Abstract
The Nucleosome Remodeling and Deacetylase (NuRD) complex is one of the major chromatin remodeling complexes found in cells. It plays an important role in regulating gene transcription, genome integrity and cell cycle progression. Through its impact on these basic cellular processes, increasing evidence indicates that alterations in the activity of this macromolecular complex can lead to developmental defects, oncogenesis and accelerated ageing. Recent genetic and biochemical studies have elucidated the mechanisms of NuRD action in modifying the chromatin landscape. These advances have the potential to lead to new therapeutic approaches to birth defects and cancer.
Introduction
Factors that regulate the biochemical state of chromatin affect gene transcription, genome integrity and cell division, basic cellular processes with broad implications for human development and disease. Increasingly, efforts are being directed towards developing therapies that target chromatin remodeling complexes. These therapeutic strategies have the potential to reprogram cells and thereby promote tissue regeneration and repair, and reverse the oncogenic phenotype of tumor cells [for review see [1]].
The Nucleosome Remodeling and Deacetylase (NuRD) complex is one of four major ATP-dependent chromatin remodeling complexes, and since its discovery almost two decades ago, there has been much progress in determining its functions (Figure 1). While initially identified as a transcriptional silencer or repressor, it is now appreciated to have more complex effects on gene transcription, including in gene activation. In addition, other significant biological functions (e.g. DNA repair) have been attributed to NuRD through its modifications of chromatin and post-translational modification of other transcription factors (e.g. p53 acetylation status, [2]), independent of regulating gene expression. Alterations in NuRD activity have been implicated in embryonic development and a broad range of human diseases, including in cancer and ageing (Tables 1 and 2). In this review, our aim is to discuss some of the major recent advances elucidating important functional roles for NuRD during embryonic development and disease. When possible, we highlight findings with the potential to lead to novel therapeutic strategies.
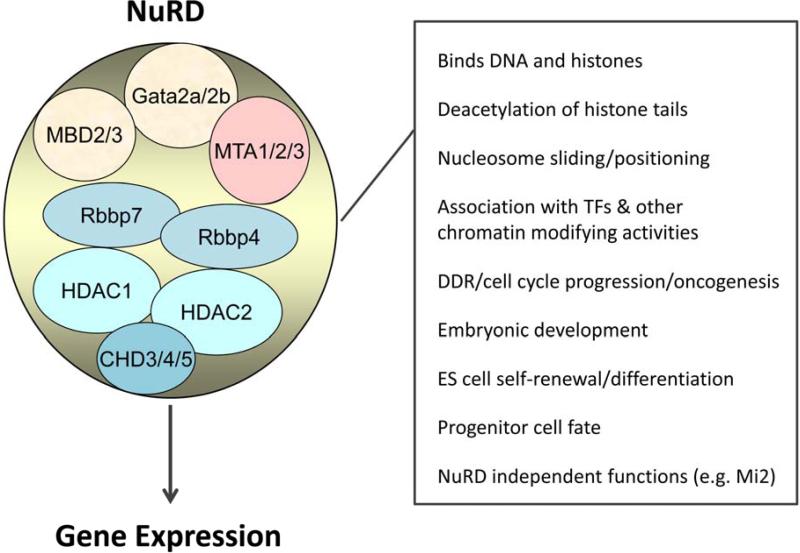
Figure 1. Functions of the Nucleosome Remodeling and Deacetylase (NuRD) Complex.
NuRD core components are shown in the illustration. NuRD regulates gene transcription through binding gene enhancers and promoters, and modifying the state of chromatin. More recent data implicate NuRD in DNA repair processes. Box on the right indicates known biochemical functions (top four on list) of NuRD components and biological roles of the complex. Increasingly, NuRD-independent functions are being described for some components (e.g., Mi2, Mta1). TF=tissue restricted transcription factors; DDR=DNA damage response
TABLE 1.
| Gene | Biochemical Function | ES Cell Phenotype | Mouse Phenotype |
|---|---|---|---|
| Hdac1/2 | Deacetylation of lysine residues on histone tails and other proteins | Hdac1−/− ES cells are viable with decreased proliferation and colony formation [102]. Hdac2−/− ES cells are viable and show no defect in proliferation or differentiation [103]. | Hdac1−/− embryos die between E9.5 and E10.5. Hdac2−/− embryos exhibit perinatal lethality due to cardiac defects [104]. Conditional deletion of Hdac1/2 indicates that these genes are redundantly required in a broad range of organs (e.g. neurons, T cells, adipocytes, kidneys) [103-105]. |
| Chd3/4/5 | Helicase/ATPase that uses the energy from ATP to remodel chromatin through histone sliding; PHD domains bind histone H3 [81] | None reported1 | Chd4 conditional mutants revealed it is required for T cell, hematopoetic stem cell, epidermal progenitor cell, and renal progenitor cell self-renewal and differentiation [19, 45, 60]. |
| Mbd2/3 | Methyl-CpG binding domain; Mbd2, but not Mbd3, binds methlylated DNA with high affinity [21] | Mbd3−/− ES cells are viable but fail to downregulate progenitor genes, and fail to differentiate [50]. | Mbd2−/− embryos are viable and fertile but the mothers have a nurturing defect. Mbd3−/− embryos are embryonic lethal between E3.5 and E8.5 [43]. |
| Mta1/2/3 | SANT and GATA DNA binding domains; directly interacts with HDAC proteins and Rbbp4/7 [41, 106] | None reported | Mta1−/− embryos are viable and exhibit cardiovascular, craniofacial, integument and skeletal defects [107] [MGI, 2010]2. Mta2−/− mutants exhibit skin lesions, bodyweight loss, glomerulonephritis, liver inflammation, and production of autoantibodies [108]. |
| Rbbp4/7 | Histone H4 binding; associates with multiple cellular proteins and thought to act as a scaffold; interacts directly with MTA1 [106] | None reported | Conditional Rbbp7 mutants have decreased CD8-positive T cell numbers, and male embryos of a gene trap allele have abnormal developmental patterning [MGI 2008, 2010]3. |
| Gata2a/2b | Nuclear zinc finger protein that binds DNA and directly interacts with Mbd2/3 [27] | None reported | Gata2a−/− embryos display anemia by E10.5, yolk sacs are devoid of red blood cells, and are embryonic lethal by E11.5 [109]. |
Databases searched for ES cell phenotype and mouse phenotype included PubMed (www.ncbi.nlm.nih.gov/pubmed), Mouse Genome Informatics (MGI, www.informatics.jax.org), and Gene Cards (www.genecards.org).
Wellcome Trust Sanger Institute, Alleles produced for the EUCOMM and EUCOMMTools projects by the Wellcome Trust Sanger Institute. MGI Direct Data Submission. 2010.
Mouse Genome Informatics (MGI) and National Center for Biotechnology Information (NCBI), Mouse Gene Trap Data Load from dbGSS. Database Download. 2008, and Wellcome Trust Sanger Institute, Alleles produced for the EUCOMM and EUCOMMTools projects by the Wellcome Trust Sanger Institute. MGI Direct Data Submission. 2010.
TABLE 2.
| Gene | Human Disease/Animal Model | Potential Translational Application |
|---|---|---|
| Hdac1/2 | Hdac2 promotes cardiac hypertrophy [110]; Hdac1/2 combined deficiency in T cells leads to neoplastic transformation [103]; acute ischemic injury and organ fibrosis reduced by Hdac inhibition [111, 112]. | Broad spectrum (e.g. TSA) and Hdac class1 specific (aminobenzamides) inhibitors [113]; valproic acid (epilepsy); two FDA approved drugs for cutaneous T cell lymphoma (Vorinostat, Romidepsin) [114]; phase 3 clinical trials in progress for other malignancies1; expansion of cord blood stem cells [115] |
| Chd3/4/5 | Mi2β auto-antibody diagnostic marker of dermatomyositis; frequent mutations of CHD4 in serous endometrial cancer [79, 80]; deletion of Chd4 in mice leads to erythroid leukemia [60]. CHD4 implicated in glioblastoma [94] CHD5 deletion and promoter methylation in CNS and other tumors [95] | Silencing of Mi2-NuRD to increase fetal Hgb as treatment of hemoglobinopathies (e.g thalassemia) [42]; calixarene-based supramolecular hosts prevent binding of CHD4 PHD2 to H3Kme3 [116]. |
| Mbd2/3 | Frequent chromosomal deletion encompassing MBD3 in serous endometrial cancer[80]; Mbd2 deficiency in mice suppresses intestinal tumor formation [117]. | Mbd3 inhibition for efficient generation of iPS cells [57, 58]; DNA methylation inhibitors to modulate Mbd2-dependent gene silencing (cancer, thalassemia) [26]; disrupt interaction with p66 (see Gata 2a/2b column below) |
| Mta1/2/3 | Increases in expression of MTAs correlate with increased metastases and poor outcome in a large variety of tumors [118]. Mta1-3 modulate pathways in breast cancer (Estrogen Receptor, Wnt, EMT); CNVs overlapping MTA3 in Li-Fraumeni syndrome with brain tumors [119]; Mta2 mouse mutant with systemic lupus-like features [108] | Potentially attractive targets for cancer and inflammatory diseases. No structures available to guide drug design; potential need for selectivity for specific MTAs. |
| Rbbp4/7 | Probable role in disease processes through its interactions with multiple cellular proteins, though specific role in pathogenesis not clearly defined2. | Structure based drug design possible but given broad/promiscuous range of protein interactions may have significant toxicity. |
| Gata2a/2b | GATA2b loss-of-function and truncating mutations associated with autosomal dominant intellectual impairment and dysmorphic facial features [69, 74] | Inhibition of Gata 2a/2b colied-coil interaction with Mbd2/3 with peptide or small molecule to disrupt NuRD [27] |
Databases searched for human disease/animal model include PubMed (www.ncbi.nlm.nih.gov/pubmed), Mouse Genome Informatics (MGI, www.informatics.jax.org), Gene Cards (www.genecards.org), and Online Mendelian Inheritance in Man (OMIM, www.ncbi.nlm.nih.gov/omim).
NuRD Biochemistry
NuRD was originally purified in 1998 [3-6] independently from different cell types and species. It is unique among chromatin remodeling complexes in having two distinct enzymatic activities, ATP- dependent nucleosome remodeling as well as histone deacetylase activity [4]. Recent studies have revealed that Lysine Demethylase 1 (Lsd1) physically and functionally interacts with NuRD in some contexts [7-9], thereby associating the complex with a third catalytic activity; however, biochemical studies that have carefully defined the stoichiometry of NuRD complex components indicate that Lsd1 is not likely to be a core NuRD component [10]. In contrast, recent quantitative proteomic studies suggest that Cyclin-Dependent Kinase 2 Associated Protein 1 (CDK2AP1), also known as Deleted in Oral Cancer 1 (DOC-1), may be a core component of NuRD [10-12] that has been overlooked in the initial purification of the complex.
NuRD is highly conserved from plants to animals and widely expressed in developing and mature tissues. The complex consists of at least 6 proteins: a) Enzymatic components, histone (protein) deacetylases HDAC1 and HDAC2 which remove acetyl groups from lysine residues on histone tails and from other proteins, such as p53 [13]. Chromodomain-helicase-DNA-binding proteins, CHD3 (Mi2-α) or CHD4 (Mi2-β), which use the energy of ATP to reposition nucleosomes on DNA [14]. A third family member, CHD5, is preferentially, though not exclusively expressed in the central nervous system and testis [15, 16]. Recent evidence indicates that Mi2-β has many NuRD independent functions, and it is possible that some of the reported Mi2-NuRD functions may be due to Mi2-β and not the complex [17-19]. b) Non-enzymatic components, methyl-CpG-binding domain protein MBD2 or MBD3. MBD2 binds cytosine-methylated DNA, however, MBD3 lacks this capacity to bind cytosine-methylated DNA with high affinity [20, 21]. Metastasis-associated proteins MTA1, MTA2, or MTA3 mediate binding to DNA and to HDAC1, and to other transcription factors and co-regulators that interact with the NuRD complex in various cell types [22-24]. Histone-binding proteins Rbbp7 and Rbbp4 (RbAp46 and RbAp48, respectively) bind histone H4 and are thought to function as scaffolding proteins or chaperones which coordinate assembly of multi-protein complexes [25]. Nuclear zinc-finger protein Gata2a and Gata2b (p66α/β) directly interact with MBD2/3 via a coiled-coiled antiparallel interface [26, 27]. While HDAC1/2 and Rbbp7/4 are present in other histone deacetylase co-repressor complexes, such as SIN3 and coREST [28, 29], Mi2, MBD3 and MTA are relatively specific to NuRD and thought to be defining components of this complex. As is evident from the above description, some of the components belong to families of related proteins encoded by gene paralogues. Consequently, different combinations of these factors can result in a diversity of NuRD complexes. For example, the three MTA family members may be found in mutually exclusive NuRD complexes with distinct and in some cases antagonistic activities [30, 31]. MBD2 and MBD3 have non-overlapping functions and these factors are also found in unique complexes [26, 32]. Thus, tissue and context specific effects of NuRD can be expected to occur via a mechanism of employing different combinations of NuRD subunits. Another mechanism by which NuRD can exert its multifaceted effects in different contexts is through its association with tissue specific transcription and co-regulatory factors that are not core NuRD components [33-35]. As described below, defining the biochemical basis for these types of interactions holds promise for targeted therapies in specific cancers and possibly other pathologic conditions.
Based on the current understanding of the biochemical and structural features of NuRD components, it is apparent that multiple subunits can interact directly with DNA, and with histones to regulate gene expression and genome integrity. At present, it is not well understood how the complex is assembled and recruited to specific sites in the genome to exert its effects. However, recent studies have provided new insight into how NuRD activity is regulated. Post-translational modifications of NuRD components play an important role in controlling its function. Compelling data in this regard are described in recent elegant work from the Kumar laboratory illustrating how post-translational modifications in a single subunit result in dynamic effects on target gene expression [36]. These investigators determined that methylation of lysine 532 on MTA1 by the methyltransferase G9A is required for formation of a NuRD repressive complex. Methylated MTA1 has increased affinity for histone H3, leading to formation of the H3K9me2 histone mark and recruitment of CHD4 (Mi2-β) to create a repressive chromatin environment. In contrast, when Lsd1 demethylates MTA1, CHD4 and the NuRD complex are displaced. De-methlyated MTA1 then associates with the NURF chromatin remodeling complex factor BPTF and the histone acetyltransferase (HAT) p300/CBP to activate gene transcription. Thus, MTA1 acts as dynamic molecular switch that can rapidly alter gene expression in opposing directions in response to specific physiological or developmental cues. It is likely that other post-translational modifications described for NuRD components [37-39], will be found to regulate activity of the complex and these could represent interesting drug targets. In addition to post-translational modifications, recent studies provide insights into how HDAC activity is regulated in the context of co-repressor complexes by investigating the structure of HDAC1 in complex with MTA1 [40]. These studies revealed that the ELM2 and SANT domains of MTA1 make extensive contacts with HDCA1, wrapping around its catalytic domain. Binding of Ins(1,4,5,6)P4 to the SANT domain of MTA1 significantly enhanced HDAC activity. A similar paradigm has been proposed for all class I HDACs and their association with SANT domains in the context of different co-repressor complexes, such as the HDAC3- containing SMRT co-repressor complex [41]. Thus, rather than being constitutively active, this model suggests that HDAC activity associated with the NuRD complex is dynamically regulated by a distinct class of inositol phosphate signaling molecules.
Structural analysis of the MBD2/p66 interaction has provided information on the importance of targeting the coiled-coil interaction to alter gene expression. Silencing of fetal γ-globin gene expression is mediated by an MBD2-containing NuRD complex [26, 42]. Assembly of Mi2-NuRD at this genomic locus depends on the p66α/MBD2 interaction which occurs through the small coiled-coil domain in p66α, forming a complex between two stable helices. Forced expression of the coiled-coil peptide functions as a competitive inhibitor that disrupts the interaction between full length MBD2 and p66α, prevents recruitment of Mi2-NuRD and results in increased fetal globin expression. Thus, development of chemical inhibitors based on this structural information could in principal be tested to treat hemoglobinopathies. The p66α coiled-coil peptide has a similar affinity for MBD3, suggesting that this strategy could also be used to target MBD3-containing NuRD complexes [27].
NuRD, ES cell pluripotency and embryonic development
One of the most interesting stories surrounding NuRD and embryonic development and ES cells involves the methyl-CpG-binding domain protein MBD3. Gene deletion of Mbd3 in mice resulted in embryonic lethality between embryonic day (E) E3.5 and E8.5, indicating that Mbd3 was required for embryonic development [43]. Deletion of Mbd2 results in viable embryos that have a maternal nurturing defect. Using gene targeting methods to knock out Mbd3 conditionally in embryonic stem (ES) cells, the Hendrich laboratory found that Mbd3 -/- mouse ES cells grew more slowly than wild type ES cells and contained a reduced amount of NuRD components Mta1, Mta2, and Rbbp7 [44]. In addition, interaction experiments showed that the NuRD complex was no longer intact in Mbd3 -/- stem cells, indicating that Mbd3 was required for stable complex formation. Mbd3 mutant embryoid bodies did not express markers of differentiated embryonic cells, and failed to downregulate expression of progenitor markers Oct4 and Nanog [44]. Instead of differentiating, Mbd3 -/- ES cells seemed to be in a persistent state of self-renewal. These studies highlight the importance of Mbd3 for NuRD complex formation, and a role for Mbd3/NuRD in the self-renewal/lineage commitment transition of embryonic stem cells. An analogous role for NuRD has been described in the skin where Mi2β was shown to mediate developmental transitions between epidermal progenitors to basal, follicular and matrix cell precursors [45].
Schübeler and colleagues investigated the functional binding sites of the MBD protein family in ES cells [46]. Using a biotin expression system they performed ChIP-seq to generate genome wide binding profiles of MBD1, 2, 3, 4, and MecP2. Genome wide in ES cells and ES cell clone derived neuronal cells they found that binding of MBD1, 2, 4, and MecP2 had increased binding at exons and promoters, and decreased binding at repetitive DNA. Moreover, binding of these factors had a linear relationship with the methylation density of chromatin; regions with higher methylation density had increased binding for MBD1, 2, 4, and MecP2. Mutations disrupting the methyl- binding domains of the biotin tagged proteins in ES Cells or conditions to prevent chromatin methylation (Dnmt1, Dnmt3a, and Dnmt3b triple knockout), abolished these findings, indicating that binding was dependent on chromatin methylation. MBD3 binding showed no enrichment at methylated islands, and genome wide binding was independent of methylation density, indicating that binding of MBD3 was independent of the methylation state of chromatin. Importantly, they found that both MBD2 and MBD3 bind a subset of targets in cooperation with NuRD components Chd4, Hdac1, and Hdac2. These genomic regions are low in CpG density and DNA methylation, but enriched for H3K4me1 and H3K27ac and are DNAse1 hypersensitive, all hallmarks of active regulatory regions. These regions are tissue specific regulatory regions, active promoters and enhancers, and account for almost all MBD3 binding sites and a subset of MBD2 methylation independent binding sites. These findings have been corroborated in other cell types by independent investigators [47-49]. In addition, they found no NuRD component binding other than MBD2 at methylated sites, which contradicts current models of MBD2-mediated targeting of NuRD to methylated sites.
Recent work from the Hendrich laboratory has further elucidated the critical role of NuRD to balance the self-renewal and differentiation of ES cells, indicating a more complex role for NuRD in this context [50]. Transcriptional heterogeneity has been suggested as one mechanism by which a population of stem cells can commit cells to different lineages and also maintain a self-renewing population of stem cells. Under conditions of self-renewal, NuRD functions to restrict the expression of a subset of pluripotency genes (Tbx3, Klf4, Klf5) in ES cells. Knockdown of Mbd3 in ES cells leads to upregulated expression of these pluripotency factors and LIF-independent self-renewal, consistent with the findings in the Mbd3 knock out mouse. The presence of Mbd3 and Mi2β at the promoters and gene bodies of these genes suggests that the effect of NuRD on their transcription is direct. These investigators go on to demonstrate that in a population of wild type ES cells there is a mixture of low and high expressing cells for these NuRD-regulated pluripotency genes. In the absence of NuRD, all cells revert to a high expressing phenotype, indicating that NuRD is required to maintain transcriptional heterogeneity. Their data also suggest that restricting the level of pluripotency gene expression is necessary for differentiation to proceed, consistent with the defective lineage commitment observed in Mbd3-deficient ES cells.
In a separate study these same authors established a link between NuRD and the Polycomb complex providing further insight into the molecular mechanisms that control the decision of stem cell self-renewal versus differentiation. Polycomb-Repressive Complex 2 (PRC2) acts to di- and trimethylate H3K27 via the methyltransferase Ezh2 as part of the mechanism by which it mediates transcriptional silencing, (for a current review of PRC2 mechanism of action see [51]). Many of the Polycomb Group (PcG) protein target genes have been extensively identified in ES cells and a number of them are bivalent, having both a transcriptional repressive mark (H3K27me3) and an active transcriptional histone mark (H3K4me3) [52, 53]. These bivalent target genes in ES cells are “primed” to be efficiently activated or repressed by modifying the histone marks during the course of progenitor self-renewal or differentiation. Transcriptional profiling of wild type and Mbd3-/- ES cells found that 839 genes were downregulated while 531 genes were upregulated [54], indicating that NuRD is likely to activate as well as silence gene transcription in ES cells. Of the genes that were upregulated, only 0.2% of genes were associated with inactive chromatin, 17% with bivalent chromatin, and 64% with active chromatin in wild type ES cells, indicating that NuRD is preferentially recruited and/or modifies chromatin of bivalent and active genes. At bivalent NuRD target genes in Mbd3-/- ES cells, chromatin was enriched for H3K27ac and depleted of H3K27me3, suggesting that NuRD and PRC2 act in concert at these target genes to maintain H3K27 in a deacetylated, trimethylated state. Furthermore, the authors showed that the presence/activity of NuRD at common bivalent target genes is required for the recruitment of the PRC2 complex.
Overall, these studies of the role of Mbd3 in ES cells suggest that NuRD plays a role in setting critical levels of gene expression in ES cells. The complex functions to restrict the level of pluripotency genes, and in maintaining transcriptional heterogeneity in a population of stem cells. In cooperation with PRC2, NuRD maintains differentiation genes in a silent, but poised state for rapid activation to allow specific cell lineages to develop in response to appropriate developmental cues. Interestingly, the cooperative role of NuRD and PRC2-mediated gene repression has also been shown to have a role in acute pro-myelocytic leukemia (APL). The abnormal PML-RARα fusion protein found in APL recruits NuRD to repress differentiation genes in hematopoetic progenitor cells, thereby promoting leukemic transformation. Morey et al. showed that this repressive of effect of NuRD required recruitment of PRC2 to silence PML-RARα -target gene promoters [55].
Additional mechanisms have been proposed to explain how NuRD inhibits pluripotency gene expression in ES cells to enable their differentiation. In mouse ES cells, Lsd1 knock down or chemical inhibition upregulated pluripotency genes; similarly, deficiency of Mbd3 resulted in a failure to downregulate pluripotency gene expression and reduced the capacity of ES cells to differentiate. NuRD and Lsd1 physically interact and co-occupy active enhancers of many genes, including pluripotency genes [9]. The data of Whyte et al. suggest that NuRD, acting through its ability to deacetylate histone H3K9, enables Lsd1 to bind and demethylate histone H3K4me1. The net effects of these coordinated histone modifications are to silence the enhancers of pluripotency genes, thereby permitting differentiation to be induced. Beyer et al recently uncovered a mechanism whereby Tgfβ can promote both maintenance of pluripotency and mesendoderm specification [56]. These investigators demonstrated that a complex including Hippo pathway components and the Tgfβ nuclear effectors SMAD2/3 cooperate with NuRD to dampen expression of pluripotency genes and to silence mesendoderm differentiation genes. This study reveals a role for NuRD in interpreting a morphogen signal (Tgfβ) in balancing the self-renewal and differentiation decision.
An exciting development related to the function of NuRD in stem cell biology comes from a recent study revealing that Mbd3/NuRD plays a crucial role in the efficient (100%) reprogramming of induced pluripotent stem (iPS) cells [57]. This represents a highly significant enhancement, as reprogramming efficiency of fibroblasts into iPS cells has generally been very low (<1%). These investigators used a siRNA screen to identify epigenetic repressor factors that may boost the efficiency of primed pluripotent epiblast stem cells (EpiSCs) to a native pluripotent state. Out of a list of 15 proteins involved in DNA repression (ex. Dnmt 1-3, Suz12, Hdac 1-3, Sin3a) only Mbd3 increased the efficiency of reprogramming from ~20% to >90%. Secondary reprogramming from somatic cells harvested from Mbd3-/- transgenic embryos containing an inducible Oct4/Sox2/Klf4/c-Myc (OSKM) cassette showed reprogramming efficiencies of 100% after 8 days compared to 20% in Mbd3 +/+ fibroblasts. The authors also demonstrated efficient reprogramming of human fibroblast cells into iPS cells using MBD3 siRNA knockdown two days after induction of OSKM factors. Not only was the reprogramming more efficient, the reprogramming was synchronized, or deterministic and not stochastic, which will allow future studies to focus on the molecular events leading toward iPS cells. Maybe most surprisingly was that the removal of one protein caused reprogramming rates to soar to 100%. The mechanism by which Mbd3/NuRD normally silences the pluripotency program is not known, however the authors showed that Mbd3 interacted with the OSKM factors in HEK293 cells using overexpression, Chd4 (Mi2-β) interacted with OSKM factors in fibroblasts undergoing reprogramming, and both Mbd3 and Chd4 were recruited to pluripotency target genes only after induction of OSKM factors . Knock down of Mbd3 also significantly reduced the time required to convert somatic cells to iPS cells. A study by Luo et al. also supports the conclusion that NuRD blocks reprogramming and knockdown of Mbd3 improves the efficiency of iPS cell generation [58]. If these dramatic effects on reprogramming efficiency are substantiated by additional independent studies, identification of Mbd3-NuRD as a critical barrier to iPS cell generation would be a major step forward in this field. By using Mbd3 RNAi it would then be possible to more readily develop iPS cell lines from patients from a variety of disease states, thereby accelerating the potential for translational studies in complex or rare diseases.
A common theme is beginning to emerge regarding the biological role of NuRD in embryonic development. Analogous to its role in ES cells, several studies point to a key role for this complex in multipotent progenitors in different tissues, though potentially acting via distinct molecular mechanisms. Among the best well studied is the hematopoetic system. In multipotent hematopoetic progenitors, Mi2β promotes commitment to the lymphoid lineage [59]. Conditional deletion of Mi2β in bone marrow leads to an initial expansion of hematopoetic stem cells that can differentiate into the erythroid, but not the lymphoid or myeloid lineages [60]. Hematopoetic progenitor cells are depleted as differentiated proerythrobalsts with features of erthyroid leukemia accumulate. The lymphoid-restricted DNA binding factor Ikaros tethers NuRD to active genes involved in lymphoid maturation, often in close proximity to RNA polymerase II [49]. In vitro studies suggest that the presence of Ikaros at NuRD bound genes inhibits the nucleosome sliding activity of Mi2β. Zhang et al. propose that loss of Ikaros provides NuRD more accessibility to histones, thereby enabling the complex to inhibit expression of lymphoid maturation genes. Interestingly, loss of Ikaros also resulted in increased access of Mi2β to genes that generally promote cell proliferation and metabolism. Paradoxically, at these sites Mi2-NuRD activated gene expression, suggesting a mechanism that could drive leukemogenesis. In contrast to the findings in ES cells, NuRD seems to function antagonistically to PRC2 at a subset of these ectopically activated genes. Thus, in different contexts, the interplay between NuRD and other chromatin modifying complexes can vary, further highlighting the potential for paradoxical effects of NuRD in cancer, other diseases and development.
Studies in several model organisms have also revealed that NuRD is required for development of erythroid progenitors, and this effect is mediated in part through the association of Friend of Gata (FOG) with the NuRD complex [61-64]. FOG interacts with NuRD via a conserved 12 amino acid motif that binds MTA and Rbbp4/7 NuRD subunits. Elegant studies from the Blobel lab demonstrated that disruption of the Fog1-NuRD interaction in knock-in mice resulted in production of fewer and less mature erythroid and megakaryocyte colonies, indicating the physiological significance of Fog1-NuRD association in vivo [65, 66]. In the developing kidney, a different zinc finger transcription factor, Sall1, contains the same NuRD binding motif as FOG. Recent studies demonstrate that, similar to Sall1, Mi2-NuRD is required to maintain nephron progenitors during embryonic kidney formation [67, 68]. Disruption of the Sall1-NuRD interaction in vivo in knock-in mice also leads to depletion of nephron progenitors, and consequently congenital renal hypoplasia (D. Denner, J. Basta, M.Rauchman unpublished observations). Collectively, these studies highlight the role of NuRD in maintaining progenitor cell populations in different contexts by associating with tissue restricted transcription factors that are not core NuRD components. In some cases, disruption of these NuRD-co-factor interactions is thought to contribute to phenotypes in several human birth defect syndromes associated with intellectual impairment, dysmorphic facies and anomalies of various other organs [69-74].
Cancer and Ageing
NuRD has been shown to have opposing effects in cancer, both promoting and inhibiting tumor growth and metastasis in different tissues. To some extent, these paradoxical effects might be explained by the ability of NuRD to associate with or modulate the activity of both tumor suppressors (e.g. p53) and oncogenic factors (e.g. Bcl-6) [13, 33]. In addition, differences in NuRD complex composition with regard to the MTA subunits display antagonistic effects in the same tissue. MTA1 expression is increased in breast and other tumors, and correlates with an increased risk of metastasis and poor outcomes [75]. Inhibition of Estrogen Receptor (ER) target gene expression by MTA1 is one likely mechanism to mediate this effect [76]. Additionally, MTA1 is thought to be a downstream effector of the MYC oncogene, which could explain why increased levels of MTA1 are associated with high tumor grade and invasiveness in a variety of cancers [77]. In contrast, MTA3 is normally expressed in breast ductal epithelium and lost in breast cancer. MTA3 inhibits the epithelial-to-mesenchymal transition (EMT) by repressing Snail gene expression [30]. Thus, loss of MTA3 in breast cancer promotes EMT, thereby leading to reduced cell adhesion and a migratory cell phenotype that predisposes to tumor invasiveness and metastases. Wang et al. showed that Lsd1 directly interacts with MTA2 and co-purifies with NuRD in breast cancer cells [8]. Their data suggest that a NuRD complex that requires Lsd1 activity suppresses EMT by acting on the TGF signaling pathway. MTA3 has been shown to directly repress Wnt4 gene transcription, suggesting yet another potential pathway by which its reduced activity could promote breast cancer [78]. Overexpression of MTA1 or MTA3 in transgenic mice has opposing effects on ductal branching of breast epithelium and on cell proliferation, further underscoring the notion that these subunits of NuRD are not redundant and can have opposing effects.
Although the role of NuRD in cancer is complex and may differ significantly according to tumor type, recent studies have increasingly implicated alterations in its function in cancer. Two recent papers described whole exome sequencing of serous uterine tumors, a highly aggressive form of endometrial cancer [79, 80]. In addition to finding disruption of known cancer genes, both groups identified frequent (17-19%) somatic mutations in CHD4 (Mi2-β). A number of these heterozygous mutations occurred in domains that are predicted to disrupt key functions of the protein, including in the catalytic core of the helicase domain and in the second plant homeodomain (PHD) finger that binds methylated histone H3K9 [81-83]. Copy number variants (CNVs) involving genomic regions that include CDH4 and MBD3 were also relatively common [80], suggesting that the NuRD complex is playing an important pathogenic role. Consistent with this observation, MTA1 is upregulated in endometrial cancer and its knockdown in tumor cell lines inhibited cell migration and proliferation [84]. This study also found that miR-30c, a micro RNA whose expression is down-regulated in uterine cancer, binds the 3’UTR and inhibits MTA expression. The molecular mechanism by which CHD4 mutations lead to uterine cancer is not understood. NuRD can suppress Wnt/beta-catenin signaling [85] which is a driver of endometrial hyperplasia [86]. EMT with loss of E-cadherin expression is thought to play an important role in the aggressiveness of uterine cancer; thus, the regulation of EMT by MTA/NuRD described in breast cancer may also be important in this context. It has also been suggested that mutations in the ATPase domain of CHD4 could predominantly impair the DNA damage response recently attributed to the NuRD complex [2, 87]. Currently, therapeutic options for this sub-type of endometrial cancer are limited and it accounts for a disproportionate number of uterine cancer deaths. These findings suggest new targets for intervention by compounds that directly modulate NuRD activity or alter expression of its subunits.
NuRD can associate with a variety of transcription factors and co-regulatory factors to mediate tissue and context specific effects. A recent study on hepatocellular cancer highlights how targeting this interface has the potential to be exploited for therapeutic purposes. An aggressive sub-type of hepatocellular carcinoma displays a progenitor cell-like phenotype. Among the onco-fetal genes that are highly expressed in this tumor is SALL4, a multi-zinc finger transcription factor [88]. The SALL family of transcription factors contains a conserved 12 amino acid domain that mediates direct interaction with NuRD [35]. Yong et al. were able to show that a peptide with the same sequence as this NuRD interacting domain in SALL4 can compete for its interaction with NuRD. This peptide could relieve SALL4-NuRD mediated repression of PTEN and reduce tumor growth in an animal model. A similar paradigm was shown for the role of SALL4 in promoting leukemogenesis, where use of the same peptide that disrupts NuRD –dependent transcriptional repression reduced tumor cell viability and engraftment in an animal model [89]. Similar effects were noted with HDAC inhibitors, suggesting that the tumor suppressing effect of abrogating the SALL-NuRD interaction is dependent on reduction of NuRD complex associated HDAC activity. SALL4 overexpression has been noted in other tumors [90] and thus this strategy may have broader application as a new therapeutic target for specific cancers. An excellent candidate is uterine cancer. As noted above, alterations in NuRD are directly implicated in this tumor. Similar to liver cancer, SALL4 overexpression is found in endometrial cancer where it correlates with poor outcome [91]. Thus, it is plausible that disruption of SALL4-NuRD interaction in this tumor may also be beneficial. SALL family proteins are overexpressed or mutated in various cancers and its interaction with NuRD may emerge as an important mechanism for tumorogenesis or metastasis [90, 92, 93].
An RNAi screen performed to identify genes that contribute to therapy resistant tumor initiating cells in gliobalstoma also implicates a key role for NuRD in association with specific tissue restricted DNA binding proteins [94]. These authors discovered that ZFHX4, a transcription factor that associates with CHD4-NuRD, inhibits neural differentiation genes. While this function serves to promote the neural precursor state during development, it also promotes tumor formation. ZFHX4 and NuRD have overlapping patterns of genomic localization and control similar gene expression programs. Silencing of ZFHX4 reduced tumor cell proliferation, promoted differentiation and reduced tumor burden in xenograft mouse models of glioblastoma. Because CHD5 is preferentially expressed in the CNS and its deletion has been implicated in CNS tumors (see [95] for review), it is also possible that a NuRD complex containing CHD5 is involved in promoting gliobalastoma. Interestingly, the screen by Chudnovsky et al. also identified Sall3, a transcription factor that is also known to associate with NuRD through the same motif found in Sall1 and Sall4 [35]; Sall3 was shown to regulate proliferation/differentiation of neural precursors [96]. Together, these findings implicate NuRD as a regulator of neural precursor cell differentiation and CNS tumor formation, potentially acting through different DNA binding co-factors.
Several studies have now reported that NuRD plays a role in the DNA damage response and maintenance of genome integrity. Cells depleted of NuRD components MTA2 or Mi2β displayed increased sensitivity to ionizing radiation and spontaneous DNA damage [97-99]. Rescue of these defects depends on the ATPase activity of Mi2β, indicating that its helicase activity is important for maintaining genomic integrity [99]. Two potential mechanisms have been invoked for NuRD in this process, recruitment of CHD4-NuRD to sites of DNA damage by PARP1 or the RING finger ubiquitin ligase RNF8 [2, 97, 99]. These emerging functions for NuRD in maintaining genome integrity will likely turn out to contribute to its role in oncogenesis. In addition to cancer, there is an accumulation of DNA damage with ageing. Pegoraro et al. found a reduction of NuRD component expression and HDAC activity in physiological ageing and premature ageing associated with Progeria [100]. Knock down of NuRD components was accompanied by chromosomal defects typically seen in ageing cells. In contrast to these findings, a recent study found that loss of the Mi2 orthologue LET-418, but not depletion of other NuRD components, increased longevity in C. elegans and Drosphila [101]. Therefore, the discrepancy between these results and those in mammalian cells could be explained by species differences, and the fact that Mi2 may have both NuRD-dependent and independent effects on ageing. Thus, the role of NuRD in ageing is complex, similar to what we have learned thus far about its role in cancer.
Conclusion
NuRD has emerged as an important regulator of gene expression that controls the fate of stem cells in many contexts. This is accomplished through the diversity of NuRD subunits, its association with a wide range of tissue restricted co-factors, and its ability to cooperate with other chromatin modifying complexes. Recent discoveries highlight the important role of NuRD in cancer, ES and progenitor cell fate, and embryonic development. Because of its broad expression and multifaceted effects on chromatin, it is likely that future studies will identify key roles for this macromolecular complex in other disease processes. Advances in understanding of NuRD complex assembly, the molecular basis for its association with specific co-factors, and signals that regulate its activity have the potential to lead to new therapeutics.
Acknowlegements
The authors would like to acknowledge support from NIDDK, the American Heart Association and the March of Dimes. All the authors have read the journal's authorship agreement and the manuscript has been reviewed by and approved by all authors.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors have read the journal's policy on disclosure of conflict of interest. The authors have no conflict of interest disclosures.
References
- 1.Suva ML, Riggi N, Bernstein BE. Epigenetic reprogramming in cancer. Science. 2013 Mar 29;339(6127):1567–70. doi: 10.1126/science.1230184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Polo SE, Kaidi A, Baskcomb L, Galanty Y, Jackson SP. Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. The EMBO journal. 2010 Sep 15;29(18):3130–9. doi: 10.1038/emboj.2010.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Y, LeRoy G, Seelig HP, Lane WS, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998 Oct 16;95(2):279–89. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- 4.Xue Y, Wong J, Moreno GT, Young MK, Cote J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998 Dec;2(6):851–61. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- 5.Wade PA, Jones PL, Vermaak D, Wolffe AP. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr Biol. 1998 Jul 2;8(14):843–6. doi: 10.1016/s0960-9822(98)70328-8. [DOI] [PubMed] [Google Scholar]
- 6.Tong JK, Hassig CA, Schnitzler GR, Kingston RE, Schreiber SL. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature. 1998 Oct 29;395(6705):917–21. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- 7.Adamo A, Sese B, Boue S, Castano J, Paramonov I, Barrero MJ, et al. LSD1 regulates the balance between self-renewal and differentiation in human embryonic stem cells. Nat Cell Biol. 2011 Jun;13(6):652–9. doi: 10.1038/ncb2246. [DOI] [PubMed] [Google Scholar]
- 8.Wang Y, Zhang H, Chen Y, Sun Y, Yang F, Yu W, et al. LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell. 2009 Aug 21;138(4):660–72. doi: 10.1016/j.cell.2009.05.050. [DOI] [PubMed] [Google Scholar]
- 9.Whyte WA, Bilodeau S, Orlando DA, Hoke HA, Frampton GM, Foster CT, et al. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature. 2012 Feb 9;482(7384):221–5. doi: 10.1038/nature10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smits AH, Jansen PW, Poser I, Hyman AA, Vermeulen M. Stoichiometry of chromatin-associated protein complexes revealed by label-free quantitative mass spectrometry-based proteomics. Nucleic acids research. 2013 Jan 7;41(1):e28. doi: 10.1093/nar/gks941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim JJ, Khalid O, Vo S, Sun HH, Wong DT, Kim Y. A novel regulatory factor recruits the nucleosome remodeling complex to wingless integrated (Wnt) signaling gene promoters in mouse embryonic stem cells. The Journal of biological chemistry. 2012 Nov 30;287(49):41103–17. doi: 10.1074/jbc.M112.416545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spruijt CG, Bartels SJ, Brinkman AB, Tjeertes JV, Poser I, Stunnenberg HG, et al. CDK2AP1/DOC-1 is a bona fide subunit of the Mi-2/NuRD complex. Molecular bioSystems. 2010 Sep;6(9):1700–6. doi: 10.1039/c004108d. [DOI] [PubMed] [Google Scholar]
- 13.Gururaj AE, Singh RR, Rayala SK, Holm C, den Hollander P, Zhang H, et al. MTA1, a transcriptional activator of breast cancer amplified sequence 3. Proceedings of the National Academy of Sciences of the United States of America. 2006 Apr 25;103(17):6670–5. doi: 10.1073/pnas.0601989103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bouazoune K, Mitterweger A, Langst G, Imhof A, Akhtar A, Becker PB, et al. The dMi-2 chromodomains are DNA binding modules important for ATP-dependent nucleosome mobilization. The EMBO journal. 2002 May 15;21(10):2430–40. doi: 10.1093/emboj/21.10.2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thompson PM, Gotoh T, Kok M, White PS, Brodeur GM. CHD5, a new member of the chromodomain gene family, is preferentially expressed in the nervous system. Oncogene. 2003 Feb 20;22(7):1002–11. doi: 10.1038/sj.onc.1206211. [DOI] [PubMed] [Google Scholar]
- 16.Zhuang T, Hess RA, Kolla V, Higashi M, Raabe TD, Brodeur GM. CHD5 is required for spermiogenesis and chromatin condensation. Mech Dev. 2014 Feb;131:35–46. doi: 10.1016/j.mod.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amaya M, Desai M, Gnanapragasam MN, Wang SZ, Zu Zhu S, Williams DC, Jr., et al. Mi2beta-mediated silencing of the fetal gamma-globin gene in adult erythroid cells. Blood. 2013 Apr 25;121(17):3493–501. doi: 10.1182/blood-2012-11-466227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kunert N, Wagner E, Murawska M, Klinker H, Kremmer E, Brehm A. dMec: a novel Mi-2 chromatin remodelling complex involved in transcriptional repression. The EMBO journal. 2009 Mar 4;28(5):533–44. doi: 10.1038/emboj.2009.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williams CJ, Naito T, Arco PG, Seavitt JR, Cashman SM, De Souza B, et al. The chromatin remodeler Mi-2beta is required for CD4 expression and T cell development. Immunity. 2004 Jun;20(6):719–33. doi: 10.1016/j.immuni.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 20.Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol. 1998 Nov;18(11):6538–47. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hashimoto H, Liu Y, Upadhyay AK, Chang Y, Howerton SB, Vertino PM, et al. Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic acids research. 2012 Jun;40(11):4841–9. doi: 10.1093/nar/gks155. PubMed PMID: 22362737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hong W, Nakazawa M, Chen YY, Kori R, Vakoc CR, Rakowski C, et al. FOG-1 recruits the NuRD repressor complex to mediate transcriptional repression by GATA-1. The EMBO journal. 2005 Jul 6;24(13):2367–78. doi: 10.1038/sj.emboj.7600703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Humphrey GW, Wang Y, Russanova VR, Hirai T, Qin J, Nakatani Y, et al. Stable histone deacetylase complexes distinguished by the presence of SANT domain proteins CoREST/kiaa0071 and Mta-L1. The Journal of biological chemistry. 2001 Mar 2;276(9):6817–24. doi: 10.1074/jbc.M007372200. [DOI] [PubMed] [Google Scholar]
- 24.Roche AE, Bassett BJ, Samant SA, Hong W, Blobel GA, Svensson EC. The zinc finger and C-terminal domains of MTA proteins are required for FOG-2-mediated transcriptional repression via the NuRD complex. J Mol Cell Cardiol. 2008 Feb;44(2):352–60. doi: 10.1016/j.yjmcc.2007.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murzina NV, Pei XY, Zhang W, Sparkes M, Vicente-Garcia J, Pratap JV, et al. Structural basis for the recognition of histone H4 by the histone-chaperone RbAp46. Structure. 2008 Jul;16(7):1077–85. doi: 10.1016/j.str.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gnanapragasam MN, Scarsdale JN, Amaya ML, Webb HD, Desai MA, Walavalkar NM, et al. p66Alpha-MBD2 coiled-coil interaction and recruitment of Mi-2 are critical for globin gene silencing by the MBD2-NuRD complex. Proceedings of the National Academy of Sciences of the United States of America. 2011 May 3;108(18):7487–92. doi: 10.1073/pnas.1015341108. PubMed PMID: 21490301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walavalkar NM, Gordon N, Williams DC., Jr Unique features of the anti-parallel, heterodimeric coiled-coil interaction between methyl-cytosine binding domain 2 (MBD2) homologues and GATA zinc finger domain containing 2A (GATAD2A/p66alpha). The Journal of biological chemistry. 2013 Feb 1;288(5):3419–27. doi: 10.1074/jbc.M112.431346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ahringer J. NuRD and SIN3 histone deacetylase complexes in development. Trends Genet. 2000;16(8):351–6. doi: 10.1016/s0168-9525(00)02066-7. [DOI] [PubMed] [Google Scholar]
- 29.Kelly RD, Cowley SM. The physiological roles of histone deacetylase (HDAC) 1 and 2: complex co-stars with multiple leading parts. Biochem Soc Trans. 2013 Jun;41(3):741–9. doi: 10.1042/BST20130010. [DOI] [PubMed] [Google Scholar]
- 30.Fujita N, Jaye DL, Kajita M, Geigerman C, Moreno CS, Wade PA. MTA3, a Mi-2/NuRD complex subunit, regulates an invasive growth pathway in breast cancer. Cell. 2003 Apr 18;113(2):207–19. doi: 10.1016/s0092-8674(03)00234-4. [DOI] [PubMed] [Google Scholar]
- 31.Zhang H, Stephens LC, Kumar R. Metastasis tumor antigen family proteins during breast cancer progression and metastasis in a reliable mouse model for human breast cancer. Clin Cancer Res. 2006 Mar 1;12(5):1479–86. doi: 10.1158/1078-0432.CCR-05-1519. [DOI] [PubMed] [Google Scholar]
- 32.Le Guezennec X, Vermeulen M, Brinkman AB, Hoeijmakers WA, Cohen A, Lasonder E, et al. MBD2/NuRD and MBD3/NuRD, two distinct complexes with different biochemical and functional properties. Mol Cell Biol. 2006 Feb;26(3):843–51. doi: 10.1128/MCB.26.3.843-851.2006. PubMed PMID: 16428440. Pubmed Central PMCID: 1347035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fujita N, Jaye DL, Geigerman C, Akyildiz A, Mooney MR, Boss JM, et al. MTA3 and the Mi-2/NuRD complex regulate cell fate during B lymphocyte differentiation. Cell. 2004 Oct 1;119(1):75–86. doi: 10.1016/j.cell.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 34.Kim J, Sif S, Jones B, Jackson A, Koipally J, Heller E, et al. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity. 1999 Mar;10(3):345–55. doi: 10.1016/s1074-7613(00)80034-5. [DOI] [PubMed] [Google Scholar]
- 35.Lauberth SM, Rauchman M. A conserved 12-amino acid motif in Sall1 recruits the nucleosome remodeling and deacetylase corepressor complex. The Journal of biological chemistry. 2006 Aug 18;281(33):23922–31. doi: 10.1074/jbc.M513461200. [DOI] [PubMed] [Google Scholar]
- 36.Nair SS, Li DQ, Kumar R. A core chromatin remodeling factor instructs global chromatin signaling through multivalent reading of nucleosome codes. Mol Cell. 2013 Feb 21;49(4):704–18. doi: 10.1016/j.molcel.2012.12.016. PubMed PMID: 23352453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007 May 25;316(5828):1160–6. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 38.Segre CV, Chiocca S. Regulating the regulators: the post-translational code of class I HDAC1 and HDAC2. J Biomed Biotechnol. 2011;2011:690848. doi: 10.1155/2011/690848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang T, Jian W, Luo Y, Fu X, Noguchi C, Bungert J, et al. Acetylation of histone deacetylase 1 regulates NuRD corepressor complex activity. The Journal of biological chemistry. 2012 Nov 23;287(48):40279–91. doi: 10.1074/jbc.M112.349704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Millard CJ, Watson PJ, Celardo I, Gordiyenko Y, Cowley SM, Robinson CV, et al. Class I HDACs share a common mechanism of regulation by inositol phosphates. Mol Cell. 2013 Jul 11;51(1):57–67. doi: 10.1016/j.molcel.2013.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Watson PJ, Fairall L, Santos GM, Schwabe JW. Structure of HDAC3 bound to co-repressor and inositol tetraphosphate. Nature. 2012 Jan 19;481(7381):335–40. doi: 10.1038/nature10728. PubMed PMID: 22230954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Costa FC, Fedosyuk H, Chazelle AM, Neades RY, Peterson KR. Mi2beta is required for gamma-globin gene silencing: temporal assembly of a GATA-1-FOG-1-Mi2 repressor complex in beta-YAC transgenic mice. PLoS genetics. 2012;8(12):e1003155. doi: 10.1371/journal.pgen.1003155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hendrich B, Guy J, Ramsahoye B, Wilson VA, Bird A. Closely related proteins MBD2 and MBD3 play distinctive but interacting roles in mouse development. Genes & development. 2001 Mar 15;15(6):710–23. doi: 10.1101/gad.194101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaji K, Caballero IM, MacLeod R, Nichols J, Wilson VA, Hendrich B. The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nature cell biology. 2006 Mar;8(3):285–92. doi: 10.1038/ncb1372. [DOI] [PubMed] [Google Scholar]
- 45.Kashiwagi M, Morgan BA, Georgopoulos K. The chromatin remodeler Mi-2beta is required for establishment of the basal epidermis and normal differentiation of its progeny. Development. 2007 Apr;134(8):1571–82. doi: 10.1242/dev.001750. [DOI] [PubMed] [Google Scholar]
- 46.Baubec T, Ivanek R, Lienert F, Schubeler D. Methylation-dependent and -independent genomic targeting principles of the MBD protein family. Cell. 2013 Apr 11;153(2):480–92. doi: 10.1016/j.cell.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 47.Gunther K, Rust M, Leers J, Boettger T, Scharfe M, Jarek M, et al. Differential roles for MBD2 and MBD3 at methylated CpG islands, active promoters and binding to exon sequences. Nucleic acids research. 2013 Mar 1;41(5):3010–21. doi: 10.1093/nar/gkt035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shimbo T, Du Y, Grimm SA, Dhasarathy A, Mav D, Shah RR, et al. MBD3 localizes at promoters, gene bodies and enhancers of active genes. PLoS genetics. 2013 Dec;9(12):e1004028. doi: 10.1371/journal.pgen.1004028. PubMed PMID: 24385926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang J, Jackson AF, Naito T, Dose M, Seavitt J, Liu F, et al. Harnessing of the nucleosome-remodeling-deacetylase complex controls lymphocyte development and prevents leukemogenesis. Nat Immunol. 2012 Jan;13(1):86–94. doi: 10.1038/ni.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reynolds N, Latos P, Hynes-Allen A, Loos R, Leaford D, O'Shaughnessy A, et al. NuRD suppresses pluripotency gene expression to promote transcriptional heterogeneity and lineage commitment. Cell Stem Cell. 2012 May 4;10(5):583–94. doi: 10.1016/j.stem.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011 Jan 20;469(7330):343–9. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS genetics. 2008 Oct;4(10):e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007 Aug 2;448(7153):553–60. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reynolds N, Salmon-Divon M, Dvinge H, Hynes-Allen A, Balasooriya G, Leaford D, et al. NuRD-mediated deacetylation of H3K27 facilitates recruitment of Polycomb Repressive Complex 2 to direct gene repression. The EMBO journal. 2012 Feb 1;31(3):593–605. doi: 10.1038/emboj.2011.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morey L, Brenner C, Fazi F, Villa R, Gutierrez A, Buschbeck M, et al. MBD3, a component of the NuRD complex, facilitates chromatin alteration and deposition of epigenetic marks. Mol Cell Biol. 2008 Oct;28(19):5912–23. doi: 10.1128/MCB.00467-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beyer TA, Weiss A, Khomchuk Y, Huang K, Ogunjimi AA, Varelas X, et al. Switch enhancers interpret TGF-beta and Hippo signaling to control cell fate in human embryonic stem cells. Cell Rep. 2013 Dec 26;5(6):1611–24. doi: 10.1016/j.celrep.2013.11.021. [DOI] [PubMed] [Google Scholar]
- 57.Rais Y, Zviran A, Geula S, Gafni O, Chomsky E, Viukov S, et al. Deterministic direct reprogramming of somatic cells to pluripotency. Nature. 2013 Oct 3;502(7469):65–70. doi: 10.1038/nature12587. [DOI] [PubMed] [Google Scholar]
- 58.Luo M, Ling T, Xie W, Sun H, Zhou Y, Zhu Q, et al. NuRD blocks reprogramming of mouse somatic cells into pluripotent stem cells. Stem cells. 2013 Jul;31(7):1278–86. doi: 10.1002/stem.1374. [DOI] [PubMed] [Google Scholar]
- 59.Ng SY, Yoshida T, Zhang J, Georgopoulos K. Genome-wide lineage-specific transcriptional networks underscore Ikaros-dependent lymphoid priming in hematopoietic stem cells. Immunity. 2009 Apr 17;30(4):493–507. doi: 10.1016/j.immuni.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yoshida T, Hazan I, Zhang J, Ng SY, Naito T, Snippert HJ, et al. The role of the chromatin remodeler Mi-2beta in hematopoietic stem cell self-renewal and multilineage differentiation. Genes & development. 2008 May 1;22(9):1174–89. doi: 10.1101/gad.1642808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang HT, Kathrein KL, Barton A, Gitlin Z, Huang YH, Ward TP, et al. A network of epigenetic regulators guides developmental haematopoiesis in vivo. Nat Cell Biol. 2013 Dec;15(12):1516–25. doi: 10.1038/ncb2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li X, Jia S, Wang S, Wang Y, Meng A. Mta3-NuRD complex is a master regulator for initiation of primitive hematopoiesis in vertebrate embryos. Blood. 2009 Dec 24;114(27):5464–72. doi: 10.1182/blood-2009-06-227777. [DOI] [PubMed] [Google Scholar]
- 63.Miccio A, Blobel GA. Role of the GATA-1/FOG-1/NuRD pathway in the expression of human beta-like globin genes. Mol Cell Biol. 2010 Jul;30(14):3460–70. doi: 10.1128/MCB.00001-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mimoto MS, Christian JL. Friend of GATA (FOG) interacts with the nucleosome remodeling and deacetylase complex (NuRD) to support primitive erythropoiesis in Xenopus laevis. PLoS One. 2012;7(1):e29882. doi: 10.1371/journal.pone.0029882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gregory GD, Miccio A, Bersenev A, Wang Y, Hong W, Zhang Z, et al. FOG1 requires NuRD to promote hematopoiesis and maintain lineage fidelity within the megakaryocytic-erythroid compartment. Blood. 2010 Mar 18;115(11):2156–66. doi: 10.1182/blood-2009-10-251280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang Y, Meng R, Hayes V, Fuentes R, Yu X, Abrams CS, et al. Pleiotropic platelet defects in mice with disrupted FOG1-NuRD interaction. Blood. 2011 Dec 1;118(23):6183–91. doi: 10.1182/blood-2011-06-363580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Basta JM, Robbins L, Kiefer SM, Dorsett D, Rauchman M. Sall1 balances self-renewal and differentiation of renal progenitor cells. Development. 2014 Mar;141(5):1047–58. doi: 10.1242/dev.095851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Denner DR, Rauchman M. Mi-2/NuRD is required in renal progenitor cells during embryonic kidney development. Dev Biol. 2013 Mar 15;375(2):105–16. doi: 10.1016/j.ydbio.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.de Ligt J, Willemsen MH, van Bon BW, Kleefstra T, Yntema HG, Kroes T, et al. Diagnostic exome sequencing in persons with severe intellectual disability. The New England journal of medicine. 2012 Nov 15;367(20):1921–9. doi: 10.1056/NEJMoa1206524. [DOI] [PubMed] [Google Scholar]
- 70.Kiefer SM, Robbins L, Barina A, Zhang Z, Rauchman M. SALL1 truncated protein expression in Townes-Brocks syndrome leads to ectopic expression of downstream genes. Hum Mutat. 2008 Sep;29(9):1133–40. doi: 10.1002/humu.20759. [DOI] [PubMed] [Google Scholar]
- 71.Liu ZLF, Ruan K, Zhang J, Mej Y, Wu J, Shi Y. Structural and Functional Insights into the Human Borjeson-Forssman-Lehmann Syndrome Associated Protein PHF6. The Journal of biological chemistry. 2014 doi: 10.1074/jbc.M113.535351. Epub Feb 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Verstappen G, van Grunsven LA, Michiels C, Van de Putte T, Souopgui J, Van Damme J, et al. Atypical Mowat-Wilson patient confirms the importance of the novel association between ZFHX1B/SIP1 and NuRD corepressor complex. Hum Mol Genet. 2008 Apr 15;17(8):1175–83. doi: 10.1093/hmg/ddn007. [DOI] [PubMed] [Google Scholar]
- 73.Wieczorek D, Bogershausen N, Beleggia F, Steiner-Haldenstatt S, Pohl E, Li Y, et al. A comprehensive molecular study on Coffin-Siris and Nicolaides-Baraitser syndromes identifies a broad molecular and clinical spectrum converging on altered chromatin remodeling. Hum Mol Genet. 2013 Dec 20;22(25):5121–35. doi: 10.1093/hmg/ddt366. [DOI] [PubMed] [Google Scholar]
- 74.Willemsen MH, Nijhof B, Fenckova M, Nillesen WM, Bongers EM, Castells-Nobau A, et al. GATAD2B loss-of-function mutations cause a recognisable syndrome with intellectual disability and are associated with learning deficits and synaptic undergrowth in Drosophila. Journal of medical genetics. 2013 Aug;50(8):507–14. doi: 10.1136/jmedgenet-2012-101490. [DOI] [PubMed] [Google Scholar]
- 75.Nicolson GL, Nawa A, Toh Y, Taniguchi S, Nishimori K, Moustafa A. Tumor metastasis-associated human MTA1 gene and its MTA1 protein product: role in epithelial cancer cell invasion, proliferation and nuclear regulation. Clin Exp Metastasis. 2003;20(1):19–24. doi: 10.1023/a:1022534217769. [DOI] [PubMed] [Google Scholar]
- 76.Mazumdar A, Wang RA, Mishra SK, Adam L, Bagheri-Yarmand R, Mandal M, et al. Transcriptional repression of oestrogen receptor by metastasis-associated protein 1 corepressor. Nat Cell Biol. 2001 Jan;3(1):30–7. doi: 10.1038/35050532. [DOI] [PubMed] [Google Scholar]
- 77.Zhang XY, DeSalle LM, Patel JH, Capobianco AJ, Yu D, Thomas-Tikhonenko A, et al. Metastasis-associated protein 1 (MTA1) is an essential downstream effector of the c-MYC oncoprotein. Proceedings of the National Academy of Sciences of the United States of America. 2005 Sep 27;102(39):13968–73. doi: 10.1073/pnas.0502330102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang H, Singh RR, Talukder AH, Kumar R. Metastatic tumor antigen 3 is a direct corepressor of the Wnt4 pathway. Genes & development. 2006 Nov 1;20(21):2943–8. doi: 10.1101/gad.1461706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Le Gallo M, O'Hara AJ, Rudd ML, Urick ME, Hansen NF, O'Neil NJ, et al. Exome sequencing of serous endometrial tumors identifies recurrent somatic mutations in chromatin-remodeling and ubiquitin ligase complex genes. Nature genetics. 2012 Dec;44(12):1310–5. doi: 10.1038/ng.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhao S, Choi M, Overton JD, Bellone S, Roque DM, Cocco E, et al. Landscape of somatic single-nucleotide and copy-number mutations in uterine serous carcinoma. Proceedings of the National Academy of Sciences of the United States of America. 2013 Feb 19;110(8):2916–21. doi: 10.1073/pnas.1222577110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mansfield RE, Musselman CA, Kwan AH, Oliver SS, Garske AL, Davrazou F, et al. Plant homeodomain (PHD) fingers of CHD4 are histone H3-binding modules with preference for unmodified H3K4 and methylated H3K9. The Journal of biological chemistry. 2011 Apr 1;286(13):11779–91. doi: 10.1074/jbc.M110.208207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Musselman CA, Ramirez J, Sims JK, Mansfield RE, Oliver SS, Denu JM, et al. Bivalent recognition of nucleosomes by the tandem PHD fingers of the CHD4 ATPase is required for CHD4-mediated repression. Proceedings of the National Academy of Sciences of the United States of America. 2012 Jan 17;109(3):787–92. doi: 10.1073/pnas.1113655109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Watson AA, Mahajan P, Mertens HD, Deery MJ, Zhang W, Pham P, et al. The PHD and chromo domains regulate the ATPase activity of the human chromatin remodeler CHD4. J Mol Biol. 2012 Sep 7;422(1):3–17. doi: 10.1016/j.jmb.2012.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kong X, Xu X, Yan Y, Guo F, Li J, Hu Y, et al. Estrogen Regulates the Tumour Suppressor MiRNA-30c and Its Target Gene, MTA-1, in Endometrial Cancer. PLoS One. 2014;9(3):e90810. doi: 10.1371/journal.pone.0090810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Major MB, Roberts BS, Berndt JD, Marine S, Anastas J, Chung N, et al. New regulators of Wnt/beta-catenin signaling revealed by integrative molecular screening. Sci Signal. 2008;1(45):ra12. doi: 10.1126/scisignal.2000037. [DOI] [PubMed] [Google Scholar]
- 86.Villacorte M, Suzuki K, Hirasawa A, Ohkawa Y, Suyama M, Maruyama T, et al. beta-Catenin signaling regulates Foxa2 expression during endometrial hyperplasia formation. Oncogene. 2013 Jul 18;32(29):3477–82. doi: 10.1038/onc.2012.376. [DOI] [PubMed] [Google Scholar]
- 87.O'Shaughnessy A, Hendrich B. CHD4 in the DNA-damage response and cell cycle progression: not so NuRDy now. Biochem Soc Trans. 2013 Jun;41(3):777–82. doi: 10.1042/BST20130027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yong KJ, Gao C, Lim JS, Yan B, Yang H, Dimitrov T, et al. Oncofetal gene SALL4 in aggressive hepatocellular carcinoma. The New England journal of medicine. 2013 Jun 13;368(24):2266–76. doi: 10.1056/NEJMoa1300297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gao C, Dimitrov T, Yong KJ, Tatetsu H, Jeong HW, Luo HR, et al. Targeting transcription factor SALL4 in acute myeloid leukemia by interrupting its interaction with an epigenetic complex. Blood. 2013 Feb 21;121(8):1413–21. doi: 10.1182/blood-2012-04-424275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Miettinen M, Wang Z, McCue PA, Sarlomo-Rikala M, Rys J, Biernat W, et al. SALL4 Expression in Germ Cell and Non-Germ Cell Tumors: A Systematic Immunohistochemical Study of 3215 Cases. Am J Surg Pathol. 2014 Mar;38(3):410–20. doi: 10.1097/PAS.0000000000000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li A, Jiao Y, Yong KJ, Wang F, Gao C, Yan B, et al. SALL4 is a new target in endometrial cancer. Oncogene. 2013 Dec 16; doi: 10.1038/onc.2013.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Love C, Sun Z, Jima D, Li G, Zhang J, Miles R, et al. The genetic landscape of mutations in Burkitt lymphoma. Nature genetics. 2012 Dec;44(12):1321–5. doi: 10.1038/ng.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wolf J, Muller-Decker K, Flechtenmacher C, Zhang F, Shahmoradgoli M, Mills GB, et al. An in vivo RNAi screen identifies SALL1 as a tumor suppressor in human breast cancer with a role in CDH1 regulation. Oncogene. 2013 Dec 2; doi: 10.1038/onc.2013.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chudnovsky Y, Kim D, Zheng S, Whyte WA, Bansal M, Bray MA, et al. ZFHX4 Interacts with the NuRD Core Member CHD4 and Regulates the Glioblastoma Tumor-Initiating Cell State. Cell Rep. 2014 Jan 30;6(2):313–24. doi: 10.1016/j.celrep.2013.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kolla V, Zhuang T, Higashi M, Naraparaju K, Brodeur GM. Role of CHD5 in Human Cancers: 10 Years Later. Cancer research. 2014 Feb 1;74(3):652–8. doi: 10.1158/0008-5472.CAN-13-3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Harrison SJPM, Managhan AP. Sall3 is required for the terminal maturation of olfactory glomerular interneurons. J Comp Neurol. 2008 Apr 10;507(5):1780–94. doi: 10.1002/cne.21650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Larsen DH, Poinsignon C, Gudjonsson T, Dinant C, Payne MR, Hari FJ, et al. The chromatin-remodeling factor CHD4 coordinates signaling and repair after DNA damage. J Cell Biol. 2010 Sep 6;190(5):731–40. doi: 10.1083/jcb.200912135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sims JK, Wade PA. Mi-2/NuRD complex function is required for normal S phase progression and assembly of pericentric heterochromatin. Mol Biol Cell. 2011 Sep;22(17):3094–102. doi: 10.1091/mbc.E11-03-0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Smeenk G, Wiegant WW, Vrolijk H, Solari AP, Pastink A, van Attikum H. The NuRD chromatin-remodeling complex regulates signaling and repair of DNA damage. J Cell Biol. 2010 Sep 6;190(5):741–9. doi: 10.1083/jcb.201001048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pegoraro G, Kubben N, Wickert U, Gohler H, Hoffmann K, Misteli T. Ageing-related chromatin defects through loss of the NURD complex. Nat Cell Biol. 2009 Oct;11(10):1261–7. doi: 10.1038/ncb1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.De Vaux V, Pfefferli C, Passannante M, Belhaj K, von Essen A, Sprecher SG, et al. The Caenorhabditis elegans LET-418/Mi2 plays a conserved role in lifespan regulation. Aging Cell. 2013 Dec;12(6):1012–20. doi: 10.1111/acel.12129. [DOI] [PubMed] [Google Scholar]
- 102.Lagger G, O'Carroll D, Rembold M, Khier H, Tischler J, Weitzer G, et al. Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. The EMBO journal. 2002 Jun 3;21(11):2672–81. doi: 10.1093/emboj/21.11.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dovey OM, Foster CT, Conte N, Edwards SA, Edwards JM, Singh R, et al. Histone deacetylase 1 and 2 are essential for normal T-cell development and genomic stability in mice. Blood. 2013 Feb 21;121(8):1335–44. doi: 10.1182/blood-2012-07-441949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Montgomery RL, Davis CA, Potthoff MJ, Haberland M, Fielitz J, Qi X, et al. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes & development. 2007 Jul 15;21(14):1790–802. doi: 10.1101/gad.1563807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Montgomery RL, Hsieh J, Barbosa AC, Richardson JA, Olson EN. Histone deacetylases 1 and 2 control the progression of neural precursors to neurons during brain development. Proceedings of the National Academy of Sciences of the United States of America. 2009 May 12;106(19):7876–81. doi: 10.1073/pnas.0902750106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lejon S, Thong SY, Murthy A, AlQarni S, Murzina NV, Blobel GA, et al. Insights into association of the NuRD complex with FOG-1 from the crystal structure of an RbAp48.FOG-1 complex. The Journal of biological chemistry. 2011 Jan 14;286(2):1196–203. doi: 10.1074/jbc.M110.195842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Manavathi B, Peng S, Rayala SK, Talukder AH, Wang MH, Wang RA, et al. Repression of Six3 by a corepressor regulates rhodopsin expression. Proceedings of the National Academy of Sciences of the United States of America. 2007 Aug 7;104(32):13128–33. doi: 10.1073/pnas.0705878104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lu X, Kovalev GI, Chang H, Kallin E, Knudsen G, Xia L, et al. Inactivation of NuRD component Mta2 causes abnormal T cell activation and lupus-like autoimmune disease in mice. The Journal of biological chemistry. 2008 May 16;283(20):13825–33. doi: 10.1074/jbc.M801275200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Charles MA, Saunders TL, Wood WM, Owens K, Parlow AF, Camper SA, et al. Pituitary-specific Gata2 knockout: effects on gonadotrope and thyrotrope function. Molecular endocrinology. 2006 Jun;20(6):1366–77. doi: 10.1210/me.2005-0378. [DOI] [PubMed] [Google Scholar]
- 110.Trivedi CM, Luo Y, Yin Z, Zhang M, Zhu W, Wang T, et al. Hdac2 regulates the cardiac hypertrophic response by modulating Gsk3 beta activity. Nature medicine. 2007 Mar;13(3):324–31. doi: 10.1038/nm1552. [DOI] [PubMed] [Google Scholar]
- 111.Cianciolo Cosentino C, Skrypnyk NI, Brilli LL, Chiba T, Novitskaya T, Woods C, et al. Histone deacetylase inhibitor enhances recovery after AKI. Journal of the American Society of Nephrology : JASN. 2013 May;24(6):943–53. doi: 10.1681/ASN.2012111055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Novitskaya T, McDermott L, Zhang KX, Chiba T, Paueksakon P, Hukriede NA, et al. A PTBA small molecule enhances recovery and reduces postinjury fibrosis after aristolochic acid-induced kidney injury. American journal of physiology Renal physiology. 2014 Mar 1;306(5):F496–504. doi: 10.1152/ajprenal.00534.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Brilli LL, Swanhart LM, de Caestecker MP, Hukriede NA. HDAC inhibitors in kidney development and disease. Pediatric nephrology. 2013 Oct;28(10):1909–21. doi: 10.1007/s00467-012-2320-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Slingerland M, Guchelaar HJ, Gelderblom H. Histone deacetylase inhibitors: an overview of the clinical studies in solid tumors. Anti-cancer drugs. 2014 Feb;25(2):140–9. doi: 10.1097/CAD.0000000000000040. [DOI] [PubMed] [Google Scholar]
- 115.Chaurasia P, Gajzer DC, Schaniel C, D'Souza S, Hoffman R. Epigenetic reprogramming induces the expansion of cord blood stem cells. The Journal of clinical investigation. 2014 Apr 24; doi: 10.1172/JCI70313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Allen HF, Daze KD, Shimbo T, Lai A, Musselman CA, Sims JK, et al. Inhibition of histone binding by supramolecular hosts. The Biochemical journal. 2014 May 1;459(3):505–12. doi: 10.1042/BJ20140145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sansom OJ, Berger J, Bishop SM, Hendrich B, Bird A, Clarke AR. Deficiency of Mbd2 suppresses intestinal tumorigenesis. Nature genetics. 2003 Jun;34(2):145–7. doi: 10.1038/ng1155. [DOI] [PubMed] [Google Scholar]
- 118.Li DQ, Pakala SB, Nair SS, Eswaran J, Kumar R. Metastasis-associated protein 1/nucleosome remodeling and histone deacetylase complex in cancer. Cancer research. 2012 Jan 15;72(2):387–94. doi: 10.1158/0008-5472.CAN-11-2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Aury-Landas J, Bougeard G, Castel H, Hernandez-Vargas H, Drouet A, Latouche JB, et al. Germline copy number variation of genes involved in chromatin remodelling in families suggestive of Li-Fraumeni syndrome with brain tumours. European journal of human genetics : EJHG. 2013 Dec;21(12):1369–76. doi: 10.1038/ejhg.2013.68. [DOI] [PMC free article] [PubMed] [Google Scholar]