Abstract
Fluorescent conjugated DNA oligonucleotides for antisense targeting of mRNA has the potential of improving tumor/normal tissue ratios over that achievable by nuclear antisense imaging. By conjugating the Cy5.5 emitter to the 3′ equivalent end of a 25 mer phosphorothioate (PS) antisense major DNA and hybridizing with a shorter 18 mer phosphodiester (PO) complementary minor DNA (cDNA) with the Black Hole inhibitor BHQ3 on its 5′ end (i.e., PS DNA25-Cy5.5/PO cDNA18-BHQ3), we previously achieved antisense optical imaging in mice as a proof of this concept. In a process of optimization, we have now evaluated the stability of a small series of duplexes with variable-length minor strands. From these results, a new study anti-mdr1 antisense duplex was selected with a 10 mer minor strand (i.e., PS DNA25-Cy5.5/PO cDNA10-BHQ3). The new study duplex shows stability in serum environments at 37 °C and provides a dramatically enhanced fluorescence in KB-G2 (pgp++) cells when compared with KB-31 (pgp±) as evidence of antisense dissociation at its mdr1 mRNA target. The duplex was also administered to KB-G2 tumor bearing mice, and when compared to the duplex used previously, the fluorescence from the tumor thigh was more obvious and the tumor-to-background fluorescence ratio was improved. In conclusion, by a process designed to optimize the duplex for optical antisense tumor targeting, the fluorescence signal was improved both in cells and in tumored mice.
INTRODUCTION
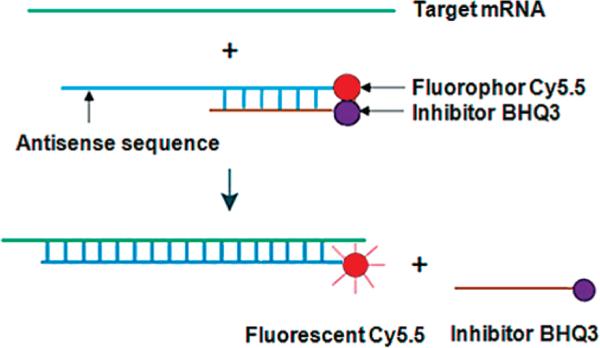
Oligomers such as DNA for antisense imaging of mRNA are potentially powerful candidates for detection of endogenous gene expression in vivo (1-5). In conventional imaging of mRNA expression in vivo, antisense oligonucleotides are usually radiolabeled with gamma ray emitting radionuclides. However, our experience and that of other laboratories suggest that achieving adequate tumor/normal radioactivity ratios by anti-sense nuclear imaging is often problematic (6). As an alternative to nuclear imaging, optical imaging has the potential to improve tumor/normal tissue ratio by modulating the detectable signal in the target through the judicious use of fluorescence resonance energy transfer (FRET) (7-11). We have been developing an approach to optical antisense imaging that in principle should provide fluorescence images in tumored subjects by an antisense mechanism that are free of background fluorescence (12). In this method, linear fluorophor-conjugated oligomer duplexes are arranged to inhibit fluorescence and to dissociate only in the presence of their target mRNA. Thus, the fluorescence signal should in principle be inhibited everywhere except in the target cell. The approach is illustrated in Scheme 1.
Scheme 1.
Schematic Diagram Illustrating the Principle of Antisense Optical Imaging
Recently, we reported on our first optical DNA duplex consisting of a Cy5.5 conjugated 25 mer phosphorothioate (PS) DNA hybridized to a BHQ3 conjugated complementary 18 mer phosphodiester (PO) cDNA for optical antisense imaging (i.e., PS DNA25-Cy5.5/PO cDNA18-BHQ3) (12). Although fluorescence in the tumor was clearly evident, this duplex also provided a fluorescence background making obvious the need for duplex optimization. In a process of optimization, we now report that five DNA duplexes differing in the length of the complementary minor strand were compared for stability using a simple thiazole orange (TO) fluorescence assay (13). On the basis of the screening results, the shortest minor strand (10 mer) was selected and, after hybridization to the 25 mer PS antisense major strand, was studied in cells and in tumored animals in comparison to that of the 18 mer major strand used previously.
MATERIALS AND METHODS
As in our earlier investigation, all duplexes used the 25 mer anti-mdr1 uniform PS antisense (5′-AAG-ATC-CAT-CCC GAC-CTC-GCG-CTC-C-3′) major strand (14-16), either native or conjugated with the Cy5.5 fluorophor on the 3′ end attached via a three carbon linker. The native duplexes were formed by hybridization with a complementary uniform native PO minor strand with one of five chain lengths. The study conjugated duplexes were formed by hybridization of the same major strand with either a 10 or 18 mer PO minor strand conjugated with a Black Hole Quencher 3 (BHQ3) quencher on the 5′ end attached via a two carbon linker. Controls for the two study conjugated duplexes were identical except that the two fluorophors were reversed:
Native Duplexes.
PS-DNA25/PO-cDNA25
PS-DNA25/PO-cDNA22
PS-DNA25/PO-cDNA18
PS-DNA25/PO-cDNA14
PS-DNA25/PO-cDNA10
Conjugated Study and Control Duplexes.
PS-DNA25-Cy5.5/PO-cDNA10-BHQ3 (study duplex)
PS-DNA25-BHQ3/PO-cDNA10-Cy5.5 (control)
PS-DNA25-Cy5.5/PO-cDNA18-BHQ3 (comparison duplex)
PS-DNA25-BHQ3/PO-cDNA18-Cy5.5 (control)
The native DNAs (Integrated DNA Technologies, Coralville, IA) and the conjugated DNAs (Biosearch Technologies, Novato, CA) were purchased HPLC purified. The TO dye was obtained from Sigma-Aldrich (St Louis, MO) and was used as received. The KB-G2 (pgp++) and KB-31 (pgp±) human epidermoid carcinoma cell lines were a gift from Isamu Sugawara (Research Institute of Tuberculosis, Tokyo, Japan) and were cultured overnight at 37 °C, 5% CO2, in Dulbecco's modified Eagle's medium (DMEM, Invitrogen, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS). The cells were incubated with DNAs in 1% FBS/DMEM medium.
Measurement of Native Duplexes
The five native duplexes were screened for stability in serum using a simple TO assay developed in this laboratory (13). Each duplex was added to a 96 well microplate at 100 μL per well and incubated at 500 nM in 70% normal mouse serum (Jackson Immuno Research Laboratories, Inc., West Grove, PA) at 37 °C for different times before the addition of the indicator TO at 6 μM. Fluorescence intensities were measured at room temperature on a Fluorescence Microplate Reader (Safire, Tecan Group Ltd.). In this case, 504 nm was used for excitation and 527 nm for detection of TO fluorescence. The TO concentration that was just sufficient to saturate the duplexes was determined by trial and error to be 6 μM. In the screening assay, decreased fluorescence was an indication of TO release and therefore of duplex instability due to dissociation and/or degradation.
Measurement of Conjugated Duplexes in Solution
Both the 10 mer and 18 mer conjugated study duplexes were prepared at different molar ratios by mixing the 10 or 18 mer cDNA with the DNA25-Cy5.5 at a molar ratio of 1:1, 2:1, 3:1, 5:1, 8:1, and 10:1 in wells of a 96 well microplate before the fluorescence was measured on the Microplate Reader. In this case, 685 nm was used for excitation and 706 nm for detection of Cy5.5 fluorescence. The DNA25-Cy5.5 concentration was 50 nM in all cases to provide a convenient fluorescence intensity.
The stability in serum of the DNA25-Cy5.5/cDNA10-BHQ3 duplex was measured by adding 100 μL of each duplex solution in a 96 well plate in quadruplicate to a final duplex concentration of 50 nM in 70% normal mouse serum (Jackson Immuno Research Laboratories, Inc., West Grove, PA) and incubated at 37 °C for up to 25 h. As control, the singlet DNA25-Cy5.5 was added to serum at the identical molar concentration in the identical manner. The fluorescence intensity was measured at each time point on the Microplate Reader.
Measurement of Conjugated Duplexes in Cells
The cell studies were performed in KB-G2 and KB-31 cells in quadruplicate in 96 well plates seeded at 10 000 cells per well and incubated with 10% FBS/DMEM culture medium overnight. In one experiment, both study and control duplexes were incubated in KB-G2 cells in 1% FBS/DMEM medium at 50, 100, 300, and 500 nM for 3 h at 37 °C. In another experiment, all four conjugated duplexes were incubated with both KB-G2 and KB-31 cells in 1% FBS/DMEM medium at 500 nM for 3 h at 37 °C. In all cases, the cells were washed twice with 1% FBS/DMEM culture medium and twice with PBS. To improve the reproducibility of the measurement, the cells were lysed before measuring the fluorescence. The lysis buffer (150 mM PBS, pH 7.2, 1% Triton X-100) was added at 100 μL per well, and the cells were incubated for 1 h at 37 °C. The fluorescence intensity of each lysis solution was measured on the Microplate Reader.
Animal Studies
All animal studies were performed with the approval of the UMMS Institutional Animal Care and Use Committee. Male nude mice (NIH Swiss, Taconic Farms, Germantown, NY) at 7 weeks of age were each injected subcutaneously in the left thigh with 100 μL suspension containing 106 KB-G2 cells with greater than 95% viability. Mice were used for imaging studies when tumors were about 0.7 cm in any dimension and after having been on a chlorophyll-free diet (AIN-93G Purified Diet, Harlan Teklad, Madison, WI) for 5 days prior to imaging. Mice bearing KB-G2 tumor were administered 3 nmol of DNA25-Cy5.5/cDNA10-BHQ3 duplex in 100 μL of PBS via a tail vein. In vivo fluorescence imaging was performed on an IVIS 100 small animal imaging system (Xenogen, Alameda, CA) using a Cy5.5 filter set. During imaging, the animals were lightly anesthetized with 1.5% isoflurane in 2 L/min oxygen. Fluorescence images were acquired at 20 min, 2 h, 7 h, and 23 h after injection using a 0.5 s exposure time (f/stop 4). Image presentation and quantification were performed using Living Image 2.5 software (Xenogen). Ellipsoid regions of interest (ROI) of equal area were drawn on the tumor and on the contralateral muscle in the normal thigh.
RESULTS
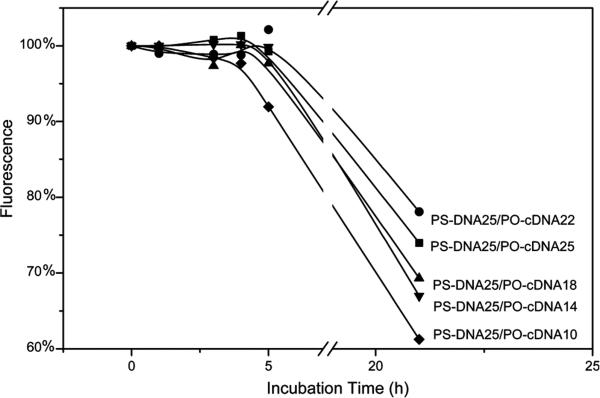
Measurement of Native Duplexes
The five native DNA duplexes, each with the same 25 mer major strand but different length of minor strand, were incubated at 37 °C in 70% normal mouse serum at 500 nM for different times before the addition of the TO indicator. The fluorescence was measured immediately after TO addition with shaking. Instability of the DNA duplex will result in a decreased fluorescence signal as denaturation of the duplex will reduce TO intercalation. Figure 1 presents the change of fluorescence intensity as a function of the incubation time in 37 °C serum and shows in all cases no change in fluorescence for at least 5 h of incubation. Thus, all duplexes appear to be stable in serum for a period sufficiently long for tumor targeting in vivo. Because of the similar stabilities, the duplex with the shortest minor strand (i.e., PS-DNA25/PO-cDNA10) was selected for further study on the assumption that it would exhibit the least stability in the presence of its mRNA target. The duplex of our earlier investigation (i.e., PS-DNA25/PO-cDNA18) was used for comparison.
Figure 1.
Percent fluorescence change for five duplexes in 70% serum at 37 °C as a function of incubation time. In each case, the fluorescence at time zero was set as 100%.
Measurement of Conjugated Duplexes in Solution
Figure 2 presents the fluorescence intensity of DNA-Cy5.5 when different molar ratios of cDNA10-BHQ3 or cDNA18-BHQ3 were added. The results show that about 90% of the fluorescence signal of Cy5.5 is quenched when hybridized with either cDNA at a 1:1 molar ratio.
Figure 2.
Fluorescence intensity of DNA25-Cy5.5 in PBS solution alone and after hybridized with increasing molar ratios of cDNA10-BHQ3 or cDNA18-BHQ3. Error bars represent standard deviations for three replicates and are too small to be visible on most data points.
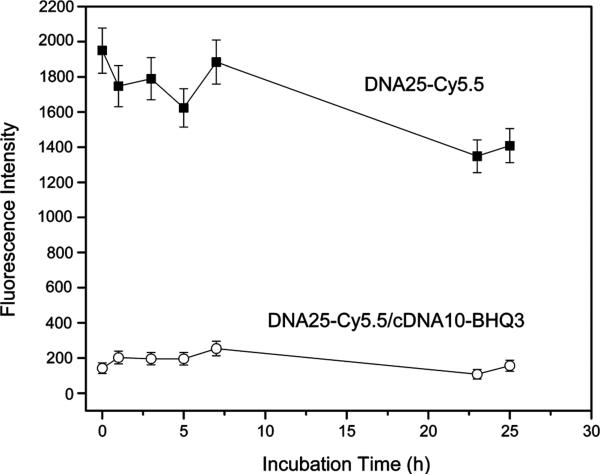
As mentioned above, the duplex with the shortest minor strand (i.e., PS-DNA25/PO-cDNA10) was selected as the study duplex on the assumption that it would exhibit the least stability in the presence of its mRNA target. The stability of this conjugated duplex was therefore evaluated over 25 h, again at 100 nM in 70% normal mouse serum at 37 °C, and the results compared to that of the singlet DNA25-Cy5.5. As shown in Figure 3, the study duplex did not show a substantial change in fluorescence over 25 h, suggesting duplex stability over this period.
Figure 3.
Fluorescence intensity showing stability of DNA25-Cy5.5/cDNA10-BHQ3 duplex compared to the DNA25-Cy5.5 single strand in 70% normal mouse serum at 37 °C over 25 h. Error bars represent standard deviations for three replicates.
Measurement of Conjugated Duplexes in Cells
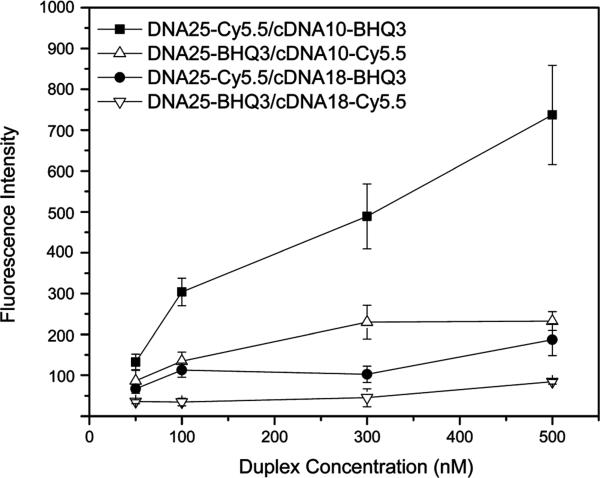
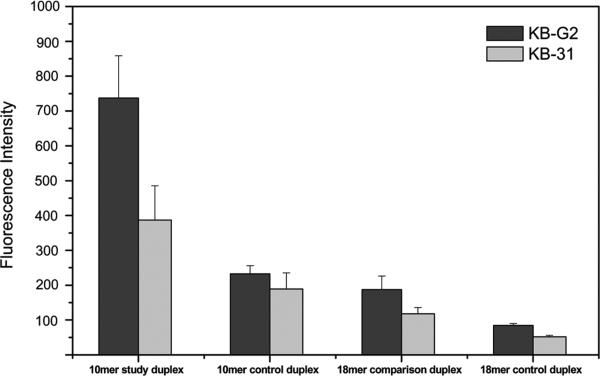
The KB-G2 cells were incubated with the study anti mdr1 mRNA antisense DNA25-Cy5.5/cDNA10-BHQ3 duplex and its control duplex in which the fluorophors were reversed. For comparison, the antisense DNA25-Cy5.5/cDNA18-BHQ3 duplex of our previous investigation (12) was also studied as a comparison duplex along with its similar control. Incubations were for 3 h at 37 °C at concentrations between 50 nM and 500 nM. Figure 4 presents the cellular fluorescence intensity vs duplex concentration and shows that the fluorescence signal of the cells is in all cases significantly higher (p < 0.05) when incubated with the 10 mer study or 18 mer comparison duplex compared to their respective control duplexes that differ only in reversed fluorophors. However, the fluorescence of cells incubated with the 10 mer study duplex is about 4-fold higher than cells incubated with the 18 mer comparison duplex as evidence of the decreased stability of the 10 mer compared to the 18 mer duplex, most probably due to antisense targeting.
Figure 4.
Cellular fluorescence of KB-G2 cells after incubation for 3 h with the 10 mer anti-mdr1 study duplex (DNA25-Cy5.5/cDNA10-BHQ3), its 10 mer control duplex with fluorophors reversed (DNA25-BHQ3/cDNA10-Cy5.5) and the 18 mer comparison duplex and its control at four concentrations. Error bars represent standard deviations for four replicates.
If the higher fluorescence of KB-G2 cells incubated with the study compared to the 18 mer comparison duplex was due to antisense dissociation as is likely, then the fluorescence should be reduced in KB-31 cells because of its limited mdr1 expression. Figure 5 presents the results of incubating both the KB-G2 and KB-31 cells with the same four duplexes as above at 500 nM. No significant differences in fluorescence were observed between the two cell lines when incubated with either the 10 mer or 18 mer control duplexes presumably because the Cy5.5 fluorophor is, in these cases, on the minor strand of each duplex that are either not dissociated (presumably in KB-31 cells) or are dissociated and released from the cell if binding does occur (presumably in KB-G2 cells). In both cases, the results would be the same: an absence of fluorescence. By contrast, the fluorescence of KB-G2 cells was statistically higher (p < 0.05) compared to the KB-31 cells when incubated with either the study 10 mer or 18 mer comparison duplex presumably because of the limited expression of the mRNA targets in KB-31. However, compared to the 18 mer duplex, the shorter 10 mer study duplex produces a much stronger fluorescence signal.
Figure 5.
Cellular fluorescence of KB-G2 and KB-31 cells after incubation for 3 h with the 10 mer anti-mdr1 study duplex (DNA25-Cy5.5/cDNA10-BHQ3), its 10 mer control duplex with fluorophors reversed (DNA25-BHQ3/cDNA10-Cy5.5), and the corresponding 18 mer comparison duplex and its control duplex at 500 nM. (Error bars represent standard deviations for four replicates.)
Animal Study
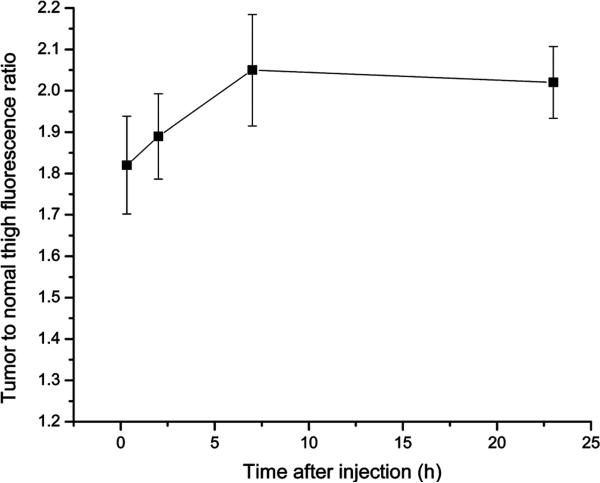
Three nude mice each bearing KB-G2 tumors in the left thigh received an injection of 3 nmol DNA25-Cy5.5/cDNA10-BHQ3 study duplex and were imaged at different times over 2 days. Images of one mouse obtained without contrast manipulation are shown in Figure 6. Tumor fluorescence is evident at all times. The fluorescence intensity ratios at four postinjection time points were calculated for the three animals, and the averages are shown in Figure 7. A positive tumor/normal thigh ratio was achieved as early as 20 min postinjection and was followed by a constant increase until a maximum at 2.05 was reached at 7 h postinjection and was constant thereafter.
Figure 6.
Whole-body dorsal fluorescence images of one mouse bearing a KB-G2 tumor in the left thigh at (A) 20 min; (B) 2 h; (C) 7 h; and (D) 23 h postinjection of 3 nmol study duplex (DNA25-Cy5.5/cDNA10-BHQ3).
Figure 7.
Average tumor-to-normal thigh fluorescence ratios vs time after injection of the study duplex. Error bars represent standard deviations for three animals. Fluorescence quantitation was accomplished using Living Image 2.5 software (Xenogen).
Compared to the images obtained earlier in the same mouse tumor model following administration of the comparison DNA25-Cy5.5/cDNA18-BHQ3 duplex (12), the study DNA25-Cy5.5/cDNA10-BHQ3 duplex of this investigation provided images with a more intense fluorescence in the tumor thigh and an averaged tumor-to-background ratio superior to the 1.6 ratio observed by us previously (12). The most likely explanation for the improved results is increased instability of the duplex with the shorter minor strand in the presence of the target mRNA.
DISCUSSION
Optical imaging is a rapidly developing biomedical technology that enables the investigation of biological/molecular events in both cell culture and small live animals and is already practical in the clinic for imaging surface cancers and those accessible by endoscopy. However, self-absorption of the optical signal can seriously complicate optical imaging even when employing near-infrared fluorescence (NIRF) imaging probes with fluorescence emission in the near-infrared where tissue hemoglobin and water, the major absorbers of visible and infrared light, respectively, have their lowest absorption coefficients (17-19). While self-absorption of γ rays in connection with nuclear imaging is much more limited, nuclear imaging, unlike optical imaging, does not offer the possibility of modulating the signal at its target site. Through the judicious use of fluorescence resonance energy transfer (FRET), optical imaging has the potential to improve the tumor/normal tissue ratio and thus become an attractive alternative to nuclear imaging (20-23).
Previously, we reported on a successful optical antisense imaging study using a Cy5.5 conjugated 25 mer antisense PS–DNA hybridized to a BHQ3 conjugated complementary 18 mer PO–DNA showing fluorescence only upon dissociation (12). The duplex was designed to resist dissociation except by an antisense mechanism in the presence of its target mRNA in tumor. Although there was obvious tumor fluorescence obtained with this duplex, there was also a background in normal tissue and therefore a less than optimal tumor/nontarget ratio. The low tumor/nontarget ratio was attributed to a combination of excessive duplex stability in the presence of the mRNA target and excessive duplex instability in circulation and in tissues and led to the conclusion that optimization was required. In this investigation, optimization was focused on varying the DNA chain length rather than changing the backbone of the phosphorothioate and phosphdiester DNAs to that of second generation oligomer analogues such as phosphodiamidate morpholinos. While keeping the antisense major strand at 25 mer, the complementary sense minor strand was varied from 25 to 10 mer and the stabilities of the duplexes measured in 37 °C serum by the TO assay. Complementary chain lengths of less than 10 mer were not considered due to concerns of low duplex binding specificity and low stability (24, 25). The results, shown in Figure 1, demonstrate that all five native duplexes were sufficiently stable by this assay in serum, and therefore, the shortest 10 mer chain length was selected for further investigation.
The results presented in Figures 4 and 5 both provide evidence of antisense targeting for both the 10 and 18 mer study duplexes. In the first instance, a significantly higher fluorescence was observed in the mdr1 expressing KB-G2 cells for the study duplexes compared to the control duplexes with fluorophors reversed, a result that would not be expected by nonspecific accumulation. In the case of the control duplexes, presumably after dissociation, the Cy5.5 PO cDNA is rapidly degraded and the fluorophor cleared from the cell. In the second instance, the fluorescence intensity of the mdr1 expressing KB-G2 cells was significantly higher than that of the control low level mdr1 expressing KB-31 cells especially in the case of the 10 mer study duplex, again providing evidence of antisense targeting.
Furthermore, the shorter DNA25-Cy5.5/cDNA10-BHQ3 study duplex provided a much higher fluorescence in cells compared to the 18 mer DNA25-Cy5.5/cDNA18-BHQ3 comparison duplex, presumably due to a lower stability toward dissociation of the study duplex in the presence of its target mRNA. Finally, the whole-body fluorescence images in KB-G2 tumor bearing mice, one of which is presented in Figure 6, show obvious fluorescence in tumor at all time points, and an improvement in the average tumor/background fluorescence ratio of 2.05 for this duplex compared to an average value of 1.6 obtained previously in the same mouse model with the comparison duplex (12).
Taken together, the work presented here provides further evidence that optical antisense imaging with fluorescent duplexes is possible and our results have been improved by the process described herein designed to optimize duplex stabilities for this application. However, it is noteworthy that there is still some unexpected fluorescence background in animals administered the optimized duplex. Obviously, the Cy5.5 in these animals is not quenched completely by the BHQ3 inhibitor, indicating that the duplex has partly dissociated in some nontarget tissues such as liver due to instability of the fluorophors and/or duplex instability to nucleases therein. A better understanding of the properties of these duplex probes in vivo will be required to further optimize this approach.
ACKNOWLEDGMENT
We thank Dr. Isamu Sugawara (Research Institute of Tuberculosis, Tokyo, Japan) for providing the KB-G2 and KB-31 cell lines for this investigation. This investigation was funded by the NIH (CA-12959).
LITERATURE CITED
- 1.Lewis MR, Jia F. Antisense imaging: and miles to go before we sleep? J. Cell. Biochem. 2003;90:464–472. doi: 10.1002/jcb.10641. [DOI] [PubMed] [Google Scholar]
- 2.Rosi NL, Giljohann DA, Thaxton CS, Lytton-Jean AKR, Han MS, Mirkin CA. Oligonucleotidemodified gold nanoparticles for intracellular gene regulation. Science. 2006;312:1027–1030. doi: 10.1126/science.1125559. [DOI] [PubMed] [Google Scholar]
- 3.Hnatowich DJ, Nakamura K. Antisense targeting in cell culture with radiolabeled DNAs-a brief review of recent progress. Ann. Nucl. Med. 2004;18:363–368. doi: 10.1007/BF02984478. [DOI] [PubMed] [Google Scholar]
- 4.Hnatowich DJ, Nakamura K. The influence of chemical structure of DNA and other oligomer radiopharmaceuticals on tumor delivery. Curr. Opin. Mol. Ther. 2006;8:136–143. [PubMed] [Google Scholar]
- 5.Dewanjee MK, Ghafouripour AK, Kapadvanjwala M, Dewanjee S, Seafini AN, Lopez DM, Sfakianakis GN. Noninvasive imaging of c-myc oncogene messenger RNA with Indium-111-antisense probes in a mammary tumor-bearing mouse model. J. Nucl. Med. 1994;35:1054–1061. [PubMed] [Google Scholar]
- 6.Nakamura K, Fan C, Liu G, Gupta S, He J, Dou S, Kubo A, Rusckowski M, Hnatowich DJ. Evidence of antisense tumor targeting in mice. Bioconjugate Chem. 2004;15:1475–1480. doi: 10.1021/bc0499073. [DOI] [PubMed] [Google Scholar]
- 7.Becker A, Hessenius C, Licha K, Ebert B, Sukowski U, Semmler W, Wiedenmann B, Grotzinger C. Receptor-targeted optical imaging of tumors with near-infrared fluorescent ligands. Nat. Biotechnol. 2001;19:327–331. doi: 10.1038/86707. [DOI] [PubMed] [Google Scholar]
- 8.Weissleder R, Tung CH, Mahmood U, Bogdanov A. In vivo imaging of tumors with protease-activated near-infrared fluorescent probes. Nat. Biotechnol. 1999;17:375–378. doi: 10.1038/7933. [DOI] [PubMed] [Google Scholar]
- 9.Ogawa M, Kosaka N, Choyke PL, Kobayashi H. In vivo molecular imaging of cancer with a quenching near-Infrared fluorescent probe using conjugates of monoclonal antibodies and indocyanine green. Cancer Res. 2009;69:1268–1272. doi: 10.1158/0008-5472.CAN-08-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ogawa M, Kosaka N, Choyke PL. Tumor-specific detection of an optically targeted antibody combined with a quencher-conjugated neutravidin “quencher-chaser”: a dual “quench and chase” strategy to improve target to nontarget ratios for molecular imaging of cancer. Bioconjugate Chem. 2009;20:147–154. doi: 10.1021/bc8003765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Silverman AP, Kool ET. Quenched probes for highly specific detection of cellular RNAs. Trends Biotechnol. 2005;23:225–230. doi: 10.1016/j.tibtech.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 12.Liu X, Wang Y, Nakamura K, Liu G, Dou S, Kubo A, Rusckowski M, Hnatowich DJ. Optical antisense imaging of tumor with fluorescent DNA duplexes. Bioconjugate Chem. 2007;18:1905–1911. doi: 10.1021/bc700221d. [DOI] [PubMed] [Google Scholar]
- 13.Liang M, Liu X, Nakamura K, Chen X, Cheng D, Liu G, Dou S, Wang Y, Rusckowski M, Hnatowich DJ. A convenient thiazole orange fluorescence assay for the evaluation of DNA duplex hybridization stability. Mol. Imaging Biol. 2009 doi: 10.1007/s11307-009-0221-4. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakamura K, Wang Y, Liu X, Kubo A, Hnatowich DJ. Cell culture and xenograft-bearing animal studies of radiolabeled antisense DNA carrier nanoparticles with streptavidin as a linker. J. Nucl. Med. 2007;48:1845–1852. doi: 10.2967/jnumed.106.039339. [DOI] [PubMed] [Google Scholar]
- 15.Nakamura K, Kubo A, Hnatowich DJ. Antisense targeting of P-glycoprotein expression in tissue culture. J. Nucl. Med. 2005;46:509–513. [PubMed] [Google Scholar]
- 16.Liu X, Nakamura K, Wang Y, Zhang S, He J, Liu G, Dou S, Kubo A, Rusckowski M, Hnatowich DJ. Improved delivery in cell culture of radiolabeled antisense DNAs by duplex formation. Mol. Imaging Biol. 2006;8:278–283. doi: 10.1007/s11307-006-0050-7. [DOI] [PubMed] [Google Scholar]
- 17.Cheng Z, Levi J, Xiong Z, Gheysens O, Keren S, Chen X, Gambhir SS. Near-infrared fluorescent deoxyglucose analogue for tumor optical imaging in cell culture and living mice. Bioconjugate Chem. 2006;17:662–669. doi: 10.1021/bc050345c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ntziachristos V, Bremer C, Weissleder R. Fluorescence imaging with near-infrared light: new technological advances that enable in vivo molecular imaging. Eur. Radiol. 2003;13:195–208. doi: 10.1007/s00330-002-1524-x. [DOI] [PubMed] [Google Scholar]
- 19.Kaijzel EL, van der Pluijm G, Lowik CWGM. Whole-body optical imaging in animal models to assess cancer development and progression. Clin. Cancer Res. 2007;13:3490–3497. doi: 10.1158/1078-0432.CCR-07-0402. [DOI] [PubMed] [Google Scholar]
- 20.Jares-Erijman E, Jovin T. FRET imaging. Nat. Biotechnol. 2003;21:1387–1395. doi: 10.1038/nbt896. [DOI] [PubMed] [Google Scholar]
- 21.Kim S, Lim YT, Soltesz EG, Grand De A. M., Lee J, Nakayama A, Parker JA, Mihaljevic T, Laurence RG, Dor DM, Cohn LH, Bawendi MG, Frangioni JV. Near-infrared fluorescent type II quantum dots for sentinel lymph node mapping. Nat. Biotechnol. 2004;22:93–97. doi: 10.1038/nbt920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao X, Cui Y, Levenson RM, Chung LWK, Nie S. In vivo cancer targeting and imaging with semiconductor quantum dots. Nat. Biotechnol. 2004;22:969–976. doi: 10.1038/nbt994. [DOI] [PubMed] [Google Scholar]
- 23.Montet X, Ntziachristos V, Grimm J, Weissleder R. Tomographic fluorescence mapping of tumor targets. Cancer Res. 2005;65:6330–6336. doi: 10.1158/0008-5472.CAN-05-0382. [DOI] [PubMed] [Google Scholar]
- 24.Sugimoto N, Nakano S, Katoh M, Matsumura A, Nakamuta H, Ohmichi T, Yoneyama M, Sasaki M. Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochemistry. 1995;34:11211–11216. doi: 10.1021/bi00035a029. [DOI] [PubMed] [Google Scholar]
- 25.Wetmur JG. DNA probes: applications of the principles of nucleic acid hybridization. Crit. ReV. Biochem. Mol. Biol. 1991;26:227–259. doi: 10.3109/10409239109114069. [DOI] [PubMed] [Google Scholar]