Abstract
Background
Ivory Coast is a West African country with the highest reported cases of Buruli ulcer, a disabling subcutaneous infection due to Mycobacterium ulcerans. However, the prevalence of environmental M. ulcerans is poorly known in this country.
Methods
We collected 496 environmental specimens consisting of soil (n = 100), stagnant water (n = 200), plants (n = 100) and animal feces (n = 96) in Ivory Coast over five months in the dry and wet seasons in regions which are free of Buruli ulcer (control group A; 250 specimens) and in regions where the Buruli ulcer is endemic (group B; 246 specimens). After appropriate total DNA extraction incorporating an internal control, the M. ulcerans IS2404 and KR-B gene were amplified by real-time PCR in samples. In parallel, a calibration curve was done for M. ulcerans Agy99 IS2404 and KR-B gene.
Results
Of 460 samples free of PCR inhibition, a positive real-time PCR detection of insertion sequence IS2404 and KR-B gene was observed in 1/230 specimens in control group A versus 9/230 specimens in group B (P = 0.02; Fisher exact test). Positive specimens comprised seven stagnant water specimens, two feces specimens confirmed to be of Thryonomys swinderianus (agouti) origin by real-time PCR of the cytb gene; and one soil specimen. Extrapolation from the calibration curves indicated low inoculums ranging from 1 to 102 mycobacteria/mL.
Conclusion
This study confirms the presence of M. ulcerans in the watery environment surrounding patients with Buruli ulcer in Ivory Coast. It suggests that the agouti, which is in close contacts with populations, could play a role in the environmental cycle of M. ulcerans, as previously suggested for the closely related possums in Australia.
Introduction
Ivory Coast is the country most affected by Buruli ulcer, a WHO-reportable disabling subcutaneous infection caused by to Mycobacterium ulcerans [1]. Indeed, more than 1,000 cases are reported every year in this country, despite the efforts of public health units [1]. The reservoir of M. ulcerans is unknown but the absence of evidence of human-to-human transmission suggests an environmental reservoir. Accordingly, several epidemiological publications in West Africa have pointed to contact with stagnant water as a significant risk factor for contracting Buruli ulcer [2–5]. The slow growth of M. ulcerans makes it easily overgrown by contaminant microorganisms; therefore, the search for this pathogen in the environment has been mainly based on the molecular detection of M. ulcerans DNA, i.e. chromosomal and plasmidic sequence insertion IS2404 and the KR-B plasmidic gene encoding a polyketide synthase participating into the biosynthesis of the mycolactone toxin [6,7].
In Ivory Coast, the use of these molecular methods confirmed the detection of M. ulcerans in freshwater insects [7–10]. Two isolates, Nau CI 001 and Nau CI 002, have also been cultivated from Naucoris spp. water insects [8] but this observation has been disputed [11,12]. So far, only one environmental isolate has been firmly sequence-confirmed from an aquatic Hemiptera (in Benin, another West African country) [11].
In this study, we launched a new environmental sampling campaign, investigating soil, stagnant and running water, plants and feces of an endemic small mammal, Thryonomys swinderianus (agouti), in four regions of Ivory Coast. The aim was to develop understanding about the ecological and geographical distribution of the pathogen in this country for Buruli ulcer.
Materials and Methods
Environmental samples
We carried-out a campaign of environmental samplings between June and October 2014 (June-July is the wet season and August-October is the dry season) in four regions of Ivory Coast comprising regions of low incidence of Buruli ulcer (control group A) and regions of high incidence of Buruli ulcer (group B) as documented by official records of active surveillance by the “Programme National de Lutte contre l’Ulcer de Buruli” (National Program against Buruli Ulcer) [1] and confirmed to one of us (Tian B.D.R.) by interviewing nurses and village leaders (Fig 1). This work was carried out with the approval of the national program against Buruli ulcer in Ivory Coast (PNLCI) and no further permissions were required for the environmental sampling of common goods. This study did not involve any endangered or protected species, and no vertebrates.
Fig 1. Map of Ivory Coast (Based on OCHA/ReliefWeb) showing the Buruli ulcer endemic areas based on World Health Organization data (2013) and location of studied environmental samples.
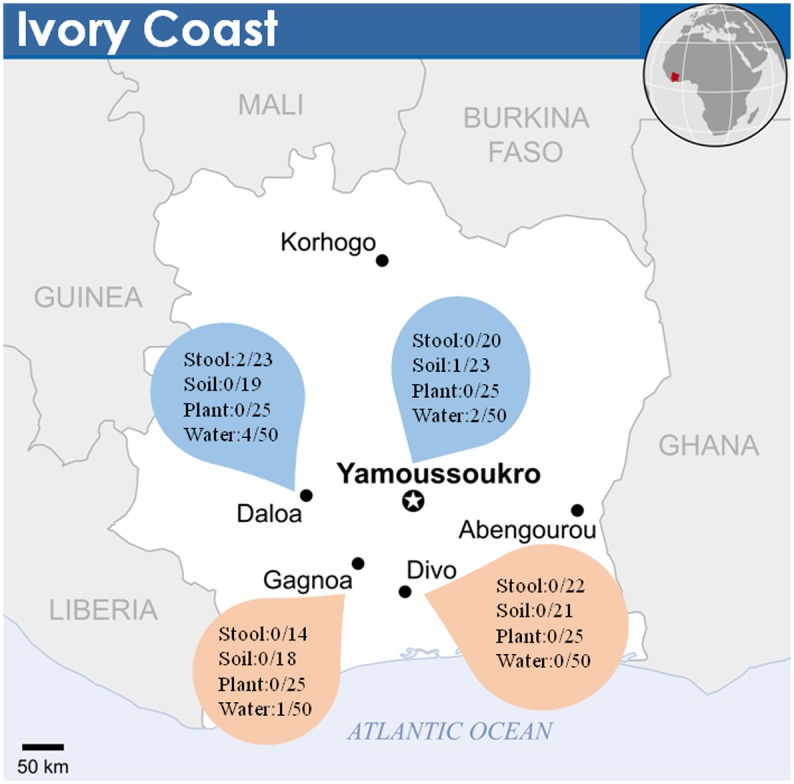
Each circle figures the number of positive samples over collected samples. Blue teardrops, endemic areas; tan teardrops, non-endemic areas.
A series of water samples (n = 200) were collected in endemic areas (n = 100) and in non-endemic areas (n = 100) at the edge and middle of stagnant waters at an average 50-cm depth. A 10 mL volume of water was collected in a 15 mL sterile tube (Sarstedt, Nümbrecht, Germany) which was immediately closed, sealed and placed in a cooler at 4°C. Non-identified plants were collected mainly in permanent and temporary stagnant waters and rivers in endemic areas (n = 50) and in non-endemic areas (n = 50). One hundred plant fragments from endemic plants in Ivory Coast including roots, stems and leaves were harvested and each sample placed into a sterile 15 mL Falcon tube stored as described above. Soil samples (n = 100; endemic areas, n = 50; non-endemic areas n = 50) were collected at half a meter or a meter from the water (river, standing water). Finally, feces fragments collected from the ground (n = 96; endemic areas, n = 48; non-endemic areas, n = 48), morphologically identified by a traditional hunter as Thryonomys swinderianus (agouti) stools, were put into a sterile tube as described above. Molecular confirmation of T. swinderianus feces was done by cytochrome b gene standard PCR. All feces specimens were confirmed to be T. swinderianus (agouti) by exhibiting a cytb amplification with Ct values of 25. All samples were stored at 4°C in our laboratory for analysis. Collected plant fragments and fecal samples did not come from species listed as endangered.
DNA extraction
Total soil DNA was extracted using a soil DNA extraction kit according to the manufacturer’s instructions (NucleoSpin®Soil, Macherey-Nagel, Düren, Germany). Total stool DNA was extracted using a stool extraction kit according to the manufacturer’s instructions (QIAmp®, DNA Stool, Qiagen, Stochach, Germany). Total DNA was extracted from plants and sedimented water using the NucleoSpin Tissue Kit (Macherey—Nagel). M. ulcerans Agy99 DNA was extracted using a commercial kit (Tissue Kit, Macherey-Nagel) as a positive control. PCR inhibition was assessed by adding 10μL of internal control into 190μL of sample, as previously described [13]. Further, efficiency of DNA extraction for M. ulcerans in feces was assessed by spiking a final inoculum of 106 M. ulcerans Agy99 colony-forming units (cfu)/mL agouti feces. These artificially infected feces were extracted as described above. In each extraction batch of 25 samples, two negative controls consisting of distilled water were included. All stool or soil specimens demonstrating PCR inhibition were diluted 1:10 and 1:100 in PBS before being tested again.
Real-time PCR amplifications
In the first step, a calibration curve was made for each one of the two M. ulcerans DNA targets by using real-time PCR as described below, in a series of 10-fold M. ulcerans Agy99 suspensions from 106 cfu/mL to 1 cfu/mL; suspensions were extracted as described above in triplicate. Then, for each specimen, IS2404 and KR-B were tentatively amplified using real-time PCR (RT-PCR) performed incorporating RT-PCR reagents (Takyon, Eurogentec, Liege, Belgium) and primers and probes as previously described [6], in a CFX 96™ real time PCR thermocycler and detection system (BIO-Rad, Marnes-la-Coquette, France). PCR was conducted under a 20 μL volume containing 5 μL of DNA, 0.5 μL of each primer, 0.5 μL of probe, 3.5 μL of sterile water and 10 μL of mastermix (Eurogentec). The RT-PCR program comprised one cycle at 50°C for two minutes and 40 cycles at 95°C for 15 seconds and 60°C for one minute. Two negative controls for 25 samples were incorporated into each PCR run. All samples were tested in duplicate. A specimen was considered as positive for the detection of M. ulcerans when both the insertion sequences IS2404 and the KR-B gene detection was positive (Ct ≤ 40 cycles) in replicates. The Ct value cut-off was chosen in order to increase the sensitivity of the detection.
Statistical analysis
According to the low effectives, frequencies of quantitative data were compared using the Fisher’s exact test. A difference was considered significant when the P value < 0.05.
Results
Between June and October 2004, a total of 496 environmental specimens were collected in Ivory Coast, consisting of 100 soil specimens, 200 stagnant water specimens, 100 plant specimens and 96 feces specimens. A total of 246 specimens were collected in control group A negative control geographic areas and a total of 250 specimens were collected in Buruli ulcer endemic geographic group B areas.
In preliminary RT-PCR experiments, the negative controls remained negative and the M. ulcerans positive control yielded consistent calibration curves for IS2404 and KR-B gene (S1 Table). As for the samples, a total of 36/496 (7.2%) samples including 17 stool samples and 19 soil samples did not yield amplification of the internal control, suggesting PCR inhibition. None of these 36 samples (pure and diluted 1:10 and 1:100) yielded any amplification for M. ulcerans and they were deleted from further analysis. Of 460 PCR inhibition-free samples, 43 (9.3%) yielded at least one positive RT-PCR detection of IS2404 or KR (Table 1). Of these 43 specimens, only 10/460 (2.1%) specimens were positive for IS2404 and KR and fulfilled our definition of a PCR- positive specimen. Positive samples yielded M. ulcerans inoculums estimated to be of 1 cfu/mL to 102 cfu/mL. The prevalence of the positive detection of IS2404 was significantly higher in endemic group B geographic areas (19/230, 8.3%) than in negative control group A geographic areas (4/230, 1.7%)(P = 0.002; OR = 5.1 [1.7–15.2]. The prevalence of the positive detection of M. ulcerans was significantly higher in endemic group B geographic areas (9/230, 3.9%) than in negative control group A geographic areas (1/230, 0.04%) (P = 0.02, Fisher’s exact test; OR = 9.33 [1.17–74.22]. These 10 M. ulcerans-positive specimens consisted of seven water specimens collected from permanent puddles in the Haut-Sassandra region, the Yamoussoukro district and the Gôh region in June-October, two T. swinderianus (agouti) feces specimens collected in the Haut-Sassandra region in October and one soil specimen collected in Yamoussoukro district in October. Altogether, 6/281(2.5%) specimens collected in the dry season were positive versus 4/215 (1.9%) specimens collected in the wet season (P = 0.76).
Table 1. Molecular detection of M. ulcerans in environment, Ivory Coast and number of environmental samples free of PCR inhibition and positive for the RT-PCR detection of M. ulcerans (CFU, colony-forming units, Ct cycle threshold).
| IS2404 | KR-B | IS2404 and KR-B True positive | Estimated CFU/mL | |||||
|---|---|---|---|---|---|---|---|---|
| positive | Ct | CFU/mL | positive | Ct | CFU/mL | |||
| Soil (n = 81) | 2 | 33 | 10 | 1 | 35 | 10 | 1 | 10 |
| 37 | 10 | |||||||
| Plant (n = 100) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Stool (n = 79) | 5 | 33 | 10 | 2 | 35 | 102 | 2 | 102 |
| 37 | 10 | 38 | 10 | 10 | ||||
| 35 | 10 | - | ||||||
| 38 | 100 | - | ||||||
| 34 | 10 | - | ||||||
| 39 | 10 | - | ||||||
| Water (n = 200) | 16 | 36 | 10 | 7 | 38 | 10 | 7 | 10 |
| 31 | 10 | 33 | 102 | 102 | ||||
| 30 | 102 | 32 | 102 | 102 | ||||
| 33 | 10 | 35 | 10 | 10 | ||||
| 32 | 10 | 34 | 10 | 10 | ||||
| 35 | 10 | 36 | 10 | 10 | ||||
| 31 | 10 | 33 | 102 | 102 | ||||
| 38 | 100 | - | ||||||
| 39 | 100 | - | ||||||
| 35 | 10 | - | ||||||
| 36 | 10 | - | ||||||
| 37 | 10 | - | ||||||
| 39 | 100 | - | ||||||
| 36 | 10 | - | ||||||
| 38 | 100 | - | ||||||
| 37 | 10 | - | ||||||
| Total (n = 460) | 23 | 10 | 10 | |||||
Discussion
Ivory Coast is the country most affected by Buruli ulcer and many efforts have been undertaken to clarify risk factors, significantly linking cases with proximity to dams used for irrigation and aquaculture [2]. However, surprisingly, few data have been reported from the direct search for the causative agent M. ulcerans in these environments in Ivory Coast. Here we report the results of the first systematic investigation in this West African country. Based on data reported in similar investigations conducted in Benin [5], Ghana [14] and Australia [15], we looked for M. ulcerans DNA in soil, stagnant water, aquatic plants and feces of the most prevalent endemic mammal in Ivory coast, T. swinderianus. We voluntarily neglected the investigation of water insects as they have been extensively studied in Ivory Coast and have previously shown to be infected by M. ulcerans [7–10]. In order to ensure the specificity of RT-PCR detection, we retained as positive only those specimens exhibiting a positive amplification for one insertion sequence IS2404 plus the KR-B gene, in the presence of negative controls according to standard interpretation. Indeed, none of these three markers is specific by itself for M. ulcerans, since IS2404 has been also detected in Mycobacterium liflandii [16], and IS2404 and KR-B gene in some isolates of Mycobacterium liflandii [17], of Mycobacterium marinum [18] and Mycobacterium pseudoshottsii [19].
Using these stringent parameters, we detected M. ulcerans DNA in only 2.1% environmental specimens. Apart from one positive stagnant water specimen in one non-endemic region, it is significant to note that all positive specimens were collected in areas endemic for Buruli ulcer. This observation agrees with the current knowledge that M. ulcerans is an environmental organism of as yet uncertain sources [4–7]. Alternatively, this difference could be due to the difference in the composition of sources between the endemic and non-endemic areas here investigated. Interestingly, 7/10 (70%) positive specimens had been collected during the dry season [3, 20–22]. The detection of M. ulcerans in seven stagnant water specimens was not surprising, as similar observations have been done in several studies in neighboring African countries [5] as well as in Australia [14] and South America [20]. We detected M. ulcerans inocula 100 times much lower than those previously reported in Benin; however, different methodology between the two studies may account for this difference [5]. The proximity to dams and stagnant water resulting from irrigation has been demonstrated as a risk factor for Buruli ulcer in Ivory Coast [2] and other countries [5, 15, 20]. The role of water however remains unknown as it could be a reservoir by itself or could support a reservoir for M. ulcerans but does not resume the cycle of this organism which is presumably inoculated to cause Buruli ulcer [5, 23–25]. More unexpected was the observation of two soil specimens collected during the dry season at 50 cm and one meter from stagnant water. Soil has rarely been found positive for M. ulcerans, except in Benin [5]. In fact, we recently confirmed this field observation, by observing a four-month survival of M. ulcerans in experimentally contaminated soil [26].
The detection of M. ulcerans in two feces specimens collected 21 days apart in two different villages (Daloa, Haut-Sassandra Region) from T. swinderianus (agouti) mammals was also unexpected. This observation is very interesting as the agouti is known to be endemic in watery areas resulting from irrigation efforts in Ivory Coast where it destroys rice fields and is in close contacts with populations as it is hunted for consumption of its meat. Moreover, T. swinderianus was shown to develop Buruli ulcer after experimental subcutaneous inoculation of M. ulcerans [27]. This observation recalls the detection of M. ulcerans in the feces of possums in Australia in geographic regions which are endemic for Buruli ulcer [28]. Both animals are small mammals. Combined with earlier observations in Ivory Coast [12], the observations reported here support a natural cycle of M. ulcerans which is more complex than previously anticipated. In Ivory Coast, additional field and experimental studies will need to be conducted in order to interpret our observations and to determine whether the digestive tract of the agouti is merely contaminated by bypassing M. ulcerans organisms from contaminated water; or if the agouti plays any role in the environmental cycle of M. ulcerans, through fecal contamination of soil and direct contamination of populations.
Supporting Information
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information file.
Funding Statement
These authors have no support or funding to report.
References
- 1.WHO Global health observatory data. Available: http://www.who.int/gho/neglected_diseases/buruli_ulcer/en/. Accessed 26 June 2015.
- 2.Brou T, Broutin H, Elguero E, Asse H, Guegan JF. Landscape diversity related to Buruli ulcer disease in Côte d'Ivoire. PLOS Negl Trop Dis. 2008; 2:e271 10.1371/journal.pntd.0000271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marion E, Eyangoh S, Yeramian E, Doannio J, Landier J, Aubry J, et al. Seasonal and regional dynamics of M. ulcerans transmission in environmental context: deciphering the role of water bugs as hosts and vectors. PLOS Negl Trop Dis. 2010; 4:e731 10.1371/journal.pntd.0000731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Williamson HR, Benbow ME, Nguyen KD, Beachboard DC, Kimbirauskas RK, McIntosh MD, et al. Distribution of Mycobacterium ulcerans in Buruli Ulcer endemic and non-endemic aquatic sites in Ghana. PLOS Negl Trop Dis. 2008; 2:e205 10.1371/journal.pntd.0000205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williamson HR, Benbow ME, Campell LP, Johnson CR, Sopoh G, Baroqui Y, et al. Detection of Mycobacterium ulcerans in the environment predicts prevalence of Buruli ulcer in Benin. PLOS Negl Trop Dis. 2012; 6:e1506 10.1371/journal.pntd.0001506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fyfe JAM, Lavender CJ, Johnson PDR, Globan M, Sievers A, Azuolas J, et al. Development and application of two multiplex real-time PCR assays for the detection of Mycobacterium ulcerans in clinical and environmental samples. Appl Environ Microbiol. 2007; 73:4733–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ngazoa-Kakou ES, Ekaza E, Aka N, Coulibaly-N’Golo D, Coulibaly B, Dosso M. Evaluation of real-time PCR for Mycobacterium ulcerans in endemic region in Côte d’Ivoire. Af J Microbiol Res. 2011; 5:2211–6. [Google Scholar]
- 8.Marsollier L, Robert R, Aubry J, Saint Andre JP, Kouakou H, Legras P, et al. Aquatic insects as a vector for Mycobacterium ulcerans. Appl Environ Microbiol. 2002; 68:4623–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doannio JM, Konan KL, Dosso FN, Koné AB, Konan YL, Sankaré Y, et al. Micronecta sp (Corixidae) and Diplonychus sp (Belostomatidae), two aquatic Hemiptera hosts and/or potential vectors of Mycobacterium ulcerans (pathogenic agent of Buruli ulcer) in Côte d'Ivoire. Med Trop. 2011; 71:53–7. [PubMed] [Google Scholar]
- 10.Konan KL, Doannio JM, Coulibaly NG, Ekaza E, Marion E, Assé H, et al. Detection of the IS2404 insertion sequence and ketoreductase produced by Mycobacterium ulcerans in the aquatic Heteroptera in the health districts of Dabou and Tiassalé in Côte d'Ivoire. Med Sante Trop. 2015; 25:44–51. 10.1684/mst.2014.0363 [DOI] [PubMed] [Google Scholar]
- 11.Portaels F, Meyers WM, Ablordey A, Castro AG, Chemlal K, de Rijk P, et al. First cultivation and characterization of Mycobacterium ulcerans from the environment. PLOS Negl Trop Dis. 2008; 2:e178 10.1371/journal.pntd.0000178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marsollier L, Aubry J, Saint-André JP, Robert R, Legras P, Manceau AL, et al. Ecology and transmission of Mycobacterium ulcerans. Pathol Biol (Paris). 2003; 51:490–5. [DOI] [PubMed] [Google Scholar]
- 13.Ninove L, Nougairede A, Gazin C, Thirion L, Delogu I, Zandotti C, et al. RNA and DNA bacteriophages as molecular diagnosis controls in clinical virology: a comprehensive study of more than 45,000 routine PCR tests. PLOS One. 2011; 6:e16142 10.1371/journal.pone.0016142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Narh CA, Mosi L, Quaye C, Dassi C, Konan DO, Tay SCK, et al. Source tracking Mycobacterium ulcerans infections in the Ashanti region, Ghana. PLOS Negl Trop Dis. 2015; 9:e0003437 10.1371/journal.pntd.0003437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roberts B, Hirts B. Immunomagnetic separation and PCR for detection of Mycobacterium ulcerans. J Clin Microbiol. 1997; 35:2709–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tobias NJ, Doig KD, Medema MH, Chen H, Haring V, Moore R, et al. Complete genome sequence of the frog pathogen Mycobacterium ulcerans ecovar Liflandii. J Bacteriol. 2013; 195:556–64. 10.1128/JB.02132-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mve-Obiang A, Lee RE, Umstot ES, Trott KA, Grammer TC, Parker JM, et al. A newly discovered mycobacterial pathogen isolated from laboratory colonies of Xenopus species with lethal infections produces a novel form of mycolactone, the Mycobacterium ulcerans macrolide toxin. Infect Immun. 2005; 73:3307–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stinear TP, Seemann T, Harrison PF, Jenkin GA, Davies JK, Johnson PD, et al. Insights from the complete genome sequence of Mycobacterium marinum on the evolution of Mycobacterium tuberculosis. Genome Res. 2008; 18:729–41. 10.1101/gr.075069.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ranger BS, Mahrous EA, Mosi L, Adusumilli S, Lee RE, Colorni A, et al. Globally distributed mycobacterial fish pathogens produce a novel plasmid-encoded toxic macrolide, mycolactone F. Infect Immun. 2006; 74:6037–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morris A, Gozlan R, Marion E, Marsollier L, Andreou D, Sanhueza D, et al. First detection of Mycobacterium ulcerans DNA in environmental samples from South America. PLOS Negl Trop Dis. 2014; 8:e2660 10.1371/journal.pntd.0002660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Landier J, Constantin de Magny G, Garchitorena A, Guégan JF, Gaudart J, Marsollier L, et al. Seasonal Patterns of Buruli Ulcer Incidence, Central Africa, 2002–2012. Emerg Infect Dis. 2015; 21:1414–7. 10.3201/eid2108.141336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garchitorena A, Guégan JF, Léger L, Eyangoh S, Marsollier L, Roche B. Mycobacterium ulcerans dynamics in aquatic ecosystems are driven by a complex interplay of abiotic and biotic factors. Elife. 2015. 4:e07616 10.7554/eLife.07616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Merritt RW, Walker ED, Small PLC, Wallace JR, Johnson PDR, Benbow ME, et al. Ecology and transmission of Buruli ulcer disease: a systematic review. PLOS Negl Trop Dis. 2010; 4:e911 10.1371/journal.pntd.0000911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marion E, Deshayes C, Chauty A, Cassisa V, Tchibozo S, Cottin J, et al. Detection of Mycobacterium ulcerans DNA in water bugs collected outside the aquatic environment in Benin. Med Trop. 2011; 71:169–72. [PubMed] [Google Scholar]
- 25.Gryseels S, Amissah D, Durnez L, Vandelannoote K, Leirs H, De Jonckheere J, et al. Amoebae as Potential Environmental Hosts for Mycobacterium ulcerans and Other Mycobacteria, but Doubtful Actors in Buruli Ulcer Epidemiology. PLOS Negl Trop Dis. 2012; 6:e1764 10.1371/journal.pntd.0001764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tian RDB, Lepidi H, Nappez C, Drancourt M. Experimental survival of Mycobacterium ulcerans in watery soil, a potential source of Buruli ulcer. Am J Trop Med Hyg. 2015; pii 15–0568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Addo P, Adu-Addai B, Quartey M, Abbas M, Okang I, Owusu E, et al. Clinical and histopathological presentation of Buruli Ulcer in experimentally infected grasscutters (Thryonomys swinderianus). Internet J Trop Med. 2006; volume 2. [Google Scholar]
- 28.Fyfe JAM, Lavender CJ, Handasyde KA, Legione AR, O'Brien CR, Stinear TP, et al. A major role for mammals in the ecology of Mycobacterium ulcerans. PLOS Negl Trop Dis. 2010; 4:e791 10.1371/journal.pntd.0000791 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information file.


