Abstract
The modern sequencing technologies are generating large volumes of information at the transcriptome and genome level. Translation of this information into a biological meaning is far behind the race due to which a significant portion of proteins discovered remain as proteins of unknown function (PUFs). Attempts to uncover the functional significance of PUFs are limited due to lack of easy and high throughput functional annotation tools. Here, we report an approach to assign putative functions to PUFs, identified in the transcriptome of mulberry, a perennial tree commonly cultivated as host of silkworm. We utilized the mulberry PUFs generated from leaf tissues exposed to drought stress at whole plant level. A sequence and structure based computational analysis predicted the probable function of the PUFs. For rapid and easy annotation of PUFs, we developed an automated pipeline by integrating diverse bioinformatics tools, designated as PUFs Annotation Server (PUFAS), which also provides a web service API (Application Programming Interface) for a large-scale analysis up to a genome. The expression analysis of three selected PUFs annotated by the pipeline revealed abiotic stress responsiveness of the genes, and hence their potential role in stress acclimation pathways. The automated pipeline developed here could be extended to assign functions to PUFs from any organism in general. PUFAS web server is available at http://caps.ncbs.res.in/pufas/ and the web service is accessible at http://capservices.ncbs.res.in/help/pufas.
Introduction
Advances in sequencing technologies have opened up unlimited opportunities for better understanding of the molecular events occurring spatially and temporally during the growth and development of an organism. Large volumes of genomic and transcriptomic information have been developed and a broad spectrum of bioinformatic tools as well as experimental strategies have been adopted for their annotation. However, linking such huge gene sequence information with a biological meaning remains a challenge, leaving behind a major portion of the identified proteins as proteins of unknown function (PUFs) in public databases. About 16 and 30% of proteins are unannotated in bacteria and yeast genomes [1][2]. In eukaryotes, over 40% of the proteins encoded by genomes is reported to lack functional annotation [3][4]. In a model plant system Arabidopsis thaliana, approximately 30–34% of the total genome is composed of PUFs [5]. Several attempts have been reported in diverse organisms, to uncover the biological role of PUFs, enumerating their functional significance in growth, development, survival and response to adverse environmental conditions [6][7][8]. There is a need to assign function to PUFs for prospecting interesting genes until then, our understanding on the complexities in the growth and development of an organism and its interaction with the biotic and abiotic environment remains unclear. The functional annotation of all the PUFs based on laboratory experiments would be time consuming and expensive. Hence, several bioinformatic tools focusing on sequence similarity, co-expression, interactions, protein structures etc., have been widely used [9][3][10][11]. However, as evident from the prominent existence of PUFs in the genomes of many organisms, high throughput pipelines and methodologies to rapidly annotate PUFs and to elucidate their biological roles would be useful. In this study, we are reporting a strategy to enumerate the functions of the PUFs generated through any sequencing platforms.
We attempted to annotate the PUFs identified from expressed sequence tag (EST) library of mulberry leaf tissue exposed to drought stress. The preliminary functional annotation of the library yielded diverse proteins of known functions (PKFs), where as several genes have been identified as PUFs. For analysis of PUFs, we developed a pipeline using freely available bio-informatics tools and attempted to assign putative functions to many mulberry PUFs. Further, for rapid and high-throughput annotation of PUFs, we developed an automated pipeline and tested its application. We also examined the relevance of three annotated PUFs by in-vivo gene expression in mulberry. The stress-responsive PUFs identified in this study could be subjected for further functional characterization to elucidate their significance in plant growth and development, as well as in abiotic stress tolerance. This approach for the annotation of PUFs would be useful in assigning functions to many uncharacterized proteins identified from diverse transcriptome and genome datasets, irrespective of species of the organism and their growth, development and environmental conditions.
Materials and Methods
Ethics statement: In this study, we used mulberry (Morus alba L.) genotype, Dudia white, which is being maintained in the Department of Crop Physiology, University of Agricultural Sciences, GKVK, Bengaluru, India. There is no need of formal approval for this type of study, as the research is carried out in a public sector (non-profit) organization, and the study does not involve any genetic modification of the genotype used.
Plant material, RNA extraction, cDNA library preparation and sequencing
Stem cuttings of mulberry (Morus alba L.) genotype, Dudia white were used to generate healthy plants in pots (30kg capacity) filled with potting mixture. The plants were grown in the garden of the Department of Crop Physiology, University of Agricultural Sciences, GKVK, Bengaluru, India. Two months old healthy plants were subjected to different levels of drought stress (70–80% mild, 55–65% moderate and 40–50% severe) at soil field capacity (FC), imposed by gravimetric approach [12].
Leaf tissue was collected from the plants experiencing different levels of drought stress and total RNA was isolated according to a modified protocol [13]. From the total RNA, messenger RNA (mRNA) was extracted using mRNA isolation kit (Oligotex mRNA Mini kit Qiagen, CA, USA) and equal amounts of mRNA were pooled. The mRNA enriched fraction was converted to 454 barcoded cDNA library as reported [14]. In brief, from mRNA (200ng), cDNA was synthesized using cDNA synthesis kit (SuperScript Double-Stranded cDNA Synthesis Kit, Invitrogen, CA, USA) with 100mM random hexamer primers (Fermentas, USA). The double-stranded cDNA synthesized was purified and nebulized using kit supplied with the GS Titanium Library Preparation kit (454 Life Sciences, CT, USA) following their recommendations (30psi for 1minute) and purified with a QIAquick PCR minelute column (Qiagen, CA, USA) and eluted in 50μL elution buffer (EB). The sample library prepared was analysed using a Qubit fluorometer (Invitrogen, CA) and average fragment sizes were determined on the Bioanalyzer (Agilent, CA, USA). The process of library preparation, emulsion-based clonal amplification and sequencing on the 454 Genome Sequencer FLX Titanium system were performed according to the manufacturer’s instructions (454 Life Sciences, CT, USA; M/s. Sasya Gentech, Bangalore, India). Signal processing and base calling were performed using bundled 454 data analysis software v2.6.
De novo assembly and annotation
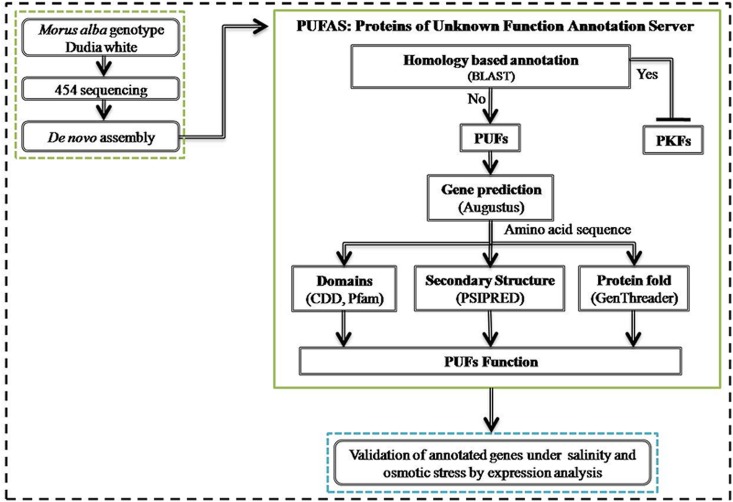
The DNA sequences obtained were processed and contigs were assembled using de novo Roche 454’s Newbler from a non-normalized mulberry cDNA library [14]. The transcriptome data was submitted to the National Center for Biotechnology Information’s (NCBI) Sequence Read Archive (SRA) with the study accession number of SRP047446. The contigs were annotated using blastx against NCBI-nr (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and broadly classified as PKFs and PUFs. The PUFs were selected for function prediction by bioinformatic approaches and, the randomly selected PUFs were used for experimental validation. The schematic representation of the events followed to process the ESTs is depicted in Fig 1.
Fig 1. The schematic representation of different events staring from library generation by 454 sequencing, assembly, annotation and other analysis performed.
Gene prediction from contigs
The gene prediction was carried out using available online tools like Softberry’s HMM-based ab-initio gene structure prediction by FGENESH [15] with Populus trichocarpa as reference genome and AUGUSTUS (A. thaliana), a HMM-based eukaryotic gene prediction server [16] and the longest gene length prediction was accepted as the gene model.
Computational annotation of PUFs
Searches were performed in Pfam database [17] and Conserved Domain Database (CDD) of the NCBI [18] for annotating the PUFs for the presence of domains and protein family in the targeted protein sequences. Fold prediction tools, like GenTHREADER [19], PHYRE2 [20] and 3DPSSM [21] were used to predict the compatible folds and associate to the function.
Development of Server for Function Annotation of Proteins of Unknown Function (PUFAS)
PUFAS web interface is developed using Javascript, HTML and CSS. Input for PUFAS is contigs, from that it predicts the possible function of an input sequence. In PUFAS, analysis was performed using the NCBI blastx for finding the homologous. Based on the preliminary annotation, the PUFs were further taken for gene prediction using AUGUSTUS, and from this the amino acid sequence was used as an input for tools like Pfam and CDD for identification of domains associated with query sequence. The GenThreader was used for the fold prediction and all these tools implemented with options to choose user-defined statistics values. Function could be assigned on the basis of the predicted domains and fold. User can download the output as a batch file.
Phylogenetic analysis
To reveal the divergence of one of the unknown genes, PUF39, in other plant genomes, a BLAST search was performed to identify the homologous genes (blastp with default parameters) against the NCBI-nr database. All hits below an E-value of 0.001 were retrieved as homologous sequences from other genomes in the GenBank database. Multiple sequence alignment was performed using ClustalW, and the alignment edited manually, tree was constructed using neighbour-joining (NJ) method in MEGA5.0 at a bootstrap value of 1000 [22].
Protein-protein interactions
Selected PUFs were queried for protein-protein interactions using the STRING database by applying a conservative score threshold of 0.7 [23].
Expression analysis of selected PUFs
Stress treatments
To study the expression pattern of the selected PUFs under other abiotic stresses, experiments were conducted under controlled laboratory conditions. Salinity and oxidative stresses were simulated by exposing the freshly collected intact twigs of mulberry to 250mM sodium chloride (NaCl) and 15μM methyl viologen (MV), respectively. Leaf tissues were collected at 6, 12, 24 and 48 hours after the stress imposition, while water treated twigs served as control.
Quantitative Real-Time PCR (qRT-PCR)
The total RNA was isolated from 100mg of the leaf tissue collected from the respective stress treatments according to a modified protocol [13]. About 3μg of total RNA was reverse transcribed to synthesize first strand cDNA using the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, USA). The cDNA was used as the template for expression analysis and the house keeping gene, elongation factor (elf) was used as the internal control. The qRT-PCR was performed in a q-PCR machine (Opticon2, MJ Research, USA), with the fluorescent dye SYBR-green (SYBR Premix Ex Taq, Perfect Real Time, Takara, Japan) under standardized PCR conditions using target specific primers as listed (S1 File). The relative transcript level was calculated from three independent replications; calculated using comparative ΔCt method [24] and student t-test was performed (p = 0.05).
Results
Sequencing and annotation of the drought specific transcriptome of mulberry
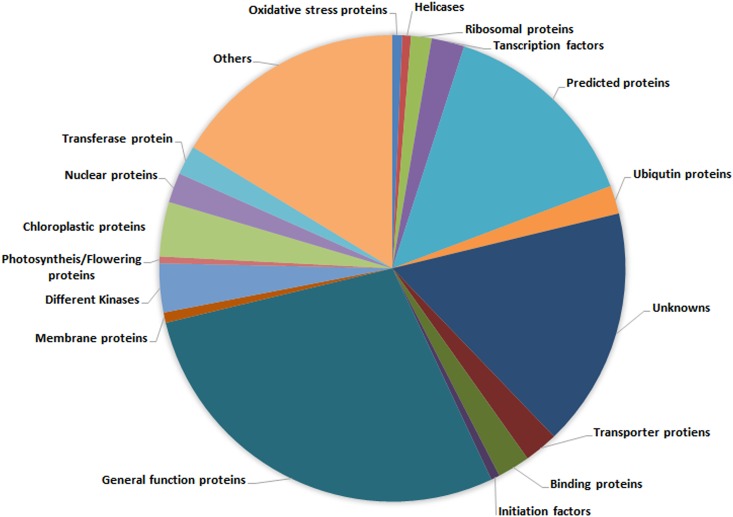
The cDNA library developed from drought stressed leaf tissues of mulberry yielded 10,190 ESTs. As a preliminary stage of library analysis, all the ESTs were searched at NCBI against nr database with a stringent E-value of 1e-5, from which 5319 ESTs were annotated and classified into PKFs and PUFs. The PKFs belong to various functional as well as regulatory proteins such as kinases, ribosomal proteins, membrane proteins, transporters, transcription factors (TFs), etc. (Fig 2). Detailed GO annotation information is provided as S2 File. The remaining ESTs were annotated as uncharacterized, hypothetical and unknown proteins which we considered as PUFs, as they lacked an experimental backup for function prediction. In our study, which was initiated in January 2014, we considered some of the PUFs, which were above 500bp, for functional annotation.
Fig 2. Distribution of selected GO terms based on NCBI-BLAST analysis of mulberry transcriptome generated from drought stressed leaf tissue.
Gene prediction from contigs
In order to confirm the gene boundaries, we employed FGENESH and AUGUSTUS, the prediction which gave longest open reading frame (ORF) were selected for further annotation and experimental study (S3 File).
Annotation of PUFs using computational tools
Sequence and structure based computational tools were utilized for the annotation of PUFs. Pfam and CDD search could identify the domains and families present in selected PUFs. The sequence analyses and domain association for all PUFs considered in the study are presented (S3 File). Following this, PUFs were also subjected to structure based functional analysis tools (S3 File). Integrating the sequence and structure based analysis, gene functions were predicted and data generated on some of the PUFs are presented in Table 1. For high throughput and rapid annotation of PUFs, we developed a web server PUFAS and implemented a RESTFul web service API for the programmatic access to the tool that can enable large-scale annotation of PUFs from a genome.
Table 1. Annotated PUFs using the computational tools.
| Name of the contig | Annotation from NCBI | Predicted function |
|---|---|---|
| contig00286 | Unknown | Thioredoxin like protein |
| contig00529 | Unknown | ACT domain containing protein |
| contig01194 | Unknown | Universal stress protein like |
| contig01391 | Unknown | Ankyrin repeat protein |
| contig01796 | Unknown | Transferase like |
| contig01963 | Unknown | CBS domain containing protein |
| contig02017 | Unknown | GroEL-like chaperone |
| contig02101 | Unknown | Oxidoreductase like |
| contig02328 | Unknown | RNA binding protein |
| contig02670 | Unknown | Aldo-keto reductase like |
| contig04385 | Unknown | Major intrinsic protein |
| contig04437 | Unknown | Transketolase like |
| contig04699 | Unknown | GDP-fucose protein O-fucosyltransferase |
| contig04820 | Unknown | Aldehyde reductase |
| contig04823 | Unknown | GDP-fucose protein O-fucosyltransferase |
| contig04921 | Unknown | Ankyrin repeat protein |
| contig5180 | Unknown | Dehydrogenase like |
| contig05224 | Unknown | Protein phosphatase like |
| contig05320 | Unknown | Dehydrogenase like |
| contig05347 | Unknown | Ferritin like protein |
| contig05421 | Unknown | Dehydrogensase/ reductase |
| contig05454 | Unknown | Late embryogenesis abundant protein like |
| contig05496 | Unknown | Acyl esterase like |
| contig05505 | Unknown | Metallophosphatase |
| contig05537 | Unknown | Ubiquitin like protein |
| contig05592 | Unknown | Dehydrogenase like |
| contig05650 | Unknown | ATP synthase |
| contig05740 | Unknown | Transferase like |
| contig05864 | Unknown | Hydrolase like |
| contig05991 | Unknown | TPR protein |
| contig06030 | Unknown | Sulphite exporter like |
| contig06333 | Unknown | Chlorophyll a-b binding protein |
| contig06433 | Unknown | Chlorophyll a-b binding protein |
| contig06595 | Unknown | Trios phosphate isomerase |
| contig06596 | Unknown | Rab5 like protein |
| contig06640 | Unknown | Phosphoglucomutase |
| contig06735 | Unknown | PLATZ like transcription factor |
| contig06750 | Unknown | Elongation factor protein |
| contig06773 | Unknown | Protein kinase like |
| contig06932 | Unknown | RNA binding protein like |
| contig07145 | Unknown | Rab like protein |
| contig07540 | Unknown | Dormancy/ auxin associated protein |
| contig07570 | Unknown | Ras related protein |
| contig07599 | Unknown | Cytochrome c oxidase |
| contig07639 | Unknown | Proteosome regulatory complex subunit like |
| contig08042 | Unknown | Aldolase like |
| contig08184 | Unknown | Ubiquitin conjugating enzyme like |
| contig08330 | Unknown | Rab like protein |
| contig08474 | Unknown | Ribosomal protein like |
| contig08640 | Unknown | Hydrolase like |
| contig08772 | Unknown | Metal transport protein |
| contig08856 | Unknown | Pholem protein like |
| contig08939 | Unknown | Ubiquinol-cytochrome c reductase complex subunit like |
| contig09315 | Unknown | Dehalogenase like |
| contig09355 | Unknown | Elongation factor like protein |
| contig09421 | Unknown | Ribosomal protein like |
| contig00062 | Hypothetical | Metallothionein like |
| contig00355 | Hypothetical | Pepsin like |
| contig00754 | Hypothetical | Ca binding epidermal growth factor like protein |
| contig01002 | Hypothetical | Late embryogenesis abundunt protein like |
| contig01413 | Hypothetical | Heavy metal binding protein like |
| contig01734 | Hypothetical | Leucine rich repeat protein |
| contig01852 | Hypothetical | Esterase like |
| contig01866 | Hypothetical | KH RNA binding protein |
| contig01896 | Hypothetical | Protein kinase like |
| contig01986 | Hypothetical | Armadillo repeat protein |
| contig02310 | Hypothetical | Perforin like |
| contig02336 | Hypothetical | Ribosomal protein like |
| contig02443 | Hypothetical | SKP1 like protein |
| contig02598 | Hypothetical | Dehydrogense lke |
| contig02665 | Hypothetical | Hydrolase like |
| contig02872 | Hypothetical | Plant retinoblastoma associated protein |
| contig03421 | Hypothetical | Transport protein like |
| contig03453 | Hypothetical | Phosphoglucomutase |
| contig04442 | Hypothetical | WD repeat protein |
| contig04444 | Hypothetical | Acid protease like |
| contig04469 | Hypothetical | SART 1 family protein |
| contig04481 | Hypothetical | Tubulin associated protein |
| contig04536 | Hypothetical | Myb like protein |
| contig04717 | Hypothetical | Transporter like |
| contig04834 | Hypothetical | Vacuolar sorting associated protein |
| contig04885 | Hypothetical | PH domain containing protein |
| contig04956 | Hypothetical | Transferase like |
| contig04977 | Hypothetical | Phosphoprotein like |
| contig00200 | Uncharacterized | DNA binding protein |
| contig00467 | Uncharacterized | Late embryogenesis abundant protein like |
| contig00638 | Uncharacterized | Transferase like |
| contig00917 | Uncharacterized | Ribosomal protein like |
| contig01190 | Uncharacterized | Zinc finger protein |
| contig01205 | Uncharacterized | Transferase like |
| contig01222 | Uncharacterized | Pseudouridine synthase like |
| contig01234 | Uncharacterized | Transferase like |
| contig01406 | Uncharacterized | Peroxidase like |
| contig01408 | Uncharacterized | Peroxidase like |
| contig01644 | Uncharacterized | Hydrolase like |
| contig01646 | Uncharacterized | Transferase like |
| contig01671 | Uncharacterized | RNA binding protein |
| contig01710 | Uncharacterized | Hydrolase like |
| contig01711 | Uncharacterized | Hydrolase like |
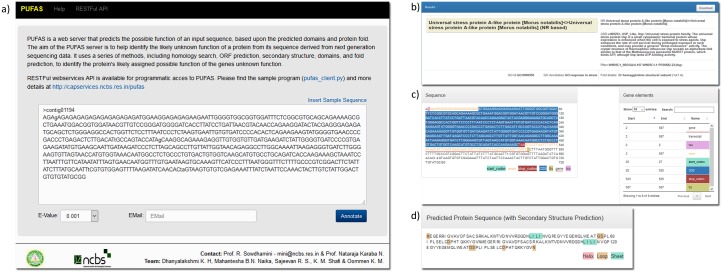
PUFAS provides a web platform for performing PUFs analysis of next generation sequencing data. A user can submit a single or list of contigs from genomics or transcriptomics experiments, and can select statistical values to perform the annotation analyses. A successful PUFAS run provides batch file contains predicted gene boundaries, associated domains, secondary structure and fold predicted. The downloadable files can be used to filter significance level association of annotation based on user need. The automated server showed similar results as in case of manual annotation. This has been illustrated with the help of one of the PUFs from our study (Fig 3a–3d). The web service API enables the programmatic access of the tool, submit the sequence and save the result on the local desktop or laptop independent of the geographical location, the programming language or the computer platform. Sample python program to access the PUFAS web service is available in the website and as S4 File.
Fig 3. Web interface of PUFAS.
a) Option to input of contigs identified from a genome by using next-generation sequencing, b) Output of Pfam and CDD, c) Gene identification and its elements, d) Amino acid sequence of a given contig.
From the analysis, the result of three most promising PUFs, where domain association was strong, is presented in detail (Table 2). The stress-responsive nature of the selected genes was validated by qRT-PCR. As per the sequence and fold predictions, one of the genes, the PUF3 is predicted to have an adenine nucleotide-binding domain-like fold and belongs to the USP-like protein family. PUF39 is predicted to be a PLATZ1-like TF having a plant AT-rich binding region by sequence-based domain association studies while the structure based analysis could not derive an annotation, within acceptable limits of E-value. PUF42 was predicted to be an RNA-binding protein with a conserved RRM1 motif, but did not find a significant fold in the 3DPSSM and GenTHREADER. However, using PHYRE2, we could associate this protein to retain an RNA binding domain with coverage of 56% of the amino acid residues with 99.8% confidence. The salient results of these three PUFs are presented in Table 2.
Table 2. Sequence and structure based annotation of selected PUFs.
| Contig ID | PUFs | Sequence based annotation | Structure based annotation | Annotation | |||
|---|---|---|---|---|---|---|---|
| CDD | PFam | 3DPSSM | PHYRE | GenTHREADER | |||
| Contig 01194 | PUF3 | Universal stress protein family | Universal stress protein family | ETFP adenine nucleotide-binding domain-like | Adenine nucleotide alpha hydrolase-like | Adenine nucleotide alpha hydrolase-like | MaUSP-like |
| Contig 06735 | PUF39 | PLATZ1 transcription factor | PLATZ1 transcription factor | Immunoglobulin-like beta-sandwich | Gene regulation, hydrolase | DNA clamp | MaPLATZ1- like |
| Contig 06932 | PUF42 | RNA recognition motif | RNA recognition motif | No Prediction | RNA binding protein | Ferredoxin-like | MaRRM1- like |
Phylogenetic analysis
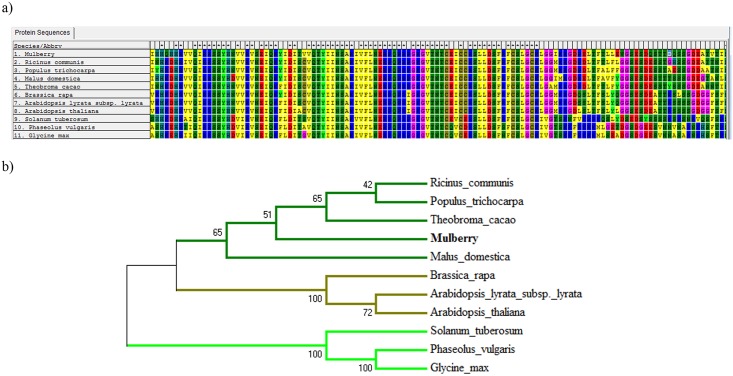
Computational methods to analyze the phylogenetic relationships have been instrumental in annotation of protein functions [25] for which MEGA5.0 was used. We extracted protein sequence from the genomes having homologues to one of the mulberry PUFs i.e., PUF39 to understand the features of the similar genes and sequence conservation in other genomes (Fig 4a) and an unrooted phylogenetic tree was constructed (Fig 4b). The average sequence identity among species is 53% and the PUF39 was observed to cluster with other tree species; notably homologues from Leguminace and Brassicacae family members clustered together (Fig 4b).
Fig 4. Phylogenetic analysis of selected PUF.
a) Multiple sequence alignment of mulberry PUF39 with homologous genes from other plant genomes. b) Unrooted phylogenetic tree of PUF39. The Genbank accession numbers of the sequences are Ricinus communis (XP_002510750.1), Populus trichocarpa (XP_002301890.1), Malus domestica (XP_008381650.1), Theobroma cacao (XP_007018633.1), Brassica rapa (XP_009149581.1), Arabidopsis lyrata subsp. Lyrata (XP_002890421.1), Arabidopsis thaliana (NP_001117322.1), Solanum tuberosum (XP_006352862.1), Phaseolus vulgaris (XP_007131503.1) and Glycine max (XP_003540084.1).
Expression analysis
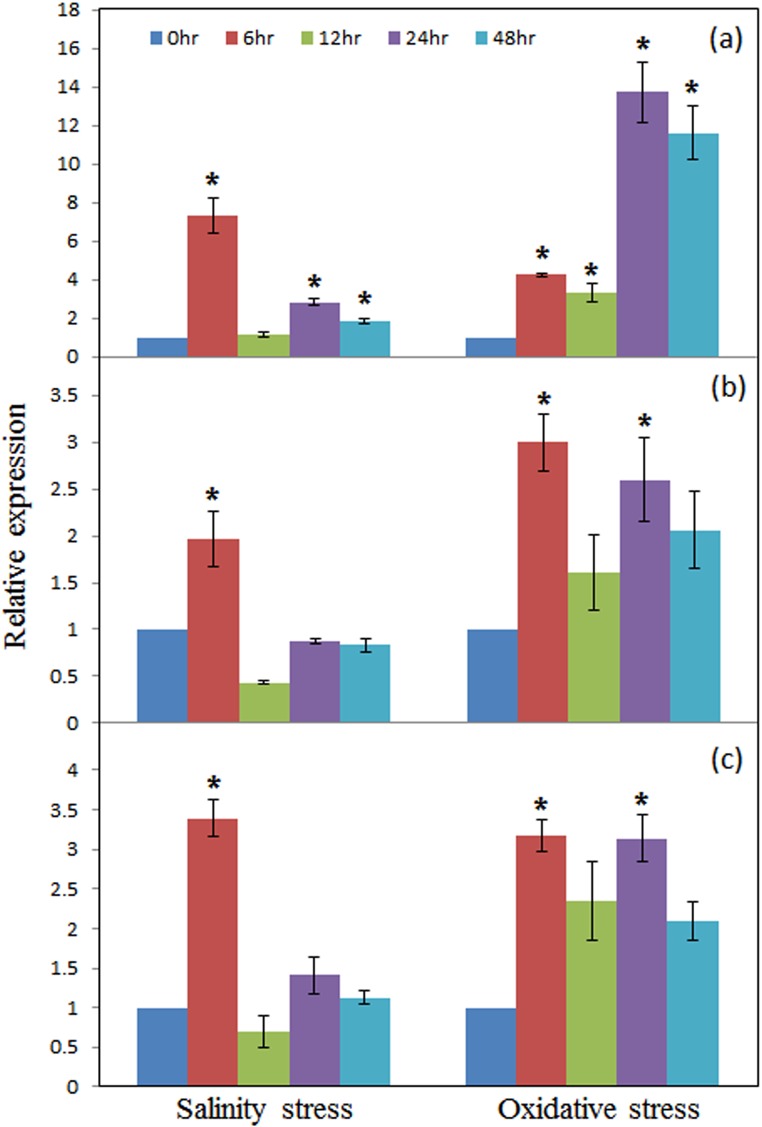
Analysis of gene expression is one of the most important approaches to highlight the functional aspect of genes. Expression pattern of the above discussed computationally annotated PUFs identified from drought specific transcriptome were studied under other abiotic stresses. Significant increase (p = 0.05) in transcript levels of PUF3 was observed under NaCl induced salt stress as well as methyl viologen induced oxidative stress (Fig 5a). The relative transcript level of PUF39 (designated as MaPLATZ1-like protein) was significantly up-regulated at six hours under salinity as well as oxidative stress (Fig 5b), followed by down-regulation during subsequent exposure to stress suggesting that the gene is early stress-responsive. The relative expression levels of MaRRM1-like genes studied under simulated salinity and oxidative stress conditions indicated significant increase at six hours after stress imposition (Fig 5c).
Fig 5. Expression analysis of selected PUFs by qRT-PCR in leaf tissue of mulberry genotype, Dudia white.
Relative transcript levels of MaUSP-like (a), MaPLATZ1-like (b), and MaRRM1-like (c) genes under salinity and oxidative stresses. Total RNA was isolated from the leaf tissues of mulberry twigs exposed to 250mM NaCl (salinity stress) and 15μM methyl viologen (oxidative stress) at 6, 12, 24, and 48 hours after stress imposition. Transcripts were normalized to the expression of the elongation factor gene. The data shown are mean ± SE from three independent experiments. Asterisk indicates the significant difference between control and treatments at p = 0.05.
Protein-protein interactions
Functional role of proteins can also be inferred by their interactions in biological networks. We derived the information from STRING database to support functional annotation using the closely related Arabidopsis proteins. USP-like protein from A. thaliana (AT3G17020) shares 69% identity with that of the MaUSP-like protein, and AT3G17020 has been shown to interact with AT3G48750 (cyclin-dependent kinase-A CDKA-1), well known for their crucial role in cell cycle regulation (S1a Fig). MaPLATZ1-like protein from Arabidopsis (AT1G21000) interacts with AT1G72830 (NF-YA3) (S1b Fig). The mulberry MaRRM-like protein has 65% identity with the A. thaliana gene AT4G17720. The A. thaliana protein has been reported to have direct interaction with AT5G58750 (wound responsive protein related), AT5G58690 (Phosphoinositide specific phospholipase C family protein), AT3G46060 (ATRAB8A), AT3G311730 (ATFP8), AT1G56330 (ATSAR1B), AT1G74620 (GAMMA CA2) based on experimental evidence and also with the genes like AT4G25680 and AT4G25660 (S1c Fig).
Discussion
As a result of the revolutionary expansion in NGS technologies, a large volume of data in genome and transcriptome level has been developed, while their interpretation and generation of biological significance has been a challenge. Since, PUFs have come up as a significant portion of many genomes, we assume that their functional annotation can reveal the presence of novel candidate genes associated with growth and developmental pathways. We made an attempt to address the functional annotation of PUFs, by developing a pipeline and validating it using the drought stressed leaf transcriptome of mulberry, as an example. The pipeline involves an initial screening of the ESTs into known and unknown function against NCBI-BLAST analysis with a stringent cut off value, followed by functional annotation using computational tools.
Pipeline of annotation
All in silico tools used in our pipeline (Table 3) are publicly accessible. The sequence analysis tools are efficient in identifying the presence of conserved domains, if any, in the given sequence which could be correlated to the probable function of the gene. In addition to this, the results predicted by the fold recognition tools could improve our annotation. The phylogenetic analysis as well as protein-protein interaction studies provide additional support to our function prediction. Several attempts have been made for the annotation of proteins lacking experimental supports [26][27], utilizing online computational tools in diverse organisms. Here we propose that, in addition to the in silico annotation, the functional relevance of annotated PUFs could be better understood, if they are taken forward to simple laboratory level experimental setup. Hence, our approach holds an improvisation over the existing ones by analyzing their expression pattern too. The expression pattern of the PUFs derived from a drought stressed library, analyzed under multiple abiotic stress conditions suggests they are stress responsive and hence may have a role in stress acclimation in mulberry.
Table 3. Tools used in the study.
| Sl.No | Tool | Purpose | URL |
|---|---|---|---|
| I. Transcriptome assembly | |||
| 1 | Newbler | De novo assembly | - |
| II. Annotation tool | |||
| 1 | NCBI BLAST | Preliminary annotation of transcripts | www.ncbi.nlm.nih.gov/ |
| III. Gene prediction | |||
| 1 | FGENESH | Prediction of open reading frame | http://www.softberry.com/berry.phtml?topic=fgenesh&group=programs&subgroup=gfind |
| 2 | AUGUSTUS | Prediction of open reading frame | http://bioinf.uni-greifswald.de/augustus/submission.php |
| IV. Sequence analysis | |||
| 1 | CDD | Identification of conserved domain | http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi |
| 2 | Pfam | Protein family classification | http://pfam.xfam.org/search |
| V. Fold analysis | |||
| 1 | PSIPRED | Fold recognition | http://bioinf.cs.ucl.ac.uk/psipred/ |
| 2 | PHYRE2 | Fold recognition | http://www.sbg.bio.ic.ac.uk/phyre2/ |
| 3 | GenTHREADER | Fold recognition | http://bioinf.cs.ucl.ac.uk/psipred/ |
| VI. Gene Ontology | |||
| 1 | Gene Ontology | Annotation | http://geneontology.org/ |
| VII. Phylogenetic analysis | |||
| 1 | MEGA5 | http://www.megasoftware.net/ | |
| VIII. Protein-protein interaction | |||
| 1 | STRING | To find out interacting partners | http://string-db.org/ |
| IX. Expression analysis | |||
| 1 | qRT-PCR | To analyze the functional significance | - |
Annotation of PUFs using computational tools and development of PUFAS
Generally, annotation of proteins has been approached in various directions using existing computational tools. One of the simplest methods followed is relating the protein to sequence conservation and domain association. The presence of a conserved region or functional domain could be an indication of the probable functional role of the protein [28]. The PUFs that have at least one previously defined motif or domain can be called as proteins of defined features (PDFs) [28][29][4]. Fold recognition methods like threading and hybrid threading/sequence fold recognition have been widely used in assigning functions to proteins as they can often recognize even the most distant homologues. In some cases, even distantly related proteins with similar structure could be identified [30]. Hence, in addition to the sequence based prediction, we used fold recognition tools like GenTHREADER, PHYRE2 and 3DPSSM which have been extensively used in other studies [31][32][33]. We also adopted Gene Ontology (GO) annotations for identifying the function of proteins, as GO is known for giving hints to the function at various levels [3][34]. By integrating these sequence and structure based approaches we could annotate some of the PUFs from the drought specific transcriptome of mulberry (Table 1). As per our annotation, the PUFs belong to various structural/functional as well as regulatory proteins like enzymes, chaperons, signaling molecules, ribosomal proteins, TFs, etc. The PUFAS server is capable of processing the PUFs with satisfactory output in a user friendly way. Although, there are pipelines and automated servers available, which rely on well defined protein sequences [35][36], PUFAS has additional features required for NGS data analysis. The server can accept outputs from various NGS platforms, process and predict gene function.
Analysis of expression pattern of the selected PUFs
We tried to highlight the relevance of the annotated PUFs by a random selection of three genes, one belonging to category of regulatory protein (MaRRM1-like), another upstream TF (MaPLATZ1-like) and third downstream functional protein (MaUSP-like) for expression analysis by qRT-PCR. From other reports, USP domain containing proteins have been up-regulated under different stress signals in plants [37]. In Arabidopsis, there are 44 putative homologues of USPs which are either ATP binding or non-ATP binding type [38] and the exact function of these proteins are yet to be known. PUF3 annotated as MaUSP-like protein, which was significantly up-regulated under multiple stresses in our study could be one of the potential candidates for imparting cellular level tolerance to abiotic stress in mulberry. PLATZ1 proteins are zinc dependent DNA binding proteins binding to AT rich regions of nucleotide sequence to bring about transcriptional repression with a possible involvement in cell cycle regulation [39]. Earlier studies report the involvement of DNA binding PLATZ1-like TF in embryo development [40] and tendril and inflorescence development [41]. In addition to the above, we propose that this PLATZ1-like gene identified from mulberry has a role in abiotic stress response, which need to be tested furthur. The PUF42 has a conserved RNA recognition motif, RRM1 which is one of the most common characteristic features of RNA binding proteins (RBPs) in plants. A wide variety of roles have been implicated for RBPs involved in abiotic stress-response in plants [42][43]. Light, salinity and abscisic acid are known to induce rapid alteration of the RBPs [41][44] as well as modulate the stress induced gene expression within minutes to hours after stress imposition [44] suggesting their involvement in early stress response. Similar to these, MaRRM1-like is suggested be a stress-responsive protein that might modify the stress response of the mulberry plants through mechanisms that are yet to be studied.
Large-scale analysis of PUFs and testing of PUFAS
We selected 100 transcript sequences of known functions from A. thaliana and performed a test using the tool, PUFAS. A script, that can access the PUFAS API service, was used to perform the analysis. The results from the server were compared against the known function and identified that PUFAS could predict the same function in 93% cases (S5 File). This proves that the PUFAS API Service and the web interface can be of a great help in annotating the entire transcriptome of an organism.
Prediction of protein-protein interactions
Protein-protein interaction is the hallmark of all living organisms [45]. More than 70% of the physically interacting proteins share similar functional annotations [46] suggesting their joint recruitment in a biological function. Hence, the functional role of proteins can also be revealed by the study of its interactions in a biological network and can be used for providing leads to functional roles of the PUFs [27]. In Arabidopsis, the interacting partners of USP-like protein (PUF3) i.e., CDKs have also been well related to stress perception and responses in plants there by regulating their strategies for growth, development and adaptation under biotic and abiotic cues [47][48]. Our analysis suggests that the MaUSP-like protein can be possibly associated with mechanisms that are related to cell cycle regulation during stress response. Recent reports suggest that NF-Y TFs have been interacting with other TF families also to influence various plant responses [49]. In this view, the MaPLATZ1-like protein from mulberry could also be a possible interacting partner for NF-YA3. The RRM1-like protein (PUF42) from A. thaliana has been associated with multiple pathways in growth, development and stress-response. The interaction study was attempted only to get a lead towards the probable function. However, there is a need to confirm the interactions.
Conclusion
The strategies used in our pipeline to annotate PUFs are simple and can be used by a wide spectrum of computational as well as experimental biologists. In the present study, sequence analysis and structure-based fold predictions have been used to uncover putative biochemical functions of a few hitherto uncharacterized genes of interest. The associated biochemical functions and domains could be extended to assign possible biological functions by deriving knowledge of their homologues in other model organisms. The study of PUFs can answer some of the unanswered questions regarding the interacting partners of many proteins in many biological systems and hence can reveal additional players in the transcriptional and translational regulatory events at molecular level. The approach followed in this study can pave way for validation of many PUFs in diverse organisms. The ultimate test would be to functionally validate the selected PUFs in model or test systems by down-regulation or over expression studies. The advantage of the approach is that all the computational tools used in this study are freely accessible. The strategy demonstrated would also be adopted for annotation of PUFs at whole genome level across diverse organisms.
Supporting Information
a) Protein-protein interaction between AT3G17020 and CDC2. b) Protein-protein interaction between AT1G2100 and NF-YA3. c) Protein-protein interaction between AT4G17720 and AT5G58750, AT5G58690 (Phosphoinositide specific phospholipase C family protein), AT3G46060 (ATRAB8A), AT3G311730 (ATFP8), AT1G56330 (ATSAR1B), AT1G74620 (GAMMA CA2), AT4G25680 and AT4G25660.
(TIF)
(XLSX)
(XLSX)
(XLSX)
(PY)
(XLS)
Acknowledgments
The research is supported by the Department of Biotechnology, Government of India grant No. BT/PR13457/PBD/19/214/2010 to KNN. KHD would like to thank Department of Science and Technology, (DST), Government of India for DST-INSPIRE Fellowship. MBNN and RS thank the National Center for Biological Sciences (NCBS), Bengaluru for infrastructure, MBNN also thank NCBS for Bridge Postdoctoral Fellowship. KNN would like to thank Department of Science and Technology-Fund for Improvement of Science and Technology (DST-FIST) for the infrastructure facilities. KHD, MBNN, OKM, KMS, RSS and KNN thank NCBS and University of Agricultural Sciences (UAS), Bengaluru for the general facilities.
Data Availability
All relevant data are within the paper and its Supporting Information files. Transcriptome raw data is available from the NCBI-SRA database (accession number SRP047446).
Funding Statement
The research is supported by the Department of Biotechnology, Government of India grant no. BT/PR13457/PBD/19/214/2010 to KNN. KHD would like to thank Department of Science and Technology (DST), Government of India for DST-INSPIRE Fellowship.
References
- 1.Pena-Castillo L, Hughes TR. Why are there still over 1000 uncharacterized yeast genes? Genetics. 2007;176: 7–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meier M, Sit RV, Quake SR. Proteome-wide protein interaction measurements of bacterial proteins of unknown function. Proc Natl Acad Sci U S A. 2013;110: 477–482. 10.1073/pnas.1210634110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Horan K, Jang C, Bailey-Serres J, Mittler R, Shelton C, Harper JF, et al. Annotating genes of known and unknown function by large-scale coexpression analysis. Plant Physiol. 2008;147: 41–57. 10.1104/pp.108.117366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luhua S, Ciftci-Yilmaz S, Harper J, Cushman J, Mittler R. Enhanced tolerance to oxidative stress in transgenic Arabidopsis plants expressing proteins of unknown function. Plant Physiol. 2008;148: 280–292. 10.1104/pp.108.124875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, Sasidharan R, et al. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 2012;40 (Database issue): D1202–10. 10.1093/nar/gkr1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guengerich FP, Tang Z, Salamanca-Pinzón SG, Cheng Q. Characterizing proteins of unknown function: orphan cytochrome p450 enzymes as a paradigm. Mol Interv. 2010;10: 153–163. 10.1124/mi.10.3.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pandey SP, Minesinger BK, Kumar J, Walker GC. A highly conserved protein of unknown function in Sinorhizobium meliloti affects sRNA regulation similar to Hfq. Nucleic Acids Res. 2011;39: 4691–4708. 10.1093/nar/gkr060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellison CE, Kowbel D, Glass NL, Taylor JW, Brem RB. Discovering functions of unannotated genes from a transcriptome survey of wild fungal isolates. MBio. 2014;5: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo F, Yang Y, Zhong J, Gao H, Khan L, Thompson DK, et al. Constructing gene co-expression networks and predicting functions of unknown genes by random matrix theory. BMC Bioinformatics. 2007;8: 299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doerks T, van Noort V, Minguez P, Bork P. Annotation of the M. tuberculosis hypothetical orfeome: Adding functional information to more than half of the uncharacterized proteins. PLoS One. 2012;7: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schuller A, Slater AW, Norambuena T, Cifuentes JJ, Almonacid LI, Melo F. Computer-based annotation of putative AraC/XylS-family transcription factors of known structure but unknown function. J Biomed Biotechnol. 2012. 10.1155/2012/103132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karaba A, Dixit S, Greco R, Aharoni A, Trijatmiko KR, Marsch-Martinez N, et al. Improvement of water use efficiency in rice by expression of HARDY, an Arabidopsis drought and salt tolerance gene. Proc Natl Acad Sci U S A. 2007;104: 15270–15275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sajeevan RS, Shivanna MB, Nataraja KN. An efficient protocol for total RNA isolation from healthy and stressed tissues of mulberry (Morus sp.) and other species: Am J Plant Sci. 2014; 2057–2065. [Google Scholar]
- 14.Lambert JD, Chan XY, Spiecker B, Sweet HC. Characterizing the embryonic transcriptome of the snail Ilyanassa. Integr Comp Biol. 2010;50: 768–777. 10.1093/icb/icq121 [DOI] [PubMed] [Google Scholar]
- 15.Solovyev V, Kosarev P, Seledsov I, Vorobyev D. Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol. 2006;77 Suppl 1: S10 1–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stanke M, Steinkamp R, Waack S, Morgenstern B. AUGUSTUS: a web server for gene finding in eukaryotes. Nucleic Acids Res. 2004;32: W309–W312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40: D290–D301. 10.1093/nar/gkr1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marchler-Bauer A, Zheng C, Chitsaz F, Derbyshire MK, Geer LY, Geer RC, et al. CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res. 2013;41: D348–D352. 10.1093/nar/gks1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones DT. GenTHREADER: An efficient and reliable protein fold recognition method for genomic sequences. J Mol Biol. 1999;287: 797–815. [DOI] [PubMed] [Google Scholar]
- 20.Bennett-lovsey RM, Herbert AD, Sternberg MJE, Kelley LA. Exploring the extremes of sequence/ structure space with ensemble fold recognition in the program Phyre. Proteins. 2008;70: 611–25. [DOI] [PubMed] [Google Scholar]
- 21.Kelley LA, Maccallum RM, Sternberg MJE. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J Mol Biol. 2000;299: 499–520. [DOI] [PubMed] [Google Scholar]
- 22.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28: 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.von-Mering C, Huynen M, Jaeggi D, Schmidt S, Bork P, Snel B. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31: 258–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3: 1101–1108. [DOI] [PubMed] [Google Scholar]
- 25.Wu F, Mueller LA, Crouzillat D, Pétiard V, Tanksley SD. Combining bioinformatics and phylogenetics to identify large sets of single-copy orthologous genes (COSII) for comparative, evolutionary and systematic studies: a test case in the euasterid plant clade. Genetics. 2006;174: 1407–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar K, Prakash A, Tasleem M, Islam A, Ahmad F, Hassan MI. Functional annotation of putative hypothetical proteins from Candida dubliniensis. Gene. 2014;543: 93–100. 10.1016/j.gene.2014.03.060 [DOI] [PubMed] [Google Scholar]
- 27.Shahbaaz M, Hassan MI, Ahmad F. Functional annotation of conserved hypothetical proteins from Haemophilus influenzae Rd KW20. PLoS One. 2013;8: e84263 10.1371/journal.pone.0084263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gollery M, Harper J, Cushman J, Mittler T, Girke T, Zhu J, et al. What makes species unique? The contribution of proteins with obscure features. Genome Biol. 2006;7: R57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gollery M, Harper J, Cushman J, Mittler T, Mittler R. POFs: what we don’t know can hurt us. Trends Plant Sci. 2007;12: 492–496. [DOI] [PubMed] [Google Scholar]
- 30.Ye Y, Godzik A. Database searching by flexible protein structure alignment. Protein Sci. 2004,13: 1841–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang C, Tesar C, Gu M, Babnigg G, Joachimiak A, Pokkuluri PR, et al. Extracytoplasmic PAS-like domains are common in signal transduction proteins. J Bacteriol. 2010;192: 1156–1159. 10.1128/JB.01508-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aguilera F, McDougall C, Degnan BM. Origin, evolution and classification of type-3 copper proteins: lineage-specific gene expansions and losses across the Metazoa. BMC Evol Biol. 2013;13: 96 10.1186/1471-2148-13-96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naika M, Shameer K, Sowdhamini R. Comparative analyses of stress-responsive genes in Arabidopsis thaliana: insight from genomic data mining, functional enrichment, pathway analysis and phenomics. Mol Biosyst. 2013;9: 1888–1908. 10.1039/c3mb70072k [DOI] [PubMed] [Google Scholar]
- 34.Xu Y, Zhou W, Zhou Y, Wu J, Zhou X. Transcriptome and Comparative Gene Expression Analysis of Sogatella furcifera (Horváth) in Response to Southern Rice Black-Streaked Dwarf Virus. PLoS One. 2012; 7: e36238 10.1371/journal.pone.0036238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nokso-koivisto J. PANNZER: high-throughput functional annotation of uncharacterized proteins in an error-prone environment. 2015;31: 1544–1552. [DOI] [PubMed] [Google Scholar]
- 36.Mckay T, Hart K, Horn A, Kessler H, Mills JL, Bardhi K, et al. Annotation of proteins of unknown function : initial enzyme results: 2015;43–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loukehaich R, Wang T, Ouyang B, Ziaf K, Li H, Zhang J, et al. SpUSP, an annexin-interacting universal stress protein, enhances drought tolerance in tomato. J Exp Bot. 2012;63: 5593–606. 10.1093/jxb/ers220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kerk D, Bulgrien J, Smith DW, Gribskov M. Arabidopsis proteins containing similarity to the universal stress protein domain of bacteria. Plant Physiol. 2003;131: 1209–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagano Y, Furuhashi H, Inaba T, Sasaki Y. A novel class of plant-specific zinc-dependent DNA-binding protein that binds to A/T-rich DNA sequences. Nucleic Acids Res. 2011;29: 4097–4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Vega-Bartol JJ, Simoes M, Lorenz WW, Rodrigues AS, Alba R, Dean JFD, et al. Transcriptomic analysis highlights epigenetic and transcriptional regulation during zygotic embryo development of Pinus pinaster. BMC Plant Biol. 2013;13: 123 10.1186/1471-2229-13-123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Díaz-Riquelme J, Martínez-Zapater JM, Carmona MJ. Transcriptional analysis of tendril and inflorescence development in grapevine (Vitis vinifera L.). PLoS One. 2014;9: e92339 10.1371/journal.pone.0092339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kwak KJ, Kim YO, Kang H. Characterization of transgenic Arabidopsis plants overexpressing GR-RBP4 under high salinity, dehydration, or cold stress. J Exp Bot. 2005;56: 3007–3016. [DOI] [PubMed] [Google Scholar]
- 43.Ambrosone A, Costa A, Leone A, Grillo S. Beyond transcription: RNA-binding proteins as emerging regulators of plant response to environmental constraints. Plant Sci. 2012;182: 12–18. 10.1016/j.plantsci.2011.02.004 [DOI] [PubMed] [Google Scholar]
- 44.Jiang J, Wang B, Shen Y, Wang H, Feng Q, Shi H. The Arabidopsis RNA binding protein with K homology motifs, SHINY1, interacts with the C-terminal domain phosphatase-like 1 (CPL1) to repress stress-inducible gene expression. PLoS Genet. 2013;9: e1003625 10.1371/journal.pgen.1003625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kelly W, Stumpf M. Protein-protein interactions: from global to local analyses. Curr Opin Biotechnol. 2008;19: 396–403. 10.1016/j.copbio.2008.06.010 [DOI] [PubMed] [Google Scholar]
- 46.Mayer ML, Hieter P. Protein networks-built by association. Nat Biotechnol. 2000;18: 1242–1243. [DOI] [PubMed] [Google Scholar]
- 47.Schuppler U, He P, John P, Munns R. Effect of water stress on cell division and cell-division-cycle 2-like cell-cycle kinase activity in wheat leaves. Plant Physiol. 1998;117: 667–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kitsios G, Doonan JH. Cyclin dependent protein kinases and stress responses in plants. Plant Signal Behav. 2011;6: 204–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu JX, Howell SH. bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell. 2010;22: 782–96. 10.1105/tpc.109.072173 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
a) Protein-protein interaction between AT3G17020 and CDC2. b) Protein-protein interaction between AT1G2100 and NF-YA3. c) Protein-protein interaction between AT4G17720 and AT5G58750, AT5G58690 (Phosphoinositide specific phospholipase C family protein), AT3G46060 (ATRAB8A), AT3G311730 (ATFP8), AT1G56330 (ATSAR1B), AT1G74620 (GAMMA CA2), AT4G25680 and AT4G25660.
(TIF)
(XLSX)
(XLSX)
(XLSX)
(PY)
(XLS)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. Transcriptome raw data is available from the NCBI-SRA database (accession number SRP047446).