Abstract
Single-stranded RNA viruses package their genomes into capsids enclosing fixed volumes. We assayed the ability of bacteriophage MS2 coat protein to package large, defined fragments of its genomic, single-stranded RNA. We show that the efficiency of packaging into a T=3 capsid in vitro is inversely proportional to RNA length, implying that there is a free-energy barrier to be overcome during assembly. All the RNAs examined have greater solution persistence lengths than the internal diameter of the capsid into which they become packaged, suggesting that protein-mediated RNA compaction must occur during assembly. Binding ethidium bromide to one of these RNA fragments, which would be expected to reduce its flexibility, severely inhibited packaging, consistent with this idea. Cryo-EM structures of the capsids assembled in these experiments with the sub-genomic RNAs show a layer of RNA density beneath the coat protein shell but lack density for the inner RNA shell seen in the wild-type virion. The inner layer is restored when full-length virion RNA is used in the assembly reaction, implying that it becomes ordered only when the capsid is filled, presumably because of the effects of steric and/or electrostatic repulsions. The cryo-EM results explain the length dependence of packaging. In addition, they show that for the sub-genomic fragments the strongest ordered RNA density occurs below the coat protein dimers forming the icosahedral 5-fold axes of the capsid. There is little such density beneath the proteins at the 2-fold axes, consistent with our model in which coat protein dimers binding to RNA stem–loops located at sites throughout the genome leads to switching of their preferred conformations, thus regulating the placement of the quasi-conformers needed to build the T=3 capsid. The data are consistent with mutual chaperoning of both RNA and coat protein conformations, partially explaining the ability of such viruses to assemble so rapidly and accurately.
Keywords: viral RNA, assembly efficiency, RNA folding, cryo-electron microscopy, RNA phage
Introduction
Viruses with single-stranded RNA (ssRNA) genomes are a large fraction of all known viruses and include major pathogens of many organisms including humans.1 Many of these viruses construct highly symmetrical protein shells that self-assemble around inherently asymmetric genomic RNAs. Such assembly mechanisms pose generically interesting structural puzzles that remain largely unsolved.2 How do these viruses preferentially package their genomic RNAs from a large background of cellular RNA? How do they regulate the formation of their protein capsids, maximising the formation of a correctly self-assembled capsid rather than non-specific aggregates? This is an especially interesting problem for viruses that use the principles of quasi-equivalence to construct their capsids. Such viruses must simultaneously regulate the formation of different quasi-conformers of the same protein sequence in the proportions needed to build a capsid of the required size and symmetry.2,3
Recently, we showed for the ssRNA bacteriophage MS2 (Fig. 1) that quasi-equivalent conformer switching can be controlled by binding of short RNA stem–loops to the basic building block of the MS2 capsid, a coat protein dimer (CP2).4,5 For MS2, there is a well characterised sequence-specific interaction between CP2 and a specific RNA stem–loop (TR) of just 19 nt in length that can fold from within the genomic RNA (Fig. 1a).6–11 The TR sequence encompasses the start codon of the viral replicase, and coat protein binding in vivo results in translational repression of the replicase cistron. The interaction between TR and CP2 is believed to function as an assembly initiation site in vivo, ensuring in this case cognate packaging of MS2 RNA even in the presence of competing RNA phages.12 In vitro, TR:CP2 confers packaging specificity on RNAs but this effect is modest in longer RNAs.13,14
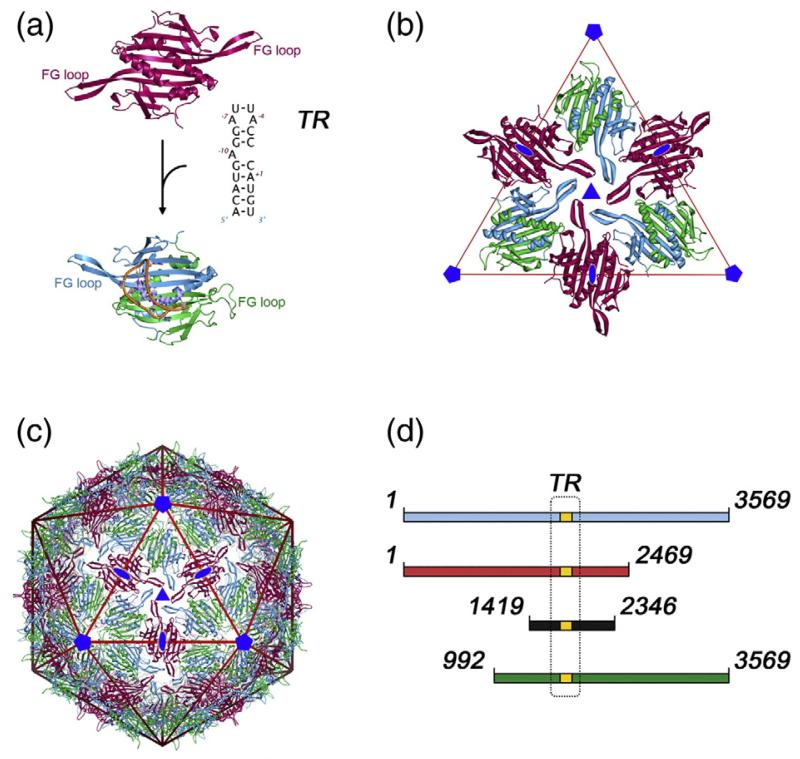
Fig. 1.
The architecture of the T=3 MS2 capsid and sub-genomic RNA fragments. (a) In the T=3 capsid, the MS2 coat protein subunit (CP) is found in three quasi-equivalent conformers: A (blue), B (green) and C (deep red). These conformers form two types of dimer (CP2), an asymmetric A/B dimer, and a symmetric C/C dimer, which differ primarily in the orientation of the loops between the F and G β-strands (FG loop). Conformational switching between symmetric and asymmetric conformations is promoted by the binding of a 19 nt RNA stem–loop (TR). (b) The A/B and C/C dimers are the basic building block of the capsid.5 The extended FG loops of the A and C conformers pack around the icosahedral 3-fold axis and the compact loops of the B conformer pack around the icosahedral 5-fold axis. (c) Such packing leads to the T=3 caspid, containing 60 A/B and 30 C/C dimers. (d) The RNAs used in this study. The position of the TR assembly-initiation sequence is in gold and boxed, and its sequence is described in (a). The RNAs used for assembly reactions are shown in red (5′RNA), black (iRNA) and green (3′RNA). The full-length genomic RNA extracted from virions is shown in blue. The numbering corresponds to the nucleotide sequence of the genomic RNA (GenBank accession number NC_001417; Table 2). Figures 1 and 5 were produced using UCSF Chimera68 and PyMOL (www.pymol.org).
The T=3 protein capsid of MS2 (Fig. 1b and c) has an icosahedral surface lattice consisting of 180 coat protein subunits packaged as 90 non-covalent dimers.15,16 The dimers define the quasi-conformers in this structure (A/B and C/C), which differ primarily in the conformations of the loop of polypeptide connecting the F and G β-strands, the FG-loop. Using a combination of chromatography, mass spectrometry and NMR to assay the effects of TR binding by protein dimers, we showed that in the absence of RNA the proteins are symmetric and C/C-like.5 Stopped-flow fluorescence experiments showed that TR:CP2 interaction is very rapid and essentially diffusion-controlled, suggesting that the initial RNA complex occurs with a C/C-like dimer.17 NMR shows, however, that TR binding promotes asymmetry within the dimer caused by conformational changes within the FG-loops, i.e. the TR:CP2 rapidly becomes A/B-like (Fig. 1a). The results are consistent with the RNA displacing a conformational equilibrium towards the asymmetric dimer, which in the absence of RNA lies largely on the side of the symmetric structure. A recent all atom normal mode analysis of the TR:CP2 complex suggests that the conformer conversion may occur via a process of dynamic allostery as a result of asymmetric binding of an A-type RNA duplex.18 Such a mechanism is consistent with the lack of RNA sequence-specificity of the conformational change.4
Neither the symmetric nor the asymmetric species by themselves is competent for rapid assembly. RNA-free protein dimers do not self-aggregate significantly over many hours, as judged by mass spectrometry. Similarly, TR:CP2 complexes in which the RNA is present at saturation are also relatively stable, forming minor aggregates only after several hours.5 If both coat protein dimer species are present, however, assembly to higher order intermediates and the T=3 capsid is extremely rapid,5,69 suggesting that the RNA-free and RNA-bound forms of the protein are the only building blocks needed to make the capsid, i.e. that they correspond to C/C-like and A/B-like dimers. These results do not imply that capsids will not form in the absence of RNA or that they will not package non-specific RNAs. At high concentrations of coat protein or over long periods of time, capsids are formed slowly in low yield in the absence of RNA, consistent with the conformational equilibrium described above. Here, we address the implications of the simple RNA allosteric conformational shift model, based on assembly triggered by small RNA stem–loops, on capsid assembly with the genomic RNA, where the TR sequence occurs only once.
The roles that genomic RNAs play in the assembly of ssRNA viruses have not been studied extensively and are still poorly understood. In part, this may reflect the fact that we have few data on the structures of the RNAs within the virion.1 There are now many high-resolution X-ray structures of ssRNA viruses but in most cases there is little or no electron density corresponding to the RNA. A range of technical factors19 contribute to this, including the symmetry averaging that is routinely applied to improve the density of the highly symmetric protein capsids. The genomes are fundamentally asymmetric objects, so this averaging tends to remove density for genomic RNA unless it is packaged with strict icosahedral symmetry.20 This effect is exacerbated because the middle to low-resolution diffraction data contribute most to structure determination of nucleic acids.19 Recently, the available structural information has increased due to the determination of more structures that include low-resolution data, and the growing use of cryo-electron microscopy (cryo-EM) to investigate virus structures.21-26 In a number of cases, density has been found directly below the capsid protein layers that can be confidently ascribed to RNA.20,22-24,27
The X-ray structure of the MS2 virion16 does not contain any significant density that could be the genomic RNA, even though the full-length 3569 nt ssRNA is present. Our recent work, inspired by previous cryo-EM structures presented by van Duin and colleagues,23,24 gives a dramatically different insight into the RNA structure of this particle. In contrast to the high-resolution X-ray structure, the icosahedrally-averaged cryo-EM map at sub-nanometre (~9 Å) resolution,22 contains significant amounts of ordered density, which we estimate may account for up to 85% of the genome. These contrasting results, in two structures both of which were icosahedrally averaged, imply that although the genome is not ordered at atomic resolution, it is nevertheless ordered at intermediate resolution and is packaged with pseudo-icosahedral symmetry. The RNA density consists of two concentric shells connected by columns of density at the icosahedral 5-fold symmetry axes. The most distal density in the outer shell of RNA is located directly below the protein subunits within the capsid, suggesting that multiple RNA–protein contacts occur during assembly. Note that the internal density must include the RNA-binding domain(s) of the single copy of maturation protein that is present in mature virions.
In the averaged cryo-EM structure there is RNA density below all the protein dimers in the capsid, i.e. at both C/C and A/B positions, although there are differences in the density below each type of dimer. A similar reconstruction of T=3 capsids reassembled in the presence of multiple TR stem–loops also shows density below both types of dimer, consistent with the stopped-flow result suggesting that TR can bind C/C dimers, as well as A/Bs. In principle, therefore, two types of RNA–protein interaction occur with the coat proteins of the capsid. Since RNA–protein dimer interactions are highly favourable, with a binding free energy (ΔG) of ≈ −50 kJ mol−1,28 these contacts are a significant driving force for capsid assembly. Although the averaging in the cryo-EM structures makes unambiguous interpretation difficult, the differences in RNA density below the two types of protein dimer suggest that appropriately positioned RNA stem–loops within the genome could actively contribute to coat protein conformer switching throughout capsid assembly. This would be advantageous for cognate RNA, because it would make capsid formation both efficient and rapid at a time when the host cell is about to lyse by providing additional co-operative inter-molecular contacts.29 It would also favour specific encapsidation in the presence of cellular RNA.
In order to test these ideas, we have carried out in vitro assembly experiments with large genomic RNA fragments and determined the cryo-EM structures of the resulting assembly products. The results suggest that the RNAs are packaged co-operatively by the coat proteins in a reaction that involves RNA compaction. Sub-genomic fragments show significantly stronger RNA density below 5-fold than at 2-fold positions, consistent with the expectations of the multiple conformer switching model.
Results
The dependence of assembly efficiency on RNA length
The cryo-EM reconstruction of the MS2 virion shows density for the majority of its RNA genome organised into two concentric shells.22 Approximately two-thirds of the observed RNA density lies directly beneath the lattice of RNA binding sites presented on the inner surface of coat protein subunits, with the remainder located in an inner shell connected along the 5-fold axes. In order to explore how this complex RNA structure forms during capsid assembly, we created three DNA templates for RNA polymerase T7 (see Materials and Methods) encompassing different lengths of the MS2 genome (Fig. 1d). Two of the transcripts encompass approximately two-thirds of the MS2 genomic sequence from either the 5′ (5′RNA) or the 3′ (3′RNA) end. The third, smaller transcript (iRNA) encompasses the central section of the RNA genome and represents approximately one-third of the total. All three fragments include the sequence corresponding to the TR assembly initiation site, where we assume assembly would initiate in each case, allowing direct comparison of the effects of RNA length and structure on capsid assembly. These RNA sub-genomic fragments were used in assembly assays, together with phenol-extracted, full-length virion RNA (vRNA).
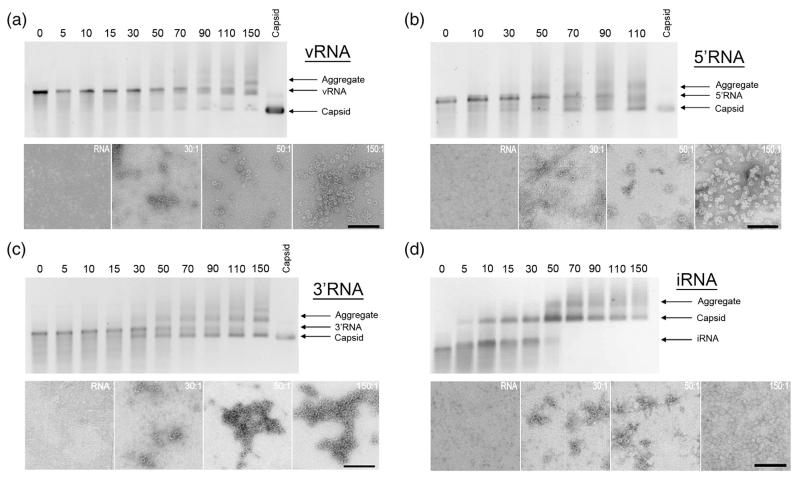
The RNAs were used to trigger capsid assembly in the presence of dissociated coat protein dimers (CP2). The RNAs, at a concentration of 40 nM, were mixed with various concentrations of CP2 encompassing a range of CP2:RNA stoichiometry from sub-stoichiometric to values in excess of those required to allow complete assembly of the T=3 shell. Under these conditions, high-affinity coat protein–TR interactions with every RNA would be expected to initiate assembly at the same site. Reaction mixtures were incubated at 24°C for 3 h and the products were monitored by gel mobility assays and negative stain transmission electron microscopy (Fig. 2). For the vRNA and 5′ RNA, the protein-free nucleic acid migrated more slowly than a sample of recombinant T=3 MS2 capsid,5,30 the mobility of the 3′RNA was very similar to that of the capsid and iRNA migrated more rapidly than the capsid. For each RNA assembly reaction, there are bands in the gels migrating more slowly than the capsid and protein-free RNA, which might be aggregates. A band of protein-free RNA remained throughout most of the titrations, implying that some RNAs proceeded to assemble whilst others did not bind protein. This is consistent with the appearance of T=3 capsids in transmission electron micrographs and on the gels at sub-stoichiometric ratios of CP2: RNA. These results suggest that partially assembled capsids compete successfully with protein-free RNA for the available free CP2. The efficiency of capsid formation was clearly dependent on both coat protein concentration and the RNA fragment being packaged.
Fig. 2.
Gel mobility and electron microscopy assays of MS2 capsid assembly. Assembly reactions with (a) vRNA, (b) 5′RNA, (c) 3′RNA, and (d) iRNA. The top half of (a – d) shows a native agarose gel of capsid assembly reactions induced with the respective RNA. The number above each lane indicates the CP2:RNA stoichiometry; i.e. coat protein dimer:RNA, of the assembly reaction in that lane. The migration positions of the RNA fragment used in each panel, the recombinant MS2 capsid and aggregated material are indicated. Selected reactions were negatively stained with 2% (w/v) uranyl acetate and imaged by electron microscopy. The scale bars represent 200 nm.
For some RNAs, there is little difference between the mobility of the free RNA and that of the T=3 capsid, which means that gel mobility assays are not ideal for these studies. We therefore switched to using sedimentation velocity assays, which allowed us to characterise all the components present in the assembly mixtures simultaneously (Fig. 3). Under the assembly conditions, the MS2 virion, the recombinant T = 3 capsid and the starting CP2 are all well resolved. These components appeared homogeneous with sedimentation coefficients very similar to those reported earlier (Table 1).31-35 Note that the recombinant capsid30 lacks both MS2 genomic RNA and the single copy of the maturation (A) protein present in the virion. From absorption measurements, it appears to contain significant amounts of non-specifically encapsidated Escherichia coli RNA, which could include the recombinant coat protein mRNA. The sedimentation of the different RNA fragments suggested that they were conformationally heterogeneous, although each was dominated by a major species with an s20,w value proportional to its length. A fraction of each sample sedimented more slowly, perhaps corresponding to an unfolded or partly folded form. The sedimentation coefficients of the faster-migrating components are similar to data in the literature for full-length MS2 RNA,34,36 and those for the sub-genomic fragments are comparable to values for similarly sized ribosomal RNAs (Table 1).37 The amount of tertiary structure in these RNAs was assessed by removing divalent metal ions with EDTA.38 This treatment resulted in a 10~15% reduction in the observed s20,w values for the longer RNAs, suggesting that the fragments are at least partially folded. The smallest fragment (iRNA) showed no significant change in its s20,w value, implying that its structure is not dependent on divalent metal ions. The calculated frictional coefficients show that all the RNAs are highly asymmetric. Estimates of the Stokes radii suggest that all have persistence lengths that are larger than the internal diameter of the T=3 capsid (Table 1). Assembly must therefore involve a rearrangement of these RNA solution conformations to permit packaging within the protein container.
Fig. 3.
Assays of packaging efficiency with different lengths of RNA. The figure shows the results of sedimentation velocity assays of the different assembly reactions. The two top-most panels show C(S) versus S plots of capsid assembly reaction components; i.e. coat protein dimer (CP2); T = 3 capsid;30 vRNA and the unliganded RNA transcripts, all at a concentration of 40 nM. Note, the coat protein dimer sample is at the same concentration as the 90:1 reaction in the lower panel, is in assembly buffer and has been incubated similarly to the RNA–coat protein mixtures; i.e. it is the negative control for the effect of RNA on assembly. Its sedimentation does not differ from coat protein dimer starting material during the course of the experiment. The remaining panels show similar plots for titrations of each RNA with increasing concentrations of CP2 after 4 h at 20 °C in 0.04 M ammonium acetate, 1 mM magnesium acetate, pH 7.2.
Table 1.
Hydrodynamic properties of MS2 capsid assembly components
| S exp | S 20,w | f/f0 | R S | |
|---|---|---|---|---|
| MS2 virion | 74.6±0.5 | 75.1 | 1.1 | 11.6 |
| MS2 capsids | 65.4±2.0 | 65.8 | 1.2 | 11.7 |
| CP2 | 2.3±0.2 | 2.3 | 1.4 | 2.8 |
| Virion RNA | 25.7±1.5 | 25.8 | 3.0 | 18.4 |
| Virion RNA + EDTA | 22.1±1.8 | 22.3 | 3.4 | 21.3 |
| 5′RNA | 20.4±1.3 | 20.5 | 2.9 | 15.9 |
| 5′RNA + EDTA | 17.3±1.2 | 17.4 | 3.4 | 18.4 |
| 3′RNA | 21.2±1.3 | 21.3 | 2.9 | 16.1 |
| 3′RNA + EDTA | 18.5±1.6 | 18.6 | 3.5 | 19.5 |
| iRNA | 9.7±0.9 | 9.8 | 3.2 | 12.8 |
| iRNA + EDTA | 10.6±1.8 | 10.7 | 3.0 | 11.8 |
Sexp is the experimental sedimentation coefficient in 40 mM ammonium acetate, 1 mM magnesium acetate at 20 °C.
S20,w is the sedimentation coefficient corrected for buffer conditions. f/f0 The frictional coefficient, an indication of how spherical a particle is. f/f0 is 1 for a spherical particle.
RS=Stokes radius.
We then analysed capsid assembly with each RNA fragment in the presence of increasing ratios of CP2:RNA, from a stoichiometry of 10:1 to 110:1 (Fig. 3). The results are consistent with the conclusions from the gel mobility experiments; as the protein concentration increases there is an increase in the amount of RNA sedimenting with an s20,w value similar (62–66 S) to that of the recombinant T=3 capsid (65 S). There are only small differences in s20,w values between capsids assembled around the different RNAs. These are difficult to interpret unambiguously because RNA and protein have significantly different partial specific volumes, making it difficult to predict the s20,w values of each capsid product. The importance of this effect can be judged by comparing the s20,w values of capsids assembled around short (≤19 nt in length) RNA stem–loops, in which the RNAs are confined to the layer adjacent to the protein shell. These values range from 53–63 S, significantly lower than the values for the larger RNAs even though some of these contain a larger mass of RNA. Note that the sedimentation of CP2 alone in the upper panel is for the protein incubated at the same concentration as the 90:1 reaction in incubation buffer. Its behaviour is identical with that of the starting material, emphasising the role(s) of the RNA in promoting efficient assembly. Few, if any, RNA species sediment with s20,w values intermediate between the starting materials and the T=3 products. The stoichiometry of these particles is hard to estimate but the molar excess of CP2 is such that, in principle, each T=3 shell should contain only one RNA. At higher CP2:RNA ratios, in addition to assembled T=3 shells, small amounts of faster sedimenting material were also present, perhaps corresponding to the slower moving material seen in gel assays. The clear result is that not all RNAs promote assembly with the same efficiency, as judged by their ability to assemble into species with s20,w values similar to that of the T=3 capsid. The efficiency of encapsidation is inversely proportional to the length of the RNA, implying that there are energy barriers that must be overcome to package these RNAs into the capsid. This would be consistent with the need to reduce the Stokes radius of each RNA molecule. As a consequence of this property, vRNA is the least efficiently packaged RNA.
RNA-binding ligands prevent capsid assembly
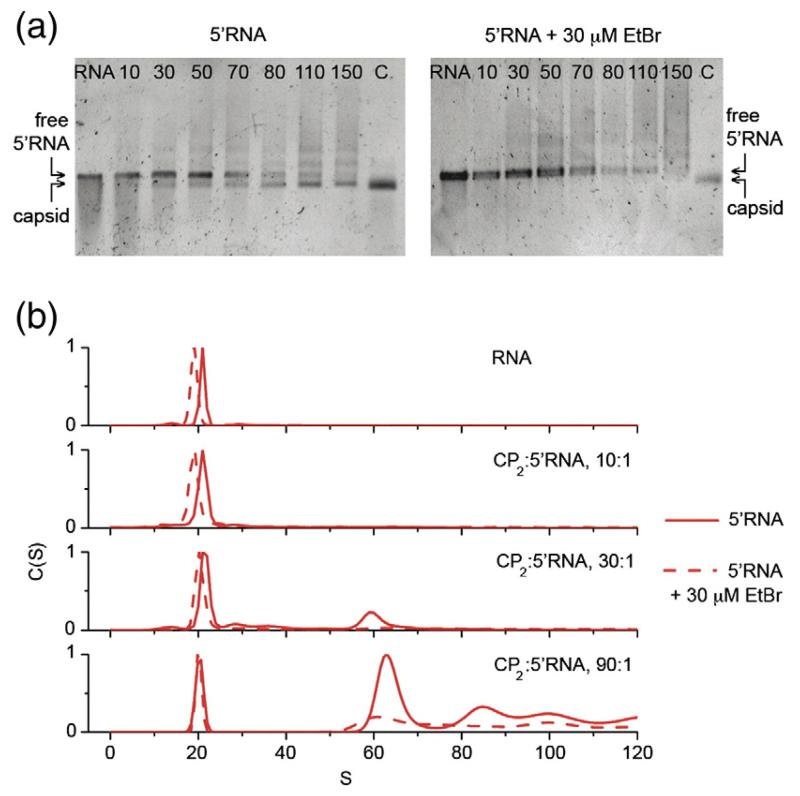
One consequence of the apparent structural rearrangement of RNA during assembly is that reagents that prevent or inhibit RNA folding should reduce the efficiency of packaging. To test this idea, we carried out similar assembly reactions with the 5′RNA in the presence of ethidium bromide (EtBr), which intercalates between the nucleotide bases thereby stiffening and distorting RNA structure.39,40 EtBr was added to the reaction at a stoichiometry of 1 EtBr molecule/8 nt of RNA, which is close to saturation.41,42 The RNA sediments more slowly in the presence of EtBr (Fig. 4), consistent with our expectation of the effect of the ligand on the conformation and flexibility of the RNA in solution. Very little capsid is formed in this reaction and most of the RNA remains protein-free as judged by its mobility in gels and its sedimentation in the centrifuge. These results might arise if EtBr competes with coat protein for binding the TR site within the fragments, preventing assembly initiation. Mass spectra of assembly reactions driven by TR stem–loops, however, show no evidence that EtBr binds to TR or reduces binding by coat protein.69 This result suggests that the EtBr effects arise due to increased resistance of the longer RNAs to compaction.
Fig. 4.
The effect(s) of ethidium bromide on the efficiency of capsid assembly. Capsid assembly was monitored at increasing stoichiometric ratios of CP2:5′RNA in the presence and in the absence of 30 μM EtBr. (a) Using native agarose gel electrophoresis, no signal corresponding to MS2 capsids was detected in the presence of EtBr after 4 h. (b) Sedimentation velocity analysis confirms that assembly is restricted in the presence of EtBr. 5′ RNA sediments slower in the presence of EtBr, suggesting that EtBr intercalation results in expansion of RNA structure and inhibits folding of the RNA into a structure compatible with the capsid.
Cryo-EM reconstruction of the sub-genomic assembly products
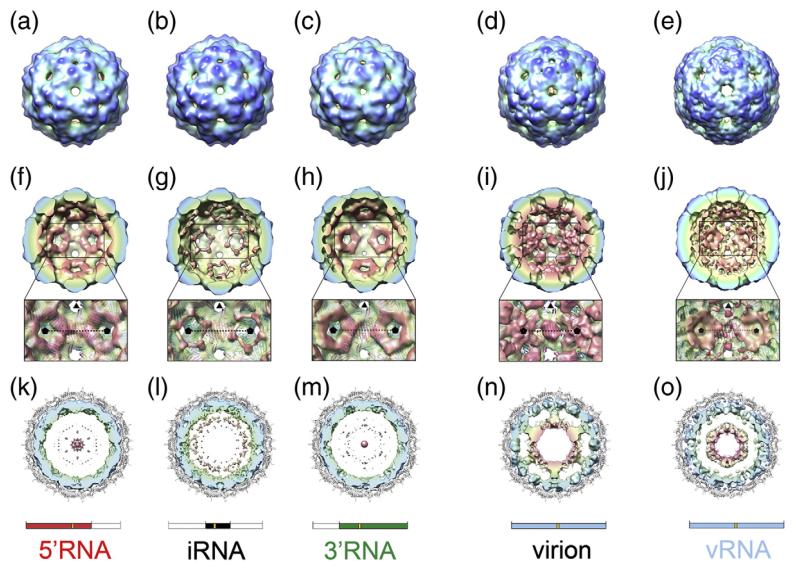
To assess the effect of RNA length on the structure of the packaged RNA, assembly reactions were carried out with each of the sub-genomic RNAs and the products were examined by cryo-EM. Three-dimensional (3D) reconstructions were then determined for the resultant T=3 virus-like particles (Fig. 5). The icosahedrally averaged cryo-EM maps (resolution 16–18 Å; Supplementary Data Fig. S1) are all similar in the protein layer to both the crystal structure for the MS2 virion at this resolution (Fig. 5a–c; Supplementary Data Fig. S2b–d) and our previous cryo-EM reconstruction (Fig. 5d).22 A shell of density corresponding to RNA is present in all three structures, lying immediately beneath the protein capsid layer (Fig. 5f–h). However, there is little if any ordered RNA density at shorter radii (Fig. 5k–m); i.e. towards the centre of the particle where the second shell of RNA is seen in the wild-type virion (Fig. 5n). The RNA density that is observed in these structures is primarily located around the icosahedral 5-fold axes, the sites where the A/B dimers are located in the T=3 capsid. Much less density is present at the 2-fold axes; i.e. beneath the positions of the C/C dimers (Fig. 5f–h). The fact that all of these 3-D structures are icosahedrally averaged complicates the interpretation of the density. However, these results clearly imply that, for each of the sub-genomic RNAs, there is high occupancy of RNA in the 60 binding sites below A/B dimers (surrounding the icosahedral 5-fold vertices), and a much lower occupancy beneath the 30 C/C dimers (on the icosahedral 2-fold axes). This disparity in occupancy is consistent with the idea that multiple RNA-mediated switching of quasi-conformers occurs throughout the assembly of the T=3 shell by RNA stem–loops of differing sequence. It appears that the genomic RNA acts to guide the conformation of protein dimers throughout the assembly of capsids.
Fig. 5.
Cryo-EM structures of MS2 capsids. (a–e) Surface representation of the cryo-EM structures of MS2 capsids reassembled in the presence of 5′RNA (a), iRNA (b), 3′RNA (c) and vRNA (e). For comparison, the cryo-EM structure of the native virion is also shown (d).22 All structures have been Fourier filtered at ~15 Å resolution, and are radially coloured from blue at high radius to red at low radius. (f–j) The rear halves of the 3-D structures of 5′RNA (f), iRNA (g), 3′RNA (h), wt MS2 virion (i) and vRNA (j) with the same radial colouring scheme. The view for each structure is looking down an icosahedral 2-fold axis. The inset is an expanded portion of each structure showing details of the RNA density surrounding the 5-fold axes (indicated by black pentagons). Atomic coordinates (cartoon representation and coloured as in Fig. 1) are fit into a now semi-transparent density. There is little or no density for RNA beneath the C/C dimers (on the 2-fold axes). (k–o) Central, 40 Å thick cross-section views of 5′RNA (k), iRNA (l), 3′RNA (m), the wt MS2 virion (n) and vRNA (o). In each case, the view is along a 3-fold axis, and the density for CP has been masked away. The fitted atomic coordinates are shown as a grey cartoon. The unfiltered density for packaged RNA is coloured radially. The virion (n) shows an outer shell of RNA immediately beneath the protein capsid and a second shell of RNA at lower radii, connected to the outer shell along 5-fold axes. The inner-shell density is not seen in the sub-genomic RNA cryo-EM structures at an equivalent threshold level, and only very weak features are seen in the maps at lower radii (k–m). The inner shell reappears when the vRNA is packaged (o). Schematics for the RNA present in the structures shown in each column are shown for clarity.
An assembly reaction was also carried out with vRNA but it was difficult to obtain sufficient data for a 3-D reconstruction because the majority of capsids appeared to have low RNA occupancy (Supplementary Data Fig. S3), compounding the low assembly efficiency (see above). In order to complete a reconstruction of an authentic vRNA containing capsid, “empty” particles (see Materials and Methods) were excluded from the dataset. The resulting reconstruction at ~30 Å resolution was calculated from just 80 individual particles (from 65 of the 288 micrographs initially recorded), and is clearly biased towards showing a ‘full’ structure. It does, however, allow a direct comparison between the vRNA capsid and the sub-genomic RNA fragment capsids (and the native virion), albeit one that is difficult because of the large differences between the respective resolutions of their electron density maps. However, a qualitative comparison suggests that in the protein capsid region, the vRNA map is again consistent with the known crystallo-graphic structure at this resolution (Fig. 5e; Supplementary Data Fig. S2d). The arrangement in the outer shell of RNA is similar to that seen earlier with the sub-genomic RNA maps; a ring of density surrounds the 5-fold axes below the A/B dimers with weaker density beneath the C/C dimers (Fig. 5j). This again implies a higher RNA occupancy at A/B than at C/C dimers when the virion RNA is packaged in vitro.
In marked contrast to the structures with subgenomic fragments, additional vRNA density is seen in a second shell at lower radius (Fig. 5o). The disparity in resolution again makes comparison to our ~9 Å reconstruction of the virion difficult, but this additional density is broadly similar in size and radius to that seen in the wild-type virion (Fig. 5n). In the latter map, the RNA shells are connected by columns of density extending along the 5-fold axes. This connection is absent from the vRNA structure; weak densities corresponding to the inner ends of these columns are visible but they do not extend outwards to join up with the outer shell (Fig. 5o). The similarities between vRNA and virion structures are consistent with the idea that the RNA tertiary structures within virions assembled in vivo and within particles assembled in vitro with vRNA are similar, albeit with reduced order in the vRNA particle, and the presence of maturation protein in the virion. We believe that the most probable explanation for the structural data presented here is that all these genomic RNAs of various lengths adopt similar folds in the capsid with respect to the coat protein layer. The features that form the inner shell are present at least in part but are not observed with the shorter RNAs. It appears they only become well ordered, and thus visible in an icosahedrally averaged cryo-EM structure, when the capsid packages RNA to the extent that it experiences electrostatic or steric constraints. This is consistent with the energy barriers inferred from the gel and centrifugation assembly experiments.
Discussion
Earlier, we showed that the MS2 coat protein dimer can be switched between quasi-conformers by binding to short RNA stem–loops and that there is little if any sequence specificity to this effect.4,18 When both RNA-free and RNA-bound forms of the coat protein dimer are present simultaneously, rapid self-assembly occurs towards the T=3 capsid. In contrast, each isolated conformation of the coat protein dimer is relatively stable, forming higher order species and eventually capsids only very inefficiently and slowly. These observations have led us to propose an assembly mechanism in which coat protein dimers of differing conformation bind alternately to a growing capsid shell.69 The experiments described above were aimed at clarifying whether this assembly mechanism applies when assembly is driven by genomic RNA–protein interactions.
From our results, it appears that large, subgenomic RNAs are encapsidated in vitro in a process that is highly co-operative. We assume that assembly initiates via the interaction of a CP2 dimer and the TR sequence present in each test RNA as a stem–loop.6 This should be the highest affinity RNA–protein contact, and would result in the formation of a CP2:RNA(TR) complex that would yield the first A/B protein conformer. Additional coat protein dimers in solution, which are C/C-like, will then bind to this complex creating intermediates on the pathway to the T=3 capsid. At particle 5-fold axes incoming protein subunits must adopt the A/B conformation in order to generate the correct curvature of the final capsid. If non-TR stem–loops throughout the large RNAs are able to engage with these dimers in the same way that TR does, then multiple RNA-mediated conformational switching events will contribute to the efficient assembly of the T=3 capsid. No such repeated RNA–protein interaction is required at the particle 2-fold sites, i.e. underneath the C/C dimers. Although RNA can bind there it either does so in a way that does not trigger the conformational change, which we have shown is likely to be caused by differential contacts to one polypeptide loop in one half of the protein dimer,18 or is unable to cause such a change because a dimer is already locked into a C/C site. The cooperativity of assembly would then arise because assembly intermediates would present a number of growth points for the addition of coat proteins dimers (protein–protein interfaces), which would also be adjacent to RNA fragments able to bind incoming subunits. Even though there is little sequence specificity to the latter interaction, there are likely to be many stem–loops within the genomic RNA that could promote conformer switching.43,44 RNA binding would also make positive contributions to the free energy of assembly. This mechanism is consistent with our previous results using smaller RNA fragments because we have shown that short natural sequence extensions of the TR stem–loop at either the 5′ or 3′ ends increase the rates of T=3 capsid assembly; the longer the RNA the faster the reaction.4 Such an assembly mechanism would be highly efficient,45 allowing the virus to complete specific encapsidation of its cognate RNA rapidly and before cell lysis. Heterologous RNA encapsidation would also be possible but would be less efficient if it did not initiate via a TR site.
This model makes several predictions, a number of which have been tested here. Multiple switching events require multiple RNA–protein interactions, in this case via RNA stem–loop structures. The required stem–loops need not necessarily be accessible in the solution conformation of the large RNAs used in these experiments, which might thus have to undergo conformational rearrangements of their own to expose the coat protein-binding sites. The centrifugation data are consistent with such a process (Figs. 3 and 4). All the RNAs tested need to be compacted during assembly. If they all initiate assembly at the TR site then we would expect there to be a defined relationship between the RNA sequence/structure and the lattice of RNA binding sites presented by the protein capsid. In addition, the assembly efficiency of longer RNAs under experimental conditions is lower than that for the shorter ones, presumably due to the need to rearrange more elements of secondary/tertiary structure. The fact that the shortest RNA (iRNA, 928nt) is most efficiently encapsidated suggests that the switching effect is not absolutely required throughout assembly because that fragment is not large enough to switch all 60 A/B dimers within the T=3 capsid (60× 20 nt). The stoichiometry of the experiments shown in Fig. 3 is not consistent with capsids containing multiple copies of the iRNA. Therefore RNA-mediated switching must occur for only a fraction of the assembly process, the remainder of the capsid being controlled by protein–protein interactions. These phases of assembly might occur with significantly different efficiency, with the RNA-mediated phase being the most efficient and including RNA conformational condensation. If it is more difficult for the RNA to rearrange its conformation, e.g. in the presence of a ligand such as EtBr or due to steric/electrostatic clashes for vRNA, assembly is either completely inhibited or less efficient. Obviously, assembly efficiency in vivo must depend on the starting conformation of the RNA being packaged, which might be different from that of the solution transcripts used here. A second prediction of the assembly model is that the RNA interactions at A/B and C/C dimers, and hence the cryo-EM density, will be different. Although the icosahedral-averaging makes unambiguous interpretation of the virion and the T=3 capsids assembled in vitro difficult, the results are consistent with this view. This is especially true of the sub-genomic fragments (Fig. 5 f–h and k–m) because the RNA density at 5-fold axes is so much stronger than at 2-fold axes.
The inner shell of RNA is largely absent from the particles assembled in vitro except when the full-length vRNA is packaged (Fig. 5j and o). This might reflect altered RNA folds in packing the different RNAs, or simply the lack of the steric/electrostatic constraints required to order the RNA features forming the inner ring if the capsid is not “full”. We favour the latter explanation because the alternative implies that the RNA and coat proteins can sense the global status of the RNA, i.e. whether it is full-length or not, at the start of assembly. It seems more likely that assembly is driven by local protein–protein and protein–RNA interactions. An obvious inference from the 3-D structures presented here is that there is a limit to the amount of RNA that can be packaged into the MS2 capsid. This is perhaps surprising, given that vRNA density in the virion, including the RNA-binding domain of the maturation protein, occupies only about one-third of the available internal volume.22 The nucleic acid packing density here is significantly less than that achieved when procapsids of double-stranded DNA bacteriophage are filled by packaging motors.46
The compaction of ssRNA genomes during assembly may well be a common property of these pathogens. Using atomic force microscopy, McPherson and colleagues showed that rapid removal of the protein coats from other ssRNA viruses allows at least a partial reversal of this event, the genomic RNAs expanding over time or upon mild heating.47,48 Gelbart, Knobler and colleagues showed that many ssRNA viruses have genomes that are smaller in solution than non-viral RNAs of similar length.49 The assembly mechanism described here for MS2 would generate an evolutionary constraint on its genomic RNA sequence, and thereby its size and structure. The idea that there is a repeated and functionally important RNA–protein contact in the T=3 shell seems at odds with the fact that no such ordered structure is seen in the X-ray density map. However, most of the RNA is ordered at intermediate resolution.22 In addition, many RNA stem–loops with significant sequence or functional group differences from TR will bind to all the CP2 in the context of the T=3 shell.50–53 Presumably, when each of the RNA stem–loops in contact with the protein layer are composed of different sequences and detailed secondary structures, the non-crystallographic symmetry averaging needed to solve the phage structure removes these features from the final electron density map.
In the MS2 virion, genomic RNA makes contact with the maturation protein that is absent from all of the in vitro assembly reactions. A single copy of the maturation protein is incorporated when the virion assembles in vivo.54,55 It is thought to make sequence-specific contacts with the genomic RNA close to both its 5′ and 3′ ends,56 as well as being the receptor binding component; the side of the F pilus in this case.57,58 Maturation protein could therefore potentially loop the RNA, substantially, reducing the conformational space that it can explore. Such a conformational restraint would limit the number of incorrect tertiary interactions that can form before assembly, potentially leading to increased assembly efficiency. Isolated maturation protein is largely insoluble and so it is not yet possible to test if this is true. Wild-type virion preparations do contain a fraction of assembled T=3 particles lacking the maturation protein. As a result, these particles are non-infectious although they contain virion RNA.59 It appears, therefore, that maturation protein–RNA binding is not an absolute pre-requisite to RNA encapsidation, even in vivo.
The prediction that there is a packaging size limit and the implication that secondary structure elements in solution are conserved in packaged RNAs are inherently testable and we are designing experiments to do so. The conclusions from the present study are that RNA binding can switch coat protein conformation throughout the assembly process, although this could be less crucial at the later stages, and that, simultaneously, protein-binding by the RNA leads to rearrangement of the RNA solution conformation that allows it to fit into the space being enclosed. The RNA and coat proteins therefore chaperone each other’s conformation during assembly.
Material and Methods
Protein and RNA preparation
MS2 coat protein was over-expressed in E. coli and purified as T=3 capsids as described.60 Recombinant capsids were treated with 60% (v/v) acetic acid followed by gel filtration and buffer exchange into 20 mM acetic acid to yield MS2 coat protein as dimers, which was confirmed by mass spectrometry and sedimentation analysis. The concentration of CP2 was calculated by measuring absorbance at 280 nm (A280) and at 260 nm (A260) and substituting the values into the formula:61
The MS2 sub-genomic RNA fragments were prepared by in vitro transcription from cDNA templates. To prepare these templates, a 2676 bp cDNA corresponding to nucleotides 1–2676 of the MS2 genome was generated by reverse transcription (RT)-PCR of MS2 vRNA using Thermoscript™ reverse transcriptase and the primers 1F and 2676R, and subsequently cloned into pGEM-T (Promega) to afford pGEMT_2676 (GenBank accession number GQ456168). Similarly, a 3528 bp cDNA corresponding to nucleotides 19–3546 of the MS2 genome was prepared using the primers 19F and 3546R, and cloned into pSMART (Lucigene) to afford pSMART_3528 (GenBank accession number GQ456167). It proved difficult to generate a full-length cDNA clone incorporating both terminal nucleotide sequences of the MS2 genome and therefore two clones were used. Sequence analysis indicated that the sequence of these clones is identical in overlapping regions.
T7 promoter-containing cDNA for use in in vitro transcription reactions was produced by PCR amplification from the cDNA plasmids using Pfu polymerase (Stratagene). The 5′ cDNA was amplified from pGEMT_2676 using the primers 1F_T7 and 2469R. The 3′ cDNA was amplified from pSMART_3528 using the primers 992F and 3569R. The iRNA was amplified from pSMART_3528 using the primers 1419F and 2346R. The primer sequences are given in Table 2.
Table 2.
Sequences of primers used to generate MS2 cDNA clones and T7 promoter containing cDNA for in vitro transcription
| Name | Primer |
|---|---|
| 1.F | GGGTGGGACCCCTTTCGG |
| 1.F_T7 | GATAATACGACTCACTATAGGGTGGGACCCCTTTCGG |
| 19F | GGTCCTGCTCAACTTCCTGTCG |
| 992F | GGGAAAAGGTGCCTTTCTCATT |
| 992F_T7 | GATAATACGACTCACTATAGGGAAAAGGTGCCTTTCTCATT |
| 1419F_T7 | GATAATACGACTCACTATAGGGGTCGCTGAATGGATCAGC |
| 2346R | CTGTAAACACTCCGTTCCCTACA |
| 2676R | TTGTGGAAAATAGTTCCCATCG |
| 2469R | CATTCAGGTCTATACCAACGG |
| 3546R | TAGTTACCAAATCGGGAGAATCC |
| 3569R | TGGGTGGTAACTAGCCAAGCAGCTAGTTACCAAATCGGGAGAATCC |
The MS2 sub-genomic RNA fragments, encompassing nucleotides 1–2469 (5′RNA), 991–3569 (3′RNA) and 1419–2346 (iRNA) of the MS2 RNA genome, were prepared by in vitro transcription using a MEGAscript® T7 transcription kit (Ambion) according to the manufacturer’s instructions. The MS2 virion RNA was isolated from wild type MS2 phage (a gift from Professor David S. Peabody, University of New Mexico) by extraction with phenol/chloroform followed by precipitation with 0.1 M sodium acetate and two volumes of EtOH. The integrity of the all RNA fragments was verified by denaturing agarose gel electrophoresis before use in capsid assembly reactions and their concentrations were determined from measurement of A260.
Assembly reactions
Capsid assembly was done by titrating the RNA with increasing amounts of CP2 in assembly buffer (40 mM ammonium acetate, 1 mM magnesium acetate, pH 7.2). The assembly reactions used a fixed concentration of RNA (40 nM) with 5–150 molar excess of CP2 in 20 μl and 320 μl final reaction volumes for gel mobility-shift analysis and sedimentation analysis, respectively, and allowed to proceed for 3 h at 24 °C.
Gel mobility-shift analysis
Loading buffer (5 μl of 40% (w/v) sucrose, 0.25% (w/v) bromophenol blue) was added to 20 μl of each assembly reaction before electrophoresis in a 1.0% (w/v) agarose gel (100 ml) at 80 V for 90 min and then stained in TBE running buffer containing 5 μg/ml EtBr and visualised at 254 nm.
Sedimentation velocity analysis
The assembly reactions were monitored using a Beckman Xl-1 analytical ultracentrifuge at 17,000 rpm for 6–12 h at 20 °C. Sedimentation was monitored by measuring A260 at intervals of 12 min. For the RNAs, the 50 first scans were used for fitting the sedimentation profiles with the program Sedfit with a continuous distribution c(s) Lamm equation model.62 RNAs were examined in the absence of divalent metal ions using a buffer containing 30 μM EDTA. The buffer viscosity and density were calculated with the program Sednterp (Dr Thomas Laue, Department of Biochemistry, University of New Hampshire, Durham, NH) and were estimated at 0.001008 poise (1 poise=0.1 Pa s) and η=0.99835 g/ml respectively. The RNA partial specific volume was set at 0.53 ml/g.35 Stokes radii for capsid assembly components were obtained directly from Sedfit. The partial specific volumes of the virion and capsid were set at 0.73 ml/g and at 0.68 ml/g for the CP2.63
Assembly reactions for cryo-EM
Capsid assembly was done by mixing RNA with CP2 in 50 mM Tris–acetate, 1 mM magnesium acetate, 40 mM ammonium acetate, pH 7.6, on ice. Assembly with virion RNA, 5′RNA and iRNA was done at a 1:60 molar ratio of RNA and CP2 for 13 h and then a CP2 chase was added to bring the molar ratio to 1:100 and assembly continued for a further 3 h. Assembly with 3′RNA was done at a 1:268 molar ratio of RNA to CP2 for 16 h. These conditions were varied slightly to get the best yields with the 3′RNA. T=3 shells were then picked for the cryo-EM dataset. After the set assembly time, all samples were flash-frozen by plunging into liquid nitrogen-cooled liquid ethane.
Cryo-EM data collection and image pre-processing
Sample preparation, data collection and image preprocessing were carried out as described for the wild-type MS2 sample,22 with the exception that micrographs were digitized using an Imacon Flextight 848 scanner (Hasselblad A/S, Copenhagen, Demark) at a final object sampling of 1.87 Å/pixel. Unless stated otherwise, all image processing steps were carried out using SPIDER.64
Particle selection of virion RNA assembled capsids
Particles recognizable as assembled capsids were selected interactively using the program Boxer,65 and corrected for the effects of the microscope contrast transfer function by computational flipping of image phases in SPIDER. The particles were centred and then subjected to multivariate statistical analysis (MSA) using IMAGIC.66 The classes showed two populations of capsids described below (and see Supplementary Data Fig. S3a). The entire data set was manually sorted into full and empty particles, and only the full capsids, which appeared to have packaged the entire virion RNA, were used in 3-D reconstruction. The reconstructions from the empty particles contained very little density for RNA in either inner or outer shells, and presumably represent virus-like particles (VLPs) assembled around shorter RNA fragments.
3-D reconstruction
3-D reconstruction and refinement were done as described,22 with the exception that the low-resolution starting model was projected across 53 orientations (angular sampling of 3.5°) for the sub-genomic RNA assembly reactions and across 19 orientations (angular sampling of 5.5°) for assembly with vRNA. The resolutions of the final reconstructions, calculated from 80, 1538, 1697 and 1740 molecular views for the vRNA, 5′RNA, iRNA and 3′RNA structures, respectively, were ~23–32 Å, 17.5 Å, 16 Å and 16 Å, respectively, using the 0.5 Fourier shell correlation criterion (Supplementary Data Fig. S1).
Atomic structure fitting and masking
Fitted atomic coordinates, described earlier,22 for the MS2 capsid (PDB ID 1AQ367) were docked into each EM map. To remove the density corresponding to the protein shell from the EM maps, the atomic coordinates for the capsid were converted to density, filtered to the calculated resolution of each map and normalised. This density was then subtracted from each map.
Data bank accession codes
The cryo-EM maps are deposited in the EMDB as EMD6792 (5′RNA), EMD6793 (iRNA), EMD6794 (3′RNA), and EMD6795 (vRNA).
Supplementary Material
Acknowledgements
We thank Dr Sean Killen and Mike Wallis for computing support. We thank Dr Arthur Olsen (TRSI, La Jolla), for suggesting the ethidium bromide experiments, Dr David Scott, University of Nottingham for discussions of the sedimentation results, and Professor Reidun Twarock, University of York, and the members of the P.G.S., N.A.R. and Twarock groups, for helpful discussions of the MS2 system. O.R. and K.T. thank the Wellcome Trust and the Leeds University Interdisciplinary Institute for Bionanotechnology, respectively, for PhD studentship support. N.A.R. and P.G.S. are grateful to The Leverhulme Trust for the project grant support that funded part of this work.
Abbreviations used
- ssRNA
single-stranded RNA
- cryo-EM
cryo-electron microscopy
- vRNA
virion RNA
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.jmb.2010.05.058
References
- 1.Schneemann A. The structural and functional role of RNA in icosahedral virus assembly. Annu. Rev. Microbiol. 2006;60:51–67. doi: 10.1146/annurev.micro.60.080805.142304. [DOI] [PubMed] [Google Scholar]
- 2.Harrison SC, Olson AJ, Schutt CE, Winkler FK. Tomato bushy stunt virus at 2.9 Å resolution. Nature. 1978;276:368–373. doi: 10.1038/276368a0. [DOI] [PubMed] [Google Scholar]
- 3.Caspar DL, Klug A. Physical principles in the construction of regular viruses. Cold Spring Harbor Symp. Quant. Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- 4.Basnak G, Morton VL, Rolfsson O, Stonehouse NJ, Ashcroft AE, Stockley PG. Viral genomic single-stranded RNA directs the pathway toward a T = 3 capsid. J. Mol. Biol. 2010;395:924–936. doi: 10.1016/j.jmb.2009.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stockley PG, Rolfsson O, Thompson GS, Basnak G, Francese S, Stonehouse NJ, et al. A simple, RNA-mediated allosteric switch controls the pathway to formation of a T = 3 viral capsid. J. Mol. Biol. 2007;369:541–552. doi: 10.1016/j.jmb.2007.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gell C, Sabir T, Westwood J, Rashid A, Smith DAM, Harris SA, Stockley PG. Single-molecule fluorescence resonance energy transfer assays reveal heterogeneous folding ensembles in a simple RNA stem-loop. J. Mol. Biol. 2008;384:264–278. doi: 10.1016/j.jmb.2008.08.088. [DOI] [PubMed] [Google Scholar]
- 7.Lago H, Fonseca SA, Murray JB, Stonehouse NJ, Stockley PG. Dissecting the key recognition features of the MS2 bacteriophage translational repression complex. Nucleic Acids Res. 1998;26:1337–1344. doi: 10.1093/nar/26.5.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Valegård K, Murray JB, Stonehouse NJ, van den Worm S, Stockley PG, Liljas L. The three-dimensional structures of two complexes between recombinant MS2 capsids and RNA operator fragments reveal sequence-specific protein–RNA interactions. J. Mol. Biol. 1997;270:724–738. doi: 10.1006/jmbi.1997.1144. [DOI] [PubMed] [Google Scholar]
- 9.Valegård K, Murray JB, Stockley PG, Stonehouse NJ, Liljas L. Crystal structure of an RNA bacteriophage coat protein-operator complex. Nature. 1994;371:623–626. doi: 10.1038/371623a0. [DOI] [PubMed] [Google Scholar]
- 10.Romaniuk PJ, Lowary P, Wu HN, Stormo G, Uhlenbeck OC. RNA binding site of R17 coat protein. Biochemistry. 1987;26:1563–1568. doi: 10.1021/bi00380a011. [DOI] [PubMed] [Google Scholar]
- 11.Gralla J, Steitz JA, Crothers DM. Direct physical evidence for secondary structure in an isolated fragment of R17 bacteriophage mRNA. Nature. 1974;248:204–208. doi: 10.1038/248204a0. [DOI] [PubMed] [Google Scholar]
- 12.Ling CM, Hung PP, Overby LR. Independent assembly of Qbeta and MS2 phages in doubly infected Escherichia coli. Virology. 1970;40:920–929. doi: 10.1016/0042-6822(70)90138-8. [DOI] [PubMed] [Google Scholar]
- 13.Beckett D, Wu HN, Uhlenbeck OC. Roles of operator and non-operator RNA sequences in bacteriophage R17 capsid assembly. J. Mol. Biol. 1988;204:939–947. doi: 10.1016/0022-2836(88)90053-8. [DOI] [PubMed] [Google Scholar]
- 14.Beckett D, Uhlenbeck OC. Ribonucleo-protein complexes of R17 coat protein and a translational operator analog. J. Mol. Biol. 1988;204:927–938. doi: 10.1016/0022-2836(88)90052-6. [DOI] [PubMed] [Google Scholar]
- 15.Golmohammadi R, Valegård K, Fridborg K, Liljas L. The refined structure of bacteriophage MS2 at 2.8 Å resolution. J. Mol. Biol. 1993;234:620–639. doi: 10.1006/jmbi.1993.1616. [DOI] [PubMed] [Google Scholar]
- 16.Valegård K, Liljas L, Fridborg K, Unge T. The three-dimensional structure of the bacterial virus MS2. Nature. 1990;345:36–41. doi: 10.1038/345036a0. [DOI] [PubMed] [Google Scholar]
- 17.Lago H, Parrott AM, Moss T, Stonehouse NJ, Stockley PG. Probing the kinetics of formation of the bacteriophage MS2 translational operator complex: identification of a protein conformer unable to bind RNA. J. Mol. Biol. 2001;305:1131–1144. doi: 10.1006/jmbi.2000.4355. [DOI] [PubMed] [Google Scholar]
- 18.Dykeman EC, Stockley PG, Twarock R. Dynamic allostery controls coat protein conformer switching during MS2 phage assembly. J. Mol. Biol. 2010;395:916–923. doi: 10.1016/j.jmb.2009.11.016. [DOI] [PubMed] [Google Scholar]
- 19.Tsuruta H, Reddy VS, Wikoff WR, Johnson JE. Imaging RNA and dynamic protein segments with low-resolution virus crystallography: experimental design, data processing and implications of electron density maps. J. Mol. Biol. 1998;284:1439–1452. doi: 10.1006/jmbi.1998.2231. [DOI] [PubMed] [Google Scholar]
- 20.Tang L, Johnson KN, Ball LA, Lin T, Yeager M, Johnson JE. The structure of pariacoto virus reveals a dodecahedral cage of duplex RNA. Nat. Struct. Biol. 2001;8:77–83. doi: 10.1038/83089. [DOI] [PubMed] [Google Scholar]
- 21.Opalka N, Beckmann R, Boisset N, Simon MN, Russel M, Darst SA. Structure of the filamentous phage pIV multimer by cryo-electron microscopy. J. Mol. Biol. 2003;325:461–470. doi: 10.1016/s0022-2836(02)01246-9. [DOI] [PubMed] [Google Scholar]
- 22.Toropova K, Basnak G, Twarock R, Stockley PG, Ranson NA. The three-dimensional structure of genomic RNA in bacteriophage MS2: implications for assembly. J. Mol. Biol. 2008;375:824–836. doi: 10.1016/j.jmb.2007.08.067. [DOI] [PubMed] [Google Scholar]
- 23.van den Worm SHE, Koning RI, Warmenhoven HJ, Koerten HK, van Duin J. Cryo electron microscopy reconstructions of the Leviviridae unveil the densest icosahedral RNA packing possible. J. Mol. Biol. 2006;363:858–865. doi: 10.1016/j.jmb.2006.08.053. [DOI] [PubMed] [Google Scholar]
- 24.Koning R, van den Worm S, Plaisier JR, van Duin J, Pieter Abrahams J, Koerten H. Visualization by cryo-electron microscopy of genomic RNA that binds to the protein capsid inside bacteriophage MS2. J. Mol. Biol. 2003;332:415–422. doi: 10.1016/s0022-2836(03)00846-5. [DOI] [PubMed] [Google Scholar]
- 25.Lee KK, Johnson JE. Complementary approaches to structure determination of icosahedral viruses. Curr. Opin. Struct. Biol. 2003;13:558–569. doi: 10.1016/j.sbi.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 26.Venkataraman S, Reddy SP, Loo J, Idamakanti N, Hallenbeck PL, Reddy VS. Structure of Seneca Valley Virus-001: an oncolytic picornavirus representing a new genus. Structure. 2008;16:1555–1561. doi: 10.1016/j.str.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tihova M, Dryden KA, Le T. v. L., Harvey SC, Johnson JE, Yeager M, Schneemann A. Nodavirus coat protein imposes dodecahedral RNA structure independent of nucleotide sequence and length. J. Virol. 2004;78:2897–2905. doi: 10.1128/JVI.78.6.2897-2905.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carey J, Uhlenbeck OC. Kinetic and thermodynamic characterization of the R17 coat protein-ribonucleic acid interaction. Biochemistry. 1983;22:2610–2615. doi: 10.1021/bi00280a003. [DOI] [PubMed] [Google Scholar]
- 29.Meyvisch C, Teuchy H, van Montagu M. Electron microscopy of the intracellular development of bacteriophage MS2 in Escherichia coli. J. Virol. 1974;13:1356–1367. doi: 10.1128/jvi.13.6.1356-1367.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mastico RA, Talbot SJ, Stockley PG. Multiple presentation of foreign peptides on the surface of an RNA-free spherical bacteriophage capsid. J. Gen. Virol. 1993;74:541–548. doi: 10.1099/0022-1317-74-4-541. [DOI] [PubMed] [Google Scholar]
- 31.Hohn T. Role of RNA in the assembly process of bacteriophage fr. J. Mol. Biol. 1969;400:935–947. doi: 10.1016/0022-2836(69)90088-6. [DOI] [PubMed] [Google Scholar]
- 32.Hohn T. The assembly of protein particles of the RNA bacteriophage fr in absence of RNA. Biochem. Biophys. Res. Commun. 1969;36:7–17. doi: 10.1016/0006-291x(69)90641-x. [DOI] [PubMed] [Google Scholar]
- 33.Hohn T. Selfassembly of defective particles of the bacteriophage fr. Eur. J. Biochem. 1967;2:152–155. doi: 10.1111/j.1432-1033.1967.tb00119.x. [DOI] [PubMed] [Google Scholar]
- 34.Sugiyama T, Hebert RR, Hartman KA. Ribonucleoprotein complexes formed between bacteriophage MS2 RNA and MS2 protein in vitro. J. Mol. Biol. 1967;25:455–463. doi: 10.1016/0022-2836(67)90198-2. [DOI] [PubMed] [Google Scholar]
- 35.Enger MD, Stubbs EA, Mitra S, Kaesberg P. Biophysical characteristics of the RNA-containing bacterial virus R17. Proc. Natl Acad. Sci. USA. 1963;49:857–860. doi: 10.1073/pnas.49.6.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fiers W. Studies on the bacteriophage MS2 III. Sedimentation heterogeneity of viral RNA preparations. Virology. 1967;33:413–424. doi: 10.1016/0042-6822(67)90117-1. [DOI] [PubMed] [Google Scholar]
- 37.Yi QM, Wong KP. The effects of magnesium ions on the hydrodynamic shape, conformation, and stability of the ribosomal 23S RNA from E. coli. Biochem. Biophys. Res. Commun. 1982;104:733–739. doi: 10.1016/0006-291x(82)90698-2. [DOI] [PubMed] [Google Scholar]
- 38.Leipply D, Draper DE. Dependence of RNA tertiary structural stability on mg(2+) concentration: interpretation of the hill equation and coefficient. Biochemistry. 2010;49:1843–1853. doi: 10.1021/bi902036j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsai CC, Jain SC, Sobell HM. Visualization of drug-nucleic acid interactions at atomic resolution. I. Structure of an ethidium/dinucleoside monophosphate crystalline complex, ethidium:5-iodouridylyl (3′-5′) adenosine. J. Mol. Biol. 1977;114:301–315. doi: 10.1016/0022-2836(77)90252-2. [DOI] [PubMed] [Google Scholar]
- 40.Waring MJ. Complex formation between ethidium bromide and nucleic acids. J. Mol. Biol. 1965;13:269–282. doi: 10.1016/s0022-2836(65)80096-1. [DOI] [PubMed] [Google Scholar]
- 41.LePecq JB, Paoletti C. A fluorescent complex between ethidium bromide and nucleic acids. Physical–chemical characterization. J. Mol. Biol. 1967;27:87–106. doi: 10.1016/0022-2836(67)90353-1. [DOI] [PubMed] [Google Scholar]
- 42.Gray PN, Saunders GF. Binding of ethidium bromide to 5-S ribosomal RNA. Biochim. Biophys. Acta. 1971;254:60–77. doi: 10.1016/0005-2787(71)90114-6. [DOI] [PubMed] [Google Scholar]
- 43.Skripkin EA, Adhin MR, de Smit MH, van Duin J. Secondary structure of the central region of bacteriophage MS2 RNA. Conservation and biological significance. J. Mol. Biol. 1990;211:447–463. doi: 10.1016/0022-2836(90)90364-R. [DOI] [PubMed] [Google Scholar]
- 44.Fiers W, Contreras R, Duerinck F, Haegeman G, Iserentant D, Merregaert J, et al. Complete nucleotide sequence of bacteriophage MS2 RNA: primary and secondary structure of the replicase gene. Nature. 1976;260:500–507. doi: 10.1038/260500a0. [DOI] [PubMed] [Google Scholar]
- 45.Williamson JR. Cooperativity in macromolecular assembly. Nat. Chem. Biol. 2008;4:458–465. doi: 10.1038/nchembio.102. [DOI] [PubMed] [Google Scholar]
- 46.Earnshaw WC, Casjens SR. DNA packaging by the double-stranded DNA bacteriophages. Cell. 1980;21:319–331. doi: 10.1016/0092-8674(80)90468-7. [DOI] [PubMed] [Google Scholar]
- 47.Kuznetsov YG, McPherson A. Atomic force microscopy investigation of Turnip Yellow Mosaic Virus capsid disruption and RNA extrusion. Virology. 2006;352:329–337. doi: 10.1016/j.virol.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 48.Kuznetsov YG, Daijogo S, Zhou J, Semler BL, McPherson A. Atomic force microscopy analysis of icosahedral virus RNA. J. Mol. Biol. 2005;347:41–52. doi: 10.1016/j.jmb.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 49.Yoffe AM, Prinsen P, Gopal A, Knobler CM, Gelbart WM, Ben-Shaul A. Predicting the sizes of large RNA molecules. Proc. Natl Acad. Sci. USA. 2008;105:16153–16158. doi: 10.1073/pnas.0808089105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Horn WT, Convery MA, Stonehouse NJ, Adams CJ, Liljas L, Phillips SEV, Stockley PG. The crystal structure of a high affinity RNA stem-loop complexed with the bacteriophage MS2 capsid: further challenges in the modeling of ligand-RNA interactions. RNA. 2004;10:1776–1782. doi: 10.1261/rna.7710304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Helgstrand C, Grahn E, Moss T, Stonehouse NJ, Tars K, Stockley PG, Liljas L. Investigating the structural basis of purine specificity in the structures of MS2 coat protein RNA translational operator hairpins. Nucleic Acids Res. 2002;30:2678–2685. doi: 10.1093/nar/gkf371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grahn E, Stonehouse NJ, Adams CJ, Fridborg K, Beigelman L, Matulic-Adamic J, Warriner SL, Stockley PG, Liljas L. Deletion of a single hydrogen bonding atom from the MS2 RNA operator leads to dramatic rearrangements at the RNA-coat protein interface. Nucleic Acids Res. 2000;28:4611–4616. doi: 10.1093/nar/28.23.4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grahn E, Moss T, Helgstrand C, Fridborg K, Sundaram M, Tars K, et al. Structural basis of pyrimidine specificity in the MS2 RNA hairpin-coat-protein complex. RNA. 2001;7:1616–1627. [PMC free article] [PubMed] [Google Scholar]
- 54.Steitz JA. Identification of the A protein as a structural component of bacteriophage R17. J. Mol. Biol. 1968;33:923–936. doi: 10.1016/0022-2836(68)90328-8. [DOI] [PubMed] [Google Scholar]
- 55.Steitz JA. Isolation of the A protein from bacteriphage R17. J. Mol. Biol. 1968;33:937–945. doi: 10.1016/0022-2836(68)90329-x. [DOI] [PubMed] [Google Scholar]
- 56.Shiba T, Suzuki Y. Localization of A protein in the RNA-A protein complex of RNA phage MS2. Biochim. Biophys. Acta. 1981;654:249–255. doi: 10.1016/0005-2787(81)90179-9. [DOI] [PubMed] [Google Scholar]
- 57.Brinton CC, Gemski P, Carnahan J. A new type of bacterial pilus genetically controlled by the fertility factor of E. coli K 12 and its role in chromosome transfer. Proc. Natl Acad. Sci. USA. 1964;52:776–783. doi: 10.1073/pnas.52.3.776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Crawford EM, Gesteland RF. The adsorption of bacteriophage R-17. Virol. J. 1964;22:165–167. [Google Scholar]
- 59.Krahn PM, Paranchych W. Heterogeneous distribution of A protein in R17 phage preparations. Virology. 1971;43:533–535. doi: 10.1016/0042-6822(71)90332-1. [DOI] [PubMed] [Google Scholar]
- 60.Stonehouse NJ, Stockley PG. Effects of amino acid substitution on the thermal stability of MS2 capsids lacking genomic RNA. FEBS Lett. 1993;334:355–359. doi: 10.1016/0014-5793(93)80711-3. [DOI] [PubMed] [Google Scholar]
- 61.Layne E. Spectrophotometric and turbidimetric methods for measuring proteins. Methods Enzymol. 1957;3:447–454. [Google Scholar]
- 62.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Strauss JH, Sinsheimer RL. Purification and properties of bacteriophage MS2 and of its ribonucleic acid. J. Mol. Biol. 1963;7:43–54. doi: 10.1016/s0022-2836(63)80017-0. [DOI] [PubMed] [Google Scholar]
- 64.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 65.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 66.van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J. Struct. Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- 67.van den Worm SH, Stonehouse NJ, Valegård K, Murray JB, Walton C, et al. Crystal structures of MS2 coat protein mutants in complex with wild-type RNA operator fragments. Nucleic Acids Res. 1998;26:1345–1351. doi: 10.1093/nar/26.5.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera – a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 69.Victoria L, Morton VL, Dykeman EC, Stonehouse NJ, Ashcroft AE, Twarock R, Stockley PG. The impact of viral RNA on assembly pathway selection. J. Mol. Biol. 2010;401:298–308. doi: 10.1016/j.jmb.2010.05.059. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.