Introduction
Where did I come from? Where am I going? And why does it matter? We might all have felt the existential angst of these questions at some point in our lives, but it is really from the point of view, if such existed, of a cell in a mammalian embryo that I want to ponder this question and to consider how, over time, imaging and transgenics have honed our ability to trace the lineage of cells during development with increasing precision and allowed us to better understand why and how it matters. As technology has opened new windows into the process of embryogenesis, questions have been framed regarding the interplay between genetics, lineage and cell behavior, and our concept of lineage analysis has expanded to include determining a mechanistic explanation for lineage patterns. Where things come from can to some extent explain how they got there, but only if we understand the mechanisms through which they reached their destinations. Cell lineage is the framework for understanding causes and mechanisms of cellular diversity, unification of the whole organism, cellular cooperation, stability of the phenotype and its relationship to pluripotency. It is the thread that holds development together and is arguably the most important concept in developmental biology.
Thus, cell lineage is something every card-carrying developmental biologist thinks about and incorporates into their work. My own pedigree as a developmental biologist includes many cell lineage studies starting with my earliest work in Richard L. Gardner's laboratory. In the best tradition of experimental embryology, Richard had developed a method of introducing cells into preimplantation mammalian embryos that, combined with genetic differences between the introduced cells and the embryo, allowed him to trace the lineage of the introduced cells in a chimeric embryo (Gardner, 1968). Using this method and even more elaborate cut-and-paste micromanipulation procedures in which embryos were taken apart and put back together in different combinations, we carried out studies aimed at tracing the lineage of the earliest differentiated cell types of the mouse blastocyst, the inner cell mass, primitive endoderm and trophectoderm, in an attempt to determine the origins of postimplantation tissues (Gardner and Papaioannou, 1975; Gardner et al., 1973; Papaioannou, 1982). Until that time, knowledge of cell lineage in mammalian embryos was inferred from histological studies or by analogy with other experimental organisms like the chick, which were much more straightforward to manipulate. These early studies were far from ideal lineage tracing studies: For one thing, the genetic markers available (isozyme variants) were ubiquitously expressed and developmentally neutral but were impossible to detect at the cellular/spatial level as the tissue was destroyed in the analysis; for another, the micromanipulation method was extremely invasive and the success of the introduction of foreign cells depended entirely on the mammalian embryo's inherent and immense tolerance of abuse and capacity for regulation. In spite of these limitations, the lineage relationships discerned and the fate maps inferred from these experiments provided a foundation of embryonic lineages that has been further refined over the years, but has essentially stood the test of time. This essay chronicles how experiments even with severe limitations can provide useful approximations of lineage relationships, and how the refining of those approximations with methodological innovations has broadened our concept of lineage relationships in the complex mammalian embryo.
How to trace cell lineage
Prior to the late 18th century, preformationist ideas of embryonic development, in which body parts were preformed in the ovum and only needed to grow larger, did not require any concept of cell lineage. However, long before these ideas were rampant, Aristotle had formulated a theory that the adult existed only potentially in the embryo and that a process of differentiation occurred as development unfolded. It eventually became clear, with the invention and use of the microscope, that there was no homunculus lurking in the sperm and that body parts of a fetus or adult did indeed come from undifferentiated parts of an earlier stage embryo by a process of differentiation, proliferation and cell movement, hence the need to trace lineage to understand how the final form came into being. Fate maps, the representation of the future fate of cells or tissues projected onto any given embryonic stage, became standard tools for thinking about and describing the dynamic process of differentiation and the destiny of cells during development.
In theory, tracing cell lineage in an embryo is simple: find some means of marking a cell without disturbing the embryo and then keep track of the marked cell and its descendents as the embryo develops to see where they go. In practice, however, this simple goal is fraught with difficulties, particularly in mammalian embryos that are protected from view by the mother's body, presenting a challenge for ‘seeing where they go’. Similarly, the condition ‘without disturbing the embryo’ is tricky. Either the act of marking the cell or the marker itself could potentially disturb the embryo. As for the marker, Anne McLaren proclaimed the attributes of the ideal genetic cell marker for tracking cell lineage in chimaeras in her book “Mammalian Chimaeras” (McLaren, 1976). Similar criteria are still applicable to any marker used in cell lineage studies. Ideally a marker would be 1) ubiquitous, so that any cell population could be marked and traced, 2) cell localized, marking cells and not, for instance, extracellular structures, 3) cell autonomous, so that the mark stayed with the original cell, 4) heritable and 5) stable in mitotic progeny, so that it was not diluted out or lost by cell division, 6) easy to detect, for obvious reasons, and 7) developmentally neutral, such that the presence of the marker did not alter the course of development. Furthermore, for genetic markers, multiple distinguishable forms or alleles would be valuable for tracking more than one cell at a time. McLaren claimed that the ideal marker did not exist, but fortunately that did not stop her or anyone since from developing lineage tracing markers with closer and closer approximations to the ideal.
The classic experimental embryology approach
We can think of mammalian lineage tracing studies as belonging to two different eras: the experimental embryology era and the genetic era. Although the early chimaera experiments starting in the 1960's depended on genetic differences to distinguish cells, the methodology was in the tradition of pure experimental embryology – add a cell, take a cell away, move cells around and see what happens. The goal of many studies was to test not only the fate of cells but also their potential, such that cells might be moved heterotopically (to a different location) and heterochronically (to a different aged embryo), as well as orthotopically (as close as possible to the original location). Even in orthotopic transfers where the goal was to elucidate a cell's normal fate, the extent to which development was disturbed by the procedures, including the time outside the mother's reproductive tract, was impossible to judge as the mammalian embryo's ability to regulate for abuse is staggering and the disturbed course of events may not be the same as the undisturbed course of events even though the end result is a normal embryo. Nonetheless, multiple approaches leading to similar results lent credibility to fate maps that were drawn. As an example, a study by Rosa Beddington (Beddington, 1982) probed the fate and potential of different regions of the epiblast by mechanically transferring clumps of marked cells orthotopically and heterotopically in embryos maintained in culture. The cells were labeled with tritiated thymidine, a marker that did not meet the criterion of ‘easy to detect’ as it could only be visualized by autoradiography of sectioned embryos, and it had the disadvantage of dilution with mitosis, thus not meeting the ‘stable in mitotic progeny’ criterion. The culture period no doubt also lent experimental variability, but in spite of these limitations, there was consistency in the results and general agreement with the fate maps of other well-studied amniotes such as chick.
A variation on the cut-and-paste chimeric approach to lineage studies, but still within the experimental embryology tradition, is the application of markers to cells in intact embryos. By leaving the tissues in situ, this method reduces disturbance to the embryos, although application of a marker could still affect the behavior of the marked cells. An example of this approach is a series of experiments by Kirstie Lawson and Roger Pederson (e.g. (Lawson and Pedersen, 1987) tracing the lineage of different cell types of the early postimplantation embryo by intracellular injection of horseradish peroxidase and locating the descendents of the cell several days later by enzymatic assay. This particular marker did not appear to adversely affect the cells and although it diluted out with mitosis, it was still easily detectable after 5-6 cell divisions. A feature of this study that came at a high cost in labor was that a single cell was injected and thus only a single clone from a lineage was traced in each embryo. This can be an advantage for precision mapping but also a disadvantage in that even a large number of single cell clones can only provide a sample of the whole picture. Also limiting was the period of invisibility between injecting the marker and harvesting the embryos – a black box between marking and analysis. What was missing was the possibility of seeing exactly how a cell got from one place to the next.
DiI, a lipophilic dye that is incorporated into cell membranes, is an example of a marker that can be followed through many cell divisions due to its intense fluorescence. A study from the Beddington lab using DiI labeling of the pregastrulation distal visceral endoderm inferred by lineage tracing that these cells moved into the position of anterior visceral endoderm (AVE) (Thomas et al., 1998). The interest in these specific cells came from the expression pattern of the homeobox gene Hex, which was seen initially in the distal visceral endoderm and later in the AVE, giving the impression of cell movement. But, as pointed out in this paper, gene expression cannot serve as a lineage marker, hence the need for an actual lineage trace to show the relationship between distal visceral endoderm and AVE. This conjunction of lineage tracing and defining a cell population based on a specific gene expression pattern was a seminal experiment leading to the idea of the AVE, a classical extraembryonic tissue, as a patterning center for the anterior/posterior axis of the embryo and also presaging a new era of lineage tracing based on gene expression.
The rise of ‘applied’ genetic markers
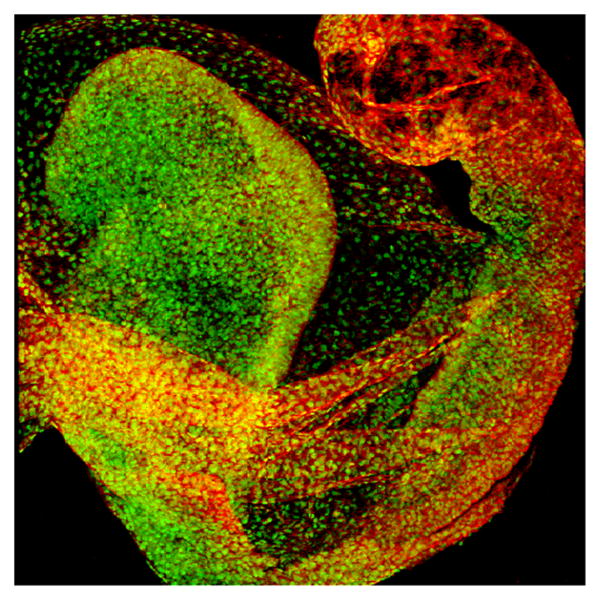
Experimental embryology and transgenic technology began to come together dramatically for cell lineage studies in the 1990s. The barriers posed by inadequate or cumbersome cell markers and the problem of following cells in real time were answered with the introduction of fluorescent markers into the germ line of mice. Miraculously, the mammalian embryo once again proved to be quite tolerant, not only of the presence of foreign fluorescent proteins in its cells, but also of the various imaging modalities required to visualize the proteins in real time in living embryos in culture. The markers, being the products of transgenes, were indelible, heritable and easy to follow. The first such transgenic marker, developed by Magda Zernicka-Goetz in Martin Evan's lab was green fluorescent protein (GFP) put under the control of a promoter active in proliferating cells (Zernicka-Goetz et al., 1997). A huge variety of transgenes and transgenic lines have since been fashioned for specific purposes, including, for example, a ubiquitous, nuclear-localized GFP line of mice produced in my lab by Kat Hadjantonakis (Hadjantonakis and Papaioannou, 2004) (Figure 1), and many other, multicolor fluorescent genes under the control of tissue- or cell-specific regulatory elements.
Figure 1.
A embryo at midgestation carrying an H2B:eGFP transgene counterstained to highlight cell membranes. Photo courtesy of Guy Eakin, Anna-Katerina Hadjantonakis, Virginia E. Papaioannou and Richard R. Behringer and reproduced with permission from Cold Spring Harbor Press (Papaioannou and Behringer, 2004).
Ubiquitously expressed markers are particularly useful for following cell lineage in chimaeras, where marked cells are added to unmarked embryos, as in the work from Patrick Tam's lab establishing fate maps of the primitive streak at different stages of gastrulation (Kinder et al., 1999). Markers that fluoresce under the control of tissue-specific promoters, however, are more useful if one wants to trace cell lineage in undisturbed embryos. But with this type of study, the concept of cell lineage shifts slightly as the definition of the cell type being traced depends on the pattern of gene expression at a particular point in time, rather than on morphology and location. Gene expression is not necessarily a constant, however, and although a particular gene may mark a given cell population at one specific point in development, this may change with time and, in addition, there may be other areas of expression. As elegantly stated in a recent paper, marker gene expression indicates the “state not fate” of cells (Viotti et al., 2014). Only continuous monitoring of individual cells can unequivocally ascertain lineage relationships as the marker will provide a dynamic picture of gene expression changes such that a population of marked cells may grow or decline as cells turn the gene on or off with developmental progress.
Nonetheless, so-called tissue-specific promoters have been extremely valuable in providing insight into cell lineage based on gene expression, with the added benefits of tracking cell behavior. For example, in a study by Shankar Srinivas, Tristan Rodriguez and others in Rosa Beddingdon's lab, a Hex:GFP transgene was used to mark cells of the distal visceral endoderm (Srinivas et al., 2004). Endogenous Hex is expressed in the distal visceral endoderm and later in the AVE, suggesting but not proving that these two areas were linked by lineage. Experiments mentioned previously using DiI as an applied lineage marker supported this idea, but with the Hex:GFP transgene, labeled distal visceral endoderm cells could be followed in real time in living embryos. The results showed active migration of the fluorescently labeled cells toward the anterior midline, confirming the lineage of the AVE and corroborating the DiI applied marker study. The potential complications of cessation of expression or of new cells beginning to express the gene were abrogated by constant monitoring of individual fluorescent cells.
Another aspect of this study illustrates the ambiguity about what exactly is being marked by a so-called tissue-specific promoter. It was noted that Hex expression in the AVE is not uniform, i.e., there is a mixture of expressing and non-expressing cells. Similarly, the Hex:GFP transgene faithfully recapitulated this salt-and-pepper expression, thus the lineage of only the expressing cells was followed in this experiment. This begs the question of whether the subset of non-expressing cells of the AVE, which is a morphologically and functionally defined area, have a similar cell lineage, or whether they are derived from another embryonic source.
Patchy gene expression was not an issue in a study from the Hadjantonakis laboratory (Kwon et al., 2008) in which an entire cell population, the visceral endoderm of the pregastrulation embryo was labeled with an Afp∷GFP transgene. This study shows the power of lineage tracing an entire population of cells, rather than individual clones or a subset of cells. The results led to an unexpected conclusion at odds with the prevailing models of endoderm development in that they revealed a persistence of the progeny of visceral endoderm cells in areas of the embryos formerly thought to consist entirely of newly formed definitive endoderm from the anterior primitive streak, and further, revealed a contribution of these so-called extraembryonic cells to the fetal gut. In this case it was the perdurance of the fluorescent protein that allowed the recognition of this lineage of cells after the endogenous Afp promoter was downregulated. Live imaging then allowed an assessment of the behavior of these cells and the discovery of the intercalation of definitive endoderm by a process of “coordinated egression” rather than displacement of an intact epithelial sheet, as was previously thought.
Further refinements for gene expression lineage tracing
Rather than putting the cell marker directly downstream of a promoter region of interest, another tactic is to drive Cre recombinase by a specific cis-regulatory element and then combine this transgene with a separate Cre-reporter transgene such that a recombination event occurs where Cre is active, causing a marker to be expressed. As this is a heritable genetic change, any cell where the recombination has occurred and all its progeny will express the marker even when gene expression stops. This allows for long term lineage tracing with the caveat that additional cells will be added to the marked lineages as long as activity of the regulatory element of interest continues and wherever else it drives Cre expression. An additional level of control can be added by making the Cre driver inducible, so that the timing of activity of Cre can be experimentally controlled. In this way, cohorts of cells expressing a particular gene (captured by the regulatory element driving that gene) can be marked within particular temporal limits.
Of course each additional experimental manipulation adds complexity and occasionally a limitation to the tracing of undisturbed cell lineages. To wit, the presence of the foreign protein Cre may not always be developmentally neutral (e.g. (Lewis et al., 2013; Naiche and Papaioannou, 2007), but when properly controlled, these experimental methods can trace cells into postnatal stages. Conceptually, however, it is important to realize that the transient expression of a particular gene may have little do with the eventual fate of the progeny cells: lineage tracing of cells from a certain location that express a particular gene will show where those cells eventually come to rest, but whether or not that gene played any role in the course of their lineage is a different question altogether. To get at this question, one useful approach is to look in mutant embryos for cell lineage changes due to the loss of the gene.
Clonal analysis within cell lineages
Analysis of the behavior of individual cell clones is related to, but distinct from, cell lineage analysis. Typically, clonal analysis follows all the progeny of a particular cell, but in the context of development, the clonal progeny of a cell from an early embryo may contribute to multiple ‘lineages’ as those lineages segregate during differentiation. In other words, clonal analysis is based simply on the mitotic progeny of a single cell, whilst cell lineage studies are based on following the fate of cells, usually a population of cells, defined by a particular anatomical location and/or gene expression pattern, most commonly after some lineage segregation has occurred. That said, the two concepts are complementary in that every cell has a clonal origin and future (if it is still dividing), and is also part of a cell lineage in terms of its position and differentiated state. Clonal analysis methods are useful for probing the fine grained behavior of individual cells within lineages composed of many cells.
Retrospective analysis of cell clones in genetic mosaics, where a somatic event creates a genetic mutation detectable at the cellular level, has been a classic technique of experimental embryology, particularly in Drosophila. Variations on this method have been applied to the mouse using rare somatic mutations in transgenes. An early example was a defective lacZ transgene that could be repaired by rare, sporadic interchromosomal recombination events to produce a functional gene that produced a detectable LacZ marker. This was used, for example, with a ubiquitous promoter to mark random clones to investigate lineage segregation during early embryogenesis and gastrulation (Tzouanacou et al., 2009). Limitations of this type of retrospective analysis are that the cell of origin is not defined and it can be difficult to distinguish redundant events, particularly when the recombination events are frequent. Thus in this example, analysis was limited to cases where the recombination events were extremely rare, necessitating a large number of specimens to obtain a reasonably comprehensive representation of the patterns of clonal growth. Nonetheless, this study was important in confirming the existence of a bipotential neuromuscular progenitor pool persisting throughout axis elongation.
Fluorescent proteins can also be used for clonal analysis with the advantages that there are many spectral variants that can be simultaneously visualized and distinguished in living embryos. Variations on the theme of clonal analysis have been developed using transgenes with the colorful names of Brainbow, Rainbow and Confetti, which, as their names imply, result in clones of different hues through recombinase-mediated excision and/or inversion of multiple fluorescent protein transgene cassettes to produce recombinant cells with many possible outcomes. Thus, stochastic recombination allows distinction among multiple clones within a population, and multiple clones can be followed within each specimen. Originally developed to study neurons in the brain, they have wide application (Weissman and Pan, 2015). Baggiolini and colleagues used the Confetti transgene, combined with inducible, tissue-specific promoters driving Cre recombinase, to answer a long controversial issue regarding the potency of neural crest cells before and after they leave the neural tube. Using rigorous statistical methods to exclude redundant recombination events, they found that most neural crest cells are multipotent both pre- and postmigration, but that their fates can be heterogeneous, as seen by variation in the range of differentiated fates in different clones (Baggiolini et al., 2015).
Mutant effects on cell lineage
Over the years, a lot of effort has gone into developing the perfect marker for lineage tracing to better understand cell behavior in undisturbed embryos. But one particular kind of disturbance forms the basis of a slightly different concept of cell lineage, that is, investigating the properties of cell with mutations by watching how they interact with normal cells during development. This has long been a staple in the analysis of chimaeras to distinguish cell autonomous from non-autonomous mutant effects (e.g. (Papaioannou and Gardner, 1979) and of course, determining the tissue-specific effects of any mutation is a standard part of mutation analysis (Papaioannou and Behringer, 2004). But the question here has more to do with competition and whether mutant cells behave differently from wild type cells in the same environment. Thus, the techniques of mosaic analysis can be modified slightly to juxtapose mutant and normal cells with a minimum of disruption to other aspects of development. A beautiful example of this combines the Mosaic Analysis with Double Markers (MADM) system with a linked mutation to mark the two daughter cells of a rare clone with different fluorescent markers and simultaneously produce a homozygous mutation in one of the daughters. This is accomplished through Cre-mediated interchromosomal recombination of two loxP-containing transgenes at homologous chromosomal locations. Originally developed and used to trace neuronal connections (Zong et al., 2005), the system has great potential for approaching questions of micro-lineage and the effect of mutations during organogenesis. Frank Costantini's laboratory, for example, has used this method to investigate branching morphogenesis of the ureteric bud during kidney development by following double marked clones of wild type and mutant ureteric bud epithelium with time-lapse imaging in vitro. By following differentially marked sister clones with one clone wild type and the other with mutations in the receptor tyrosine kinase gene Ret or its target gene, Etv4, they showed that Ret/Etv4 signaling regulates cell movements that are part of the basic mechanism of ureteric bud branch formation, independently of effects on cell proliferation (Riccio et al., 2015).
Final thoughts
One of the special challenges of studying mammalian embryology is the flexibility and regulative capacity of the embryo. Because of this ability to absorb and compensate for disruptions, it stands to reason that cell lineages even in undisturbed embryos will not be invariant but that there will be wiggle room at the margins as a tradeoff between flexibility and stability. Nonetheless, the consistency of development implies that lineage history serves as a guiding force in cell fate decisions as development unfolds, so the questions of where do cells come from and where do they go are highly relevant. Lineage studies can link morphology with gene expression patterns and cell fate, converging on the functional role of genes in development and reconciling heterogeneity of gene expression with fate maps. In the future, following the fate of cells and clones of cells with specific, multiple-gene expression profiles will provide an even finer-grained map so that cell lineage studies will not be going out of fashion any time soon.
Acknowledgments
I would like to thank Kat Hadjantonakis for helpful comments on the manuscript. This work was supported by the Eunice Kennedy Shriver National Institute of Child Health & Human Development of the National Institutes of Health (NIH) grant R37HD033082. The content is solely the responsibility of the author and does not necessarily represent the official views of the NIH.
References
- Baggiolini A, Varum S, Mateos JM, Bettosini D, John N, Bonalli M, Ziegler U, Dimou L, Clevers H, Furrer R, Sommer L. Cell Stem Cell. 2015;16(3):314–22. doi: 10.1016/j.stem.2015.02.017. 2015 Mar 5. [DOI] [PubMed] [Google Scholar]
- Beddington RS. J Embryol Exp Morphol. 1982;69:265–85. [PubMed] [Google Scholar]
- Gardner RL. Nature. 1968;220:596–597. doi: 10.1038/220596a0. [DOI] [PubMed] [Google Scholar]
- Gardner RL, Papaioannou VE. The Early Development of Mammals. 1975;2:107–132. [Google Scholar]
- Gardner RL, Papaioannou VE, Barton SC. J Embryol exp Morph. 1973;30:561–572. [PubMed] [Google Scholar]
- Hadjantonakis AK, Papaioannou VE. BMC Biotechnology. 2004;4:33. doi: 10.1186/1472-6750-4-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinder SJ, Tsang TE, Quinlan GA, Hadjantonakis AK, Nagy A, Tam PP. Development. 1999;126:4691–701. doi: 10.1242/dev.126.21.4691. [DOI] [PubMed] [Google Scholar]
- Kwon GS, Viotti M, Hadjantonakis AK. Dev Cell. 2008;15:509–20. doi: 10.1016/j.devcel.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson KA, Pedersen RA. Development. 1987;101:627–52. doi: 10.1242/dev.101.3.627. [DOI] [PubMed] [Google Scholar]
- Lewis AE, Vasudevan HN, O'Neill AK, Soriano P, Bush JO. Dev Biol. 2013;379:229–34. doi: 10.1016/j.ydbio.2013.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren. Mammalian Chimaeras. Cambridge University Press; Cambridge: 1976. p. 154. [Google Scholar]
- Naiche LA, Papaioannou VE. Genesis. 2007;5:768–775. doi: 10.1002/dvg.20353. [DOI] [PubMed] [Google Scholar]
- Papaioannou VE. J Embryol exp Morph. 1982;68:199–209. [PubMed] [Google Scholar]
- Papaioannou VE, Behringer RR. Mouse Phenotypes, A Handbook of Mutation Analysis. Cold Spring Harbor Press; Cold Spring Harbor, NY: 2004. p. 226. [Google Scholar]
- Papaioannou VE, Gardner RL. J Embryol exp Morph. 1979;52:153–163. [PubMed] [Google Scholar]
- Riccio P, Cebrian C, Zong H, Hippenmeyer S, Costantini F. PLoS Biol. 2015 doi: 10.1371/journal.pbio.1002382. in review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S, Rodriguez T, Clements M, Smith JC, Beddington RS. Development. 2004;131:1157–64. doi: 10.1242/dev.01005. [DOI] [PubMed] [Google Scholar]
- Thomas PQ, Brown A, Beddington RSP. Development. 1998;125:85–94. doi: 10.1242/dev.125.1.85. [DOI] [PubMed] [Google Scholar]
- Tzouanacou E, Wegener A, Wymeersch FJ, Wilson V, Nicolas JF. Dev Cell. 2009;17:365–76. doi: 10.1016/j.devcel.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Viotti M, Foley AC, Hadjantonakis AK. Philos Trans R Soc Lond B Biol Sci. 2014;369(1657) doi: 10.1098/rstb.2013.0547. 2014 Dec 5. pii: 20130547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman TA, Pan YA. Genetics. 2015;199(2):293–306. doi: 10.1534/genetics.114.172510. 2015 Feb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zernicka-Goetz M, Pines J, McLean Hunter S, Dixon JP, Siemering KR, Haseloff J, Evans MJ. Development. 1997;124:1133–7. doi: 10.1242/dev.124.6.1133. [DOI] [PubMed] [Google Scholar]
- Zong H, Espinosa JS, Su HH, Muzumdar MD, Luo L. Cell. 2005;121:479–92. doi: 10.1016/j.cell.2005.02.012. [DOI] [PubMed] [Google Scholar]