Abstract
Despite initial remission, approximately 60-70% of adult and 30% of pediatric patients experience relapse or refractory AML. Studies so far have identified base line gene expression profiles of pathogenic and prognostic significance in AML, however extent of change in gene expression post-initiation of treatment has not been investigated. Exposure of leukemic cells to chemotherapeutic agents such as cytarabine, a mainstay of AML chemotherapy can trigger adaptive response by influencing leukemic cell transcriptome and hence development of resistance or refractory disease. It is however challenging to perform such a study due to lack of availability of specimens post-drug treatment. In this study our primary objective was to identify in vivo cytarabine induced changes in leukemia cell transcriptome and to evaluate their impact on clinical outcome. Our results highlight genes relevant to cytarabine resistance and support the concept of targeting cytarabine-induced genes as a means of improving response.
Keywords: Cytarabine, Acute Myeloid Leukemia, Gene Expression, Event free Survival
Introduction
In treating AML, refractory disease remains the greatest challenge. Despite initial remission, approximately 60-70% of adult patients and 30% of pediatric patients will die of relapse and refractory AML.(1, 2) Moreover, standard chemotherapy treatment with cytarabine and anthracyclines cause significant adverse events including myelosuppression, high risk for infections, need for transfusions, mucositis, neurotoxicity, and cardiotoxicity.(3, 4) Anti-leukemia drugs such as cytarabine act intra-cellularly by incorporation into DNA. Inefficient cellular uptake, reduced intracellular activation, increased degradation, or expansion of the dNTP pools could result in development of cellular resistance to ara-C.(5) Although intracellular cytarabine-triphosphate levels correlate with AML response and small increases in cytarabine-triphosphate result in increased AML cell-cytotoxic effect, prior attempts in AML patients to increase cytarabine dosing did not improve clinical outcomes.(6) Together, these data suggest that cytarabine resistance involves other mechanisms.
Previously, we used gene-expression array to identify diagnostic AML cell gene-expression signatures predictive of in vitro and in vivo response to chemotherapy.(7) This study demonstrated a validated method of integrating genomic-data at diagnosis with important pharmacologic and clinical outcomes. Based on this experience, we reasoned that examining gene-expression changes in AML cells after exposure to cytarabine would provide additional information regarding intracellular response to treatment. However efforts to study in vivo leukemic cell gene expression changes induced by chemotherapy has been relatively limited. This is primarily due to technical difficulty in obtaining samples at adequate time and in enough quantity to study transcriptomic changes. Our study is unique in this aspect as it reports in vivo cytarabine induced gene expression levels in leukemic cells obtained at diagnosis and 24 hr post-cytarabine infusion. Additionally, we also report gene-expression changes corresponding to worse outcomes may explain mechanisms of refractory disease and may serve as potential targets for enhancing response to cytarabine
MATERIALS AND METHODS
Study population
Twenty-four de novo AML patients enrolled on AML02 trial [clinicaltrias.gov identifier NCT00136084] with matched specimens available at diagnosis [pre-treatment] and 24 hours after the start of first dose cytarabine were included in this study (6). Patients with acute promyelocytic leukemia or Down's syndrome were excluded. Patients were randomly assigned to receive induction 1 therapy consisting of high-dose [3 g/m2, days 1, 3 and 5] or low-dose [100 mg/m2, days 1-10] cytarabine along with daunorubicin [50mg/m2: days 2, 4 and 6] and etoposide [days 2-6] by intravenous infusion. Subsequent therapy was adapted based on diagnostic risk features and induction 1 response measured as minimal residual disease [MRD] as assessed by flow cytometry. The details of study design and clinical outcome are described elsewhere (6). St. Jude Institutional Review Board approved the study, and informed consent was obtained from parents/guardians and consents/assents from the individuals themselves when appropriate.
Specimen Collection and AML Cell Enrichment
Bone marrow was aspirated at diagnosis [pre-treatment] and 24 hours after the start of first dose cytarabine. Enrichment for AML cells was performed using Ficoll-Hypaque density-gradient centrifugation, as previously described.(8) If necessary, specimens were further enriched to achieve > 80% blasts by immunomagnetic sorting [Miltenyi Biotech, Germany].
Gene Expression Profiling
Gene expression profiling of AML cells was performed using GeneChip® Human Genome U133 Plus 2.0 Array [Affymetrix, Santa Clara, CA]. Details regarding RNA isolation, labeling of cRNA, and scanning of Affymetrix arrays have been published previously(9).
Pharmacology Measurements
AML cells from diagnostic bone marrow specimens were treated in vitro with various concentrations of cytarabine [range, 0.002 – 2.5 ng/μL] followed by MTT assays to determine the median lethal concentration [LC50] value [previously described (7, 9).
Clinical Outcomes
Patients were classified as having low-risk AML if the AML cells harbored t[8;21], inv[16], or t[9;11] chromosome abnormalities. High-risk AML included those with del[7], FLT3-ITD mutation, t[6;9], megakaryoblastic AML, treatment-related AML, or AML arising from MDS. All other patients were classified as standard-risk AML. IWG AML response criteria were used to classify clinical responses.(10) Flow cytometry was used to measure minimal residual disease [MRD at day 22], as previously described.(11) MRD was defined as 1 or more AML cells per 1000 bone marrow mononuclear cells [i.e., ≥ 0.1%]. Overall study design, patient characteristics and details of endpoints are provided in Table- 1 and Supplementary Figure-1S shows overall study schema.
Table 1.
Patient characteristics by arm.
| Feature | HDAC | LDAC | P value |
|---|---|---|---|
| Gender | 0.089 | ||
| Female | 7 | 5 | |
| Male | 2 | 10 | |
| Age | 0.089 | ||
| <10 Years | 2 | 10 | |
| >=10 Years | 7 | 5 | |
| WBC | 0.657 | ||
| <50 | 7 | 9 | |
| >=50 | 2 | 6 | |
| Race | 1 | 2 | 1 |
| Black | |||
| Other | 1 | 2 | |
| White | 7 | 11 | |
| Cytogenetics | 0.277 | ||
| 11q23 | 4 | 2 | |
| CBF | 3 | 3 | |
| Normal | 1 | 5 | |
| Other | 1 | 5 | |
| Provisional Risk | 0.666 | ||
| High | 2 | 6 | |
| Low | 3 | 3 | |
| Standard | 4 | 6 | |
Statistical Analysis
For each subject and each probe-set, the expression-change was defined as the log-transformed MAS5.0 normalized signal of the 24-hour sample minus that of the baseline sample so that a positive/negative change indicates that expression increased [decreased] from baseline to post-cytarabine infusion.
Arm-Stratified Wilcoxon Signed-Rank Test
An arm-stratified signed-rank test was used to identify probe-sets with significant expression changes for both arms, and the rank-sum test was used to identify probe-sets with expression changes that differed significantly between cytarabine treatment arms. The following statistical testing procedure was performed for each probe-set. The Wilcoxon signed-rank test statistic was computed from the expression changes separately for each arm. We also computed the expected value and variance of these statistics under the null hypothesis that the median expression change in the population equals zero. The final z-statistic was computed by subtracting the sum of the null expectations from the sum of the signed-rank statistics and dividing the result by the square root of the sum of the null variances. A final p-value was computed by comparing the final z-statistic to the standard normal distribution.
Rank-Sum Test
For each probe-set, the rank-sum test statistic was computed to test the null hypothesis that the two arms had equal median expression changes. The p-value was determined by comparing the observed statistic to a set of statistics obtained by 10,000 random permutations of the assignment of the arm label to ranks of the expression change values. The expected value and variance of the rank-sum statistics under the null hypothesis were used to z-transform the rank-sum statistic.
PROMISE Analyses
PRojection-Onto-the-Most-Interesting-Statistical-Evidence [PROMISE](12) was used to explore the association of expression changes with LC50, MRD, and EFS. Event-free survival [EFS] was defined as the time elapsed from protocol enrollment to the earliest of disease resistance, relapse, death, development of a second malignancy, or death with times for subjects living and free of these treatment failures censored at last follow-up. For this analysis, MRD was numerically represented as 0 [no detectable disease], 1 [between 0.1% and 1% of cells are leukemic], or 2 [>1% of cells are leukemic]. An arm-stratified Spearman-rank correlation statistic was used to characterize the association of expression changes with LC50 and MRD. The sign of this statistic indicates the direction of association of the expression change with these two endpoints. The statistic of Jung, Owzar, and George [2005] was used to characterize the association of expression changes with the duration of EFS (13). A positive value of this statistic indicates that an increased expression value associates with longer EFS. For each probe-set, the PROMISE statistic was defined as the sum of the LC50 association statistic and the MRD association statistic minus the EFS association statistic. A positive value of the PROMISE statistic indicates a beneficial pattern of association in the sense that a greater expression change value associates with lower LC50 [greater sensitivity to cytarabine], lower MRD [less residual disease after one course], and a lower rate of EFS treatment failures. A negative value of the PROMISE statistic indicates a detrimental pattern of association in the sense that a greater expression change value associates with greater LC50 [greater resistance to cytarabine], greater MRD [more residual disease after one course], and a greater rate of EFS treatment failures. P-values were determined by 10,000 arm-stratified permutations of the assignment of the entire expression change profile to the vector of endpoint data values. For each analysis described above, the robust FDR method of Pounds and Cheng (14) was applied to the p-values to obtain estimates of the false discovery rate. These estimates are reported as q-values.
In vitro validation by si-RNA mediated knock down of selected genes
Fourteen genes identified in PR2 analysis, 5 from PR3 analysis and 5 that were significant in both PR2 and PR3 analysis were targeted by a rapid and high-throughput siRNA-drug modifier screening in THP-1 cell lines. THP-1 cells were transfected with 3 individual siRNAs per selected gene and was tested alongside standard transfection controls (three replicates experimental plates were utilized). DCK was used a positive control-Since DCK is rate limiting enzyme in activation of cytarabine to cytarabine monophosphate its knock down should increase drug resistance. Post-siRNA transfection, cells were treated with different concentrations of cytarabine (0μM; 0.1μM-IC10; 0.8μM-IC50; and 10μM-IC90) for 48 hrs followed by multi-parametric nuclear morphometry assays using automated microscopy to document the individual and combined phenotypic effects of siRNA gene silencing and cytarabine on cell growth and proliferation. The following Definiteness parameters were used to quantify changes in nuclear morphometry: i) Number of nuclei, absolute number of nuclei per image field; indication of cell proliferative activity. ii) Condensed nuclei index, (% condensed nuclei), the percentage of nuclei classified as having condensed chromatin (defined by intensity and granularity) Serves as an indication of apoptotic and mitotic nuclei. iii) Aberrant nuclei index, (% deformed “aberrant” nuclei), the percentage of nuclei classified as misshapen (defined by circularity and elliptical fit). iv) Large nuclei index, (% large nuclei), based on a size cut-off. V) Small nuclei index, (% small nuclei), based on a size cut-off. Normalization of all well means was done using corresponding Negative controls per cell line, time point and compound concentration. For controls, median and standard deviation of 3 normalized wells per plate, determination of CV% as an expression of intra-plate variation. Median and standard deviation of individual normalized wells over 3 replicate plates, determination of CV% as an expression of inter-plate variation, which were in the acceptable range. Normalized siRNA effect on compound-treated cells (normalized well means (IC10; IC50 or IC90)/normalized well means (buffer control) was determined.
Two methods were utilized to determine siRNA mediated effect: RSA ranking: The hit selection algorithm, RSA or “redundant siRNA activity”, uses an iterative hypergeometric distribution formula to calculate the statistical significance of the siRNA phenotypic readout and ranking for individual genes (indicated by LogP value). By considering the effect of all three siRNAs for a gene, and not a single high value siRNA, this algorithm is more sophisticated in its handling of outliers. Because it uses ranks, it does not depend on an underlying data distribution (e.g., Gaussian). RSA analyses using customized scripts (adapted from König et al.(15)) were carried out on normalized siRNA effect on compound-treated cells for all readouts. For maximum flexibility of downstream analyses, RSA was run “in both directions” considering as positives either high or low values (i.e., increase or decrease of phenotypic effect under drug treatment vs. buffer control). Hit selection 1 (RSA analysis): Normalized siRNA effect on compound-treated cells was determined at all IC levels independently. Cut-off strategy: A simple way to arrive at a list of hits is to apply a cut-off threshold (2XSD of neg control) for the number of nuclei (normalized siRNA effect on compound-treated cells). Two out of three siRNAs per target gene should pass this threshold.
RESULTS
Among the patients included in this study 25% were classified as low-risk AML, 42% standard-risk, and 33% high-risk. MRD ≥0.1 was present in 33% of patients after the first cycle of cytarabine induction chemotherapy.
AML Gene Expression Changes Induced By Cytarabine
An arm-stratified signed-rank test was used to identify probe-sets with significant expression changes for both arms. We identified 51 genes with significant increase and 5 genes with significant decrease in expression after exposure to cytarabine chemotherapy [p≤0.001, q=0.34; Table-2]. Table 2 gives the z-statistic, p-value, and robust FDR estimate [q-value] for the probe-sets with p ≤ 0.001. A positive z-statistic indicates that expression of the probe-set showed a significant increase during the ara-C infusion and a negative z-statistic indicates that expression of the probe-set showed significant decrease during the ara-C infusion.
Table 2.
List of genes with significant change in expression post ara-C infusion in AML patients.
| Affy probe ID | Gene.Symbol | Gene Name | Statistics | Pvalue | BH95_q | PC06_q |
|---|---|---|---|---|---|---|
| 202284_s_at | CDKN1A | cyclin-dependent kinase inhibitor 1A [p21, Cip1] | 165 | 0.0000 | 0.1295 | 0.1203 |
| 202665_s_at | WASPIP | HIV-1 Rev binding protein | 154 | 0.0001 | 0.3179 | 0.1203 |
| 209294_x_at | TNFRSF10B | tumor necrosis factor receptor superfamily, member 10b | 153 | 0.0001 | 0.3179 | 0.1203 |
| 213316_at | KIAA1462 | 152 | 0.0001 | 0.3179 | 0.1203 | |
| 214054_at | DOK2 | docking protein 2, 56kDa | 151 | 0.0001 | 0.3179 | 0.1203 |
| 202872_at | ATP6V1C1 | ATPase, H+ transporting, lysosomal 42kDa, V1 subunit C1 | 150 | 0.0002 | 0.3179 | 0.1203 |
| 210105_s_at | FYN | FYN oncogene related to SRC, FGR, YES | 150 | 0.0002 | 0.3179 | 0.1203 |
| 212357_at | KIAA0280 | 150 | 0.0002 | 0.3179 | 0.1203 | |
| 201297_s_at | MOBK1B | MOB1, Mps One Binder kinase activator-like 1B [yeast] | 150 | 0.0002 | 0.3179 | 0.1203 |
| 221653_x_at | APOL2 | apolipoprotein L, 2 | 149 | 0.0002 | 0.3179 | 0.1203 |
| 218373_at | AKTIP | AKT Interacting Protein | 149 | 0.0002 | 0.3179 | 0.1203 |
| 205643_s_at | PPP2R2B | protein phosphatase 2, regulatory subunit B, beta isoform | 149 | 0.0002 | 0.3179 | 0.1203 |
| 212932_at | RAB3GAP1 | RAB3 GTPase activating protein subunit 1 [catalytic] | 149 | 0.0002 | 0.3179 | 0.1203 |
| 203217_s_at | ST3GAL5 | ST3 beta-galactoside alpha-2,3-sialyltransferase 5 | 149 | 0.0002 | 0.3179 | 0.1203 |
| 203409_at | DDB2 | damage-specific DNA binding protein 2, 48kDa | 148 | 0.0003 | 0.3179 | 0.1203 |
| 202270_at | GBP1 | guanylate binding protein 1, interferon-inducible, 67kDa | 148 | 0.0003 | 0.3179 | 0.1203 |
| 215350_at | SYNE1 | spectrin repeat containing, nuclear envelope 1 | 148 | 0.0003 | 0.3179 | 0.1203 |
| 214900_at | ZKSCAN1 | zinc finger with KRAB and SCAN domains 1 | 148 | 0.0003 | 0.3179 | 0.1203 |
| 219777_at | GIMAP6 | GTPase, IMAP family member 6 | 146 | 0.0004 | 0.3682 | 0.2653 |
| 211977_at | GPR107 | G protein-coupled receptor 107 | 146 | 0.0004 | 0.3682 | 0.2653 |
| 206765_at | KCNJ2 | potassium inwardly-rectifying channel, subfamily J, member 2 | 146 | 0.0004 | 0.3682 | 0.2653 |
| 216069_at | PRMT2 | protein arginine methyltransferase 2 | 146 | 0.0004 | 0.3682 | 0.2653 |
| 214006_s_at | GGCX | gamma-glutamyl carboxylase | 145 | 0.0005 | 0.3682 | 0.2834 |
| 219243_at | GIMAP4 | GTPase, IMAP family member 4 | 145 | 0.0005 | 0.3682 | 0.2834 |
| 215255_at | IGSF9B | immunoglobulin superfamily, member 9B | 145 | 0.0005 | 0.3682 | 0.2834 |
| 216841_s_at | SOD2 | superoxide dismutase 2, mitochondrial | 145 | 0.0005 | 0.3682 | 0.2834 |
| 217818_s_at | ARPC4 | actin related protein 2/3 complex, subunit 4, 20kDa | 144 | 0.0005 | 0.3682 | 0.3324 |
| 221042_s_at | CLMN | calmin [calponin-like, transmembrane] | 144 | 0.0005 | 0.3682 | 0.3324 |
| 209457_at | DUSP5 | dual specificity phosphatase 5 | 144 | 0.0005 | 0.3682 | 0.3324 |
| 216080_s_at | FADS3 | fatty acid desaturase 3 | 144 | 0.0005 | 0.3682 | 0.3324 |
| 218092_s_at | HRB | HIV-1 Rev binding protein | 144 | 0.0005 | 0.3682 | 0.3324 |
| 210734_x_at | MAX | MYC associated factor X | 144 | 0.0005 | 0.3682 | 0.3324 |
| 205456_at | CD3E | CD3e molecule, epsilon [CD3-TCR complex] | 143 | 0.0007 | 0.3682 | 0.3393 |
| 214551_s_at | CD7 | CD7 molecule | 143 | 0.0007 | 0.3682 | 0.3393 |
| 220546_at | FLJ11783 | 143 | 0.0007 | 0.3682 | 0.3393 | |
| 209969_s_at | STAT1 | signal transducer and activator of transcription 1, 91kDa | 143 | 0.0007 | 0.3682 | 0.3393 |
| 202748_at | GBP2 | guanylate binding protein 2, interferon-inducible | 142 | 0.0008 | 0.3682 | 0.3419 |
| 202595_s_at | LEPROTL1 | leptin receptor overlapping transcript-like 1 | 142 | 0.0008 | 0.3682 | 0.3419 |
| 207705_s_at | RP4-691N24.1 | 142 | 0.0008 | 0.3682 | 0.3419 | |
| 208540_x_at | S100A11 | S100 calcium binding protein A11 pseudogene | 142 | 0.0008 | 0.3682 | 0.3419 |
| 214838_at | SFT2D2 | SFT2 domain containing 2 | 142 | 0.0008 | 0.3682 | 0.3419 |
| 206511_s_at | SIX2 | SIX homeobox 2 | 142 | 0.0008 | 0.3682 | 0.3419 |
| 219423_x_at | TNFRSF25 | tumor necrosis factor receptor superfamily, member 25 | 142 | 0.0008 | 0.3682 | 0.3419 |
| 205299_s_at | BTN2A2 | butyrophilin, subfamily 2, member A2 | 141 | 0.0010 | 0.3682 | 0.3424 |
| 221724_s_at | CLEC4A | C-type lectin domain family 4, member A | 141 | 0.0010 | 0.3682 | 0.3424 |
| 210968_s_at | RTN4 | reticulon 4 | 141 | 0.0010 | 0.3682 | 0.3424 |
| 210569_s_at | SIGLEC9 | sialic acid binding Ig-like lectin 9 | 141 | 0.0010 | 0.3682 | 0.3424 |
| 208992_s_at | STAT3 | signal transducer and activator of transcription 3 | 141 | 0.0010 | 0.3682 | 0.3424 |
| 203567_s_at | TRIM38 | tripartite motif-containing 38 | 141 | 0.0010 | 0.3682 | 0.3424 |
| 203732_at | TRIP4 | thyroid hormone receptor interactor 4 | 141 | 0.0010 | 0.3682 | 0.3424 |
| 211422_at | TRPM3 | transient receptor potential cation channel, subfamily M, member 3 | 141 | 0.0010 | 0.3682 | 0.3424 |
| 221747_at | TNS1 | tensin 1 | 4 | 0.0002 | 0.3179 | 0.1203 |
| 203720_s_at | ERCC1 | excision repair cross-complementing rodent repair deficiency, complementation group 1 | 9 | 0.0005 | 0.3682 | 0.3234 |
| 205592_at | IL8 | interleukin 8 | 9 | 0.0005 | 0.3682 | 0.3234 |
| 218225_at | ECSIT | ECSIT homolog [Drosophila] | 10 | 0.0007 | 0.3682 | 0.3348 |
| 221932_s_at | GLRX5 | glutaredoxin 5 | 10 | 0.0007 | 0.3682 | 0.3348 |
The expected rank [statistic] is 76.5, a statistic greater than 76.5 indicates that there are more positive values in expression change [higher median expression at 24 hours].
Probes with decrease in expression post-araC are shaded in grey.
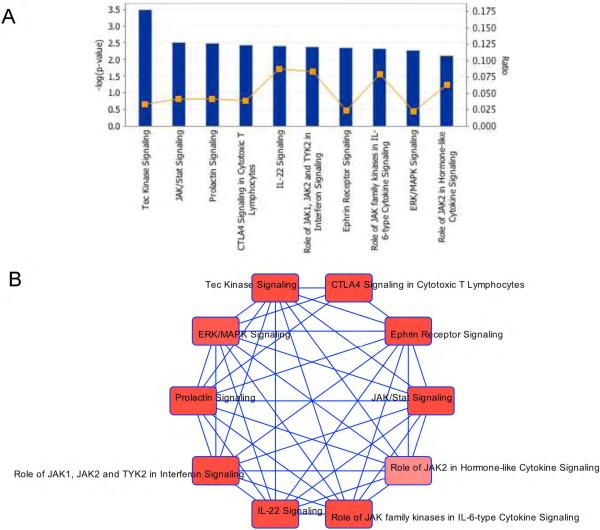
Several genes hold strong potential for biological and clinical relevance. Specifically, we found change in expression of the DNA excision repair genes DDB2 [1.9-fold increase, p=0.0003,q=0.12] and ERCC1 [1.4-fold decrease, p=0.0006,q=0.34] after cytarabine. Components of the PI3K/Akt activation pathway, including AKTIP showed increased expression after cytarabine [p=0.0002,q=0.12]. STAT1 and STAT3, signal transducer and transcription activators involved in multiple pathways [FLT3 signaling, MAPK and Jak/Stat signaling pathways etc.], were increased post cytarabine treatment [p =0.0007and 0.001, respectively]. Among other genes of significant biological/clinical interest that were increased in expression by cytarabine [p<0.001] included FYN, an member of tyrosine kinase oncogene family, MAX-MYC associated protein is an oncoprotein, CDKN1A [p21CIP1], a cyclin dependent kinase inhibitor; GTPAses-GIMAP4 and GIMAP6, transmembrane receptors [TNFRS10B, TNFRS25, CLEC4A and CD3E] involved in regulating caspases, protein phosphatases [PPP2R2B and DUSP5]; transporters [ATP6V1C1 and SLC4A1], transcription regulators [KMT2A, SIX2,TRIP4, ZKSCAN1]. Analysis by Ingenuity pathway analysis tool mapped these genes to Tec Kinase, JAK/Stat, ERK/MAPK, Prolactin and Ephrin, IL22 and CTLA4 signaling pathways [Figure 1].
Figure 1.
Pathway analysis utilizing Ingenuity pathway analysis tool of genes demonstrating significant change in expression post-cytarabine infusion in AML patients. A] Top 10 canonical pathways for genes with significant change in expression post cytarabine infusion. Y-axis indicates Log p value (calculated with the right-tailed Fisher's Exact Test) and Ratio (percentage of genes in a pathway that were also found in results). The ratio is therefore good for looking at which pathway has been affected the most based on the percentage of genes uploaded into IPA. B) The network of these 10 pathways demonstrating interactions between the pathways due to shared genes.
Cytarabine Dose did not result in significant difference in AML Gene Expression changes
To determine whether cytarabine dose impacted changes in gene-expression, patients were categorized into two groups [high dose vs. low dose] according to the cytarabine dose received during first-induction chemotherapy. The two groups did not differ in age, sex, race, or molecular-translocations [Table 1]. Changes in AML cell gene-expression were not different between the two treatment groups, after consideration of multiple testing [q = 1.0; Supplementary Table-1]. This result is congruent with the AML02 clinical trial results, which demonstrated no significant differences in day 22 MRD levels or EFS between the two randomized doses of cytarabine (6).
PROMISE analysis
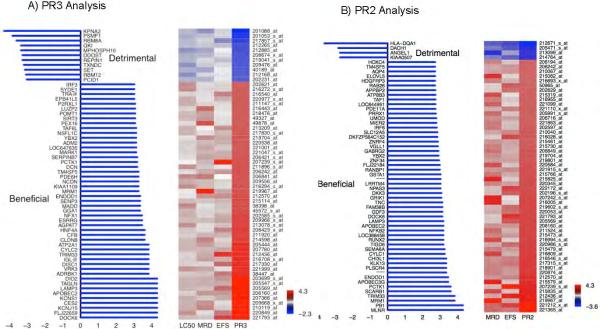
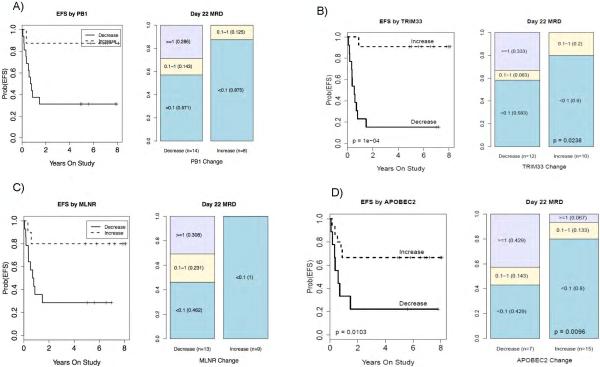
We then used the PROMISE statistical procedure to identify gene with expression changes that were associated with detrimental outcomes [higher LC50, positive MRD at day 22, and a longer event-free survival time period] OR beneficial outcomes [lower LC50, negative MRD, shorter EFS]. When analyzing for all three outcomes of interest [PR3 analysis], 65 genes were significantly [p ≤0.001, q=0.32] associated with beneficial or detrimental outcome. 13/65 [20%] of the genes were associated with detrimental response and 52/65 genes were associated with beneficial response [p<0.001, and Figure 2A]. Because LC50 data was unavailable for some study participants [n=16], we performed PROMISE analysis with two clinical outcomes of interest [MRD and EFS: PR2 analysis]. In this analysis, we identified 32 genes as significantly associated with clinical response [p ≤0.001, q=0.72, Figure 2B]. None of identified genes in this study associated with AML risk group assigned at diagnosis [p>0.05 for each gene]. Genes with significant association at q <0.3 are summarized in Table 3 and Supplementary Table 2S provides full list of genes from PR2 and PR3 analysis. Figures 3A-D illustrates the association of the expression changes of selected probe-sets [identified in PR2/ PR3 analysis] with MRD and event free survival [EFS]. PB1, polybromo1, was the top gene in PR2 analysis with expression change predictive of MRD22 and EFS [PR2 p =0.000; MRD, p=0.0026, EFS p=0.0005, Figure-3A]. PB1 is involved in transcriptional activation and repression of genes involved in chromatin remodeling and acts as a negative regulator of cell proliferation. Change in TRIM33, a transcriptional repressor with a role in cell proliferation, was associated with a favorable outcome [PR3 p=0.0001, q=0.117;EFS p=0.0001;MRD p=0.0238, Figure-3B]. TRIM33 has been shown to mediate erythroid differentiation of hematopoietic stem/progenitor in response to TGFbeta(16). Similarly MLNR expression change was also predictive of clinical outcome, [PR2 p =0.0001, MRD p=<0.0001 and EFS p=0.02, Figure 3C]. Increased expression of APOBEC2, a cytidine-deaminase family member was associated with a beneficial pattern of association [PR3 p<0.0001, q=0;EFS p=0.01;MRD p=0.009; Figure-3D]. HLA-DQA1- belonging to HLA class II alpha chain paralogues was associated with unfavorable outcome [PR2 p=0.002,EFS p=0.04 and MRD p=0.0004]; haplotypes within this and other members of the HLA family have been implicated in the risk of developing CML and ALL(17, 18). An increase in the expression of RUNX2 [AML3], a member of the RUNX family, showed a beneficial association pattern [PR2 p=0.0006, q=0.51, MRD p=0.0008]. Fusion of the RUNX family gene RUNX1 with ETO is considered a low-risk feature that is associated with a better prognosis(19). Increased expression of the nuclear oncogene SET showed a detrimental association pattern [PR3 p=0.0007,q=0.30; MRD p=0.01]. Increased expression of DKK3, a tumor suppressor that inhibits WNT oncogenic signaling and is involved in the regulation of mortalization-related gene expression(20), showed a beneficial association pattern [PR2 p=0.0008, EFS; p=0.0017, MRD p=0.03].
Figure 2.
Therapeutically beneficial and detrimental patterns of association detected by the PROMISE method. A) The three-endpoint PROMISE analysis (PR3) identified 65 probe sets with cytarabine induced changes in expression levels that showed a beneficial or detrimental pattern of association with in vitro LC50, MRD and EFS. B) The two-endpoint PROMISE analysis (PR2) identified 30 probe sets with cytarabine induced change in expression levels that showed beneficial or detrimental patterns of association with MRD and EFS. X-axis values give a log10 p-value with sign defined by the pattern (negative for detrimental and positive for beneficial). Each row represents a gene and each column represents a clinical endpoint: MRD and EFS; PR2 indicates the statistical values corresponding to the PROMISE analysis. Colors are assigned according to the signed log10 p-value. EFS: Event-free survival; MRD: Minimal residual disease; PROMISE: Projection onto the Most Interesting Statistical Evidence.
Table 3.
PROMISE analysis identified genes change in expression post ara-C to be predictive of beneficial or detrimental patterns of association with clinical endpoints in AML patients [q<0.3]
| A. PROMISE 3 [PR3] analysis utilizing 3 endpoints LC50, MRD22 and EFS | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ProbelD | Gene Symbol | LC50. stat | MRD22. stat | EFS. stat | PROMIS E. stat | LC50 p value | MRD22 p value | EFS p value | PR3 p value | BH95_q | PC06_q | Risk p value |
| 201088_at | KPNA2 | 0.708 | 0.568 | 0.290 | −0.522 | 0.044 | 0.006 | 0.136 | 0.000 | 0.000 | 0.000 | 0.171 |
| 203699_s_at | DIO2 | −0.708 | −0.468 | −0.506 | 0.561 | 0.040 | 0.026 | 0.009 | 0.000 | 0.000 | 0.000 | 0.472 |
| 205547_s_at | TAGLN | −0.773 | −0.462 | −0.447 | 0.561 | 0.030 | 0.026 | 0.020 | 0.000 | 0.000 | 0.000 | 0.662 |
| 205569_at | LAMP3 | −0.773 | −0.406 | −0.614 | 0.598 | 0.028 | 0.060 | 0.001 | 0.000 | 0.000 | 0.000 | 0.686 |
| 206160_at | APOBEC2 | −0.602 | −0.531 | −0.493 | 0.542 | 0.110 | 0.010 | 0.010 | 0.000 | 0.000 | 0.000 | 0.245 |
| 207366_at | KCNS1 | −0.773 | −0.365 | −0.448 | 0.529 | 0.024 | 0.085 | 0.020 | 0.000 | 0.000 | 0.000 | 0.879 |
| 209668_x_at | CES2 | −0.773 | −0.537 | −0.253 | 0.521 | 0.030 | 0.008 | 0.203 | 0.000 | 0.000 | 0.000 | 0.658 |
| 210119_at | KCNJ15 | −0.773 | −0.586 | −0.331 | 0.564 | 0.027 | 0.004 | 0.090 | 0.000 | 0.000 | 0.000 | 0.531 |
| 220849_at | FLJ22659 | −0.838 | −0.291 | −0.549 | 0.559 | 0.007 | 0.177 | 0.004 | 0.000 | 0.000 | 0.000 | 0.719 |
| 221793_at | DOCK6 | −0.708 | −0.400 | −0.650 | 0.586 | 0.043 | 0.064 | 0.000 | 0.000 | 0.000 | 0.000 | 0.886 |
| 201053_s_at | PSMF1 | 0.773 | 0.403 | 0.306 | −0.494 | 0.027 | 0.057 | 0.116 | 0.000 | 0.153 | 0.118 | 0.842 |
| 205444_at | ATP2A1 | −0.773 | −0.523 | −0.349 | 0.548 | 0.029 | 0.011 | 0.071 | 0.000 | 0.153 | 0.118 | 0.259 |
| 207780_at | CYLC2 | −0.708 | −0.435 | −0.460 | 0.534 | 0.042 | 0.041 | 0.017 | 0.000 | 0.153 | 0.118 | 0.858 |
| 212436_at | TRIM33 | −0.602 | −0.478 | −0.668 | 0.583 | 0.116 | 0.024 | 0.000 | 0.000 | 0.153 | 0.118 | 0.983 |
| 216708_x_at | IGL2 | −0.643 | −0.520 | −0.411 | 0.525 | 0.096 | 0.013 | 0.033 | 0.000 | 0.153 | 0.118 | 0.638 |
| 217330_at | DISC1 | −0.643 | −0.307 | −0.607 | 0.519 | 0.097 | 0.152 | 0.001 | 0.000 | 0.153 | 0.118 | 0.284 |
| 221999_at | VRK3 | −0.838 | −0.504 | −0.233 | 0.525 | 0.008 | 0.017 | 0.246 | 0.000 | 0.153 | 0.118 | 0.388 |
| 38447_at | ADRBK1 | −0.643 | −0.400 | −0.546 | 0.530 | 0.091 | 0.059 | 0.003 | 0.000 | 0.153 | 0.118 | 0.809 |
| 208429_x_at | HNF4A | −0.659 | −0.277 | −0.549 | 0.495 | 0.066 | 0.200 | 0.004 | 0.000 | 0.217 | 0.192 | 0.854 |
| 211920_at | CFB | −0.643 | −0.450 | −0.408 | 0.500 | 0.097 | 0.031 | 0.032 | 0.000 | 0.217 | 0.192 | 0.943 |
| 214598_at | CLDN8 | −0.708 | −0.579 | −0.309 | 0.532 | 0.043 | 0.004 | 0.114 | 0.000 | 0.217 | 0.192 | 0.435 |
| 217857_s_at | RBM8A | 0.708 | 0.325 | 0.530 | −0.521 | 0.042 | 0.133 | 0.005 | 0.000 | 0.217 | 0.192 | 0.476 |
| 202585_s_at | NFX1 | −0.773 | −0.326 | −0.427 | 0.509 | 0.027 | 0.132 | 0.028 | 0.000 | 0.278 | 0.254 | 0.941 |
| 209966_x_at | ESRRG | −0.667 | −0.417 | −0.472 | 0.519 | 0.050 | 0.049 | 0.013 | 0.000 | 0.278 | 0.254 | 0.355 |
| 213078_x_at | AGPAT7 | −0.838 | −0.291 | −0.372 | 0.500 | 0.008 | 0.172 | 0.052 | 0.000 | 0.278 | 0.254 | 0.498 |
| B. PROMISE 2 [PR2] analysis utilizing 2 clinical endpoints [MRD22 and EFS] | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ProbelD | Gene. Symbol | MRD22. stat | EFS. stat | PROMI SE. stat | MRD22 p value | EFS p value | PR2 p value | BH95_q | PC06_q | Risk p value | ||
| 221212_x_at | PB1 | −0.613 | −0.635 | 0.624 | 0.003 | 0.001 | 0.000 | 0.000 | 0.000 | 0.212 | ||
| 221365_at | MLNR | −0.611 | −0.529 | 0.570 | 0.002 | 0.004 | 0.000 | 0.000 | 0.000 | 0.284 | ||
Figure 3.
Association of cytarabine induced change in expression levels of A) PB1, B) TRIM33, C) MLNR and D) APOBEC2 with EFS and MRD (day 22) in AML patients.
In vitro validation of selected genes using siRNA mediated knockdown
After identifying inducible genes that were also associated with clinical importance, we next questioned whether targeting these genes could modify leukemia response to cytarabine. Fourteen genes identified in PR2 analysis, 5 from PR3 analysis and 5 that were significant in both PR2 and PR3 analysis were targeted by a rapid and high-throughput siRNA-drug modifier screening in THP-1 cell lines (Table-4). Each gene was targeted with 3 individual siRNAs and was tested alongside standard transfection controls (DCK was used a positive control). Post-siRNA transfection, cells were treated with different concentrations of cytarabine (0μM; 0.1μM-IC10; 0.8μM-IC50; and 10μM-IC90) followed by multi-parametric nuclear morphometry assays using automated microscopy to document the individual and combined phenotypic effects of siRNA gene silencing and cytarabine on cell growth and proliferation. siRNA mediated knockdown of CHI3L, NFKB2, APOBEC3G, REPIN1, or DOCK6 increased cytarabine-sensitivity; while knockdown of ADRBK1, NPAS3, SCARB1, TIGD6, or TNC, increased cytarabine-resistance (Table-4).
Table 4.
Effect of siRNA mediated knockdown of selected genes on cytarabine sensitivity in THP1 cells.
| Gene symbol | PROMISE analysis | siRNA-drug modifier GENE LEVEL effect-Based on RSA ranking | siRNA-drug modifier GENE LEVEL effect-Based on threshold (at least 2 siRNAs passing threshold) | Non-neutral genes refer to genes, which do yield a detectible RNAi phenotype in the absence of the drug (phenotype was sometimes further enhanced post-drug treatment) | Only one of three siRNA influence drug response |
|---|---|---|---|---|---|
| ADRBK1 | PR3 | Increased cytarabine resistance | Increased cytarabine resistance | ||
| APOBEC2 | PR2 and PR3 | Increased sensitivity | |||
| APOBEC3G | PR2 and PR3 | Increased cytarabine sensitivity | non-neutral | ||
| CHI3L1 | PR2 | Increased cytarabine sensitivity | |||
| CYLC1 | PR2 | Increased resistance | |||
| DCK | +ve control | Increased cytarabine resistance | Increased cytarabine resistance | ||
| DOCK6 | PR2 and PR3 | Increased cytarabine sensitivity | |||
| ENDOD1 | PR2 and PR3 | Increased resistance | |||
| GDF3 | PR2 | ||||
| GSTA1 | PR2 | Increased resistance | |||
| KLK13 | PR2 | ||||
| LRRTM4 | PR2 | Increased resistance | |||
| MARK1 | PR3 | ||||
| MLNR | PR2 | ||||
| NFKB2 | PR2 | Increased cytarabine sensitivity | |||
| NPAS3 | PR2 | Increased cytarabine resistance | non-neutral | Increased resistance | |
| QKI | PR3 | ||||
| RANBP1 | PR2 | ||||
| REPIN1 | PR3 | Increased cytarabine sensitivity | Increased cytarabine sensitivity | ||
| RUNX2 | PR2 | Increased resistance | |||
| SCARB1 | PR2 | Increased cytarabine resistance | Increased cytarabine resistance | ||
| SET | PR3 | non-neutral | |||
| TIGD6 | PR2 | Increased cytarabine resistance | Increased cytarabine resistance | ||
| TNC | PR2 | Increased cytarabine resistance | |||
| TRIM33 | PR2 and PR3 | non-neutral |
DISCUSSION
In this study we examined cytarabine induced in vivo gene expression changes in pediatric AML patients. Most of the studies in literature have focused on gene expression profiling of the of diagnostic chemo naïve tumor specimens. The knowledge gained from gene expression signature identified in diagnostic specimens have opened up opportunities for biomarker identification as well as identification of potential targets of drug development. In our previous work we have used gene expression array to identify an AML cell gene expression signature that predicts intracellular ara-CTP concentration and clinical response to chemotherapy.(7) This study demonstrated a validated method of integrating genomic data at the time of diagnosis with important pharmacologic and clinical outcomes. Based on this experience, and the gene expression changes by chemotherapy, we reasoned that examining gene expression changes in AML cells after exposure to cytarabine would provide relevant information regarding intracellular response to treatment. However there is no study to the best of our knowledge that reports in vivo gene expression changes induced by cytarabine. One of the challenges to perform such a study is technical difficulty in obtaining clinical samples especially post treatment. One of the unique and significant feature of our study was availability of bone marrow samples 24hr post initiation of cytarabine infusion, since no other chemotherapeutic agent was yet initiated samples obtained at this time point reflect gene expression differences unique to cytarabine. To the best of our knowledge, this is the first study reporting in vivo gene-expression changes that occur during cytarabine treatment in pediatric AML patients.
Our analysis of gene expression changes post cytarabine treatment identified genes of biological interest such as components of PI3K/AKT pathway such as AKTIP, genes involved in DNA repair DDB2 and ERCC1. AKTIP regulates protein kinase B [PKB]/Akt signaling [critical for cell growth, glucose-metabolism, and apoptosis] by enhancing the phosphorylation of PKB regulatory sites (14). This result confirms and extends findings from our previous work that showed significant correlation between diagnostic AML cell gene expression of PIK3C3 (involved in the PI3K/PTEN/Akt/mTOR signaling cascade) and worse clinical outcomes.(7) Identification of genes that were mapped to Tech signaling is of potential interest given the fact Bruton's Tyrosine kinase (BTK), a member of Tec kinase family and a key regulator of B-cell Receptor (BCR) is being explored as a potential target in lymphoma and leukemia(21). Pharmacological screening of ibrutinib as inhibitor of BTK kinase has shown promising results in AML warranting clinical evaluation (22) Interestingly we did not observed significant impact of cytarabine dose on gene-expression change, this was consistent with the clinical outcome that demonstrated no significant difference between the two randomized doses of cytarabine.
Additionally, gene expression changes corresponding to worse outcomes may explain mechanisms of refractory disease and may serve as potential targets for enhancing response to cytarabine. PROMISE method to identify gene expression changes that are predictive of clinically meaningful pattern of association with multiple endpoints identified 65 genes in PR3 (LC50, MRD and EFS) and 32 in PR2 (MRD and EFS] analysis [p<0.001]. Some of the genes of potential interest as novel therapeutic agents include: PB1 (aka PBRM1, BAF180), PolyBromo 1 is a bromodomain protein codes for a subunit of ATP-depend chromatin remodeling complex [SWI/SNF-A], cytarabine induced expression of PB1 to be predictive of better outcome [p<10-4]. PBRM1 mutations have been found to be frequent in cancer and in renal cell carcinoma approximately 40% of tumor samples have been shown to harbor PBRM1 mutations. Loss of PB1 expression has been associated with poor prognosis and studies in renal carcinoma suggest PBRM1 to be a tumor suppressor by acting as a targeting submit of nucleosome remodeling complex (23-25). P53 transcriptional activity has also been shown to be dependent on PBRM1, thereby resulting in onset of cancer with loss of PBRM1 (26-28). Although PBRM1 has not yet been implicated in pathology or prognosis of AML, our results show it as a potential target, therapeutic manipulation of PBRM1 is being explored in renal cell carcinoma and if successful might open up opportunities to modify treatment strategies in AML. Another gene belonging to bromo-domain family is TRIM33, a transcriptional repressor with a role in cell proliferation. TRIM33 is a multifunctional protein implicated in TGFb signaling and hematopoietic stem cell [HSC] aging by regulating the balance between lymphoid and myeloid derived HSCs (29). In mice lacking TRIM33 premature hematopoietic aging has been implicated in predisposition to myeloproliferative disease as CML. TRIM33 has also been shown to act in PARP-dependent DNA damage response by timely removal of ACL1 from damages chromatin thereby facilitating DNA repair (30). Our results are in consensus with proposed biological functions of TRIM33 and with supporting evidence form literature opens up potential opportunities to develop therapeutic strategies to module DNA repair efficiency in tumors lacking TRIM33. Among tumor suppressors another gene of significant clinical value is DKK3, which inhibits WNT oncogenic signaling and is involved in the regulation of mortalization-related gene expression(20). Down-regulation of DKK3 via promoter hyper-methylation has been associated with poor prognosis in ALL(20), which is concordant with our finding that increased expression of DKK3 associates with beneficial outcomes. Thus, combining a hypomethylating agent such as decitabine with cytarabine might be a strategy to induce expression of DKK3.
SET, which is a target of translocation in AML and is involved in tumor metastasis, chromatin remodeling, apoptosis and the MAP/ERK pathway. SET also inhibits the GZMA-activated DNase NME1, a nucleotide diphosphate kinase involved in cytarabine activation(31). SET antagonism has been implicated in overcoming drug-resistance in myeloid leukemia(32). This finding is concordant with our result that increased expression of SET is detrimental and suggests JAK2 inhibition as a potential therapeutic strategy for AML.
DIO2, belongs to family of deiodinases that is involve din thyroid metabolism, additionally have been implicated in maintaining balance between proliferation and differentiation. Recent study identified seleno-compounds that can module expression levels of DIO enzymes thereby allowing modulation of balance between proliferation and differentiation as a therapeutic strategy (33).
Overall our results for the first time report in vivo cytarabine induced gene-expression changes in AML, since cytarabine was the only drug that patients received at that time the results reflect gene expression changes specific to cytarabine. Drug induced in vivo expression changes can often trigger adaptive responses that can contribute to development of resistance or refractory disease. Key genes [such as tumor suppressors DKK3, TRIM33, PBRM1, an oncogene SET, cytidine-deaminase family members APOBEC2 and APOBEC3G] influenced by cytarabine infusion that were also predictive of response can serve as potential targets for enhancing therapeutic strategies. The results highlight genes relevant to cytarabine resistance and support the concept of targeting genes modulated by cytarabine exposure as a means of improving response.
Future in depth studies will help in understanding the interplay of these genes/pathways to better understand mechanisms of cytarabine resistance in AML. Importantly, novel agents directed at these targets may serve as potential therapeutics to improve clinical outcomes. In summary, our results identified genes of potential biological and therapeutic significance that are influenced by cytarabine treatment thereby opening up opportunities for future research to elucidate the mechanisms underlying AML response/resistance and identify targets of development of novel agents.
Supplementary Material
Acknowledgements
E. Coustan-Smith and D. Campana performed the MRD studies; we are thankful to them for sharing the MRD data. We gratefully acknowledge the technical support of E. Wuitschick, M. Griffin and S. Orwick, and the database assistance of N. Kornegay and M. Wilkinson. This work was supported in part by NIH P30CA021765 [St Jude], NIH/NCI R01CA132946 [Lamba and Pounds], American Society of Hematology Bridge funding award [Lamba] and by the American Lebanese Syrian Associated Charities [ALSAC]. The Leukemia & Lymphoma Society supported C.R.C. with a Scholar in Clinical Research award [2400-13] and a Quest for Cures grant [0725-14].
Footnotes
Conflict of Interest: Authors have no conflicts to disclose.
REFERENCES
- 1.Jabbour EJ, Estey E, Kantarjian HM. Adult acute myeloid leukemia. Mayo Clinic proceedings Mayo Clinic. 2006;81(2):247–60. doi: 10.4065/81.2.247. [DOI] [PubMed] [Google Scholar]
- 2.Rubnitz JE, Gibson B, Smith FO. Acute myeloid leukemia. Pediatr Clin North Am. 2008;55(1):21–51, ix. doi: 10.1016/j.pcl.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 3.Gaydos LA, Freireich EJ, Mantel N. The quantitative relation between platelet count and hemorrhage in patients with acute leukemia. N Engl J Med. 1962;266:905–9. doi: 10.1056/NEJM196205032661802. [DOI] [PubMed] [Google Scholar]
- 4.Bodey GP, Rodriguez V, Chang HY. Narboni. Fever and infection in leukemic patients: a study of 494 consecutive patients. Cancer. 1978;41(4):1610–22. doi: 10.1002/1097-0142(197804)41:4<1610::aid-cncr2820410452>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 5.Galmarini CM, Mackey JR, Dumontet C. Nucleoside analogues: mechanisms of drug resistance and reversal strategies. Leukemia. 2001;15(6):875–90. doi: 10.1038/sj.leu.2402114. [DOI] [PubMed] [Google Scholar]
- 6.Rubnitz JE, Inaba H, Dahl G, Ribeiro RC, Bowman WP, Taub J, et al. Minimal residual disease-directed therapy for childhood acute myeloid leukaemia: results of the AML02 multicentre trial. Lancet Oncol. 2010;11(6):543–52. doi: 10.1016/S1470-2045(10)70090-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lamba JK, Crews KR, Pounds SB, Cao X, Gandhi V, Plunkett W, et al. Identification of predictive markers of cytarabine response in AML by integrative analysis of gene-expression profiles with multiple phenotypes. Pharmacogenomics. 2011;12(3):327–39. doi: 10.2217/pgs.10.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holleman A, Cheok MH, den Boer ML, Yang W, Veerman AJ, Kazemier KM, et al. Geneexpression patterns in drug-resistant acute lymphoblastic leukemia cells and response to treatment. N Engl J Med. 2004;351(6):533–42. doi: 10.1056/NEJMoa033513. [DOI] [PubMed] [Google Scholar]
- 9.Ross ME, Mahfouz R, Onciu M, Liu HC, Zhou X, Song G, et al. Gene expression profiling of pediatric acute myelogenous leukemia. Blood. 2004;104(12):3679–87. doi: 10.1182/blood-2004-03-1154. [DOI] [PubMed] [Google Scholar]
- 10.Cheson BD, Bennett JM, Kopecky KJ, Buchner T, Willman CL, Estey EH, et al. Revised recommendations of the International Working Group for Diagnosis, Standardization of Response Criteria, Treatment Outcomes, and Reporting Standards for Therapeutic Trials in Acute Myeloid Leukemia. J Clin Oncol. 2003;21(24):4642–9. doi: 10.1200/JCO.2003.04.036. [DOI] [PubMed] [Google Scholar]
- 11.Coustan-Smith E, Ribeiro RC, Rubnitz JE, Razzouk BI, Pui CH, Pounds S, et al. Clinical significance of residual disease during treatment in childhood acute myeloid leukaemia. Br J Haematol. 2003;123(2):243–52. doi: 10.1046/j.1365-2141.2003.04610.x. [DOI] [PubMed] [Google Scholar]
- 12.Pounds S, Cheng C, Cao X, Crews KR, Plunkett W, Gandhi V, et al. PROMISE: a tool to identify genomic features with a specific biologically interesting pattern of associations with multiple endpoint variables. Bioinformatics. 2009;25(16):2013–9. doi: 10.1093/bioinformatics/btp357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jung SH, Owzar K, George SL. A multiple testing procedure to associate gene expression levels with survival. Stat Med. 2005;24(20):3077–88. doi: 10.1002/sim.2179. [DOI] [PubMed] [Google Scholar]
- 14.Pounds S, Cheng C. Robust estimation of the false discovery rate. Bioinformatics. 2006;22(16):1979–87. doi: 10.1093/bioinformatics/btl328. [DOI] [PubMed] [Google Scholar]
- 15.Konig R, Chiang CY, Tu BP, Yan SF, DeJesus PD, Romero A, et al. A probability-based approach for the analysis of large-scale RNAi screens. Nat Methods. 2007;4(10):847–9. doi: 10.1038/nmeth1089. [DOI] [PubMed] [Google Scholar]
- 16.He W, Dorn DC, Erdjument-Bromage H, Tempst P, Moore MA, Massague J. Hematopoiesis controlled by distinct TIF1gamma and Smad4 branches of the TGFbeta pathway. Cell. 2006;125(5):929–41. doi: 10.1016/j.cell.2006.03.045. [DOI] [PubMed] [Google Scholar]
- 17.Di Bernardo MC, Broderick P, Harris S, Dyer MJ, Matutes E, Dearden C, et al. Risk of developing chronic lymphocytic leukemia is influenced by HLA-A class I variation. Leukemia. 2013;27(1):255–8. doi: 10.1038/leu.2012.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Urayama KY, Thompson PD, Taylor M, Trachtenberg EA, Chokkalingam AP. Genetic Variation in the Extended Major Histocompatibility Complex and Susceptibility to Childhood Acute Lymphoblastic Leukemia: A Review of the Evidence. Frontiers in oncology. 2013;3:300. doi: 10.3389/fonc.2013.00300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rubnitz JE, Raimondi SC, Halbert AR, Tong X, Srivastava DK, Razzouk BI, et al. Characteristics and outcome of t(8;21)-positive childhood acute myeloid leukemia: a single institution's experience. Leukemia. 2002;16(10):2072–7. doi: 10.1038/sj.leu.2402633. [DOI] [PubMed] [Google Scholar]
- 20.Roman-Gomez J, Jimenez-Velasco A, Agirre X, Castillejo JA, Navarro G, Barrios M, et al. Transcriptional silencing of the Dickkopfs-3 (Dkk-3) gene by CpG hypermethylation in acute lymphoblastic leukaemia. British journal of cancer. 2004;91(4):707–13. doi: 10.1038/sj.bjc.6602008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu H, Hu C, Wang A, Weisberg EL, Chen Y, Yun CH, et al. Discovery of a BTK/MNK dual inhibitor for lymphoma and leukemia. Leukemia. 2015 doi: 10.1038/leu.2015.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rushworth SA, Murray MY, Zaitseva L, Bowles KM, MacEwan DJ. Identification of Bruton's tyrosine kinase as a therapeutic target in acute myeloid leukemia. Blood. 2014;123(8):1229–38. doi: 10.1182/blood-2013-06-511154. [DOI] [PubMed] [Google Scholar]
- 23.da Costa WH, Rezende M, Carneiro FC, Rocha RM, da Cunha IW, Carraro DM, et al. Polybromo-1 (PBRM1), a SWI/SNF complex subunit is a prognostic marker in clear cell renal cell carcinoma. BJU Int. 2014;113(5b):E157–63. doi: 10.1111/bju.12426. [DOI] [PubMed] [Google Scholar]
- 24.Pawlowski R, Muhl SM, Sulser T, Krek W, Moch H, Schraml P. Loss of PBRM1 expression is associated with renal cell carcinoma progression. Int J Cancer. 2013;132(2):E11–7. doi: 10.1002/ijc.27822. [DOI] [PubMed] [Google Scholar]
- 25.Thompson M. Polybromo-1: the chromatin targeting subunit of the PBAF complex. Biochimie. 2009;91(3):309–19. doi: 10.1016/j.biochi.2008.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brownlee PM, Chambers AL, Oliver AW, Downs JA. Cancer and the bromodomains of BAF180. Biochem Soc Trans. 2012;40(2):364–9. doi: 10.1042/BST20110754. [DOI] [PubMed] [Google Scholar]
- 27.Burrows AE, Smogorzewska A, Elledge SJ. Polybromo-associated BRG1-associated factor components BRD7 and BAF180 are critical regulators of p53 required for induction of replicative senescence. Proc Natl Acad Sci U S A. 2010;107(32):14280–5. doi: 10.1073/pnas.1009559107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niimi A, Chambers AL, Downs JA, Lehmann AR. A role for chromatin remodellers in replication of damaged DNA. Nucleic Acids Res. 2012;40(15):7393–403. doi: 10.1093/nar/gks453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quere R, Saint-Paul L, Carmignac V, Martin RZ, Chretien ML, Largeot A, et al. Tif1gamma regulates the TGF-beta1 receptor and promotes physiological aging of hematopoietic stem cells. Proc Natl Acad Sci U S A. 2014;111(29):10592–7. doi: 10.1073/pnas.1405546111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kulkarni A, Oza J, Yao M, Sohail H, Ginjala V, Tomas-Loba A, et al. Tripartite Motifcontaining 33 (TRIM33) protein functions in the poly(ADP-ribose) polymerase (PARP)-dependent DNA damage response through interaction with Amplified in Liver Cancer 1 (ALC1) protein. J Biol Chem. 2013;288(45):32357–69. doi: 10.1074/jbc.M113.459164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fan Z, Beresford PJ, Oh DY, Zhang D, Lieberman J. Tumor suppressor NM23-H1 is a granzyme A-activated DNase during CTL-mediated apoptosis, and the nucleosome assembly protein SET is its inhibitor. Cell. 2003;112(5):659–72. doi: 10.1016/s0092-8674(03)00150-8. [DOI] [PubMed] [Google Scholar]
- 32.Agarwal A, MacKenzie RJ, Pippa R, Eide CA, Oddo J, Tyner JW, et al. Antagonism of SET using OP449 enhances the efficacy of tyrosine kinase inhibitors and overcomes drug resistance in myeloid leukemia. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20(8):2092–103. doi: 10.1158/1078-0432.CCR-13-2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stoedter M, Renko K, Ibanez E, Plano D, Becker NP, Martitz J, et al. Strong induction of iodothyronine deiodinases by chemotherapeutic selenocompounds. Metallomics. 2015;7(2):347–54. doi: 10.1039/c4mt00273c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.