Abstract
Purpose
Aromatase inhibitors (AI) can exert unfavorable effects on lipid profiles; however, previous studies have reported inconsistent results. We describe the association of single-nucleotide polymorphisms (SNP) in candidate genes with lipid profiles in women treated with adjuvant AIs.
Experimental Design
We conducted a prospective observational study to test the associations between SNPs in candidate genes in estrogen signaling and AI metabolism pathways with fasting lipid profiles during the first three months of AI therapy in postmenopausal women with early breast cancer randomized to adjuvant letrozole or exemestane. We performed genetic association analysis and multivariable linear regressions using dominant, recessive, and additive models.
Results
A total of 303 women had complete genetic and lipid data and were evaluable for analysis. In letrozole-treated patients, SNPs in CYP19A1, including rs4646, rs10046, rs700518, rs749292, rs2289106, rs3759811, and rs4775936 were significantly associated with decreases in triglycerides (TG) by 20.2 mg/dL and 39.3mg/dL (p<0.00053), respectively, and with variable changes in high-density lipoprotein (HDL-C) from decreases by 4.2 mg/dL to increases by 9.8 mg/dL (p<0.00053).
Conclusion
Variants in CYP19A1 are associated with decreases in TG and variable changes in HDL-C in postmenopausal women on adjuvant AIs. Future studies are needed to validate these findings, and to identify breast cancer survivors who are at higher risk for cardiovascular disease with AI therapy.
Keywords: single nucleotide polymorphisms, lipid profiles, pharmacogenomics, breast cancer, aromatase inhibitors
Introduction
Compared to tamoxifen, adjuvant aromatase inhibitors (AI) reduce the risk of recurrence and death in postmenopausal women with hormone receptor-positive breast cancer and are an integral component of adjuvant therapy, considered to be a standard of care (1, 2). At the same time several studies are investigating the role of extended duration of endocrine therapy, particularly AI therapy in postmenopausal women, which may prolong AI associated toxicities (3).
While all three approved third generation AIs (exemestane, anastrozole, letrozole) reduce the concentrations of effective circulating estrogens by inhibiting aromatase, and improve breast cancer outcomes, individual women may experience different toxicities with different AIs (4, 5). Amongst other detrimental effects, AIs may exert unfavorable effects on lipid profiles in treated women. In contrast, tamoxifen appears to exert a favorable effect on lipid profiles (6).
Previous studies investigating the effects of different AIs on lipid profiles have demonstrated mixed results (2, 7–14). Because of the intricate biological relationship of estrogen with lipid profiles and metabolism, the mixed results from previous studies may be explained by heterogeneity in genes involved in estrogen signaling, and estrogen and AI metabolism (15, 16). For example, certain polymorphisms in the estrogen receptor alpha (ESR1) gene have been shown to be associated with increased low density lipoprotein cholesterol (LDL-C) and triglyceride concentrations in women treated with AIs (17). Developing models for cardiovascular risk factors in breast cancer survivors is particularly relevant because many women are cured after a diagnosis of early stage breast cancer and are expected to become long-term survivors (18). We report the results of a planned subset analysis of a prospective randomized trial that evaluated the association of single nucleotide polymorphisms (SNP) in candidate genes involved in estrogen and AI metabolism with lipid profiles in postmenopausal women receiving adjuvant AIs.
Materials and Methods
Study Design
The data in this report are derived from a planned sub-analysis of the Exemestane and Letrozole Pharmacogenomics (ELPh) study, a large prospective multicenter randomized observational open-label trial evaluating the effects of two years of therapy with either letrozole or exemestane in postmenopausal women with early stage breast cancer on a variety of biomarkers of estrogen activity and potential AI-related effects. The parent study has been described in detail previously (19).
Eligible participants were postmenopausal women with a biopsy-proven hormone receptor-positive ductal carcinoma in situ (DCIS) or stage I-III breast cancer either considering primary hormone therapy with AIs or in sequence with tamoxifen. Participants must have completed planned breast surgery, adjuvant or neoadjuvant chemotherapy, and adjuvant radiation therapy. Patients previously treated with AIs, with a history of bilateral mastectomy or radiation therapy to the contralateral breast, or with a history of gynecological malignancies were excluded.
Participants were randomized in a stratified fashion based on prior chemotherapy, prior tamoxifen, and bisphosphonate use. Participants were randomly assigned to receive either exemestane (Aromasin; supplied by Pfizer, Inc., 25 mg orally per day) or letrozole (Femara; supplied by Novartis Pharmaceuticals Corporation, 2.5 mg orally per day) and followed for 2 years. Whole blood was collected at baseline and deoxyribonucleic acid (DNA) was isolated for SNP genotyping in candidate genes. Serum lipid profiles were collected at baseline prior to initiating AI therapy and following 3 months of treatment.
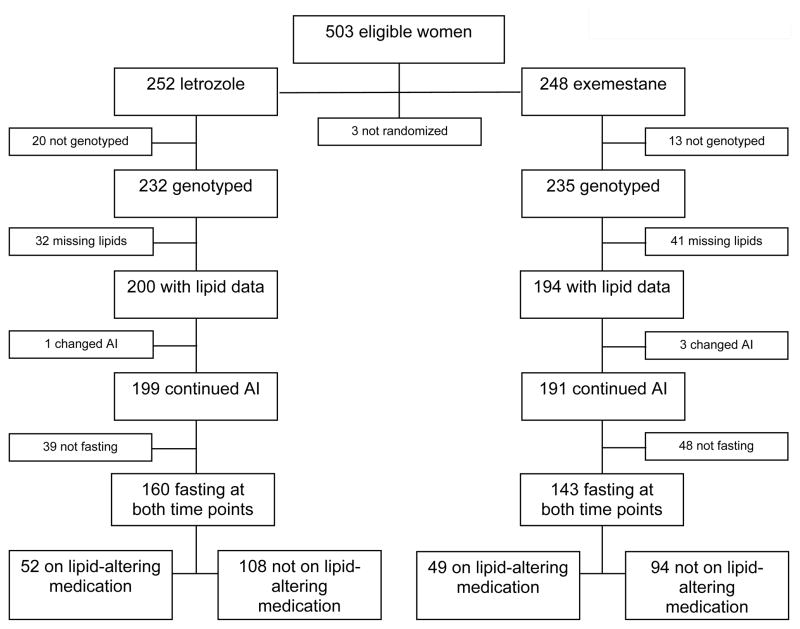
Participants were excluded from the analysis if they did not undergo genotyping, if lipid data were not available either at baseline or 3 months, if they discontinued or crossed over to a different AI during the first 3 months, or if they were not fasting at both baseline and 3 months. Participants taking lipid-altering medications (including statins, fibrates, and/or ezetimibe) during the first 3 months were excluded from the primary analysis but included in a sub-analysis. The derivation of the cohort is described in Figure 1.
Figure 1.
Consort diagram.
Participants were recruited from breast cancer clinics at participating sites including the University of Michigan Comprehensive Cancer Center, the Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins, and the Melvin and Bren Simon Cancer Center at the Indiana University School of Medicine. The study protocol was approved by Institutional Review Boards at all sites and enrolled subjects provided signed written informed consent.
Sample collection
Sample collection and measurement of lipid profiles and estradiol
Venous blood samples for lipid panel analyses were collected at baseline and 3 months after fasting overnight for at least 12 hours.
Serum total cholesterol (TC), high density lipoprotein cholesterol (HDL-C), LDL-C, and triglycerides (TG) were analyzed by standardized enzymatic methodology at Clinical Laboratory Improvement Amendments (CLIA) certified laboratories at the University of Michigan Health System, the Johns Hopkins Medical Laboratories, and the Indiana University Health Pathology Laboratory.
Plasma estradiol was analyzed using an ultrasensitive gas chromatography and tandem mass spectrometry assay, as previously described (20). The lower limits of quantification were 1.25 or 0.625 pg/mL, as determined by calibration curves run with plasma sample batches. In the cohort analyzed, none of the baseline values were below the lower limit of quantification. Since the majority of plasma estradiol concentrations at month three were below the lower limit of quantification (n=278 of the analyzed cohort), these concentrations were set to 0 for data analysis.
Sample collection and processing of candidate genes
Whole blood was collected at enrollment of each study participant. DNA was extracted from whole blood using Qiafilter Blood DNA Maxi kits (Qiagen, Inc., Valencia, CA).
Candidate genes were selected during protocol development based on their known roles in AI drug metabolism (cytochrome P450 2A6 (CYP2A6), and 3A5 (CYP3A5)), estrogen metabolism (Armadillo Repeat gene deleted in Velo-Cardio-Facial syndrome (ARVCF), Catechol-O-methyltransferase (COMT), cytochrome P450 19A1 (CYP19A1)), ESR1 and 2, progesterone receptor (PGR)), co-regulation of the estrogen receptor, (ER, E1a binding protein p300 (EP300), enhancer of zeste 2 polycomb repressive complex 2 (EZH2), nuclear receptor coactivator 1 (NCOA1), 2 (NCOA2), 3 (NCOA3), nuclear receptor corepressor 1 (NCOR1), 2 (NCOR2), nuclear receptor interaction protein (NRIP), proline-, glutamic acid-, and leucine-rich protein-1 (PELP1)), and neuropeptide signaling (5-Hydroxytryptamine Receptor 1A (HTR1A), 2A (HTR2A), serotonin transporter gene (SCL6A4), hypocretin (orexin) neuropeptide precursor (HCRT), hypocretin receptor type 1 (HCRTR1), and 2 (HCRTR2)). Genes related to neuropeptide signaling were not included in the analysis as they were not felt to be relevant to lipid metabolism; this determination was made before the statistical analysis. Genotyping for all SNPs was performed using the BioTrove OpenArray™ platform (Applied Biosystems, Inc., Foster City, CA). Genotype quality control was performed before genetic association analysis, by randomly selecting 10% of the samples and re-genotyping them to validate results. CYP2A6 and CYP3A5 were genotyped using allelic-discrimination TaqMan SNP genotyping assays (Applied Biosystems, Foster City, CA) as previously described (21).
The Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK) criteria were used for reporting these biomarker results (22).
Statistical Analysis
Characteristics of participants at baseline were summarized and described between those on lipid-altering medications and those who were not using Fisher’s exact test and Wilcoxon rank sum tests. Changes in lipid parameters from baseline to 3 months following treatment initiation were summarized overall and by treatment groups and tested for differences with paired t tests. Data analysis was performed using participants who underwent genotyping and follow-up for lipid measurements and who fulfilled criteria for this analysis as described above. For 143/145 SNPs, the minor and major alleles were identified and used to classify participants for each model type (recessive, dominant, additive). CYP2A6 and CYP3A5 were genotyped using different assays as described above, therefore these were analyzed separately. SNPs with a homozygous (defined as two copies of the minor allele) genotype frequency of less than 5 participants were excluded from analyses, leaving 92/143 SNPs for analysis.
A multivariable linear regression model was run for each combination of the143 SNPs, SNP genotype (recessive, dominant or additive), 7 treatment subgroups (all patients, all patients on letrozole, patients on letrozole +/− lipid-altering medication, all patients on exemestane, patients on exemestane +/− lipid-altering medication), and 4 lipid parameters, a multivariable linear regression model was run. There were 12,012 potential models (84 subgroups by 143 SNPs). Eliminating models where there were fewer than 5 participants with the homozygous genotype resulted in 5,796 models across 92 of the 94 SNPs, excluding CYP2A6 and CYP3A5. For each model, the dependent variable was in the difference in the lipid parameter, and independent variables were SNP genotype, participant age, body mass index (BMI), race, change in estradiol levels from baseline to 3 months, and indicators of prior hormonal therapy and tamoxifen use. For each of these models, we specifically tested for Hardy Weinberg equilibrium, calculated the minor allele frequency, and calculated the additional difference in lipids attributable to the SNP with its corresponding p-value. The same multivariable linear regression modeling approach as described for the 143 SNPs was used to analyze CYP2A6 and CYP3A5, this resulted in 84 models (3 gene classifications (CYP3A5 expressed versus not, CYP2A6 normal versus intermediate/slow, CYP2A6 normal/intermediate versus slow) by 7 patient subgroups by 4 lipid outcomes). For the models that were run on the entire cohort, we also performed a test for interaction between the SNP and treatment arm on the change in lipid parameter. To adjust for multiple comparisons, models with a resulting Bonferonni-corrected p-value < 0.00053 (=0.05/94) were considered significant.
Results
Study Population
Of the 503 evaluable participants enrolled in the ELPh trial, 303 met eligibility criteria for this analysis (Figure 1). Of these, 101 participants were taking lipid-altering medications, and were analyzed separately.
The median age of the overall randomized cohort (n=500) was 59 years (range 35 to 89), which consisted of 441 (88%) Caucasians and 46 (9%) African Americans. The median BMI was 29.0 (range 17.7 to 55.9), over the first 3 months the average change in BMI was 0.15. Of the overall randomized cohort, 228 (46%) women had previously been treated with either adjuvant or neoadjuvant chemotherapy, and 184 (37%) had been treated with adjuvant tamoxifen for a median of 2.3 years duration (range 0.1 to 12.9). Compliance data was available in 433 patients at 3 months, and 95% of patients (n=412) reported they had not missed a single dose of AI in the previous week, suggesting a highly compliant cohort. The cohort included in this analysis (n=303) was similar to the overall cohort (Table 1). Patients not taking lipid-altering medications (n=202) were slightly younger (p=0.002), had lower BMI (p=0.02), and were more likely to have received chemotherapy (p=0.01) compared to those taking lipid-altering medication (n=101), but otherwise had similar characteristics. The most common class of lipid-altering medications used by participants was statins.
Table 1.
Characteristics of patients enrolled and randomized in ELPh trial, and subdivided by those included in the analysis. P-values for Fisher’s exact test or Wilcoxon rank sum tests for differences in patients on and not on lipid-altering medication (LAM).
| Entire Cohort (n=500) | Cohort Analyzed (n=303) | Participants not on LAM (n=202) | Participants on LAM (n=101) | P-value | |
|---|---|---|---|---|---|
| Age, median (range) | 59 (35, 89) | 59 (35, 84) | 58 (38, 83) | 61 (35, 84) | 0.002 |
| BMI at diagnosis, median (range) | 29 (17.7, 55.9) | 29.3 (18.4, 55.9) | 28.6 (18.4, 53.4) | 30.5 (20.7, 55.9) | 0.02 |
| Race, n (%) | |||||
| White | 441 (88) | 263 (87) | 176 (87) | 87 (86) | 0.81 |
| African American | 46 (9) | 34 (11) | 21 (10) | 13 (13) | |
| Asian | 12 (2) | 5 (2) | 4 (2) | 1 (1) | |
| Other | 1 (0) | 1 (0) | 1 (0) | 0 (0) | |
| Treatment arm | |||||
| Exemestane | 248 (50) | 143 (47) | 94 (47) | 49 (49) | 0.81 |
| Letrozole | 252 (50) | 160 (53) | 108 (53) | 52 (51) | |
| Prior chemotherapy, n (%) | 228 (46) | 131 (43) | 98 (49) | 33 (33) | 0.01 |
| Prior tamoxifen use, n (%) | 184 (37) | 100 (33) | 70 (35) | 30 (30) | 0.44 |
| Years on tamoxifen, median (range) | 2.3 (0.1, 12.9) | 2.1 (0.1, 5.2) | 2.1 (0.1, 5.2) | 2.1 (0.2, 5) | 0.92 |
Single nucleotide polymorphism (SNP), body mass index (BMI), standard deviation (SD), number (n).
Changes in Lipid Profiles in Cohort and by AI
Key changes in lipid profiles are summarized in Table 2. After 3 months of letrozole in participants not taking lipid-altering medications, TC increased by 5.9 mg/dL (p=0.003) and LDL-C increased by 5.5 mg/dL (p<0.007). However, in participants taking lipid-altering medications, TC decreased and LDL-C remained unchanged. In participants on exemestane not taking lipid-altering medications, TC decreased by 5.9 mg/dL (0.02) and HDL-C decreased by 7.8 mg/dL (p<0.001); however, in those taking lipid-altering medications, decreases in TC were more pronounced, HDL-C remained unchanged, and TG levels were also decreased.
Table 2.
Change in lipid parameters after 3 months of aromatase inhibitor therapy in the entire cohort, and in participants treated with letrozole or exemestane whether on lipid-altering medications (LAM) or not. Patients not taking LAM are also further subdivided by treatment arm. P-values are for changes from baseline to 3 months within patient group. Interaction p-values test whether changes in lipids after 3 months differ between patients on and not on LAM, separately by treatment arm.
| Lipid Parameter | N | Baseline, mean (SD) | 3 Months, mean (SD) | Change, mean (SD) | P-value | Interaction P-value |
|---|---|---|---|---|---|---|
| Overall Cohort | ||||||
| TC, mg/dL | 422 | 199.7 (37.5) | 196.3 (36.1) | −3.4 (27.3) | 0.01 | |
| HDL, mg/dL | 422 | 58.6 (17) | 54.9 (17.3) | −3.7 (11.4) | < 0.001 | |
| LDL, mg/dL | 419 | 117.1 (33.1) | 118 (31.3) | 1 (26.6) | 0.45 | |
| TG, mg/dL | 422 | 121.1 (62.7) | 116.9 (62.4) | −4.1 (49.6) | 0.09 | |
| Letrozole (not on LAM) | ||||||
| TC, mg/dL | 147 | 201.5 (32.8) | 207.4 (33.3) | 5.9 (23.6) | 0.003 | |
| HDL, mg/dL | 147 | 60.9 (16.7) | 59.7 (17.1) | −1.3 (8.5) | 0.08 | |
| LDL, mg/dL | 147 | 119.4 (28.6) | 124.8 (30.5) | 5.5 (24) | 0.007 | |
| TG, mg/dL | 147 | 105.9 (47.1) | 108.4 (47.8) | 2.4 (38.2) | 0.44 | |
| Letrozole (on LAM) | ||||||
| TC, mg/dL | 72 | 186.3 (42.1) | 180.1 (38) | −6.2 (25.5) | 0.04 | 0.001 |
| HDL, mg/dL | 72 | 52.9 (15.6) | 51.3 (18.1) | −1.6 (13.6) | 0.33 | 0.86 |
| LDL, mg/dL | 70 | 105 (36.9) | 100.1 (31) | −4.9 (26.8) | 0.13 | 0.007 |
| TG, mg/dL | 72 | 138.8 (71.2) | 141.4 (74.4) | 2.6 (53.3) | 0.68 | 0.98 |
| Exemestane (not on LAM) | ||||||
| TC, mg/dL | 132 | 207.9 (36.4) | 201.9 (30.5) | −5.9 (28.6) | 0.02 | |
| HDL, mg/dL | 132 | 62.7 (17.8) | 54.8 (15.7) | −7.8 (8.2) | < 0.001 | |
| LDL, mg/dL | 132 | 122.8 (32.2) | 125.6 (26.1) | 2.8 (27.2) | 0.23 | |
| TG, mg/dL | 132 | 112 (54.1) | 104.6 (53.8) | −7.4 (51.7) | 0.1 | |
| Exemestane (on LAM) | ||||||
| TC, mg/dL | 71 | 194.5 (39.9) | 179.3 (37.8) | −15.3 (27.7) | < 0.001 | 0.02 |
| HDL, mg/dL | 71 | 51.9 (14) | 48.5 (17.1) | −3.5 (16.2) | 0.08 | 0.04 |
| LDL, mg/dL | 70 | 113.4 (36.5) | 107.3 (32.5) | −6.1 (28.6) | 0.08 | 0.03 |
| TG, mg/dL | 71 | 151.3 (80.6) | 132.8 (79.5) | −18.5 (59) | 0.01 | 0.19 |
Abbreviations: total cholesterol (TC), high density lipoprotein (HDL), low density lipoprotein (LDL), triglycerides (TG). Only patients with data at both time points are included.
Association of SNPs in Candidate Genes on Lipid Profiles
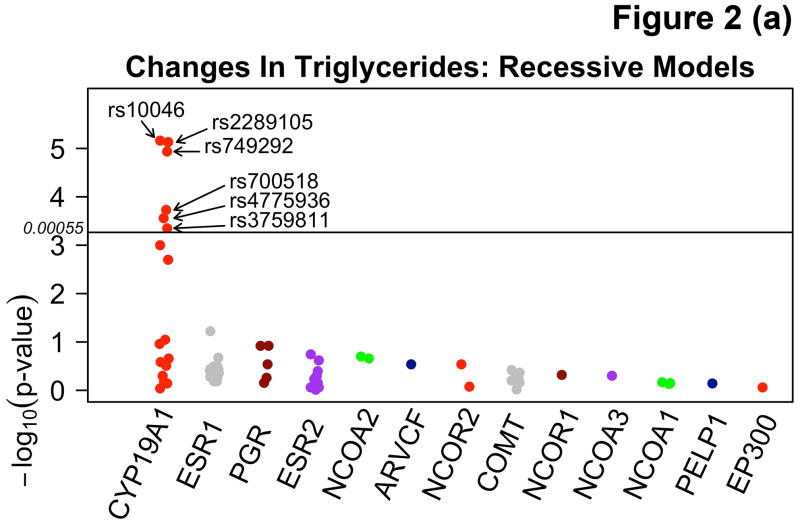
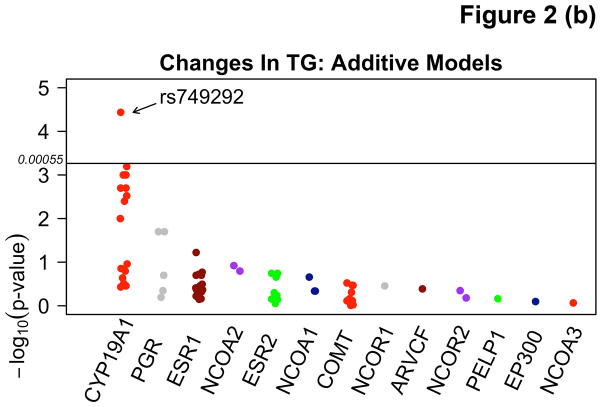
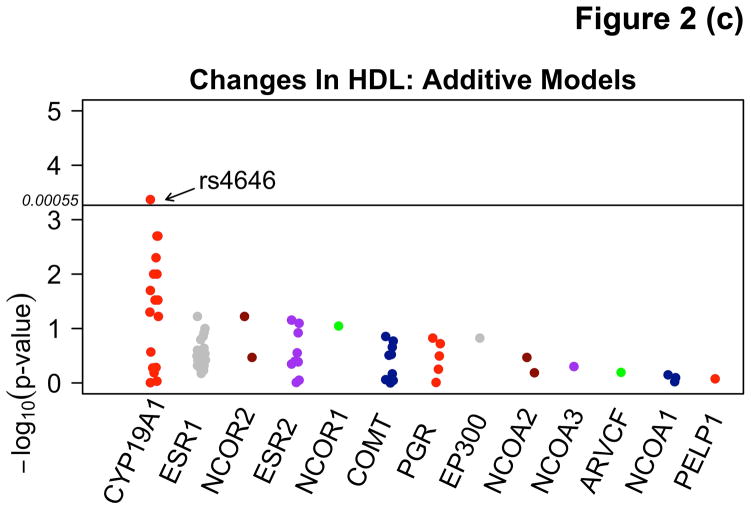
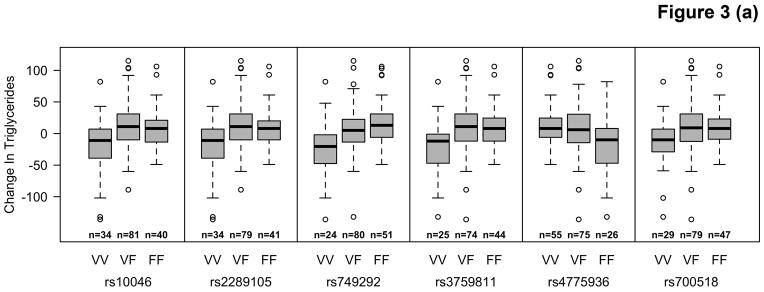
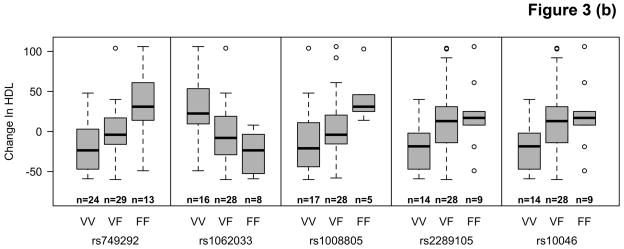
Among participants taking letrozole, variants in the CYP19A1 gene were associated with significant decreases in TG ranging from 20.2 – 39.3 mg/dL, and decreases in HDL-C (4.2 mg/dL) using additive and recessive models. Among participants taking letrozole on lipid-altering medications, some specific variants of CYP19A1 were associated with increases of 6.7 – 9.5 mg/dL in HDL-C and others with decreases of 6.2 – 6.6 mg/dL in both dominant and additive models. There were no significant associations with SNPs among patients taking letrozole not on lipid-altering medications. Statistically significant changes (p<0.00053) are summarized in Table 3 and compared to SNPs in other candidate genes not reaching statistical significance in Figure 2. Mean allele frequency (MAF) ranged from 0.27 – 0.48, and is summarized in Table 3. TG and HDL-C changes in significant SNPs were further described in recessive, dominant, and additive models (Figure 3).
Table 3.
Significant findings of multivariable linear regressions analyzing genetic associations between candidate gene SNPs and lipid profiles in AI-treated participants, adjusted for age, BMI, race, change in plasma estradiol, and prior hormonal therapy and tamoxifen use.
| Cohort | Lipid Parameter | Model | Gene (SNP) | MAF | HWE P-value | Number of HOM | Minor Allele | Mean Change, mg/dL (SE) | P-value |
|---|---|---|---|---|---|---|---|---|---|
| All Patients on Letrozole (n=160) | TG | Recessive | CYP19A1 (rs10046*) | 0.48 | 0.63 | 34 | A | −36.4 (7.8) | 0.0000069 |
| TG | Recessive | CYP19A1 (rs2289105*) | 0.48 | 0.87 | 34 | C | −36.45 (7.8) | 0.0000074 | |
| TG | Recessive | CYP19A1 (rs3759811*) | 0.43 | 0.61 | 25 | C | −32.3 (9.0) | 0.00045 | |
| TG | Recessive | CYP19A1 (rs700518*) | 0.44 | 0.75 | 29 | C | −32.3 (8.4) | 0.00019 | |
| TG | Recessive | CYP19A1 (rs4775936*) | 0.41 | 1.0 | 26 | C | −33.2 (8.9) | 0.00028 | |
| HDL | Additive | CYP19A1 (rs4646*) | 0.27 | 1.0 | 11 | A | −4.2 (1.16) | 0.00043 | |
| TG | Recessive | CYP19A1 (rs749292*) | 0.41 | 0.51 | 24 | A | −39.3 (8.6) | 0.000012 | |
| TG | Additive | CYP19A1 (rs749292*) | 0.41 | 0.51 | 24 | A | −20.2 (4.7) | 0.000037 | |
| Patients on Letrozole taking LAM (n=52) | HDL | Additive | CYP19A1 (rs749292*) | 0.47 | 0.58 | 10 | A | 6.7 (1.4) | 0.000037 |
| HDL | Additive | CYP19A1 (rs1062033*) | 0.42 | 0.58 | 8 | G | 6.9 (1.5) | 0.000043 | |
| HDL | Dominant | CYP19A1 (rs1008805*) | 0.38 | 0.24 | 5 | G | −6.6 (1.7) | 0.00037 | |
| HDL | Dominant | CYP19A1 (rs749292*) | 0.47 | 0.58 | 10 | A | 9.5 (2.4) | 0.0003 | |
| HDL | Additive | CYP19A1 (rs10046*) | 0.45 | 0.57 | 9 | G | −6.2 (1.6) | 0.00046 | |
| HDL | Additive | CYP19A1 (rs2289105*) | 0.45 | 0.57 | 9 | T | −6.2 (1.6) | 0.00046 |
Mean allele frequency (MAF), Hardy-Weinberg equilibrium (HWE), homozygotes (HOM), standard error (SE), adenine (A), cytosine (C), guanine (G), thymine (T)
Figure 2.
Manhattan plots demonstrating associations between variants in the CYP19A1 gene and absolute changes in triglycerides (TG) using recessive (A) and additive (B) models, and changes in high density lipoprotein (HDL-C) using additive models (C) in letrozole treated participants.
Figure 3.
Distribution of changes in TG in letrozole treated women not on lipid altering medications (LAM) (A) and HDL-C in letrozole treated women on LAM (B) by CYP19A1 genotype.
We did not observe significant SNP-lipid associations in exemestane treated participants overall nor by whether they were taking lipid-altering medications or not in these or other variants (Supplementary Table 1). We also did not observe any evidence of interaction between any of the SNPs and treatment arm on the changes in lipid outcomes. We performed a sensitivity analysis with only Caucasian women (Supplementary Table 2) and another analysis without adjusting for changes in plasma estradiol concentrations (Supplementary Table 3), and found similar results. All alleles from significant SNPs were found to be in Hardy Weinberg Equilibrium.
Discussion
We have prospectively demonstrated that, in women treated with letrozole, variants of CYP19A1 are associated with mostly favorable effects in TG and HDL-C. These associations were observed despite a general analysis finding no association between letrozole and changes in TG (p=0.44) and HDL-C (p=0.08), suggesting that pharmacogenetic factors play a powerful role in predicting AI-associated lipid changes. The product of the CYP19A1 gene is crucial in the conversion of pre-estrogens into estrogens, which can alter lipid concentrations; therefore, a possible mechanism explaining our results may be altered estrogen metabolism in letrozole-treated patients who have specific SNPs in CYP19A1. While we observed significant associations in all women treated with letrozole, and in a subset of those taking lipid-altering medications, we did not find an association in the subset of patients not on lipid-altering medications. This result may be due to small numbers in subset analysis, or because the effects lipid-altering medications on lipid profiles may have influenced results.
We did not observe significant associations between SNPs and lipids in exemestane-treated women, although in a general analysis we found that exemestane was associated with decreases in TC and HDL-C, and that taking lipid-altering medications improved the overall lipid profile. Failure to find any associations in exemestane-treated women in the SNP analysis may have been due to a small sample size being underpowered to detect modest pharmacogenetic effects. Alternatively, non-steroidal versus steroidal AIs have been shown to have different pharmacodynamic effects, and our results may indeed suggest different biological effects (15, 23–25).
Our general analysis is similar to results our group has previously reported in the ELPh cohort despite minor differences in cohort derivation (15). Overall, effects of both letrozole and exemestane on lipid profiles are unfavorable. Notably, while exemestane appears to decrease TC, this may be due to decreases in HDL-C also observed (TC= LDL-C + HDL-C + TG/5). While published literature of the effects on AIs has been mixed, many studies have demonstrated decreased in HDL-C with exemestane, and increased LDL-C with letrozole treatment [7–14]. However, we report that negative effects on lipid profiles by letrozole and exemestane are eliminated in patients already on lipid-altering medications. To our knowledge this is the first report finding such results.
Certain strengths and limitations should be considered when interpreting these data. A significant strength of this study is that this is a planned sub-analysis of a large prospective study. The subject population is diverse; therefore, results can be extrapolated to similar populations. Correlative study blood samples were collected in the majority of patients, with 79% (393 from 500 evaluable patients) of patients having genotype and lipid data at corresponding time points. A unique strength of our analysis is that changes in estradiol at the same times lipids were collected are adjusted for. The results of this analysis are similar whether estrogen levels are adjusted for or not, suggesting a powerful pharmacodynamic effect.
Limitations to consider when interpreting these data include the fact that allelic frequencies vary amongst different races, and both Caucasian and African American women are included in this analysis. To address this consideration, we statistically adjusted our analysis for race. Furthermore, we performed a sensitivity analysis in only Caucasian women, and found results to be similar. Another limitation to consider is that our sample was relatively small after excluding participants ineligible for this analysis, which reduces power. However, as mentioned above, this was a planned sub-analysis of a prospective randomized cohort, which adds to validity of our findings. Another consideration when interpreting these results is that we investigated only candidate genes relevant to AI metabolism and estrogen signaling; however, variants in other genes may potentially play a role in determining AI-mediated effects on lipids. While this may be so, our group has substantial experience with selected candidate genes, and similar panels have been linked to rates of AI discontinuation and letrozole concentration in AI treated women, suggesting that candidate genes selected were appropriate (21, 26, 27). Future large scale investigation should include genome-wide association studies (GWAS) to validate our findings and to explore new candidate genes. Another consideration when interpreting these results is that while obesity, defined by BMI, is associated with lipid abnormalities, central obesity, estimated by waist circumference, may be a more accurate surrogate of this relationship (28–30). We adjusted our analysis to include the effects of BMI, although a more precise surrogate may have been waist circumference, which may be particularly relevant since up to 96% of women gain weight after breast cancer diagnosis and treatment (31, 32). These results may justify investigating pharmacogenetic strategies to assess cardiovascular risk in breast cancer survivors. While the Arimidex, Tamoxifen, Alone or in Combination (ATAC) trial investigators found no difference in non-breast cancer related deaths between aromatase inhibitors and tamoxifen, a review analyzing 30,023 women found that patients treated with AIs had an increased risk of cardiovascular disease (OR=1.26, 95% CI: 1.10 – 1.43, p<0.001) compared to tamoxifen (33, 34). Cardiovascular disease risk is a significant competing comorbidity to breast cancer recurrence, and data from this same cohort of patients used in this study has demonstrated that 43% of women had a predicted 10-year cardiovascular disease risk equivalent to breast cancer recurrence risk and 37% had cardiovascular disease risk higher than breast cancer recurrence risk. Particularly, patients with stage 1 disease had a significantly increased risk of developing heart disease (OR 6.1, 95% CI: 3.4–11.2, p<0.0001) (18). Since dyslipidemias are an important cardiovascular risk factor, identifying those patients at highest risk for dyslipidemias due to AI treatment is critical. Our data suggest that women taking letrozole with variants in CYP19A1 may enjoy a favorable modulation in TG, although HDL-C levels may or may not undergo favorable changes.
This study contributes evidence that pharmacogenomic biomarkers play a role as predictors of AI toxicity (21, 27). Furthermore, pharmacogenomic biomarkers may help explain why different AIs may exert differential effects in target tissues. Additional studies investigating SNP-based models predicting specific AI toxicity may help identify patients at risk and guide management, particularly in regards to cardiovascular risk factors. This is particularly relevant because many breast cancer survivors are cured of their breast cancer; however, anti-cancer therapies in some women may place them at higher risk for cardiovascular disease.
Supplementary Material
Statement of Translational Relevance.
Aromatase inhibitors (AIs) are a cornerstone of therapy for postmenopausal women with early breast cancer. However, there is a concern that negative effects on lipid profiles can potentially increase cardiovascular risk. While this relationship has not been clear to date, pharmacogenomics may help identify which patients may or may not be at risk. This study investigates whether single nucleotide polymorphisms (SNPs) in candidate genes related to estrogen and AI signaling and metabolism predict changes to lipid profiles in AI-treated women. Our findings that SNPs in the CYP19A1 gene are associated with modulation of high-density lipoprotein and triglyercerides demonstrate the powerful effects of pharamcogenomics. Future studies may ultimately lead to personalized and improved management of cardiovascular risk factors.
Acknowledgments
Financial Support: This study was funded in part by the NIH/NIGMS Pharmacogenetics Network Grant, P30 CA06973, Pfizer, Novartis, the Breast Cancer Research Foundation (N003173, JMR, NLH), NIH 1RO1GM099143 (JMR, SO, NLH), and Fashion Footwear Charitable Foundation of New York/QVC Presents Shoes on Sale ™ (DFH). Vered Stearns holds the Breast Cancer Research Chair in Oncology.
We thank Dr. Gary Rosner for helpful assistance with the statistical analysis.
Footnotes
Conflicts of Interest: Dr. Vered Stearns has received research funding from Abbvie, Celgene, Medimmune, Merck, Novartis, Pfizer, and Puma. Dr. Daniel Hayes has received research funding from Johnson and Johnson, Puma Biotechnology, Pfizer, and Aztra Zeneca. Dr. Hayes also has personal financial interest in Oncimmune and Inbiomotion, has consulted for Pfizer and Lilly Oncology, has sponsored clinical research from Janssen R&D, Puma Biotechnology, Pfizer, AstraZeneca, and has royalties from Janssen R&D. Dr. Lynn Henry has received research funding from Astra Zeneca, Eli Lilly, Sanofi Aventis, BioMarin Pharmaceuticals, and Celldex Pharmaceuticals. Dr. David Flockhart has received research funding from Pfizer and Novartis. The rest of the authors have no conflicts of interest to declare.
References
- 1.Burstein HJ, Prestrud AA, Seidenfeld J, Anderson H, Buchholz TA, Davidson NE, et al. American Society of Clinical Oncology clinical practice guideline: update on adjuvant endocrine therapy for women with hormone receptor-positive breast cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2010;28:3784–96. doi: 10.1200/JCO.2009.26.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dowsett M, Cuzick J, Ingle J, Coates A, Forbes J, Bliss J, et al. Meta-analysis of breast cancer outcomes in adjuvant trials of aromatase inhibitors versus tamoxifen. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2010;28:509–18. doi: 10.1200/JCO.2009.23.1274. [DOI] [PubMed] [Google Scholar]
- 3.Higgins MJ, Liedke PE, Goss PE. Extended adjuvant endocrine therapy in hormone dependent breast cancer: the paradigm of the NCIC-CTG MA.17/BIG 1-97 trial. Crit Rev Oncol Hematol. 2013;86:23–32. doi: 10.1016/j.critrevonc.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 4.Goss PE, Ingle JN, Pritchard KI, Ellis MJ, Sledge GW, Budd GT, et al. Exemestane versus anastrozole in postmenopausal women with early breast cancer: NCIC CTG MA.27--a randomized controlled phase III trial. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2013;31:1398–404. doi: 10.1200/JCO.2012.44.7805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geisler J, Haynes B, Anker G, Dowsett M, Lonning PE. Influence of letrozole and anastrozole on total body aromatization and plasma estrogen levels in postmenopausal breast cancer patients evaluated in a randomized, cross-over study. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2002;20:751–7. doi: 10.1200/JCO.2002.20.3.751. [DOI] [PubMed] [Google Scholar]
- 6.Hayes DF, Skaar TC, Rae JM, Henry NL, Nguyen AT, Stearns V, et al. Estrogen receptor genotypes, menopausal status, and the effects of tamoxifen on lipid levels: revised and updated results. Clin Pharmacol Ther. 2010;88:626–9. doi: 10.1038/clpt.2010.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lonning PE, Geisler J, Krag LE, Erikstein B, Bremnes Y, Hagen AI, et al. Effects of exemestane administered for 2 years versus placebo on bone mineral density, bone biomarkers, and plasma lipids in patients with surgically resected early breast cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2005;23:5126–37. doi: 10.1200/JCO.2005.07.097. [DOI] [PubMed] [Google Scholar]
- 8.Francini G, Petrioli R, Montagnani A, Cadirni A, Campagna S, Francini E, et al. Exemestane after tamoxifen as adjuvant hormonal therapy in postmenopausal women with breast cancer: effects on body composition and lipids. Br J Cancer. 2006;95:153–8. doi: 10.1038/sj.bjc.6603258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Montagnani A, Gonnelli S, Cadirni A, Caffarelli C, Del Santo K, Pieropan C, et al. The effects on lipid serum levels of a 2-year adjuvant treatment with exemestane after tamoxifen in postmenopausal women with early breast cancer. Eur J Intern Med. 2008;19:592–7. doi: 10.1016/j.ejim.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 10.Markopoulos C, Polychronis A, Dafni U, Koukouras D, Zobolas V, Tzorakoleftherakis E, et al. Lipid changes in breast cancer patients on exemestane treatment: final results of the TEAM Greek substudy. Annals of oncology: official journal of the European Society for Medical Oncology/ESMO. 2009;20:49–55. doi: 10.1093/annonc/mdn545. [DOI] [PubMed] [Google Scholar]
- 11.Markopoulos C, Chrissochou M, Michailidou A, Tzoracoleftherakis E, Xepapadakis G, Papadiamantis J, et al. Effect of exemestane on the lipidemic profile of post-menopausal operable breast cancer patients following 5–7 years of adjuvant tamoxifen: preliminary results of the ATENA substudy. Anticancer Drugs. 2005;16:879–83. doi: 10.1097/01.cad.0000173478.12981.e1. [DOI] [PubMed] [Google Scholar]
- 12.Markopoulos C, Dafni U, Misitzis J, Zobolas V, Tzoracoleftherakis E, Koukouras D, et al. Extended adjuvant hormonal therapy with exemestane has no detrimental effect on the lipid profile of postmenopausal breast cancer patients: final results of the ATENA lipid substudy. Breast cancer research: BCR. 2009;11:R35. doi: 10.1186/bcr2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elisaf MS, Bairaktari ET, Nicolaides C, Kakaidi B, Tzallas CS, Katsaraki A, et al. Effect of letrozole on the lipid profile in postmenopausal women with breast cancer. Eur J Cancer. 2001;37:1510–3. doi: 10.1016/s0959-8049(01)00155-1. [DOI] [PubMed] [Google Scholar]
- 14.Wasan KM, Goss PE, Pritchard PH, Shepherd L, Palmer MJ, Liu S, et al. The influence of letrozole on serum lipid concentrations in postmenopausal women with primary breast cancer who have completed 5 years of adjuvant tamoxifen (NCIC CTG MA.17L) Annals of oncology: official journal of the European Society for Medical Oncology/ESMO. 2005;16:707–15. doi: 10.1093/annonc/mdi158. [DOI] [PubMed] [Google Scholar]
- 15.Bell LN, Nguyen AT, Li L, Desta Z, Henry NL, Hayes DF, et al. Comparison of changes in the lipid profile of postmenopausal women with early stage breast cancer treated with exemestane or letrozole. J Clin Pharmacol. 2012;52:1852–60. doi: 10.1177/0091270011424153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Emre A, Sahin S, Erzik C, Nurkalem Z, Oz D, Cirakoglu B, et al. Effect of hormone replacement therapy on plasma lipoproteins and apolipoproteins, endothelial function and myocardial perfusion in postmenopausal women with estrogen receptor-alpha IVS1-397 C/C genotype and established coronary artery disease. Cardiology. 2006;106:44–50. doi: 10.1159/000092598. [DOI] [PubMed] [Google Scholar]
- 17.Koukouras D, Marioli DJ, Papadopoulos K, Adonakis GL, Armeni AK, Georgopoulos NA, et al. Association of estrogen receptor alpha (ERα) gene polymorphisms with endometrial thickness and lipid profile in women with breast cancer treated with aromatase inhibitors. Gynecol Endocrinol. 2012;28:859–62. doi: 10.3109/09513590.2012.671393. [DOI] [PubMed] [Google Scholar]
- 18.Bardia A, Arieas ET, Zhang Z, Defilippis A, Tarpinian K, Jeter S, et al. Comparison of breast cancer recurrence risk and cardiovascular disease incidence risk among postmenopausal women with breast cancer. Breast cancer research and treatment. 2012;131:907–14. doi: 10.1007/s10549-011-1843-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henry NL, Giles JT, Ang D, Mohan M, Dadabhoy D, Robarge J, et al. Prospective characterization of musculoskeletal symptoms in early stage breast cancer patients treated with aromatase inhibitors. Breast cancer research and treatment. 2008;111:365–72. doi: 10.1007/s10549-007-9774-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henry NL, Xia R, Banerjee M, Gersch C, McConnell D, Giacherio D, et al. Predictors of recovery of ovarian function during aromatase inhibitor therapy. Annals of oncology: official journal of the European Society for Medical Oncology/ESMO. 2013;24:2011–6. doi: 10.1093/annonc/mdt149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Desta Z, Kreutz Y, Nguyen AT, Li L, Skaar T, Kamdem LK, et al. Plasma letrozole concentrations in postmenopausal women with breast cancer are associated with CYP2A6 genetic variants, body mass index, and age. Clin Pharmacol Ther. 2011;90:693–700. doi: 10.1038/clpt.2011.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Altman DG, McShane LM, Sauerbrei W, Taube SE. Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK): explanation and elaboration. PLoS Med. 2012;9:e1001216. doi: 10.1371/journal.pmed.1001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goss PE, Hadji P, Subar M, Abreu P, Thomsen T, Banke-Bochita J. Effects of steroidal and nonsteroidal aromatase inhibitors on markers of bone turnover in healthy postmenopausal women. Breast cancer research: BCR. 2007;9:R52. doi: 10.1186/bcr1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller WR, Bartlett J, Brodie AM, Brueggemeier RW, di Salle E, Lonning PE, et al. Aromatase inhibitors: are there differences between steroidal and nonsteroidal aromatase inhibitors and do they matter? Oncologist. 2008;13:829–37. doi: 10.1634/theoncologist.2008-0055. [DOI] [PubMed] [Google Scholar]
- 25.Goss PE, Hershman DL, Cheung AM, Ingle JN, Khosla S, Stearns V, et al. Effects of adjuvant exemestane versus anastrozole on bone mineral density for women with early breast cancer (MA.27B): a companion analysis of a randomised controlled trial. The Lancet Oncology. 2014;15:474–82. doi: 10.1016/S1470-2045(14)70035-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henry NL, Chan HP, Dantzer J, Goswami CP, Li L, Skaar TC, et al. Aromatase inhibitor-induced modulation of breast density: clinical and genetic effects. Br J Cancer. 2013;109:2331–9. doi: 10.1038/bjc.2013.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henry NL, Skaar TC, Dantzer J, Li L, Kidwell K, Gersch C, et al. Genetic associations with toxicity-related discontinuation of aromatase inhibitor therapy for breast cancer. Breast cancer research and treatment. 2013;138:807–16. doi: 10.1007/s10549-013-2504-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hubert HB, Feinleib M, McNamara PM, Castelli WP. Obesity as an independent risk factor for cardiovascular disease: a 26-year follow-up of participants in the Framingham Heart Study. Circulation. 1983;67:968–77. doi: 10.1161/01.cir.67.5.968. [DOI] [PubMed] [Google Scholar]
- 29.Grundy SM, Barnett JP. Metabolic and health complications of obesity. Dis Mon. 1990;36:641–731. [PubMed] [Google Scholar]
- 30.Pouliot MC, Despres JP, Lemieux S, Moorjani S, Bouchard C, Tremblay A, et al. Waist circumference and abdominal sagittal diameter: best simple anthropometric indexes of abdominal visceral adipose tissue accumulation and related cardiovascular risk in men and women. Am J Cardiol. 1994;73:460–8. doi: 10.1016/0002-9149(94)90676-9. [DOI] [PubMed] [Google Scholar]
- 31.Vance V, Mourtzakis M, McCargar L, Hanning R. Weight gain in breast cancer survivors: prevalence, pattern and health consequences. Obes Rev. 2011;12:282–94. doi: 10.1111/j.1467-789X.2010.00805.x. [DOI] [PubMed] [Google Scholar]
- 32.Heasman KZ, Sutherland HJ, Campbell JA, Elhakim T, Boyd NF. Weight gain during adjuvant chemotherapy for breast cancer. Breast cancer research and treatment. 1985;5:195–200. doi: 10.1007/BF01805994. [DOI] [PubMed] [Google Scholar]
- 33.Amir E, Seruga B, Niraula S, Carlsson L, Ocana A. Toxicity of adjuvant endocrine therapy in postmenopausal breast cancer patients: a systematic review and meta-analysis. Journal of the National Cancer Institute. 2011;103:1299–309. doi: 10.1093/jnci/djr242. [DOI] [PubMed] [Google Scholar]
- 34.Cuzick J, Sestak I, Baum M, Buzdar A, Howell A, Dowsett M, et al. Effect of anastrozole and tamoxifen as adjuvant treatment for early-stage breast cancer: 10-year analysis of the ATAC trial. The Lancet Oncology. 2010;11:1135–41. doi: 10.1016/S1470-2045(10)70257-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.