chain acyl-CoA dehydrogenase
- MCEE
methylmalonyl-CoA epimerase
- MCM
methylmalonyl-CoA mutase
- MTP
mitochondrial trifunctional protein
- NARP
neuropathy, ataxia, retinal pigmentosa
- nDNA
nuclear DNA
- NUBPL
nucleotide-binding protein-like
- O2
oxygen
- OXPHOS
oxidative phosphorylation
- PCC
propionyl-CoA carboxylase
- PCCA
propionyl-CoA carboxylase alpha subunit
- PCCB
propionyl-CoA carboxylase beta subunit
- PD
Parkinson's disease
- SBCAD
short/branched chain specific acyl-CoA dehydrogenase, mitochondrial
- SCAD
short chain acyl-CoA dehydrogenase
- ∆ψm
mitochondrial membrane potential
- TCA
tricarboxylic acid
- VLCAD
very long chain acyl-CoA dehydrogenase
FUNDING
This work was supported by the Australian Research Council Future Fellowship scheme [grant number FT120100459 (to M.McK.)]; the Australian Postgraduate Award scheme (to A.N.S.); the William Buckland Foundation (to M.McK.); the Australian Mitochondrial Disease Foundation (to M.McK. and A.N.S.); and the Victorian Government Infrastructure Program.
References
- 1.Nunnari J., Suomalainen A. Mitochondria: in sickness and in health. Cell. 2012;148:1145–1159. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thorburn D.R., Sugiana C., Salemi R., Kirby D.M., Worgan L., Ohtake A., Ryan M.T. Biochemical and molecular diagnosis of mitochondrial respiratory chain disorders. Biochim. Biophys. Acta. 2004;1659:121–128. doi: 10.1016/j.bbabio.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 3.Smeitink J., van den Heuvel L., DiMauro S. The genetics and pathology of oxidative phosphorylation. Nat. Rev. Genet. 2001;2:342–352. doi: 10.1038/35072063. [DOI] [PubMed] [Google Scholar]
- 4.Kompare M., Rizzo W.B. Mitochondrial fatty-acid oxidation disorders. Semin. Pediatr. Neurol. 2008;15:140–149. doi: 10.1016/j.spen.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 5.Dudkina N.V., Kouřil R., Peters K., Braun H.-P., Boekema E.J. Structure and function of mitochondrial supercomplexes. Biochim. Biophys. Acta. 2010;1797:664–670. doi: 10.1016/j.bbabio.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 6.Hackenbrock C.R., Hochli M., Chau R.M. Calorimetric and freeze fracture analysis of lipid phase transitions and lateral translational motion of intramembrane particles in mitochondrial membranes. Biochim. Biophys. Acta. 1976;455:466–484. doi: 10.1016/0005-2736(76)90318-7. [DOI] [PubMed] [Google Scholar]
- 7.Hatefi Y., Rieske J. The preparation and properties of DPNH—cytochrome c reductase (complex I–III of the respiratory chain) Methods Enzymol. 1967;10:225–231. [Google Scholar]
- 8.Ragan C., Heron C. The interaction between mitochondrial NADH-ubiquinone oxidoreductase and ubiquinol-cytochrome c oxidoreductase. Evidence for stoicheiometric association. Biochem. J. 1978;174:783–790. doi: 10.1042/bj1740783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu C.A., Yu L., King T.E. Soluble cytochrome b-c1 complex and the reconstitution of succinate-cytochrome c reductase. J. Biol. Chem. 1974;249:4905–4910. [PubMed] [Google Scholar]
- 10.Berry E.A., Trumpower B.L. Isolation of ubiquinol oxidase from Paracoccus denitrificans and resolution into cytochrome bc1 and cytochrome c-aa3 complexes. J. Biol. Chem. 1985;260:2458–2467. [PubMed] [Google Scholar]
- 11.Iwasaki T., Matsuura K., Oshima T. Resolution of the aerobic respiratory system of the thermoacidophilic archaeon, Sulfolobus sp. strain 7: I. The archaeal terminal oxidase supercomplex is a functional fusion of respiratory complexes III and IV with no c-type cytochromes. J. Biol. Chem. 1995;270:30881–30892. doi: 10.1074/jbc.270.52.30881. [DOI] [PubMed] [Google Scholar]
- 12.Sone N., Sekimachi M., Kutoh E. Identification and properties of a quinol oxidase super-complex composed of a bc1 complex and cytochrome oxidase in the thermophilic bacterium PS3. J. Biol. Chem. 1987;262:15386–15391. [PubMed] [Google Scholar]
- 13.Eubel H., Heinemeyer J., Braun H.-P. Identification and characterization of respirasomes in potato mitochondria. Plant Physiol. 2004;134:1450–1459. doi: 10.1104/pp.103.038018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eubel H., Jansch L., Braun H.P. New insights into the respiratory chain of plant mitochondria. Supercomplexes and a unique composition of complex II1. Plant Physiol. 2003;133:274–286. doi: 10.1104/pp.103.024620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Minauro-Sanmiguel F., Wilkens S., Garcia J.J. Structure of dimeric mitochondrial ATP synthase: novel F0 bridging features and the structural basis of mitochondrial cristae biogenesis. Proc. Natl. Acad. Sci. U.S.A. 2005;102:12356–12358. doi: 10.1073/pnas.0503893102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schagger H., Pfeiffer K. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 2000;19:1777–1783. doi: 10.1093/emboj/19.8.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Acin-Perez R., Bayona-Bafaluy M.P., Fernandez-Silva P., Moreno-Loshuertos R., Perez-Martos A., Bruno C., Moraes C.T., Enriquez J.A. Respiratory complex III is required to maintain complex I in mammalian mitochondria. Mol. Cell. 2004;13:805–815. doi: 10.1016/s1097-2765(04)00124-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diaz F., Fukui H., Garcia S., Moraes C.T. Cytochrome c oxidase is required for the assembly/stability of respiratory complex I in mouse fibroblasts. Mol. Cell Biol. 2006;26:4872–4881. doi: 10.1128/MCB.01767-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKenzie M., Lazarou M., Thorburn D.R., Ryan M.T. Mitochondrial respiratory chain supercomplexes are destabilized in Barth Syndrome patients. J. Mol. Biol. 2006;361:462–469. doi: 10.1016/j.jmb.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 20.Eble K.S., Coleman W.B., Hantgan R.R., Cunningham C.C. Tightly associated cardiolipin in the bovine heart mitochondrial ATP synthase as analyzed by 31P nuclear magnetic resonance spectroscopy. J. Biol. Chem. 1990;265:19434–19440. [PubMed] [Google Scholar]
- 21.Gomez B., Robinson N.C. Phospholipase digestion of bound cardiolipin reversibly inactivates bovine cytochrome bc1. Biochemistry. 1999;38:9031–9038. doi: 10.1021/bi990603r. [DOI] [PubMed] [Google Scholar]
- 22.Barth P.G., Scholte H.R., Berden J.A., Van der Klei-Van Moorsel J.M., Luyt-Houwen I.E., Van ’t Veer-Korthof E.T., Van der Harten J.J., Sobotka-Plojhar M.A. An X-linked mitochondrial disease affecting cardiac muscle, skeletal muscle and neutrophil leucocytes. J. Neurol. Sci. 1983;62:327–355. doi: 10.1016/0022-510x(83)90209-5. [DOI] [PubMed] [Google Scholar]
- 23.Barth P.G., Van den Bogert C., Bolhuis P.A., Scholte H.R., van Gennip A.H., Schutgens R.B.H., Ketel A.G. X-linked cardioskeletal myopathy and neutropenia (Barth syndrome): Respiratory-chain abnormalities in cultured fibroblasts. J. Inherit. Metab. Dis. 1996;19:157–160. doi: 10.1007/BF01799418. [DOI] [PubMed] [Google Scholar]
- 24.Boumans H., Grivell L.A., Berden J.A. The respiratory chain in yeast behaves as a single functional unit. J. Biol. Chem. 1998;273:4872–4877. doi: 10.1074/jbc.273.9.4872. [DOI] [PubMed] [Google Scholar]
- 25.Lapuente-Brun E., Moreno-Loshuertos R., Acin-Perez R., Latorre-Pellicer A., Colas C., Balsa E., Perales-Clemente E., Quiros P.M., Calvo E., Rodriguez-Hernandez M.A., et al. Supercomplex assembly determines electron flux in the mitochondrial electron transport chain. Science. 2013;340:1567–1570. doi: 10.1126/science.1230381. [DOI] [PubMed] [Google Scholar]
- 26.Maranzana E., Barbero G., Falasca A.I., Lenaz G., Genova M.L. Mitochondrial respiratory supercomplex association limits production of reactive oxygen species from complex I. Antioxid. Redox Signal. 2013;19:1469–1480. doi: 10.1089/ars.2012.4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lenaz G., Baracca A., Barbero G., Bergamini C., Dalmonte M.E., Del Sole M., Faccioli M., Falasca A., Fato R., Genova M.L., et al. Mitochondrial respiratory chain super-complex I–III in physiology and pathology. Biochim. Biophys. Acta. 2010;1797:633–640. doi: 10.1016/j.bbabio.2010.01.025. [DOI] [PubMed] [Google Scholar]
- 28.Genova M.L., Bianchi C., Lenaz G. Supercomplex organization of the mitochondrial respiratory chain and the role of the Coenzyme Q pool: pathophysiological implications. BioFactors. 2005;25:5–20. doi: 10.1002/biof.5520250103. [DOI] [PubMed] [Google Scholar]
- 29.Blaza J.N., Serreli R., Jones A.J.Y., Mohammed K., Hirst J. Kinetic evidence against partitioning of the ubiquinone pool and the catalytic relevance of respiratory-chain supercomplexes. Proc. Natl. Acad. Sci. U.S.A. 2014;111:15735–15740. doi: 10.1073/pnas.1413855111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dudkina N.V., Kudryashev M., Stahlberg H., Boekema E.J. Interaction of complexes I, III, and IV within the bovine respirasome by single particle cryoelectron tomography. Proc. Natl. Acad. Sci. U.S.A. 2011;108:15196–15200. doi: 10.1073/pnas.1107819108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Galli C., Marangoni F. Recent advances in the biology of n-6 fatty acids. Nutrition. 1997;13:978–985. doi: 10.1016/s0899-9007(97)00341-9. [DOI] [PubMed] [Google Scholar]
- 32.Yamashita A., Sugiura T., Waku K. Acyltransferases and transacylases involved in fatty acid remodeling of phospholipids and metabolism of bioactive lipids in mammalian cells. J. Biochem. 1997;122:1–16. doi: 10.1093/oxfordjournals.jbchem.a021715. [DOI] [PubMed] [Google Scholar]
- 33.van der Vusse G.J., Glatz J.F., Stam H.C., Reneman R.S. Fatty acid homeostasis in the normoxic and ischemic heart. Physiol. Rev. 1992;72:881–940. doi: 10.1152/physrev.1992.72.4.881. [DOI] [PubMed] [Google Scholar]
- 34.Jansson E., Kaijser L. Substrate utilization and enzymes in skeletal muscle of extremely endurance-trained men. J. Appl. Physiol. 1987;62:999–1005. doi: 10.1152/jappl.1987.62.3.999. [DOI] [PubMed] [Google Scholar]
- 35.Kuhajda F.P. In: Encyclopedia of Biological Chemistry. Lane W.J.L.D., editor. Waltham: Academic Press; 2013. pp. 275–280. [Google Scholar]
- 36.Houten S.M., Herrema H., Te Brinke H., Denis S., Ruiter J.P., van Dijk T.H., Argmann C.A., Ottenhoff R., Muller M., Groen A.K., et al. Impaired amino acid metabolism contributes to fasting-induced hypoglycemia in fatty acid oxidation defects. Hum. Mol. Genet. 2013;22:5249–5261. doi: 10.1093/hmg/ddt382. [DOI] [PubMed] [Google Scholar]
- 37.Houten S.M., Violante S., Ventura F.V., Wanders R.J. The biochemistry and physiology of mitochondrial fatty acid beta-oxidation and its genetic disorders. Annu. Rev. Physiol. 2016;78:23–44. doi: 10.1146/annurev-physiol-021115-105045. [DOI] [PubMed] [Google Scholar]
- 38.Hirsch D., Stahl A., Lodish H.F. A family of fatty acid transporters conserved from mycobacterium to man. Proc. Natl. Acad. Sci. U.S.A. 1998;95:8625–8629. doi: 10.1073/pnas.95.15.8625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohtake A., Murayama K., Mori M., Harashima H., Yamazaki T., Tamaru S., Yamashita Y., Kishita Y., Nakachi Y., Kohda M., et al. Diagnosis and molecular basis of mitochondrial respiratory chain disorders: exome sequencing for disease gene identification. Biochim. Biophys. Acta. 2014;1840:1355–1359. doi: 10.1016/j.bbagen.2014.01.025. [DOI] [PubMed] [Google Scholar]
- 40.Man P.Y., Turnbull D.M., Chinnery P.F. Leber hereditary optic neuropathy. J. Med. Genet. 2002;39:162–169. doi: 10.1136/jmg.39.3.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taylor R.W., Turnbull D.M. Mitochondrial DNA mutations in human disease. Nat. Rev. Genet. 2005;6:389–402. doi: 10.1038/nrg1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jun A.S., Brown M.D., Wallace D.C. A mitochondrial DNA mutation at nucleotide pair 14459 of the NADH dehydrogenase subunit 6 gene associated with maternally inherited Leber hereditary optic neuropathy and dystonia. Proc. Natl. Acad. Sci. U.S.A. 1994;91:6206–6210. doi: 10.1073/pnas.91.13.6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shoffner J.M., Brown M.D., Stugard C., Jun A.S., Pollock S., Haas R.H., Kaufman A., Koontz D., Kim Y., Graham J.R., et al. Leber's hereditary optic neuropathy plus dystonia is caused by a mitochondrial DNA point mutation. Ann. Neurol. 1995;38:163–169. doi: 10.1002/ana.410380207. [DOI] [PubMed] [Google Scholar]
- 44.Fassone E., Rahman S. Complex I deficiency: clinical features, biochemistry and molecular genetics. J. Med. Genet. 2012;49:578–590. doi: 10.1136/jmedgenet-2012-101159. [DOI] [PubMed] [Google Scholar]
- 45.Bugiani M., Invernizzi F., Alberio S., Briem E., Lamantea E., Carrara F., Moroni I., Farina L., Spada M., Donati M.A., et al. Clinical and molecular findings in children with complex I deficiency. Biochim. Biophys. Acta. 2004;1659:136–147. doi: 10.1016/j.bbabio.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 46.Andrews B., Carroll J., Ding S., Fearnley I.M., Walker J.E. Assembly factors for the membrane arm of human complex I. Proc. Natl. Acad. Sci. 2013;110:18934–18939. doi: 10.1073/pnas.1319247110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ogilvie I., Kennaway N.G., Shoubridge E.A. A molecular chaperone for mitochondrial complex I assembly is mutated in a progressive encephalopathy. J. Clin. Invest. 2005;115:2784–2792. doi: 10.1172/JCI26020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dunning C.J., McKenzie M., Sugiana C., Lazarou M., Silke J., Connelly A., Fletcher J.M., Kirby D.M., Thorburn D.R., Ryan M.T. Human CIA30 is involved in the early assembly of mitochondrial complex I and mutations in its gene cause disease. EMBO J. 2007;26:3227–3237. doi: 10.1038/sj.emboj.7601748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saada A., Vogel R.O., Hoefs S.J., van den Brand M.A., Wessels H.J., Willems P.H., Venselaar H., Shaag A., Barghuti F., Reish O., et al. Mutations in NDUFAF3 (C3ORF60), encoding an NDUFAF4 (C6ORF66)-interacting complex I assembly protein, cause fatal neonatal mitochondrial disease. Am. J. Hum. Genet. 2009;84:718–727. doi: 10.1016/j.ajhg.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saada A., Edvardson S., Rapoport M., Shaag A., Amry K., Miller C., Lorberboum-Galski H., Elpeleg O. C6ORF66 is an assembly factor of mitochondrial complex I. Am. J. Hum. Genet. 2008;82:32–38. doi: 10.1016/j.ajhg.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sugiana C., Pagliarini D.J., McKenzie M., Kirby D.M., Salemi R., Abu-Amero K.K., Dahl H.H., Hutchison W.M., Vascotto K.A., Smith S.M., et al. Mutation of C20orf7 disrupts complex I assembly and causes lethal neonatal mitochondrial disease. Am. J. Hum. Genet. 2008;83:468–478. doi: 10.1016/j.ajhg.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McKenzie M., Tucker E.J., Compton A.G., Lazarou M., George C., Thorburn D.R., Ryan M.T. Mutations in the gene encoding C8orf38 block complex I assembly by inhibiting production of the mitochondria-encoded subunit ND1. J. Mol. Biol. 2011;414:413–426. doi: 10.1016/j.jmb.2011.10.012. [DOI] [PubMed] [Google Scholar]
- 53.Calvo S.E., Tucker E.J., Compton A.G., Kirby D.M., Crawford G., Burtt N.P., Rivas M., Guiducci C., Bruno D.L., Goldberger O.A., et al. High-throughput, pooled sequencing identifies mutations in NUBPL and FOXRED1 in human complex I deficiency. Nat. Genet. 2010;42:851–858. doi: 10.1038/ng.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nouws J., Nijtmans L., Houten S.M., van den Brand M., Huynen M., Venselaar H., Hoefs S., Gloerich J., Kronick J., Hutchin T., et al. Acyl-CoA dehydrogenase 9 is required for the biogenesis of oxidative phosphorylation complex I. Cell Metab. 2010;12:283–294. doi: 10.1016/j.cmet.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 55.Mimaki M., Wang X., McKenzie M., Thorburn D.R., Ryan M.T. Understanding mitochondrial complex I assembly in health and disease. Biochim. Biophys. Acta. 2012;1817:851–862. doi: 10.1016/j.bbabio.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 56.Bénit P., Chretien D., Kadhom N., de Lonlay-Debeney P., Cormier-Daire V., Cabral A., Peudenier S., Rustin P., Munnich A., Rötig A. Large-scale deletion and point mutations of the nuclear NDUFV1 and NDUFS1 genes in mitochondrial complex I deficiency. Am. J. Hum. Genet. 2001;68:1344–1352. doi: 10.1086/320603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kevelam S.H., Rodenburg R.J., Wolf N.I., Ferreira P., Lunsing R.J., Nijtmans L.G., Mitchell A., Arroyo H.A., Rating D., Vanderver A., et al. NUBPL mutations in patients with complex I deficiency and a distinct MRI pattern. Neurology. 2013;80:1577–1583. doi: 10.1212/WNL.0b013e31828f1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McFarland R., Kirby D.M., Fowler K.J., Ohtake A., Ryan M.T., Amor D.J., Fletcher J.M., Dixon J.W., Collins F.A., Turnbull D.M., et al. De novo mutations in the mitochondrial ND3 gene as a cause of infantile mitochondrial encephalopathy and complex I deficiency. Ann. Neurol. 2004;55:58–64. doi: 10.1002/ana.10787. [DOI] [PubMed] [Google Scholar]
- 59.Komen J.C., Thorburn D.R. Turn up the power–pharmacological activation of mitochondrial biogenesis in mouse models. Br. J. Pharmacol. 2014;171:1818–1836. doi: 10.1111/bph.12413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rotig A., Munnich A. Genetic features of mitochondrial respiratory chain disorders. J. Am. Soc. Nephrol. 2003;14:2995–3007. doi: 10.1097/01.asn.0000095481.24091.c9. [DOI] [PubMed] [Google Scholar]
- 61.Moggio M., Colombo I., Peverelli L., Villa L., Xhani R., Testolin S., Di Mauro S., Sciacco M. Mitochondrial disease heterogeneity: a prognostic challenge. Acta Myol. 2014;33:86–93. [PMC free article] [PubMed] [Google Scholar]
- 62.Schapira A.H.V., Cooper J.M., Dexter D., Clark J.B., Jenner P., Marsden C.D. Mitochondrial complex I deficiency in Parkinson's disease. J. Neurochem. 1990;54:823–827. doi: 10.1111/j.1471-4159.1990.tb02325.x. [DOI] [PubMed] [Google Scholar]
- 63.Keeney P.M., Xie J., Capaldi R.A., Bennett J.P., Jr Parkinson's disease brain mitochondrial complex I has oxidatively damaged subunits and is functionally impaired and misassembled. J. Neurosci. 2006;26:5256–5264. doi: 10.1523/JNEUROSCI.0984-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Greenamyre J.T., Sherer T.B., Betarbet R., Panov A.V. Complex I and Parkinson's disease. IUBMB Life. 2001;52:135–141. doi: 10.1080/15216540152845939. [DOI] [PubMed] [Google Scholar]
- 65.Lin M.T., Cantuti-Castelvetri I., Zheng K., Jackson K.E., Tan Y.B., Arzberger T., Lees A.J., Betensky R.A., Beal M.F., Simon D.K. Somatic mitochondrial DNA mutations in early Parkinson and incidental Lewy body disease. Ann. Neurol. 2012;71:850–854. doi: 10.1002/ana.23568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rivner M.H., Shamsnia M., Swift T.R., Trefz J., Roesel R.A., Carter A.L., Yanamura W., Hommes F.A. Kearns-Sayre syndrome and complex II deficiency. Neurology. 1989;39:693–696. doi: 10.1212/wnl.39.5.693. [DOI] [PubMed] [Google Scholar]
- 67.Ghezzi D., Goffrini P., Uziel G., Horvath R., Klopstock T., Lochmuller H., D'Adamo P., Gasparini P., Strom T.M., Prokisch H., et al. SDHAF1, encoding a LYR complex-II specific assembly factor, is mutated in SDH-defective infantile leukoencephalopathy. Nat. Genet. 2009;41:654–656. doi: 10.1038/ng.378. [DOI] [PubMed] [Google Scholar]
- 68.Bugiani M., Lamantea E., Invernizzi F., Moroni I., Bizzi A., Zeviani M., Uziel G. Effects of riboflavin in children with complex II deficiency. Brain Dev. 2006;28:576–581. doi: 10.1016/j.braindev.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 69.Bourgeron T., Rustin P., Chretien D., Birch-Machin M., Bourgeois M., Viegas-Pequignot E., Munnich A., Rotig A. Mutation of a nuclear succinate dehydrogenase gene results in mitochondrial respiratory chain deficiency. Nat. Genet. 1995;11:144–149. doi: 10.1038/ng1095-144. [DOI] [PubMed] [Google Scholar]
- 70.Alston C., Ceccatelli Berti C., Blakely E., Oláhová M., He L., McMahon C., Olpin S., Hargreaves I., Nolli C., McFarland R., et al. A recessive homozygous p.Asp92Gly SDHD mutation causes prenatal cardiomyopathy and a severe mitochondrial complex II deficiency. Hum. Genet. 2015;134:869–879. doi: 10.1007/s00439-015-1568-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Birch-Machin M.A., Taylor R.W., Cochran B., Ackrell B.A., Turnbull D.M. Late-onset optic atrophy, ataxia, and myopathy associated with a mutation of a complex II gene. Ann. Neurol. 2000;48:330–335. [PubMed] [Google Scholar]
- 72.Burnichon N., Briere J.J., Libe R., Vescovo L., Riviere J., Tissier F., Jouanno E., Jeunemaitre X., Benit P., Tzagoloff A., et al. SDHA is a tumor suppressor gene causing paraganglioma. Hum. Mol. Genet. 2010;19:3011–3020. doi: 10.1093/hmg/ddq206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schimke R.N., Collins D.L., Stolle C.A. Paraganglioma, neuroblastoma, and a SDHB mutation: Resolution of a 30-year-old mystery. Am. J. Med. Genet. Part A. 2010;152A:1531–1535. doi: 10.1002/ajmg.a.33384. [DOI] [PubMed] [Google Scholar]
- 74.Schiavi F., Boedeker C.C., Bausch B., Peczkowska M., Gomez C.F., Strassburg T., Pawlu C., Buchta M., Salzmann M., Hoffmann M.M., et al. Predictors and prevalence of paraganglioma syndrome associated with mutations of the SDHC gene. JAMA. 2005;294:2057–2063. doi: 10.1001/jama.294.16.2057. [DOI] [PubMed] [Google Scholar]
- 75.Cascon A., Ruiz-Llorente S., Cebrian A., Telleria D., Rivero J.C., Diez J.J., Lopez-Ibarra P.J., Jaunsolo M.A., Benitez J., Robledo M. Identification of novel SDHD mutations in patients with phaeochromocytoma and/or paraganglioma. Eur. J. Hum. Genet. 2002;10:457–461. doi: 10.1038/sj.ejhg.5200829. [DOI] [PubMed] [Google Scholar]
- 76.Ohlenbusch A., Edvardson S., Skorpen J., Bjornstad A., Saada A., Elpeleg O., Gartner J., Brockmann K. Leukoencephalopathy with accumulated succinate is indicative of SDHAF1 related complex II deficiency. Orphanet J. Rare Dis. 2012;7:69. doi: 10.1186/1750-1172-7-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bayley J.P., Kunst H.P., Cascon A., Sampietro M.L., Gaal J., Korpershoek E., Hinojar-Gutierrez A., Timmers H.J., Hoefsloot L.H., Hermsen M.A., et al. SDHAF2 mutations in familial and sporadic paraganglioma and phaeochromocytoma. Lancet Oncol. 2010;11:366–372. doi: 10.1016/S1470-2045(10)70007-3. [DOI] [PubMed] [Google Scholar]
- 78.Baysal B.E., Ferrell R.E., Willett-Brozick J.E., Lawrence E.C., Myssiorek D., Bosch A., van der Mey A., Taschner P.E., Rubinstein W.S., Myers E.N., et al. Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science. 2000;287:848–851. doi: 10.1126/science.287.5454.848. [DOI] [PubMed] [Google Scholar]
- 79.Bénit P., Lebon S., Rustin P. Respiratory-chain diseases related to complex III deficiency. Biochim. Biophys. Acta. 2009;1793:181–185. doi: 10.1016/j.bbamcr.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 80.Brown M.D., Voljavec A.S., Lott M.T., Torroni A., Yang C.C., Wallace D.C. Mitochondrial DNA complex I and III mutations associated with Leber's hereditary optic neuropathy. Genetics. 1992;130:163–173. doi: 10.1093/genetics/130.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.de Lonlay P., Valnot I., Barrientos A., Gorbatyuk M., Tzagoloff A., Taanman J.W., Benayoun E., Chretien D., Kadhom N., Lombes A., et al. A mutant mitochondrial respiratory chain assembly protein causes complex III deficiency in patients with tubulopathy, encephalopathy and liver failure. Nat. Genet. 2001;29:57–60. doi: 10.1038/ng706. [DOI] [PubMed] [Google Scholar]
- 82.Nogueira C., Barros J., Sa M.J., Azevedo L., Taipa R., Torraco A., Meschini M.C., Verrigni D., Nesti C., Rizza T., et al. Novel TTC19 mutation in a family with severe psychiatric manifestations and complex III deficiency. Neurogenetics. 2013;14:153–160. doi: 10.1007/s10048-013-0361-1. [DOI] [PubMed] [Google Scholar]
- 83.Marin-Garcia J., Hu Y., Ananthakrishnan R., Pierpont M.E., Pierpont G.L., Goldenthal M.J. A point mutation in the cytb gene of cardiac mtDNA associated with complex III deficiency in ischemic cardiomyopathy. Biochem. Mol. Biol. Int. 1996;40:487–495. doi: 10.1080/15216549600201053. [DOI] [PubMed] [Google Scholar]
- 84.Hinson J.T., Fantin V.R., Schönberger J., Breivik N., Siem G., McDonough B., Sharma P., Keogh I., Godinho R., Santos F., et al. Missense mutations in the BCS1L gene as a cause of the Björnstad syndrome. N. Engl. J. Med. 2007;356:809–819. doi: 10.1056/NEJMoa055262. [DOI] [PubMed] [Google Scholar]
- 85.Ghezzi D., Arzuffi P., Zordan M., Da Re C., Lamperti C., Benna C., D'Adamo P., Diodato D., Costa R., Mariotti C., et al. Mutations in TTC19 cause mitochondrial complex III deficiency and neurological impairment in humans and flies. Nat. Genet. 2011;43:259–263. doi: 10.1038/ng.761. [DOI] [PubMed] [Google Scholar]
- 86.Van Biervliet J.P., Bruinvis L., Ketting D., De Bree P.K., Van der Heiden C., Wadman S.K. Hereditary mitochondrial myopathy with lactic acidemia, a De Toni-Fanconi-Debre syndrome, and a defective respiratory chain in voluntary striated muscles. Pediatr. Res. 1977;11:1088–1093. doi: 10.1203/00006450-197711100-00005. [DOI] [PubMed] [Google Scholar]
- 87.van Bon B.W., Oortveld M.A., Nijtmans L.G., Fenckova M., Nijhof B., Besseling J., Vos M., Kramer J.M., de Leeuw N., Castells-Nobau A., et al. CEP89 is required for mitochondrial metabolism and neuronal function in man and fly. Hum. Mol. Genet. 2013;22:3138–3151. doi: 10.1093/hmg/ddt170. [DOI] [PubMed] [Google Scholar]
- 88.Massa V., Fernandez-Vizarra E., Alshahwan S., Bakhsh E., Goffrini P., Ferrero I., Mereghetti P., D'Adamo P., Gasparini P., Zeviani M. Severe infantile encephalomyopathy caused by a mutation in COX6B1, a nucleus-encoded subunit of cytochrome c oxidase. Am. J. Hum. Genet. 2008;82:1281–1289. doi: 10.1016/j.ajhg.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lim S.C., Smith K.R., Stroud D.A., Compton A.G., Tucker E.J., Dasvarma A., Gandolfo L.C., Marum J.E., McKenzie M., Peters H.L., et al. A founder mutation in PET100 causes isolated complex IV deficiency in Lebanese individuals with Leigh syndrome. Am. J. Hum. Genet. 2014;94:209–222. doi: 10.1016/j.ajhg.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jonckheere A.I., Smeitink J.A.M., Rodenburg R.J.T. Mitochondrial ATP synthase: architecture, function and pathology. J. Inherit. Metab. Dis. 2012;35:211–225. doi: 10.1007/s10545-011-9382-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rodenburg R.J. Biochemical diagnosis of mitochondrial disorders. J. Inherit. Metab. Dis. 2011;34:283–292. doi: 10.1007/s10545-010-9081-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Holt I.J., Harding A.E., Petty R.K., Morgan-Hughes J.A. A new mitochondrial disease associated with mitochondrial DNA heteroplasmy. Am. J. Hum. Genet. 1990;46:428–433. [PMC free article] [PubMed] [Google Scholar]
- 93.D'Aurelio M., Vives-Bauza C., Davidson M.M., Manfredi G. Mitochondrial DNA background modifies the bioenergetics of NARP/MILS ATP6 mutant cells. Hum. Mol. Genet. 2010;19:374–386. doi: 10.1093/hmg/ddp503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Spiegel R., Khayat M., Shalev S.A., Horovitz Y., Mandel H., Hershkovitz E., Barghuti F., Shaag A., Saada A., Korman S.H., et al. TMEM70 mutations are a common cause of nuclear encoded ATP synthase assembly defect: further delineation of a new syndrome. J. Med. Genet. 2011;48:177–182. doi: 10.1136/jmg.2010.084608. [DOI] [PubMed] [Google Scholar]
- 95.De Meirleir L., Seneca S., Lissens W., De Clercq I., Eyskens F., Gerlo E., Smet J., Van Coster R. Respiratory chain complex V deficiency due to a mutation in the assembly gene ATP12. J. Med. Genet. 2004;41:120–124. doi: 10.1136/jmg.2003.012047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ionasescu V., Hug G., Hoppel C. Combined partial deficiency of muscle carnitine palmitoyltransferase and carnitine with autosomal dominant inheritance. J. Neurol. Neurosurg. Psychiatry. 1980;43:679–682. doi: 10.1136/jnnp.43.8.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hale D.E., Batshaw M.L., Coates P.M., Frerman F.E., Goodman S.I., Singh I., Stanley C.A. Long-chain acyl coenzyme A dehydrogenase deficiency: an inherited cause of nonketotic hypoglycemia. Pediatr. Res. 1985;19:666–671. doi: 10.1203/00006450-198507000-00006. [DOI] [PubMed] [Google Scholar]
- 98.Aoyama T., Uchida Y., Kelley R.I., Marble M., Hofman K., Tonsgard J.H., Rhead W.J., Hashimoto T. A novel disease with deficiency of mitochondrial very-long-chain acyl-CoA dehydrogenase. Biochem. Biophys. Res. Commun. 1993;191:1369–1372. doi: 10.1006/bbrc.1993.1368. [DOI] [PubMed] [Google Scholar]
- 99.Yang Z., Zhao Y., Bennett M.J., Strauss A.W., Ibdah J.A. Fetal genotypes and pregnancy outcomes in 35 families with mitochondrial trifunctional protein mutations. Am. J. Obstet. Gynecol. 2002;187:715–720. doi: 10.1067/mob.2002.125893. [DOI] [PubMed] [Google Scholar]
- 100.Ibdah J.A., Bennett M.J., Rinaldo P., Zhao Y., Gibson B., Sims H.F., Strauss A.W. A fetal fatty-acid oxidation disorder as a cause of liver disease in pregnant women. N. Engl. J. Med. 1999;340:1723–1731. doi: 10.1056/NEJM199906033402204. [DOI] [PubMed] [Google Scholar]
- 101.Vockley J., Whiteman D.A.H. Defects of mitochondrial β-oxidation: a growing group of disorders. Neuromuscul. Disord. 2002;12:235–246. doi: 10.1016/s0960-8966(01)00308-x. [DOI] [PubMed] [Google Scholar]
- 102.DiMauro S., DiMauro P.M. Muscle carnitine palmityltransferase deficiency and myoglobinuria. Science. 1973;182:929–931. doi: 10.1126/science.182.4115.929. [DOI] [PubMed] [Google Scholar]
- 103.Scalais E., Bottu J., Wanders R.J.A., Ferdinandusse S., Waterham H.R., De Meirleir L. Familial very long chain acyl-CoA dehydrogenase deficiency as a cause of neonatal sudden infant death: Improved survival by prompt diagnosis. Am. J. Med. Genet. 2015;167:211–214. doi: 10.1002/ajmg.a.36803. [DOI] [PubMed] [Google Scholar]
- 104.Wilcken B., Wiley V., Hammond J., Carpenter K. Screening newborns for inborn errors of metabolism by tandem mass spectrometry. N. Engl. J. Med. 2003;348:2304–2312. doi: 10.1056/NEJMoa025225. [DOI] [PubMed] [Google Scholar]
- 105.Stoler J.M., Sabry M.A., Hanley C., Hoppel C.L., Shih V.E. Successful long-term treatment of hepatic carnitine palmitoyltransferase I deficiency and a novel mutation. J. Inherit. Metab. Dis. 2004;27:679–684. doi: 10.1023/b:boli.0000042979.42120.55. [DOI] [PubMed] [Google Scholar]
- 106.Spiekerkoetter U., Lindner M., Santer R., Grotzke M., Baumgartner M.R., Boehles H., Das A., Haase C., Hennermann J.B., Karall D., et al. Treatment recommendations in long-chain fatty acid oxidation defects: consensus from a workshop. J. Inherit. Metab. Dis. 2009;32:498–505. doi: 10.1007/s10545-009-1126-8. [DOI] [PubMed] [Google Scholar]
- 107.Grosse S.D., Khoury M.J., Greene C.L., Crider K.S., Pollitt R.J. The epidemiology of medium chain acyl-CoA dehydrogenase deficiency: an update. Genet. Med. 2006;8:205–212. doi: 10.1097/01.gim.0000204472.25153.8d. [DOI] [PubMed] [Google Scholar]
- 108.Blakemore A.I., Singleton H., Pollitt R.J., Engel P.C., Kolvraa S., Gregersen N., Curtis D. Frequency of the G985 MCAD mutation in the general population. Lancet. 1991;337:298–299. doi: 10.1016/0140-6736(91)90907-7. [DOI] [PubMed] [Google Scholar]
- 109.Andresen B.S., Bross P., Udvari S., Kirk J., Gray G., Kmoch S., Chamoles N., Knudsen I., Winter V., Wilcken B., et al. The molecular basis of medium-chain acyl-CoA dehydrogenase (MCAD) deficiency in compound heterozygous patients: is there correlation between genotype and phenotype? Hum. Mol. Genet. 1997;6:695–707. doi: 10.1093/hmg/6.5.695. [DOI] [PubMed] [Google Scholar]
- 110.Millington D., Koeberl D. Metabolic screening in the newborn. Growth Genet. Horm. 2003;19:33–38. [Google Scholar]
- 111.Andresen B.S., Olpin S., Poorthuis B.J.H.M., Scholte H.R., Vianey-Saban C., Wanders R., Ijlst L., Morris A., Pourfarzam M., Bartlett K., et al. Clear correlation of genotype with disease phenotype in very-long-chain Acyl-CoA dehydrogenase deficiency. Am. J. Hum. Genet. 1999;64:479–494. doi: 10.1086/302261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Doi T., Abo W., Tateno M., Hayashi K., Hori T., Nakada T., Fukao T., Takahashi Y., Terada N. Milder childhood form of very long-chain acyl-CoA dehydrogenase deficiency in a 6-year-old Japanese boy. Eur. J. Pediatr. 2000;159:908–911. doi: 10.1007/pl00008368. [DOI] [PubMed] [Google Scholar]
- 113.Tyni T., Palotie A., Viinikka L., Valanne L., Salo M.K., von Döbeln U., Jackson S., Wanders R., Venizelos N., Pihko H. Long-chain 3-hydroxyacyl–coenzyme A dehydrogenase deficiency with the G1528C mutation: clinical presentation of thirteen patients. J. Pediatr. 1997;130:67–76. doi: 10.1016/s0022-3476(97)70312-3. [DOI] [PubMed] [Google Scholar]
- 114.Spiekerkoetter U., Sun B., Khuchua Z., Bennett M.J., Strauss A.W. Molecular and phenotypic heterogeneity in mitochondrial trifunctional protein deficiency due to beta-subunit mutations. Hum. Mutat. 2003;21:598–607. doi: 10.1002/humu.10211. [DOI] [PubMed] [Google Scholar]
- 115.Spierkerkoetter U., Khuchua Z., Yue Z., Strauss A.W. The early-onset phenotype of mitochondrial trifunctional protein deficiency: a lethal disorder with multiple tissue involvement. J. Inherit. Metab. Dis. 2004;27:294–296. doi: 10.1023/b:boli.0000028839.57386.88. [DOI] [PubMed] [Google Scholar]
- 116.Corydon M.J., Vockley J., Rinaldo P., Rhead W.J., Kjeldsen M., Winter V., Riggs C., Babovic-Vuksanovic D., Smeitink J., De Jong J., et al. Role of common gene variations in the molecular pathogenesis of short-chain acyl-CoA dehydrogenase deficiency. Pediatr. Res. 2001;49:18–23. doi: 10.1203/00006450-200101000-00008. [DOI] [PubMed] [Google Scholar]
- 117.Gregersen N., Winter V.S., Corydon M.J., Corydon T.J., Rinaldo P., Ribes A., Martinez G., Bennett M.J., Vianey-Saban C., Bhala A., et al. Identification of four new mutations in the short-chain acyl-CoA dehydrogenase (SCAD) gene in two patients: one of the variant alleles, 511C–>T, is present at an unexpectedly high frequency in the general population, as was the case for 625G–>A, together conferring susceptibility to ethylmalonic aciduria. Hum. Mol. Genet. 1998;7:619–627. doi: 10.1093/hmg/7.4.619. [DOI] [PubMed] [Google Scholar]
- 118.Gregersen N., Andresen B.S., Corydon M.J., Corydon T.J., Olsen R.K., Bolund L., Bross P. Mutation analysis in mitochondrial fatty acid oxidation defects: Exemplified by acyl-CoA dehydrogenase deficiencies, with special focus on genotype-phenotype relationship. Hum. Mutat. 2001;18:169–189. doi: 10.1002/humu.1174. [DOI] [PubMed] [Google Scholar]
- 119.Wolfe L., Jethva R., Oglesbee D., Vockley J. In: GeneReviews(R) Pagon R.A., Adam M.P., Ardinger H.H., Wallace S.E., Amemiya A., Bean L.J.H., Bird T.D., Fong C.T., Mefford H.C., Smith R.J.H., et al., editors. Seattle: University of Washington; 1993. [PubMed] [Google Scholar]
- 120.Peters H., Buck N., Wanders R., Ruiter J., Waterham H., Koster J., Yaplito-Lee J., Ferdinandusse S., Pitt J. ECHS1 mutations in Leigh disease: a new inborn error of metabolism affecting valine metabolism. Brain. 2014;137:2903–2908. doi: 10.1093/brain/awu216. [DOI] [PubMed] [Google Scholar]
- 121.Sakai C., Yamaguchi S., Sasaki M., Miyamoto Y., Matsushima Y., Goto Y. ECHS1 mutations cause combined respiratory chain deficiency resulting in Leigh syndrome. Hum. Mutat. 2015;36:232–239. doi: 10.1002/humu.22730. [DOI] [PubMed] [Google Scholar]
- 122.Ferdinandusse S., Friederich M.W., Burlina A., Ruiter J.P.N., Coughlin C.R., Dishop M.K., Gallagher R.C., Bedoyan J.K., Vaz F.M., Waterham H.R., et al. Clinical and biochemical characterization of four patients with mutations in ECHS1. Orphanet J. Rare Dis. 2015;10:79. doi: 10.1186/s13023-015-0290-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tetreault M., Fahiminiya S., Antonicka H., Mitchell G.A., Geraghty M.T., Lines M., Boycott K.M., Shoubridge E.A., Mitchell J.J., Michaud J.L., et al. Whole-exome sequencing identifies novel ECHS1 mutations in Leigh syndrome. Hum. Mutat. 2015;134:981–991. doi: 10.1007/s00439-015-1577-y. [DOI] [PubMed] [Google Scholar]
- 124.Haack T.B., Jackson C.B., Murayama K., Kremer L.S., Schaller A., Kotzaeridou U., de Vries M.C., Schottmann G., Santra S., Büchner B., et al. Deficiency of ECHS1 causes mitochondrial encephalopathy with cardiac involvement. Ann. Clin. Transl. Neurol. 2015;2:492–509. doi: 10.1002/acn3.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yamada K., Aiba K., Kitaura Y., Kondo Y., Nomura N., Nakamura Y., Fukushi D., Murayama K., Shimomura Y., Pitt J., et al. Clinical, biochemical and metabolic characterisation of a mild form of human short-chain enoyl-CoA hydratase deficiency: significance of increased N-acetyl-S-(2-carboxypropyl)cysteine excretion. J. Med. Genet. 2015;52:691–698. doi: 10.1136/jmedgenet-2015-103231. [DOI] [PubMed] [Google Scholar]
- 126.Enns G.M., Bennett M.J., Hoppel C.L., Goodman S.I., Weisiger K., Ohnstad C., Golabi M., Packman S. Mitochondrial respiratory chain complex I deficiency with clinical and biochemical features of long-chain 3-hydroxyacyl-coenzyme A dehydrogenase deficiency. J. Pediatr. 2000;136:251–254. doi: 10.1016/s0022-3476(00)70111-9. [DOI] [PubMed] [Google Scholar]
- 127.Tyni T. Pathology of skeletal muscle and impaired respiratory chain function in long-chain 3-hydroxyacyl-CoA dehydrogenase deficiency with the G1528C mutation. Neuromuscul. Disord. 1996;6:327–337. doi: 10.1016/0960-8966(96)00352-5. [DOI] [PubMed] [Google Scholar]
- 128.Nouws J., Te Brinke H., Nijtmans L.G., Houten S.M. ACAD9, a complex I assembly factor with a moonlighting function in fatty acid oxidation deficiencies. Hum. Mol. Genet. 2014;23:1311–1319. doi: 10.1093/hmg/ddt521. [DOI] [PubMed] [Google Scholar]
- 129.Gargus J.J., Boyle K., Bocian M., Roe D.S., Vianey-Saban C., Roe C.R. Respiratory complex II defect in siblings associated with a symptomatic secondary block in fatty acid oxidation. J. Inherit. Metab. Dis. 2003;26:659–670. doi: 10.1023/b:boli.0000005659.52200.c1. [DOI] [PubMed] [Google Scholar]
- 130.Arpa J., Campos Y., Gutiérrez-Molina M., Cruz-Martinez A., Arenas J., Caminero A.B., Palomo F., Morales C., Barreiro P. Benign mitochondrial myopathy with decreased succinate cytochrome C reductase activity. Acta Neurol. Scand. 1994;90:281–284. doi: 10.1111/j.1600-0404.1994.tb02722.x. [DOI] [PubMed] [Google Scholar]
- 131.Vockley J., Rinaldo P., Bennett M.J., Matern D., Vladutiu G.D. Synergistic heterozygosity: disease resulting from multiple partial defects in one or more metabolic pathways. Mol. Genet. Metab. 2000;71:10–18. doi: 10.1006/mgme.2000.3066. [DOI] [PubMed] [Google Scholar]
- 132.Sumegi B., Srere P.A. Complex I binds several mitochondrial NAD-coupled dehydrogenases. J. Biol. Chem. 1984;259:15040–15045. [PubMed] [Google Scholar]
- 133.Taylor W.A., Mejia E.M., Mitchell R.W., Choy P.C., Sparagna G.C., Hatch G.M. Human trifunctional protein alpha links cardiolipin remodeling to beta-oxidation. PloS One. 2012;7:e48628. doi: 10.1371/journal.pone.0048628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wang Y., Mohsen A.W., Mihalik S.J., Goetzman E.S., Vockley J. Evidence for physical association of mitochondrial fatty acid oxidation and oxidative phosphorylation complexes. J. Biol. Chem. 2010;285:29834–29841. doi: 10.1074/jbc.M110.139493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Parker W.D., Jr, Filley C.M., Parks J.K. Cytochrome oxidase deficiency in Alzheimer's disease. Neurology. 1990;40:1302–1303. doi: 10.1212/wnl.40.8.1302. [DOI] [PubMed] [Google Scholar]
- 136.Das A.M., Fingerhut R., Wanders R.J., Ullrich K. Secondary respiratory chain defect in a boy with long-chain 3-hydroxyacyl-CoA dehydrogenase deficiency: possible diagnostic pitfalls. Eur. J. Pediatr. 2000;159:243–246. doi: 10.1007/s004310050063. [DOI] [PubMed] [Google Scholar]
- 137.Tonin A.M., Amaral A.U., Busanello E.N., Grings M., Castilho R.F., Wajner M. Long-chain 3-hydroxy fatty acids accumulating in long-chain 3-hydroxyacyl-CoA dehydrogenase and mitochondrial trifunctional protein deficiencies uncouple oxidative phosphorylation in heart mitochondria. J. Bioenerg. Biomembr. 2013;45:47–57. doi: 10.1007/s10863-012-9481-9. [DOI] [PubMed] [Google Scholar]
- 138.Friedrich T., Bottcher B. The gross structure of the respiratory complex I: a Lego System. Biochim. Biophys. Acta. 2004;1608:1–9. doi: 10.1016/j.bbabio.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 139.Duarte M., Pópulo H., Videira A., Friedrich T., Schulte U. Disruption of iron-sulphur cluster N2 from NADH: ubiquinone oxidoreductase by site-directed mutagenesis. Biochem. J. 2002;364:833–839. doi: 10.1042/BJ20011750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Grigorieff N. Three-dimensional structure of bovine NADH:ubiquinone oxidoreductase (complex I) at 22 A in ice. J. Mol. Biol. 1998;277:1033–1046. doi: 10.1006/jmbi.1998.1668. [DOI] [PubMed] [Google Scholar]
- 141.Lazarou M., Thorburn D.R., Ryan M.T., McKenzie M. Assembly of mitochondrial complex I and defects in disease. Biochim. Biophys. Acta. 2009;1793:78–88. doi: 10.1016/j.bbamcr.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 142.Schiff M., Haberberger B., Xia C., Mohsen A.W., Goetzman E.S., Wang Y., Uppala R., Zhang Y., Karunanidhi A., Prabhu D., et al. Complex I assembly function and fatty acid oxidation enzyme activity of ACAD9 both contribute to disease severity in ACAD9 deficiency. Hum. Mol. Genet. 2015;24:3238–3247. doi: 10.1093/hmg/ddv074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.McKenzie M., Ryan M.T. Assembly factors of human mitochondrial complex I and their defects in disease. IUBMB Life. 2010;62:497–502. doi: 10.1002/iub.335. [DOI] [PubMed] [Google Scholar]
- 144.Pagliarini D.J., Calvo S.E., Chang B., Sheth S.A., Vafai S.B., Ong S.-E., Walford G.A., Sugiana C., Boneh A., Chen W.K., et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Narayan S.B., Master S.R., Sireci A.N., Bierl C., Stanley P.E., Li C., Stanley C.A., Bennett M.J. Short-chain 3-hydroxyacyl-coenzyme A dehydrogenase associates with a protein super-complex integrating multiple metabolic pathways. PloS One. 2012;7:e35048. doi: 10.1371/journal.pone.0035048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tyni T., Majander A., Kalimo H., Rapola J., Pihko H. Pathology of skeletal muscle and impaired respiratory chain function in long-chain 3-hydroxyacyl-CoA dehydrogenase deficiency with the G1528C mutation. Neuromuscul. Disord. 1996;6:327–337. doi: 10.1016/0960-8966(96)00352-5. [DOI] [PubMed] [Google Scholar]