Abstract
The redundancy of the genetic code implies that most amino acids are encoded by multiple synonymous codons. In all domains of life, a biased frequency of synonymous codons is observed at the genome level, in functionally related genes (e.g., in operons), and within single genes. Other codon bias variants include biased codon pairs and codon co-occurrence. Although translation initiation is the key step in protein synthesis, it is generally accepted that codon bias contributes to translation efficiency by tuning the elongation rate of the process. Moreover, codon bias plays an important role in controlling a multitude of cellular processes, ranging from differential protein production to protein folding. Here we review currently known types of codon bias and how they may influence translation. We discuss how understanding the principles of codon bias and translation can contribute to improved protein production and developments in synthetic biology.
The central dogma of molecular biology concerns the general principle of protein expression: DNA is transcribed to mRNA, which is translated to protein. The key molecules of translation are the set of tRNAs, each providing a direct, specific link between a triplet of nucleotides and the corresponding amino acid. Ribosomes are the engines of translation that accommodate the tRNAs and mRNA (Figure 1). Deciphering the genetic code revealed that 61 codons (triplets) encode the standard 20 amino acids, whereas the remaining 3 are translation stop signals. The genetic code is nearly universal, meaning that almost all organisms use exactly the same codons for a specific amino acid. Because 18 of 20 amino acids are encoded by multiple synonymous codons, the genetic code is called “degenerate.”
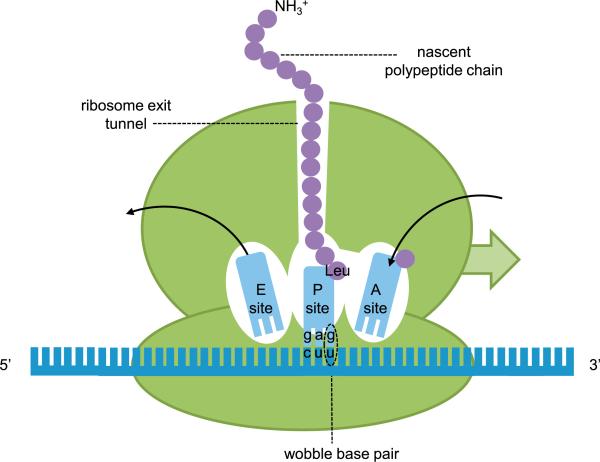
Figure 1. Translation in the Ribosome and tRNA Structure.
Cartoon of the ribosome (green) during translation of a mRNA (blue) with a wobbling codon-anticodon base pair encoding a leucine amino acid. A site, aminoacyl-tRNA site; E site, exit site; P site, peptidyl-tRNA site.
Because synonymous mutations do not affect the identity of the encoded amino acid, they were originally thought to have no consequences for protein function or organismal fitness and were therefore regarded as “silent mutations.” However, comparative sequence analysis revealed a non-random distribution of synonymous codons in genes of different organisms. Each organism seems to prefer a different set of codons over others; this phenomenon is called codon bias (Sharp and Li, 1986).
Several important variations of codon bias have recently been discovered, such as the existence of a ramp of rare, slowly translated codons at the 5′ end of protein-coding sequences (Tuller et al., 2010) and the co-occurrence of certain codons (Cannarozzi et al., 2010). Apart from directly affecting general protein expression levels, it has been established that codon bias also influences protein folding (Pechmann and Frydman, 2013) and differential regulation of protein expression (Gingold et al., 2014). In addition to in silico analyses of codon bias, the development of ribosome density profiling has allowed experimental monitoring of the translation elongation rate at single-codon resolution (Ingolia, 2014). Partly on the basis of ribosome profiling data, some studies have shown that codon bias plays an important role in translation efficiency. Alternatively, however, it is concluded that translation efficiency relies on other features of the coding sequence, such as mRNA secondary structure (Kudla et al., 2009) and the presence of Shine-Dalgarno-like sequences (Li et al., 2012).
Here we provide a comprehensive overview of distinct variations of codon bias. We discuss how codon bias can tune expression at multiple levels: genome, operon, and gene. Furthermore, we discuss how rules for codon bias may be further elucidated and applied to improve engineering projects, ranging from the biotechnological production of single proteins to more complex synthetic biology endeavors.
General Codon Bias Variants
By the end of the 1970s, the development of DNA sequencing had enabled comparisons of the rapidly growing number of gene sequences. Striking differences were observed in the preference of distinct organisms to use certain synonymous codons over others (Grantham et al., 1980). It did not take long to discover that codon usage also differs among genes within one genome (Ikemura, 1985). Soon after, metrics for the frequency of optimal codons were proposed, such as the commonly used codon adaptation index (CAI) (Sharp and Li, 1987). The CAI for a certain organism is based on the codon usage frequency in a reference set of highly expressed genes, such as the ones encoding ribosomal proteins. The CAI for a specific gene can be determined by comparing its codon usage frequency with this reference set.
Wobbling and tRNA Modifications
Analysis of the tRNA content of organisms in all domains of life showed that they never contain a full set of tRNAs with anticodons complementary to the 61 different codons; for example, 39 tRNAs with distinct anticodons are present in the bacterium Escherichia coli, 35 in the archaeon Sulfolobus solfataricus, and 45 in the eukaryote Homo sapiens (Table 1). In some Mycoplasma species and related species, the smallest sets are found, consisting of only 28 tRNAs with distinct anticodons (Grosjean et al., 2010). Translation of multiple synonymous codons by a single tRNA has been demonstrated to occur by wobble base-pairing: standard Watson-Crick base-pairing (A-U, G-C) is required at the first and second positions of a codon, and “wobbling” (e.g., G-U) is allowed at the third position of a codon (corresponding to the 5′ position of the anticodon, i.e., position 34 of a tRNA) (Crick, 1966; Söll et al., 1966). However, the affinity by which synonymous codons are recognized via wobble base-pairing is not similar. For instance, tRNAs with G in the 5′ position of the anticodon have a higher binding affinity for C-ending codons than for U-ending codons (Crick, 1966; Söll et al., 1966).
Table 1.
Codon Frequencies and tRNA Gene Copy Numbers for E. coli, S. solfataricus, and H. sapiens
| Second Base Codon |
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T |
C |
A |
G |
||||||||||||||||
| Amino Acid | Eco Codon % (tRNA #) | Sso Codon % (tRNA #) | Hsa Codon % (tRNA #) | Amino Acid | Eco Codon % (tRNA #) | Sso Codon % (tRNA #) | Hsa Codon % (tRNA #) | Amino Acid | Eco Codon % (tRNA #) | Sso Codon % (tRNA #) | Hsa Codon % (tRNA #) | Amino Acid | Eco Codon % (tRNA #) | Sso Codon % (tRNA #) | Hsa Codon % (tRNA #) | ||||
| First Base Codon |
T | Phe | 2.22 (0) |
2.63 (0) |
1.76 (0) |
Ser | 0.80 (0) |
1.50 (0) |
1.52 (11) |
Tyr | 1.60 (0) |
3.10 (0) |
1.22 (1) |
Cys | 0.50 (0) |
0.40 (0) |
1.06 (0) |
T | Third Base Codon |
| 1.65 (2) |
1.80 (1) |
2.03 (12) |
0.86 (2) |
0.76 (1) |
1.77 (0) |
1.22 (3) |
1.69 (1) |
1.53 (14) |
0.64 (1) |
0.19 (1) |
1.26 (30) |
C | |||||||
| Leu | 1.39 (1) |
4.11 (1) |
0.77 (7) |
0.71 (1) |
1.55 (1) |
1.22 (5) |
Stop | 0.21 (0) |
0.18 (0) |
0.10 (2)a |
Stop | 0.09 (1)b |
0.10 (3)b |
0.16 (3)b |
A | ||||
| 1.36 (1) |
1.59 (1) |
1.29 (7) |
0.89 (1) |
0.48 (1) |
0.44 (4) |
0.03 (0) |
0.07 (0) |
0.08 (1)a |
Trp | 1.53 (1) |
1.05 (1) |
1.32 (9) |
G | ||||||
| C | 1.10 (0) |
1.52 (0) |
1.32 (12) |
Pro | 0.70 (0) |
1.23 (0) |
1.75 (10) |
His | 1.29 (0) |
0.82 (0) |
1.09 (0) |
Arg | 2.09 (4) |
0.17 (0) |
0.45 (7) |
T | |||
| 1.11 (1) |
0.70 (1) |
1.96 (0) |
0.55 (1) |
0.55 (1) |
1.98 (0) |
0.97 (1) |
0.46 (1) |
1.51 (11) |
2.2 (0) |
0.06 (1) |
1.04 (0) |
C | |||||||
| 0.39 (1) |
1.91 (1) |
0.72 (3) |
0.84 (1) |
1.60 (1) |
1.69 (7) |
Gln | 1.54 (2) |
1.56 (1) |
1.23 (11) |
0.35 (0) |
0.13 (1) |
0.62 (6) |
A | ||||||
| 5.29 (4) |
0.55 (1) |
3.96 (10) |
2.32 (1) |
0.41 (1) |
0.69 (4) |
2.89 (2) |
0.53 (1) |
3.42 (20) |
0.54 (1) |
0.05 (1) |
1.14 (4) |
G | |||||||
| A | Ile | 3.04 (0) |
3.36 (0) |
1.60 (14) |
Thr | 0.89 (0) |
2.01 (0) |
1.31 (10) |
Asn | 1.77 (0) |
2.63 (0) |
1.70 (2) |
Ser | 0.87 (0) |
1.66 (0) |
1.21 (0) |
T | ||
| 2.52 (3) |
1.11 (1) |
2.08 (3) |
2.34 (2) |
0.68 (1) |
1.89 (0) |
2.16 (4) |
1.67 (1) |
1.91 (32) |
1.60 (1) |
0.73 (1) |
1.95 (8) |
C | |||||||
| 0.43 (0) |
4.94 (0) |
0.75 (5) |
0.70 (1) |
1.38 (1) |
1.51 (6) |
Lys | 3.37 (6) |
3.96 (1) |
2.44 (16) |
Arg | 0.20 (1) |
2.52 (1) |
1.22 (6) |
A | |||||
| Met Start |
2.78 (8) |
2.07 (3) |
2.20 (20) |
1.44 (2) |
0.64 (1) |
0.61 (6) |
1.03 (0) |
3.76 (1) |
3.19 (17) |
0.11 (1) |
1.76 (1) |
1.20 (5) |
G | ||||||
| G | Val | 1.83 (0) |
2.76 (0) |
1.10 (11) |
Ala | 1.53 (0) |
2.24 (0) |
1.84 (29) |
Asp | 3.22 (0) |
3.42 (0) |
2.18 (0) |
Gly | 2.48 (0) |
2.19 (0) |
1.08 (0) |
T | ||
| 1.53 (2) |
0.72 (1) |
1.45 (0) |
2.56 (2) |
0.72 (1) |
2.77 (0) |
1.91 (3) |
1.25 (1) |
2.51 (19) |
2.97 (4) |
0.67 (1) |
2.22 (15) |
C | |||||||
| 1.09 (5) |
2.81 (1) |
0.71 (5) |
2.02 (3) |
1.92 (1) |
1.58 (9) |
Glu | 3.96 (4) |
3.84 (1) |
2.90 (13) |
0.79 (1) |
2.58 (1) |
1.65 (9) |
A | ||||||
| 2.62 (0) |
1.22 (1) |
2.18 (16) |
3.37 (0) |
0.71 (1) |
0.74 (5) |
1.78 (0) |
2.95 (1) |
3.96 (13) |
1.11 (1) |
0.97 (1) |
1.65 (7) |
G | |||||||
Overview of codon frequency usage in all coding sequences and tRNA gene copy numbers for E. coli K12 (Eco), S. solfataricus (Sso), and H. sapiens (Hsa) (data obtained from http://gtrnadb.ucsc.edu).
Suppressor tRNAs suppressing the stop codon.
tRNAs incorporating selenocysteine amino acid into selenoproteins and suppressing the TGA stop codon.
The influence of wobble base-pairing on decoding rates of codons by the ribosome is still unresolved and complex to analyze. The translation kinetics of different codon-anticodon pairs are complex, as the following processes can play a role: (1) the diffusion kinetics of the matching tRNA, (2) the relative codon-binding affinity of matching tRNAs over mismatching tRNAs (Gromadski et al., 2006), and (3) the translocation kinetics of mRNA and tRNA through the ribosome, which are affected by anticodon-codon interactions (Khade and Joseph, 2011). Recently conflicting results were published that reported either slower (Stadler and Fire, 2011) or faster (Gardin et al., 2014) decoding of wobbling codons. This process deserves a more detailed analysis of both data sets and methods and should also take into account the effect of tRNA modifications on wobble base-pairing.
Modified nucleotides present in tRNAs further extend the range of recognized synonymous codons by affecting the ability of these tRNAs to form wobble base pairs (Agris et al., 2007). Some specific, key tRNA modifications are present in only some domains of life (Grosjean et al., 2010). First, a key tRNA modification, present in eukaryotes and to some extent in bacteria, is the modification of adenine-34 to inosine-34, which allows non-Watson-Crick pairing with adenine, cytosine, and uridine. Second, exclusively in bacteria, the key tRNA modifications of uridine-34 to hydroxy-uridine and derivatives are found, allowing wobble pairing with adenine, guanosine, and uridine. These key tRNA modifications explain many differences in the tRNA sets that are present in archaea, bacteria, and eukaryotes (Table 1) (Novoa et al., 2012).
Correlation of Codon Bias with tRNA Pools
After the discovery of codon bias, a positive correlation was found between the frequency of codons and the concentration of tRNAs with complementary anticodons (Figure 2A) (Ikemura, 1985). This fact was established for several prokaryotes and unicellular eukaryotes (Kanaya et al., 1999). However, this correlation initially could not be identified for several, mostly multicellular eukaryotes.
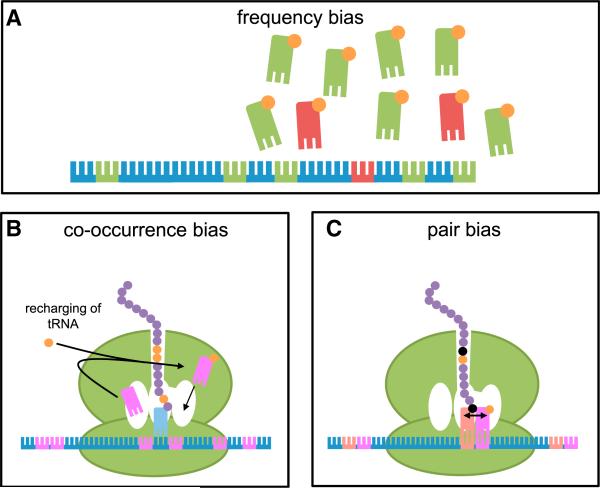
Figure 2. Different Types of Codon Bias.
(A) Frequency bias will result in effective protein production when the frequency of used codons matches the cellular tRNA population.
(B) Co-occurrence bias enhances protein expression, presumably because of tRNA recharging in the vicinity of the translating ribosome.
(C) Pair bias is probably selected because of more optimal interactions of tRNAs in the A site and the P site.
To better analyze the relation between codon frequency bias and tRNA abundance in multicellular organisms, the tRNA adaptation index (tAI) was developed (dos Reis et al., 2004). This metric is based on the copy number of tRNA genes, assumed to be correlated with tRNA abundance in cells, and also takes into account the efficiency of codon-anticodon binding, related to Crick's wobble rules (Crick, 1966). On the basis of computational analyses, it was concluded that organisms with larger genomes have higher tRNA gene redundancy, which would decrease selection for specific codons (dos Reis et al., 2004). This explained why in multicellular organisms with larger genomes, no positive correlation between codon usage and tRNA abundance could be identified in many studies. However, most studies at that time estimated tRNA abundance on the basis of tRNA gene copy numbers.
Correlations based on tRNA copy numbers do not take into account that pools of distinct tRNAs and aminoacyl-tRNA species are dynamic and can vary considerably in different conditions. For example, it was demonstrated by microarray analysis that tRNA expression abundance in humans varies widely among different tissues. This abundance could be statistically correlated to codon usage of highly expressed genes specific for those tissues (Dittmar et al., 2006). Furthermore, in bacteria, it was found that the charging levels of different tRNAs recognizing synonymous codons vary drastically in response to amino acid starvation; that is, although the pool of some synonymous tRNAs remains completely charged, the charged fraction of others can decline to zero (Dittmar et al., 2005; Elf et al., 2003).
So far, codon frequencies were correlated mostly with the total supply of tRNAs. However, when a more frequently used codon is recognized by a more abundant tRNA species, this codon will also compete for this tRNA with more codons. To take this into account, the normalized translation efficiency (nTE) metric was introduced, correcting for supply as well as demand rates of tRNAs (Pechmann and Frydman, 2013). The nTE considers codons to be more optimal if their relative tRNA abundance on the basis of gene copy number (supply) exceeds their relative cognate codon usage (demand) on the basis of codon frequencies in mRNA. Although the tAI and nTE already give good indications of the availability of tRNAs for the translation of synonymous codons, this approximation could still be improved. The actual important value is the level of mature aminoacyl-tRNAs ready for amino acid delivery in the translation process. However, because the levels of tRNA expression and charging can undergo major fluctuations on the basis of cellular condition, it is not straightforward to take these values into account. In addition, it was demonstrated that codon frequency bias can be better correlated with tRNA gene frequencies in all domains of life, if two major, domain-specific tRNA modification types are taken into account (Novoa et al., 2012).
To summarize, highly expressed proteins are generally encoded by genes that contain relatively high proportions of co-dons recognized by abundant, charged tRNAs with kinetically efficient codon-anticodon base-pairing. This explains to a large extent the observed codon frequency bias in many genes and genomes. In addition to codon frequency bias, two other general types of codon bias have been identified in recent years: codon pair bias and codon co-occurrence bias; these will be discussed hereafter.
Synonymous Codon Co-occurrence Bias
Recently it was shown that not only the overall frequency of synonymous codons but also the order in which they reside in a gene are biased. While studying all coding sequences of Saccharomyces cerevisiae, a bias was revealed of clustered synonymous codons, called codon co-occurrence bias. Instead of a random distribution of synonymous codons on a coding sequence, there is a bias to cluster those synonymous codons that are recognized by the same tRNA (i.e., identical codons and isoaccepting codons) (Cannarozzi et al., 2010) (Figure 2B). The effect of co-occurrence bias involves both frequent and rare codons and is most prominent in highly expressed genes that must be rapidly induced, such as those involved in stress response (Cannarozzi et al., 2010).
It has been suggested that tRNAs remain in proximity to the translating ribosome after their exit from the E site and that they are subsequently recharged by the corresponding amino-acyl-tRNA synthetases that somehow co-localize with the ribosome (Cannarozzi et al., 2010; Godinic-Mikulcic et al., 2014). At the next occurrence of the same or isoaccepting codon, the charged tRNA would be readily available for translation; this would have a positive effect on translation efficiency (Cannarozzi et al., 2010).
Co-occurrence bias has been demonstrated in eukaryotes, bacteria, and archaea (Cannarozzi et al., 2010; Shao et al., 2012; Zhang et al., 2013). However, co-occurrence of identical codons is strongly biased in all domains of life, whereas co-occurrence of non-identical isoaccepting codons is less prominent in prokaryotes than in eukaryotes (Shao et al., 2012; Zhang et al., 2013). The fact that co-occurrence of non-identical isoaccepting occurs more in eukaryotes most likely correlates with differences in affinity of codon-anticodon pairs between eukaryotes and prokaryotes (Shao et al., 2012). Domain-specific key modifications of tRNA result in differences in affinities of wobble base-pairing for certain synonymous codons. It has been hypothesized that only non-identical codon pairs that are recognized by a tRNA with similarly high affinity may result in co-occurrence bias (Shao et al., 2012). The described findings demonstrate that the use of identical and some isoaccepting codons in close proximity are generally advantageous for the translation process.
Non-synonymous Codon Pair Bias
In addition to codon frequency and co-occurrence, also the context in which a codon resides is under selective constraint. Nucleotides neighboring a particular codon are distributed in a non-random manner (Buchan et al., 2006; Gutman and Hatfield, 1989). This phenomenon is called codon pair bias (Figure 2C). For example, there are eight possible codon pairs to encode the adjacent amino acids alanine and glutamate. On the basis of codon frequencies, one would expect these amino acids to be encoded equally by GCC-GAA and GCA-GAG codon pairs. However, in humans, the GCC-GAA pair is heavily underrepresented compared with the expected frequency, even though it contains GCC, the most prevalent codon for alanine (Coleman et al., 2008). Some codon pairs are universally avoided or preferred; for example, nnUAnn codon pairs are usually underrepresented, whereas nnGCnn codon pairs are most preferred (Tats et al., 2008).
Although the exact mechanism by which codon pair bias might enhance translation efficiency is currently not well understood, it is assumed that tRNAs in the A and P sites of the ribosome can interact and as such influence the efficiency of the translation process (Figure 2C) (Buchan et al., 2006). Several viral genomes also contain codon pair bias, which generally matches that of their host. Modification of this codon pair usage in virulence-related genes of viruses and has been presented as an elegant strategy to produce vaccines with attenuated viruses (Coleman et al., 2008). However, it was recently suggested that this attenuation may be caused by an increased CpG and UpA dinucleo-tide bias rather than by a changed codon pair bias, because these dinucleotides are generally used at a low frequency in RNA and small DNA viruses infecting mammals and plants (Tulloch et al., 2014).
Is Translation Efficiency Correlated with Codon Bias?
The efficiency of translation, and the resulting protein production, is determined by both translation initiation and elongation rates. While the translation initiation rate controls how often a transcript is translated, the translation elongation rate controls the speed of this translation process.
Influence of the Coding Sequence on Translation Initiation
To initiate translation, ribosomes need to be sequestered on the mRNA, and the start codon must be recognized. This initiation process is facilitated by regulatory sequences. In prokaryotes, the binding strength between the Shine-Dalgarno sequence upstream of the coding sequence in the mRNA and the anti-Shine-Dalgarno sequence in the 16S rRNA regulate the efficiency of translation initiation. In eukaryotes, the Kozak sequence around the start codon is involved in interaction with the pre-initiation complex for translation (Tuller and Zur, 2015). Hence, the strength of mRNA folding around the regulatory initiation sequences and start codon can influence translation initiation efficiency; these 5′ mRNA secondary structures are also partially influenced by the 5′ end of the coding sequence. By analyzing libraries of synonymous variants of reporter genes in E. coli and S. cerevisiae, it was concluded that most of the observed variation in protein expression can be explained by differences in mRNA folding in the 5′ end of the mRNA (Bentele et al., 2013; Goodman et al., 2013; Kudla et al., 2009). However, there is a lively debate on these results, as it has been argued that the effect of mRNA secondary structures is overestimated because the aforementioned studies relied mainly on synthetic reporter gene variants with unusually strong mRNA secondary structures (Supek and Šmuc, 2010; Tuller and Zur, 2015).
Is the Translation Elongation Rate Controlled by Codon Bias?
Generally it is assumed that translation efficiency is determined mostly by translation initiation. However, at least to some extent, the translation elongation rate also appears to contribute to translation efficiency. A recent report showed that at a high translation initiation rate, a high translation elongation rate is required for optimal expression. Low expression, on the other hand, can be caused independently either by a low initiation rate or by low translation rates (Chu et al., 2014). Regarding translation elongation rates, it was assumed that more frequently used codons, recognized by abundant tRNAs, result in faster translation elongation and higher translation efficiency (Berg and Kurland, 1997). This assumption was confirmed by the observation that optimizing the overall codon sequence to more frequent codons does, at least in some cases, result in increased heterologous gene expression (Gustafsson et al., 2004).
Ribosome density profiling now allows genome-wide analysis of ribosome distribution down to a single-codon resolution (Figure 3) (Ingolia, 2014; Ingolia et al., 2009). Ribosome density profiles are very well suitable for obtaining snapshots of genome-wide gene translation (translatome); hereby translation rate profiles within single genes can be obtained and potentially be correlated with codon usage (Figure 3A). Ribosome density profiles were also used to estimate intergenic translation efficiency differences (Quax et al., 2013). However, ribosome densities can be used to compare the translation efficiency of genes only if the translation elongation rates for those genes are similar (Figure 3B) (Ingolia, 2014; Quax et al., 2013).
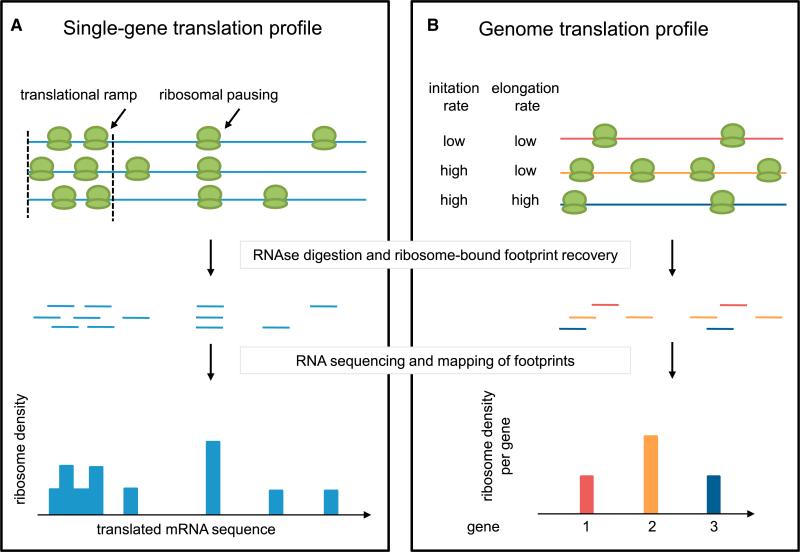
Figure 3. Ribosome Density Profiling to Measure Translation.
(A) Single-gene translation profiles based on ribosome density analysis are used to characterize intragenic fluctuations in translation elongation rates (e.g., corresponding to ramps of rare codons or pause sites at Shine-Dalgarno motifs).
(B) Genome translation profiles based on ribosome density analysis provide insight in intergenic differences in translation efficiency (translatome) by normalizing gene specific ribosome densities to their mRNA abundance. Importantly, intergenic differences in translation efficiency can be identified only if similar translation elongation rates can be assumed (e.g., on the basis of a similar codon bias in the genes to compare).
Many studies of ribosome profiling data have found that no correlations can be detected between high translation elongation rates and frequently used codons (Ingolia, 2014). On the other hand, correlations were found between strong mRNA secondary structures and a local speed reduction of translating ribosomes in yeast (Pop et al., 2014; Shah et al., 2013). Furthermore, ribosomes were found to slow down at sequences encoding specific amino acid sequences, such as consecutive proline residues (Woolstenhulme et al., 2013) or positively charged amino acids (Charneski and Hurst, 2013). In bacteria, ribosome profiling experiments identified ribosomal pausing at Shine-Dalgarno-like sequences (Li et al., 2012). These studies found no correlation between the codon usage frequency and ribosome speed. Some studies in eukaryotes reported that more frequent codons are translated at the same rate as rare codons (Pop et al., 2014; Qian et al., 2012). These studies suggest that codon bias is adapted to tRNA pools as a strategy to balance tRNA supply and demand and hence to achieve optimal translation. In contrast, two recent studies concluded that rare codons with less abundant cognate tRNAs are decoded slower, thus resulting in decreased translation elongation rates (Dana and Tuller, 2014; Gardin et al., 2014). To reach these conclusions, these studies applied novel statistical methods for analyzing ribosome profiling data, to remove bias caused either by highly expressed genes (Gardin et al., 2014) or by extreme ribosome pause events that are unrelated to codon frequency (Dana and Tuller, 2014). The latter studies also optimized and compared different experimental protocols for ribosome profiling. Only for the mouse ribosome profiling data, analyzed using the novel statistical method, still no correlation between codon decoding time and tRNA gene copy numbers (tAI) could be found (Dana and Tuller, 2014). However, this may be related to differential tRNA expression and/or charging levels in different tissues in multicellular organisms such as mice. By applying the novel statistical methods, no correlation between codon decoding times and charged tRNAs could be seen (Dana and Tuller, 2014), yet aminoacyl-tRNA synthetase levels were used as an estimator for charged tRNAs, which is not a proven method to determine aminoacyl-tRNA pools.
In summary, the translation elongation rate is tuned by the interplay of multiple features of the coding sequences, including frequent and rare codon usage with their related tRNA pools, but also mRNA secondary structures and Shine-Dalgarno-like sequences.
Intragenic Codon Landscapes and Expression
In addition to codon frequency bias on genome level, local codon bias within genes is observed. Many genes have locally biased distributions of rare and frequent codons, which results in a gene “codon landscape.” As a consequence of this codon landscape, variable translation rates may occur along a gene's mRNA. Variable local translation rates can regulate the even distribution of translating ribosomes on the mRNA (Tuller et al., 2010), tune the protein co-translational folding process (Zhang et al., 2009), and facilitate protein translocation across membranes (Pechmann et al., 2014). Different types of intragenic codon landscapes that contribute to the modulation of elongation speed are discussed here.
Rare Codon Ramps to Reduce Ribosome Jamming
Analysis of the distribution of frequent and rare codons within genes revealed a common pattern predominantly in highly expressed prokaryotic and eukaryotic genes: a ramp sequence immediately downstream of the start codon consisting of 30–50 relatively rare codons (Tuller et al., 2010) (Figure 4A). The translation rate of this codon ramp was predicted to be relatively slow, after which it would increase to reach a plateau during elongation of the remaining part of the gene. Experimental confirmation of the slower translation of the ramp came from inspection of ribosome density profiles of yeast, which revealed a higher ribosome density for the 50 5′ end codons of genes (Tuller et al., 2010). The only exception is the codon following the start codon; it is usually translated with high efficiency, possibly to support fast release of the initiator tRNAMet (Tuller et al., 2010). The presence of the ramp is thought to increase the overall efficiency of protein synthesis. The relatively slow start of the elongation process appears to evenly space the ribosomes in order to reduce ribosome traffic jams during further elongation of highly expressed proteins with accordingly high ribosome densities on the mRNA (Tuller et al., 2010).
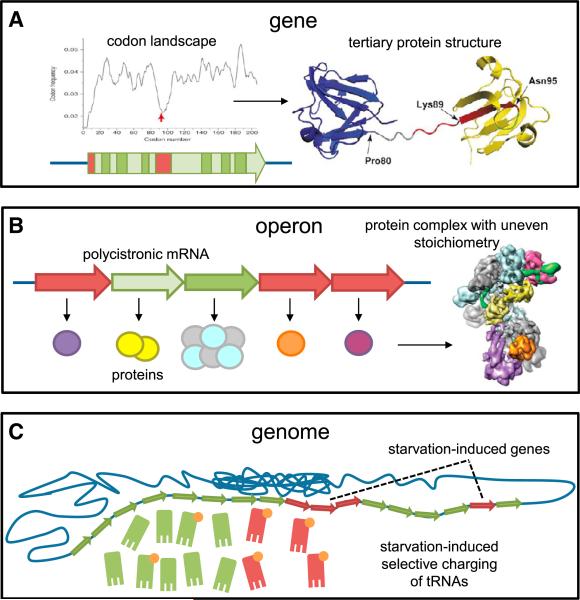
Figure 4. Codon Landscapes and Intergenic Codon Bias.
(A) The intragenic codon landscape can modulate the translation elongation rate, contributing to appropriate co-translational folding of the protein's secondary structure elements and domains. The red arrow indicates a stretch of rare codons encoding the red colored residues in the protein structure. Apart from a ramp sequence at 5′ end of the coding sequence, a valley in the codon landscape occurs between the two domains. The picture represents the bovine β-B2 crystallin and is adapted from Komar (2009).
(B) Differential expression of genes residing in an operon is controlled by differential translation. This is accomplished mainly by enhanced initiation (as reflected by a higher ribosome density) and, to some extent, by adjusted elongation (as reflected by a more optimal codon bias). An example is shown of the operon encoding the CRISPR-associated Cascade complex from E. coli with uneven stoichiometry; cryo-electron microscopic structure is from Wiedenheft et al. (2011).
(C) The expression of sets of functionally related genes can be co-regulated on the basis of their codon bias. The available pools of aminoacyl-tRNAs can specifically change under certain conditions, which lead to improved translation of genes with adapted codon bias. As an example, genes encoding amino acid biosynthesis pathways are shown, which contain many rare codons translated by tRNAs that remain highly charged during amino acid starvation.
An alternative explanation is that the apparent ramp of rare codons is a side effect of selection for reduced mRNA secondary structures at the 5′ end of coding sequences (Goodman et al., 2013; Shah et al., 2013). Some studies could not confirm slower translation elongation at the coding region's 5′ end; these investigators claimed that this earlier observed phenomenon was an artifact either of the ribosome profiling experiment (Ingolia et al., 2011) or of the data analysis, because shorter genes have higher initiation rates (Shah et al., 2013). However, different experimental protocols and data normalization for ribosome profiling did again confirm a slower translation elongation rate at the 5′ end of highly expressed genes (Tuller and Zur, 2015). To summarize, multiple features of the 5′ terminus of coding sequences play different roles in translation, which complicates efforts to separate the individual effects of each of these features on translation efficiency (Tuller and Zur, 2015).
Codon Landscapes for Protein Translation across the Membrane
Recently, rare codon clusters were identified in genes for membrane and secretory proteins; in yeast, these clusters occur 35–40 codons downstream of the binding sites for the signal recognition particle (SRP) (Pechmann et al., 2014). The SRP recognizes secretory signal sequences or transmembrane protein segments as they emerge from the ribosomal exit tunnel. The 35- to 40-codon distance between these rare codon clusters and the SRP-binding sequence spans the length of the ribosomal exit tunnel. After SRP binding to the emerging peptide chain, the nascent chain ribosome complex is transferred to the membrane translocation machinery for co-translational transport of the protein across the membrane. The local translation slowdown by the rare codon clusters would provide additional time for the SRP to associate with the nascent chain ribosome complex and hence could facilitate the subsequent membrane translocation (Pechmann et al., 2014). Translation slowdown at these sites in yeast was confirmed by analyzing ribosome profiling data for transmembrane and secretory proteins (Pechmann et al., 2014).
Furthermore, ribosomes in E. coli were recently found to slow down during membrane protein targeting. This slowdown of the translation elongation was probably caused not by rare codons but by Shine-Dalgarno-like sequences approximately 35 codons downstream of the transmembrane helices (Fluman et al., 2014). Apart from rare codons, slower translation elongation at the 5′ end of coding sequences of membranes proteins may also be caused by a bias for positively charged amino acids found at the cytoplasmic N terminus of membrane proteins (Charneski and Hurst, 2014).
Codon Landscapes and Co-translational Protein Folding
Clusters of rare codons within the codon landscape may modulate the translational rate to coordinate protein translation rate with co-translational protein folding (Purvis et al., 1987). A correlation between certain protein structures and rare or frequent codons has been demonstrated experimentally for some proteins (Spencer et al., 2012; Zhang et al., 2009) and supported computationally for larger sets of proteins (Pechmann and Frydman, 2013; Saunders and Deane, 2010). Using the nTE metric, it was concluded that frequent codons are depleted in regions that encode coils in protein structures. This may relate to the fact that coils contain loops that have keys roles in co-translational folding. For α helices, enrichment of both rare and frequent codon clusters is observed. At the start of α helices, a specific codon usage pattern is observed: rare codons before the helix and at positions 1 and 4 and frequent codons occur at positions 2 and 3; this may reflect the complex co-translational folding of α helices, which already occurs within the ribosomal exit tunnel. β sheets are encoded mainly by regions enriched in frequent codons (Pechmann and Frydman, 2013). The observed trends strongly suggest a correlation of codon bias with co-translational folding; however, complete understanding of the rules requires further analysis. In addition, it should be noted that certain structurally and/or functionally important regions (e.g., catalytic residues) may also be encoded by frequent codons not because of folding kinetics but because these frequent codons may enhance translational accuracy of these important amino acids (Drummond and Wilke, 2008). It was experimentally demonstrated that substitution of rare codons by synonymous frequent codons can cause improper folding that results in either degradation or aggregation (inclusion bodies) (Spencer et al., 2012; Zhang et al., 2009), most likely because of a distorted balance of the protein folding process. In addition, synonymous mutations may also result in more subtle changes, probably by slightly affecting the folding process and resulting in minor changes in the protein structure or in post-translational modifications. Consequently, these “silent” mutations may result in subtle but significant changes in protein functionality (Kimchi-Sarfaty et al., 2007; Zhou et al., 2013).
Intergenic Codon Bias and Differential Expression
Differences in codon bias occur not only between regions within individual genes (intragenic, as discussed in the previous section) but also between sets of genes, either clustered in operons or scattered in a genome (Figures 4B and 4C). The latter intergenic codon bias may facilitate differential expression of sets of genes. In this section, we discuss some types of intergenic codon bias and related differential gene expression.
Starvation Conditions and Codon Bias
The large number of rare codons in particular gene clusters that encode amino acid biosynthetic pathways appears counterintuitive. However, this observation makes perfect sense when the relative levels of aminoacyl-tRNA under starvation conditions are considered. These rare codons present in amino acid biosynthesis genes are recognized by tRNAs that remain relatively highly charged during starvation (Dittmar et al., 2005). Furthermore, during starvation, lowly charged tRNAs have a role in the “transcriptional attenuation control” mechanism, found in bacterial amino acid biosynthesis operons (Elf and Ehrenberg, 2005). This control mechanism relies on the competition between ribosomes translating at the leader cistron of an mRNA and the RNA polymerase transcribing the cistrons downstream on the polycistronic mRNA. In the leader cistron of the mRNA, codons recognized by tRNAs with low charging levels during starvation are present, which during starvation results in ribosome stalling during translation of the leader. This stalling affects the secondary structure of a downstream sequence in the mRNA and as such relieves transcription repression that results in further transcription of downstream cistrons encoding the biosynthetic pathway enzymes (Elf and Ehrenberg, 2005).
Cell-Cycle, Differentiation, and Stress Regulation by Codon Usage
Variations in tRNA expression during different states of a cell may enable differential expression of sets of genes with codons adapted to different tRNA pools. For example, concentrations of tRNAs and aminoacyl-tRNA synthetases have been demonstrated to oscillate during the human cell cycle (Frenkel-Morgenstern et al., 2012). As a consequence, gene sets that are expressed at different phases of the cell cycle have different codon usage (Frenkel-Morgenstern et al., 2012). This provides a codon bias strategy that supports cell-cycle regulation. In humans and other vertebrates, tRNA concentrations differ in proliferating and differentiating cell types (Gingold et al., 2014). Genes specific for proliferation and differentiation processes have a corresponding codon bias; this implies that the two distinct translational programs that operate during proliferation and differentiation are regulated by codon bias. The cyanobacterium Synechococcus elongatus uses codons to adjust its protein production to fluctuating environmental conditions. The genes encoding the circadian clock-associated oscillator proteins contain rare codons leading to low expression at low temperatures. This causes the desired suppression of circadian regulation at low temperatures (Xu et al., 2013).
Altered tRNA modification patterns present another strategy to adapt gene expression of large sets of genes to different conditions. RNA modifications can alter the codon-anticodon binding affinity of tRNA molecules. Translation of certain codons can be favored in this way, resulting in enhanced expression of gene sets that contain elevated frequencies of these codons. Environmental factors have been reported to cause changes in RNA modifications. For example, in yeast, stress induced by DNA-damaging compounds (Begley et al., 2007) or oxidative stress (Chan et al., 2012) upregulates specific tRNA-modifying enzymes. Interestingly, in the genes required for coping with these stress factors, the codons recognized by these modified tRNAs are overrepresented (Begley et al., 2007; Chan et al., 2012). Thus, fluctuations of the (aminoacyl-)tRNA pools and tRNA modifications play a role in regulating expression of genes with adapted codon usage.
Differential Expression within Operons
As outlined in the previous sections, functionally related genes generally have a similar codon bias that allows their co-regulation under specific conditions. However, significant differences in the degree of codon bias of related genes have been described as well. Even within prokaryotic operons, codon bias of individual genes may differ considerably. Within the ATP-synthase operon, genes encoding the highly abundant ATPase subunits were observed to be enriched in codons recognized by abundant tRNAs (Gouy and Gautier, 1982). In a recent comparative genomics analysis, the correlation between subunit stoichiometry and codon bias was demonstrated in many different operons in bacteria and archaea. These operons were selected because they encode protein complexes with established uneven subunit stoichiometry and included highly expressed complexes (e.g., ribosome, ATPase) as well as poorly expressed complexes (e.g., CRISPR-associated Cascade complex) (Quax et al., 2013) (Figure 4B). Translation of several polycistronic messengers encoding protein complexes with uneven stoichiometry were evaluated by ribosome density profiles (Li et al., 2012); this analysis revealed that the cistrons coding for the abundant subunits had correlating high ribosome densities (Quax et al., 2013). This indicates that translation initiation of these cistrons allows differential translation. In addition, a positive correlation was found between more highly expressed subunits and codon frequency bias and co-occurrence bias. This correlation strongly suggests a role of codon bias in tuning the elongation rate of highly expressed subunits. Differential translation is proposed as a universal control mode to tune differential production of operon-encoded protein complexes with uneven stoichiometry (Quax et al., 2013). In addition to the aforementioned protein complexes, differential translation is important for other groups of (operon-encoded) related proteins of which differential production is required, including control systems and metabolic pathways (Li et al., 2014).
Selection Pressure on Codon Usage
It is firmly established that codon usage is biased in the majority of living organisms. Two not mutually exclusive explanations on the evolution of this bias have been formulated: (1) non-randomness of mutation and (2) selection for codon bias (Hershberg and Petrov, 2008). Some nucleotides or codons may have higher mutation rates, resulting in lower frequencies of some codons and nucleotides. Some investigators have claimed that codon bias is related mainly to the non-random mutations caused by the global GC content of an organism, because this GC content seems determined for the complete genome, not only for the coding part of the genome (Chen et al., 2004; Knight et al., 2001). However, as previously discussed, different types of codon usage bias were observed to be related to translation efficiencies; therefore, this codon bias must be under selective pressure during evolution, and mutation rates alone cannot explain the various observations. Especially intragenic and intergenic codon bias cannot be explained by the mutational theory (Hershberg and Petrov, 2008).
Certain codons may be selected to achieve efficient and/or accurate translation, and both may influence cellular fitness to a certain extent (Hershberg and Petrov, 2008). Higher translation accuracy may result from using codons that are recognized by abundant tRNAs. Improved translation accuracy will avoid wasting resources and energy caused by production of nonfunctional proteins. The “accuracy theory” is supported by the aforementioned detection of more stringent selection for codon bias at crucial positions in proteins, potentially to ensure high-fidelity translation of these structurally and/or functionally important residues (Drummond and Wilke, 2008; Stoletzki and Eyre-Walker, 2007; Zhou et al., 2009). In addition, stronger codon bias was found for longer genes, most likely because of relatively higher resource costs for mistranslation of larger proteins (Stoletzki and Eyre-Walker, 2007).
Selection pressure on translation efficiency can act locally, because codon usage landscapes can affect protein folding. In addition, there will be selection for local coding sequence features such as mRNA secondary structures, because these allow tuning of gene expression. Selection pressure may also act on codon usage at a global scale, because more optimal codons may give rise to higher global translation rates, thus keeping more ribosomes available and thereby increasing cellular fitness (Andersson and Kurland, 1990; Berg and Kurland, 1997). In the struggle of life that occurs in most natural ecosystems, the avail ability of limited resources will imply that the translation efficiency will impose a strong selective pressure on an optimal codon bias. This notion is supported by an enhanced frequency of optimal codons in highly expressed genes in rapidly growing organisms (Ran and Higgs, 2012). In summary, the different types of codon bias that have evolved in all domains of life reflect an optimized combination of frequent and rare codons, which will allow appropriate translation of a certain gene, in a certain organism, under certain conditions, and eventually in a certain tissue or organelle.
Applying Codon Bias as a Means to Improve Protein Production
Codon bias has been studied intensively during the past three decades, and some of the insights gained have been widely applied in biotechnology as a strategy to optimize gene expression for improving protein production rates and yields. The two major strategies are adjusting the expressed set of tRNAs and adjusting the codon usage of a gene of interest.
Expressing Additional tRNA Genes in the Production Host
Additional copies of tRNA genes are introduced to enhance tRNA levels, aiming for the heterologous expression of genes that contain many rare codons. At present, this strategy has been used mainly in bacterial production systems. Several commercial bacterial strains expressing extra tRNAs from a plasmid are available for this purpose (e.g., E. coli Rosetta [pRARE plasmid with genes of tRNAs that recognize the following codons: AGG, AGA, AUA, CUA, CCC, GGA], E. coli BL21-CodonPlus [pRIL plasmid with genes of tRNAs that recognize the following codons: AGG, AGA, AUA, CUA, CCC]) (Gustafsson et al., 2004) (Table 1). There are many examples demonstrating that this strategy was successful, or at least resulted in improved functional protein production. However, in many other cases, this approach did not enhance protein yields, for instance, when slow translation of stretches of rare codons in a gene was required for proper protein folding (Zhang et al., 2009). The main problem with this strategy is that the protein-specific codon landscape (frequent as well as rare codons) is not taken into account.
Designing Codon-Optimized Genes
The decreasing costs of de novo DNA synthesis have allowed the synthesis of codon-optimized genes. The potential space for synonymous codon substitutions throughout a gene is extremely large; for a protein of 300 amino acids, more than 10100 different coding sequence variants are possible (Welch et al., 2009a). Therefore, automated codon optimization algorithms have been developed to design coding sequences optimized for increased expression in certain hosts (Gould et al., 2014). Most DNA synthesis companies offer codon optimization services, primarily on the basis of confidential algorithms. Many of these algorithms optimize codon usage by maximizing a gene's CAI to match that of the expression host, along with optimizing for some sequence features. Sequence features regularly taken into account are GC content and avoidance of repeats and motifs such as ribonuclease (RNase) recognition sites, transcriptional terminator sites, Shine-Dalgarno-like sequences, and sequences that lead to strong mRNA secondary structures (Table 2) (Gustafsson et al., 2012). There are many reports of successful codon optimization of coding sequences, which can lead up to 1,000-fold increased gene expression by optimizing the coding sequence (Gustafsson et al., 2012). For expression of a large set of human genes in E. coli, such algorithms were reported to be more successful than expressing additional tRNAs (Maertens et al., 2010). However, for many genes, expression was not improved by such algorithms (Gustafsson et al., 2012; Maertens et al., 2010). Thus, the output of these algorithms is not a guarantee for success, for example because synonymous mutations might interfere with protein folding by altering the codon landscape.
Table 2.
Coding Sequence Features Relevant for Synthetic Gene Design
| Global Host and Condition-Related Codon Usage |
| codon usage frequency of highly expressed genes (CAI) |
| tRNA gene copy numbers (tAI) |
| balance tRNA supply/mRNA codon demand (nTE)a |
| tRNA expression levelsa |
| charged tRNA levelsa |
| tRNA modification levelsa |
| synonymous codon co-occurrence biasa |
| non-synonymous codon pair biasa |
| Local Gene-Level Codon Usage Landscape |
| rare codon ramps at 5′ end of coding sequencesa |
| codon frequencies related to protein co-translational foldinga |
| codon frequencies related to protein translocation across a membranea |
| Features Not Directly Related to Codon Usage |
| GC content |
| mRNA folding at 5′ end of coding sequences |
| mRNA folding throughout coding sequencea |
| Shine-Dalgarno-like sequences (prokaryotes)a |
| transcriptional terminators |
| restriction sites |
| RNase E sites |
| hidden stop codons |
| sequence repeats |
| transcription factor recognition sequences |
| CpG and other dinucleotide bias (eukaryotes) |
| polyadenylation signals (eukaryotes) |
| cryptic splicing signals (eukaryotes) |
Most coding sequence features listed are discussed in this review; some sequence features not directly related to codon usage are not discussed here but have been reviewed previously (Gould et al., 2014; Gustafsson et al., 2012).
Features that deserve more specific attention for synthetic gene design and/or better elucidation of their relevance for protein expression
Alternative approaches for synonymous codon design have also been reported. The “codon harmonization algorithm” adapts the codons in a way that the original codon landscape of the gene in the original host is maintained in the expression hosts (Angov et al., 2008). Hence, this algorithm retains a larger proportion of rare codons in comparison with other optimization algorithms. A successful application of this algorithm was the improved heterologous expression of a few proteins in E. coli (Angov et al., 2008; Spencer et al., 2012). A more systematic experimental and statistical approach to optimize coding sequences for heterologous gene expression in E. coli identified several crucial amino acids and specific synonymous codons that are essential for high expression. These crucial codons generally were not the most frequently used codons in highly expressed genes in the host, but some crucial codons were related to cognate tRNAs that remain highly charged under starvation conditions (Welch et al., 2009b). Evaluating different codon optimization strategies to improve protein expression is not straightforward. First, generally only single case studies of optimizing coding sequences for different proteins have been reported and are thus difficult to evaluate (Welch et al., 2009a). Second, the many coding sequence features that influence expression (Table 2), and their uncertain hierarchy in different genes, in different hosts, and under different conditions, pose a major challenge.
Challenges Ahead
Unraveling Codon Bias and Other Factors Influencing Expression
A future challenge in studying the relation between coding sequences and protein production is to perform a thorough comparative analysis of all currently known, and yet to be discovered, features of coding sequences that influence the translation process. This may be achieved by further improvements and integration of experimental approaches and statistical analyses. Experimental RNA-sequencing data that determine mRNA and tRNA abundance, ribosome density profiling, and proteomics should be integrally analyzed. To date, most coding sequence features affecting gene expression have been derived from natural expression data or overexpression of randomly generated libraries of reporter proteins. A systematic approach designing synthetic gene variants will be more efficient than generating and testing random gene variants (Gustafsson et al., 2012). The variant design should systematically vary coding sequence features that potentially affect expression and whenever possible minimize the co-variance between individual features. Such a systematic approach would allow us to reveal the effects of several coding sequence features with a relatively limited number of variants to be tested (Gustafsson et al., 2012).
Many studies that have addressed codon bias by analyzing libraries of synthetic reporter variants have resulted mainly in the identification of coding sequence features that affect translation initiation (Goodman et al., 2013; Kudla et al., 2009). To analyze the effect of codon bias on translation elongation, it would be better to use a recently developed series of synthetic translation initiation elements that result in constant translation initiation rates independent of the 5′ end of a coding sequence (Mutalik et al., 2013). To complement studies of overexpressed synonymous reporter variants, studies of synonymous codon variants of naturally expressed genes may reveal additional rules for translation efficiency. Chromosomal genes can be replaced by synthetic, strategically designed synonymous variants. Recently, a set of highly expressed chromosomal genes in E. coli has been replaced by codon-shuffled variants (Lajoie et al., 2013). However, this study was limited to only three variants of these genes, which does not yet allow for composing solid rules on how the coding sequence affects gene expression. Further probing the hierarchy of features hidden in the coding sequence that contribute to the efficiency of protein synthesis requires the analysis of many different genes from different domains of life, cell types, and conditions.
Further Improving Synthetic Gene Design
Natural selection formed optimal codon landscapes for different types of genes and organisms, and now the main challenge is to understand the rules on how to recreate these landscapes for high-level production in heterologous production systems. The codon optimization field is gradually moving away from the common concept that synthetic genes should contain as much as possible frequent codons in order to achieve high protein production. Many features are generally accepted to be important sequence for synthetic gene design (Table 2). To date, several of these features have hardly been used in synthetic gene design and deserve more attention in future attempts. For example, a better experimental analysis of aminoacyl-tRNA abundance under protein overproduction conditions should provide the basis for better codon optimizations ensuring a balanced aminoacyl-tRNA supply for production of the protein of interest. This also implies that more accurate metrics than CAI, for example the nTE, for predicting optimal codon bias should be considered.
Given our deeper understanding of how the codon landscape influences the efficiency of translation and protein folding, novel design rules for synthetic genes should be considered. Especially for high-level expression of secreted or membrane proteins, along with adjustment of the codon bias to the heterologous expression host, landscape features such as clusters of rare codons or other coding sequence pause sites need to be incorporated in the gene design. Further refinements of the “codon harmonization” approach (Angov et al., 2008) would be promising for this purpose.
Applying Codon Bias as a Tool in Synthetic Biology
Improved rational gene design underlies the attempts in synthetic biology to create synthetic gene circuits, biosynthetic pathways, or even new genomes. In the design of circuits or pathways, expression of functionally related genes is crucial. For designing synthetic operons to express these circuits or pathways, one can learn from recently elucidated roles of differential codon bias and other factors in differentially expressed cistrons in prokaryotic operons (Li et al., 2014; Quax et al., 2013).
Synthetic biology now enables engineering at genome level. Rare codons have been replaced by synonymous codons on a genome-wide scale, and removed rare codons could be reassigned to encode non-natural amino acids (Lajoie et al., 2013; Mukai et al., 2015; Rovner et al., 2015). Furthermore, the nucleotide alphabet of life has recently been expanded by introducing two synthetic nucleotides in E. coli (Malyshev et al., 2014). For a sweeping modification of the codon alphabet, a better understanding of codon bias will be advantageous. In addition, recent advances in DNA assembly have permitted the assembly and transplantation of complete synthetic genomes (Gibson, 2014). This technology practically enables the redesign of genomes from scratch. However, a more thorough understanding of codon bias is needed for rational design of an optimally functional synthetic genome with sensible choices of codon usage, related tRNA genes, and tRNA-modifying enzymes.
ACKNOWLEDGMENTS
We are grateful to Henry Grosjean for stimulating discussions. This work was supported by the Fonds voor Wetenschappelijk Onderzoek (FWO-Belgium) by an FWO Pegasus Marie-Curie Grant to T.E.F.Q., the IP/OP program Systems Biology of Wageningen University to N.J.C., the National Institute of General Medical Sciences (grant GM22854) to D.S., and the Netherlands Organization for Scientific Research (NWO) by an ALW-TOP grant (854.10.003) to J.O.
Footnotes
AUTHOR CONTRIBUTIONS
T.E.F.Q. and N.J.C. reviewed the literature. T.E.F.Q., N.J.C., D.S., and J.O. wrote the paper.
REFERENCES
- Agris PF, Vendeix FAP, Graham WD. tRNA's wobble decoding of the genome: 40 years of modification. J. Mol. Biol. 2007;366:1–13. doi: 10.1016/j.jmb.2006.11.046. [DOI] [PubMed] [Google Scholar]
- Andersson SG, Kurland CG. Codon preferences in free-living microorganisms. Microbiol. Rev. 1990;54:198–210. doi: 10.1128/mr.54.2.198-210.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angov E, Hillier CJ, Kincaid RL, Lyon JA. Heterologous protein expression is enhanced by harmonizing the codon usage frequencies of the target gene with those of the expression host. PLoS ONE. 2008;3:e2189. doi: 10.1371/journal.pone.0002189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begley U, Dyavaiah M, Patil A, Rooney JP, DiRenzo D, Young CM, Conklin DS, Zitomer RS, Begley TJ. Trm9-catalyzed tRNA modifications link translation to the DNA damage response. Mol. Cell. 2007;28:860–870. doi: 10.1016/j.molcel.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentele K, Saffert P, Rauscher R, Ignatova Z, Blüthgen N. Efficient translation initiation dictates codon usage at gene start. Mol. Syst. Biol. 2013;9:675. doi: 10.1038/msb.2013.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg OG, Kurland CG. Growth rate-optimised tRNA abundance and codon usage. J. Mol. Biol. 1997;270:544–550. doi: 10.1006/jmbi.1997.1142. [DOI] [PubMed] [Google Scholar]
- Buchan JR, Aucott LS, Stansfield I. tRNA properties help shape codon pair preferences in open reading frames. Nucleic Acids Res. 2006;34:1015–1027. doi: 10.1093/nar/gkj488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannarozzi G, Schraudolph NN, Faty M, von Rohr P, Friberg MT, Roth AC, Gonnet P, Gonnet G, Barral Y. A role for codon order in translation dynamics. Cell. 2010;141:355–367. doi: 10.1016/j.cell.2010.02.036. [DOI] [PubMed] [Google Scholar]
- Chan CT, Pang YL, Deng W, Babu IR, Dyavaiah M, Begley TJ, Dedon PC. Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat. Commun. 2012;3:937. doi: 10.1038/ncomms1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charneski CA, Hurst LD. Positively charged residues are the major determinants of ribosomal velocity. PLoS Biol. 2013;11:e1001508. doi: 10.1371/journal.pbio.1001508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charneski CA, Hurst LD. Positive charge loading at protein termini is due to membrane protein topology, not a translational ramp. Mol. Biol. Evol. 2014;31:70–84. doi: 10.1093/molbev/mst169. [DOI] [PubMed] [Google Scholar]
- Chen SL, Lee W, Hottes AK, Shapiro L, McAdams HH. Codon usage between genomes is constrained by genome-wide mutational processes. Proc. Natl. Acad. Sci. U S A. 2004;101:3480–3485. doi: 10.1073/pnas.0307827100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu D, Kazana E, Bellanger N, Singh T, Tuite MF, von der Haar T. Translation elongation can control translation initiation on eukaryotic mRNAs. EMBO J. 2014;33:21–34. doi: 10.1002/embj.201385651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman JR, Papamichail D, Skiena S, Futcher B, Wimmer E, Mueller S. Virus attenuation by genome-scale changes in codon pair bias. Science. 2008;320:1784–1787. doi: 10.1126/science.1155761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick FH. Codon–anticodon pairing: the wobble hypothesis. J. Mol. Biol. 1966;19:548–555. doi: 10.1016/s0022-2836(66)80022-0. [DOI] [PubMed] [Google Scholar]
- Dana A, Tuller T. The effect of tRNA levels on decoding times of mRNA codons. Nucleic Acids Res. 2014;42:9171–9181. doi: 10.1093/nar/gku646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmar KA, Sørensen MA, Elf J, Ehrenberg M, Pan T, Sørensen M. Selective charging of tRNA isoacceptors induced by amino-acid starvation. EMBO Rep. 2005;6:151–157. doi: 10.1038/sj.embor.7400341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmar KA, Goodenbour JM, Pan T. Tissue-specific differences in human transfer RNA expression. PLoS Genet. 2006;2:e221. doi: 10.1371/journal.pgen.0020221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Reis M, Savva R, Wernisch L. Solving the riddle of codon usage preferences: a test for translational selection. Nucleic Acids Res. 2004;32:5036–5044. doi: 10.1093/nar/gkh834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DA, Wilke CO. Mistranslation-induced protein mis-folding as a dominant constraint on coding-sequence evolution. Cell. 2008;134:341–352. doi: 10.1016/j.cell.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elf J, Ehrenberg M. What makes ribosome-mediated transcriptional attenuation sensitive to amino acid limitation? PLoS Comput. Biol. 2005;1:e2. doi: 10.1371/journal.pcbi.0010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elf J, Nilsson D, Tenson T, Ehrenberg M. Selective charging of tRNA isoacceptors explains patterns of codon usage. Science. 2003;300:1718–1722. doi: 10.1126/science.1083811. [DOI] [PubMed] [Google Scholar]
- Fluman N, Navon S, Bibi E, Pilpel Y. mRNA-programmed translation pauses in the targeting of E. coli membrane proteins. eLife. 2014;3:e03440. doi: 10.7554/eLife.03440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkel-Morgenstern M, Danon T, Christian T, Igarashi T, Cohen L, Hou YM, Jensen LJ. Genes adopt non-optimal codon usage to generate cell cycle-dependent oscillations in protein levels. Mol. Syst. Biol. 2012;8:572. doi: 10.1038/msb.2012.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardin J, Yeasmin R, Yurovsky A, Cai Y, Skiena S, Futcher B. Measurement of average decoding rates of the 61 sense codons in vivo. eLife. 2014;3:1–20. doi: 10.7554/eLife.03735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG. Programming biological operating systems: genome design, assembly and activation. Nat. Methods. 2014;11:521–526. doi: 10.1038/nmeth.2894. [DOI] [PubMed] [Google Scholar]
- Gingold H, Tehler D, Christoffersen NR, Nielsen MM, Asmar F, Kooistra SM, Christophersen NS, Christensen LL, Borre M, Sørensen KD, et al. A dual program for translation regulation in cellular proliferation and differentiation. Cell. 2014;158:1281–1292. doi: 10.1016/j.cell.2014.08.011. [DOI] [PubMed] [Google Scholar]
- Godinic-Mikulcic V, Jaric J, Greber BJ, Franke V, Hodnik V, Anderluh G, Ban N, Weygand-Durasevic I. Archaeal aminoacyl-tRNA synthetases interact with the ribosome to recycle tRNAs. Nucleic Acids Res. 2014;42:5191–5201. doi: 10.1093/nar/gku164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman DB, Church GM, Kosuri S. Causes and effects of N-terminal codon bias in bacterial genes. Science. 2013;342:475–479. doi: 10.1126/science.1241934. [DOI] [PubMed] [Google Scholar]
- Gould N, Hendy O, Papamichail D. Computational tools and algorithms for designing customized synthetic genes. Front. Bioeng. Biotechnol. 2014;2:41. doi: 10.3389/fbioe.2014.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M, Gautier C. Codon usage in bacteria: correlation with gene expressivity. Nucleic Acids Res. 1982;10:7055–7074. doi: 10.1093/nar/10.22.7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantham R, Gautier C, Gouy M, Mercier R, Pavé A. Codon catalog usage and the genome hypothesis. Nucleic Acids Res. 1980;8:r49–r62. doi: 10.1093/nar/8.1.197-c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromadski KB, Daviter T, Rodnina MV. A uniform response to mismatches in codon-anticodon complexes ensures ribosomal fidelity. Mol. Cell. 2006;21:369–377. doi: 10.1016/j.molcel.2005.12.018. [DOI] [PubMed] [Google Scholar]
- Grosjean H, de Crécy-Lagard V, Marck C. Deciphering synonymous codons in the three domains of life: co-evolution with specific tRNA modification enzymes. FEBS Lett. 2010;584:252–264. doi: 10.1016/j.febslet.2009.11.052. [DOI] [PubMed] [Google Scholar]
- Gustafsson C, Govindarajan S, Minshull J. Codon bias and heterologous protein expression. Trends Biotechnol. 2004;22:346–353. doi: 10.1016/j.tibtech.2004.04.006. [DOI] [PubMed] [Google Scholar]
- Gustafsson C, Minshull J, Govindarajan S, Ness J, Villalobos A, Welch M, Drive OB, Suite A, Park M. Engineering genes for predictable protein expression. Protein Expr. Purif. 2012;83:37–46. doi: 10.1016/j.pep.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutman GA, Hatfield GW. Nonrandom utilization of codon pairs in Escherichia coli. Proc. Natl. Acad. Sci. U S A. 1989;86:3699–3703. doi: 10.1073/pnas.86.10.3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberg R, Petrov DA. Selection on codon bias. Annu. Rev. Genet. 2008;42:287–299. doi: 10.1146/annurev.genet.42.110807.091442. [DOI] [PubMed] [Google Scholar]
- Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms. Mol. Biol. Evol. 1985;2:13–34. doi: 10.1093/oxfordjournals.molbev.a040335. [DOI] [PubMed] [Google Scholar]
- Ingolia NT. Ribosome profiling: new views of translation, from single codons to genome scale. Nat. Rev. Genet. 2014;15:205–213. doi: 10.1038/nrg3645. [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaya S, Yamada Y, Kudo Y, Ikemura T. Studies of codon usage and tRNA genes of 18 unicellular organisms and quantification of Bacillus subtilis tRNAs: gene expression level and species-specific diversity of codon usage based on multivariate analysis. Gene. 1999;238:143–155. doi: 10.1016/s0378-1119(99)00225-5. [DOI] [PubMed] [Google Scholar]
- Khade PK, Joseph S. Messenger RNA interactions in the decoding center control the rate of translocation. Nat. Struct. Mol. Biol. 2011;18:1300–1302. doi: 10.1038/nsmb.2140. [DOI] [PubMed] [Google Scholar]
- Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, Ambudkar SV, Gottesman MM. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- Knight RD, Freeland SJ, Landweber LF. A simple model based on mutation and selection explains trends in codon and amino-acid usage and GC composition within and across genomes. Genome Biol. 2001;2:RESEARCH0010. doi: 10.1186/gb-2001-2-4-research0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komar AA. A pause for thought along the co-translational folding pathway. Trends Biochem. Sci. 2009;34:16–24. doi: 10.1016/j.tibs.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Kudla G, Murray AW, Tollervey D, Plotkin JB. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009;324:255–258. doi: 10.1126/science.1170160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lajoie MJ, Kosuri S, Mosberg JA, Gregg CJ, Zhang D, Church GM. Probing the limits of genetic recoding in essential genes. Science. 2013;342:361–363. doi: 10.1126/science.1241460. [DOI] [PubMed] [Google Scholar]
- Li G-WW, Oh E, Weissman JS. The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature. 2012;484:538–541. doi: 10.1038/nature10965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li GW, Burkhardt D, Gross C, Weissman JS. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell. 2014;157:624–635. doi: 10.1016/j.cell.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maertens B, Spriestersbach A, von Groll U, Roth U, Kubicek J, Gerrits M, Graf M, Liss M, Daubert D, Wagner R, Schäfer F. Gene optimization mechanisms: a multi-gene study reveals a high success rate of full-length human proteins expressed in Escherichia coli. Protein Sci. 2010;19:1312–1326. doi: 10.1002/pro.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malyshev DA, Dhami K, Lavergne T, Chen T, Dai N, Foster JM, Corrêa IR, Jr., Romesberg FE. A semi-synthetic organism with an expanded genetic alphabet. Nature. 2014;509:385–388. doi: 10.1038/nature13314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai T, Hoshi H, Ohtake K, Takahashi M, Yamaguchi A, Hayashi A, Yokoyama S, Sakamoto K. Highly reproductive Escherichia coli cells with no specific assignment to the UAG codon. Sci. Rep. 2015;5:9699. doi: 10.1038/srep09699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutalik VK, Guimaraes JC, Cambray G, Lam C, Christoffersen MJ, Mai Q-A, Tran AB, Paull M, Keasling JD, Arkin AP, Endy D. Precise and reliable gene expression via standard transcription and translation initiation elements. Nat. Methods. 2013;10:354–360. doi: 10.1038/nmeth.2404. [DOI] [PubMed] [Google Scholar]
- Novoa EM, Pavon-Eternod M, Pan T, Ribas de Pouplana L. A role for tRNA modifications in genome structure and codon usage. Cell. 2012;149:202–213. doi: 10.1016/j.cell.2012.01.050. [DOI] [PubMed] [Google Scholar]
- Pechmann S, Frydman J. Evolutionary conservation of codon optimality reveals hidden signatures of cotranslational folding. Nat. Struct. Mol. Biol. 2013;20:237–243. doi: 10.1038/nsmb.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechmann S, Chartron JW, Frydman J. Local slowdown of translation by nonoptimal codons promotes nascent-chain recognition by SRP in vivo. Nat. Struct. Mol. Biol. 2014;21:1100–1105. doi: 10.1038/nsmb.2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pop C, Rouskin S, Ingolia NT, Han L, Phizicky EM, Weissman JS, Koller D. Causal signals between codon bias, mRNA structure, and the efficiency of translation and elongation. Mol. Syst. Biol. 2014;10:770. doi: 10.15252/msb.20145524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purvis IJ, Bettany AJ, Santiago TC, Coggins JR, Duncan K, Eason R, Brown AJ. The efficiency of folding of some proteins is increased by controlled rates of translation in vivo. A hypothesis. J. Mol. Biol. 1987;193:413–417. doi: 10.1016/0022-2836(87)90230-0. [DOI] [PubMed] [Google Scholar]
- Qian W, Yang JR, Pearson NM, Maclean C, Zhang J. Balanced codon usage optimizes eukaryotic translational efficiency. PLoS Genet. 2012;8:e1002603. doi: 10.1371/journal.pgen.1002603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quax TE, Wolf YI, Koehorst JJ, Wurtzel O, van der Oost R, Ran W, Blombach F, Makarova KS, Brouns SJ, Forster AC, et al. Differential translation tunes uneven production of operon-encoded proteins. Cell Rep. 2013;4:938–944. doi: 10.1016/j.celrep.2013.07.049. [DOI] [PubMed] [Google Scholar]
- Ran W, Higgs PG. Contributions of speed and accuracy to translational selection in bacteria. PLoS ONE. 2012;7:e51652. doi: 10.1371/journal.pone.0051652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovner AJ, Haimovich AD, Katz SR, Li Z, Grome MW, Gassaway BM, Amiram M, Patel JR, Gallagher RR, Rinehart J, Isaacs FJ. Recoded organisms engineered to depend on synthetic amino acids. Nature. 2015;518:89–93. doi: 10.1038/nature14095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders R, Deane CM. Synonymous codon usage influences the local protein structure observed. Nucleic Acids Res. 2010;38:6719–6728. doi: 10.1093/nar/gkq495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P, Ding Y, Niemczyk M, Kudla G, Plotkin JB. Rate-limiting steps in yeast protein translation. Cell. 2013;153:1589–1601. doi: 10.1016/j.cell.2013.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao ZQ, Zhang YM, Feng XY, Wang B, Chen JQ. Synonymous codon ordering: a subtle but prevalent strategy of bacteria to improve translational efficiency. PLoS ONE. 2012;7:e33547. doi: 10.1371/journal.pone.0033547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Li WH. An evolutionary perspective on synonymous codon usage in unicellular organisms. J. Mol. Evol. 1986;24:28–38. doi: 10.1007/BF02099948. [DOI] [PubMed] [Google Scholar]
- Sharp PM, Li WH. The codon Adaptation Index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söll D, Jones DS, Ohtsuka E, Faulkner RD, Lohrmann R, Hayatsu H, Khorana HG. Specificity of sRNA for recognition of codons as studied by the ribosomal binding technique. J. Mol. Biol. 1966;19:556–573. doi: 10.1016/s0022-2836(66)80023-2. [DOI] [PubMed] [Google Scholar]
- Spencer PS, Siller E, Anderson JF, Barral JM. Silent substitutions predictably alter translation elongation rates and protein folding efficiencies. J. Mol. Biol. 2012;422:328–335. doi: 10.1016/j.jmb.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler M, Fire A. Wobble base-pairing slows in vivo translation elongation in metazoans. RNA. 2011;17:2063–2073. doi: 10.1261/rna.02890211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoletzki N, Eyre-Walker A. Synonymous codon usage in Escherichia coli: selection for translational accuracy. Mol. Biol. Evol. 2007;24:374–381. doi: 10.1093/molbev/msl166. [DOI] [PubMed] [Google Scholar]
- Supek F, Šmuc T. On relevance of codon usage to expression of synthetic and natural genes in Escherichia coli. Genetics. 2010;185:1129–1134. doi: 10.1534/genetics.110.115477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tats A, Tenson T, Remm M. Preferred and avoided codon pairs in three domains of life. BMC Genomics. 2008;9:463. doi: 10.1186/1471-2164-9-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuller T, Zur H. Multiple roles of the coding sequence 5′ end in gene expression regulation. Nucleic Acids Res. 2015;43:13–28. doi: 10.1093/nar/gku1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuller T, Carmi A, Vestsigian K, Navon S, Dorfan Y, Zaborske J, Pan T, Dahan O, Furman I, Pilpel Y. An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell. 2010;141:344–354. doi: 10.1016/j.cell.2010.03.031. [DOI] [PubMed] [Google Scholar]
- Tulloch F, Atkinson NJ, Evans DJ, Ryan MD, Simmonds P. RNA virus attenuation by codon pair deoptimisation is an artefact of increases in CpG/UpA dinucleotide frequencies. eLife. 2014;3:e04531. doi: 10.7554/eLife.04531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch M, Villalobos A, Gustafsson C, Minshull J. You’re one in a googol: optimizing genes for protein expression. J. R. Soc. Interface. 2009a;6(Suppl 4):S467–S476. doi: 10.1098/rsif.2008.0520.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch M, Govindarajan S, Ness JE, Villalobos A, Gurney A, Minshull J, Gustafsson C. Design parameters to control synthetic gene expression in Escherichia coli. PLoS ONE. 2009b;4:e7002. doi: 10.1371/journal.pone.0007002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, Lander GC, Zhou K, Jore MM, Brouns SJJ, van der Oost J, Doudna JA, Nogales E. Structures of the RNA- guided surveillance complex from a bacterial immune system. Nature. 2011;477:486–489. doi: 10.1038/nature10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolstenhulme CJ, Parajuli S, Healey DW, Valverde DP, Petersen EN, Starosta AL, Guydosh NR, Johnson WE, Wilson DN, Buskirk AR. Nascent peptides that block protein synthesis in bacteria. Proc. Natl. Acad. Sci. U S A. 2013;110:E878–E887. doi: 10.1073/pnas.1219536110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Ma P, Shah P, Rokas A, Liu Y, Johnson CH. Non-optimal codon usage is a mechanism to achieve circadian clock conditionality. Nature. 2013;495:116–120. doi: 10.1038/nature11942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Hubalewska M, Ignatova Z. Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat. Struct. Mol. Biol. 2009;16:274–280. doi: 10.1038/nsmb.1554. [DOI] [PubMed] [Google Scholar]
- Zhang YM, Shao ZQ, Yang LT, Sun XQ, Mao YF, Chen JQ, Wang B. Non-random arrangement of synonymous codons in archaea coding sequences. Genomics. 2013;101:362–367. doi: 10.1016/j.ygeno.2013.04.008. [DOI] [PubMed] [Google Scholar]
- Zhou T, Weems M, Wilke CO. Translationally optimal codons associate with structurally sensitive sites in proteins. Mol. Biol. Evol. 2009;26:1571–1580. doi: 10.1093/molbev/msp070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Guo J, Cha J, Chae M, Chen S, Barral JM, Sachs MS, Liu Y. Non-optimal codon usage affects expression, structure and function of clock protein FRQ. Nature. 2013;495:111–115. doi: 10.1038/nature11833. [DOI] [PMC free article] [PubMed] [Google Scholar]