Abstract
Despite advances in treatment approaches for head and neck squamous cell carcinoma (HNSCC), survival rates have remained stagnant due to the paucity of preclinical models that accurately reflect the human tumor. Patient-derived xenografts (PDXs) are an emerging model system where patient tumors are implanted directly into mice. Increased understanding of the application and limitations of PDXs will facilitate their rational use. Studies to date have not reported protein profiles of PDXs. Therefore, we developed a large cohort of HNSCC PDXs and found that tumor take rate was not influenced by the clinical, pathologic or processing features. Protein expression profiles, from a subset of the PDXs, were characterized by reverse phase protein array (RPPA) and the data was compared to The Cancer Genome Atlas (TCGA) HNSCC data. Cluster analysis revealed that HNSCC PDXs were more similar to primary HNSCC than to any other tumor type. Interestingly, while a significant fraction of proteins were expressed similarly in both primary HNSCC and PDXs, a subset of proteins/phosphoproteins were expressed at higher (or lower) levels in PDXs compared to primary HNSCC. These findings indicate that the proteome is generally conserved in PDXs, but mechanisms for both positive and negative model selection and/or differences in the stromal components exist.
Implications
Proteomic characterization of HNSCC PDXs demonstrates potential drivers for model selection and provides a framework for improved utilization of this expanding model system.
Keywords: RPPA, PDX, HNSCC, in vivo model systems, proteomic characterization
INTRODUCTION
Head and neck squamous cell carcinoma (HNSCC) is the sixth leading cancer worldwide, with more than 600,000 incident cases per year (1, 2). Despite advances in multimodality therapy, HNSCC is frequently lethal, and five-year overall survival (OS) has increased minimally since 1990 (3). As of 2015, only 6 drugs are approved by the U.S. Food and Drug Administration (FDA) for the systemic treatment of HNSCC. The two most recent approvals, for the anti-microtubule chemotherapy docetaxel and the epidermal growth factor receptor (EGFR) antibody cetuximab, occurred in 2006 with no subsequent advancements in systemic therapy (2). Ongoing efforts to identify the subset of HNSCC patients who will respond to cytotoxic chemotherapy or EGFR targeting have been limited, in part, by the use of immortalized cell lines and xenografts derived from these cell lines for preclinical experiments. We recently reported that the genetic alterations found in HNSCC tumors often bear little resemblance to the changes that characterize immortalized HNSCC cell lines, and vice versa (1). The development of more relevant preclinical models is essential to facilitate more effective clinical translation. For example, the vast majority of HNSCC cell line-derived xenografts respond to cetuximab treatment, unlike most patients with HNSCC (3).
Patient-derived xenografts (PDXs) are emerging as a model platform that may better reflect human cancer compared with xenografts derived from immortalized cell lines that have been propagated indefinitely in culture (4, 5). Characterization of PDXs derived from a variety of cancers to date has included genomic sequencing, transcriptional profiling and immunohistochemical staining (6, 7). HNSCC PDXs specifically have been evaluated for mutations, gene expression and candidate proteins (8–13). Protein expression profiling of PDXs has been extremely limited (14–16).
In the present study we developed a collection of HNSCC-derived PDX models, assessed potential clinical and pathologic features that may influence take rates, assessed cryopreservation feasibility, and characterized protein expression of a subset of models for comparison with human HNSCC tumors. We were able to successfully establish and characterize HNSCC PDXs in NOD SCID gamma, but not athymic nude mice. We found that 40% (76/190) of the proteins analyzed by reverse phase protein array (RPPA), including phosphoproteins indicative of pathway activity, were conserved between HNSCC primary tumors and PDXs. Of the proteins that were differentially expressed between human HNSCC tumors and PDXs, 53% (34/64) were expressed at higher levels in the human tumors compared with PDXs, implicating negative model selection and 47% (30/64) were expressed at higher levels in the PDXs compared with human tumors, suggesting positive model selection. Different tumor/stroma ratios and/or reactivity of antibodies with murine stromal compartments likely also contributes to dissimilar protein levels observed.
MATERIALS AND METHODS
HNSCC patient-derived xenograft propagation
Tissues were collected under the auspices of a tissue bank protocol approved by the University of Pittsburgh Institutional Review Board. Following HNSCC tumor resection, de-identified patient samples underwent quality control to ensure 70% tumor composition. Samples were delivered to the laboratory in antibiotic/antimycotic solution and the time from resection to implantation was recorded. Tumor samples were cut into 25 mg pieces and directly implanted into mice. NOD/SCID gamma mice (Jackson Laboratories; Bar Harbor, ME) were anesthetized using isofluorane and a small (<10 mm) incision in the flank. 25 mg of tumor was placed in the pocket of the incision site and the wound closed with surgical adhesive. Analgesic was administered and the animals monitored until fully ambulatory. Mice were kept in isolation for 7–10 days and checked regularly for wound healing. Subsequently, mice were checked weekly for tumor formation. All animal studies were performed under an approved IACUC protocol at the University of Pittsburgh.
PDX cryopreservation
When tumors reached 1 cm maximum diameter, mice were sacrificed and the tumors were harvested. Tumors were cut into ~3 mm3 fragments and placed in 4°C RPMI-1640 media with 10% fetal bovine serum, 1% penicillin and streptomycin, and 5% DMSO (Life Technologies, Grand Island, NY). Tumors were placed at −20°C for 2 hours, at −80°C for 24 hours, and subsequently transferred to liquid nitrogen for long term storage. To transplant viably frozen tumors the tumors were thawed at 37°C and washed with 2 volumes of warm RPMI-1640 to remove DMSO. Tumor fragments were suspended in 100 uL BD Matrigel Matrix (BD Bioscience, USA) and kept on ice for immediate implantation.
Reverse-phase protein array (RPPA)
Samples were prepared as described previously (17, 18). RPPA was performed by the RPPA core facility at University of Texas MD Anderson Cancer Center. For human cancers, protein expression data were generated by RPPA for 4,778 patient tumors and 13 HNSCC PDXs using 190 antibodies. Patient tumor samples were profiled under the auspices of The Cancer Genomics Atlas and The Cancer Proteome Atlas (TCGA, http://cancergenome.nih.gov; TCPA TCPAportal.org) and included 127 bladder urothelial carcinomas (BLCA), 752 breast cancers (BRCA), 464 colon and rectal adenocarcinomas (COAD and READ), 299 gastric cancers, 215 glioblastoma multiforme (GBM), 212 head and neck squamous cell carcinomas (HNSC), 454 renal clear cell carcinomas (KIRC), 260 low grade gliomas (LGG), 237 lung adenocarcinomas (LUAD), 195 lung squamous cell carcinomas (LUSC), 208 melanomas, 412 high-grade serous ovarian cystadenocarcinomas (OVCA), 164 prostate cancers, 375 thyroid cancers, and 404 uterine corpus endometrial carcinomas (UCEC). RPPA slides were quantified using ArrayPro (Media Cybernetics Washington DC) to generate signal intensities that were further processed by SuperCurve (19) to estimate relative protein levels (in log2 scale). RPPA slide quality was monitored by a QC classifier (20) and only the slides whose QC scores were above 0.8 (on a 0–1 scale) were used for further analysis. The TCGA samples were run in a total of 7 batches, and merged via a replicates based normalization (RBN) method (21) which uses replicate samples run across multiple batches to adjust the data for batch effects.
Statistical analysis
We first applied unsupervised clustering analysis using normalized protein expression data and identified a branch with the majority of HNSCC samples from TCGA that were also comparable to HNSCC PDX samples. Comparison of protein expression of primary HNSCC tumors with HNSCC PDX samples was conducted using a Student’s t-test for each protein. False discovery rate (FDR) and Bonferroni corrected p-values were used to identify statistically significant and non-significant differences in protein expression patterns. All data analysis was conducted using R statistical package version 3.1.2 (https://www.r-project.org/).
RESULTS
Predictors of HNSCC PDX outgrowth
The factors that influence PDX engraftment are incompletely understood. Establishment and maintenance of PDXs require substantial resources, thus identification of clinical, pathologic or processing features that could increase successful in vivo establishment would facilitate translational research. We first implanted tumors in athymic nude mice and found that only 4/26 (15%) developed tumors so we abandoned this mouse strain. Between September 2009 and March of 2014 we implanted 76 tumors (from 71 specimens) into NOD SCID mice and 61 demonstrated growth in vivo (~80% take rate, Table 1). Similar to prior studies we found that these tumors could be cryopreserved through 24 months and passaged serially. Mean time to first tumor outgrowth was 3.5 months (Median 3.0 months; Range 0.97 to 11.2 months) with outgrowth upon passaging in mice being faster (Average 1.5 months; Median 1.5 months; Range 0.5 to 10 months). Passaging the PDX in mice at least once before cryopreservation enhanced the viability of the model as approximately 25% of primary HNSCC that engrafted successfully could not be subsequently passaged. There was no significant difference in take rates according to patient age, sex, tumor site, including metastatic lymph nodes or time from resection to implantation, or HPV status (χ2test; Table 2). All tumors were implanted less than 4 hours following excision. We received only 5 tumors that were HPV positive and 4 of these were successfully grown in mice. Of the 76 implanted tumors, 66 (87%) were primary-untreated HNSCC, 6 (8%) were post-treatment residual tumors, and 4 (5%) were recurrent tumors. We also tested the viability of 4 PDXs after cryopreservation, and 3 of 4 tumors could be regrown following 24 months in liquid nitrogen. We continue to track these tumors for sustained viability.
Table 1.
PDX Parameters
| Total specimens | PDX Formation | No growth | Take Rate | P-value | |
|---|---|---|---|---|---|
| Specimen Weight (g) | |||||
| Lowest 25th percentile (<=0.145) | 17 | 14 | 3 | 82.4% | 0.11 |
| 25–50th percentile (0.145–0.226) | 17 | 14 | 3 | 82.4% | |
| 50–75th percentile (0.226–0.342) | 15 | 15 | 0 | 100.0% | |
| Highest 25th percentile (>0.342) | 16 | 13 | 3 | 81.3% | |
| Mouse strain | |||||
| Nude | 26 | 4 | 22 | 15.4% | |
| NOD SCID | 76 | 61 | 15 | 80.3% | |
| Time to implantation (minutes) | |||||
| <100 | 48 | 38 | 10 | 79.2% | 0.43 |
| >=100 | 14 | 13 | 1 | 92.9% | |
| Unknown | 9 |
Table 2.
PDX donor clinical and pathologic information
| Total specimens | PDX Formation | No growth | Take Rate | P-value | |
|---|---|---|---|---|---|
| Site | |||||
| Oropharynx | 9 | 8 | 1 | 88.9% | 0.80 |
| Pharynx | 3 | 3 | 0 | 100.0% | |
| Oral cavity | 39 | 32 | 7 | 82.1% | |
| Lymph node | 11 | 8 | 3 | 72.7% | |
| Larynx | 9 | 7 | 2 | 77.8% | |
| AJCC Stage | |||||
| Stage I/II | 11 | 8 | 3 | 72.7% | 0.37 |
| Stage III | 13 | 9 | 4 | 69.2% | |
| Stage IV | 35 | 30 | 5 | 85.7% | |
| Unknown | 12 | 11 | 1 | 91.7% | |
| Age (years) | |||||
| years=<55 | 26 | 23 | 2 | 88.5% | 0.36 |
| 56–60 | 17 | 16 | 1 | 94.1% | |
| 61–67 | 15 | 10 | 5 | 66.7% | |
| >=68 | 13 | 11 | 2 | 84.6% | |
| Gender | |||||
| Male | 58 | 47 | 11 | 81.0% | 1 |
| Female | 13 | 11 | 2 | 84.6% | |
| HPV Status | |||||
| Positive | 5 | 4 | 1 | 80.0% | NA |
| Negative | 66 | 54 | 12 | 81.8% |
HNSCC protein expression is not age or site specific
To determine if overall primary HNSCC protein expression was associated with age or tumor site, we were able to obtain age and tumor site information for 55 primary HNSCC with TCPA data. Of the 55 specimens 2 were HPV positive and 2 had unknown HPV status. Protein profiles did not cluster by age or anatomic site of the primary HNSCC (Supplemental Figure 1). This finding suggests that head and neck tumors that arise in different anatomic sites may not represent distinct etiological cancers.
HNSCC-derived PDXs are more similar to HNSCC than any other tumor type
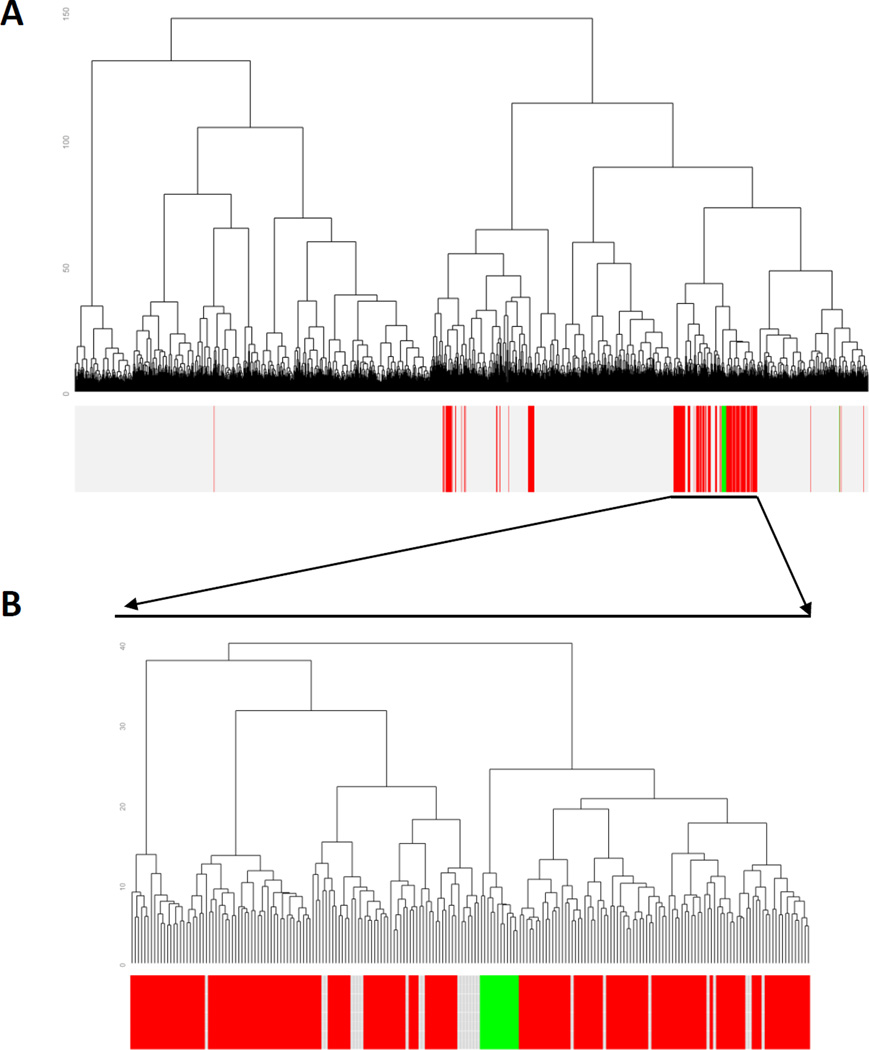
A subset of our HNSCC PDX collection (n=13) were analyzed at the early passages (10 of the 13 were at the first passage and 3 of 13 were at the second passage) by reverse phase protein array (RPPA), which assessed expression levels of 190 candidate total and phosphorylated proteins generally involved in signal transduction. A total of 4,778 tumor specimens from 15 different cancer types previously analyzed by RPPA were then compared to HNSCC-derived PDXs to determine the concordance of protein expression patterns (Figure 1a). The majority of human HNSCC tumors (173/212) clustered together and nearly all of HNSCC-derived PDXs (12/13) clustered within the major branch of HNSCC specimens in unsupervised clustering (Figure 1b). To our knowledge, this is the first comparison of RPPA/proteomic analysis of PDXs and primary human tumors in HNSCC.
Figure 1. HNSCC PDXs cluster with HNSCC patient tumors.
A) RPPA data from 4778 specimens in the TCPA database comprising 14 different types of cancer (GBM, OVCA, LUAD, LUSC, BRCA, KIRC, UCEC, COAD, Gastric, HNSC, Melanoma, Thyroid, BLCA, Prostate, LGG) were compared to 13 HNSCC PDX specimens by unsupervised clustering. A total of 190 proteins were utilized for this comparison. HNSCC is depicted in red, PDX in green, other tumor types are shown in light gray. B) Magnification of the sub-branch containing the primary HNSCC region of the cluster.
Subsets of proteins are similar between primary HNSCC and PDXs
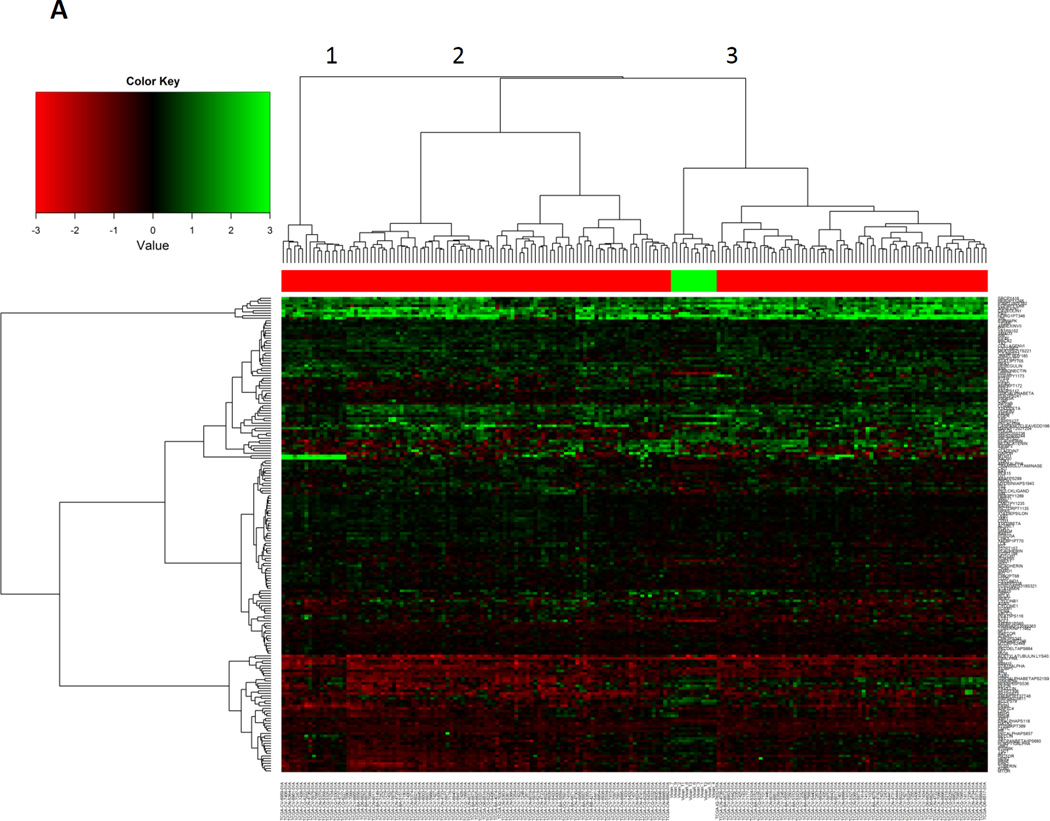
We then compared the 12 HNSCC-derived PDXs that clustered with the main branch of primary HNSCC to the primary HNSCC specimens. We found that the PDXs clustered together within HNSCC samples, demonstrating that the PDX are more similar to each other than to primary HNSCC. This indicates that despite similarities there are also changes in the PDX protein expression compared with primary HNSCC, likely due to both murine stromal components and changes in the HNSCC component (Figure 2a). Primary HNSCC is also somewhat heterogeneous with 3 subgroups noted in Figure 2a. The PDXs clustered with the largest subgroup of primary HNSCC indicating that the PDXs are more representative of one of the three subgroups of HNSCC (Figure 2a).
Figure 2. HNSCC PDXs cluster together when compared to HNSCC alone.
A) 173 HNSCC specimens and 12 HNSCC PDX specimens in the major HNSCC sub-branch cluster from Figure 1B. The three major sub-branches are denoted (numbers 1–3). A total of 190 proteins for each specimen were analyzed. B) Clustering analysis of 76 proteins expressed at similar levels in both primary HNSCC and HNSCC PDXs. T-test was applied to compare these 173 HNSCC and 12 PDX specimens. A false discovery rate > 0.05 was deemed as not significantly different. C) A more conservative p-value adjustment was used (Bonferroni Correction) and the adjusted p-value < 0.05 was applied to identify differentially expressed proteins (n=64).
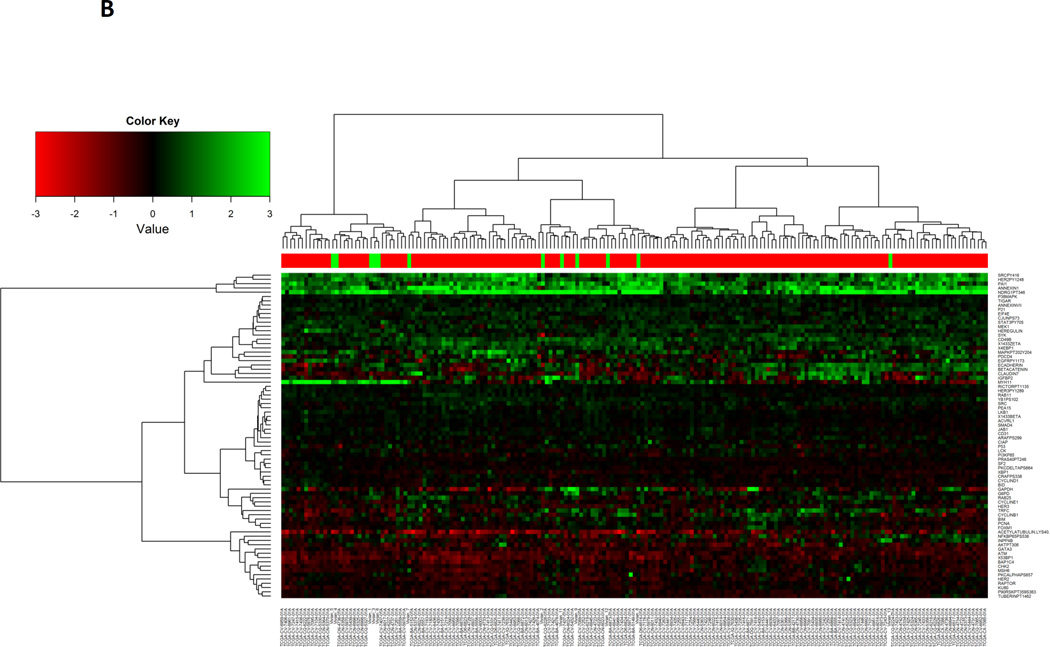
To determine the concordance and discordance of levels of protein expression between HNSCC human tumors and PDXs, we compared expression levels of 190 individual proteins from 173 HNSCC tumors and 12 PDXs. When an adjusted p-value exceeded the pre-specified FDR (≥ 0.05), the expression of a specific protein was classified as similar between HNSCC tumors and PDXs. We identified 76 proteins expressed at similar levels in both primary HNSCC and PDXs, which are depicted with unsupervised clustering (Figure 2b, Supplemental Table 1, Supplemental Figure 2a). The PDX samples intermingle with the primary HNSCC, indicating that these 76 proteins are likely to be preserved in a PDX.
Subsets of proteins are expressed at different levels in primary HNSCC compared with PDXs
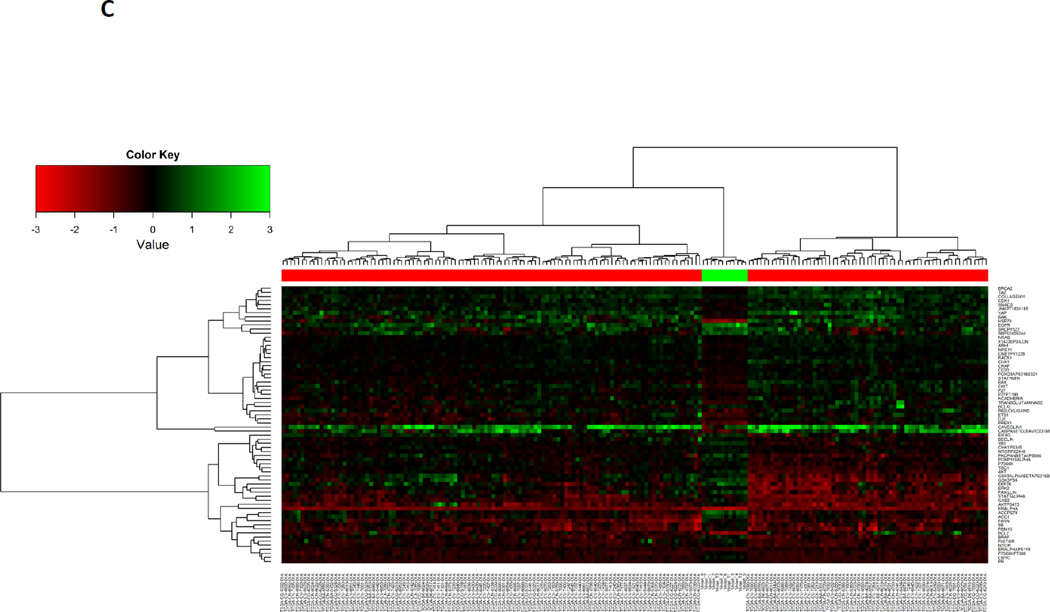
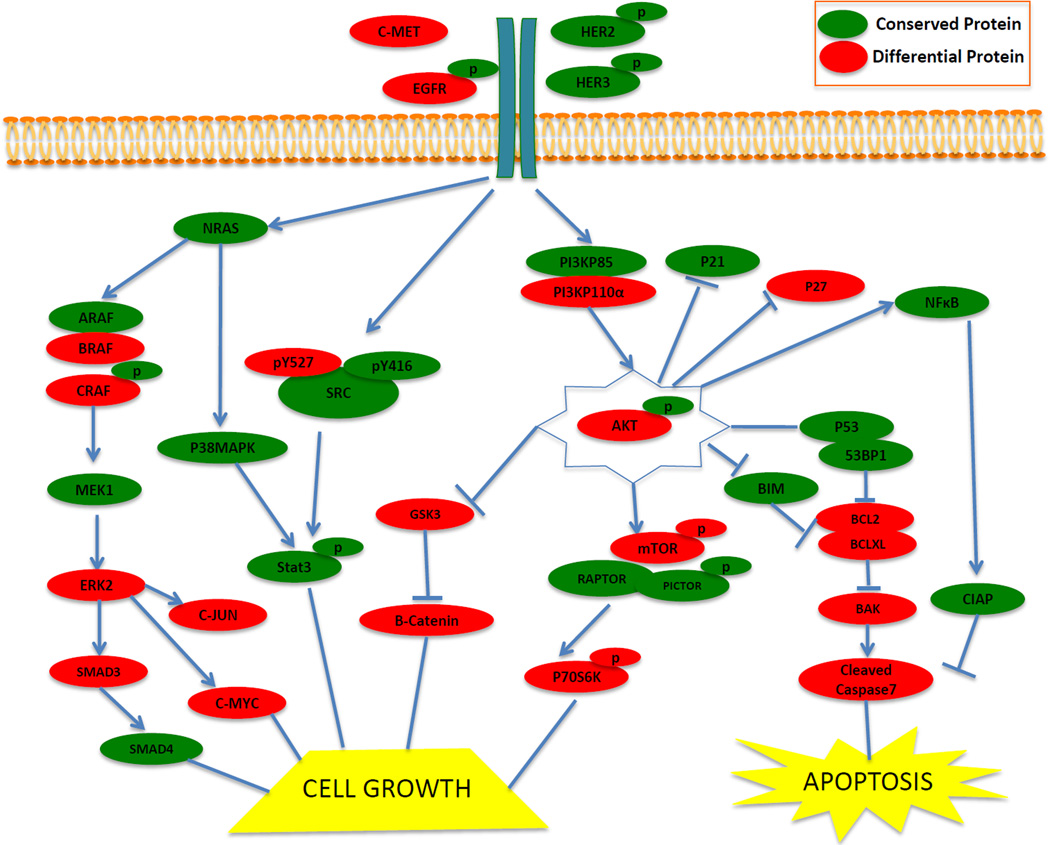
To define the proteins that were differentially expressed between primary HNSCC and PDXs, Bonferroni correction was used to stringently adjust p-values for multiple testing; an adjusted p-value ≤ 0.05 was considered significant. We found that 64 proteins were either significantly increased (n=30) or decreased (n=34) in PDXs compared with primary HNSCC tumors. Unsupervised clustering analysis of expression levels of these differentially expressed proteins demonstrated that PDXs clustered separately from the primary HNSCC (Figure 2c, Supplemental Table 2, Supplemental Figure 2b). Manual pathway matching did not find evidence that the majority of the proteins in any given signaling pathway were either preserved or differentially expressed in PDXs compared with the human HNSCC (Figure 3).
Figure 3. Conserved and differentially expressed proteins in major HNSCC signaling pathways.
Representative differences and similarities between HNSCC PDXs and primary human tumors are depicted in cell growth and apoptosis pathways. Proteins that were not significantly different between HNSCC PDXs and human tumors are depicted in green (subset of the n=76 proteins). Proteins that were significantly different between HNSCC PDXs and human tumors are depicted in red (subset of the n=64 proteins).
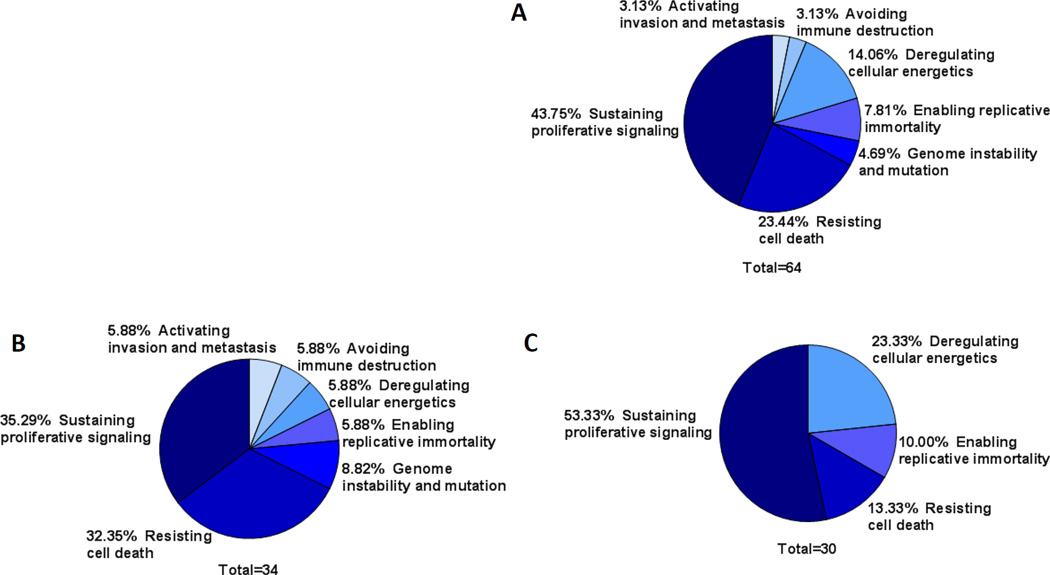
Of the 64 differentially expressed proteins, 30 demonstrated increased expression in PDXs (positive model selection) while 34 demonstrated decreased expression in PDXs (negative model selection). We categorized all 64 proteins to one of the cancer hallmark phenotypes (sustaining proliferative signaling, evading growth suppressors, avoiding immune destruction, enabling replicative immortality, tumor-promoting inflammation, activating invasion and metastasis, inducing angiogenesis, genome instability and mutation, resisting cell death, deregulating cellular energetics) proposed by Hanahan and Weinberg (Figure 4a) (22) and found that negative model selection (decreased protein expression) was represented by many different categories of proteins (Figure 4b), demonstrating no preferred pathway of negative selection. The positive model selection (increased protein expression) was comprised of fewer categories and primarily involved proteins that sustain proliferative signaling (Figure 4c). Expression levels of the proteins that were differentially expressed in HNSCC PDXs were generally not outside the range of expression in the primary HNSCC, but rather the PDXs clustered to high or low within the primary HNSCC range (Supplemental Figure 2b). There were only 6 proteins with more than half of the PDX specimens outside the expression range of primary HNSCC. AKT, c-Myc and PR were increased in PDXs above the range of primary HNSCC, while BCL2, c-Kit, and HSP70 were decreased in PDXs below the range of primary HNSCC. These cumulative findings suggest that while most proteins are conserved in PDXs, there is evidence that proteins associated with proliferative signaling may be preferentially selected during the process of creating PDX models.
Figure 4. HNSCC PDX protein expression clustered by cancer hallmarks is different between positive and negative selection model proteins.
A) Proteins that are expressed at different levels in HNSCC PDXs compared to primary HNSCC were categorized according to the hallmarks of cancer (31). B) Proteins that were negatively selected in HNSCC PDXs (lower expression in PDXs compared to primary HNSCC) were categorized by the hallmarks of cancer. C) Proteins that were positively selected in PDXs (high expression in PDXs compared to primary HNSCC) were categorized by the hallmarks of cancer.
DISCUSSION
The need for improved preclinical model systems to develop cancer therapeutics is supported by the high failure rate in the clinic of agents that eradicate tumors in mice (23). The heterogeneous nature of cancer and the known changes that arise when tumors are immortalized in cell culture contribute to the limitations of cell line-derived xenograft model systems (24–26). We developed patient-derived xenografts from primary HNSCC specimens in an effort to better understand the capacity and limitations of these models as well as to test discrete hypotheses regarding response to molecular targeted therapies (3, 27, 28). Some studies have demonstrated no difference in engraftment with tumor biology or clinical characteristics (8), while other studies have demonstrated some increase in engraftment with poorly differentiated primary HNSCC or the presence of nodal disease (29). We did not identify a clinical, pathologic or tumor processing variable, including time from resection to implantation, that affected the PDX take rate (Table 2). Although hypoxia and devascularization likely impact tumor take rate, our HNSCC samples were all implanted within 4 hours of surgical resection. Published take rates for HNSCC PDXs range from 30% to 70% and for PDXs generally range from 10% to 90% (6, 7). Our overall take rate was 80% and we developed the capability of cryopreservation following of established PDXs, thereby allowing the development of a robust collection that can be used over time.
PDX models have been developed in a number of cancers with evidence that these models more accurately reflect the human tumor compared with cell line-derived xenografts (3, 9, 24, 30, 31). Characterization of these PDXs so far has focused on genomic and transcriptomic profiling with most studies reporting a general concordance between gene amplification, mutation, and gene expression in the primary and PDX tumor (6, 7). The limitations of PDXs noted to date include evidence that there are molecular and structural changes in PDXs both at early and late passages, particularly noted in endothelial cells (32, 33). Lack of concordance has arisen in some models due to significant intratumoral heterogeneity (31, 34, 35). Additionally, several tumors known to metastasize in the patient do not metastasize in PDXs (36). Human tumor microenvironment, drug kinetics, and the lack of an immune system are ongoing challenges that are being addressed slowly with solutions such as humanized mice and in silico methods (6, 37–39). It is possible that limitations of PDXs have been under-reported to date due to the real improvements of these models compared with cell line xenografts coupled with the natural selection of publications focused on positive data.
Studies on HNSCC PDXs are limited with each publication validating specific aspects of the PDX model or a small number of therapeutic options. To date studies in HNSCC PDXs and PDXs in general have not compared the PDXs to larger control groups of primary tumors to determine if the PDX is a good representation of the disease; rather, publications have favored a parent primary to PDX comparison. While this is important for initial validation of a new PDX, such comparisons are inadequate to inform about the effectiveness of these PDXs in modeling the broader disease. Without comparison to a larger primary tumor dataset like the TCGA, it is possible to have a high level of primary to PDX concordance with a significant engraftment bias for a subtype of the disease. HNSCC PDXs appear to have a reasonable concordance with their matched primary tumor for: promoter methylation (9), histology (10, 12, 29), limited protein markers by IHC (10, 12, 29), and some gene expression (29). The availability of TCGA data on primary human tumors, including HNSCC, provides an opportunity to compare preclinical models systems to human cancer. To our knowledge, this is the first study to compare protein profiling in HNSCC PDXs to a repository of comprehensively profiled HNSCC human tumors. We found that HNSCC PDXs express an overall protein profile that is largely similar to primary HNSCCs (when compared to other tumor types), suggesting that they represent a promising preclinical model system for mechanistic and therapeutic studies. Analysis of the HNSCC PDXs compared to HNSCC primary tumors alone also revealed that the HNSCC PDXs were more similar to one another than to primary HNSCC tumors, suggesting that model selection factors may limit the ability of PDXs to reflect human tumor heterogeneity. In addition, since the stroma of established and passaged PDX is murine, these models cannot currently be used to study human stroma or the impact of stroma on proteomic profiling. By comparing the proteins that were significantly different or similar between HNSCC PDXs and human tumors we anticipated finding that common stromal pathways were different and common tumor/epithelial pathways were similar. Instead, we found significant heterogeneity among expression of canonical proteins and phosphorylated proteins in well-established signaling pathways implicated in HNSCC. This was particularly surprising given that prior genomic and transcriptomic analyses have demonstrated preservation of these features in HNSCC PDXs compared with human tumors (9, 29). This underlines the importance of protein analysis in the characterization of these model systems. Ideally, an unbiased proteomic approach could be employed, but the cost and time limitations of mass spectrometry-based proteomics coupled with the absence of these data systematically associated with TCGA cohorts, suggests that alternative strategies must be considered. One of the strengths of the present study was the large control cohort from the TCPA (40), which allowed us to analyze the HNSCC PDX protein profiles in the setting of primary HNSCC heterogeneity and to evaluate them as overall models of HNSCC. We did not have access to matched HNSCC primary and PDX RPPA data. A matched primary analysis, as seen in the prior nucleic acid analyses, is valuable for comparing individual preservation profiles but inherently selects for a subgroup of HNSCC and may not provide sufficient information about modeling the heterogeneity of HNSCC.
We found evidence for both positive and negative model selection among the differentially expressed proteins in the PDXs compared to primary HNSCC. Increased expression of AKT and c-Myc in the HNSCC PDXs is consistent with the roles of AKT in cell growth and proliferation and c-Myc in cell cycle progression and apoptosis. Increased expression of PR was surprising; differences in expression of these hormone signaling proteins possibly reflect the sexual dimorphism between the predominately male primary HNSCC tissue and female mice used for these model systems. Decreased expression of BCL2 in PDXs below the range of primary HNSCC supports the apoptotic role of BCL2, while decreased expression of c-Kit in PDXs may be due to the loss of the cytokines involved in signaling through this receptor. The negative selection and further downregulation of BCL2 and c-Kit expression in HNSCC PDXs underscores the plasticity of tumor protein expression and the importance of characterization of model systems beyond genomics. HSP70 is also decreased in PDXs. HSP70 is a chaperone protein that is upregulated by various cellular stressors. HSP70 may be higher in primary tumors due to upregulation during the often lengthy primary tumor resection process compared to the rapid resection of established PDXs with flash freezing, which may cause little to no upregulation of this protein due to stress. Categorizing these proteins according to the previously defined hallmarks of cancer suggested that proteins that sustain proliferative signaling comprised a larger component of positive model selection proteins than any other category, while negative selection proteins were more heterogeneous and included proteins in all categories (22). This indicates that proteins that aid in cell proliferation are more likely to be increased in PDXs overall, and care should be taken when studying therapeutic targeting of these important oncogenic pathways as the upregulation of these proteins may make these PDXs more reliant on these pathways than the original tumors. In the absence of clear signaling pathway similarities or differences between HNSCC PDXs and primary tumors, studies on targeted treatments for HNSCC in PDXs are limited. The protein expression profile may help to determine the use of these models to guide precision medicine
Patient tissue is a valuable tool in the study of cancer and the ability to propagate that tool using PDX models (compared to traditional cell culture) holds promise. We demonstrate here that HNSCC PDXs have limitations that must be considered in the context of preclinical modeling studies. An individual PDX may harbor the mutations identified in the primary human tumor but relative protein expression may be altered by the process of implanting and propagating these tumors in mice. Changes in protein expression indicate a need for careful characterization of protein expression in PDXs for studies that are predicated on an understanding of expression of total and phosphorylated proteins implicated in oncogenic signaling. Additionally, a single PDX may recapitulate the response of the primary tumor to a specific therapy, but such a finding may not be extrapolated to conclude that the primary tumor is reflective of HNSCC drug responses in general. Analysis of paired primary and PDX genomic, transcriptomic, proteomic and metabolomic profiles for PDXs at each passage would provide additional insights into this model system.
Our results suggest the limitations of utilizing only genomic and transcriptomic data to characterize a PDX model. It is important to recognize that protein may provide us with more information about the functional differences affecting therapeutic and mechanistic studies. Despite the concordance of genomic and transcriptomic data between primary human tumors and PDXs (6, 7) we found some significant differences at the protein level. These differences are likely attributable to changes in the stromal component of the tumor that subsequently alters protein expression in the tumor cells in conjunction with factors that mediate both positive and negative model selection. PDXs represent a new and important tool for preclinical modeling of cancer therapeutics. Understanding both the capacity and limitations of these models should guide their rational selection for specific studies.
Supplementary Material
Acknowledgments
NIH P50CA097190 and the American Cancer Society (JRG)
NIH K07 CA137140 (AME)
Department of Veterans Affairs BLR&D. This work does not reflect the views of the US Government or the Department of Veterans Affairs. (UD)
RPPA work was performed in the MDACC CCSG supported core NCI CA16672
JRG has research funding from Novartis and BMS.
Footnotes
The remaining authors declare that they have no conflicts of interest.
REFERENCES
- 1.Li H, Wawrose JS, Gooding WE, Garraway LA, Lui VW, Peyser ND, et al. Genomic analysis of head and neck squamous cell carcinoma cell lines and human tumors: a rational approach to preclinical model selection. Molecular cancer research : MCR. 2014;12(4):571–582. doi: 10.1158/1541-7786.MCR-13-0396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vermorken JB, Trigo J, Hitt R, Koralewski P, Diaz-Rubio E, Rolland F, et al. Open-label, uncontrolled, multicenter phase II study to evaluate the efficacy and toxicity of cetuximab as a single agent in patients with recurrent and/or metastatic squamous cell carcinoma of the head and neck who failed to respond to platinum-based therapy. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2007;25(16):2171–2177. doi: 10.1200/JCO.2006.06.7447. [DOI] [PubMed] [Google Scholar]
- 3.Quesnelle KM, Wheeler SE, Ratay MK, Grandis JR. Preclinical modeling of EGFR inhibitor resistance in head and neck cancer. Cancer biology & therapy. 2012;13(10):935–945. doi: 10.4161/cbt.20846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lodhia KA, Hadley AM, Haluska P, Scott CL. Prioritizing therapeutic targets using patient-derived xenograft models. Biochimica et biophysica acta. 2015;1855(2):223–234. doi: 10.1016/j.bbcan.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siolas D, Hannon GJ. Patient-derived tumor xenografts: transforming clinical samples into mouse models. Cancer research. 2013;73(17):5315–5319. doi: 10.1158/0008-5472.CAN-13-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malaney P, Nicosia SV, Dave V. One mouse, one patient paradigm: New avatars of personalized cancer therapy. Cancer letters. 2014;344(1):1–12. doi: 10.1016/j.canlet.2013.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tentler JJ, Tan AC, Weekes CD, Jimeno A, Leong S, Pitts TM, et al. Patient-derived tumour xenografts as models for oncology drug development. Nature reviews Clinical oncology. 2012;9(6):338–350. doi: 10.1038/nrclinonc.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen J, Milo GE, Shuler CF, Schuller DE. Xenograft growth and histodifferentiation of squamous cell carcinomas of the pharynx and larynx. Oral surgery, oral medicine, oral pathology, oral radiology, and endodontics. 1996;81(2):197–202. doi: 10.1016/s1079-2104(96)80415-x. [DOI] [PubMed] [Google Scholar]
- 9.Hennessey PT, Ochs MF, Mydlarz WW, Hsueh W, Cope L, Yu W, et al. Promoter methylation in head and neck squamous cell carcinoma cell lines is significantly different than methylation in primary tumors and xenografts. PloS one. 2011;6(5):e20584. doi: 10.1371/journal.pone.0020584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kimple RJ, Harari PM, Torres AD, Yang RZ, Soriano BJ, Yu M, et al. Development and characterization of HPV-positive and HPV-negative head and neck squamous cell carcinoma tumorgrafts. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19(4):855–864. doi: 10.1158/1078-0432.CCR-12-2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prince ME, Sivanandan R, Kaczorowski A, Wolf GT, Kaplan MJ, Dalerba P, et al. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(3):973–978. doi: 10.1073/pnas.0610117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seshadri M, Merzianu M, Tang H, Rigual NR, Sullivan M, Loree TR, et al. Establishment and characterization of patient tumor-derived head and neck squamous cell carcinoma xenografts. Cancer biology & therapy. 2009;8(23):2275–2283. doi: 10.4161/cbt.8.23.10137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stein AP, Saha S, Liu CZ, Hartig GK, Lambert PF, Kimple RJ. Influence of handling conditions on the establishment and propagation of head and neck cancer patient derived xenografts. PloS one. 2014;9(6):e100995. doi: 10.1371/journal.pone.0100995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown KE, Chagoya G, Kwatra SG, Yen T, Keir ST, Cooter M, et al. Proteomic profiling of patient-derived glioblastoma xenografts identifies a subset with activated EGFR: implications for drug development. Journal of neurochemistry. 2015;133(5):730–738. doi: 10.1111/jnc.13032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li S, Shen D, Shao J, Crowder R, Liu W, Prat A, et al. Endocrine-therapy-resistant ESR1 variants revealed by genomic characterization of breast-cancer-derived xenografts. Cell reports. 2013;4(6):1116–1130. doi: 10.1016/j.celrep.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang X, Claerhout S, Prat A, Dobrolecki LE, Petrovic I, Lai Q, et al. A renewable tissue resource of phenotypically stable, biologically and ethnically diverse, patient-derived human breast cancer xenograft models. Cancer research. 2013;73(15):4885–4897. doi: 10.1158/0008-5472.CAN-12-4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carey MS, Agarwal R, Gilks B, Swenerton K, Kalloger S, Santos J, et al. Functional proteomic analysis of advanced serous ovarian cancer using reverse phase protein array: TGF-beta pathway signaling indicates response to primary chemotherapy. Clinical cancer research : an official journal of the American Association for Cancer Research. 2010;16(10):2852–2860. doi: 10.1158/1078-0432.CCR-09-2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tibes R, Qiu Y, Lu Y, Hennessy B, Andreeff M, Mills GB, et al. Reverse phase protein array: validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Molecular cancer therapeutics. 2006;5(10):2512–2521. doi: 10.1158/1535-7163.MCT-06-0334. [DOI] [PubMed] [Google Scholar]
- 19.Hu J, He X, Baggerly KA, Coombes KR, Hennessy BT, Mills GB. Non-parametric quantification of protein lysate arrays. Bioinformatics. 2007;23(15):1986–1994. doi: 10.1093/bioinformatics/btm283. [DOI] [PubMed] [Google Scholar]
- 20.Ju Z, Liu W, Roebuck PL, Siwak DR, Zhang N, Lu Y, et al. Development of a robust classifier for quality control of reverse-phase protein arrays. Bioinformatics. 2015;31(6):912–918. doi: 10.1093/bioinformatics/btu736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akbani R, Ng PK, Werner HM, Shahmoradgoli M, Zhang F, Ju Z, et al. A pan-cancer proteomic perspective on The Cancer Genome Atlas. Nature communications. 2014;5:3887. doi: 10.1038/ncomms4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 23.Johnson JI, Decker S, Zaharevitz D, Rubinstein LV, Venditti JM, Schepartz S, et al. Relationships between drug activity in NCI preclinical in vitro and in vivo models and early clinical trials. British journal of cancer. 2001;84(10):1424–1431. doi: 10.1054/bjoc.2001.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daniel VC, Marchionni L, Hierman JS, Rhodes JT, Devereux WL, Rudin CM, et al. A primary xenograft model of small-cell lung cancer reveals irreversible changes in gene expression imposed by culture in vitro. Cancer research. 2009;69(8):3364–3373. doi: 10.1158/0008-5472.CAN-08-4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lawrence MS, Sougnez C, Lichtenstein L, Cibulskisl K, Lander E, Gabriel SB, et al. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517(7536):576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333(6046):1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lui VW, Hedberg ML, Li H, Vangara BS, Pendleton K, Zeng Y, et al. Frequent mutation of the PI3K pathway in head and neck cancer defines predictive biomarkers. Cancer discovery. 2013;3(7):761–769. doi: 10.1158/2159-8290.CD-13-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sen M, Pollock NI, Black J, DeGrave KA, Wheeler S, Freilino ML, et al. JAK kinase inhibition abrogates STAT3 activation and head and neck squamous cell carcinoma tumor growth. Neoplasia. 2015;17(3):256–264. doi: 10.1016/j.neo.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peng S, Creighton CJ, Zhang Y, Sen B, Mazumdar T, Myers JN, et al. Tumor grafts derived from patients with head and neck squamous carcinoma authentically maintain the molecular and histologic characteristics of human cancers. Journal of translational medicine. 2013;11:198. doi: 10.1186/1479-5876-11-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fichtner I, Rolff J, Soong R, Hoffmann J, Hammer S, Sommer A, et al. Establishment of patient-derived non-small cell lung cancer xenografts as models for the identification of predictive biomarkers. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14(20):6456–6468. doi: 10.1158/1078-0432.CCR-08-0138. [DOI] [PubMed] [Google Scholar]
- 31.Rubio-Viqueira B, Jimeno A, Cusatis G, Zhang X, Iacobuzio-Donahue C, Karikari C, et al. An in vivo platform for translational drug development in pancreatic cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2006;12(15):4652–4661. doi: 10.1158/1078-0432.CCR-06-0113. [DOI] [PubMed] [Google Scholar]
- 32.Gray DR, Huss WJ, Yau JM, Durham LE, Werdin ES, Funkhouser WK, Jr, et al. Short-term human prostate primary xenografts: an in vivo model of human prostate cancer vasculature and angiogenesis. Cancer research. 2004;64(5):1712–1721. doi: 10.1158/0008-5472.can-03-2700. [DOI] [PubMed] [Google Scholar]
- 33.Sanz L, Cuesta AM, Salas C, Corbacho C, Bellas C, Alvarez-Vallina L. Differential transplantability of human endothelial cells in colorectal cancer and renal cell carcinoma primary xenografts. Laboratory investigation; a journal of technical methods and pathology. 2009;89(1):91–97. doi: 10.1038/labinvest.2008.108. [DOI] [PubMed] [Google Scholar]
- 34.Fu X, Guadagni F, Hoffman RM. A metastatic nude-mouse model of human pancreatic cancer constructed orthotopically with histologically intact patient specimens. Proceedings of the National Academy of Sciences of the United States of America. 1992;89(12):5645–5649. doi: 10.1073/pnas.89.12.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim MP, Evans DB, Wang H, Abbruzzese JL, Fleming JB, Gallick GE. Generation of orthotopic and heterotopic human pancreatic cancer xenografts in immunodeficient mice. Nature protocols. 2009;4(11):1670–1680. doi: 10.1038/nprot.2009.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mueller BM, Reisfeld RA. Potential of the scid mouse as a host for human tumors. Cancer metastasis reviews. 1991;10(3):193–200. doi: 10.1007/BF00050791. [DOI] [PubMed] [Google Scholar]
- 37.Eckhardt SG. Challenges to Pdtx Models in Drug Development. Annals of Oncology. 2013;24:15. [Google Scholar]
- 38.Kopetz S, Lemos R, Powis G. The promise of patient-derived xenografts: the best laid plans of mice and men. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18(19):5160–5162. doi: 10.1158/1078-0432.CCR-12-2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuperwasser C, Chavarria T, Wu M, Magrane G, Gray JW, Carey L, et al. Reconstruction of functionally normal and malignant human breast tissues in mice. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(14):4966–4971. doi: 10.1073/pnas.0401064101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li J, Lu Y, Akbani R, Ju Z, Roebuck PL, Liu W, et al. TCPA: a resource for cancer functional proteomics data. Nature methods. 2013;10(11):1046–1047. doi: 10.1038/nmeth.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.