Abstract
In the production of fermented foods, microbes play an important role. Optimization of fermentation processes or starter culture production traditionally was a trial-and-error approach inspired by expert knowledge of the fermentation process. Current developments in high-throughput ‘omics’ technologies allow developing more rational approaches to improve fermentation processes both from the food functionality as well as from the food safety perspective. Here, the authors thematically review typical bioinformatics techniques and approaches to improve various aspects of the microbial production of fermented food products and food safety.
Keywords: bioinformatics; microorganisms; food, genomics; predictive models
Background
Food is an indispensable part of our daily life. Many food products undergo some form of processing before they reach the consumer, ranging from fermentation to packaging. In many of these processes, microorganisms play important roles, either in transforming the food into the desired end product (e.g. fermentation of olives, rice, bread, alcoholic beverages such as beer and wine, fermented meat, kimchi and various fermented dairy products such as cheese and yogurt) or in spoiling or contaminating the food.
The type of microorganisms used in a fermentation process greatly influences the properties of the fermented product [1]. For example, yeasts produce ethanol as the main fermentation product, whereas the main fermentation product of lactic acid bacteria is lactic acid. The food industry is very active in optimizing strain performance with respect to diversification of product properties such as flavour and texture and with respect to controlling fermentation, by using defined starter cultures to initiate the fermentation process [1].
Strain optimization is an expert-knowledge-guided process involving trial-and-error approaches that are nowadays increasingly backed up by recent high-throughput ‘omics’ developments to improve fermentation processes [2] and to assess safety of food products [3].
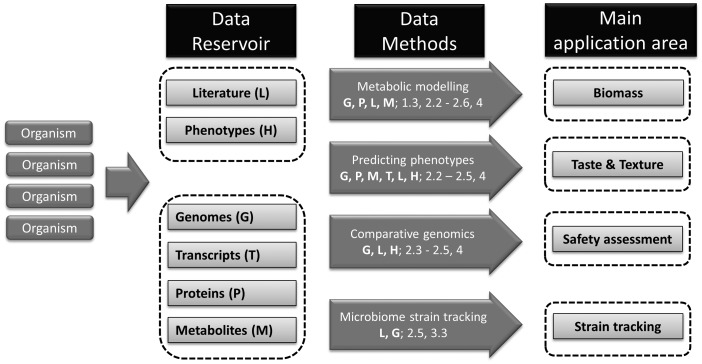
Bioinformatics plays an increasing role in predicting and assessing the desired and undesired effects of microorganisms on food [4]. A combination of bioinformatics with laboratory verification of selected findings is particularly powerful. In this review, we focus on bioinformatics methods that can be used to improve the microbial production of fermented food products. These include genomics-based functional predictions, the creation of genome-scale metabolic models and prediction of complex food properties, such as taste and texture, and properties of complex fermentations. All application areas (outlined in the paragraphs below) and their relation to data streams and bioinformatics are described in Figure 1. A glossary of the bioinformatics concepts, methods and tools is provided in Table 1.
Figure 1.
Data and bioinformatics applied in food application areas. Central in this figure are the food application areas (right panel). From organisms, different data sets can be obtained (data reservoir); their abbreviation is given within parentheses. Middle panel: one (of many important) methods and other methods/data sources (see Table 1 for an explanation) relevant for a main application area shown. Interpretation example: for safety assessment, genomes (G), literature (L) and phenotypes (H) are used with the gene function annotation (2.3), orthology (2.4), comparative genomics (2.5) and predicting phenotypes (4) techniques (see Table 1).
Table 1.
Glossary of food bioinformatics concepts and techniques, their explanation and their application
| Term | Description and examples of tools |
|---|---|
| 1. Big data/grid/cloud/ | With the increasing volume and heterogeneity of data sets (often referred to as “Big Data”), high performance computing is needed for analysis of the data. Many bioinformatics methods have been adapted to run on clusters of multiple computers (grid computing) and on large remotely located servers (cloud computing) [5]. Galaxy [6] and KNIME [7] are two popular software solutions to integrate and distribute larger data analysis tasks to the grid/cloud. Examples of cloud-based bioinformatics applications: HBLAST (BLAST, the most used bioinformatics sequence alignment tool) [8]; TPP, a proteomic analysis tool [9]; HIPPIE: promoter analysis provided as Amazon Machine Image [10]; BG7: bacterial genome annotation based on Amazon Web Services [11]. |
| 1.1 Data mining | Statistical and machine learning techniques to determine trends in typically large data sets. Unsupervised techniques (sample grouping is not explicitly used in the analysis) include: principal component analysis (PCA) and clustering algorithms (e.g. K-means, hierarchical). Supervised techniques (sample grouping is taken into account) include: ANOVA, Mann–Whitney U test, partial least squares analysis (PLS), machine learning (e.g. by support vector machines (SVM) [12] and random forest [13]): a computational model is trained to use properties derived from samples to predict the status of samples. See term 4 for examples. |
| 1.2 Virtual machines (VM) | A large computer file (disk image) that consists of an operating system (e.g. Linux), software tools and data. The image can be run on an actual computer using virtual machine software that emulates an actual computer. In other words, a computer in a computer. The advantage of VMs is that they are portable (can be run on many different types of computer hardware), easy to backup and more straight-forward to maintain. Examples of the use of VMs are the generic bioinformatics tools in the NEBC Bio-Linux distribution [14] and the 16s analysis suite Qiime (see term 3.1). |
| 1.3 Databases | Databases are organized collections of biological data. Bioinformatics is only successful if databases with high-quality data are available, together with structured vocabularies that describe the content of the data sets. An updated overview of relevant biological databases can be found here: http://www.oxfordjournals.org/our_journals/nar/database/cap/. |
| 2. Genome sequencing | Determining the complete genome sequence of a microbial strain of interest. Next-generation sequencing (NGS) techniques allow for high-throughput and high-quality sequencing results. Especially the combination if different techniques (e.g. Illumina and Pacific Biosystems or PacBio) result in high-quality (circular) genomes [15]. |
| 2.1 Sequencing data (FASTQ) | Sequencing data are represented in FASTQ format. These files provide, next to the raw sequence data, additional information regarding the quality of the reads. In this manner, quality control and trimming can be applied. |
| 2.2 Assembly | Raw sequence reads of different NGS technologies can be assembled into contigs, long stretches of DNA sequence representing part of the genome. Most of the assembly methods are based on alignment of sequence reads with each other (de novo assembly) or against a reference genome (mapping assembly), thereby generating long DNA sequences from the fragments generated by the sequencing. Some examples of assembly tools are Ray [16], MIRA [17] and IDBA [18]. |
| 2.2.1 Scaffolding | Organizing the contigs from the assembly (2.2) to larger, gapped, DNA sequences. Some NGS techniques (e.g. Illumina) allow the synthesis of paired end (PE) or mate pair (MP) libraries; libraries with a fixed insert size that are sequenced at both ends. As reads span a larger DNA fragment, the matched reads pairs can be used to order contigs, even if the sequence in between the contigs has not been assembled. In general, most assembly tools allow for scaffolding, but also dedicated tools exist, such as SSPACE [19]. |
| 2.2.2 Gap closure strategies | After scaffolding, genome sequences will most often contain gaps. Common strategies to fill these gaps are generating new sequencing data using, for example, PacBio’s long reads [15], or predicting the most likely order and orientation of the contigs using bioinformatics tools like Projector2 [20] or Mauve [21]. These tools infer contig order by comparing them to one or more reference sequences. |
| 2.3 Gene function annotation | Gene function is typically inferred from similarity in amino acid sequence. Gene functions can be predicted by comparing sequences to databases containing genes with known functions with tools like RAST [22] and Prokka [23]. |
| 2.4 Orthology | Genes in different organisms are orthologous when they were the same gene in the last common ancestor. Reconstructing the evolutionary history of genes allows the prediction of functional equivalence (i.e. orthologous genes are likely to have similar functions). Tools are OrthoMCL [24] and Orthagogue [25]. |
| 2.5 Comparative genomics | All analyses in which genome sequences or genome content of multiple organisms are compared. |
| 2.6 Metabolic modelling | Prediction of growth, and recruitment of metabolic pathways, of microbes by using the genome sequence as an inventory of all possible metabolic reactions. Genome-scale metabolic models can be constructed using automated [26] or comparative genomics analyses [27]. Once constructed, the models can be used to simulate growth by, for example, flux balance analysis (FBA) and to determine the boundaries of fluxes by flux variability analysis (FVA). Tools for modelling are Pysces [28], the SEED [26] and VANTED [29]. |
| 3 Microbiome analysis | All microbes present in a particular niche are termed a microbiome. Analysis of microbiomes can be done using different next-generation sequencing-based techniques (see below). |
| 3.1 16s rRNA sequencing | 16s amplicon sequencing is the generation of sequence reads from conserved regions of the 16s gene. Amplicon sequencing (e.g. by Illumina) is used to identify the bacterial (and sometimes archaeal) component of microbial communities. Examples of software to infer community composition from sequencing data are Qiime [30] and Mothur [31]. 16s sequencing is a relatively cheap and well-established technique and as such an ideal starting point for characterization of complex cultures for which limited prior knowledge is available. |
| 3.1.1 Functional prediction | 16s sequences derived from a particular ecological niche indicate the taxa present and their relative abundance. From these data, presence of gene functions in those taxa can be performed using, e.g., PICRUSt [32]. It infers the presence of gene functions in given taxa using already sequenced genomes part of the same taxa. |
| 3.2 Shotgun metagenomics and metatranscriptomics | Random fragments of the DNA or (enriched) mRNA of a given microbiome are sequenced with next-generation sequencing [33, 34]. Metagenomics and metatranscriptomics techniques are powerful, as they allow circumventing growing microbes while still determining their gene content or gene expression. This provides insight into the molecular functions encoded by the DNA, taxonomic assignment of that DNA fragments or inferring similar information for expressed mRNAs. This method can be used in addition to sequencing of individual isolates from complex cultures. Sequencing of individual isolates, however, has the advantage that comparative genomics can be done and metabolic models can be built more straight-forwardly, provided that the isolates under study are representative of the biodiversity present in the complex culture. |
| 3.2.1 Assembly | Using the sequence overlap, the DNA/RNA-derived sequences can be assembled into larger contigs, see [35] for a recent comparison of tools. Functional annotation of these larger fragments is more straight-forward, but the fraction of reads that can be assembled into contigs depends on both the complexity of the microbiome (many different microbes with varying abundances) as well as the presence of microdiversity (many different microbes with similar genome sequences). |
| 3.2.2 Annotation | Similar to the genome of a single bacterium, the sequences of a metagenome can be functionally and taxonomically annotated by comparing (assembled) sequences or predicted gene products against one or more reference databases with sequences with known functions from known taxonomic origin. Gene context such as operons are, however, primarily missing in shotgun metagenomics reads/contigs. A few tools are: PhymmBL [36] (taxonomic classification using sequence-based models), MG-RAST [37] (functional and taxonomic classification using alignment to reference databases) and MetaPhlAn [38] (taxonomy prediction using taxon-specific marker genes). |
| 3.3 Strain typing and tracking | Pinpointing the presence of a particular microbe (strain) in a biological sample. Using MLST markers [39] (multi-locus sequence typing), PCR based on unique DNA fragments [40] or strain-specific markers [41], the abundance of particular strains can be followed during the course of a fermentation. Potential downside of these techniques is that only known biodiversity can be traced. Therefore, the performance must be evaluated on new strains. New biodiversity can be uncovered, provided that a genomic target is well-designed (e.g. targeting a gene that is single copy with sufficient resolution to distinguish between strains). |
| 4 Predicting phenotypes | Gene–trait matching: machine learning or statistics methods are used to predict the phenotype of a bacterial strain based on the presence/absence of particular genes [42], (parts of) metabolic pathways [43, 44] or classifications from experts [45]. Transcriptome–trait matching: gene expression data (based on microarray or RNAseq) instead of gene presence are used. Transcriptome data from multiple strains grown under the same condition [46] or the same strain grown under different conditions can be used [47–49]. |
| 5 Metabolomics | The simultaneous measurement of multiple metabolites in biological samples [50]. Metabolomics is a technique that can be applied to describe reaction products of microorganisms in defined media and in food samples. Its data are very suitable to be associated to results from sensory measurements [51, 52]. |
Translating genome information into functional predictions
The prediction of function from sequence information is one of the fundamental roles of bioinformatics. The large variety of sequencing techniques generates a large amount of genomics data. Harnessing the power of these data requires careful identification of functional elements in these data and associating the sequence information with function, for example by comparing predicted protein sequences to sequences with known functions. This type of analysis can identify functions for genes (crucial information for metabolic modelling; see below), e.g. prediction of laccases [53]; predict functions for most genes in a bacterial genome [23, 54]; and suggest properties for specific strains of bacteria by projecting the predicted functions of all its genes on pathway databases [55, 56], predicting properties of, e.g., Bifidobacteria in the gut environment [57] or even predict functionalities of complex microbial communities [22, 32]. For genes where a sequence similarity search does not yield a good prediction, their function may be deduced by correlating the presence and absence of the gene in organisms with the presence and absence of a certain phenotypic trait in the same set of organisms (also referred to as gene–trait matching; GTM) [42, 58]. For example, a set of proteins was predicted to be involved in the degradation of plant (oligo-) saccharides by linking isolation source of bacteria to gene presence/absence [59]. Comparative analysis of the genome sequences of a species where some strains have a positive impact (e.g. flavour enhancement) while others are detrimental (e.g. spoilage) can be used to identify genetic elements potentially underlying these differences, as was done for the yeast Brettanomyces bruxellensis [60]. Tools that can be used to link -omics data to phenotypes are PhenoLink [58] and DuctApe [43]. These approaches require a genome sequence, which might be relatively difficult to obtain for microbes that are difficult to grow in culture. Techniques like multiple displacement amplification [61] can be used to amplify DNA from a single cell, and a range of genome assembly tools can be used to assemble the reads obtained from single-cell sequencing [62].
Mobile elements such as transposons, plasmids or phages can carry functionality from one bacterial strain to another. An example is the galactose utilization operon transfer between Lactococcus lactis strains studied by next-generation sequencing and bioinformatics techniques [63]. Identifying potential transposon insertion sites is crucial to this end and can be facilitated by bioinformatics tools such as transposon insertion finder [64].
Improving metabolite production and biomass
Improvement of the food production process by optimizing biomass yield is a topic of continuous attention. A technique to rationally improve fermentation yield is genome-scale metabolic modelling [65]. In this process, the genome sequence of the organism is used as an inventory of the metabolic potential of the strain of interest. Metabolic models have been made for many microbes, including several of food-relevant microorganisms [66–69]. Although the quality of a genome sequence can be a limiting factor (e.g. missed gene due to low sequencing coverage), the metabolic model can be completed by identifying metabolic reactions that are missing in the model, but likely present due to the fact that they are part of metabolic reaction cascades or ‘pathways’ [70]. Complete genome-scale metabolic models together with algorithms such as flux balance analysis allow the in silico simulation of growth of the organism under the (metabolic) restrictions provided by the substrate availability in the medium. These growth simulations can then be used to optimize medium composition to better fit the organism requirements [71]. In addition, the models can suggest alternative or cheaper substrates for fermentation [69], and improve the production of compounds such as amino acids [72] or succinic acid [73], taking into account possible changes in activity with respect to flavour or texture activity of the strain. These models have also been implemented in complex (multistrain) fermentation processes, providing insight in the interactions between different species/strains in a complex fermentation [74].
A second factor that improves the overall yield is the robustness of strains after harvesting. Also, this factor can significantly be influenced by changing fermentation conditions under which starter cultures are prepared. By correlating gene expression levels to the survival of L. lactis, an application of transcriptome–trait matching (TTM), a number of genes that were potentially causative related to survival were identified. Subsequent knock-out of the genes proved that these genes were indeed important for the strains’ phenotype. This shows that not only gene content but also expression of genes is important for a given phenotype. In other words, preconditioning L. lactis strains, followed by GTM and TTM, allows improving their survival to heat and oxidative stresses, typically encountered during spray drying [46, 47].
Improving texture and flavour
The fermentation process also influences the texture and flavour properties of the food product. These characteristics are microorganism-specific [75] and can be changed by fermentation, e.g. the production of flavours by adding adjunct strains to cheese fermentations [76], or the addition of exopolysaccharide-producing organisms to improve the texture of yoghurt [77, 78]. Also, flavour profiles of wine can be modified by either altering fermentation conditions or changing the wine starter cultures [79]. Whereas improvements can be made by testing a variety of experimental settings, bioinformatics and data analytics may be used to optimize the experimental designs [80–82].
The performance of a microorganism under particular fermentation conditions may be deduced from gene content of these microorganisms. Using a metabolic model, L. lactis MG1363 flavour formation could be predicted and was subsequently experimentally verified [67]. Likewise, the genomic sequence of Lactobacillus delbrueckii subsp. bulgaricus revealed how this organism is adapted to for the fermentation of milk and the production of yoghurt [83]. Similar analyses have been carried out for Oenococcus oeni [84] and yeast genomes [85] and their relation to wine fermentation. Due to the larger complexity of yeast genomes, this analysis is more challenging [86].
Using GTM growth on various sugars can relatively well be predicted based on gene content, e.g. for L. lactis, Lactobacillus plantarum, Lactobacillus paracasei and Bifidobacterium breve [58, 87–89]. In the same studies it became apparent that predicting more complex phenotype such as stress tolerance is less straight-forward to predict based on gene content alone [58, 87]. Information on the transcript levels of the genes (see above) might be taken into account to better predict these phenotypes. TTM can similarly be used to associate the expression of microorganism genes to texture and flavour characteristics of a product, such as improving the production of organic acids by knowledge-based altering fermentation conditions [48].
The effects on taste and texture are mainly caused by the metabolites that are produced or converted during fermentations. Rather than associating gene content with effects on taste texture, metabolite patterns may be used directly to predict final sensory characteristics. The golden standard test for sensory characteristics of a fermented product is a quantitative descriptive analysis by a trained sensory panel. These tests are elaborate and require production of substantial amounts of the product. The results are dependent on the panel experience and the attributes that are used to describe the product properties [51]. With metabolomics profiling techniques, it is now possible to simultaneously measure hundreds of metabolites in food samples [50]. This, together with the development of small-scale product screening methods [90], has led to the development of many new statistical methods to associate instrumental data, such as, for example, gas chromatography–mass spectrometry, to sensory data [51, 52, 91–94].
Risk assessment
Rather than predicting functions for all genes in a bacterial genome, selectively screening microbial genome sequences for genes with specific functionalities can be a highly sensitive and computationally efficient way of identifying potential health or safety risks of microbial strains present in a sample. The potential of a specific bacterium for antibiotic resistance or virulence can be investigated by comparing its genome sequence to a reference database containing known resistance genes and virulence factors [95]. Similar approaches have been described for the identification of persistence of bacteria in food products [45], anaerobic spore-forming organisms in food [96] and potential pathogens in metagenomics data [97]. This (meta)genomics-based methodology can be extended to a wide range of functionalities, e.g. production of antimicrobial peptides [98–100] and resistance to cleaning procedures commonly used in food production settings [101, 102]. A requirement for getting useful results out of metagenomics experiments is a dedicated database with gene–function relations and access to domain knowledge on the specific functionality to specify gene functions.
Mixed culture fermentations characterization
Complex fermentations involve an (un)defined (wild) starter culture with different microbes (bacteria, yeasts and fungi) that together ferment a substrate to the product. Examples are cheese, malolactic wine, soy and seafood fermentations [103, 104]. In these fermentations, strong succession of microbes can occur, for instance in wine fermentation, the microbes Saccharomyces cerevisiae and Oenococcus oenii [105, 106]. Similar to the above-described GTM and TTM approaches to associate (transcription of) genes to phenotypes, presence and absence of (combinations of) microorganisms (or their functionality) can be associated to fermentation product characteristics.
The first step in characterizing a fermentation is to determine what microorganisms are present at the different stages of the fermentation and to correlate these to other measurements such as metabolomics [107] or the presence of phages [108]. The properties of microbial consortia are determined by the functional potential encoded in all microbial genomes. Metagenomics has an advantage over conventional sequencing of single isolates from consortia because it also reveals DNA of otherwise unculturable organisms. Based on the sequences found in a consortium, functionalities of the microbes can be predicted. Due to the succession of microbes in a fermentation, it is important to omit DNA from dead microbes before building predictive models based on sequences. One way to sequester ‘dead’ DNA, and therefore not sequencing it, is the use of propidium mono azide [109]. Next-generation sequencing techniques that profile, e.g., the 16S gene present in all bacteria are increasingly used over molecular biology techniques, e.g. gel-based methods [110, 111]. The bioinformatics analysis of 16S data from food fermentations is quite well-established (Table 1), resulting in descriptions of the taxa present in a particular fermentation at best at the species level, but for some taxa, the genus level is challenging to obtain [112].
There is a large biodiversity beyond the species level that is not taken into account with, e.g., 16S sequencing. Even within a bacterial species, there is considerable biodiversity. For example, all genes present in strains of the Lactobacillus genus (its pan-genome) comprise over 14 000 gene families, with a single genome encoding ∼3 000 proteins [113]. A gene family typically consists of genes that are evolutionary conserved, but that might have different enzymatic functions depending on the specific protein sequence [114]. Comparative genomics, in combination with molecular strain typing, techniques have been used to uncover strain-level diversity in complex, yet relatively defined, fermentations in general [41] and specifically for L. lactis and Leuconostoc mesenteroides from cheese [108], Lactobacillus sakei from meat fermentations [115], Lactobacillus sanfranciscensis in sourdough fermentations [116] and wine yeasts [86].
With shotgun metagenomics, the DNA in the mixed-culture fermentation is profiled, but strain-level diversity is extremely difficult to deduce from shotgun metagenomics sequence fragments [108]. On the other hand, due to the enormous biodiversity, the actual presence of any strain isolate thought to be of importance in a particular mixed-culture fermentation should be established. The combination of shotgun metagenomics and comparative genomics could prove to be particularly powerful, as the shotgun metagenome DNA sequences can be aligned to the genomes of isolates in order to prove that the functionality present in the isolates covers that of the metagenome [41, 108].
Metatranscriptomics approaches allow profiling the mRNA-derived sequences of a complex fermentation. An advantage of metatranscriptomics over metagenomics approaches is that the gene expression measurement allows determining what genes are actually expressed in a mixed culture. Application of ‘metatranscriptomics’ using microarrays with the genomes of several species to determine global gene expression across species has been reported for Kimchi [117]. Only recently, metagenome and metatranscriptome sequencing of bacterial communities involved in cheese rind fermentations has been reported [118]. The strength of this study is that the metagenomics and metatranscriptomics profiles were traced to their likely sources (genome sequences of isolates from the rind cheese fermentation). Using experimental setups like the latter in combination with metabolomics measurements and appropriate follow-up studies should strengthen the point to use metagenomics/metatranscriptomics techniques to characterize and potentially optimize fermentations.
Bacteriophages play an important role in industrial fermentations due to the phenomenon of maintaining biodiversity through phage predation [119], but also because phage sweeps disrupt fermentation processes [120, 121]. Currently, however, predicting the specificity of bacteriophages and the interactions between microbes in mixed-culture fermentation are time-consuming tasks [108, 121–123].
Bioinformatics techniques that analyse the interaction of microbes and bacteriophages, and in-depth knowledge of the metabolic requirements of the microbial consortia present during fermentation could in the future lead to knowledge-based improvements of fermentation stability. This could be achieved by performing experiments with synthetic microbial consortia. The design of these consortia is currently being developed [81], and cross-kingdom interactions are being studied [124]. In a study where cheese rind bacterial communities were created based on various -omics data, knowledge of the fermentation and dedicated follow-up experiments, the potential of predicting properties of complex fermentations [118] was demonstrated. This study did not explicitly describe whether the selected strains (or close relatives) were actually present in a real fermentation. This has been described for representative L. lactis and L. mesenteroides strains of a complex cheese fermentation [108] and an L. lactis strain from a defined consortium [125].
Branding, tracing and detection
Food production and food consumption take place in complex environments in which next to the microorganisms present in the natural environment, many other sources of proteins, fat and carbohydrates are present. The presence of the endogenous flora as well as the macromolecular structures of the food can cause a lot of difficulty in detection and tracing of specific microorganisms, such as potential food pathogens or probiotic strains added to the food product for enhanced functionality.
Next to classical detection DNA-based techniques such as (q)PCR [126], new methods based on genomic data have been developed that allow for a fast and accurate tracking or detection of specific species or even strains among the natural microflora. By specific amplification and sequencing of a locus that was identified to be discriminatory between different L. plantarum strains, it was shown that one could quantify the relative presence of different strains through the passage of the gastrointestinal tract [40]. This same approach can also be followed to design specific primers to discriminate between pathogenic and non-pathogenic populations of specific species [127] and to detect a strain of interest in food products, allowing dedicated branding of a specific product.
Next to dedicated tracing of a single strain, metagenome approaches as described for studying complex fermented products, for example in cheese [118] and fermented foods of plant origin [128, 129], will also have their benefit in the detection of spoilage bacteria. Especially as these methods allow for direct profiling of the product, and do not require a culture step that could create bias in the results, they could very well prove to be more specific to detect spoilage bacteria from a product. Culturing steps will always have their merit due to limited costs and requirement of limited amounts of material. Especially in fermented products, 16s community profiling approaches will allow detecting low abundant microbes that might be overgrown in culture-dependent detection methods.
Perspectives
Bioinformatics is increasingly applied in food fermentation and safety. Below we describe some new and exciting developments in this field.
Sequence-based prediction of microbial functionality is just starting. An inventory is needed of which functionality for which bacteria can reliably be determined using sequence data. New publicly available data sets with genotype/phenotype/transcriptome such as those available for L. lactis and L. plantarum could help to develop new sequence-based functional prediction strategies such as further specified protein domains to more specifically screen for, e.g., carbohydrate active enzymes [130] and relating promoters or regulatory binding sites to phenotype [42].
By consolidating the above information, a knowledge-based in silico screening of culture collections for desired traits can be established. This would require databases that use controlled vocabularies to integrate data from genomics, systems biology, phenotypes, ingredient information, properties of batches of foods, on-line measuring of parameters during the food making process and ‘biomarkers’ for functionality in specific taxa (based on, e.g., GTM). Specific emphasis should be put in propagating the FAIR (findable, accessible, interoperable, re-usable; http://datafairport.org/) principle in storing data. Given that analyses will become more standardized and computer resource-intensive, the software and databases could be set up in a virtual machine that can subsequently be run on computer clusters or in the cloud. First steps towards data consolidation are being made in the EU-funded project GenoBox (www.genobox.eu) that aims to create a database that consolidates genotype and phenotype data that allow screening microbial genomes for functionality and safety risk factors.
Similarly, IBM and MARS have established a consortium that aims to sequence the food supply chain (http://www.research.ibm.com/client-programs/foodsafety/). Their aim is to determine nominal levels of microbial components in many food products across the globe. The resulting database can be used to assess risks of the presence of certain microbes/functionality in a given food product. Given that sufficient biodiversity has been recorded into this database, it could also be used for branding products based on unique microbiota signatures present in fermented products or foods that contain a microbiome.
Another important factor to consider in steering the performance of fermentations is the interactions between microbes and their environment. This new layer of complexity has been studied, for instance, for microbe–plant interactions for rice or coconut [131, 132] and the use of systems biology beyond genome-scale metabolic models by using kinetic models to describe interactions between microbes and their matrix [133]. These studies require a substantial knowledge base on both the properties of the microorganisms and the physical properties of the matrix in which the organism operate.
In conclusion, the increasing amount of data on food fermentation and safety encourages consolidating this information in databases that with the right experimental design, algorithms, expertise and follow-up experiments should allow enhancing the prediction of fermentation performance and safety.
Key Points.
Exploiting the vast biodiversity to create new food products or to optimize existing ones is gaining momentum.
Sequence-based prediction of microbial functionality is a powerful tool, with a clear application in screening biobanks.
Increased availability of public data sets of fermentations will allow developing better predictive models for microbial functionality.
Detection of spoilage strains on the basis of genotype.
Funding
This work was supported by a project from the Top Institute Food and Nutrition, Wageningen, the Netherlands and the Kluyver Center for Genomics of Industrial Fermentation, Delft, the Netherlands.
Biographies
Wynand Alkema is group leader fermentation & health at NIZO food research and head of the Radboudumc bioinformatics technology center. His main interests are large-scale data analytics and literature mining.
Jos Boekhorst is a bioinformatician at NIZO food research. He uses computational tools to unravel links between microbes, food, health and disease.
Michiel Wels is group leader bioinformatics and food safety at NIZO food research and is involved in applying bioinformatics approaches to different food-related research questions.
Sacha van Hijum is associate professor and leads the bacterial (meta)genomics group at the Radboudumc. He is principal scientist bioinformatics at NIZO food research and leads the bioinformatics platform of TI Food and Nutrition.
References
- 1.Smid EJ, Kleerebezem M. Production of aroma compounds in lactic fermentations. Annu Rev Food Sci Technol 2014;5:313–26. [DOI] [PubMed] [Google Scholar]
- 2.Cifuentes A. Foodomics: the necessary route to boost quality, safety and bioactivity of foods. Electrophoresis 2014;35:1517–18. [DOI] [PubMed] [Google Scholar]
- 3.Bergholz TM, Moreno Switt AI, Wiedmann M. Omics approaches in food safety: fulfilling the promise? Trends Microbiol 2014;22:275–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garrigues C, Johansen E, Crittenden R. Pangenomics–an avenue to improved industrial starter cultures and probiotics. Curr Opin Biotechnol 2013;24:187–91. [DOI] [PubMed] [Google Scholar]
- 5.Talukdar V, Konar A, Datta A, et al. Changing from computing grid to knowledge grid in life-science grid. Biotechnol J 2009;4:1244–52. [DOI] [PubMed] [Google Scholar]
- 6.Goecks J, Nekrutenko A, Taylor J, et al. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol 2010;11:R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tiwari A, Sekhar AK. Workflow based framework for life science informatics. Comput Biol Chem 2007;31:305–19. [DOI] [PubMed] [Google Scholar]
- 8.O'Driscoll A, Belogrudov V, Carroll J, et al. HBLAST: parallelised sequence similarity - a Hadoop MapReducable basic local alignment search tool. J Biomed Inform 2015. [DOI] [PubMed] [Google Scholar]
- 9.Deutsch EW, Mendoza L, Shteynberg D, et al. Trans-Proteomic Pipeline, a standardized data processing pipeline for large-scale reproducible proteomics informatics. Proteomics Clin Appl 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hwang Y, Lin C, Valladares O, et al. HIPPIE: a high-throughput identification pipeline for promoter interacting enhancer elements. Bioinformatics 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tobes R, Pareja-Tobes P, Manrique M, et al. Gene calling and bacterial genome annotation with BG7. Methods Mol Biol 2015;1231:177–89. [DOI] [PubMed] [Google Scholar]
- 12.Cortes C, Vapnik V. Support-vector networks. Mach Learn 1995;20:273–97. [Google Scholar]
- 13.Breiman L. Random forests. Mach Learn 2001;45:5–32. [Google Scholar]
- 14.Field D, Tiwari B, Booth T, et al. Open software for biologists: from famine to feast. Nat Biotechnol 2006;24:801–3. [DOI] [PubMed] [Google Scholar]
- 15.Koren S, Phillippy AM. One chromosome, one contig: complete microbial genomes from long-read sequencing and assembly. Curr Opin Microbiol 2015;23:110–20. [DOI] [PubMed] [Google Scholar]
- 16.Boisvert S, Raymond F, Godzaridis E, et al. Ray Meta: scalable de novo metagenome assembly and profiling. Genome Biol 2012;13:R122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chevreux B, Pfisterer T, Drescher B, et al. Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res 2004;14:1147–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peng Y, Leung HC, Yiu SM, et al. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 2012;28:1420–8. [DOI] [PubMed] [Google Scholar]
- 19.Boetzer M, Pirovano W. SSPACE-LongRead: scaffolding bacterial draft genomes using long read sequence information. BMC Bioinformatics 2014;15:211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Hijum SA, Zomer AL, Kuipers OP, et al. Projector 2: contig mapping for efficient gap-closure of prokaryotic genome sequence assemblies. Nucleic Acids Res 2005;33:W560–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Darling AC, Mau B, Blattner FR, et al. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 2004;14:1394–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aziz RK, Bartels D, Best AA, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 2008;9:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 2014;30:2068–9. [DOI] [PubMed] [Google Scholar]
- 24.Li L, Stoeckert CJ, Jr, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 2003;13:2178–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ekseth OK, Kuiper M, Mironov V. orthAgogue: an agile tool for the rapid prediction of orthology relations. Bioinformatics 2014;30:734–6. [DOI] [PubMed] [Google Scholar]
- 26.Aziz RK, Devoid S, Disz T, et al. SEED servers: high-performance access to the SEED genomes, annotations, and metabolic models. PLoS One 2012;7:e48053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Notebaart RA, van Enckevort FH, Francke C, et al. Accelerating the reconstruction of genome-scale metabolic networks. BMC Bioinformatics 2006;7:296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olivier BG, Rohwer JM, Hofmeyr JH. Modelling cellular systems with PySCeS. Bioinformatics 2005;21:560–1. [DOI] [PubMed] [Google Scholar]
- 29.Krach C, Junker A, Rohn H, et al. Flux visualization using VANTED/FluxMap. Methods Mol Biol 2014;1191:225–33. [DOI] [PubMed] [Google Scholar]
- 30.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010;7:335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schloss PD, Westcott SL, Ryabin T, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 2009;75:7537–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langille MG, Zaneveld J, Caporaso JG, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 2013;31:814–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Di Bella JM, Bao Y, Gloor GB, et al. High throughput sequencing methods and analysis for microbiome research. J Microbiol Methods 2013;95:401–14. [DOI] [PubMed] [Google Scholar]
- 34.Gilbert JA, Hughes M. Gene expression profiling: metatranscriptomics. Methods Mol Biol 2011;733:195–205. [DOI] [PubMed] [Google Scholar]
- 35.Celaj A, Markle J, Danska J, et al. Comparison of assembly algorithms for improving rate of metatranscriptomic functional annotation. Microbiome 2014;2:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brady A, Salzberg SL. Phymm and PhymmBL: metagenomic phylogenetic classification with interpolated Markov models. Nat Methods 2009;6:673–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Glass EM, Wilkening J, Wilke A, et al. Using the metagenomics RAST server (MG-RAST) for analyzing shotgun metagenomes. Cold Spring Harb Protoc 2010;2010:pdb prot5368. [DOI] [PubMed] [Google Scholar]
- 38.Segata N, Waldron L, Ballarini A, et al. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Methods 2012;9:811–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andreani NA, Martino ME, Fasolato L, et al. Reprint of ‘Tracking the blue: a MLST approach to characterise the Pseudomonas fluorescens group’. Food Microbiol 2015;45:148–58. [DOI] [PubMed] [Google Scholar]
- 40.van Bokhorst-van de Veen H, Smelt MJ, Wels M, et al. Genotypic adaptations associated with prolonged persistence of Lactobacillus plantarum in the murine digestive tract. Biotechnol J 2013;8:895–904. [DOI] [PubMed] [Google Scholar]
- 41.Johansen P, Vindelov J, Arneborg N, et al. Development of quantitative PCR and metagenomics-based approaches for strain quantification of a defined mixed-strain starter culture. Syst Appl Microbiol 2014;37:186–93. [DOI] [PubMed] [Google Scholar]
- 42.Dutilh BE, Backus L, Edwards RA, et al. Explaining microbial phenotypes on a genomic scale: GWAS for microbes. Brief Funct Genomics 2013;12:366–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Galardini M, Mengoni A, Biondi EG, et al. DuctApe: a suite for the analysis and correlation of genomic and OmniLog Phenotype Microarray data. Genomics 2014;103:1–10. [DOI] [PubMed] [Google Scholar]
- 44.Galardini M, Mengoni A, Mocali S. From pangenome to panphenome and back. Methods Mol Biol 2015;1231:257–70. [DOI] [PubMed] [Google Scholar]
- 45.Vangay P, Steingrimsson J, Wiedmann M, et al. Classification of Listeria monocytogenes persistence in retail delicatessen environments using expert elicitation and machine learning. Risk Anal 2014;34:1830–45. [DOI] [PubMed] [Google Scholar]
- 46.Dijkstra AR, Setyawati MC, Bayjanov JR, et al. Diversity in robustness of Lactococcus lactis strains during heat stress, oxidative stress, and spray drying stress. Appl Environ Microbiol 2014;80:603–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dijkstra AR, Alkema W, Starrenburg M, et al. Fermentation-induced variation in heat and oxidative stress phenotypes of Lactococcus lactis MG1363 reveals transcriptome signatures for robustness. Microb Cell Fact 2014;13:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bron PA, Wels M, Bongers RS, et al. Transcriptomes reveal genetic signatures underlying physiological variations imposed by different fermentation conditions in Lactobacillus plantarum. PLoS One 2012;7:e38720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Bokhorst-van de Veen H, Lee IC, Marco ML, et al. Modulation of Lactobacillus plantarum gastrointestinal robustness by fermentation conditions enables identification of bacterial robustness markers. PLoS One 2012;7:e39053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alonso A, Marsal S, Julia A. Analytical methods in untargeted metabolomics: state of the art in 2015. Front Bioeng Biotechnol 2015;3:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmidtke LM, Blackman JW, Clark AC, et al. Wine metabolomics: objective measures of sensory properties of semillon from GC-MS profiles. J Agric Food Chem 2013;61:11957–67. [DOI] [PubMed] [Google Scholar]
- 52.Procida G, Cichelli A, Lagazio C, et al. Relationships between volatile compounds and sensory characteristics in virgin olive oil by analytical and chemometric approach. J Sci Food Agric 2015. [DOI] [PubMed] [Google Scholar]
- 53.Weirick T, Sahu SS, Mahalingam R, et al. LacSubPred: predicting subtypes of Laccases, an important lignin metabolism-related enzyme class, using in silico approaches. BMC Bioinformatics 2014;15(Suppl 11):S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Overbeek R, Olson R, Pusch GD, et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res 2014;42:D206–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yamada T, Letunic I, Okuda S, et al. iPath2.0: interactive pathway explorer. Nucleic Acids Res 2011;39:W412–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kanehisa M, Goto S, Sato Y, et al. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res 2012;40:D109–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ventura M, Turroni F, van Sinderen D. Probiogenomics as a tool to obtain genetic insights into adaptation of probiotic bacteria to the human gut. Bioeng Bugs 2012;3:73–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bayjanov JR, Molenaar D, Tzeneva V, et al. PhenoLink–a web-tool for linking phenotype to ∼omics data for bacteria: application to gene-trait matching for Lactobacillus plantarum strains. BMC Genomics 2012;13:170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Siezen R, Boekhorst J, Muscariello L, et al. Lactobacillus plantarum gene clusters encoding putative cell-surface protein complexes for carbohydrate utilization are conserved in specific Gram-positive bacteria. BMC Genomics 2006;7:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Crauwels S, Zhu B, Steensels J, et al. Assessing genetic diversity among Brettanomyces yeasts by DNA fingerprinting and whole-genome sequencing. Appl Environ Microbiol 2014;80:4398–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lasken RS. Genomic DNA amplification by the multiple displacement amplification (MDA) method. Biochem Soc Trans 2009;37:450–53. [DOI] [PubMed] [Google Scholar]
- 62.Gurevich A, Saveliev V, Vyahhi N, et al. QUAST: quality assessment tool for genome assemblies. Bioinformatics 2013;29:1072–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Machielsen R, Siezen RJ, van Hijum SA, et al. Molecular description and industrial potential of Tn6098 conjugative transfer conferring alpha-galactoside metabolism in Lactococcus lactis. Appl Environ Microbiol 2011;77:555–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nakagome M, Solovieva E, Takahashi A, et al. Transposon Insertion Finder (TIF): a novel program for detection of de novo transpositions of transposable elements. BMC Bioinformatics 2014;15:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Maarleveld TR, Khandelwal RA, Olivier BG, et al. Basic concepts and principles of stoichiometric modeling of metabolic networks. Biotechnol J 2013;8:997-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Heavner BD, Smallbone K, Price ND, et al. Version 6 of the consensus yeast metabolic network refines biochemical coverage and improves model performance. Database (Oxford) 2013;2013:bat059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Flahaut NA, Wiersma A, van de Bunt B, et al. Genome-scale metabolic model for Lactococcus lactis MG1363 and its application to the analysis of flavor formation. Appl Microbiol Biotechnol 2013;97:8729–39. [DOI] [PubMed] [Google Scholar]
- 68.Pastink MI, Teusink B, Hols P, et al. Genome-scale model of Streptococcus thermophilus LMG18311 for metabolic comparison of lactic acid bacteria. Appl Environ Microbiol 2009;75:3627–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Teusink B, Wiersma A, Jacobs L, et al. Understanding the adaptive growth strategy of Lactobacillus plantarum by in silico optimisation. PLoS Comput Biol 2009;5:e1000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Benedict MN, Mundy MB, Henry CS, et al. Likelihood-based gene annotations for gap filling and quality assessment in genome-scale metabolic models. PLoS Comput Biol 2014;10:e1003882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wegkamp A, Teusink B, de Vos WM, et al. Development of a minimal growth medium for Lactobacillus plantarum. Lett Appl Microbiol 2010;50:57–64. [DOI] [PubMed] [Google Scholar]
- 72.Park SH, Kim HU, Kim TY, et al. Metabolic engineering of Corynebacterium glutamicum for L-arginine production. Nat Commun 2014;5:4618. [DOI] [PubMed] [Google Scholar]
- 73.Otero JM, Cimini D, Patil KR, et al. Industrial systems biology of Saccharomyces cerevisiae enables novel succinic acid cell factory. PLoS One 2013;8:e54144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Branco dos Santos F, de Vos WM, Teusink B. Towards metagenome-scale models for industrial applications–the case of Lactic Acid Bacteria. Curr Opin Biotechnol 2013;24:200–6. [DOI] [PubMed] [Google Scholar]
- 75.Deetae P, Bonnarme P, Spinnler HE, et al. Production of volatile aroma compounds by bacterial strains isolated from different surface-ripened French cheeses. Appl Microbiol Biotechnol 2007;76:1161–71. [DOI] [PubMed] [Google Scholar]
- 76.Whetstine ME, Drake MA, Broadbent JR, et al. Enhanced nutty flavor formation in cheddar cheese made with a malty Lactococcus lactis adjunct culture. J Dairy Sci 2006;89:3277–84. [DOI] [PubMed] [Google Scholar]
- 77.Robitaille G, Tremblay A, Moineau S, et al. Fat-free yogurt made using a galactose-positive exopolysaccharide-producing recombinant strain of Streptococcus thermophilus. J Dairy Sci 2009;92:477–82. [DOI] [PubMed] [Google Scholar]
- 78.Yilmaz MT, Dertli E, Toker OS, et al. Effect of in situ exopolysaccharide production on physicochemical, rheological, sensory, and microstructural properties of the yogurt drink ayran: an optimization study based on fermentation kinetics. J Dairy Sci 2015;98:1604–24. [DOI] [PubMed] [Google Scholar]
- 79.Carrau F, Gaggero C, Aguilar PS. Yeast diversity and native vigor for flavor phenotypes. Trends Biotechnol 2015;33:148–54. [DOI] [PubMed] [Google Scholar]
- 80.Settachaimongkon S, Nout MJ, Antunes Fernandes EC, et al. Influence of different proteolytic strains of Streptococcus thermophilus in co-culture with Lactobacillus delbrueckii subsp. bulgaricus on the metabolite profile of set-yoghurt. Int J Food Microbiol 2014;177:29–36. [DOI] [PubMed] [Google Scholar]
- 81.Bernstein HC, Carlson RP. Design, construction, and characterization methodologies for synthetic microbial consortia. Methods Mol Biol 2014;1151:49–68. [DOI] [PubMed] [Google Scholar]
- 82.Seo SW, Yang J, Min BE, et al. Synthetic biology: tools to design microbes for the production of chemicals and fuels. Biotechnol Adv 2013;31:811–17. [DOI] [PubMed] [Google Scholar]
- 83.Hao P, Zheng H, Yu Y, et al. Complete sequencing and pan-genomic analysis of Lactobacillus delbrueckii subsp. bulgaricus reveal its genetic basis for industrial yogurt production. PLoS One 2011;6:e15964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Borneman AR, McCarthy JM, Chambers PJ, et al. Comparative analysis of the Oenococcus oeni pan genome reveals genetic diversity in industrially-relevant pathways. BMC Genomics 2012;13:373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang H, Richards KD, Wilson S, et al. Genetic characterization of strains of Saccharomyces uvarum from New Zealand wineries. Food Microbiol 2015;46:92–9. [DOI] [PubMed] [Google Scholar]
- 86.Borneman AR, Pretorius IS, Chambers PJ. Comparative genomics: a revolutionary tool for wine yeast strain development. Curr Opin Biotechnol 2013;24:192–9. [DOI] [PubMed] [Google Scholar]
- 87.Bayjanov JR, Starrenburg MJ, van der Sijde MR, et al. Genotype-phenotype matching analysis of 38 Lactococcus lactis strains using random forest methods. BMC Microbiol 2013;13:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Smokvina T, Wels M, Polka J, et al. Lactobacillus paracasei comparative genomics: towards species pan-genome definition and exploitation of diversity. PLoS One 2013;8:e68731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bottacini F, O'Connell Motherway M, Kuczynski J, et al. Comparative genomics of the Bifidobacterium breve taxon. BMC Genomics 2014;15:170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bachmann H, Kruijswijk Z, Molenaar D, et al. A high-throughput cheese manufacturing model for effective cheese starter culture screening. J Dairy Sci 2009;92:5868–82. [DOI] [PubMed] [Google Scholar]
- 91.Zhu J, Chen F, Wang L, et al. Comparison of aroma-active compounds and sensory characteristics of durian (Durio zibethinus L.) wines using strains of Saccharomyces cerevisiae with odor activity values and partial least-squares regression. J Agric Food Chem 2015;63:1939–47. [DOI] [PubMed] [Google Scholar]
- 92.Xiao ZB, Liu JH, Chen F, et al. Comparison of aroma-active volatiles and their sensory characteristics of mangosteen wines prepared by Saccharomyces cerevisiae with GC-olfactometry and principal component analysis. Nat Prod Res 2015;29:656–62. [DOI] [PubMed] [Google Scholar]
- 93.Ochi H, Bamba T, Naito H, et al. Metabolic fingerprinting of hard and semi-hard natural cheeses using gas chromatography with flame ionization detector for practical sensory prediction modeling. J Biosci Bioeng 2012;114:506–11. [DOI] [PubMed] [Google Scholar]
- 94.Ochi H, Naito H, Iwatsuki K, et al. Metabolomics-based component profiling of hard and semi-hard natural cheeses with gas chromatography/time-of-flight-mass spectrometry, and its application to sensory predictive modeling. J Biosci Bioeng 2012;113:751–8. [DOI] [PubMed] [Google Scholar]
- 95.Bennedsen M, Stuer-Lauridsen B, Danielsen M, et al. Screening for antimicrobial resistance genes and virulence factors via genome sequencing. Appl Environ Microbiol 2011;77:2785–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Doyle CJ, Gleeson D, Jordan K, et al. Anaerobic sporeformers and their significance with respect to milk and dairy products. Int J Food Microbiol 2015;197C:77–87. [DOI] [PubMed] [Google Scholar]
- 97.Naccache SN, Federman S, Veeraraghavan N, et al. A cloud-compatible bioinformatics pipeline for ultrarapid pathogen identification from next-generation sequencing of clinical samples. Genome Res 2014;24:1180–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.van Heel AJ, de Jong A, Montalban-Lopez M, et al. BAGEL3: automated identification of genes encoding bacteriocins and (non-)bactericidal posttranslationally modified peptides. Nucleic Acids Res 2013;41:W448–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lira F, Perez PS, Baranauskas JA, et al. Prediction of antimicrobial activity of synthetic peptides by a decision tree model. Appl Environ Microbiol 2013;79:3156–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lata S, Mishra NK, Raghava GP. AntiBP2: improved version of antibacterial peptide prediction. BMC Bioinformatics 2010;11(Suppl 1):S19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bore E, Langsrud S. Characterization of micro-organisms isolated from dairy industry after cleaning and fogging disinfection with alkyl amine and peracetic acid. J Appl Microbiol 2005;98:96–105. [DOI] [PubMed] [Google Scholar]
- 102.da Silva Fernandes M, Kabuki DY, Kuaye AY. Biofilms of Enterococcus faecalis and Enterococcus faecium isolated from the processing of ricotta and the control of these pathogens through cleaning and sanitization procedures. Int J Food Microbiol 2015;200C:97–103. [DOI] [PubMed] [Google Scholar]
- 103.Bokulich NA, Mills DA. Next-generation approaches to the microbial ecology of food fermentations. BMB Rep 2012;45:377–89. [DOI] [PubMed] [Google Scholar]
- 104.Bokulich NA, Thorngate JH, Richardson PM, et al. Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc Natl Acad Sci USA 2014;111:E139–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Alexandre H, Costello PJ, Remize F, et al. Saccharomyces cerevisiae-Oenococcus oeni interactions in wine: current knowledge and perspectives. Int J Food Microbiol 2004;93:141–54. [DOI] [PubMed] [Google Scholar]
- 106.Jolly NP, Varela C, Pretorius IS. Not your ordinary yeast: non-Saccharomyces yeasts in wine production uncovered. FEMS Yeast Res 2014;14:215–37. [DOI] [PubMed] [Google Scholar]
- 107.Ponnusamy K, Lee S, Lee CH. Time-dependent correlation of the microbial community and the metabolomics of traditional barley nuruk starter fermentation. Biosci Biotechnol Biochem 2013;77:683–90. [DOI] [PubMed] [Google Scholar]
- 108.Erkus O, de Jager VC, Spus M, et al. Multifactorial diversity sustains microbial community stability. ISME J 2013;7:2126–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nocker A, Richter-Heitmann T, Montijn R, et al. Discrimination between live and dead cellsin bacterial communities from environmental water samples analyzed by 454 pyrosequencing. Int Microbiol 2010;13:59–65. [DOI] [PubMed] [Google Scholar]
- 110.van Hijum SA, Vaughan EE, Vogel RF. Application of state-of-art sequencing technologies to indigenous food fermentations. Curr Opin Biotechnol 2013;24:178–86. [DOI] [PubMed] [Google Scholar]
- 111.Xie G, Wang L, Gao Q, et al. Microbial community structure in fermentation process of Shaoxing rice wine by Illumina-based metagenomic sequencing. J Sci Food Agric 2013;93:3121–5. [DOI] [PubMed] [Google Scholar]
- 112.Janda JM, Abbott SL. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J Clin Microbiol 2007;45:2761–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kant R, Blom J, Palva A, et al. Comparative genomics of Lactobacillus. Microb Biotechnol 2011;4:323–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ponting CP. Issues in predicting protein function from sequence. Brief Bioinform 2001;2:19–29. [DOI] [PubMed] [Google Scholar]
- 115.Nyquist OL, McLeod A, Brede DA, et al. Comparative genomics of Lactobacillus sakei with emphasis on strains from meat. Mol Genet Genomics 2011;285:297–311. [DOI] [PubMed] [Google Scholar]
- 116.Venturi M, Guerrini S, Granchi L, et al. Typing of Lactobacillus sanfranciscensis isolates from traditional sourdoughs by combining conventional and multiplex RAPD-PCR profiles. Int J Food Microbiol 2012;156:122–6. [DOI] [PubMed] [Google Scholar]
- 117.Nam YD, Chang HW, Kim KH, et al. Metatranscriptome analysis of lactic acid bacteria during kimchi fermentation with genome-probing microarrays. Int J Food Microbiol 2009;130:140–6. [DOI] [PubMed] [Google Scholar]
- 118.Wolfe BE, Button JE, Santarelli M, et al. Cheese rind communities provide tractable systems for in situ and in vitro studies of microbial diversity. Cell 2014;158:422–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rodriguez-Valera F, Martin-Cuadrado AB, Rodriguez-Brito B, et al. Explaining microbial population genomics through phage predation. Nat Rev Microbiol 2009;7:828–36. [DOI] [PubMed] [Google Scholar]
- 120.McGrath S, Fitzgerald GF, van Sinderen D. The impact of bacteriophage genomics. Curr Opin Biotechnol 2004;15:94–99. [DOI] [PubMed] [Google Scholar]
- 121.Mahony J, McAuliffe O, Ross RP, et al. Bacteriophages as biocontrol agents of food pathogens. Curr Opin Biotechnol 2011;22:157–63. [DOI] [PubMed] [Google Scholar]
- 122.Kelly WJ, Altermann E, Lambie SC, et al. Interaction between the genomes of Lactococcus lactis and phages of the P335 species. Front Microbiol 2013;4:257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Denes T, Wiedmann M. Environmental responses and phage susceptibility in foodborne pathogens: implications for improving applications in food safety. Curr Opin Biotechnol 2014;26:45–9. [DOI] [PubMed] [Google Scholar]
- 124.Mansour S, Bailly J, Landaud S, et al. Investigation of associations of Yarrowia lipolytica, Staphylococcus xylosus, and Lactococcus lactis in culture as a first step in microbial interaction analysis. Appl Environ Microbiol 2009;75:6422–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lambie SC, Altermann E, Leahy SC, et al. Draft Genome Sequence of Lactococcus lactis subsp. cremoris HPT, the First Defined-Strain Dairy Starter Culture Bacterium. Genome Announc 2014;2:pii: e00107-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Prol MJ, Bruhn JB, Pintado J, et al. Real-time PCR detection and quantification of fish probiotic Phaeobacter strain 27-4 and fish pathogenic Vibrio in microalgae, rotifer, Artemia and first feeding turbot (Psetta maxima) larvae. J Appl Microbiol 2009;106:1292–303. [DOI] [PubMed] [Google Scholar]
- 127.Tsen HY, Chen ML, Hsieh YM, et al. Bacillus cereus group strains, their hemolysin BL activity, and their detection in foods using a 16S RNA and hemolysin BL gene-targeted multiplex polymerase chain reaction system. J Food Prot 2000;63:1496–502. [DOI] [PubMed] [Google Scholar]
- 128.Humblot C, Guyot JP. Pyrosequencing of tagged 16S rRNA gene amplicons for rapid deciphering of the microbiomes of fermented foods such as pearl millet slurries. Appl Environ Microbiol 2009;75:4354–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Botta C, Cocolin L. Microbial dynamics and biodiversity in table olive fermentation: culture-dependent and -independent approaches. Front Microbiol 2012;3:245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cantarel BL, Coutinho PM, Rancurel C, et al. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res 2009;37:D233–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Guttman DS, McHardy AC, Schulze-Lefert P. Microbial genome-enabled insights into plant-microorganism interactions. Nat Rev Genet 2014;15:797–813. [DOI] [PubMed] [Google Scholar]
- 132.Gupta A, Gopal M, Thomas GV, et al. Whole genome sequencing and analysis of plant growth promoting bacteria isolated from the rhizosphere of plantation crops coconut, cocoa and arecanut. PLoS One 2014;9:e104259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Almquist J, Cvijovic M, Hatzimanikatis V, et al. Kinetic models in industrial biotechnology - Improving cell factory performance. Metab Eng 2014;24:38–60. [DOI] [PubMed] [Google Scholar]