Abstract
The prospective isolation of purified stem cell populations has dramatically altered the field of stem cell biology and has been a major focus of research across tissues in different organisms. Muscle stem cells are now among the most intensely studied stem cell populations in mammalian systems and the prospective isolation of these cells has allowed cellular and molecular characterizations not dreamed of a decade ago. In this protocol, we describe how to isolate muscle stem cells from limb muscles of adult mice by fluorescence-activated cell sorting (FACS). We provide a detailed description of the physical and enzymatic dissociation of mononucleated cells from limb muscles, a procedure that is essential to maximize cell yield. We then describe a FACS-based method for obtaining exquisitely pure populations of either quiescent or activated muscle stem cells (VCAM+/CD31−/CD45−/Sca1−). The protocol also allows for the isolation of endothelial cells, hematopoietic cells, and mesenchymal stem cells from muscle tissue.
Keywords: satellite cell, stem cell, quiescence
INTRODUCTION
Skeletal muscle possesses remarkable regenerative ability owning to the resident stem cells1,2. In adult animals, the primary muscle stem cells (MuSCs), also termed satellite cells, are characterized by their localization underneath the basal lamina of post-mitotic myofibers and the expression of the Pax7 transcription factor3,4. In the absence of muscle injury or pathology, post-mitotic myofibers are extremely stable and exhibit a very low turnover rate1. As a result, MuSCs predominantly reside in a quiescent state under physiological conditions and therefore provide a good model system for studying cellular quiescence and chronological stem cell aging5,6. In response to muscle injury or pathological conditions that cause damage to muscle fibers, MuSCs are activated, giving rise to progeny that undergo proliferative expansion and either differentiate into mature myofibers or return to quiescence to replenish the MuSC pool. Because MuSCs possess the ability to contribute to muscle repair and regeneration upon transplantation, there is much interest in the potential of MuSCs to treat traumatic injuries and degenerative diseases of muscle7,8.
In a previously published issue of Nature Protocols, the technique of serial plating, which relies on the differential adhesion of distinct types of cells to purify myogenic cells from other mononucleated cells in skeletal muscle, was reported9. Because this approach relies on culture conditions that inevitably lead to a selection of cells with specific growth and survival characteristics in vitro, it does not necessarily (and in fact is unlikely to) yield cells that possess the characteristics of quiescent MuSCs in vivo. The myogenic cells derived from this protocol are progeny of activated MuSCs. Furthermore, in general, the expansion of muscle progenitor populations in vitro leads to a marked reduction in their efficacy to engraft and repair muscle upon transplantation10,11. Therefore, the ability to prospectively isolate quiescent MuSCs is required for the characterization and analysis of those cells, which may in turn provide clues as to how to maintain that state, perhaps through the use of protein or small molecule modulators, even as the cells are expanded in vitro.
Given the importance of prospective isolation techniques, several laboratories have described the isolation of MuSCs from skeletal muscles of adult mice by fluorescence-activated cell sorting (FACS)11–19 (Table 1). All protocols rely on both positive and negative selection by cell surface markers, as well as physical properties of cells such as forward and side scatter characteristics. Conboy et al. provide detailed FACS plots that are valuable for sorting quiescent MuSCs based on the positive markers CXCR4 and β1-integrin18. The protocol by Yi and Rossi is accompanied by a useful video in which the handling of the tissue is presented visually20. No head-to-head comparisons have been performed, but the general consensus is that most of these various protocols are roughly equally efficient and selective for isolation of quiescent MuSCs. However, expression of some of these surface makers, such as CD34, declines upon muscle injury and therefore cannot be used to isolate activated MuSCs and their progeny. To date, none of the protocols have presented evidence of the prospective isolation with equal efficacy and at equal purity of both quiescent and activated MuSCs (defined here as MuSCs that either have responded to tissue injury with morphological and biochemical changes prior to cell cycle entry or have entered the cell cycle and are actively proliferating). A few mouse strains have been developed to label MuSCs by genetic expression of fluorescent reporter genes, and these allow the isolation of both quiescent and activated MuSCs 11,17,21,22, but of course are not applicable to mice that do not bear the reporter transgene.
Table 1.
Summary of FACS protocols to isolate MuSCs based on the expression of cell surface antigens.
| Positive Markers | Negative Markers | Types of MuSCs Isolated | Type of Muscles Tested | References |
|---|---|---|---|---|
| VCAM-1 | CD31, CD45, Sca1 | quiescent and activated MuSCs | hindlimb muscles, diaphragm | Liu et.al.17 |
| SM/C2.6* | CD45 | quiescent MuSCs | hindlimb muscles | Fukada et.al.13 |
| CXCR4, β1- integrin | CD45, Mac1, Sca1 | quiescent MuSCs | hindlimb muscles | Sherwood et.al.12 |
| CD34 | CD45,Sca1¶ | quiescent MuSCs | hindlimb muscles, diaphragm | Montarras et.al.11 |
| CD34 | CD31, CD45, Sca1 | quiescent MuSCs | hindlimb muscles | Joe et.al.14 |
| α7-integrin | CD31, CD45, Sca1 | activated MuSCs | hindlimb muscles | Joe et.al.14 |
| CD34, α7- integrin | CD31, CD11b, CD45, Sca1 | quiescent MuSCs | tibialis anterior | Sacco et.al.15 Pasut et.al.31 |
This antibody is not commercially available.
Due to the lack of endothelial markers, this protocol does not discriminate MuSCs from endothelial cells.
The purpose of this article is to present a detailed protocol for the prospective isolation of MuSCs by mechanical and enzymatic dissociation followed by FACS based upon methods we have optimized. The primary advantages of the current protocol, as detailed in subsequent sections, in comparison to published protocols include the fact that we are able to sort both quiescent MuSCs and their proliferating progeny with one protocol, we have optimized the preparation so as to increase the yield of MuSCs 2–4 fold over previously published methods, we have validated the use of the protocol in mice of different ages, we have validated, using a genetic lineage model, that the protocol not only yields highly pure (>98%) populations of myogenic cells but also that it is also extremely effective in capturing virtually all of the myogenic cells in the population, and we have used this protocol to purify MuSCs from a variety of muscle groups and mouse strains, as well as from diseased muscle. We isolate quiescent and activated MuSCs from adult mice according to their expression of vascular cell adhesion molecule 1 (VCAM1; also known as Cluster of Differentiation 106 (CD106)) on the cell surface13. Of particular importance, we have performed lineage tracing studies to validate VCAM1 as a marker of MuSCs and their progeny by FACS (Supplemental Figure 1). Due to the specific expression of the Pax7 transcription factor in MuSCs in adult animals, MuSCs and their progeny are genetically labeled by YFP expression in the mouse strain Pax7CreER/+; ROSA26eYFP/+ in which the expression of Cre recombinase can be induced from the Pax7 locus without disrupting normal Pax7 expression23. We injected these mice with tamoxifen at 2 months of age to induce the expression of YFP in MuSCs. We then used FACS analysis to test for YFP expression among populations of mononucleated cells typically found in adult skeletal muscle, including (and identified by the associated cell surface marker) MuSCs (VCAM1+), endothelial cells (CD31+), hematopoietic cells (CD45+), and mesenchymal stem cells (Sca1+). Our analysis revealed that all YFP-expressing cells were positive for VCAM1 expression and negative for CD31, CD45 and Sca1. Furthermore, YFP-expressing cells could be found only in VCAM1+/CD31−/CD45−/Sca1− cells prior to and following muscle injury in both young (3 months of age) and old (23 months of age) mice. These analyses confirm that VCAM1 is a sensitive and highly specific FACS marker of MuSCs from both young and old animals, and that it is as effective for isolating activated, proliferating MuSC progeny as for quiescent MuSCs.
We have used this protocol to successfully purify MuSCs from different types of muscle, including limb and diaphragm muscles, and from adult mice of all ages, of various strains and genetic background, and with various disease conditions16,17,24–26. The high yield and purity of MuSCs from this protocol has allowed us to perform not only classic stem cell experiments in tissue culture and upon transplantation, but also biochemical and molecular analyses that often require large numbers of cells17. Furthermore, this protocol allows simultaneous isolation of mesenchymal stem cells (Sca1+), which have the potential to differentiate into fibroblasts, adipocytes and osteoblasts14, as well as endothelial cells from the CD31+ population and hematopoietic cells from the CD45+ population, from limb muscle. We have not tested the efficiency of this protocol in isolating myogenic progenitors from mice younger than 6 weeks of age.
Skeletal muscle is a dense tissue composed primarily of multinucleated myofibers. Efficient release of mononucleated cells from the tissue and removal of fiber debris are the most critical steps to obtain a large number of pure MuSCs. In this protocol, limb muscles are subjected to a series of physical and enzymatic dissociation steps to release resident mononucleated cells. Cells are then immediately stained with a cocktail of antibodies to allow the discrimination of MuSCs from endothelial cells, hematopoietic cells and mesenchymal stem cells, as well as other less well-characterized cells, by FACS. The successful execution of this protocol requires basic knowledge of muscle biology, mouse anatomy, tissue culture and FACS.
MATERIALS
REAGENTS
Mice older than 2 months of age (any strains are appropriate for this protocol.)
Ham’s F-10 media with L-glutamate (Hyclone)
Horse serum (Life Technologies)
Penicillin/streptomycin mixtures (100X, Omega Scientific)
Collagenase II (Worthington)
Dispase (Life Technologies)
Propidium Iodine (Life Technologies catalog number P3566)
*APC anti-mouse CD31 (clone MEC13.3; BioLegend catalog number 102510)
*APC anti-mouse CD45 (clone 30-F11; BioLegend catalog number 103112)
Pacific Blue anti-mouse Ly-6A/E (anti- Sca1, clone D7; BioLegend catalog number 108120)
Biotin anti-mouse CD106 (anti-VCAM1, clone 429; BioLegend catalog number 105704)
PE/Cy7 Streptavidin (BioLegend catalog number 405206)
APC Rat IgG2a, κ Isotype Control (BioLegend catalog number 400511)
APC Rat IgG2b, κ Isotype Control (BioLegend catalog number 400611)
Pacific Blue IgG2a, κ Isotype Control (BioLegend catalog number 400527)
Biotin IgG2a, κ Isotype Control (BioLegend catalog number 400503)
1x phosphate buffer saline (PBS) pH 7.4 (Life Technologies)
37% formaldehyde (Sigma F8775)
2.5 M Glycine solution
Poly-D lysine solution, 1 mg/ml (EMD Millipore A-003-E)
ECM gel from Engelbreth-Holm-Swarm murine sarcoma (Sigma E1270)
Recombinant Fibroblast Growth Factor-basic (bFGF) (PeproTech 100-18B)
DMEM (Cellgro)
Anti-Pax7 (Developmental Studies Hybridoma Bank)
Anti-MyoD (Dako M3512)
Anti-Myogenin (BD Pharmingen 556358)
Anti-MHC (clone MF 20, Developmental Studies Hybridoma Bank)
EQUIPMENT
Dumont forceps with straight tips
Dissection scissors
Sterile surgical blade size 11 (Fisher Scientific 08-915-13 or equivalent)
10-cm petri dishes
50-ml conical tubes
37°C shaking water bath (Fisher Scientific 15-453-205 or equivalent)
Clinical centrifuge
10-ml syringes, point style: Luer Lok (Fisher Scientific 14-823-2A or equivalent)
20 G 1-inch needles (Fisher Scientific 14-826-D or equivalent)
Falcon 0.40 μm cell strainer (Fisher Scientific 08-771-1 or equivalent)
2 ml round bottom microcentrifuge tube (USA Scientific 1620-2700 or equivalent)
Nutating rocker (Fisher Scientific 22-363-152 or equivalent)
Refrigerated centrifuge
Falcon 5-ml round bottom tubes with strainer cap (Fisher Scientific 08-771-23 or equivalent)
Falcon 5-ml round bottom tubes (Fisher Scientific 14-959-2A or equivalent)
Sterile hood for cell culture
BD FACSAria II or III cell sorter (BD Biosciences)
8-well chamber slides
0.1–10, 2–20, 20–200, 100–1000 μl pipets and matching tips
Sterile 5 and 10 ml serological pipets and a pipet controller
REAGENT SETUP
Wash Medium (WM)
WM is Ham’s F-10 supplemented with 10% Horse Serum and 1X penicillin/streptomycin. Keep cold (4–8°C) throughout the protocol.
Muscle Dissociation Buffer (MDB)
MDB is 700–800 U/ml Collagenase II solution prepared in WM. Prepare 10 ml for each mouse. This reagent should be prepared freshly by dissolving Collagenase II powder in WM before the dissection and kept on ice until needed. Sterilize with 0.22 μm filter if necessary.
Stock Collagenase II solution
This solution is prepared by dissolving Collagenase II powder in 1X PBS so that the final concentration is 1000 U/ml. Sterilize with 0.22 μm filter and store as 1-ml aliquots at −20°C. Thaw before use.
Stock Dispase solution
This solution is prepared by dissolving Dispase powder in 1X PBS so that the final concentration is 11 U/ml. Sterilize with 0.22 μm filter and store as 1-ml aliquots at −20°C. Thaw and centrifuge at >10,000 rpm for 1 min before use. Avoid taking the pellet after centrifugation for digest.
ECM coating solution
This solution is prepared by diluting ECM gel 1:100 in cold DMEM. Prepare freshly before use. It is important to keep the reagents cold (about 4°C) during preparation as ECM gel solidifies at higher temperature.
Stock basic fibroblast growth factor (bFGF)
The stock solution is made by dissolving bFGF in sterile 1× PBS containing 0.1% bovine serum albumin so that the concentration is 25 μg/ml. Aliquot into small vials and store at −20°C. Avoid repeated freeze and thaw cycles. Once thawed, bFGF can be stored at 4°C for up to a month.
Proliferation medium (PM)
PM is Ham’s F-10 supplemented with 10% horse serum, 1× penicillin/streptomycin, and 2.5 ng/ml bFGF. Store the medium at 4°C until ready to use. It is recommended that bFGF be added freshly.
Differentiation medium (DM)
DM is DMEM supplemented with 5% horse serum and 1× penicillin/streptomycin. Store the medium at 4°C until ready to use.
EQUIPMENT SETUP
BD FACSAria cell sorter
The 405 nm, 488 nm and 633 nm lasers are required for the detection of the fluorochromes recommended in this protocol. Using antibodies conjugated to these fluorochromes requires no compensation between the channels, thus maximizing yield and purity. These fluorochromes can also be used in conjunction with GFP or similar fluorescent proteins without compensation. A 70-μm nozzle and threshold rate of 2500–3500 events/second are recommended for better yield, purity and cell survival. Higher threshold rate compromises both yield and purity. While lower threshold rate does not impact yield or purity, it is neither time-saving nor cost-effective. We recommend using a flow rate lower than 3.0 as cell viability decreases with higher flow rates.
ECM-coated plastic tissue culture dishes and plates
Add a sufficient volume of freshly prepared ECM coating solution to cover the bottom of the vessels. Place on the rocker at 4°C and incubate for a minimum of 6 hr. The dishes or plates can then be stored at 4°C for up to a week with the coating solution on them. Aspirate the coating solution and seed cells immediately.
Poly-D-Lysine- and ECM-coated glass chamber slides
Coating of the two materials is performed sequentially. Glass chamber slides are incubated with freshly prepared 0.1 mg/ml poly-D-lysine solution (1:10 dilution of stock solution with Sterile MilliQ water) at room temperature for from 8–24 hours. Aspirate the solution and rinse the slides with sterile MilliQ water three times. After the last wash, aspirate all traces of liquid and let the slides dry in the tissue culture hood for 6–8 hr (optional: the slides can also be sterilized by UV while they are drying). Poly-D-lysine coated slides can be stored at 4°C for a few months. These slides can then be coated with freshly prepared ECM coating solution as previously described.
PROCEDURE
Dissection of limb muscles | TIMING 30 minutes per mouse
-
1
Prepare 10-cm petri dishes with 10 ml wash medium (one dish per mouse).
-
2
Euthanize mice by CO2 asphyxiation followed by cervical dislocation.
-
3
Spray the mouse liberally with 70% ethanol and place on its back in the dissection area.
-
4
Lift the skin around one ankle with forceps and make a small incision with scissors.
-
5
Extend the incision up to the knee. Retract the skin to expose all the hindlimb muscles.
-
6
Collect the hindlimb muscles in the petri dish. We usually start by cutting all tendons at the ankle. Detach the tibialis anterior, extensor digitorum longus, gastrocnemius and soleus muscles from the tibia and fibula. Cut around the knee and transfer these muscles to the petri dish. Insert a scalpel between the quadriceps and the femur at the joint above the knee and cut the muscles. Hold the quadriceps at the distal end with forceps, separate it from the femur and the rest of the muscle by pulling it toward the proximal end. Cut at the proximal end and transfer the quadriceps in the petri dish. Detach the hamstrings and remaining muscles by carefully cutting along the femur. Collect these muscles in the petri dish and trim away any visible fat and tendon.
CRITICAL STEP: It is important to collect all muscles to maximize the yield of MuSCs. Avoid damaging vessels whenever possible. If bleeding occurs, absorb blood immediately with Kimwipes. Although blood-derived cells do not interfere with the remaining steps of this protocol, the presence of large amounts of blood makes it difficult to visualize the anatomy. At the end of this step, the tibia, fibula and femur should be stripped clean without any attaching muscles. The most prominent intermuscular fat is found between the hamstring muscles and at the proximal end of the quadriceps. Remove the fat during the dissection.
-
7
Collect all muscles from the other hindlimb. Triceps of the forelimbs can also be collected if the analysis is not restricted to hindlimb muscles.
-
8
Transfer all muscles of a mouse to a fresh petri dish (or use the lid of the initial dish) and mince the muscles for about 10 minutes.
CRITICAL STEP: We recommend two ways to mince the muscles, both of which require holding one end of a piece of muscle with forceps. Then, either use scissors to cut the muscle into small pieces or use a scalpel to slice the muscle. Finish cutting all of the muscle pieces. Gather all muscles at the center of the dish and cut with scissors for an additional 1–2 minutes. At the end of this step, the preparation should yield a slurry of well-minced tissue.
Isolation of mononucleated cells | TIMING 2.5 hours
-
9
Transfer the minced muscle from each mouse to an individual 50-ml conical tube with 10 ml MDB. Muscle from 2 mice can be pooled into one 50-ml tube with 20 ml MDB. We do not recommend pooling muscle from more than 2 mice per tube as it often leads to insufficient digestion after Step 10.
-
10
Seal the tube(s) well with Parafilm and incubate in a 37°C water bath with agitation (60–70 rpm on the recommended model of shaking water bath) for 1 hr. Tubes should be positioned horizontally along the shaking path and completely submerged in water. Use weights to keep the tubes submerged.
The following steps should be performed in a sterile tissue culture hood if cells will be used for culture or transplantation.
-
11
Fill each tube to 50 ml with cold WM. Gently invert a few times to mix.
-
12
Centrifuge in a swinging bucket rotor at 500g for 5 min. Aspirate supernatant down to about 8 ml.
CRITICAL STEP: The pellet at the bottom after centrifugation is loose and can be easily disturbed. Control the aspiration strength and always aspirate only from the surface of the supernatant.
-
13
Add 1 ml stock Collagenase II solution and 1 ml stock Dispase solution.
-
14
Resuspend the pellet with a 5-ml serological pipet. Triturate 10–15 times or until the suspension travels up and down the pipet smoothly without clogging.
-
15
Seal the tube(s) well with Parafilm and incubate in a 37°C water bath with agitation (60–70 rpm on the recommended model of shaking water bath) for 30 min. Tubes should be positioned as in step 10.
-
16
Using a 10-ml syringe and 20-gauge needle, aspirate and eject the muscle suspension in and out of the syringe 10 times. CRITICAL STEP: The needle may be clogged by small pieces of tendon or undigested muscle during the first few attempts. If clogging occurs during trituration, remove the clog from the tip of the needle. If clogging occurs during ejection, pull the plunger up and down to move the clog out of position. Eject towards the wall of the tube (as opposed to the bottom) to avoid foaming.
-
17
Fill each tube to 50 ml with WM. Gently invert a few times to mix.
-
18
Centrifuge at 500g for 5 min. Aspirate supernatant down to about 10 ml.
-
19
Place a 40-μm Nylon cell strainer on a fresh 50-ml conical tube.
-
20
Resuspend the pellet with a 10-ml pipet. Transfer to the cell strainer and allow to filter by gravity.
-
21
Add 10 ml WM to the original tube. Swirl to rinse, and transfer to the same cell strainer.
-
22
Rinse the cell strainer with another 10 ml WM. Use a 20–200 μl pipet to collect any remaining liquid on the underside of the strainer.
CRITICAL STEP: The strainer can be clogged by fiber debris when two mice are processed in one sample. Make sure to recover all traces of liquid from the clogged strainer before changing to a new one. Make sure to collect everything that remains associated with the outside and underside of the strainer after filtration to maximize yield.
-
23
Fill each tube to 50 ml with WM. Gently invert a few times to mix.
-
24
Centrifuge at 500g for 5 min. Remove all supernatant by aspiration immediately after centrifugation as the pellet will loosen over time. Be very careful not to aspirate cells from the pellet.
-
25
Resuspend cell pellet in 600 μl WM.
CRITICAL STEP: We typically obtain 8–12 × 106 cells per mouse and the concentration of the cell suspension is usually 10–20 × 106 cells/ml. Quantify the number of cells and adjust the amount of antibody in the following Antibody Staining step accordingly if the number of cells falls out of this range.
Antibody staining | TIMING 1.5 hr
-
26
Transfer 10 μl of the cell suspension to a 5-ml FACS tube containing 190 μl WM and leave on ice. This is the unstained control sample.
-
27
Label nine 2-ml tubes to prepare for the single channel and fluorescence minus one (FMO) staining controls for each individual channel, APC, Pacific Blue and PE-Cy7, and antibody isotype controls. Add 190 μl WM into each tube and 10 μl of the cell suspension. Add PI so that the final concentration is 0.3 μg/ml, add 0.25 μl CD31-APC and CD45-APC, Sca1-Pacific Blue or VCAM1-Biotin antibodies to the appropriately labeled tubes. If the concentration of isotype control antibody differs from a given specific antibody, calculate the volume before applying so that the same amount of isotype control antibody is used.
Antibody Tube # 1 2 3 4 5 6 7 8 9 10 PI + CD31-APC + + + + + CD45-APC + + + + + Sca1-Pacific Blue + + + + + VCAM1-Biotin + + + + + IgG2a-APC + IgG2b-APC + IgG2a-Pacific Blue + IgG2a-Biotin + -
28
Transfer the rest of the cell suspension (about 500 μl) into a labeled 2-ml tube. For cells from each mouse, add 5 μl of each of the following antibodies: CD31-APC, CD45-APC, Sca1-Pacific Blue and VCAM1-Biotin.
CRITICAL STEP: Assuming a typical yield, all antibodies are used at about 1 μg per 107 cells except for VCAM1-Biotin which is used at about 2.5 μg per 107 cells. Titration of antibodies is usually not required when using reagents suggested by this protocol. However, optimization may be necessary for antibodies from other vendors.
-
29
Place all staining samples on a nutating rocker at 4°C and incubate for 40 min.
-
30
Fill all tubes to 2 ml with WM. Gently invert the tubes a few times to mix. Centrifuge at 250g with a fixed angle rotor for 5 min at 4°C.
-
31
Aspirate supernatant completely. Resuspend cells in control tubes in 200 μl WM and the sort sample in 500 μl WM. CRITICAL STEP: Be careful not to aspirate cells. It helps to aspirate with low-volume pipet tips with narrow openings, e.g. gel-loading tips or 1–10 μl tips.
-
32
Add 0.5 μl PE-Cy7 Streptavidin into the control tubes 3–5 and 7–9.
-
33
Add 5 μl PE-Cy7 Streptavidin and PI to a final concentration to 0.3 μg/ml into the sort sample. Place all tubes on a nutating rocker at 4°C and incubate for 20 min.
-
34
Transfer the cells in control tubes 1, 2 and 4 to labeled 5-ml FACS tubes. Leave the tubes on ice and protect from light.
-
35
Fill all tubes to 2 ml with WM. Gently invert the tubes a few times to mix. Centrifuge at 250g with a fixed angle rotor for 5 min at 4°C. Aspirate supernatant completely. CRITICAL STEP: Be careful not to aspirate cells. It helps to aspirate with low-volume pipet tips with narrow openings, e.g. gel-loading tips or 1–10 μl tips.
-
36
Resuspend cells in control tubes in 200 μl WM and transfer to labeled 5-ml FACS tubes.
-
37
Resuspend cells in the sort sample in 500 μl WM. Transfer to the strainer cap attached to a 5-ml FACS tube. Gently tap the tube on the bench to facilitate flow through.
-
38
Rinse the 2-ml tube used to stain the sort sample with another 500 μl WM and pass through the same strainer cap. Use a 20–200 μl pipet to collect any remaining liquid in the strainer cap from the underside. Add another 1 ml of WM to the sort sample and mix by gentle vortex.
CRITICAL STEP: It is best to move immediately to cell sorting for the highest yield and viability.
Cell sorting | TIMING 1–1.5 hr per mouse
-
39
Set up the cell sorter in accordance with the manufacturer’s specifications with the 70-μm nozzle.
-
40
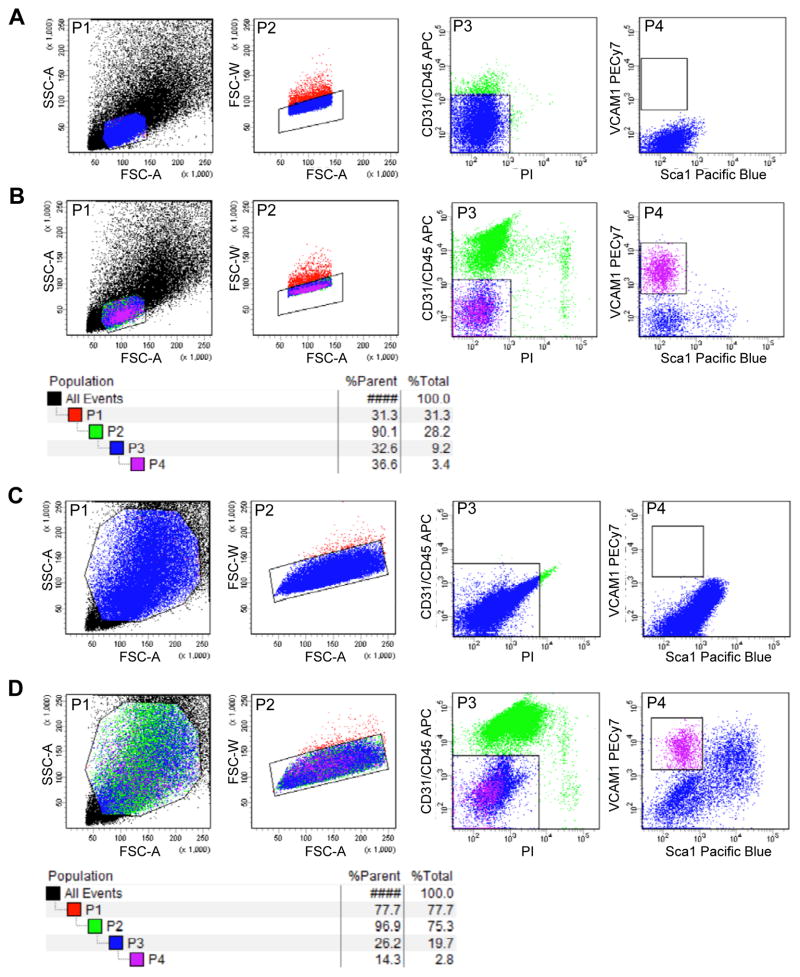
At the lowest flow rate, run the unstained sample to ensure that the voltages are appropriately set and that the cell population is properly positioned in the FSC-A and SSC plot and exhibits low background fluorescence in all channels (Figure 1A and 1C).
-
41
Run all controls to ensure that positive populations can be distinctly separated from background.
-
42
Create a gate on the FSC-A and SSC plot that excludes debris. Debris from uninjured muscle is often distinct from cells on this plot and debris from injured muscle may not be apparent (P1, Figure 1, Supplemental Figure 2 and 3A).
-
43
Create a gate on the FSC-W plot for intact single cells only (P2, Figure 1 and Supplemental Figure 2).
CRITICAL STEP: In our experience, this gate excludes dead cells and fragmented fibers, both of which exhibit irregular dimensions when isolating quiescent MuSCs from uninjured muscle. This gate can also exclude doublets of cells when isolating activated MuSCs and their proliferating progeny after acute muscle injury.
-
44
Create gates to collect the VCAM1+CD31−CD45−Sca1− population (P3 and P4, Figure 1 and Supplemental Figure 2) into 1 ml WM (or any desired media) in a 5-ml round-bottom tube. If mesenchymal stem cells are desired, collect the Sca1+CD31−CD45− population (P5, Supplemental Figure 3B). Hematopoietic and endothelial cells can also be collected if desired when CD31 and CD45 are stained with antibodies conjugated with different fluorochromes (P6 and P7, Supplemental Figure 3C).
CRITICAL STEP: We suggest running 70% ethanol through the system before sorting to minimize the risk of contamination.
Figure 1. Representative FACS profiles of quiescent and activated MuSCs.
The digested tissue preparations, as described in this protocol, from uninjured or injured limb muscles were stained with PI, APC anti-mouse CD31, APC anti-mouse CD45, Pacific Blue anti-mouse Ly-6A/E (Sca1) and Biotin anti-mouse CD106 (VCAM1) followed by PE/Cy7 Streptavidin. Cells in the P4 gate are MuSCs. (A) Profiles of the unstained quiescent MuSCs. (B) Profiles of quiescent MuSCs after antibody staining. The population hierarchy is shown under the plots. (C) Profiles of the unstained activated MuSCs and their progeny. (D) Profiles of activated MuSCs and their progeny after antibody staining. The population hierarchy is shown under the plots.
Freezing MuSCs for RNA isolation | TIMING 10 min
-
45
Transfer sorted cells to an RNase-free 1.5-ml microcentrifuge tube. If the sorted cell sample exceeds 1.5 ml, use 2 tubes or centrifuge sequentially to accommodate all cells in one tube. We do not recommend using bigger tubes as it is difficult to visualize the cell pellet in these tubes after Step 45.
-
46
Centrifuge in a fixed angle rotor at 2,500g for 5 min at 4°C.
-
47
Aspirate supernatant completely.
-
48
Snap-freeze the cell pellet in liquid nitrogen and store in liquid nitrogen or at −80°C.
Freezing MuSCs for protein extraction | TIMING 15 min
-
49
Transfer sorted cells to a clean 1.5-ml microcentrifuge tube. If the sorted cell sample exceeds 1.5 ml, use 2 tubes or centrifuge sequentially to accommodate all cells in one tube. We do not recommend using bigger tubes as it is difficult to visualize the cell pellet in these tubes after Step 49.
-
50
Centrifuge in a fixed angle rotor at 2,500g for 5 min at 4°C.
-
51
Aspirate supernatant completely.
-
52
Fill the tube with 1.5 ml 1X PBS.
-
53
Centrifuge in a fixed angle rotor at 2,500g for 5 min at 4°C and aspirate supernatant completely.
-
54
Snap-freeze the cell pellet in liquid nitrogen and store in liquid nitrogen or at −80°C.
Preparation of MuSCs for chromatin immunoprecipitation (ChIP) | TIMING 30–40 min
-
55
Transfer sorted cells to a 1.5-ml microcentrifuge tube. Measure the volume while transferring. If the sorted cell sample exceeds 1.5 ml, use 2 tubes. We do not recommend using bigger tubes as it is difficult to visualize the cell pellet in these tubes after Step 57.
-
56
Add formaldehyde to a final concentration of 1% and gently invert the tubes a few times to mix. Incubate at room temperature with gentle agitation for 10 min.
-
57
Add glycine to a final concentration of 0.125 M and gently invert the tubes a few times to mix. Incubate at room temperature with gentle agitation for 5 min.
-
58
Centrifuge in a fixed angle rotor at 5,500g for 5 min at 4°C.
-
59
Aspirate supernatant completely.
-
60
Fill the tube with 1.5 ml ice-cold 1X PBS.
-
61
Centrifuge in a fixed angle rotor at 5,500g for 5 min at 4°C and aspirate supernatant completely.
-
62
Repeat the PBS wash one more time.
-
63
Snap-freeze the cell pellet in liquid nitrogen and store in liquid nitrogen or at −80°C.
Cell culture
-
64
Aspirate the ECM coating solution from the tissue culture dishes and seed cells immediately in appropriate medium. For in vitro activation followed by spontaneous myogenic differentiation, follow option A. For in vitro expansion of myogenic progenitors, follow option B. To differentiate cultured myogenic progenitors, continue to option C after option B.
Activation of MuSCs: Seed cells in WM and incubate in standard culture conditions at 5% CO2. Change medium every 3–4 days.
Expansion of myogenic progeny derived from MuSCs: grow sorted MuSCs in PM in standard culture conditions at 5% CO2. Change medium every 2 days. Make sure cells do not grow to confluence. Cells can be passaged with trypsin with standard tissue culture practice. Use fresh ECM coated dishes or plates each time cells are passaged.
Allow cells to become nearly confluent in PM. Rinse cells 3 times with serum-free DMEM and culture cells in DM. Cells will elongate and begin to fuse within 1 day of culture in DM. The peak of differentiation and maximal fusion occurs between 2–3 days, at which point cells should be harvested or processed for analysis. Mature myotubes attach poorly and will spontaneously detach from the plate.
TIMING
MuSC isolation
Dissection of hindlimb muscles: 30 min per mouse
Isolation of mononucleated cells: 2.5 hr
Antibody staining: 1.5 hr
Cell sorting: 1–1.5 hr per mouse
Preparation of MuSCs for molecular analysis
Freezing MuSCs for RNA isolation: 10 min
Freezing MuSCs for protein extraction: 15 min
Preparation of MuSCs for chromatin immunoprecipitation (ChIP): 30–40 min
Cell culture: 30–60 minutes to set up
TROUBLSHOOTING
Trouble shooting advice can be found in Table 3.
Table 3.
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 6 | Poor cell yield | Not all muscles are collected. | Collect all hindlimb muscles until the femur, tibia and fibula are completely stripped of attaching muscles. |
| 8 | Poor cell yield | Muscles are over-minced or under-minced. Over-minced muscle makes the suspension very dense after the first Collagenase digest. Satellite cells that are already released into the suspension or attached to small fiber debris do not settle well after centrifugation and are easily aspirated at Step 12. Under-minced muscle cannot be fully digested and interfere with the dissociation at Step 16. | Mince the muscles properly. |
| 12 | Poor cell yield | Cells are aspirated. | Do not over-mince the muscles at Step 8. Aspirate slowly and always from the surface of the suspension. |
| 16 | Poor cell yield and/or purity | Cells are not dissociated from the basal lamina. | Mince the muscles properly at Step 8. Use needles of the correct gauge. Triturate properly with the syringe. Only count the complete up-and-down motion. To avoid foaming, set the needle against the wall of the tube at an angle to eject liquid. |
| 22 | Poor cell yield | Cell suspension is retained on the strainer. | Change strainer if it is clogged and recover all liquid in retention. Recover all liquid from underside of the strainer. |
| 25 | Poor staining Poor cell yield and/or purity | Too much fiber debris is present in the suspension. This can be determined by visualizing the cell suspension with a hemacytometer (dilute 10–50 times if necessary). Fiber debris appears rod- or thread- like using a 10x objective. | Follow the isolation procedures closely to optimize the yield of total mononucleated cells. Excessive fibers can be removed by density gradient centrifugation using pure fetal bovine serum, which preserves cell viability better than sucrose or sucrose polymers such as Ficoll. |
| 28 | Poor staining Poor cell yield and purity | Total mononucleated cells are not sufficiently dissociated from the muscle and/or fiber debris are not sufficiently removed. The ratio between antibody and cell number is not optimized. |
Follow the isolation procedures closely to optimize the yield of total mononucleated cells. Pay attention to all CRITICAL STEPS. The amount of each antibody may need to be optimized if different lots are used. Perform serial dilution of each antibody to identify the optimal μg of antibody to use for a fixed number of cells. Set up controls according to Step 27 to make sure positive population can be distinctly separated from background. |
| 31, 35 | Poor cell yield | Cells are aspirated. | Aspirate supernatant slowly or remove the supernatant by pipeting. |
| 38 | Poor cell yield | Cell suspension is retained on the strainer. Too much fiber debris is present in the cell suspension. Cells have aggregated. |
Change strainer if it is clogged and recover all liquid in retention. Recover all liquid from underside of the strainer. Optimize the isolation process and pay special attention to Steps 8 and 16. Filter the cell suspension through a strainer cap one more time. Proceed to sorting immediately after staining. Gently pipet up and down to resuspend cells and filter through a strainer cap if necessary. |
| 43 | Poor purity | Dead cells and cells of different size are not excluded. | Make sure a stringent FSC-W gate is drawn. Only about 1% CD31−/CD45−/Sca1−/VCAM1+ cells should be PI positive when an optimal FSC-W gate is applied. |
| 44 | Frequent sorter clogs Poor purity |
Excessive fiber debris are present in the sample. Cells do not separate well from background in the plots. |
Optimize the isolation process. Pay special attention to Steps 8 and 16 to minimize the amount of debris in the pellets at step 25. Optimize the isolation process as above. Optimize the ratio between antibody and cell number as in Step 28. Draw stringent gates in all channels to exclude contaminants. |
| 38, 44 | Contamination | Lack of attention to sterile cell sorting practice | Cell sorter should be decontaminated periodically. Run 70% ethanol in the system for at least 5 minutes before sorting. |
ANTICIPATED RESULTS
With this protocol, we routinely isolate 250,000 to 350,000 quiescent MuSCs from hindlimb muscles of a healthy, wildtype C57BL6 mouse of 2–6 months of age. This is approximately 2–4 fold greater than previously published protocols. Conboy et al reported a yield of approximately 40,000 cells per gram of muscle18,19, which translates to approximately 80,000–100,000 cells per mouse assuming a mass of approximately 2–2.5 grams of hindlimb muscle per mouse. Yi et al reported a yield of approximately 150,000 MuSCs per mouse20. The yield of MuSCs that we obtain represents about 3–4% of the total population of mononucleated cells after antibody staining. The background signal in each channel when sorting for MuSCs from mice within this age range is low, thus allowing distinct separation of the VCAM1+Sca1−CD31−CD45− population (Figure 1). When plated in culture conditions that promote differentiation, greater than 99% of the sorted cells are myogenic as determined by the presence of molecular markers, and greater than 96% undergo terminal differentiation and fusion (Figure 2). The yield of MuSCs decreases with older mice. At 22–24 months of age, hindlimb muscles of a wildtype C57BL/6 mouse that appears phenotypically healthy typically produces 150,000–200,000 MuSCs. Occasionally the yield can be lower than 100,000. This lower yield in old mice is a combined result of the reduction of MuSC number with age27–29 and the higher background signal after antibody staining. The gate to obtain the VCAM1+Sca1−CD31−CD45− cells (P4, Figure 1) from aged mice often needs to be adjusted to avoid contaminating cells or debris with high background fluorescence and thus compromises yield for purity. We have isolated MuSCs from a variety of mouse strains and found little variation in the yield and purity within a particular age range. These strains include SV129, BALB/c, C57BL/10, and transgenic mice of mixed strain backgrounds that do not exhibit defects in muscle physiology. This protocol can also be used to isolate myogenic cells in mouse models of muscular dystrophy, such as mdx or SJL/J.
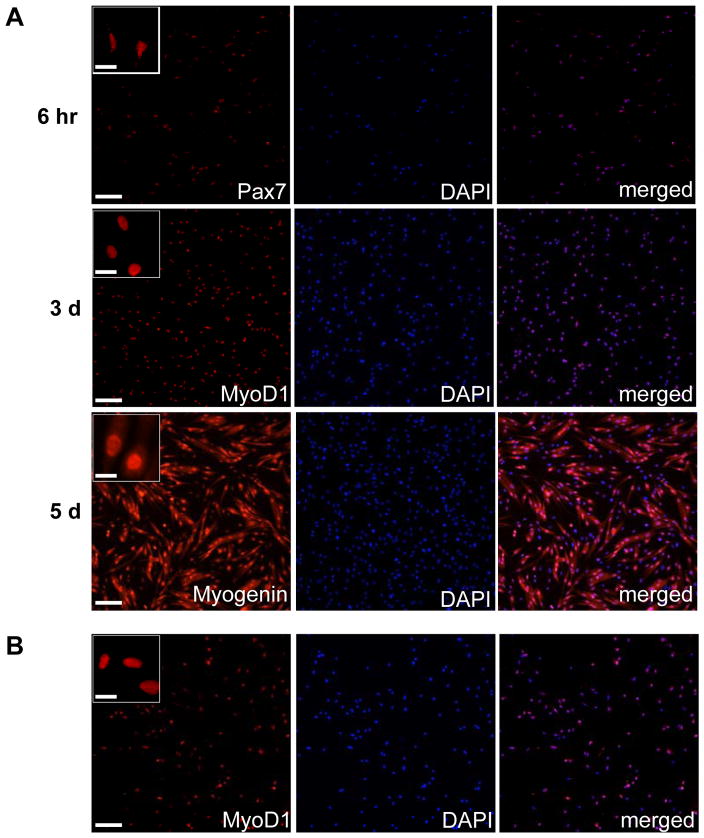
Figure 2. Confirmation of the myogenicity of isolated MuSCs.
(A) Immediately following FACS isolation, 5,000 quiescent MuSCs were plated in a well of an 8-well chamber slide that had been coated with Poly-D lysine and ECM. Cells were fixed with 4% paraformaldehyde 6 hours (6 hr), 3 days (3 d) and 5 days (5 d) following plating, and stained with antibodies against Pax7, MyoD1 and Myogenin, respectively. DAPI stains all nuclei. Note the changes in cell size and morphology. Scale bars represent 70 μm in the images and 10 μm in the inserts. (B) Immediately following FACS isolation, 5,000 activated MuSCs or their progeny from limb muscles that had been injured three days earlier were plated in a well of an 8-well chamber slide that had been coated with Poly-D lysine and ECM. Cells were fixed with 4% paraformaldehyde 6 hours later and stained with an anti-MyoD1 antibody. DAPI stains all nuclei. Scale bars represent 70 μm in the image and 10 μm in the insert.
In general, we find the physical and enzymatic isolation step to be the most important in maximizing both the yield and purity of MuSCs. An important aspect to achieve the distinct separation of different cell populations during sorting is the effective removal of fiber debris. We therefore recommend that the most attention be paid to steps 8 to 25 when learning this protocol or working with mice of different ages or disease models. Defining FACS gates is another important element of the protocol to ensure isolation of a pure population of MuSCs. We therefore have listed in this protocol a number of FACS controls that may help in determining the boundary of gates and the fidelity of the sorting antibodies (Supplemental Figures 4–6). For cost- and time-effectiveness, these controls can be eliminated without compromising yield and purity as long as the cell isolation steps have been optimized. When the cell isolation steps have been optimized, populations of cells expressing or not expressing a given surface marker can be readily distinguished (Figure 1 and Supplemental Figure 2), enabling appropriate gating without the need for additional controls. However, we recommend utilizing single channel, FMO and antibody isotype controls when attempting this protocol for the first time or when validating new antibodies. With experience, less than 1% of quiescent MuSCs isolated by this protocol are dead cells (Supplemental Figure 2C). Therefore, although the addition of a live-dead dye such as PI helps in determining whether the cell isolation steps are properly performed and whether the FACS gates are optimized, this additional step has only a marginal effect on the purity of the sorted population. It should be noted that the F10 media contains low amount of Biotin, which may have an effect on staining with the Biotin-conjugated VCAM1 antibody. While we find the F10 media appears to promote cell viability during and after sorting, it is possible to substitute it with other types of media. The recommended concentration of the VCAM1-Biotin antibody in this protocol works well with F10 media and may be decreased if using Biotin-free media.
MuSCs isolated by our protocol can be directly processed for molecular analysis, ex vivo culture, or transplantation studies. For investigators learning this protocol, we recommend assessing the purity of the isolated population by plating a small number of cells followed by staining of myogenic markers. Plated in WM, cells express high levels of Pax7 protein for up to 2 days in culture. Greater than 70% of cells express MyoD protein within 24 hours and all cells should express MyoD by 48 hours. These progeny of MuSCs will continue to differentiate and express late myogenic markers such as Myogenin and Myosin Heavy Chain (MHC). The extent of fusion depends on the density of the cells. Cells cultured at higher density result in larger myotubes with a greater number of nuclei. Isolated MuSCs proliferate rapidly in PM and all progeny should express MyoD but not Myogenin protein. If expansion is desired to obtain undifferentiated progeny of MuSCs, it is important not to grow the cells to confluence as the cells spontaneously differentiate when confluent.
We also routinely isolate activated MuSCs and their myogenic progeny from a variety of mice following intramuscular BaCl2 or cardiotoxin injections17,25,26. In our experience, the peak of myogenic proliferation in vivo occurs between day 3 and day 4 after the injection. Typically, distal (i.e., below the knees) hindlimb muscles from a 3-month-old mouse yield about one million proliferating MuSC progeny 3 days after the injection of 50 μl of a 1.2% BaCl2 solution. As MuSCs activate, they increase significantly in size26,30. Therefore in order to isolate these cells by FACS, it is important to adjust the FSC-A and SSC parameters to position these cells in the center.
A note on the issue of the sorting of quiescent MuSCs is in order because it often creates confusion. The goal of sorting MuSCs from uninjured muscle is to isolate MuSCs for analysis or study while they are still in the quiescent state. This is unequivocally the case for all protocols referenced, including ours. Once isolated, the purified cells will not enter the cell cycle, as judged by BrdU incorporation, for at least 36 hours. Thus, as functionally defined, the isolated cells are certainly quiescent. Having entered the cell cycle, the MuSC progeny will then proceed through the cell cycle in approximately 12 hours, far more rapidly than it takes them to enter the very first cell cycle from the quiescent state. That is not to say that there are not changes that occur during the isolation procedure; surely there are. As such, the quiescent cells obtained following sorting likely differ from the quiescent cells in vivo. On the other hand, we have performed detailed molecular analysis of the freshly isolated quiescent cells, and those studies suggest that the isolated cells have characteristics that would be expected of the quiescent cells in vivo and are vastly different from the proliferating progeny and even from SCs activated for 24 hours in vivo following injury, prior to entry into the cell cycle17.
Supplementary Material
Table 2.
Summary of FACS protocols to isolate MuSCs based on the genetic expression of lineage markers.
| Genotype | Genetic Manipulation | References |
|---|---|---|
| Pax7CreER; ROSA26eYFP | Conditional expression of eYFP from the ROSA26 locus upon the induction of Cre recombinase knocked into the Pax7 3′ UTR | Liu et.al.17 |
| Pax3GFP/+ | Genetic knock-in: Exon 1 of Pax3 has been replaced by sequence encoding enhanced GFP | Montarras et.al.11 |
| Tg: Pax7-nGFP | Transgenic expression of nuclear localized GFP under the regulatory element of the mouse Pax7 gene | Rocheteau et.al.18 |
| Tg: Pax7-ZsGreen | Transgenic expression of ZsGreen under the regulatory element of the mouse Pax7 gene | Bosnakovski et.al.19 |
Acknowledgments
We thank Polly Huang for technical assistance of dissection, and Lusijah Rott for technical assistance of FACS. This work was supported by grants from The Glenn Foundation for Medical Research, the National Institutes of Health (P01 AG036695, R37 AG023806, and R01 AR062185), and Department of Veterans Affairs to T.A.R.
Footnotes
Note: these antibodies can be substituted with FITC anti-mouse CD31 and CD45 (BioLegend catalog number 102506 and 103108, respectively) when sorting cells from wildtype mice or any mouse strains that do not express green fluorescent protein (GFP) or similar reporter genes in cells in skeletal muscle. Matching FITC isotype controls are BioLegend catalog numbers 400505 and 400605. In general, FITC-conjugated antibodies are more economical than APC-conjugated ones. For simplicity, APC-conjugated antibodies are described in this protocol unless stated otherwise.
Author Contribution
All authors contribute to the design and interpretation of the experiments; L.L., T.H.C. and G.W.C. conducted the experiments; L.L. and T.A.R. wrote the manuscript.
Competing Financial Interest
The authors declare no competing financial interests.
References
- 1.Morgan JE, Partridge TA. Muscle satellite cells. Int J Biochem Cell Biol. 2003;35:1151–6. doi: 10.1016/s1357-2725(03)00042-6. [DOI] [PubMed] [Google Scholar]
- 2.Rando TA. Stem cells, ageing and the quest for immortality. Nature. 2006;441:1080–6. doi: 10.1038/nature04958. [DOI] [PubMed] [Google Scholar]
- 3.Mauro A. Satellite cell of skeletal muscle fibers. J Biophys Biochem Cytol. 1961;9:493–5. doi: 10.1083/jcb.9.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seale P, Sabourin LA, Girgis-Gabardo A, Mansouri A, Gruss P, Rudnicki MA. Pax7 is required for the specification of myogenic satellite cells. Cell. 2000;102:777–86. doi: 10.1016/s0092-8674(00)00066-0. [DOI] [PubMed] [Google Scholar]
- 5.Cheung TH, Rando TA. Molecular regulation of stem cell quiescence. Nat Rev Mol Cell Biol. 2013;14:329–40. doi: 10.1038/nrm3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu L, Rando TA. Manifestations and mechanisms of stem cell aging. J Cell Biol. 2011;193:257–66. doi: 10.1083/jcb.201010131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rinaldi F, Perlingeiro RC. Stem cells for skeletal muscle regeneration: therapeutic potential and roadblocks. Transl Res. 2014;163:409–17. doi: 10.1016/j.trsl.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tedesco FS, Cossu G. Stem cell therapies for muscle disorders. Curr Opin Neurol. 2012;25:597–603. doi: 10.1097/WCO.0b013e328357f288. [DOI] [PubMed] [Google Scholar]
- 9.Gharaibeh B, Lu A, Tebbets J, Zheng B, Feduska J, Crisan M, Peault B, Cummins J, Huard J. Isolation of a slowly adhering cell fraction containing stem cells from murine skeletal muscle by the preplate technique. Nat Protoc. 2008;3:1501–9. doi: 10.1038/nprot.2008.142. [DOI] [PubMed] [Google Scholar]
- 10.Cosgrove BD, Gilbert PM, Porpiglia E, Mourkioti F, Lee SP, Corbel SY, Llewellyn ME, Delp SL, Blau HM. Rejuvenation of the muscle stem cell population restores strength to injured aged muscles. Nat Med. 2014;20:255–64. doi: 10.1038/nm.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Montarras D, Morgan J, Collins C, Relaix F, Zaffran S, Cumano A, Partridge T, Buckingham M. Direct isolation of satellite cells for skeletal muscle regeneration. Science. 2005;309:2064–7. doi: 10.1126/science.1114758. [DOI] [PubMed] [Google Scholar]
- 12.Sherwood RI, Christensen JL, Conboy IM, Conboy MJ, Rando TA, Weissman IL, Wagers AJ. Isolation of adult mouse myogenic progenitors: functional heterogeneity of cells within and engrafting skeletal muscle. Cell. 2004;119:543–54. doi: 10.1016/j.cell.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 13.Fukada S, Uezumi A, Ikemoto M, Masuda S, Segawa M, Tanimura N, Yamamoto H, Miyagoe-Suzuki Y, Takeda S. Molecular signature of quiescent satellite cells in adult skeletal muscle. Stem Cells. 2007;25:2448–59. doi: 10.1634/stemcells.2007-0019. [DOI] [PubMed] [Google Scholar]
- 14.Joe AW, Yi L, Natarajan A, Le Grand F, So L, Wang J, Rudnicki MA, Rossi FM. Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nat Cell Biol. 2010;12:153–63. doi: 10.1038/ncb2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sacco A, Doyonnas R, Kraft P, Vitorovic S, Blau HM. Self-renewal and expansion of single transplanted muscle stem cells. Nature. 2008;456:502–6. doi: 10.1038/nature07384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boutet SC, Cheung TH, Quach NL, Liu L, Prescott SL, Edalati A, Iori K, Rando TA. Alternative polyadenylation mediates microRNA regulation of muscle stem cell function. Cell Stem Cell. 2012;10:327–36. doi: 10.1016/j.stem.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu L, Cheung TH, Charville GW, Hurgo BM, Leavitt T, Shih J, Brunet A, Rando TA. Chromatin modifications as determinants of muscle stem cell quiescence and chronological aging. Cell Rep. 2013;4:189–204. doi: 10.1016/j.celrep.2013.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conboy MJ, Cerletti M, Wagers AJ, Conboy IM. Immuno-analysis and FACS sorting of adult muscle fiber-associated stem/precursor cells. Methods Mol Biol. 2010;621:165–73. doi: 10.1007/978-1-60761-063-2_11. [DOI] [PubMed] [Google Scholar]
- 19.Conboy MJ, Conboy IM. Preparation of adult muscle fiber-associated stem/precursor cells. Methods Mol Biol. 2010;621:149–63. doi: 10.1007/978-1-60761-063-2_10. [DOI] [PubMed] [Google Scholar]
- 20.Yi L, Rossi F. Purification of progenitors from skeletal muscle. J Vis Exp. 2011 doi: 10.3791/2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rocheteau P, Gayraud-Morel B, Siegl-Cachedenier I, Blasco MA, Tajbakhsh S. A subpopulation of adult skeletal muscle stem cells retains all template DNA strands after cell division. Cell. 2012;148:112–25. doi: 10.1016/j.cell.2011.11.049. [DOI] [PubMed] [Google Scholar]
- 22.Bosnakovski D, Xu Z, Li W, Thet S, Cleaver O, Perlingeiro RC, Kyba M. Prospective isolation of skeletal muscle stem cells with a Pax7 reporter. Stem Cells. 2008;26:3194–204. doi: 10.1634/stemcells.2007-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishijo K, Hosoyama T, Bjornson CR, Schaffer BS, Prajapati SI, Bahadur AN, Hansen MS, Blandford MC, McCleish AT, Rubin BP, Epstein JA, Rando TA, Capecchi MR, Keller C. Biomarker system for studying muscle, stem cells, and cancer in vivo. FASEB J. 2009;23:2681–90. doi: 10.1096/fj.08-128116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bjornson CR, Cheung TH, Liu L, Tripathi PV, Steeper KM, Rando TA. Notch signaling is necessary to maintain quiescence in adult muscle stem cells. Stem Cells. 2012;30:232–42. doi: 10.1002/stem.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheung TH, Quach NL, Charville GW, Liu L, Park L, Edalati A, Yoo B, Hoang P, Rando TA. Maintenance of muscle stem-cell quiescence by microRNA-489. Nature. 2012;482:524–8. doi: 10.1038/nature10834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodgers JT, King KY, Brett JO, Cromie MJ, Charville GW, Maguire KK, Brunson C, Mastey N, Liu L, Tsai CR, Goodell MA, Rando TA. mTORC1 controls the adaptive transition of quiescent stem cells from G0 to G(Alert) Nature. 2014;510:393–6. doi: 10.1038/nature13255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conboy IM, Conboy MJ, Wagers AJ, Girma ER, Weissman IL, Rando TA. Rejuvenation of aged progenitor cells by exposure to a young systemic environment. Nature. 2005;433:760–4. doi: 10.1038/nature03260. [DOI] [PubMed] [Google Scholar]
- 28.Gibson MC, Schultz E. Age-related differences in absolute numbers of skeletal muscle satellite cells. Muscle Nerve. 1983;6:574–80. doi: 10.1002/mus.880060807. [DOI] [PubMed] [Google Scholar]
- 29.Sajko S, Kubinova L, Cvetko E, Kreft M, Wernig A, Erzen I. Frequency of M-cadherin-stained satellite cells declines in human muscles during aging. J Histochem Cytochem. 2004;52:179–85. doi: 10.1177/002215540405200205. [DOI] [PubMed] [Google Scholar]
- 30.Tang AH, Rando TA. Induction of autophagy supports the bioenergetic demands of quiescent muscle stem cell activation. EMBO J. 2014 doi: 10.15252/embj.201488278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pasut A, Oleynik P, Rudnicki MA. Isolation of muscle stem cells by fluorescence activated cell sorting cytometry. Methods Mol Biol. 2012;798:53–64. doi: 10.1007/978-1-61779-343-1_3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.