Abstract
Defining the impacts of anthropogenic climate change on biodiversity and species distributions is currently a high priority. Niche models focus primarily on predicted changes in abiotic factors; however, species interactions and adaptive evolution will impact the ability of species to persist in the face of changing climate. Our review focuses on the use of hybrid zones to monitor species' responses to contemporary climate change. Monitoring hybrid zones provides insight into how range boundaries shift in response to climate change by illuminating the combined effects of species interactions and physiological sensitivity. At the same time, the semi-permeable nature of species boundaries allows us to document adaptive introgression of alleles associated with response to climate change.
Keywords: hybridization, distribution, gene flow, range limits, adaptive introgression
Climate change and biodiversity
Changes in the distributions of species have been and will continue to be an obvious and important consequence of climate change [1]. Over geological time, and most recently during the Pleistocene, climates have fluctuated greatly, and species distributions have changed in response. In the present day, anthropogenic impacts on global climate have become a major source of concern. Primarily because of the rapidity of the change, anthropogenic contributions to climate change have generally negative impacts on biodiversity.
Over the past decade, the ability to predict the influence of anthropogenic climate change on species' distributions and persistence has improved [2-4]. However, niche models focused on abiotic factors have dominated the literature, and these models rarely incorporate species interactions or adaptive evolution, two major factors that undoubtedly impact the ability of species to persist in the face of changing climate [2,5]. Tracking how species respond to climate change should involve a multifaceted approach – one that includes examining changes in distributions and interspecific interactions, together with analysis of adaptive evolution – in order to gain a comprehensive understanding of the impacts of climate change on contemporary biodiversity. Long term monitoring of hybrid zones is one important way to document distributional changes and observe interspecific interactions under dynamic climate scenarios; in addition hybrid zones provide an opportunity to observe and characterize patterns of adaptive introgression [6].
Hybrid zones as sensitive markers of environmental change
Hybrid zones occur where the ranges of species or other genetically distinct entities overlap; in these areas individuals from two “species” hybridize and produce offspring of mixed ancestry. The locations of many hybrid zones (e.g. black-capped (Poecile atricapillus) and Carolina (P. carolinensis) chickadees [7-9]; northern (Glaucomys sabrinus) and southern (G. volans) flying squirrels [10-12]; Papilio butterflies, [13,14]; Allonemobious crickets [15]) are influenced by climate, and change in climate can lead to hybrid zone movement (e.g., [7]), changes in range overlap, and the origin of new hybrid zones [16] (Table S1, Figure 1). It is important to note that hybrid zones can also move due to factors other than climate. Under the tension zone model [17], in which a hybrid zone is maintained by selection against individuals of mixed ancestry, (see below) hybrid zones are free to move. Tension zone position is influenced by the density of the interacting species, with hybrid zones coming to rest in density troughs. A hybrid zone can, just by chance, experience movement under this model (e.g. Mus musculus hybrid zone [18]), which could mistakenly be attributed to climate change or other co-varying environmental factors. When hybrid zone movement is detected, additional evidence is always necessary to support a role for climate-mediated factors [7,19] (see Box 1).
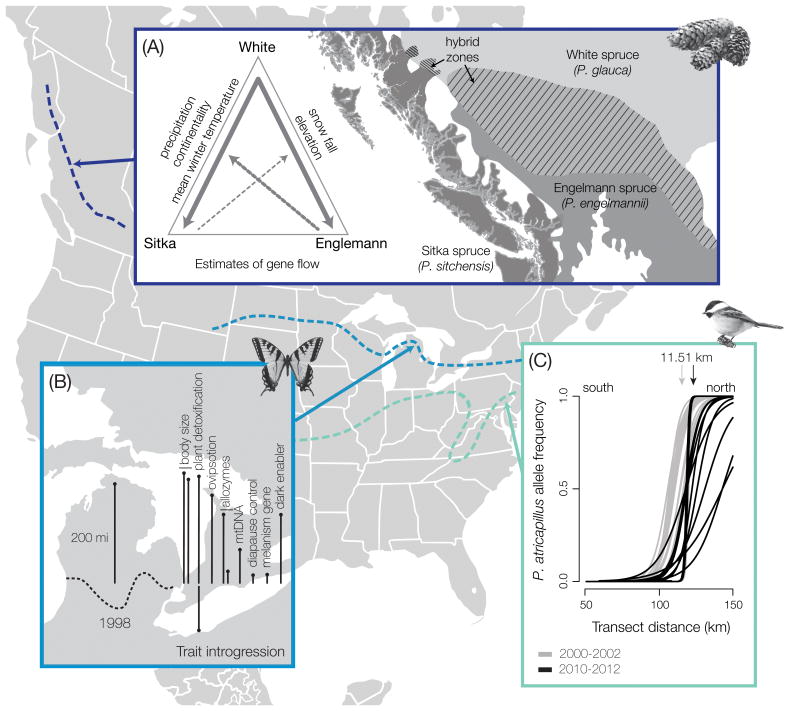
Figure 1. Hybrid zone case studies.
(A) Spruce hybrid zones (Picea glauca × P. engelmannii and P. glauca × P. sitchensis) [90]. The map to the right shows species distributions (indicated by dark gray (P. sitchensis), medium gray (P. engelmannii) and light gray (P. glauca)) and locations of the two hybrid zones. The triangle on the left summarizes estimated gene flow between the three species. Line width roughly corresponds to the percentage of shared alleles. Dashed lines indicate potential gene flow between two parental species and admixed individuals from the other hybrid zone. Climatic variables that are associated with each hybrid zone are indicated on the outside of the triangle. (B) Swallowtail butterfly hybrid zone (Papilio glaucus and P. canadensis) [13]. The map indicates the hybrid zone (dashed line) running east to west across Wisconsin. Lines represent the extent and direction (north or south) of species-specific trait introgression from 1998 to 2011. (C) Chickadee hybrid zone (Poecile atricapillus and P. carolinensis) [7]. Locus specific allele frequencies are plotted against distance along a linear transect (geographic clines) from historical (gray) and contemporary (black) sampling. Average shift north of 11.5 km in 10 years.
BOX 1. Sampling hybrid zones in a changing climate.
TEXT: Appropriate sampling is critical for understanding patterns of variation within hybrid zones and for inferring ecological and evolutionary processes from those patterns. This is particularly true when species ranges and hybrid zones shift as a result of climate change.
Markers- Molecular analyses of hybrid zones tend to focus on regions of the genome that have private alleles or highly differentiated allele frequencies between species. Genome-wide markers are now readily available for any taxon, and diagnostic markers can be identified through genome-wide sequencing of multiple individuals from several allopatric populations. These markers can be used to classify individuals within the hybrid zone as a parental type or as individuals of mixed ancestry (e.g., F1 hybrid, 1st generation backcross etc). With appropriate sampling of allopatric populations, diagnostic makers allow us to be confident that shared alleles within the hybrid zone are a result of hybridization and gene flow as opposed to ancestral polymorphism.
Time points – To capture hybrid zone movement and species range shifts, populations must be sampled at multiple time points. The time scale for repeated sampling will depend on the strength of selection, generation time and individual dispersal distances. Combining bioclimatic modeling with spatial demographic models can provide insights into expected rates of movement and therefore sampling periodicity. The best strategy will be to simultaneously track environmental and ecological variables (e.g., temperature and precipitation). Hybrid zones can also move stochastically and for reasons other than environmental change [17,21]; therefore, establishing a clear link between climate and hybrid zone movement is critically important.
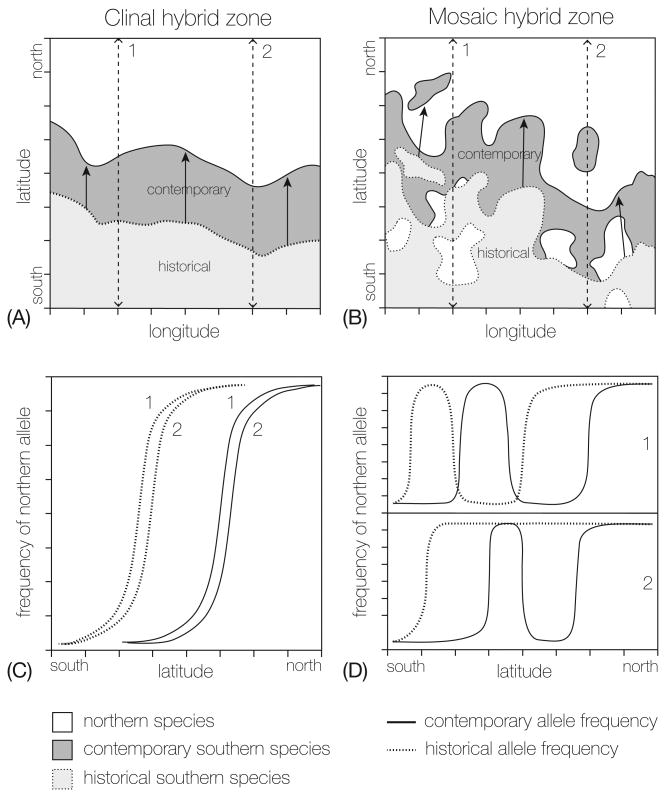
Geography – Hybrid zones should be sampled broadly to characterize the structure (e.g. clinal vs mosaic) and capture the full area of interaction between species. This is particularly true for hybrid zones that are shifting. Sampling allopatric populations of each species is essential for identifying diagnostic markers and localizing the hybrid zone. Sampling of hybrid zones can be in linear transects or over large areas (e.g.[92]). However, linear transects can make clines look broader or sharper depending on the whether the transect is orthogonal to the hybrid zone (see [18]). Restricted geographic sampling of a hybrid zone can obscure patterns of variation, particularly in mosaic hybrid zones where overlap can be extensive and occupied habitat patches can appear and disappear with shifting species distributions. If sampled only at a fine scale, the area of contact or shifts in ranges can be missed. Note: data analyses will differ depending on the type of hybrid zone sampled, see Box 3).
BOX 1, Figure I. Temporal and geographic sampling of hybrid zones in a changing climate.
The top panels represent northward shifts of a southern species (gray) and a northern species (white) for (A) clinal and (B) mosaic hybrid zones. The gray shading represents the northern range edge of the southern species' historical (light gray, dotted line) and contemporary (dark gray, solid line) distributions. Arrows highlight the shift in the species distribution. Lines 1 and 2 represent north-south transects across the hybrid zone. The change in the northern species' allele frequency along each transect for historical (dotted) and contemporary (solid) samples are plotted for the clinal hybrid zone (C) and the mosaic hybrid zone (D). The clinal hybrid zone forms an extensive and narrow zone of contact that extends east and west. In the mosaic hybrid zone, parental forms occupy distinct habitat patches in a heterogeneous landscape and hybridization occurs across patch boundaries. The patterns of variation in allele frequency differ depending on the transect. As the range of the southern species shifts north, habitat patches alter; patches disappear, new patches form and the area of each patch changes.
It has long been recognized that hybrid zones are important windows on the evolutionary process, because the outcomes of many generations of hybridization and recombination allow insights into the genetics of local adaptation, reproductive barriers and speciation [17,20-24] . Hybridization can also provide an important source of genetic variation that contributes to the evolution of novel phenotypes or adaptation to new environments [25-32]. Hybrid speciation is well documented in plants [24], but remains controversial in animals [33-36]; adaptive introgression is now a well-established phenomenon in many organisms. Studying how hybrid zones move in response to climate change will allow a more holistic understanding of the influence of both abiotic and biotic factors on range limits and how interacting species respond to climate change. Hybrid zones represent excellent systems for monitoring distributional changes both across continents and along elevational gradients; they also can provide insights into the role of adaptive introgression as a response to changing climate. Thus, hybrid zones have the potential to act as windows on anthropogenic climate change.
Here we discuss the utility of hybrid zone research for determining how species respond to contemporary climate change, either through changes in geographic distributions [7], or through adaptive introgression of alleles that facilitate species persistence [13]. Long-term monitoring of hybrid zones has enormous potential for informing (1) how species respond to climate change [37], and (2) how climate change might alter patterns of introgression [13]. This potential remains largely unrealized.
Monitoring changes in geographic distributions using hybrid zones
Understanding factors that limit species' ranges has, for many years, been a focus for ecologists and evolutionary biologists [38-44]. Defining range boundaries is inherently difficult, because population density declines towards range margins, making range boundaries diffuse and difficult to map. When competing species overlap at range edges, the increase in abundance of one species and concomitant decrease in abundance of the other species can occur over large and relatively undefined geographic areas. Only when competition between parapatric species or subspecies is intense will changes in their distributions be relatively straightforward to document. As a consequence, tracking responses to climate change can be difficult. Factors that determine range limits can be abiotic or biotic and include (but are not limited to) temperature, precipitation, and presence or absence of competitors, predators and parasites. Temperature is often implicated as an important abiotic factor determining range boundaries, and many northern and high elevation range boundaries are currently shifting northwards or skywards (in the case of mountain dwelling species) in response to climate change [45-49].
Hybrid zones can be broadly categorized as either clinal or mosaic in structure (Box 2). Clinal hybrid zones can be maintained by several different selection regimes. These include (1) endogenous selection against hybrid genotypes (tension zones) [17], (2) selection favoring different parental types at either end of an environmental gradient [50], or (3) selection in intermediate habitats favoring individuals of mixed ancestry (bounded hybrid superiority model)[51]. When selection against hybrids is strong, changes in the range of one species will be closely tied to the changes in the other species' range [37]. Further, when hybridizing species are distributed along an environmental gradient, hybridization can narrow the region in which both species co-occur, due to ecological and/or reproductive character displacement across the zone of hybridization [52-53]. A narrow zone can facilitate hybrid zone tracking by any method discussed here. In some cases, the hybridizing species will differ in their sensitivity to changing climate, in which case hybrid zone movement will presumably reflect “retreat” of the more vulnerable species.
BOX 2. Citizen science.
TEXT: Citizen science refers to data collection (and sometimes analysis) by members of the general public. These data are often related to natural systems (e.g., wildlife observations) and are collected in collaboration with scientists. A recent review highlighted 56 articles in the primary literature that dealt explicitly with citizen science, but suggested that the field was experiencing rapid growth [93]. Since that time, citizen science and use of citizen generated data has grown exponentially. We suggest that citizen science could be an important avenue of data collection for those interested in following distributional changes in hybrid zones. For example, the data used to generate a distribution map of the contact zone between black-capped and Carolina chickadees in Taylor et al. (2014) came from eBird [94], which is currently the largest citizen science database in the world. From citizen science data the authors were able to generate a high-resolution map of the species distributions and contact zone over a time period that coincided with the authors population sampling. This allowed the authors to combine information on the temporal changes in the chickadee hybrid zone with a genomic dataset and establish the link between hybrid zone movement and climate change. Numerous other citizen science databases exist [93] or are being initiated, that could serve to inform hybrid zone studies and facilitate the tracking of distributional changes over time. Even when hybrids are difficult to identify from morphology (as in the chickadee hybrid zone), participant training and rigorous data filtering will provide data suitable for high-level scientific inquiry. At the same time, citizen science best practices are being developed and made available to those interested in initiating large-scale data collection by public participants [95]. Given that we are suggesting that many hybrid zones should be monitored over the largest geographic region possible, citizen science might be one of the only ways researchers will be able to collect appropriate data. We acknowledge that there are some organisms that might lack public attention, or be inaccessible, but citizen science data collecting programs could generate the necessary involvement in many cases. Public involvement in scientific endeavors, especially when they relate to detecting the impacts of climate change on biodiversity, is increasingly important. Indeed, most scientific granting agencies require that grant proposals indicate how the proposed research will be extended into the public sphere.
Mosaic hybrid zones are also useful for monitoring range changes, but for these zones the sampling scheme is critical (Box1)[54]. In mosaic hybrid zones, parental types are distributed on a patchy landscape and hybridization most often occurs across patch boundaries. The distribution of habitat or resource patches can shift with changing climate. To monitor such shifts, mosaic hybrid zones must be sampled at appropriate spatial scales. It is important to note that hybrid zone width can change, which might or might not be related to overall movement, but can also be a result of environmental change (e.g., changes in the underlying habitat mosaic) [55]. The most useful hybrid zones for monitoring range changes in response to climate change will be those that occur between species with broad continental distributions and those that occur along elevational gradients (e.g. [15, 56]).
The utility of hybrid zones for understanding range changes has not gone unnoticed [57,58]. Indeed, the work of Godfrey Hewitt and others clearly shows that we can use the contemporary positions of many hybrid zones in both Europe and North America to better understand historical biogeography and species' responses to climate change throughout the Late Miocene to Late Pleistocene [58]. Post-glacial colonization routes have been mapped for a diversity of animal and plant taxa, using the contemporary distribution of hybrid zones, identified from morphology, genetics, or a combination [22,58,59-63]. Distributional changes in plants have also been inferred from pollen cores (e.g., [64,65]); these cores reveal rapid northward movement of many tree species over the past 10-15K years. Most of the aforementioned work focused on how historical distributions changed in response to changing climate. We propose to use the same approach to examine ongoing range changes and to make predictions about future distributions.
Determining if a hybrid zone is moving can be accomplished using distributional data in combination with phenotypic and/or genetic data (e.g., [66]). Direct measurements of hybrid zone movement require long-term sampling of any of these data types (e.g., [67-69]). Movement can also be inferred indirectly from patterns of allelic variation, but interpreting the genetic signature of hybrid zone movement is complicated (e.g., [55,68,70-72]). Consistent asymmetric introgression of genetic markers has been interpreted as evidence that these markers have been “left behind” as a hybrid zone boundary moved (e.g., [70,71]).
Classic geographic cline analysis allows us to estimate movement by examining allele or phenotype frequency changes across the landscapes at two (or more) time points (See Box 1). For each time point we can estimate the average cline center from a series of locus specific estimates and then determine the difference in cline center between time points to gain an understanding of how far a hybrid zone has moved (e.g., distance moved north [7]). The ideal phenotypes or genotypes used to measure hybrid zone movement will be fixed or nearly fixed for different morphs or alleles between the sampled populations, resulting in stepped clines across the hybrid zone and high power to assign cline parameters (e.g., center and width) (See Box 1). Given that ancestral polymorphism is not expected to leave a biogeographic signature, changes in allele frequencies between non-diagnostic markers are also useful for assessing hybrid zone movement, but power to assign cline parameters in such instances will be low. When available, diagnostic (or nearly diagnostic) differences between species allow us to be confident that alleles that are shared in the hybrid zone are the result of hybridization and introgression, as opposed to shared ancestral polymorphism (see Box 1). In a similar way, we can use distribution data and records of hybrid individuals, or of the overlap between parental species, to determine hybrid zone locations through time (e.g., [66]). This approach can be problematic when identification of hybrids in the field is difficult, or when the parental species have similar morphology.
Advances in DNA sequencing technology allow us to follow changes in hybrid zone positions in real time. The ability to generate molecular markers across the genome for non-model organisms allows detection of divergent genomic regions between closely related species (e.g., [73, 74]). The size and chromosomal distribution of these regions vary across taxa, and mechanisms for their origin and maintenance might vary as well. However, large, multi-locus datasets from broad scale sampling of species ranges and hybrid zones are now relatively easy to obtain (e.g. see Table 2 in [75]). At the same time, new target enrichment sequencing methods (e.g., exon capture) that are appropriate for the degraded DNA obtained from museum samples can extend our temporal sampling and potentially allow analysis of samples that pre-date human mediated climate change [76]. Finally, the field of citizen science has developed rapidly, and some of the largest citizen science databases are now providing scientists with the raw data necessary to track distributional changes across large geographic regions over relatively short timescales (e.g.,a decade) [7] (See Box 2). Combining molecular data from broad geographic sampling with geographic cline analyses allows the close tracking of hybrid zone centers through time. Using these resources, we can record and interpret genetic and distributional changes of hybridizing species in response to climate change.
Introgression and Adaptation
Natural hybridization has the potential to generate novel genetic combinations, either through hybrid speciation or introgression [16,24,26,34,77]. Compelling evidence for speciation via hybridization remains rare in animals [33] but is more common in plants [24]. In contrast, adaptive introgression is well documented in both plants and animals [78,79]. In fact, recent evidence for genome-wide introgression in the Anopheles species complex raises the intriguing possibility that in some cases introgression can be more extensive and more difficult to detect than previously believed [32]. There is growing evidence that introgression can lead to the acquisition of favorable alleles in response to climate change ([77], see Papilio example below). Repeated genomic sampling of hybrid zones responding to climate change will be one of the best avenues for determining the importance of adaptive introgression as a response to climate change.
In addition to using hybrid zones to monitor distributional changes of interacting species, we can also use hybrid zone data to document how climate change alters selection pressures. In this context, we must recognize that genomes are not coherent and that genome regions can have independent evolutionary histories. Using this framework, range boundaries might be defined in terms of individual genes or gene regions; that is, introgression represents a (possibly adaptive) range change for a particular genome region. This approach will allow identification of genes that affect thermal tolerances and climate-relevant physiology (i.e., genes that affect phenotypes relevant to climate change), as well as adaptation to altered environments and/or communities. In some cases alleles at these loci are highly diverged between parental species and their patterns of introgression can be tracked using cline analyses (e.g., [7,13] (Box 3). Alternatively, natural variation in hybrid zones can be used to associate particular genotypes with adaptation to climatic niches and to monitor changes in these traits as climate changes [80-83]. Hybrid zones are essentially natural mapping experiments; generations of recombination break down linkage disequilibrium in hybrid genomes and divergent alleles can be associated with environmental variables or phenotypes [84]. This approach will be particularly useful in cases where large, highly admixed or stable hybrid populations exist (e.g. spruce, poplar, house mouse).
BOX 3. Detecting admixture and introgression.
TEXT: Hybridization and recombination shuffle divergent genomes, thereby allowing regions of the genome to behave independently, depending on linkage relationships and functional interactions (epistasis). Genes or gene regions that function in the genomic background of both of the hybridizing species can be exchanged (introgress). In contrast, regions that contribute to local adaptation, premating barriers, or hybrid unfitness will exhibit restricted introgression. Therefore, patterns of introgression can help identify genomic regions that affect these phenotypes.
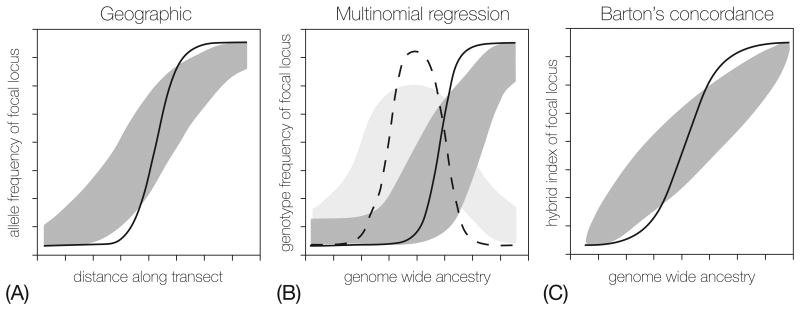
A classic approach to estimating introgression is to plot the change in allele frequencies along a transect across the hybrid zone (geographic cline) (Figure IA). Alleles that contribute to species boundaries or local adaptation will show abrupt changes in frequency across the zone, whereas alleles that introgress will show more gradual changes. Geography is a proxy for admixture; the center of the zone has the greatest admixture and hybrid individuals become less common away from the center. Geographic clines perform well when a hybrid zone is independent of the environment or occurs along an environmental gradient, but in cases where species have patchy distributions allele frequencies can change abruptly between adjacent habitat patches (see Box 1). Mosaic hybrid zones can be characterized using geographic clines but must be sampled appropriately (i.e., across single habitat patch boundaries [54,92].
An alternative is a “genomic cline” approach that plots the change in allele frequency for a locus against an estimate of genome-wide admixture. Individuals within a hybrid zone are each evaluated for the proportion of their genome derived from one parental species (hybrid index). The observed change in allele frequency at the focal locus is then plotted against an expected change based on the mean hybrid index. Significant deviations suggest genes are experiencing selection. This approach, originally introduced by Szyumra and Barton (1986), has been implemented in several new forms (see [74,96], Figures IB and IC).
Geographic and genomic cline analyses are complimentary, and each has its own application. However, in cases where hybrid zones are shifting as a result of climate change, the implementation of these approaches is critical. Geographic clines are an obvious way to document shifting species distributions, and as long as care is taken in sampling the hybrid zone (See Box 1), they can be very informative (See [7]). Genomic clines can also be used to document shifting species distributions. In this case, the change in species ranges can be evaluated by comparing the proportion of individuals with different hybrid indices at different time points or geographic locations (see [91]).
BOX 3, Figure I.
Clinal analyses for estimating introgression across a hybrid zone. Each panel depicts expected clines for a hybrid zone that is maintained by local adaptation, premating barriers that prevent the formation of hybrids, or selection against hybrids. The black lines depict a locus under selection; one that contributes to local adaptation or reproductive barriers. The gray represents the expected range of cline shapes for unlinked neutral markers. A) Classic geographic clines. B) Genomic clines modeled using multinomial regression [80, 96]. C) Genomic clines modeled using either Barton's concordance method [97], Bayesian genomic clines [98] or the log-logistic method[74].
Hybrid zone case studies
Here we describe three case studies that provide insight into the influence of climate change on species distributions and introgression. We recognize that not every hybrid zone will be amenable to this approach, and suggest other avenues of research that will further the role of hybrid zones in the study of climate change.
The widely distributed white spruce (Picea glauca), hybridizes with two western spruce species, Engelmann spruce (P. engelmannii) [85-88] and Sitka spruce (P. sitchensis) [89,90] (Figure 1A). Each spruce species occupies a unique climatic niche and remains genetically, morphologically and ecologically distinct. Yet in areas of contact, there is weak reproductive isolation and extensive gene flow. Species distributions are largely influenced by temperature, precipitation (Sitka and White spruce), continentality and mean winter temperature, all of which have the potential to change rapidly with anthropogenic climate change. Hybrid genotypes appear to form stable populations along environmental gradients between parental habitats, suggesting that combinations of parental alleles can allow hybrids to be locally adapted to conditions at the edge or outside of the parent species range [86,90]. As a result, gene flow among spruce populations might serve as a conduit for adaptive introgression of alleles that enables species to cope with changing environmental conditions [82]. At the same time, monitoring the distribution of hybrid individuals on the landscape will provide insight into the speed with which tree species can respond to climate change. Necessarily, such studies will need to be long-term, given the generation times of these long-lived species.
North American tiger swallowtail butterflies in the genus Papilio (P. glaucus and P. canadensis) hybridize in a narrow band that stretches across much of North America [13] (Figure 1B). Decades of research on this hybrid zone in the Great Lakes region of North America (>30 years) have provided unique insights into interspecific gene flow, local selection, ecological divergence, incipient speciation, and hybrid zone movement. The occurrence and movement of this hybrid zone has been attributed to climate change [13,14]. Perhaps more than any other North American hybrid zone, the swallowtail hybrid zone has provided examples of introgression of species-specific alleles in response to climate change. For example, alleles related to the ability of Papilio larvae to detoxify host plants have introgressed northwards 200km over the past 15 years, potentially driven by strong selection on novel phenotypes produced within the hybrid zone (Figure 1B). Alleles related to diapause control have also introgressed northwards, but to a lesser extent (Figure 1B). Expanded sampling along this hybrid zone to determine the geographic consistency of these patterns, and using citizen scientists to accurately map the distribution of each species through time, are two natural extensions of the work carried out thus far on Papilio.
Black-capped (Poecile atricapillus) and Carolina (P. carolinensis) chickadees hybridize in a narrow band that stretches from New Jersey to Kansas. The zone of contact appears to be defined by temperature (Figure 1C). A recent examination of the hybrid zone that combined genomic, distributional (from a citizen science database), and climatic data provided clear evidence of hybrid zone movement northwards over the past decade, and showed that the recorded movement was closely tied to warming winter temperatures [7]. This investigation was limited in geographic scope compared to the extent of the hybrid zone. Additional insight into the influence of climate change on the distribution of both species could be gained by expanding both the genetic and distributional studies. A subset of the loci from the genomic dataset used in this study were fixed for different alleles in the parental species, and preliminary analyses suggest that some of these are located in regions of the genome related to oxidative phosphorylation and metabolism [91]. Additional work on the hybrid zone could provide interesting insight into species barriers and the influence of climate change on the permanence of those barriers, as well as insight into the nature of reproductive isolation as it relates to differences in thermal tolerance in a changing climate.
The highlighted studies are just three examples of species that hybridize where they overlap; these examples are restricted to North America, and for each we already possess significant data. These hybrid zones are easy to map and monitor and have already provided insight into the effects of climate change on species' distributions and the influence of climate on local adaptation and adaptive introgression. Other hybrid zones exist both within North America and elsewhere that have the potential to inform our understanding of the influence of contemporary climate change on species distributions, and on the role of climate change in adaptive introgression (Table S1).
Species interactions and climate-driven distributional changes
Species interactions can confound our interpretation of climate-driven impacts on distribution, but close monitoring of hybrid zones should improve our understanding of these interactions and their influence. Hybrid zone studies have the potential to identify cases in which expansion of one species is limited by biotic interaction with another species; competitive interactions are rarely included as a factor in contemporary climate niche modeling. It is important to note that ecological interactions (but not introgression) can occur in the absence of hybridization. This means that parapatrically distributed species or subspecies will also offer important opportunities for understanding distributional changes in response to climate change.
Concluding remarks: The utility of monitoring hybrid zones in a changing world
For nearly half a century biologists have been using morphological and molecular markers to characterize species interactions within hybrid zones. During that time, the pace of anthropogenic climate change has accelerated with obvious consequences for species' distributions. Range expansions and contractions, and local extinctions are becoming commonplace, but we are not always well prepared to document these changes, or predict them. We are now poised to utilize both historical and current sampling of hybrid zones to understand how species interactions and adaptive responses are being transformed by our changing climate. We emphasize that hybrid zones should be sampled with this change in mind; they should not be viewed as equilibrium situations. We also highlight the utility of new genomic methods (increased genomic and geographic representation, use of historical DNA samples) and public databases (citizen science) that will facilitate studies of hybrid zone movement. As the approaches we suggest are adopted, hybrid zones will continue to act as windows on evolution and ecology in our changing world.
Supplementary Material
Acknowledgments
SAT is supported by a Banting Postdoctoral Fellowship from the Natural Sciences and Engineering Research Council of Canada, and in part by postdoctoral fellowships from the Cornell Center for Comparative and Population Genomics as well as the Fuller Evolutionary Biology Program at the Cornell Lab of Ornithology. ELL is supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the National Institutes of Health under award number R01HD073439 to J.M. Good. RGH's work on hybrid zones has been supported by NSF. Thanks to J. Hamilton for supplying species range maps and helpful comments. Thanks to L. Campagna, D. Toews, N. Mason, P. Deane-Coe, J. Berve, and I. Lovette for helpful discussion.
Glossary
- Admixture
The mixing of individuals from two (or more) source populations that have different allele, genotype, or phenotype frequencies
- Biogeographic signature
distribution of allelic or haplotypic variation that reflects species distributions in geographic space and through geological time
- Clinal hybrid zone
A hybrid zone in which character states change in clinal fashion across the zone. Clinal zones can be “tension zones,” maintained by a balance between dispersal and selection against hybrids, or they can reflect selection along an environmental gradient
- Bounded hybrid superiority model
A model that suggests that hybrid zones are maintained by selection for hybrids in intermediate habitats. Hybrids have higher fitness than either parent species in these habitats, but their fitness is lower in either parental species habitat [51]
- Hybridization
The interbreeding of individuals from two populations, or groups of populations, which are distinguishable on the basis of one or more heritable characters. [20,99]
- Hybrid zones
Regions where genetically distinct groups of individuals meet and mate resulting in at least some offspring of mixed ancestry [19,20]
- Hybrid speciation (homoploid)
Origin of new species through the production of a stable population of recombinant individuals that are reproductively isolated from the parent species
- Introgression
The incorporation of alleles of one entity into the gene pool of another, typically through hybridization and backcrossing [100]. Adaptive introgression refers to the introgression of advantageous alleles
- Mosaic hybrid zone
Hybrid zones that occur when genetically distinct groups of individuals that differ in habitat or resource use interact in a patchy environment [20]
- Species boundaries
The phenotype, genes or genome regions that remain differentiated in the face of potential hybridization and introgression [75]
- Tension zone
Clinal hybrid zones maintained by a balance between dispersal into the hybrid zone and selection against hybrids [20]
Footnotes
Scott A. Taylor – Email: sat235@cornell.edu, Phone: 6072271374, Twitter: @Dr_Scott_Taylor
Erica L. Larson, Email: ericalarson@gmail.com, Twitter: @ericallarson
Richard G. Harrison, Email: rgh4@cornell.edu
References
- 1.Rosenzweig C, et al. Attributing physical and biological impacts to anthropogenic climate change. Nature. 2008;453:353–357. doi: 10.1038/nature06937. [DOI] [PubMed] [Google Scholar]
- 2.Lavergne S, et al. Biodiversity and climate change: integrating evolutionary and ecological responses of species and communities. Ann Rev Ecol Evol Syst. 2010;41:321–350. [Google Scholar]
- 3.Bellard C, et al. Impacts of climate change on the future of biodiversity. Ecol Lett. 2012;15:365–377. doi: 10.1111/j.1461-0248.2011.01736.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quintero I, Wiens JJ. Rates of projected climate change dramatically exceed past rates of climatic niche evolution among vertebrate species. Ecol Lett. 2013;16:1095–1103. doi: 10.1111/ele.12144. [DOI] [PubMed] [Google Scholar]
- 5.Norberg J, et al. Eco-evolutionary responses of biodiversity to climate change. Nature Clim Change. 2012;2:747–751. [Google Scholar]
- 6.Chown SL, et al. Biological invasions, climate change and genomics. Evol Appl. 2015;8:23–46. doi: 10.1111/eva.12234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taylor SA, et al. Climate-Mediated Movement of an Avian Hybrid Zone. Curr Biol. 2014;24:671–676. doi: 10.1016/j.cub.2014.01.069. [DOI] [PubMed] [Google Scholar]
- 8.Curry RL. Hybridization in chickadees: much to learn from familiar birds. The Auk. 2005;122:747–758. [Google Scholar]
- 9.Reudink MW, et al. Structure and dynamics of the hybrid zone between black-capped chickadee (Poecile atricapillus) and Carolina chickadee (P. carolinensis) in southeastern Pennsylvania. The Auk. 2007;124:463–478. [Google Scholar]
- 10.Garroway CJ, et al. Climate change induced hybridization in flying squirrels. Glob Change Biol. 2010;16:113–121. [Google Scholar]
- 11.Garroway CJ, et al. The genetic signature of rapid range expansion by flying squirrels in response to contemporary climate warming. Glob Change Biol. 2011;17:1760–1769. [Google Scholar]
- 12.Bowman J, et al. Northern range boundary dynamics of southern flying squirrels: evidence of an energetic bottleneck. Can J Zool. 2005;83:1486–1494. [Google Scholar]
- 13.Scriber JM. Impacts of climate warming on hybrid zone movement: Geographically diffuse and biologically porous “species borders”. Insect Sci. 2011;18:121–159. [Google Scholar]
- 14.Scriber JM, et al. Adaptations to “thermal time” constraints in Papilio: latitudinal and local size clines differ in response to regional climate change. Insects. 2014;5:199–226. doi: 10.3390/insects5010199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Britch SC, et al. Spatio-temporal dynamics of the Allonemobius fasciatus- A. socius mosaic hybrid zone: a 14-year perspective. Mol Ecol. 2001;10:627–638. doi: 10.1046/j.1365-294x.2001.01215.x. [DOI] [PubMed] [Google Scholar]
- 16.Chunco AJ. Hybridization in a warmer world. Ecol Evol. 2014;4:2019–2031. doi: 10.1002/ece3.1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barton NH, Hewitt GM. Analysis of hybrid zones. Ann Rev Ecol Syst. 1985;16:113–148. [Google Scholar]
- 18.Macholán M, et al. Assessing multilocus introgression patterns: a case study on the mouse X chromosome in central Europe. Evolution. 2011;65:1428–1446. doi: 10.1111/j.1558-5646.2011.01228.x. [DOI] [PubMed] [Google Scholar]
- 19.Kohlmann B, et al. Environmental predictions and distributional limits of chromosomal taxa in the Australian grasshopper Caledia captiva (F.) Oecologia. 1988;75:483–493. doi: 10.1007/BF00776409. [DOI] [PubMed] [Google Scholar]
- 20.Harrison RG. Hybrid Zones and the Evolutionary Process. Oxford University Press; 1993. [Google Scholar]
- 21.Harrison RG. Hybrid zones: windows on evolutionary process. Oxford surveys in evolutionary biology 1990 [Google Scholar]
- 22.Barton NH, Hewitt GM. Adaptation, speciation and hybrid zones. Nature. 1989;341:497–503. doi: 10.1038/341497a0. [DOI] [PubMed] [Google Scholar]
- 23.Hewitt GM. Hybrid zones-natural laboratories for evolutionary studies. Trends Ecol Evol. 1988;3:158–167. doi: 10.1016/0169-5347(88)90033-X. [DOI] [PubMed] [Google Scholar]
- 24.Abbott R, et al. Hybridization and speciation. J Evol Biol. 2013;26:229–246. doi: 10.1111/j.1420-9101.2012.02599.x. [DOI] [PubMed] [Google Scholar]
- 25.Brelsford A, et al. Hybrid origin of Audubon's warbler. Mol Ecol. 2011;20:2380–2389. doi: 10.1111/j.1365-294X.2011.05055.x. [DOI] [PubMed] [Google Scholar]
- 26.Trier CN, et al. Evidence for mito-nuclear and sex-linked reproductive barriers between the hybrid Italian sparrow and its parent species. PLoS genetics. 2014 doi: 10.1371/journal.pgen.1004075. e1004075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hermansen JS, et al. Hybrid speciation in sparrows I: phenotypic intermediacy, genetic admixture and barriers to gene flow. Mol Ecol. 2011;20:3812–3822. doi: 10.1111/j.1365-294X.2011.05183.x. [DOI] [PubMed] [Google Scholar]
- 28.Elgvin TO, et al. Hybrid speciation in sparrows II: a role for sex chromosomes? Mol Ecol. 2011;20:3823–3937. doi: 10.1111/j.1365-294X.2011.05182.x. [DOI] [PubMed] [Google Scholar]
- 29.Selz OM, et al. Behavioural isolation may facilitate homoploid hybrid speciation in cichlid fish. J Evol Biol. 2014;27:275–289. doi: 10.1111/jeb.12287. [DOI] [PubMed] [Google Scholar]
- 30.Toews D, et al. Migration, mitochondria, and the yellow-rumped warbler. Evolution. 2013;68:241–255. doi: 10.1111/evo.12260. [DOI] [PubMed] [Google Scholar]
- 31.Fujita MK, et al. Introgression and phenotypic assimilation in Zimmerius flycatchers (Tyrannidae): population genetic and phylogenetic inferences from genome-wide SNPs. Sys Biol. 2014;63:134–152. doi: 10.1093/sysbio/syt070. [DOI] [PubMed] [Google Scholar]
- 32.Fontaine MC, et al. Extensive introgression in a malaria vector species complex revealed by phylogenomics. Science. 2015;347 doi: 10.1126/science.1258524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schumer M, et al. How common is homoploid hybrid speciation? Evolution. 2014;68:1553–1560. doi: 10.1111/evo.12399. [DOI] [PubMed] [Google Scholar]
- 34.Barton NH. Does hybridization influence speciation? J Evol Biol. 2013;26:267–269. doi: 10.1111/jeb.12015. [DOI] [PubMed] [Google Scholar]
- 35.Mallet J. Hybrid speciation. Nature. 2007;446:279–283. doi: 10.1038/nature05706. [DOI] [PubMed] [Google Scholar]
- 36.Barton NH. The role of hybridization in evolution. Mol Ecol. 2001;10:551–568. doi: 10.1046/j.1365-294x.2001.01216.x. [DOI] [PubMed] [Google Scholar]
- 37.Harr B, Price T. Climate change: A hybrid zone moves north. Curr Biol. 2014;24:R230–R232. doi: 10.1016/j.cub.2014.02.023. [DOI] [PubMed] [Google Scholar]
- 38.Hargreaves AL, et al. Are species' range limits simply niche limits writ large? A review of transplant experiments beyond the range. Am Nat. 2014;183:157–173. doi: 10.1086/674525. [DOI] [PubMed] [Google Scholar]
- 39.Sexton JP, et al. Evolution and ecology of species range limits. Ann Rev Ecol Evol Syst. 2009;40:415–436. [Google Scholar]
- 40.Eckert CG, et al. Genetic variation across species' geographical ranges: the central–marginal hypothesis and beyond. Mol Ecol. 2008;17:1170–1188. doi: 10.1111/j.1365-294X.2007.03659.x. [DOI] [PubMed] [Google Scholar]
- 41.Gaston KJ. Geographic range limits: achieving synthesis. Proc R Soc B: Biological Sciences. 2009;276:1395–1406. doi: 10.1098/rspb.2008.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sagarin RD, et al. Moving beyond assumptions to understand abundance distributions across the ranges of species. Trends Ecol Evol. 2006;21:524–530. doi: 10.1016/j.tree.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 43.Sagarin RD, Gaines SD. The “abundant centre”distribution: to what extent is it a biogeographical rule? Ecol Lett. 2002;5:137–147. [Google Scholar]
- 44.Hoffmann AA, Blows MW. Species borders: ecological and evolutionary perspectives. Trends Ecol Evol. 1994;9:223–227. doi: 10.1016/0169-5347(94)90248-8. [DOI] [PubMed] [Google Scholar]
- 45.Chen IC, et al. Rapid range shifts of species associated with high levels of climate warming. Science. 2011;333:1024–1026. doi: 10.1126/science.1206432. [DOI] [PubMed] [Google Scholar]
- 46.Parmesan C. Ecological and evolutionary responses to recent climate change. Ann Rev Ecol Evol Syst. 2006;37:637–669. [Google Scholar]
- 47.Abbott RJ, Brennan AC. Altitudinal gradients, plant hybrid zones and evolutionary novelty. Phil Trans R Soc. 2014;369:20130346. doi: 10.1098/rstb.2013.0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Freeman BG, Freeman A. Rapid upslope shifts in New Guinean birds illustrate strong distributional responses of tropical montane species to global warming. PNAS. 2014;111:4490–4494. doi: 10.1073/pnas.1318190111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moritz C, et al. Impact of a century of climate change on small-mammal communities in Yosemite National Park, USA. Science. 2008;322:261–264. doi: 10.1126/science.1163428. [DOI] [PubMed] [Google Scholar]
- 50.Endler JA. Geographic variation, speciation, and clines. Monogr Popul Biol. 1977;10:1–246. [PubMed] [Google Scholar]
- 51.Moore WS. An evaluation of narrow hybrid zones in vertebrates. Q Rev Biol. 1977;52:263. [Google Scholar]
- 52.Goldberg EE, Lande R. Ecological and reproductive character displacement on an environmental gradient. Evolution. 2006;60:1344–1357. [PubMed] [Google Scholar]
- 53.Goldberg EE, Lande R. Species' borders and dispersal barriers. Am Nat. 2007;170:297–304. doi: 10.1086/518946. [DOI] [PubMed] [Google Scholar]
- 54.Ross CL, Harrison RG. A fine-scale spatial analysis of the mosaic hybrid zone between Gryllus firmus and Gryllus pennsylvanicus. Evolution. 2002;56:2296–2312. doi: 10.1111/j.0014-3820.2002.tb00153.x. [DOI] [PubMed] [Google Scholar]
- 55.Buggs R. Empirical study of hybrid zone movement. Heredity. 2007;99:301–312. doi: 10.1038/sj.hdy.6800997. [DOI] [PubMed] [Google Scholar]
- 56.Brennan AC, et al. Adaptation and selection in the Senecio (Asteraceae) hybrid zone on Mount Etna, Sicily. New Phytol. 2009;183:702–717. doi: 10.1111/j.1469-8137.2009.02944.x. [DOI] [PubMed] [Google Scholar]
- 57.Remington CL. Evolutionary biology. Springer; US: 1968. Suture-zones of hybrid interaction between recently joined biotas; p. 321. [Google Scholar]
- 58.Hewitt GM. Quaternary phylogeography: the roots of hybrid zones. Genetica. 2011;139:617–638. doi: 10.1007/s10709-011-9547-3. [DOI] [PubMed] [Google Scholar]
- 59.Hewitt GM. Genetic consequences of climatic oscillations in the Quaternary. Phil Trans R Soc B. 2004;359:183–195. doi: 10.1098/rstb.2003.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hewitt GM. Speciation, hybrid zones and phylogeography—or seeing genes in space and time. Mol Ecol. 2001;10:537–549. doi: 10.1046/j.1365-294x.2001.01202.x. [DOI] [PubMed] [Google Scholar]
- 61.Hewitt G. The genetic legacy of the Quaternary ice ages. Nature. 2000;405:907–913. doi: 10.1038/35016000. [DOI] [PubMed] [Google Scholar]
- 62.Ibrahim KM, et al. Spatial patterns of genetic variation generated by different forms of dispersal during range expansion. Heredity. 1996;77:282–291. [Google Scholar]
- 63.Comes HP, Kadereit JW. The effect of Quaternary climatic changes on plant distribution and evolution. Trends in plant science. 1998;3:432–438. [Google Scholar]
- 64.Davis MB, Botkin DB. Sensitivity of cool-temperate forests and their fossil pollen record to rapid temperature change. Quat Res. 1985;23:327–340. [Google Scholar]
- 65.Williams JW, et al. Rapid and widespread vegetation responses to past climate change in the North Atlantic region. Geology. 2002;30:971–974. [Google Scholar]
- 66.Dasmahapatra KK, et al. Inferences from a rapidly moving hybrid zone. Evolution. 2002;56:741–753. doi: 10.1111/j.0014-3820.2002.tb01385.x. [DOI] [PubMed] [Google Scholar]
- 67.Rising JD. Current ornithology. Springer; US: 1983. The great plains hybrid zones; pp. 131–157. [Google Scholar]
- 68.Arntzen JW, Wallis GP. Restricted gene flow in a moving hybrid zone of the newts Triturus cristatus and T. marmoratus in western France. Evolution. 1991;45:805–816. doi: 10.1111/j.1558-5646.1991.tb04352.x. [DOI] [PubMed] [Google Scholar]
- 69.Yang SY, Selander RK. Hybridization in the grackle Quiscalus quiscula in Louisiana. Sys Biol. 1968;17:107–143. [PubMed] [Google Scholar]
- 70.Marchant AD. Apparent introgression of mitochondrial DNA across a narrow hybrid zone in the Caledia captiva species-complex. Heredity. 1988;60:39–46. [Google Scholar]
- 71.Marchant AD, et al. Gene flow across a chromosomal tension zone. I. Relicts of ancient hybridization. Heredity. 1988;61:321–328. [Google Scholar]
- 72.Shaw DD, et al. The control of gene flow across a narrow hybrid zone: a selective role for chromosomal rearrangement? Can J Zool. 1990;68:1761–1769. [Google Scholar]
- 73.Wagner CE, et al. Genome-wide RAD sequence data provide unprecedented resolution of species boundaries and relationships in the Lake Victoria cichlid adaptive radiation. Mol Ecol. 2012;22:787–798. doi: 10.1111/mec.12023. [DOI] [PubMed] [Google Scholar]
- 74.Fitzpatrick BM. Alternative forms for genomic clines. Ecol Evol. 2013;3:1951–1966. doi: 10.1002/ece3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Harrison RG, Larson EL. Hybridization, introgression, and the nature of species boundaries. J hered. 2014;105:795–809. doi: 10.1093/jhered/esu033. [DOI] [PubMed] [Google Scholar]
- 76.Bi K, et al. Unlocking the vault: next-generation museum population genomics. Mol Ecol. 2013;22:6018–6031. doi: 10.1111/mec.12516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lewontin RC, Birch LC. Hybridization as a source of variation for adaptation to new environments. Evolution. 1966;20:315–336. doi: 10.1111/j.1558-5646.1966.tb03369.x. [DOI] [PubMed] [Google Scholar]
- 78.Song Y, et al. Adaptive introgression of anticoagulant rodent poison resistance by hybridization between old world mice. Curr Biol. 2011;21:1296–1301. doi: 10.1016/j.cub.2011.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ellstrand NC, et al. Introgression of crop alleles into wild or weedy populations. Ann Rev Ecol Evol Syst. 2013;44:325–345. [Google Scholar]
- 80.Lexer C, et al. Admixture in European Populus hybrid zones makes feasible the mapping of loci that contribute to reproductive isolation and trait differences. Heredity. 2007;98:74–84. doi: 10.1038/sj.hdy.6800898. [DOI] [PubMed] [Google Scholar]
- 81.Buerkle CA, Lexer C. Admixture as the basis for genetic mapping. Trends Ecol Evol. 2008;23:686–694. doi: 10.1016/j.tree.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 82.Aitken SN, et al. Adaptation, migration or extirpation: climate change outcomes for tree populations. 2008;1:95–111. doi: 10.1111/j.1752-4571.2007.00013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lexer C, et al. Candidate gene polymorphisms associated with salt tolerance in wild sunflower hybrids: implications for the origin of Helianthus paradoxus, a diploid hybrid. New Phytologist. 2004;161:225–233. doi: 10.1046/j.1469-8137.2003.00925.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Crawford JE, Nielsen R. Detecting adaptive trait loci in nonmodel systems: divergence or admixture mapping? Mol Ecol. 2013;22:6131–6148. doi: 10.1111/mec.12562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.De La Torre AR, et al. Adaptation and exogenous selection in a Picea glauca× Picea engelmannii hybrid zone: implications for forest management under climate change. New Phytol. 2014;201:687–699. doi: 10.1111/nph.12540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.De La Torre AR, et al. Genome-wide admixture and ecological niche modelling reveal the maintenance of species boundaries despite long history of interspecific gene flow. Mol Ecol. 2014;23:2046–2059. doi: 10.1111/mec.12710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Haselhorst M, Buerkle CA. Population genetic structure of Picea engelmannii, P. glauca and their previously unrecognized hybrids in the central Rocky Mountains. Tree Genet Genome. 2013;9:669–681. [Google Scholar]
- 88.De La Torre, et al. Genetic architecture and genomic patterns of gene flow between hybridizing species of. Picea. 2015 doi: 10.1038/hdy.2015.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hamilton JA, et al. Genomic and phenotypic architecture of a spruce hybrid zone (Picea sitchensis × P. glauca) Mol Ecol. 2013;22:827–841. doi: 10.1111/mec.12007. [DOI] [PubMed] [Google Scholar]
- 90.Hamilton JA, et al. Fine-scale environmental variation contributes to introgression in a three-species spruce hybrid complex. Tree Genet Genome. 2014;11:1–14. [Google Scholar]
- 91.Taylor SA, et al. Spatiotemporally consistent genomic signatures of reproductive isolation in a moving hybrid zone. Evolution. 2014;68:3066–3081. doi: 10.1111/evo.12510. [DOI] [PubMed] [Google Scholar]
- 92.Larson EL, et al. Gene flow and the maintenance of species boundaries. Mol Ecol. 2014;23:1668–1678. doi: 10.1111/mec.12601. [DOI] [PubMed] [Google Scholar]
- 93.Silvertown J. A new dawn for citizen science. Trends Ecol Evol. 2009;24:467–471. doi: 10.1016/j.tree.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 94.Sullivan BL, et al. eBird: A citizen-based bird observation network in the biological sciences. Biol Cons. 2009;142:2282–2292. [Google Scholar]
- 95.Bonney R, et al. Citizen science: a developing tool for expanding science knowledge and scientific literacy. BioScience. 2009;59:977–984. [Google Scholar]
- 96.Gompert Z, Buerkle CA. A powerful regression-based method for admixture mapping of isolation across the genome of hybrids. Mol Ecol. 2009;18:1207–1224. doi: 10.1111/j.1365-294X.2009.04098.x. [DOI] [PubMed] [Google Scholar]
- 97.Szymura JM, Barton NH. Genetic analysis of a hybrid zone between the fire-bellied toads, Bombina bombina and B. variegata, near Cracow in southern Poland. Evolution. 1986;40:1141–1159. doi: 10.1111/j.1558-5646.1986.tb05740.x. [DOI] [PubMed] [Google Scholar]
- 98.Gompert Z, Buerkle CA. A hierarchical bayesian model for next-generation population genomics. Genetics. 2011;187:903–917. doi: 10.1534/genetics.110.124693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Woodruff DS, Gould SJ. Fifty years of interspecific hybridization: genetics and morphometrics of a controlled experiment on the land snail Cerion in the Florida Keys. Evolution. 1987;41:1022–1045. doi: 10.1111/j.1558-5646.1987.tb05874.x. [DOI] [PubMed] [Google Scholar]
- 100.Anderson E, Hubricht L. Hybridization in Tradescantia. III. The evidence for introgressive hybridization. Am J Bot. 1938;25:396–402. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.