Abstract
In the past decade, significant progress has been made in understanding both microRNA function and cellular pluripotency. Here we review the intersection of these two exciting fields. While microRNAs are not required for the establishment and maintenance of pluripotency in early development and cell culture, respectively, they are critically important in the regulation of the cell cycle structure of pluripotent stem cells as well as the silencing of the pluripotency program upon differentiation. Pluripotent cells, both in vivo and in vitro, dominantly express a single family of microRNAs, which can promote the reprogramming of a somatic cell back to a pluripotent state. Here, we review the known mechanisms by which these and other microRNAs regulate the different aspects of the pluripotent stem cell program in both mouse and human.
Keywords: embryonic stem cells, reprogramming, epiblast, ESCC, induced pluripotent stem cells
INTRODUCTION
microRNAs (miRNAs) are 20–21-base noncoding RNAs that bind target messenger RNAs (mRNAs), leading to destabilization and translational inhibition of the transcripts. This function is supported by a family of proteins called the Argonaute proteins, which, together with the miRNA, make up the RNA-induced silencing complex (RISC) (Bartel 2009, Fabian & Sonenberg 2012). Individual miRNAs can suppress hundreds of different mRNA targets simultaneously, placing them in a large, coregulated network (Farh et al. 2005). Extensive discussions of miRNA evolution, biogenesis, and mechanisms of gene repression have been provided in other excellent reviews (Ebert & Sharp 2012, Fabian & Sonenberg 2012, Peterson et al. 2009). Here we focus on the biological impact of miRNAs, specifically on pluripotent cells both in vivo and in vitro.
Cells are considered pluripotent if they are able to transition into any cell state in the adult organism. This review focuses on three manifestations of pluripotent cells where the roles of miRNAs have been most extensively characterized in recent years: (a) in vivo pluripotent cells located in the inner cell mass (ICM) and epiblast during early mouse development; (b) pluripotent stem cell (PSC) lines derived from these early stages of mouse and human embryogenesis that are stably cultured in vitro, including embryonic stem cells (ESCs) and epiblast stem cells (EpiSCs); and (c) PSCs arising from the dedifferentiation of somatic cells through in vitro manipulation, called induced pluripotent stem cells (iPSCs). Excellent reviews of miRNA regulation in other mammalian stem cell populations during later development and disease (most notably, adult stem cells, primordial germ cells, and cancer stem cells) have been published recently and are not discussed here (Cook & Blelloch 2013, Ivey & Srivastava 2010, Martignani et al. 2011).
microRNA EXPRESSION AND FUNCTION IN PLURIPOTENT CELLS DURING EARLY MOUSE DEVELOPMENT
Pluripotency in Early Mouse Development
The zygote, resulting from the fusion of an egg and a sperm, is considered totipotent because it is able to form all cell states of both embryonic and extraembryonic tissue. At approximately embryonic day (E) 3.5, the cells of the developing mouse embryo self-organize into a structure called a blastocyst, comprised of two cellular compartments: the ICM and the trophectoderm. Cells of the ICM are considered pluripotent because they give rise to all cells of the embryo proper. The ICM differentiates into the primitive endoderm and epiblast cells (Rossant & Tam 2009). Epiblast cells will go on to form all embryonic lineages and, therefore, are also considered pluripotent. Remarkably, stable PSC lines can be derived as late as E8.0, long after the initiation of gastrulation (Osorno et al. 2012). Whether these cells represent dedifferentiation of the E8.0 embryonic cells in culture or remnant pluripotent cells in vivo is unclear.
Global microRNA Function in Early Development
Early efforts to determine the importance of miRNAs during mouse development relied on genetic deletion of Dicer, which encodes an enzyme required for the biogenesis of all miRNAs (see sidebar, microRNA Biogenesis) (Figure 1) (Bernstein et al. 2003). Homozygotic loss of Dicer is embryonic lethal. Embryos are grossly abnormal by E7.5 and are ubiquitously reabsorbed by E9.5 (Bernstein et al. 2003). Follow-up studies confirmed that several genes associated with gastrulation and definitive endoderm were absent or aberrantly expressed between E6.5 and E7.5 (Bernstein et al. 2003, Spruce et al. 2010). Interestingly, at E5.5, Dicer knockout epiblasts were morphologically normal and displayed no defects in the expression of pluripotency markers or in cell proliferation. However, apoptosis within the epiblast was increased dramatically. Further, the trophectoderm exhibited abnormal gene expression and reduced cell proliferation. These data demonstrate that, in the absence of Dicer, naive pluripotent cells of the ICM are established, but embryonic development arrests shortly after implantation, likely owing to a combination of defects in the cells of the epiblast and extraembryonic tissues.
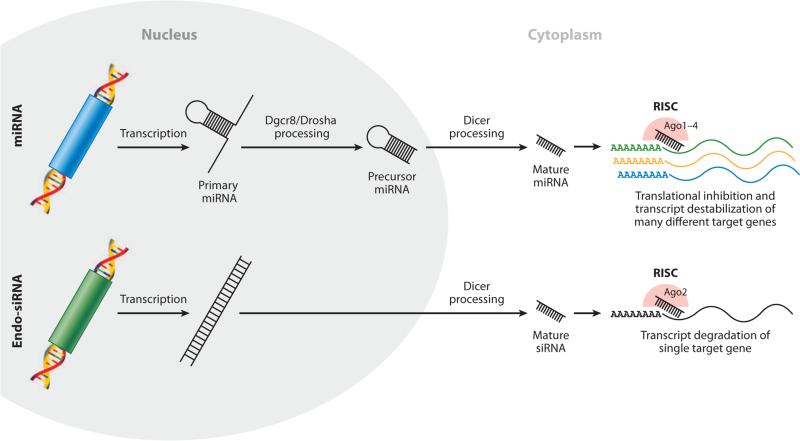
Figure 1.
Schematic of microRNA (miRNA) (top) and endogenous small interfering RNA (endo-siRNA) (bottom) biogenesis (see sidebar, microRNA Biogenesis). Abbreviation: RISC, RNA-induced silencing complex.
Although pioneering, studies based upon constitutive Dicer deletion have been limited in interpretation for two reasons. First, as Dicer−/− embryos can be generated only from crossing Dicer+/− parents, the resulting knockout embryos may be partially rescued by the contribution of a maternal Dicer transcript or enzyme. Indeed, it has been clearly shown in zebrafish that maternal Dicer does rescue early development, with an absence of any phenotypes until 10 days postfertilization, whereas maternal-zygotic loss causes major defects much earlier, beginning with the onset of gastrulation (Giraldez et al. 2005, Wienholds et al. 2003). Second, although critical for miRNA biogenesis, Dicer is also a key regulator in the processing of other noncoding small RNAs, notably endogenous small interfering RNAs (siRNAs) and noncanonical miRNAs in the mouse (see sidebar, microRNA Biogenesis) (Figure 1); thus, the phenotypes could be attributed to these other small RNA populations (Babiarz et al. 2008, Tam et al. 2008, Watanabe et al. 2008). Indeed, a study using a maternal-zygotic deletion of another miRNA biogenesis protein, Dgcr8, showed a dramatically different phenotype from Dicer loss (Suh et al. 2010). Unlike Dicer, Dgcr8 is not involved in the processing of other known classes of small RNAs (Babiarz & Blelloch 2009). Loss of maternal Dicer results in oocyte arrest (Tang et al. 2007). In contrast, loss of maternal Dgcr8 results in normal-appearing functional oocytes, showing that the Dicer phenotype is secondary to production of other classes of small RNAs, likely endo-siRNAs (Figure 2) (Suh et al. 2010). Importantly, maternal-zygotic-null Dgcr8 embryos are normal up to implantation, with intact ICMs, comparable cells numbers, and proper expression of pluripotency markers (Suh et al. 2010). Careful characterization of the postimplantation phenotypes remains to be described. Together, these studies show that miRNAs, as a class of molecules, are not required for preimplantation development, or the establishment of embryonic pluripotent cells in vivo, but are critical for postimplantation embryonic development. In the absence of a more careful analysis, it remains unclear exactly when embryogenesis is blocked in the absence of all miRNAs.
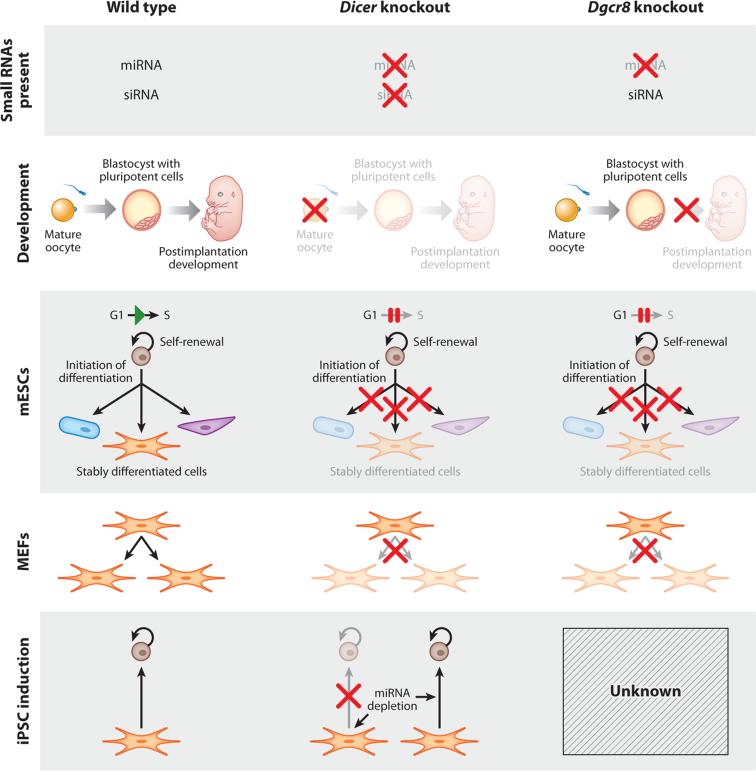
Figure 2.
Phenotypes of Dicer and Dgcr8 knockout. (Row 1) Small RNA populations present in each knockout. (Row 2) Effect of maternal or maternal-zygotic deletion of Dicer and Dgcr8 on development. Maternal Dicer knockout oocytes do not complete maturation, whereas maternal-zygotic knockout Dgcr8 oocytes mature and develop to the blastocyst stage. (Row 3) Effect of knockouts on mouse embryonic stem cell (mESC) biology. mESCs lacking Dicer or Dgcr8 retain their ability to self-renew and can initiate differentiation, but they fail to silence pluripotency factors and acquire a delayed G1-to-S transition. (Row 4) Knockout of either Dicer or Dgcr8 in mouse embryonic fibroblasts (MEFs) results in cell cycle arrest and senescence. (Row 5) Cell cycle arrest in Dicer knockout MEFs prevents reprogramming, but Dicer knockout after initiation of reprogramming is permissive for induction of pluripotent stem cells (PSCs). Abbreviations: microRNA, miRNA; siRNA, small interfering RNA.
Individual microRNA Function in Early Development
Little is known about the expression or role of specific miRNAs during early mouse development. Although the field has inferred much from profiling stable cultured PSC lines derived from ICMs and epiblasts (see below), studies that directly compare these cell lines with their isolated in vivo counterparts find significant differences in the expression of several of the most dominant miRNA families (Tang et al. 2010). One exception to this is the miR-290-295 cluster, originally discovered as an ESC-specific miRNA locus (Houbaviy et al. 2003). The miR-290 cluster is expressed at comparable levels in isolated single blastomeres and ESCs (Tang et al. 2010). Single-cell and whole-embryo analyses show that miR-290 is initially activated during the 4–8-cell stage and is repressed after E6.5, which suggests that it is expressed in the pluripotent cells of the ICM (Medeiros et al. 2011, Tang et al. 2010). Despite its dominant expression in pluripotent cells in vivo, the miR-290 cluster is not required for the establishment of pluripotency, as miR-290-cluster knockout blastocysts develop normally and can even grow to adulthood (Medeiros et al. 2011). Interestingly, partially penetrant abnormalities occur starting at E8.5, after PSCs can be derived from the epiblast, which suggests non–pluripotent cell–related effects.
A second major cluster associated with cultured PSC lines is the miR-302 cluster. Profiling of whole embryos revealed that in vivo, the miR-302 cluster is not expressed at E3.5, is highly expressed by E6.5, and is silenced again by E8.5 (Card et al. 2008). This finding is consistent with expression in the pluripotent cells of the epiblast, although the cell types that express this cluster in vivo have yet to be determined.
Studies using locked nucleic acid–based in situ hybridization concluded that members of the related miR-17-92, miR-106a-363, and miR-106b-25 clusters were also expressed in the ICM of blastocysts (Foshay & Gallicano 2009). However, similar to the miR-290 cluster, triple knockout of the three loci has no effect on early embryogenesis and instead induces late E15 lethality secondary to cardiac defects (Ventura et al. 2008). Of the more than 40 other reported genetic deletions of miRNAs, none have demonstrated embryonic defects prior to E14.5 (Kuhnert et al. 2008; Liu et al. 2008; Park et al. 2010, 2012; Zhao et al. 2007). Thus, the early embryonic-lethal phenotype of the miRNA-null Dicer and Dgcr8 knockout embryos remains unexplained. However, considering the frequent functional redundancy of miRNAs, multiple clusters likely must be deleted to recapitulate the global knockout phenotype.
microRNA EXPRESSION AND FUNCTION IN CULTURED PLURIPOTENT STEM CELL LINES
microRNA Expression in Pluripotent Stem Cell Lines
Several types of stable PSC lines can be derived from mouse and human embryos. These include mouse ESCs (mESCs), mouse EpiSCs (mEpiSCs), and human ESCs (hESCs). These lines can be passaged indefinitely in culture and differentiated into all embryonic germ layers. mESCs are derived from the ICM of blastocyst-stage embryos (Evans & Kaufman 1981, Martin 1981). They express a well-characterized circuitry of self-reinforcing transcription factors that allow for both long-term self-renewal and retention of pluripotency (see below). When injected into blastocysts, mESCs integrate into the ICM and contribute to all tissues of the developing embryo, generating chimeric animals. Interestingly, although both mESCs and hESCs are isolated from cultured blastocyst-stage preimplantation embryos, they appear to represent different steps in embryonic development, and their cellular programs are inherently different (Brons et al. 2007, Tesar et al. 2007, Thomson et al. 1998). Of note, the growth conditions, morphology, X-chromosomal activation status, and molecular profiles of hESCs are more akin to mEpiSC lines, which can be derived from postimplantation (e.g., E5.5) mouse embryos or by differentiating mESCs in the presence of Activin and Fgf. Unlike mESCs, mEpiSCs express markers of early differentiation and cannot contribute to chimeras. They are assumed to share a primed pluripotent state with hESCs, whereas mESCs are considered to be a naive cell population (Brons et al. 2007, Guo et al. 2009, Tesar et al. 2007).
The differences observed between mESCs, mEpiSCs, and hESCs extend into the miRNA expression profile (see sidebar, Pluripotency microRNA Clusters and Families) (Figure 3) (Table 1). Whereas in mESCs the polycistronic clusters mmu-miR-290-295 and mmu-miR-17-92b are the dominant contributors to the miRNA profile (Marson et al. 2008), mEpiSCs and hESCs mainly express miRNAs from the miR-302-367 cluster (Bar et al. 2008, Jouneau et al. 2012). The murine miR-290-295 cluster, as well as its human analog, miR-371-373, are at low levels in mEpiSCs and hESCs, respectively (Bar et al. 2008, Jouneau et al. 2012). Furthermore, consistent with the primed pluripotent states of mEpiSCs, there is also an upregulation of miRNAs commonly found in somatic tissue (Chiang et al. 2010, Jouneau et al. 2012).
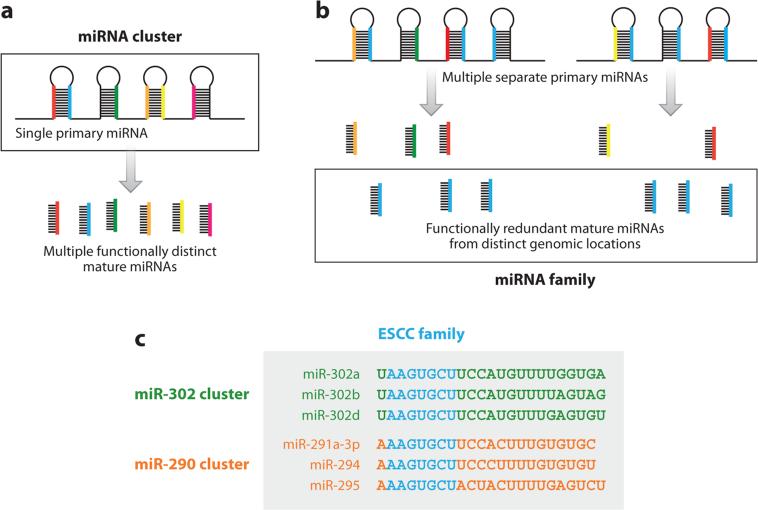
Figure 3.
microRNA (miRNA) classification. miRNAs can be classified as both (a) clusters and (b) families. miRNAs that are adjacent in the genome and encoded in the same transcript are in a cluster, regardless of the sequence of each mature miRNA. miRNAs that have the same seed sequence have overlapping targets and, therefore, are grouped into families, regardless of their location in the genome. (c) Examples of miRNAs from two different clusters that are part of the same family, the ESCC family. The shared seed sequence is highlighted in blue.
Table 1.
microRNA profile of pluripotent stem cells
| Percent of total microRNA profile | Conservation | Other tissues | |||||
|---|---|---|---|---|---|---|---|
| Human embryonic stem cell | Mouse embryonic stem cell | Mouse epiblast stem cell | |||||
| Min. | Max. | Min. | Max. | Single study | |||
| Clusters with ESCC microRNAs | |||||||
| miR-371-373 | 0.00 | 0.67 | – | – | – | hsa, mml | Germ cell tumor |
| miR-290-295 | – | – | 14 | 63 | 4 | mmu, rno | Placenta, germ cell tumor |
| miR-302-367 | 15.00 | 31.00 | 0.1 | 0.9 | 23.5 | hsa, mml, mmu, xtr | Germ cell tumor |
| miR-17-92b | 5.00 | 13.00 | 6 | 17.5 | 2.7 | hsa, mml, mmu, rno, xtr, dre | Germ cell tumor, lymphoma |
| miR-106a-363 | 0.50 | 1.50 | 2.3 | 4.8 | 2.9 | hsa, mml, mmu, rno, xtr, dre | Germ cell tumor, lymphoma |
| miR-106b-25 | 0.25 | 6.00 | 1.9 | 3.1 | 1.8 | hsa, mml, mmu, rno, xtr, dre | Germ cell tumor, lymphoma |
| Other microRNA clusters highly expressed in pluripotent cells | |||||||
| miR-200b-429 | 0.05 | 0.08 | 0.5 | 1.5 | 0.6 | hsa, mml, mmu, rno, xtr, dre | Adult tissue |
| miR-200c-141 | 0.20 | 0.40 | 0.2 | 1 | 0 | hsa, mml, mmu, rno, xtr, dre | Placenta, adult tissue |
| miR-183-182 | 0.01 | 0.09 | 1.5 | 20.5 | 14.8 | hsa, mml, mmu, rno, xtr, dre | Adult tissue, germ cell tumor, B-cell lymphoma |
| Ubiquitous and somatic microRNAs | |||||||
| miR-21 | 8.50 | 22.00 | 1.4 | 12.4 | 5.6 | hsa, mml, mmu, rno, dre | Ubiquitous |
| Combined ESCC microRNAs | |||||||
| 16.00 | 36.00 | 19 | 44 | 32 | |||
| Citations | |||||||
| Morin et al. 2008 | Bar et al. 2008 | Jouneau et al. 2012 | Marson et al. 2008 | Jouneau et al. 2012 | Kiezun et al. 2012 | Landgraf et al. 2007 | |
Abbreviations: dre, Danio rerio; ESCC, embryonic stem cell–specific cell cycle–regulating; hsa, Homo sapiens; mml, Macaca mulatta; mmu, Mus musculus; rno, Rattus norvegicus; xtr, Xenopus tropicalis.
Interestingly, despite the different dominating miRNA clusters in mESCs and mEpiSCs/hESCs, the miRNA profiles of all three of these PSC lines are dominated by a single family of conserved miRNAs, sharing an AAGUGC seed sequence, called the ESC-specific cell cycle–regulating (ESCC) family of miRNAs (see below) (Kiezun et al. 2012). The seed sequence consists of six to eight bases in the 5′ region of a miRNA and is predominantly responsible for mRNA targeting (Bartel 2009). Thus, miRNAs with the same or similar seed sequences are thought to have the same or similar targets (see sidebar, Pluripotency microRNA Clusters and Families) (Figure 3). All three of the dominant PSC miRNA clusters—miR-290-295/miR-371-373, miR-302, and miR-17-92b—contain ESCC or ESCC-like (shifted by one base) miRNAs. As a result, although expressed from different genomic loci, this single miRNA superfamily contributes to between 20% and 50% of the miRNA population in all PSC lines (Table 1). This is in contrast to somatic cells, which generally do not express the ESCC miRNAs, although the ESCC-like miRNAs of the miR-17-92b cluster are expressed in various somatic contexts (Landgraf et al. 2007). The high proportion of this common ESC seed sequence suggests a crucial contribution to PSC function and maintenance.
There are also miRNA families that are broadly expressed across somatic tissues but are absent or expressed at very low levels in PSCs. Most notable is the let-7 family of miRNAs, which is activated only upon PSC differentiation (see below) (Bar et al. 2008; Houbaviy et al. 2003, 2005; Laurent et al. 2008; Marson et al. 2008; Stadler et al. 2010; Suh et al. 2004). Interestingly, prilet-7 is present in PSCs but is not further processed into pre- and mature miRNA forms (see sidebar, microRNA Biogenesis) (Figure 1). Maturation is inhibited by the RNA-binding protein Lin28. Details of this process have been extensively reviewed elsewhere (Lehrbach & Miska 2010, Thornton & Gregory 2012).
Effect of Dicer and Dgcr8 Knockout on Embryonic Stem Cell Behavior In Vitro
Studies conducted in mESCs lacking either Dicer or Dgcr8 corroborate the in vivo observation that, globally, miRNAs are required not for establishment of pluripotent cells but rather for differentiation. Both Dicer−/− and Dgcr8−/− mESCs exhibit normal mESC morphology and express the pluripotency circuitry (Kanellopoulou et al. 2005, Wang et al. 2007). However, upon induction of differentiation, both genotypes fail to silence pluripotency markers.
Dgcr8−/− mESCs exhibit analogous, but less severe, phenotypes when compared with Dicer−/− mESCs. Whereas in differentiating conditions, Dicer−/− mESCs show a complete lack of differentiation and concomitant marker expression (such as Hnf4a, Bmp4, and Gata1), Dgcr8−/− mESCs demonstrate only reduced expression of differentiation markers like Fgf5, Brachyury, and Hnf4. Furthermore, Dgcr8−/− mESCs continue to grow even after day 16 of embryoid body formation, whereas Dicer−/− embryoid bodies stop growing after day 8 (Kanellopoulou et al. 2005, Wang et al. 2007). Furthermore, Dicer knockout ESCs are more difficult to derive and likely represent cells that manage to escape an initial proliferation arrest (Murchison et al. 2005). In contrast, Dgcr8 knockout cells are easy to derive following conditionally induced deletion (Wang et al. 2007). These phenotypic differences suggest additional functional roles for Dicer in mESCs, possibly secondary to other classes of small RNAs, such as endo-siRNAs and noncanonical miRNAs (Babiarz et al. 2008), similar to the different phenotypes associated with maternal loss of these enzymes, as described above. It will be important to dissect the roles of members of these additional classes of small RNAs in mESCs.
Unlike the ICM in Dicer knockout embryos, the Dicer and Dgcr8 knockout mESCs display a proliferation and cell cycle defect and accumulate in the G1 phase of the cell cycle (Kanellopoulou et al. 2005, Wang et al. 2008). Once again, the difference between embryos and culture may reflect persistent miRNAs in vivo resulting from maternally contributed Dicer. Consistent with the phenotypes observed in murine ESCs, knockdown of DICER in human ESCs leads to comparable defects in proliferation and self-renewal (Qi et al. 2009).
Together, these reports suggest that, though important for proliferation and cell cycle structure, miRNAs are not required for the maintenance or self-renewal of PSC lines. Rather, they are essential for proper differentiation and silencing of the pluripotency network (Figure 2). The miRNA-null Dicer−/− and Dgcr8−/− mESC lines provide a powerful tool for the discovery of specific interactions and functions of miRNA families.
microRNAs Regulating the Pluripotent Stem Cell Cycle
One of the most well-characterized miRNA-regulated PSC programs is the maintenance of the distinct ESC cell cycle structure. The typical cell cycle depends on external mitogenic cues to overcome a G1-to-S restriction point. These mitogens promote cell cycle progression by regulating a combination of factors that normally inhibit the formation of various Cyclin/Cdk complexes (Blagosklonny & Pardee 2002). Specific G1 inhibitors include members of the Ink4 family (p15, p16, p18, p19), which inhibit mainly Cdk4 and Cdk6 complexes, and members of the Cip/Kip family (p21, p27, p57), which inhibit mainly Cdk2 complexes. The Cyclin/Cdk complexes phosphor-ylate the retinoblastoma (Rb) family proteins, including Rb1, Rbl1, and Rbl2. Unphosphorylated Rb proteins inhibit E2f-mediated transcription, whereas their phosphorylation by the Cyclin/Cdk complexes releases this inhibition. E2f targets then drive exit from the restriction point and entry into S phase. Remarkably, the G1-to-S restriction point is absent in PSCs (Schratt et al. 2001). Also, CyclinE/Cdk2 activity is constitutively elevated, leading to constant Rb phosphorylation and transcription of E2f targets even in the absence of CyclinD/Cdk4,6 activity (Savatier et al. 1994, Stead 2002, White et al. 2005). The constitutive Cdk2 activity is presumably responsible for the absence of the restriction point and, at least in part, the rapid cell cycle of PSCs (reviewed in Orford & Scadden 2008).
One of the main defects in miRNA-null ESCs is a reduced proliferation rate. Part of this phenotype can be explained by changes in the cell cycle structure. miRNA-deficient ESCs accumulate in G1, which implies a crucial role of ESC-specific miRNAs in the rapid G1-to-S transition (Wang et al. 2008). Consistent with this conclusion, Dgcr8−/− ESCs exhibit high levels of Rb1 and Rbl2 as well as of cell cycle inhibitors Lats2 and p21, both of which regulate the CyclinE/Cdk2 complex (Wang et al. 2008). Furthermore, the Dgcr8−/− ESCs have decreased levels of c-Myc, a promoter of the G1-S transition (Melton et al. 2010).
A functional screen performed in Dgcr8−/− mESCs identified several miRNAs able to rescue this defect. These miRNAs possess the ESCC miRNA seed sequence (Table 1) and are mainly transcribed from the PSC-dominant miR-290-295, miR-302-367, and miR-17-92 clusters (Wang et al. 2008). miRNAs containing the ESCC seed appear to be functionally redundant in this role. The ESCC miRNAs directly target known inhibitors of the G1-to-S transition: p21, Lats2, and Rbl2 in both human and mouse (Qi et al. 2009, Subramanyam et al. 2011, Wang et al. 2008). The ESCC miRNAs also upregulate c-Myc, through an unknown intermediate (Melton et al. 2010).
Consistent with the mouse observations, knockdown of Drosha or Dicer in hESCs results in similar cell cycle defects that are restored by hsa-miR-372, as well as members of the hsa-302-367 and the hsa-miR106b-25 cluster. Also, similar to those in mouse, these miRNAs target P21 (Qi et al. 2009). Additionally, hsa-miR-195 also enhances cell cycle progression in the knockdown cells by targeting WEE1, which regulates the G2-to-M transition (Qi et al. 2009). In another study, Sengupta et al. (2009) showed that miR-92b (hsa-miR-17-92 cluster) promotes the G1-to-S transition by directly targeting the CDK inhibitor P57KIP2. These data show how several miRNAs cooperatively counteract inhibitors of the cell cycle to establish the unique ESC cell cycle profile (Figure 4).
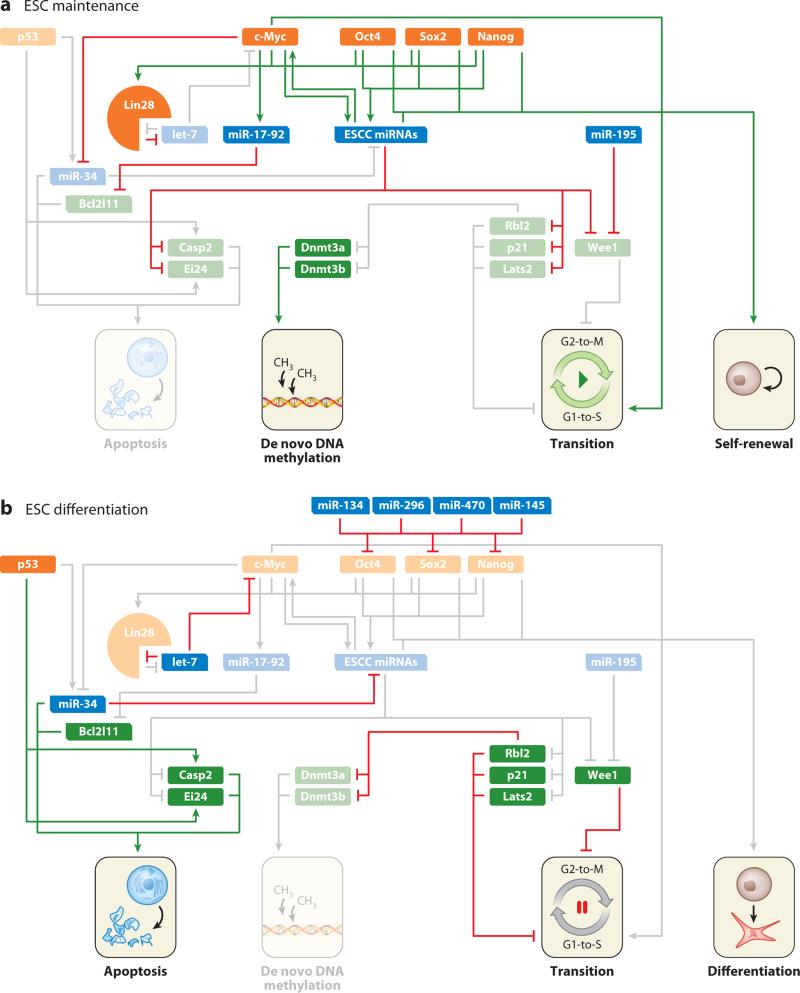
Figure 4.
Interactions between microRNAs (miRNAs) and pluripotency regulatory factors (a) in the embryonic stem cell (ESC) maintenance state and (b) upon induction of differentiation. The ESC-specific cell cycle (ESCC) miRNAs and Oct4/Nanog/Sox2 confer ESC maintenance. In contrast, let-7 supports the differentiated somatic cell state. Green lines indicate activation, red lines indicate repression, and gray lines indicate interactions that are not present in the indicated cell state.
microRNAs Regulating Pluripotent Stem Cell Differentiation
The second major defect in miRNA-null mESCs is their inability to silence the pluripotency network when cultured in differentiating conditions. This defect can be rescued by the let-7 family of miRNAs. Although widely expressed in various somatic tissues and upregulated in mESCs grown in differentiation conditions, mature let-7 miRNA is absent in mESCs (Landgraf et al. 2007; Melton et al. 2010; Thomson et al. 2004, 2006). Introduction of let-7 into Dgcr8−/− mESCs downregulates pluripotency markers, including alkaline phosphatase activity and Oct2, Sox2, and Nanog levels (Melton et al. 2010). However, let-7 is not able to shut down the self-renewal program in wild-type ESCs or when cointroduced with ESCC miRNAs in Dgcr8−/− mESCs (Melton et al. 2010), which suggests an antagonistic relationship between these miRNA families. A global analysis of let-7 targets in mESCs revealed that this family targets hundreds of members of the pluripotency network, such as Sall4, n-Myc, and Lin28 (Melton et al. 2010). The ESCC miRNAs indirectly upregulate many of these same let-7 targets (Melton et al. 2010). Exactly how the ESCC miRNAs are functionally dominant over let-7 remains to be determined, although its ability to upregulate Lin28, which inhibits let-7 processing, is likely part of the mechanism (see below).
The differentiation defect in miRNA-deficient Dicer−/− mESCs may also been linked to regulation of the Rb protein Rbl2 by ESCC miRNAs. During normal differentiation, Oct4 is silenced at the level of the promoter—first transiently by repressors and then permanently by promoter methylation—the latter process being achieved through DNA methyltransferases Dnmt3a and Dnmt3b. The initial transient silencing of the Oct4 locus occurs in both wild-type and Dicer−/− mESCs during differentiation, whereas permanent silencing through promoter methylation does not occur in Dicer−/− mESCs, leading to reactivation of Oct4. Overexpression of Dnmt3a and Dnmt3b was shown to rescue this phenotype. Dicer−/− mESCs have abnormally low levels of these enzymes, resulting in a global loss of DNA methylation (Benetti et al. 2008) and the inability to permanently silence the Oct4 promoter. Interestingly, the ESCC target Rbl2 is an inhibitor of Dnmt3a and Dnmt3b. Therefore, it was proposed that the ESCC miRNAs may indirectly enable early differentiation by suppressing Rbl2 and hence maintaining high levels of the de novo methyltransferases (Figure 4) (Benetti et al. 2008, Sinkkonen et al. 2008). However, overexpression of the miR-290 cluster prevents differentiation in wild-type mESCs (Lichner et al. 2011), and ESCC miRNAs greatly enhance the dedifferentiation of somatic cells into PSCs (see below) (Judson et al. 2009). Further exploration of the combinatorial effects of all the targets of ESCC miRNAs is needed to elucidate these apparently contradictory findings.
microRNAs in the Main Pluripotency Circuitry
In recent years, the circuitry of transcription factors that maintain pluripotency in PSCs has been well defined. Oct4, Sox2, and Nanog were identified to be at the top level of the hierarchy that controls pluripotency in the ICM as well as in ESCs (Avilion et al. 2003, Chambers et al. 2003, Mitsui et al. 2003, Nichols et al. 1998, Niwa et al. 2000). Oct4, Sox2, and Nanog, often together with other pluripotency factors, co-occupy promoters of a myriad of miRNAs and protein-coding genes in ESCs. They bind and activate promoters of genes involved in ESC maintenance, including their own promoters (Boyer et al. 2005, Marson et al. 2008). The Oct4-Sox2-Nanog triumvirate also represses genes involved in lineage commitment co-occupying these promoters with Polycomb-group proteins (PcG), potentially priming them for differentiation (Bernstein et al. 2006, Boyer et al. 2006).
In 2008, three groups were able to positively associate Oct4, Sox2, and Nanog with the dominant PSC-specific miRNA clusters: the mouse miR-290-295 cluster, the human miR-371-372 cluster, and the conserved miR-302-367 and miR-106a-363 clusters (Barroso-delJesus et al. 2008, Card et al. 2008, Marson et al. 2008). As mentioned above, these clusters each contain members with the ESCC miRNA seed sequence. Interestingly, Oct4-Sox2-Nanog also binds to the let-7g promoter, a miRNA that is known to drive differentiation, in mESCs (Marson et al. 2008). Indeed, pri-let-7g transcripts are present at high levels (Thomson et al. 2006). However, in mESCs, processing of primary let-7 transcript into mature let-7 is inhibited by Lin28 (Thornton & Gregory 2012). Lin28 itself is under transcriptional control of Oct4, Sox2, Nanog, and Tcf3 in mESCs (Marson et al. 2008); is highly expressed in ESCs; and is downregulated upon differentiation (Moss & Tang 2003).
Four reports showed that Lin28 inhibits processing of pri- and pre-let-7 transcript at the nuclear (Newman et al. 2008, Viswanathan et al. 2008) and cytoplasmic levels (Heo et al. 2008, Rybak et al. 2008). In turn, mature let-7 directly targets Lin28 mRNA, forming a self-reinforcing feedback loop (Rybak et al. 2008). In accordance with its role in maintaining low levels of prodifferentiative let-7, Lin28 overexpression was also shown to improve reprogramming efficiency of somatic cells to human iPSCs (Yu et al. 2007) (see below). High levels of immature let-7 transcripts suggested a mechanism in which loss of Lin28 with differentiation provides a fast molecular switch, which enables upregulation of mature let-7 even in the absence of transcription. Presumably, such a mechanism accelerates differentiation. Lin28 can also directly target mRNAs and likely has additional, let-7-independent roles in regulating pluripotency (Cho et al. 2012, Polesskaya et al. 2007, Qiu et al. 2010, Xu et al. 2009a). Therefore, Lin28 appears to have multiple mechanisms for promoting pluripotency that involve both miRNA and mRNA regulation.
Apart from let-7, several tissue-specific miRNAs have been shown to target and downregulate the main pluripotency factors upon differentiation: Tay et al. (2008) showed that in mPSCs, miR-134, miR-470, and miR-296 are induced upon differentiation and downregulate Oct4, Sox2, and Nanog. In hPSCs, miR-145 is induced upon differentiation and was reported to repress Oct4, Sox2, and Klf4 (Xu et al. 2009b). Although present at high levels in hPSCs, miR-21 may be upregulated upon differentiation and may target Oct4, Sox2, and Nanog, although these data have been challenged (Buckley et al. 2009, Jorgensen et al. 2009, Singh et al. 2008).
Another important pluripotency factor is c-Myc, which is a well-known oncoprotein and regulator of cell proliferation (Cartwright et al. 2005, Dang 2012). In PSCs, c-Myc co-occupies one third of the promoters of actively transcribed genes with Oct4, Sox2, and Nanog (Young 2011). These coregulated genes are enriched for metabolic and cell cycle regulators. Deletion of c-Myc, along with its family member n-Myc, triggers cell cycle arrest and early lineage commitment, which shows that they are partially redundant and contribute to maintenance of the ESC phenotype in growth conditions that contain both serum and leukemia inhibitory factor (LIF) (Davis 1993, Schulte et al. 2009, Smith et al. 2010). They also inhibit differentiation through repression of the primitive endoderm regulator Gata6 (Smith et al. 2010). Interestingly, ESCs cultured with Mapk inhibitor do not require Myc activity for self-renewal, demonstrating that this dependency relies on the growth conditions (Hishida et al. 2011).
Consistent with the previously described effects on ESC phenotypes, c-Myc and n-Myc were shown to bind the miR-290-295 promoter (Chen et al. 2008, Judson et al. 2009), and c-Myc was furthermore shown to induce the miR-302-367 cluster through an indirect mechanism (Lin et al. 2009). In turn, miR-294 indirectly activates c-Myc expression (Melton et al. 2010), implying a positive feedback loop. In human tumor cells, C-MYC and N-MYC have also been shown to induce expression of hsa-miR-17-92 and hsa-106a-363 (Northcott et al. 2009, O'Donnell et al. 2005, Schulte et al. 2008), two clusters that are highly enriched in ESCs and contain miRNAs closely related to the ESCC miRNAs. Furthermore, c-Myc binds to promoters and induces expression of other regulators of pluripotency, miR-141, miR-200, and miR-429. Reintroduction of these miRNAs into ESCs mitigates differentiation (Lin et al. 2009).
In addition to the regulation of pluripotency associated miRNAs, Myc is also involved in a negative feedback loop with let-7: let-7 targets c-Myc and n-Myc (Kumar et al. 2007, Melton et al. 2010), and the Myc proteins, in turn, inhibit biogenesis of mature let-7 through activation of Lin28 (Chang et al. 2009) (Figure 4). Furthermore, in lymphoma cells, Myc may directly inhibit let-7 expression through promoter binding (Chang et al. 2008).
microRNAs in Apoptosis
Apoptosis is a process crucial to tissue development and homeostasis as well as to the removal of damaged cells, often through the activation of the p53 pathway. Several miRNAs in the miR-290 cluster inhibit apoptosis through targeting of Casp2 and Ei24 (Zheng et al. 2011), both of which are known proapoptotic factors that are activated by p53 (Li & Yuan 2008, Zhao et al. 2005). In combination with p21 and Lats2, these proteins expand the group of p53 downstream effectors that are targeted by ESCC miRNAs (Zheng et al. 2011). Another important effector of p53-mediated DNA damage response is miR-34, which promotes both apoptosis (Chang et al. 2007) and cell cycle arrest (Tarasov et al. 2007). Interestingly, miR-34 members target C-MYC (Wei et al. 2008), and C-MYC, in turn, can negatively regulate hsa-miR-34b, at least in human lymphoma cells (Craig et al. 2011).
In human cancer cells, other pluripotency-associated miRNAs were also shown to target P53 effectors: Members of the miR-17-92 and miR-106b-25 clusters have been shown to repress apoptosis by targeting P21 (Gibcus et al. 2011) and BCL2L11 (Ho et al. 2011). Therefore, miRNAs highly expressed in ESCs, including the ESCC family of miRNAs and the miR-17/20/106 miRNAs, likely work together to repress apoptosis (Figure 4).
microRNAS IN REPROGRAMMING
An important area of study that has received much attention recently for its potential clinical application is the in vitro derivation of iPSCs from somatic cells in a process called directed dedifferentiation or reprogramming (Takahashi & Yamanaka 2006). Typically, a set of defined transcription factors is introduced into somatic cells in the form of DNA vectors. Remarkably, expression of these transcription factors induces a small fraction of these cells to adopt the morphology and molecular profile of ESCs, functionally reestablishing both self-renewal and pluripotency. Because miRNAs can be overexpressed or suppressed without the risk of genomic alterations, several groups have investigated the potential of miRNAs in the generation of iPSCs.
microRNA Expression During Reprogramming
Multiple layers of heterogeneity make studying gene expression during directed dedifferentiation a difficult endeavor. For example, the reprogramming process itself is gradual, inefficient, and stochastic, such that in a single population of reprogramming cells, relatively few individual cells are in the same transcriptional state (Buganim et al. 2012, Hanna et al. 2009, Polo et al. 2012, Samavarchi-Tehrani et al. 2010). Furthermore, many different combinations of reprogramming factors have been used, and these factors have been introduced with different technologies and supplemented with different media conditions. Each one of these changes has drastic effects on gene expression and reprogramming efficiency (Esteban et al. 2010, Hanna et al. 2009, Lee et al. 2012a). It is therefore remarkable that, over several independent studies, numerous miRNAs have been found to have relatively consistent changes in expression during this process.
The let-7 family, miR-10a/b, miR-21, miR-26a/b, miR-29a/b, the miR-30 family, miR-34b-5p, miR-34c, miR-99a/b, miR-100, miR-125a/b-5p, miR-136, miR-145, miR-146a/b, miR-199a-3p, miR-210, miR-301, and miR-434-3p have all been identified by multiple groups as miRNAs downregulated during the reprogramming of mouse embryonic fibroblasts (MEFs) with either Oct4, Sox2, and Klf4 (OSK) or Oct4, Sox2, Klf4, and c-Myc (OSKM) (Chen et al. 2012, Polo et al. 2012, Yang et al. 2011). Fewer miRNAs have been found to be consistently upregulated during reprogramming. Several groups have found that members of the miR-17-92, miR-106a-363, and miR-106b-25 clusters, as well as miR-183, are activated early in OSKM reprogramming of MEFs (Chen et al. 2012, Li et al. 2011, Polo et al. 2012). c-Myc directly binds the promoters of the miR-17-92, miR-106a-363, and miR-106b-25 clusters and is largely responsible for their activation (Liao et al. 2011). In contrast, the miR-290 cluster has been consistently found to reactivate late in reprogramming, along with other core pluripotency transcription factors (Chen et al. 2012, Judson et al. 2009, Polo et al. 2012). Interestingly, the promoter of this cluster is bound by all four reprogramming factors but is activated only following epigenetic remodeling (Judson et al. 2009, Marson et al. 2008). Other miRNAs with known function in PSCs, including miR-200, miR-130/301/721, and the miR-302 cluster, have been reported to be activated during reprogramming, but the reported degree and timing of activation have been inconsistent, likely reflecting the stochasticity of the assay (Chen et al. 2012, Li et al. 2011, Liao et al. 2011, Polo et al. 2012, Ryul Lee et al. 2012, Samavarchi-Tehrani et al. 2010, Wang et al. 2011). Indeed, miR-302 cluster reactivation during reprogramming reportedly requires not only Oct4 binding but also expression of the histone demethylase Jhdm1b and media supplementation with vitamin C, highlighting the variability introduced by both the reprogramming factors and media conditions on miRNA expression during this assay (Wang et al. 2011).
microRNAs that Regulate Reprogramming
Exogenous overexpression of more than a dozen miRNAs has been reported to enhance OSK or OSKM reprogramming of MEFs (Table 2). The first study focused specifically on the miR-290 and miR-302 clusters owing to their known high expression and function in PSC lines. Interestingly, within these clusters, only the ESCC miRNAs, including miR-294 and miR-302a, enhance OSK iPSC colony formation in a dose-dependent manner, reaching 70-fold increases in efficiency (Judson et al. 2009). Furthermore, the ESCC miRNAs increase the quality of this transition, bypassing partially reprogrammed or transformed colonies (Judson et al. 2009). This effect of the ESCC miRNAs on reprogramming has proven to be extraordinarily robust (Liao et al. 2011). Nearly all miRNAs reported to enhance reprogramming possess an ESCC miRNA seed sequence (Table 2). Variants in the sequence reduce, but do not demolish, the enhancement (Li et al. 2011, Pfaff et al. 2011). Importantly, the positive effect of ESCC miRNAs on reprogramming efficiency and specificity is conserved in human reprogramming (Subramanyam et al. 2011). Additionally, several groups have reported successful reprogramming in the absence of exogenous transcription factors by using different combinations of miRNAs, with the ESCC miRNAs being the only common component of each combination (see below) (Anokye-Danso et al. 2011; Lin et al. 2008, 2011; Miyoshi et al. 2011).
Table 2.
| Category | microRNA | Species | Seed region | Fold change | Factors | Verified functional targets | Citations |
|---|---|---|---|---|---|---|---|
| ESCC microRNA enhancers | miR-372 | hsa | -aaagugcugc- | 4–12× | osk, oskm | SMARCC2, MDB2, RBL2, CDC2L6, RHOC, MECP2, TGFBR2 | Subramanyam et al. (2011) |
| miR-302 | hsa | -uaagugcuuc- | 4–12× | osk, oksm | SMARCC2, MDB2, RBL2, CDC2L6, RHOC, MECP2, TGFBR2, P21, P130, NR2C2, AOF1, AOF2, MECP1-p66, MECP2 | Subramanyam et al. (2011), Banito et al. (2009), Hu et al. (2012), Li et al. (2011), Lin et al. (2011) | |
| mmu | -uaagugcuuc- | 10× | osk | Tgfbr2 | Judson et al. (2009), Liao et al. (2011) | ||
| miR-294 | mmu | -aaagugcuuc- | 8–70× | osk | Judson et al. (2009) | ||
| miR-295 | mmu | -aaagugcuac- | 5× | osk | Judson et al. (2009) | ||
| miR-291-3p | mmu | -aaagugcuuc- | 5× | osk | Judson et al. (2009) | ||
| ESCC-like microRNA enhancers | miR-93 | mmu | caaagugcugu- | 2–5× | osk, oskm | Tgfbr2, p21 | Li et al. (2011), Pfaff et al. (2011) |
| miR-106a/b | mmu | caaagugcuaa- | 2–5× | osk, oskm | Tgfbr2, p21 | Li et al. (2011), Liao et al. (2011), Pfaff et al. (2011) | |
| miR-17 | mmu | caaagugcuua- | 3–4× | oskm | Tgfbr2, p21 | Liao et al. (2011), Pfaff et al. (2011) | |
| miR-20 | mmu | uaaagugcuua- | 1.5–5× | osk | Liao et al. (2011), Pfaff et al. (2011) | ||
| ESCC-variant microRNA enhancers | miR-130 | mmu | -cagugcaaug- | 2–5× | osk | Meox2 | Pfaff et al. (2011) |
| miR-301 | mmu | -cagugcaaua- | 2–4× | osk | Meox2 | Pfaff et al. (2011) | |
| miR-721 | mmu | -cagugcaauu- | 2–5× | osk | Meox2 | Pfaff et al. (2011) | |
| miR-148 | mmu | -ucagugcacua- | 4× | osk | Pfaff et al. (2011) | ||
| Non-ESCC microRNA enhancers | miR-138 | mmu | agcugguguug- | 2.5× | oskm | p53 | Ye et al. (2012) |
| miR-29b | mmu | uagcaccauuu- | 2× | oskm, osk | Dnmt3a, Dnmt3b | Guo et al. (2013) | |
| microRNA inhibitors | Let-7 | mmu | ugagguaguag- | 0.25×, 0.5× | osk, oskm | n-Myc, Sall4, Lin28 | Melton et al. (2010) |
| miR-21 | mmu | uagcuuaucag- | 0.4×, 0.7× | oskm, osk | p85α, Spry1 | Yang et al. (2011) | |
| miR-29a | mmu | uagcaccaucu- | 0.4× | oskm | Cdc42, p85α, Spry1 | Yang et al. (2011) | |
| miR-34a/b | mmu | uggcagugucu- | 0.2–0.3× | osk, oskm | Nanog, Sox2, n-Myc | Choi et al. (2011) | |
| miR-199a-3p | mmu | acaguagucug- | 0.3× | osk, oskm | Wang et al. (2012) |
Bold indicates embryonic stem cell–specific cell cycle–regulating (ESCC) microRNA seed sequence.
Abbreviations: hsa, Homo sapiens; mmu, Mus musculus; osk, Oct4, Sox2, and Klf4; oskm, Oct4, Sox2, Klf4, and c-Myc.
Given the potent effect of ESCC miRNA enhancement on reprogramming, the study of ESCC miRNA targets during this process could prove extraordinarily fruitful in the identification of genes that normally prevent the reacquisition of pluripotency in somatic cells. Indeed, the predicted mechanisms of ESCC-miRNA-enhanced reprogramming have been the subject of several recent reviews (Kuo et al. 2012, Lipchina et al. 2012). Though such postulation is valuable for directing future experiments, we stress caution in making such predictions without experimental validation, as miRNA target-prediction programs exhibit highly variable sensitivity and specificity (Baek et al. 2008, Pfaff et al. 2011, Selbach et al. 2008). Furthermore, it has been well established that miRNAs exhibit cell-context specific targeting, dependent on the presence and nature of transcripts in any particular cell (Alonso 2012, Karreth et al. 2011, Subramanyam et al. 2011). For these reasons, we restrict this summary of the mechanism of ESCC miRNAs during reprogramming to those genes shown to be direct targets of the miRNAs in relevant cell types and to where the manipulation of these genes has been experimentally shown to regulate reprogramming.
Surprisingly, despite the known role of ESCC miRNAs in regulating the ESC cell cycle, ESCC miRNA reprogramming enhancement does not appear to be the result of increasing proliferation of the starting somatic population (Judson et al. 2009, Lin et al. 2011, Pfaff et al. 2011). Overexpression of ESCC miRNAs in non-PSCs can actually induce cell cycle arrest (Lin et al. 2011). In the search for mechanism, 14 human ESCC miRNA targets and 2 mouse ESCC miRNA targets have been identified that functionally regulate reprogramming (Table 2). In both mouse and human, the ESCC miRNAs have been shown to directly target Tgfbr2 and p21 (Banito et al. 2009, Li et al. 2011, Liao et al. 2011, Subramanyam et al. 2011, Wang et al. 2008). Inhibition of both Tgf-β signaling and p21-induced senescence enhances reprogramming (Banito et al. 2009, Ichida et al. 2009, Maherali & Hochedlinger 2009, Subramanyam et al. 2011). Reprogramming of fibroblasts is initiated by a mesenchymal-to-epithelial transition (MET), and further progression is inhibited by a reversal epithelial-to-mesenchymal transition (EMT) (Samavarchi-Tehrani et al. 2010). Tgf-β and Rho signaling are both well-established drivers of EMT, and the ESCC miRNAs inhibit these pathways and this transition (Liao et al. 2011, Subramanyam et al. 2011). Thus, the ESCC miRNAs likely enhance reprogramming, in part, through either EMT inhibition or MET enhancement via direct inhibition of the Tgf-β and Rho signaling pathways. Indeed, the ESCC miRNAs cause an acceleration of MET during reprogramming (Liao et al. 2011, Subramanyam et al. 2011). Interestingly, the miR-200 family is also a well-established regulator of MET and enhances MET during reprogramming (Samavarchi-Tehrani et al. 2010). Unlike the ESCC miRNAs, however, the miR-200 family does not enhance the number of eventual iPSC colonies that form, which suggests that the ESCC miRNAs must have additional mechanisms to enhance reprogramming (Liao et al. 2011, Samavarchi-Tehrani et al. 2010). Indeed, the ESCC miRNAs have been shown in human to reduce cellular senescence through direct targeting of P21 and P130; to induce OCT4 expression through inhibition of NR2C2; and to inhibit a host of epigenetic modifiers, including MBD2, MECP2, SMARCC2, AOF2, AOF1, and MECP1-p66 (Banito et al. 2009, Hu et al. 2012, Lin et al. 2011, Ryul Lee et al. 2012, Subramanyam et al. 2011). In mouse, the ESCC variant miR-130/301/721 targets Meox2, a transcript that encodes for a protein that inhibits reprogramming, presumably via p21 induction (Pfaff et al. 2011). In addition to the ESCCs, a few miRNAs with independent seed sequences have been identified to have subtle positive effects on reprogramming (Table 2). Of these, the only elucidated mechanism has been for miR-138, which inhibits p53 (Ye et al. 2012). Because miRNAs have been reported to have hundreds of targets, it will be interesting to see whether these few genes are responsible for miRNA enhancement of reprogramming or whether a more significant subset of interacting target genes will be revealed.
In addition to the miRNAs that enhance reprogramming, many of the miRNAs that are highly expressed in MEFs and downregulated upon dedifferentiation are also active inhibitors of the reprogramming process. Knockdowns of the let-7 family, miR-21, miR-29a, miR-34a/b, and miR-199a-3p all enhance mouse OSK or OSKM reprogramming two- to fivefold (Choi et al. 2011, Lee et al. 2012b, Melton et al. 2010, Wang et al. 2012, Yang et al. 2011). Interestingly, each of these miRNAs appears to be critically integrated into the Myc-p53-p21 axis. Both miR-29a and miR-21 directly target p85α, and miR-29a additionally targets Cdc42 (Yang et al. 2011). Each of these genes inhibits activation of p53 signaling—a well-established barrier to reprogramming. p53 activation inhibits reprogramming through p21-induced senescence, miR-34a activation, and miR-199a-3p activation (Banito et al. 2009, Choi et al. 2011, Wang et al. 2012). miR-199a-3p appears to also be involved in senescence, but miR-34a functions independently of p21-effects, targeting members of the pluripotency network (Nanog and Sox2) (Choi et al. 2011). Similarly, let-7 targets Sall4 and Lin28, both of which are known enhancers of reprogramming, and the latter of which inhibits let-7 (Heo et al. 2008, Melton et al. 2010, Newman et al. 2008, Rybak et al. 2008, Tsubooka et al. 2009, Viswanathan et al. 2008, Yu et al. 2007). Both miR-34a and let-7 target n-Myc, which can bind and inhibit miR-29a and miR-21 expression, completing the circuit (Choi et al. 2011, Melton et al. 2010, Yang et al. 2011). miR-29a and miR-21 also directly inhibit Spry1, thus activating Erk signaling, yet another barrier to reprogramming (Yang et al. 2011).
In contrast to these results, a recent report also identified miR-29b, which has the same seed sequence as miR-29a, as a mild enhancer of reprogramming—overexpression of the miRNA causes a twofold increase in iPSC colonies, and inhibition causes a twofold decrease in iPSC colonies (Guo et al. 2013). The mechanism of action was determined to be direct binding and inhibition of Dnmt3a and Dnmt3b, both of which inhibit reprogramming. These conflicting results concerning the role of the miR-29 family in reprogramming once again may reflect the heterogeneous and stochastic nature of the assay.
This complex network of inhibiting and enhancing miRNAs has led several groups to investigate whether miRNAs, as a class of gene regulators, are required for reprogramming. Indeed, given that miRNAs are not required for pluripotent cell viability in vivo or in vitro, but are required for differentiation, global loss of miRNA function likely would enhance reprogramming efficiency. This interesting question remains unanswered, because knockdown or knockout of Dicer or Dgcr8 causes senescence in MEFs (Kim et al. 2012, Li et al. 2011). Interestingly, when Dicer is conditionally knocked out directly before OSKM introduction, resulting in loss of miRNAs by day 5 of reprogramming, Dicer knockout iPSCs form, which suggests that miRNAs are required not for the reprogramming process but rather for somatic cell viability (Figure 2) (Kim et al. 2012).
microRNA-Only Reprogramming
Lin et al. (2008) first reported that miRNAs might be able to independently induce pluripotency in the absence of transcription factors. The authors used a retroviral construct to introduce miR-302a–d into human melanoma and prostate cancer lines. miR-302 expression caused 95% of the cells to die, but the surviving cells expressed markers of PSCs. However, these iPSC-like cells were restricted in their differentiation to the neuronal lineage. Given the significant amount of cell death, the speed of OCT4 induction, and the negative effect that miR-302 can have on somatic cell viability, the miRNAs likely were selecting for a more stem-like population.
However, in 2011, three groups reported the generation of fully reprogrammed iPSCs from normal mouse and human tissue by using miRNAs exclusively. Each group used different combinations of miRNAs and different technologies of overexpression. The ESCC miRNA miR-302 is the only common factor. Wu and colleagues used electroporation of a doxycyclinemiR-302a–d-expressing construct into normal human hair follicle cells. Approximately 5–6 days after miR-302 expression was induced, more than 90% of the cells reprogrammed, expressing OCT4, SOX2, and NANOG and demonstrating some characteristics of PSCs (Lin et al. 2011). In a second study using a lentiviral construct overexpressing miR-302a– d and miR-367, Morrisey and colleagues reported the full reprogramming of normal mouse or human fibroblasts into iPSCs (Anokye-Danso et al. 2011). The efficiency of mouse and human reprogramming was 80% in 8 days and 10% in 14 days, respectively—both efficiencies two orders of magnitude higher than those found using the traditional OSKM. The reprogramming required the expression of miR-367. Additionally, Mori and colleagues reported full reprogramming of normal mouse and human fibroblasts through transfection of synthesized mature miRNA mimics (Miyoshi et al. 2011). The authors serially transfected a mixture of miR-302a–d, miR-200, miR-369-3p, and miR-369-5p every 48 h. Using this method, researchers obtained 1–3 iPSC colonies after 14 and 20 days for mouse and human cells, respectively.
Interestingly, controversy exists over these data, as reports from independent groups have failed to reproduce miRNA-only reprogramming. Further attempts of the Wu method have been reported by three independent groups; two concluded the results could not be reproduced, and the other successfully created human iPSCs but from transformed starting populations (Hu et al. 2012, Koide et al. 2012, Ryul Lee et al. 2012). Likewise, viral overexpression of the miR-302 cluster with miR-367, as in the Morrisey method, has been reported by two independent groups, neither of which observed transcription factor–independent reprogramming (Liao et al. 2011, Ryul Lee et al. 2012). Further, both groups found miR-367 to be dispensable for the reprogramming-enhancing effects of the cluster.
Which variables allow for, in some cases, miRNA-only mediated reprogramming with stunning efficiency, and in others, complete lack thereof? The levels of the miRNAs are likely critically important, as oversaturation causes cell cycle arrest in somatic populations (Lin et al. 2011). In addition to expression levels, a recent study has demonstrated that the technical methods of factor introduction play a role in cellular reprogramming and gene activation, which suggests that the method of miRNA introduction is important (Lee et al. 2012a). Further, Koide and colleagues found that, even with transformed cells, the Wu method requires passaging onto MEFs, which suggests that specific culturing conditions are also critical (Koide et al. 2012). miRNA-only reprogramming promises an integration-free, highly efficient, and highly specific form of reprogramming with significant clinical value. Pinpointing and deciphering the unknown variables that are currently compromising the consistent reproducibility of these methods should be a priority of the field.
CONCLUSIONS AND FUTURE DIRECTIONS
Studies over the past decade have revealed significant insights into the role miRNAs play in pluripotent cell biology. Interestingly, many of the observations made in different systems are, at face value, paradoxical. For example, the dominant miRNAs expressed in PSCs, namely, the ESCC miRNAs, appear to be unnecessary for achieving and maintaining pluripotency both during early mammalian development and during in vitro culturing of PSC lines. However, these same miRNAs are powerful inducers of pluripotency during somatic cell reprogramming. Furthermore, these miRNAs are required for proper maintenance of the ESC cell cycle in vitro, but their role in cell cycle regulation in vivo remains unclear. The role of miRNAs in establishing somatic lineages is more consistent, as miRNAs are essential both in vivo for postimplantation development, with embryos arresting prior to or early in gastrulation, and in vitro for PSC differentiation.
One possible explanation for differences in the establishment versus induction of pluripotency is that miRNAs serve as a buffer against alternative cell fates. Introduction of a PSC-dominant miRNA into a somatic cell might increase iPSC production through the inhibition of somatic cell programs, which would not be seen during normal establishment of PSCs. Furthermore, that miRNA-null mESCs do not differentiate might suggest that pluripotency is a default cellular state, which somatic cells are locked out of, in part, through lineage-specific miRNA expression. This notion is quite different from Waddington's classic epigenetic landscape, which suggests that pluripotency is a higher energy state and is more difficult to achieve than somatic states. Rather, this model predicts that, without miRNAs, somatic cells might have a greater tendency to revert back to pluripotency. Similarly, the role of the ESCC miRNAs would be to prevent aberrant transitions of PSCs into more differentiated cell types through destabilization of somatic cell programs—consistent with the observations that ESCC miRNAs both stabilize (but are not required for) pluripotency and enhance reprogramming.
Future experiments can address these paradoxes by focusing on the molecular mechanisms of the major miRNA-related phenotypes described here. Which miRNAs and targets are responsible for embryonic lethality? Why do ESCC miRNAs have opposing effects on the cell cycle in mESCs and somatic cells? If the cell cycle arrest induced by miRNA-processing-machinery knockout is overridden, does total miRNA knockdown enhance or inhibit reprogramming? By what mechanisms do the ESCC miRNAs enhance reprogramming? Answering these questions will give significant insight into the role miRNAs play in the establishment, maintenance, and destabilization of both pluripotent cells and the diversity of cell types found in somatic tissue.
microRNA BIOGENESIS.
Most miRNAs are generated from longer transcripts transcribed by RNA-polymerase II. These transcripts are called primary miRNA (pri-miRNA) (Lee et al. 2004). Pri-miRNAs are processed by a protein complex consisting of the RNAse III enzyme Drosha (Lee et al. 2003) and the RNA-binding protein Dgcr8 (Denli et al. 2004, Gregory et al. 2004, Han et al. 2004, Landthaler et al. 2004) into ~70-nt hairpin precursor miRNAs (pre-miRNAs) (Figure 1). After exportation to the cytoplasm, the pre-miRNAs are cleaved by the RNAse III enzyme Dicer into 19–25-nt double-stranded mature miRNA molecules (Hammond et al. 2000). One strand of the double-stranded RNA is then bound by an Argonaute family RNA-binding protein (Ago1–4), forming the RNA-induced silencing complex (RISC) (Khvorova et al. 2003, Schwarz et al. 2003). In contrast, endogenous small interfering RNAs (endo-siRNAs) are derived from long double-stranded RNAs from various genomic locations, which bypass Dgcr8/Drosha processing but are cleaved by Dicer before being incorporating into RISC (Elbashir et al. 2001, Zamore et al. 2000). RNA-loaded RISC localizes predominantly to the 3′ untranslated region of mRNAs through complementary binding (reviewed in Bartel 2004). Depending on the free energy of the base pairing between the small miRNA and the mRNA target, RISC can favor degradation, destabilization, or translational inhibition (Baek et al. 2008, Bazzini et al. 2012, Djuranovic et al. 2012, Guo et al. 2010). Only Ago2 has slicer activity and, therefore, is capable of target degradation (Liu et al. 2004). miRNA-loaded RISCs predominantly function through translational inhibition and target destabilization, while siRNAs result in target degradation. However, there are rare examples of mature miRNAs directly cleaving targets (Yekta et al. 2004), and siRNAs can behave like miRNAs (Doensh et al. 2003).
PLURIPOTENCY microRNA CLUSTERS AND FAMILIES.
microRNAs (miRNAs) are organized by cluster and family. A cluster of miRNAs is expressed from the same locus in the genome and can contain individual miRNAs from several different families. A family of miRNA is defined by a common seed sequence—six to eight bases near the 5′ end of the miRNA—which is largely responsible for target identification (Bartel 2009). Thus, miRNAs with the same seeds often bind the same mRNA targets, whereas miRNAs from the same cluster are transcriptionally coregulated (Friedman et al. 2009).
The miRNA profile of pluripotent stem cell (PSC) lines has been well documented (Bar et al. 2008, Calabrese et al. 2007, Jouneau et al. 2012, Marson et al. 2008, Morin et al. 2008). Remarkably, unlike most somatic cells, which express a diversity of different miRNA species, a single miRNA family, known as ESC-specific cell cycle–regulating (ESCC) miRNAs, dominates PSC expression. The individual ESCC miRNAs, however, are expressed from different clusters, depending on the cell line (Table 1). These clusters contain both ESCC and non-ESCC miRNAs. Consequentially, each PSC line expresses high levels of functionally redundant ESCC miRNAs but differs in its non-ESCC miRNA profile. Generally, each of the ESCC miRNA–containing clusters is silenced during differentiation, and reactivation is associated with tumorigenesis.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- Alonso CR. A complex ‘mRNA degradation code’ controls gene expression during animal development. Trends Genet. 2012;28:78–88. doi: 10.1016/j.tig.2011.10.005. [DOI] [PubMed] [Google Scholar]
- Anokye-Danso F, Trivedi CM, Juhr D, Gupta M, Cui Z, et al. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8:376–88. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17:126–40. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiarz JE, Blelloch R. Small RNAs—their biogenesis, regulation and function in embryonic stem cells. In: StemBook, editor. Stem Cell Res. Community. Harv. Stem Cell Inst; Cambridge, MA: 2009. [PubMed] [Google Scholar]
- Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–85. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banito A, Rashid ST, Acosta JC, Li S, Pereira CF, et al. Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev. 2009;23:2134–39. doi: 10.1101/gad.1811609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar M, Wyman SK, Fritz BR, Qi J, Garg KS, et al. MicroRNA discovery and profiling in human embryonic stem cells by deep sequencing of small RNA libraries. Stem Cells. 2008;26:2496–505. doi: 10.1634/stemcells.2008-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barroso-delJesus A, Romero-López C, Lucena-Aguilar G, Melen GJ, Sanchez L, et al. Embryonic stem cell-specific miR302-367 cluster: human gene structure and functional characterization of its core promoter. Mol. Cell. Biol. 2008;28:6609–19. doi: 10.1128/MCB.00398-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzini AA, Lee MT, Giraldez AJ. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science. 2012;336:233–37. doi: 10.1126/science.1215704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benetti R, Gonzalo S, Jaco I, Muñoz P, Gonzalez S, et al. A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl2-dependent regulation of DNA methyltransferases. Nat. Struct. Mol. Biol. 2008;15:998. doi: 10.1038/nsmb0908-998b. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–26. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, et al. Dicer is essential for mouse development. Nat. Genet. 2003;35:215–17. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- Blagosklonny MV, Pardee AB. The restriction point of the cell cycle. Cell Cycle. 2002;1:103–10. [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–56. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–53. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–95. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- Buckley NJ, Johnson R, Sun YM, Stanton LW. Is REST a regulator of pluripotency? Nature. 2009;457:E5–6. doi: 10.1038/nature07784. discussion E7. [DOI] [PubMed] [Google Scholar]
- Buganim Y, Faddah DA, Cheng AW, Itskovich E, Markoulaki S, et al. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell. 2012;150:1209–22. doi: 10.1016/j.cell.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese JM, Seila AC, Yeo GW, Sharp PA. RNA sequence analysis defines Dicer's role in mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2007;104:18097–102. doi: 10.1073/pnas.0709193104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Card DA, Hebbar PB, Li L, Trotter KW, Komatsu Y, et al. Oct4/Sox2-regulated miR-302 targets cyclin D1 in human embryonic stem cells. Mol. Cell. Biol. 2008;28:6426–38. doi: 10.1128/MCB.00359-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright P, McLean C, Sheppard A, Rivett D, Jones K, Dalton S. LIF/STAT3 controls ES cell self-renewal and pluripotency by a Myc-dependent mechanism. Development. 2005;132:885–96. doi: 10.1242/dev.01670. [DOI] [PubMed] [Google Scholar]
- Chambers I, Colby D, Robertson M, Nichols J, Lee S, et al. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–55. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- Chang T-C, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol. Cell. 2007;26:745–52. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T-C, Yu D, Lee YS, Wentzel EA, Arking DE, et al. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat. Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T-C, Zeitels LR, Hwang H-W, Chivukula RR, Wentzel EA, et al. Lin-28B transactivation is necessary for Myc-mediated let-7 repression and proliferation. Proc. Natl. Acad. Sci. USA. 2009;106:3384–89. doi: 10.1073/pnas.0808300106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Wang G, Lu C, Guo X, Hong W, et al. Synergetic cooperation of microRNAs with transcription factors in iPS cell generation. PLoS ONE. 2012;7:e40849. doi: 10.1371/journal.pone.0040849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133(6):1106–17. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Chiang HR, Schoenfeld LW, Ruby JG, Auyeung VC, Spies N, et al. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev. 2010;24:992–1009. doi: 10.1101/gad.1884710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J, Chang H, Kwon SC, Kim B, Kim Y, et al. LIN28A is a suppressor of ER-associated translation in embryonic stem cells. Cell. 2012;151:765–77. doi: 10.1016/j.cell.2012.10.019. [DOI] [PubMed] [Google Scholar]
- Choi YJ, Lin CP, Ho JJ, He X, Okada N, et al. miR-34 miRNAs provide a barrier for somatic cell reprogramming. Nat. Cell Biol. 2011;13:1353–60. doi: 10.1038/ncb2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook MS, Blelloch R. Small RNAs in germline development. Curr. Top. Dev. Biol. 2013;102:159–205. doi: 10.1016/B978-0-12-416024-8.00006-4. [DOI] [PubMed] [Google Scholar]
- Craig VJ, Cogliatti SB, Imig J, Renner C, Neuenschwander S, et al. Myc-mediated repression of microRNA-34a promotes high-grade transformation of B-cell lymphoma by dysregulation of FoxP1. Blood. 2011;117:6227–36. doi: 10.1182/blood-2010-10-312231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AC. A null c-myc mutation causes lethality before 10.5 days of gestation in homozygotes and reduced fertility in heterozygous female mice. Genes Dev. 1993;7(4):671–82. doi: 10.1101/gad.7.4.671. [DOI] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–35. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- Djuranovic S, Nahvi A, Green R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 2012;336:237–40. doi: 10.1126/science.1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Petersen CP, Sharp PA. siRNAs can function as miRNAs. Genes Dev. 2003;17:438–42. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–24. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban MA, Wang T, Qin B, Yang J, Qin D, et al. Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell Stem Cell. 2010;6:71–79. doi: 10.1016/j.stem.2009.12.001. [DOI] [PubMed] [Google Scholar]
- Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–56. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat. Struct. Mol. Biol. 2012;19:586–93. doi: 10.1038/nsmb.2296. [DOI] [PubMed] [Google Scholar]
- Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, et al. The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science. 2005;310:1817–21. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- Foshay KM, Gallicano GI. miR-17 family miRNAs are expressed during early mammalian development and regulate stem cell differentiation. Dev. Biol. 2009;326:431–43. doi: 10.1016/j.ydbio.2008.11.016. [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibcus JH, Kroesen B-J, Koster R, Halsema N, de Jong D, et al. MiR-17/106b seed family regulates p21 in Hodgkin's lymphoma. J. Pathol. 2011;225:609–17. doi: 10.1002/path.2958. [DOI] [PubMed] [Google Scholar]
- Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, et al. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–38. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, et al. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–40. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- Guo G, Yang J, Nichols J, Hall JS, Eyres I, et al. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development. 2009;136:1063–69. doi: 10.1242/dev.030957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–40. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Liu Q, Wang G, Zhu S, Gao L, et al. microRNA-29b is a novel mediator of Sox2 function in the regulation of somatic cell reprogramming. Cell Res. 2013;23:142–56. doi: 10.1038/cr.2012.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–96. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–50. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–27. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Saha K, Pando B, van Zon J, Lengner CJ, et al. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol. Cell. 2008;32:276–84. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- Hishida T, Nozaki Y, Nakachi Y, Mizuno Y, Okazaki Y, et al. Indefinite self-renewal of ESCs through Myc/Max transcriptional complex-independent mechanisms. Cell Stem Cell. 2011;9:37–49. doi: 10.1016/j.stem.2011.04.020. [DOI] [PubMed] [Google Scholar]
- Ho J, Pandey P, Schatton T, Sims-Lucas S, Khalid M, et al. The pro-apoptotic protein Bim is a microRNA target in kidney progenitors. J. Am. Soc. Nephrol. 2011;22:1053–63. doi: 10.1681/ASN.2010080841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbaviy HB, Dennis L, Jaenisch R, Sharp PA. Characterization of a highly variable eutherian microRNA gene. RNA. 2005;11:1245–57. doi: 10.1261/rna.2890305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific microRNAs. Dev. Cell. 2003;5:351–58. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- Hu S, Wilson KD, Ghosh Z, Han L, Wang Y, et al. MicroRNA-302 increases reprogramming efficiency via repression of NR2F2. Stem Cells. 2012;31:259–68. doi: 10.1002/stem.1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichida JK, Blanchard J, Lam K, Son EY, Chung JE, et al. A small-molecule inhibitor of Tgf-βsignaling replaces Sox2 in reprogramming by inducing Nanog. Cell Stem Cell. 2009;5:491–503. doi: 10.1016/j.stem.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivey KN, Srivastava D. MicroRNAs as regulators of differentiation and cell fate decisions. Cell Stem Cell. 2010;7:36–41. doi: 10.1016/j.stem.2010.06.012. [DOI] [PubMed] [Google Scholar]
- Jorgensen HF, Chen ZF, Merkenschlager M, Fisher AG. Is REST required for ESC pluripotency? Nature. 2009;457:E4–5. doi: 10.1038/nature07783. discussion E7. [DOI] [PubMed] [Google Scholar]
- Jouneau A, Ciaudo C, Sismeiro O, Brochard V, Jouneau L, et al. Naive and primed murine pluripotent stem cells have distinct miRNA expression profiles. RNA. 2012;18:253–64. doi: 10.1261/rna.028878.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson RL, Babiarz JE, Venere M, Blelloch R. Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat. Biotechnol. 2009;27:459–61. doi: 10.1038/nbt.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, et al. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karreth FA, Tay Y, Perna D, Ala U, Tan SM, et al. In vivo identification of tumor-suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell. 2011;147:382–95. doi: 10.1016/j.cell.2011.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–16. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- Kiezun A, Artzi S, Modai S, Volk N, Isakov O, Shomron N. miRviewer: a multispecies microRNA homologous viewer. BMC Res. Notes. 2012;5:92. doi: 10.1186/1756-0500-5-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BM, Thier MC, Oh S, Sherwood R, Kanellopoulou C, et al. MicroRNAs are indispensable for reprogramming mouse embryonic fibroblasts into induced stem cell-like cells. PLoS ONE. 2012;7:e39239. doi: 10.1371/journal.pone.0039239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koide N, Yasuda K, Kadomatsu K, Takei Y. Establishment and optimal culture conditions of microRNA-induced pluripotent stem cells generated from HEK293 cells via transfection of microRNA-302s expression vector. Nagoya J. Med. Sci. 2012;74:157–65. [PMC free article] [PubMed] [Google Scholar]
- Kuhnert F, Mancuso MR, Hampton J, Stankunas K, Asano T, et al. Attribution of vascular phenotypes of the murine Egfl7 locus to the microRNA miR-126. Development. 2008;135:3989–93. doi: 10.1242/dev.029736. [DOI] [PubMed] [Google Scholar]
- Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat. Genet. 2007;39:673–77. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- Kuo CH, Deng JH, Deng Q, Ying SY. A novel role of miR-302/367 in reprogramming. Biochem. Biophys. Res. Commun. 2012;417:11–16. doi: 10.1016/j.bbrc.2011.11.058. [DOI] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–14. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and its D. melanogaster homolog are required for miRNA biogenesis. Curr. Biol. 2004;14:2162–67. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Laurent LC, Chen J, Ulitsky I, Mueller FJ, Lu C, et al. Comprehensive microRNA profiling reveals a unique human embryonic stem cell signature dominated by a single seed sequence. Stem Cells. 2008;26:1506–16. doi: 10.1634/stemcells.2007-1081. [DOI] [PubMed] [Google Scholar]
- Lee J, Sayed N, Hunter A, Au KF, Wong WH, et al. Activation of innate immunity is required for efficient nuclear reprogramming. Cell. 2012a;151:547–58. doi: 10.1016/j.cell.2012.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–19. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lee Y, Kim M, Han J, Yeom KH, Lee S, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–60. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YL, Peng Q, Fong SW, Chen AC, Lee KF, et al. Sirtuin 1 facilitates generation of induced pluripotent stem cells from mouse embryonic fibroblasts through the miR-34a and p53 pathways. PLoS ONE. 2012b;7:e45633. doi: 10.1371/journal.pone.0045633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrbach NJ, Miska EA. Regulation of pre-miRNA processing. Adv. Exp. Med. Biol. 2010;700:67–75. [PubMed] [Google Scholar]
- Li J, Yuan J. Caspases in apoptosis and beyond. Oncogene. 2008;27:6194–206. doi: 10.1038/onc.2008.297. [DOI] [PubMed] [Google Scholar]
- Li Z, Yang CS, Nakashima K, Rana TM. Small RNA-mediated regulation of iPS cell generation. EMBO J. 2011;30:823–34. doi: 10.1038/emboj.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao B, Bao X, Liu L, Feng S, Zovoilis A, et al. MicroRNA cluster 302–367 enhances somatic cell reprogramming by accelerating a mesenchymal-to-epithelial transition. J. Biol. Chem. 2011;286:17359–64. doi: 10.1074/jbc.C111.235960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichner Z, Páll E, Kerekes A, Pállinger E, Maraghechi P, et al. The miR-290–295 cluster promotes pluripotency maintenance by regulating cell cycle phase distribution in mouse embryonic stem cells. Differentiation. 2011;81:11–24. doi: 10.1016/j.diff.2010.08.002. [DOI] [PubMed] [Google Scholar]
- Lin C-H, Jackson AL, Guo J, Linsley PS, Eisenman RN. Myc-regulated microRNAs attenuate embryonic stem cell differentiation. EMBO J. 2009;28:3157–70. doi: 10.1038/emboj.2009.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SL, Chang DC, Chang-Lin S, Lin CH, Wu DT, et al. Mir-302 reprograms human skin cancer cells into a pluripotent ES-cell-like state. RNA. 2008;14:2115–24. doi: 10.1261/rna.1162708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SL, Chang DC, Lin CH, Ying SY, Leu D, Wu DT. Regulation of somatic cell reprogramming through inducible mir-302 expression. Nucleic Acids Res. 2011;39:1054–65. doi: 10.1093/nar/gkq850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipchina I, Studer L, Betel D. The expanding role of miR-302–367 in pluripotency and reprogramming. Cell Cycle. 2012;11:1517–23. doi: 10.4161/cc.19846. [DOI] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–41. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- Liu N, Bezprozvannaya S, Williams AH, Qi X, Richardson JA, et al. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 2008;22:3242–54. doi: 10.1101/gad.1738708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali N, Hochedlinger K. Tgfβ signal inhibition cooperates in the induction of iPSCs and replaces Sox2 and cMyc. Curr. Biol. 2009;19:1718–23. doi: 10.1016/j.cub.2009.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–33. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martignani E, Miretti S, Accornero P, Baratta M. miRNAs highlights in stem and cancer cells. Mini Rev. Med. Chem. 2011;11:1165–82. doi: 10.2174/138955711797655371. [DOI] [PubMed] [Google Scholar]
- Martin G. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. USA. 1981;78(12):7634–38. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros LA, Dennis LM, Gill ME, Houbaviy H, Markoulaki S, et al. Mir-290–295 deficiency in mice results in partially penetrant embryonic lethality and germ cell defects. Proc. Natl. Acad. Sci. USA. 2011;108:14163–68. doi: 10.1073/pnas.1111241108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton C, Judson RL, Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 2010;463:621–26. doi: 10.1038/nature08725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, et al. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–42. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- Miyoshi N, Ishii H, Nagano H, Haraguchi N, Dewi DL, et al. Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell. 2011;8:633–38. doi: 10.1016/j.stem.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Morin RD, O'Connor MD, Griffith M, Kuchenbauer F, Delaney A, et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18:610–21. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss EG, Tang L. Conservation of the heterochronic regulator Lin-28, its developmental expression and microRNA complementary sites. Dev. Biol. 2003;258:432–42. doi: 10.1016/s0012-1606(03)00126-x. [DOI] [PubMed] [Google Scholar]
- Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2005;102:12135–40. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–49. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–91. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 2000;24:372–76. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- Northcott PA, Fernandez-L A, Hagan JP, Ellison DW, Grajkowska W, et al. The miR-17/92 polycistron is up-regulated in sonic hedgehog-driven medulloblastomas and induced by N-myc in sonic hedgehog-treated cerebellar neural precursors. Cancer Res. 2009;69:3249–55. doi: 10.1158/0008-5472.CAN-08-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–43. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- Orford KW, Scadden DT. Deconstructing stem cell self-renewal: genetic insights into cell-cycle regulation. Nat. Rev. Genet. 2008;9:115–28. doi: 10.1038/nrg2269. [DOI] [PubMed] [Google Scholar]
- Osorno R, Tsakiridis A, Wong F, Cambray N, Economou C, et al. The developmental dismantling of pluripotency is reversed by ectopic Oct4 expression. Development. 2012;139:2288–98. doi: 10.1242/dev.078071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CY, Choi YS, McManus MT. Analysis of microRNA knockouts in mice. Hum. Mol. Genet. 2010;19:R169–75. doi: 10.1093/hmg/ddq367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CY, Jeker LT, Carver-Moore K, Oh A, Liu HJ, et al. A resource for the conditional ablation of microRNAs in the mouse. Cell Rep. 2012;1:385–91. doi: 10.1016/j.celrep.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson KJ, Dietrich MR, McPeek MA. MicroRNAs and metazoan macroevolution: insights into canalization, complexity, and the Cambrian explosion. BioEssays. 2009;31:736–47. doi: 10.1002/bies.200900033. [DOI] [PubMed] [Google Scholar]
- Pfaff N, Fiedler J, Holzmann A, Schambach A, Moritz T, et al. miRNA screening reveals a new miRNA family stimulating iPS cell generation via regulation of Meox2. EMBO Rep. 2011;12:1153–59. doi: 10.1038/embor.2011.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polesskaya A, Cuvellier S, Naguibneva I, Duquet A, Moss EG, Harel-Bellan A. Lin-28 binds IGF-2 mRNA and participates in skeletal myogenesis by increasing translation efficiency. Genes Dev. 2007;21:1125–38. doi: 10.1101/gad.415007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo JM, Anderssen E, Walsh RM, Schwarz BA, Nefzger CM, et al. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell. 2012;151:1617–32. doi: 10.1016/j.cell.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J, Yu JY, Shcherbata HR, Mathieu J, Wang AJ, et al. microRNAs regulate human embryonic stem cell division. Cell Cycle. 2009;8:3729–41. doi: 10.4161/cc.8.22.10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu C, Ma Y, Wang J, Peng S, Huang Y. Lin28-mediated post-transcriptional regulation of Oct4 expression in human embryonic stem cells. Nucleic Acids Res. 2010;38:1240–48. doi: 10.1093/nar/gkp1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossant J, Tam PP. Blastocyst lineage formation, early embryonic asymmetries and axis patterning in the mouse. Development. 2009;136:701–13. doi: 10.1242/dev.017178. [DOI] [PubMed] [Google Scholar]
- Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, et al. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat. Cell Biol. 2008;10:987–93. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- Ryul Lee M, Prasain N, Chae HD, Kim YJ, Mantel C, et al. Epigenetic regulation of nanog by miR-302 cluster-MBD2 completes iPS cell reprogramming. Stem Cells. 2012;31:666–81. doi: 10.1002/stem.1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samavarchi-Tehrani P, Golipour A, David L, Sung HK, Beyer TA, et al. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- Savatier P, Huang S, Szekely L, Wiman KG, Samarut J. Contrasting patterns of retinoblastoma protein expression in mouse embryonic stem cells and embryonic fibroblasts. Oncogene. 1994;9:809–18. [PubMed] [Google Scholar]
- Schratt G, Weinhold B, Lundberg AS, Schuck S, Berger J, et al. Serum response factor is required for immediate-early gene activation yet is dispensable for proliferation of embryonic stem cells. Mol. Cell. Biol. 2001;21:2933–43. doi: 10.1128/MCB.21.8.2933-2943.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Schulte JH, Horn S, Otto T, Samans B, Heukamp LC, et al. MYCN regulates oncogenic MicroRNAs in neuroblastoma. Int. J. Cancer. 2008;122:699–704. doi: 10.1002/ijc.23153. [DOI] [PubMed] [Google Scholar]
- Schulte JH, Horn S, Schlierf S, Schramm A, Heukamp LC, et al. MicroRNAs in the pathogenesis of neuroblastoma. Cancer Lett. 2009;274:10–15. doi: 10.1016/j.canlet.2008.06.010. [DOI] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Sengupta S, Nie J, Wagner RJ, Yang C, Stewart R, Thomson JA. MicroRNA 92b controls the G1/S checkpoint gene p57 in human embryonic stem cells. Stem Cells. 2009;27:1524–28. doi: 10.1002/stem.84. [DOI] [PubMed] [Google Scholar]
- Singh SK, Kagalwala MN, Parker-Thornburg J, Adams H, Majumder S. REST maintains self-renewal and pluripotency of embryonic stem cells. Nature. 2008;453:223–27. doi: 10.1038/nature06863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkkonen L, Hugenschmidt T, Berninger P, Gaidatzis D, Mohn F, et al. MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 2008;15:259–67. doi: 10.1038/nsmb.1391. [DOI] [PubMed] [Google Scholar]
- Smith KN, Singh AM, Dalton S. Myc represses primitive endoderm differentiation in pluripotent stem cells. Cell Stem Cell. 2010;7:343–54. doi: 10.1016/j.stem.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruce T, Pernaute B, Di-Gregorio A, Cobb BS, Merkenschlager M, et al. An early developmental role for miRNAs in the maintenance of extraembryonic stem cells in the mouse embryo. Dev. Cell. 2010;19:207–19. doi: 10.1016/j.devcel.2010.07.014. [DOI] [PubMed] [Google Scholar]
- Stadler B, Ivanovska I, Mehta K, Song S, Nelson A, et al. Characterization of microRNAs involved in embryonic stem cell states. Stem Cells Dev. 2010;19:935–50. doi: 10.1089/scd.2009.0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stead E. Pluripotent cell division cycles are driven by ectopic Cdk2: cyclin A/E and E2F activities. Oncogene. 2002;21(54):8320–33. doi: 10.1038/sj.onc.1206015. [DOI] [PubMed] [Google Scholar]
- Subramanyam D, Lamouille S, Judson RL, Liu JY, Bucay N, et al. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat. Biotechnol. 2011;29:443–48. doi: 10.1038/nbt.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh MR, Lee Y, Kim JY, Kim SK, Moon SH, et al. Human embryonic stem cells express a unique set of microRNAs. Dev. Biol. 2004;270:488–98. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- Suh N, Baehner L, Moltzahn F, Melton C, Shenoy A, et al. MicroRNA function is globally suppressed in mouse oocytes and early embryos. Curr. Biol. 2010;20:271–77. doi: 10.1016/j.cub.2009.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–38. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F, Kaneda M, O'Carroll D, Hajkova P, Barton SC, et al. Maternal microRNAs are essential for mouse zygotic development. Genes Dev. 2007;21:644–48. doi: 10.1101/gad.418707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F, Barbacioru C, Bao S, Lee C, Nordman E, et al. Tracing the derivation of embryonic stem cells from the inner cell mass by single-cell RNA-Seq analysis. Cell Stem Cell. 2010;6:468–78. doi: 10.1016/j.stem.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarasov V, Jung P, Verdoodt B, Lodygin D, Epanchintsev A, et al. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;6:1586–93. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–28. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, et al. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–99. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–47. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20:2202–7. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JM, Parker J, Perou CM, Hammond SM. A custom microarray platform for analysis of microRNA gene expression. Nat. Methods. 2004;1:47–53. doi: 10.1038/nmeth704. [DOI] [PubMed] [Google Scholar]
- Thornton JE, Gregory RI. How does Lin28 let-7 control development and disease? Trends Cell Biol. 2012;22:474–82. doi: 10.1016/j.tcb.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubooka N, Ichisaka T, Okita K, Takahashi K, Nakagawa M, Yamanaka S. Roles of Sall4 in the generation of pluripotent stem cells from blastocysts and fibroblasts. Genes Cells. 2009;14:683–94. doi: 10.1111/j.1365-2443.2009.01301.x. [DOI] [PubMed] [Google Scholar]
- Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–86. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, He Q, Han C, Gu H, Jin L, et al. p53-facilitated miR-199a-3p regulates somatic cell reprogramming. Stem Cells. 2012;30:1405–13. doi: 10.1002/stem.1121. [DOI] [PubMed] [Google Scholar]
- Wang T, Chen K, Zeng X, Yang J, Wu Y, et al. The histone demethylases Jhdm1a/1b enhance somatic cell reprogramming in a vitamin-C-dependent manner. Cell Stem Cell. 2011;9:575–87. doi: 10.1016/j.stem.2011.10.005. [DOI] [PubMed] [Google Scholar]
- Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, Blelloch R. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat. Genet. 2008;40:1478–83. doi: 10.1038/ng.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat. Genet. 2007;39:380–85. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–43. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- Wei JS, Song YK, Durinck S, Chen QR, Cheuk AT, et al. The MYCN oncogene is a direct target of miR-34a. Oncogene. 2008;27:5204–13. doi: 10.1038/onc.2008.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J, Stead E, Faast R, Conn S, Cartwright P, Dalton S. Developmental activation of the Rb-E2F pathway and establishment of cell cycle-regulated cyclin-dependent kinase activity during embryonic stem cell differentiation. Mol. Biol. Cell. 2005;16:2018–27. doi: 10.1091/mbc.E04-12-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienholds E, Koudijs MJ, van Eeden FJM, Cuppen E, Plasterk RH. The microRNA-producing enzyme Dicer1 is essential for zebrafish development. Nat. Genet. 2003;35:217–18. doi: 10.1038/ng1251. [DOI] [PubMed] [Google Scholar]
- Xu B, Zhang K, Huang Y. Lin28 modulates cell growth and associates with a subset of cell cycle regulator mRNAs in mouse embryonic stem cells. RNA. 2009a;15:357–61. doi: 10.1261/rna.1368009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009b;137:647–58. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- Yang CS, Li Z, Rana TM. microRNAs modulate iPS cell generation. RNA. 2011;17:1451–60. doi: 10.1261/rna.2664111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye D, Wang G, Liu Y, Huang W, Wu M, et al. MiR-138 promotes induced pluripotent stem cell generation through the regulation of the p53 signaling. Stem Cells. 2012;30:1645–54. doi: 10.1002/stem.1149. [DOI] [PubMed] [Google Scholar]
- Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–96. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- Young RA. Control of the embryonic stem cell state. Cell. 2011;144:940–54. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–20. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- Zhao X, Ayer RE, Davis SL, Ames SJ, Florence B, et al. Apoptosis factor EI24/PIG8 is a novel endoplasmic reticulum-localized Bcl-2-binding protein which is associated with suppression of breast cancer invasiveness. Cancer Res. 2005;65:2125–29. doi: 10.1158/0008-5472.CAN-04-3377. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, et al. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1–2. Cell. 2007;129:303–17. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- Zheng GX, Ravi A, Calabrese JM, Medeiros LA, Kirak O, et al. A latent pro-survival function for the mir-290-295 cluster in mouse embryonic stem cells. PLoS Genet. 2011;7:e1002054. doi: 10.1371/journal.pgen.1002054. [DOI] [PMC free article] [PubMed] [Google Scholar]