Abstract
Non-lethal exposure to ionizing radiation (IR) is a public concern due to its known carcinogenic effects. Although latency periods for IR-induced neoplasms are relatively long, the ability to detect cancer as early as possible is highly advantageous for effective therapeutic intervention. Therefore, we hypothesized that metabolites in the urine from mice exposed to total body radiation (TBI) would predict for the presence of cancer before a palpable mass was detected. In this study, we exposed mice to 0 or 5.4 Gy TBI, collected urine samples periodically over one year, and assayed urine metabolites by using mass spectrometry. Longitudinal data analysis within the first year post-TBI revealed that cancers, including hematopoietic, solid, and benign neoplasms, could be distinguished by unique urinary signatures as early as 3 months post-TBI. Furthermore, a distinction among different types of malignancies could be clearly delineated as early as 3 months post-TBI for hematopoietic neoplasms, 6 months for solid neoplasms, and by 1 year for benign neoplasms. Moreover, the feature profile for radiation-exposed mice 6 months post-TBI was found to be similar to non-irradiated control mice at 18 months, suggesting that TBI accelerates aging. These results demonstrate that urine feature profiles following TBI can identify cancers prior to macroscopic detection, with important implications for the early diagnosis and treatment.
Keywords: Radiation, carcinogenesis, metabolites, metabolomics, aging
Introduction
While there are numerous beneficial properties of ionizing radiation (IR) including diagnostic and therapeutic applications, there are also detrimental realities such as lethality, acute and chronic tissue damage, and carcinogenesis. Concern regarding these toxicities has been magnified by terrorist events or nuclear accidents over the past few years raising the possibility of exposure of large populations of people to IR. To address these concerns the U.S. government established and funded the Radiation Nuclear Countermeasures Program in 2004 to foster research and the development of radiation medical countermeasures that protect against IR-induced lethality or mitigates tissue/organ damage (1). Predominantly this program focuses on determining means of IR protection/mitigation for acute effects as opposed to late effects. IR-induced late effects become most important in the setting of non-lethal doses of IR. An example of an untoward late effect of non-lethal IR exposure is increased risk for cancer. There has been little research conducted with respect to early screening or identification of biomarkers for IR-induced cancer. A reason for this is that IR is both an initiator and promoter requiring a relatively long latency period for the development of neoplasms. The neoplasm most associated with IR exposure is hematopoietic with latency periods of 5-7 years in humans (2). Solid neoplasms induced by IR generally require longer periods of time post-treatment (10-20 years) (3). Since it is well established that early diagnosis of cancer leads to more effective therapy, means of detecting it early would be highly advantageous (4). Refined instrumentation such as ultra-high pressure liquid chromatography (UHPLC) coupled with quadrupole time-of-flight mass spectrometry (QTOFMS) has facilitated the interrogation and identification of large numbers of metabolites from easily obtainable body fluids (e.g., urine, blood) and tissue (5, 6). This has enabled the emergence of the field of metabolomics, which allows for the study of metabolites that might be associated with disease processes. Metabolic profiles have been studied in IR treated animals, but predominantly at time periods shortly after IR exposure (<30 days) (7-11).
In the current study the hypothesis was tested that metabolites in the urine from mice exposed to non-lethal total body radiation (TBI) would predict for the presence of neoplasms before it becomes visibly detected. To test this hypothesis, individually ear-tagged female C3H mice were exposed to 0 or 5.4 Gy TBI. Urine samples were routinely collected and stored after IR. Animals were euthanized when a tumor mass appeared or for humane reasons and complete necropsy and pathology was performed on each mouse. Metabolite profiles of over 700 urine samples taken from control and IR-treated mice were determined by UHPLC-QTOFMS-based metabolomics. Distinguishable feature profile (mass-to-charge ratio, retention time, and peak area) patterns could be discerned from IR-treated mice as early as 3 months post-IR compared to control mice. Further, distinct feature patterns were associated with different neoplasm histology in IR-treated mice suggesting that these profiles may be prognostic of future cancer development.
Materials and Methods
Mice
Female C3H/HenTac−MTV− (NCI Animal Production Area; Frederick, MD) mice were housed in a specific pathogen free facility on a 12 hr day/night cycle with standard laboratory chow and water provided ad libitum. When the mice were 8-9 weeks of age, they were randomly divided into two groups; 0 Gy control (n = 25) and 5.4 Gy total body radiation (TBI) (n = 75). Each mouse was ear-tagged for identification. Mice were exposed to 5.4 Gy TBI (Cesium Gamma Cell 40, Atomic Energy of Canada LTD, Ottawa, Canada) at a dose rate of 0.75 Gy/min. Mice were placed in specially designed acrylic containers that were well ventilated allowing 5 mice/container. Radiation dosimetry was confirmed by both thermoluminescent and ferrous sulfate dosimetry. Immediately following TBI the chow for both groups was switched to bacon-flavored control chow (BioServ, Frenchtown, NJ, USA) (to maintain continuity with previous studies) and water provided ad libitum. The animals were maintained for their lifespan in a climate controlled, circadian rhythm-adjusted rooms (5 mice/cage). Periodic weight assessments were made approximately monthly post-TBI. Beginning at ~3 months post-TBI urine samples were collected from 1/3 of the mice (control and irradiated) every two weeks (each mouse was sampled every 6 weeks). Spot urine samples were collected at the same time of day (during the morning hours) from mice by holding them over a sterile 100 mm petri dish. Once urine was expressed the sample was placed into a sterile Eppendorf tube, sealed, labeled with ear tag number and date, and stored at −80°C. All experiments were carried out under a protocol approved by the National Cancer Institute Animal Care and Use Committee and were in compliance with the Guide for the Care and Use Of Laboratory Animal Resource, (2011) National Research Council.
Assessment, Necropsy, and Pathology
Animals were carefully monitored a minimum of three times per week for their entire lifespan. The endpoint for the study was tumor formation (not to exceed 2 cm diameter) or until the animal reached a humane endpoint (rapid weight loss, debilitating diarrhea, rough hair coat, hunched posture, labored breathing, lethargy, persistent recumbence, jaundice, significantly abnormal neurological signs, bleeding from any orifice, proptosis or abnormal appearance of eyes, impaired mobility, or inability to obtain food or water) at which time the animal was euthanized and evaluated for the presence of tumor and cause of death. Mice were euthanized by CO2 asphyxiation and blood collected from the thoracic aorta for a complete blood count. A comprehensive necropsy examination was performed on each mouse with descriptions of gross lesions, collection of all major organs, tissues and lesions and fixation of pathology materials in 10% buffered neutral formalin. Tissues were processed and stained with hematoxylin and eosin (H&E). A board-certified veterinary pathologist performed pathology evaluation. The number of tumors, their phenotypes, tumor burden and cause of death of each animal were determined at its death. Samples were excluded if an animal died of unrelated causes. TBI exposed mice were broadly categorized as three phenotypes that developed hematopoietic neoplasms (IRH), solid neoplasms (IRS) and benign neoplasms (IRB) (Supplementary Table S1). This is a gross classification since some animals had multiple tumor phenotypes. Animals were called as IRH if they had hematopoietic neoplasms irrespective of the presence of solid and benign tumors, IRS type had solid neoplasms irrespective of the presence of benign tumors but did not have hematopoietic neoplasms, and the remaining were called IRB type irrespective of the presence benign neoplasms. All the controls (C) were treated as one category and sub-classes within C were defined as those that developed solid neoplasms or hematopoietic neoplasms (CT) and the remaining (C-) (Supplementary Table S1, S2). Given the scope of the study we were limited to studying female mice only.
UHPLC-ESI-QTOFMS Analysis of Urine Samples
Urine samples were prepared by adding 20 μL of urine to 180 μL 50% aqueous acetonitrile (50:50 water:acetonitrile). Samples were vortexed for 5 min and centrifuged at 14,000 rpm for 20 min at 4 °C to remove particulates and precipitate protein. The supernatant was transferred to an autosampler vial for analysis. A 5 μL aliquot of the supernatant was chromatographed via ultra-high pressure liquid chromatography (Acquity H-Class, Waters Corp, Milford, MA) using a 2.1×50mm Waters BEH C18 1.7 μm column and introduced via electrospray into a quadrupole time-of-flight mass spectrometer (QTOF Premier, Waters Corp., Milford, MA). The gradient mobile phase consisted of 0.1% formic acid solution (A) and acetonitrile containing 0.1% formic acid solution (B). The gradient was maintained at 100% A for 0.5 min, increased to 100% B over the next 7.5 min and returning to 100% A in the last 2 min. Data were collected operating the mass spectrometer in positive (ESI+) and negative (ESI−) electrospray ionizations in centroid mode, in full-scan range from 50 to 850 m/z. Nitrogen was used as both cone gas (50 liters/h) and desolvation gas (600 liters/h). Source temperature and desolvation temperature were set at 120°C and 350°C, respectively. The capillary and cone voltages were 3000 and 20 V, respectively. Chlorpropamide (5 μM) was added as an internal standard and was checked in 6 arbitrarily picked. Mean values of retention time (RT) and m/z ratios in ESI+ mode were 5.47 ±0.007 and 277.0411 ± (0.4 ppm), respectively. In addition, a blank followed by a MetMix standard were randomly injected at an average interval of 17 samples). Three peaks of MetMix standard were monitored at mean RT = 1.84 (± 0.012), 2.44 (± 0.01), and 5.47 (± 0.01) min and mean m/z ratios = 181.0728 (± 1 ppm), 195.0891(± 2 ppm), and 264.1751 (± 1 ppm), respectively (Supplementary Table S3).
Chemometrics and Data Deconvolution
UHPLC-ESI-QTOFMS data were acquired for 728 urine samples in randomized order. Centroided and integrated raw mass spectrometric data were processed using MarkerLynx software (Waters Crop., Milford, MA) to align the chromatograms and to obtain a data matrix consisting of unique retention time (RT), m/z, and integrated peak areas. There are 9075 and 8173 unique RT and m/z combinations respectively in ESI+ and ESI− modes resulting in over 12.5 million feature assays. Additionally, 44 blanks and 44 MetMix standards were included to ascertain instrument performance.
Intensity data were normalized to account for the variations of endogenous metabolite concentrations of urine due to water consumption of the animals and their physiological and pathophysiological factors (12). Normalization by (a) endogenous creatinine level and (b) total ion current (TIC) using ESI+ mode peak intensities indicated that either method is better than no normalization at all. It is known that creatinine levels vary with age (13). Hence, we chose TIC normalization. Total intensity of each sample was scaled to 10000 in ESI+ and ESI− modes. The complete normalized peak intensity data matrix contained 17248 rows uniquely identified by ESI mode, RT and m/z values and 728 columns each representing one urine sample. Rows are referred hereafter as features for convenience although it is possible that multiple peaks may be observed from a single metabolite due to fragmentation and/or adduct formation in the mass spectrometer.
The histogram of non-trivial peaks of an individual sample is normally distributed on logarithmic scale and the intensities of successive temporal points of a single mouse are highly correlated (Pearson correlation coefficient ~ 0.8, data not shown). Samples having poor correlations within an animal were treated as outliers. Principal component analysis and multidimensional scaling graphs were used to detect any outliers within classes. After excluding all outliers, 525 samples from 68 animals remained. In these, 313 samples were collected within 365 days from 62 animals (Supplementary Table S2). To study the gross changes among classes, an intensity threshold filter was applied to the rows in data matrix. The features having 50% or more samples above 0.1 threshold intensity level were included (about 6000 – 7500). Since a linear decrease of intensities with m/z increase was observed, the threshold was linearly varied from 0.1 to 0.0167 for m/z ratios 0 to 850 and referred as 0.1 threshold intensity level (Supplementary methods). A data set including ~11,900 features at 19% of the samples above thresholds was used for gross comparison of IR versus C.
Multivariate Data Analysis
The filtered data matrix was analyzed by projection to latent structures discriminant analysis (PLS-DA) using mixOmics software package (mixOmics package ver. 4.0-2; http://www.math.univ-toulouse.fr/~biostat/mixOmics/) in the R environment [4-5]. Data were centered and scaled in PLS-DA giving equal weights to all the features passed through the intensity filter. A wrapper R-script was used to accomplish orthogonal projections to latent structures (OPLS) which offers more easily interpretable component projection graphs. The variable importance for projection (VIP) value was determined for each feature. A subset of features having the highest VIP scores was sufficient to demonstrate the contrasts between classes. However, every feature in this subset may not be statistically significant. Class comparisons were made by Wilcoxon rank sum tests and the features found at p < 0.001 (unless explicitly mentioned otherwise) were examined in further detail. Hierarchical clustering of significant features was performed on scaled and mean or median centered data by average linkage algorithm and using 1-correlation coefficient (ρ) as distance metric. The relative abundances of the features were displayed in Red-Black-Green heat map.
A set of 960 features were chosen for classification including 811 found significant in class comparison tests between 0 and 5 Gy at p < 0.001, 61 previously reported in literature (7, 10, 11, 14), 80 having relatively high abundances (a peak intensity > 100) and 6 selected based on temporal profiles. Random Forests (RF) analysis (5, 7, 15, 16) was done using the random forest package (ver 4.6-10), in R environment to evaluate classification performance. The class prediction (out of bag) errors were computed using 10000 trees where each tree was constructed using a subset of samples drawn randomly. The nodes were split by 30 randomly drawn features and the samples that were not included in the tree were used to test classification error (see supplementary methods, Random Forest Analysis). The features were sorted by Gini importance scores and those having high scores were determined. For each time point, 9 different classifiers (C-IR, C-IRB, C-IRH, C-IRS, IRB-IRH, IRB-IRS, IRH-IRS, IRB-IRH-IRS and C-IRB-IRHIRS) were evaluated (Supplementary Table S4) and the ranks of Gini importance scores were used to choose the features for further evaluation.
Putative Metabolite Identification through Interpretation of Tandem MS Data
In order to facilitate feature identities, two approaches were taken. First, public databases including METLIN (17) and HMDB (18) were searched using accurate mass and a mass error window of 10 ppm. Second, tandem MS data was obtained for the four important features and compared with available spectra or interpreted manually.
Results
Kaplan-Meier curves for the two groups of mice are shown in Supplementary Figure S1. As expected, TBI (5.4 Gy) shortened the lifespan of C3H mice (IR (ave) curve). Individual survival curves are also shown for mice with different types of neoplasms: IRH, IRS, and IRB. As previously observed (19), survival was shortened most for irradiated mice presenting with hematopoietic neoplasms, which was the first neoplasm to emerge following TBI (data not shown).
We first questioned whether there was a difference in the feature profile of 0 Gy versus 5.4 Gy treated animals at 1 year post-TBI. At this time point there was a significant number of 5.4 Gy mice with hematopoietic neoplasms (Supplementary Figure S1); however, there were no neoplasms evident in the 0 Gy control group. The number of mice evaluable for metabolomic analysis, which had both individual pathology data and multiple urine samples were 17 controls (0 Gy) and 51 irradiated mice (5.4 Gy) (Supplementary Table S1). As shown in Supplementary Figure S2, PLS-DA analysis clearly separated the two groups.
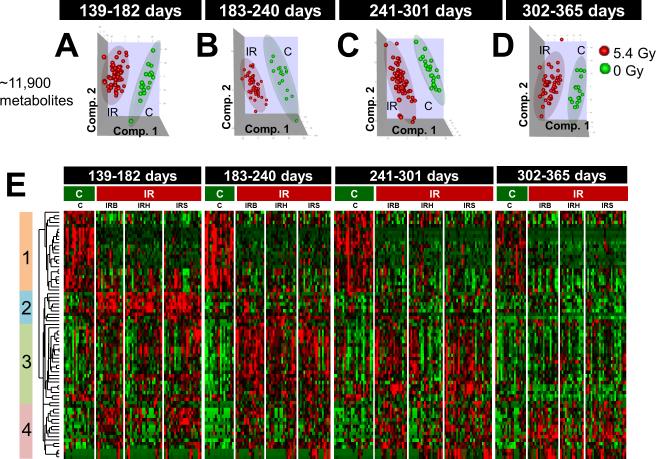
Since a major objective of this study was to identify features early post-IR that might predict for cancer, we choose to evaluate four different time periods to 1-year post-IR (Supplementary Table S2). As can be seen in Supplementary Figure S1 days 139-182 covers a period where one death was recorded; whereas, for days 302-365 approximately 30% of the 5.4 Gy mice had died. Using 11,900 features, PLS-DA analysis showed a significant difference between 0 Gy and 5.4 Gy mice in all four time periods (Figure 1A-D). The heat map is shown in Figure 1E for the 4 time periods (control and IR treated mice) using 73 features having significant differences between IR and C at p < 0.0001 in at least one of the four time groups (Supplementary Table S5). Further, the IR columns show the 3 types of tumors induced (IRB, IRH, and IRS). On the left side of the map are 4 clusters showing pattern differences between control and IR mice. It is clear that even during the first time period (139-182 days) there were significant differences in the feature profile pattern in clusters 1 and 2 between control and IR mice. Interestingly, the features in cluster 1 remained depressed compared to controls for up to 1 year post-IR. Cluster 3 shows differences in the first two-time periods; whereas, cluster 4 shows a heterogeneous response. It is clear that IR causes changes to the feature profile patterns that are clearly associated with cancer induction at early time periods before a macroscopic tumor forms (Supplementary Figures S1). Further analysis of the data was conducted to determine biomarkers that could be indentified that were predictive of IR exposure. Using 525 samples for PLS-DA analysis (up to 2 years of urine collection), four features having relatively high intensities were chosen for validation using MS/MS fragmentation analysis. As can be seen in Supplementary Figure S3A-C three of the metabolites were significantly different at a number of time points between the control and IR treated groups. One metabolite, putatively assigned as indoxyl sulfate (Supplementary Figure S3D), showed no difference between the control and IR samples over first year. A putatively identified taurine conjugated steroid (Supplementary Figure S3B), was significantly different over the entire time period examined and also was identified as one of the 73 features in the heat map analysis from Figure 1. This putatively identified new biomarker has not been reported in previous IR metabolomics studies to date (6-11). MHPG sulfate levels gradually decrease with time in IR exposed mice while it remained fairly constant in controls. The temporal changes were examined by PLSR analysis that revealed time dependent variance similar to the contrasts between C and IR (data not shown). However, attempts of modeling time as a continuous variable were unsuccessful in bringing out the contrast between 0 and 5 Gy because of complex time dependence of the profiles. Therefore, the one year time range was divided into 4 time groups to simplify the contrasts, yet preserving the temporal information. This procedure efficiently demonstrated the contrasts between the temporal profiles of C and IR, e.g., the difference of C and IR is apparent in dihydroxyquinoline levels (Supplementary Figure S3 C), while the difference in indoxyl sulfate levels are much smaller.
Figure 1.
Comparison of feature profiles of 5.4 Gy (IR) vs. 0 Gy (C). A total of 313 samples (C = 76 from 14 mice, IR = 237 from 48 mice) were grouped by the age of the animals in days as follows: (i) 139-182, (ii) 183-240, (iii) 241-301, and (iv) 302-365. (A – D): Scores plots of the first three components of OPLS-DA using ~11900 features that passed intensity filter. E. Heat map of 73 features at p < 0.0001 in at least one of the four time groups. Relative changes are shown after centering and scaling. Green – Black – Red scale is from −0.1 to +0.1 where black is mean value of the feature. Heat map indicates break up of C and IR animals including division by the phenotype of the animals developed benign tumors (IRB), hematopoietic neoplasms (IRH) and solid tumors (IRS). The vertical color bars on the left of heat map indicate clusters of features having similar patterns.
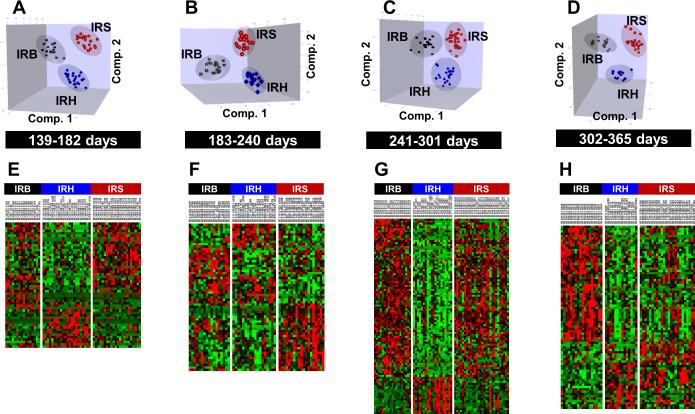
We next asked whether there were differences in feature profile patterns among the IR-induced neoplasm types. For example, in Figure 1 there were clusters where a distinct difference between the tumor types was evident (cluster 4, 1st time period, cluster 3, 3rd time period). Initially, using unsupervised clustering analysis we were unable to clearly separate the three neoplasm types. However, using PLS-DA analysis, in which each tumor type for each animal was known, it was possible to discern distinct patterns of features which could clearly separate the three neoplasm types as shown in Figure 2A-D. These data emphasize the importance of long-term lifespan studies to identify feature profiles that could indicate which animals were irradiated, but also the type of neoplasm induced. Shown in Figure 2E-H are heat maps of the top selected features obtained by template matching to distinguish IRB, IRH, and IRS (Supplementary Table S6). A striking feature was the emergence of a cluster of features, which clearly identified mice with hematopoietic neoplasms prior to any deaths caused by the tumor (Figure 2A, E). Further, both benign and solid neoplasms exhibited different cluster patterns over the time periods. These feature patterns occurred before any deaths were recorded due to these neoplasms.
Figure 2.
(A-D) Comparison of feature profiles in mice exposed to 5.4Gy radiation over four defined time periods within 1 year. The 3D graphs are the scores plots of OPLS-DA using ~11,900 features that passed the intensity filter in arbitrary orientations. (E-H) Heat maps of the top selected features by template matching to distinguish IRB, IRH and IRS (52, 61, 100, 79 features respectively over four defined time periods). Relative changes are shown after centering and scaling. Green – Black – Red scale is from −0.2 to +0.2 where black is mean value of the feature.
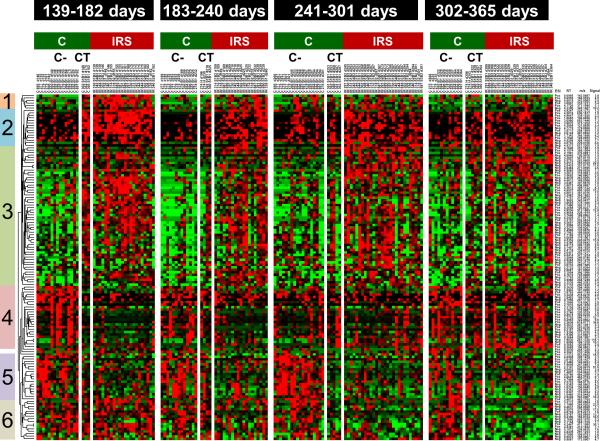
Additional analyses were conducted to determine if the feature profile obtained in control mice with solid tumors was the same or similar to IR treated mice with solid tumors. The analysis was limited due to low numbers of mice in the control group. As a result, the three control mice that died of solid neoplasms were combined with one mouse, which had a hematopoietic neoplasm, indicated as CT on the map in Figure 3. Using the features in Figure 2, we re-analyzed these features comparing only the IR solid tumor mice with controls with or without solid tumors. Despite the low number of control mice with neoplasms (n = 4), the feature profiles in the various clusters indicated that the features from control solid neoplasms mice were similar to the IR solid neoplasm mice. The data suggest that regardless of how a solid neoplasm forms (either spontaneously or induced by IR), the feature profiles are similar and indicative of the carcinogenesis process.
Figure 3.
Animals that developed solid tumors or hematopoietic neoplasms without the exposure to radiation (CT) also have feature profiles similar to those developed solid tumors by exposure to 5.4 Gy radiation (IRS). Wilcoxon rank sum tests indicated 139 features at p < 0.02 in at least a time group and OPLS-DA variable importance in projection (VIP) >2.5. Heat map indicates break up of C animals as having no-neoplasms (C-) and CT within each time group. Relative changes are shown by mean centering and scaling. Green – Black – Red scale is from −0.1 to +0.1 where black is mean value of the feature. Note the clusters having CT levels similar to IRS.
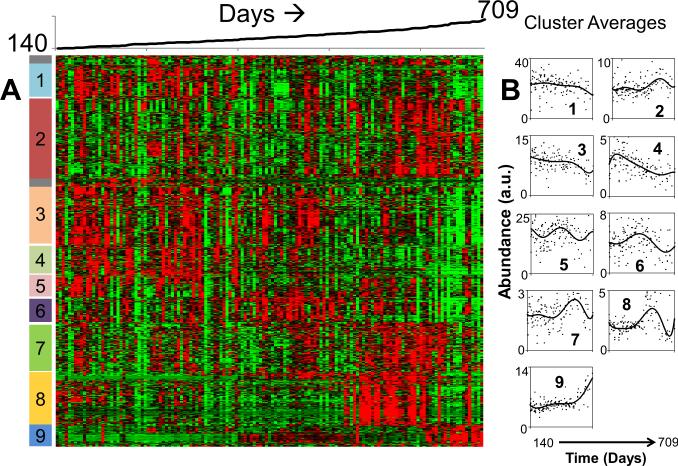
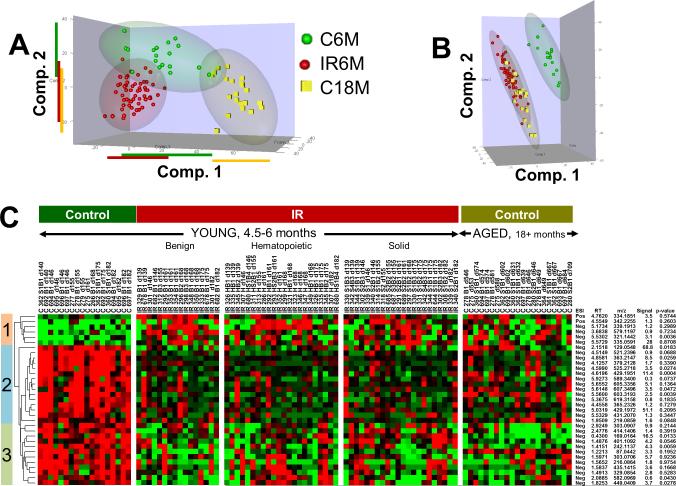
One of the hallmarks of IR exposure to mice is “life-shortening” or accelerated aging (20, 21, 21). In the current study temporal urine feature profiles were determined for 17 untreated mice over their lifespan, thus enabling an assessment of age-dependent changes in metabolism. Hierarchical clustering of about 6600 features above 0.1 intensity threshold level in at least 50% of 141 samples from all 17 control animals indicated distinct temporal profiles in the time period of 140-709 days. The heat map of a subset of 719 features having distinct temporal changes is shown in Figure 4A. Nine clusters were delineated that exhibited distinct changes over time. Cluster averages are shown in the plots to the right of the heat map a variety of patterns where features decreased, increased, or cycled over time (Figure 4B). These changes in urine features are not static, but in fact, vary considerably as the mice age. We next evaluated the feature profiles of young mice (control at 6 months), IR mice at 6 months, and old mice (control at 18 months) as shown in Figure 5. The PLS-DA analysis in Figure 5A shows a clear difference among the young control mice, young mice with IR treatment, and the older mice for the features selected. Further, the young IR mice are more closely associated with the old control mice as shown in Figure 5B. Both of these findings can be seen more clearly in the cluster map (Figure 5C) as shown for clusters 1, 2 and 3. These data suggest that IR amplifies “age-related” features similar to that seen in older mice.
Figure 4.
A) Heat map of 719 features selected from 6600 features measured in 17 control mice (0 Gy) having distinct temporal feature changes over days 140 to 709. Nine separate patterns were identified and marked on the right side of the heat map (red: high; green: low). B) Scatter plots of the mean intensities of each cluster as a function of time (cluster numbers are indicated on each plot).
Figure 5.
Relationship between 5.4 Gy radiation exposure and aging. Young animals are 139-182 days old, and Aged animals are 545-730 days old. A. Scores plot of the first three components of OPLS-DA using ~11900 features. B. Scores plot of OPLS-DA considering 5.4 Gy radiation exposed young animals and Control aged animals as one class. C. There are 33 features similar between Aged Controls and 5.4 Gy exposed Young animals. Heat maps indicate relative changes after mean centering and scaling. Green – Black – Red scale is from −0.1 to +0.1 where black is mean value of the feature. The p-values were calculated by Levene’s test for homogeneity of variance between aged animals and radiation exposed young animals. Signal column indicates maximum signal value among samples.
Random forest (RF) analysis was carried out on over 3500 features (selected by PLS-DA analysis) to select a smaller number of predictor features that classify the 9 separate control (C), IR, and the IR tumor groups over 4 time periods (36 groups total) (Supplementary Table S4). The first round of RF analysis using 250-274 features to split the node of each tree did produce a good probability (Prob 1) of classification for 4 of the groups (C vs IR (97% T1), C vs IRH (95% T1), C vs IRB (91% T1), and C versus IRS (97% T1)) but were less successful in classifying the IR tumor types from each other. However, the first RF analysis produced good classification for these groups in the other time periods as well (T2, T3, and T4). PLS-DA was performed by taking the first 275 features with the lowest Gini scores and was able to separate the 4 groups into individual clusters (data not shown). A second round of RF analysis using these 275 features improved the classification probabilities (Prob 2) for the other tumor groups (IRH, IRB, and IRS) (Supplementary Table S4). Table 1 lists the top 16 features which were selected by RF in all time periods analyzed and having low p-values (C versus IR groups) and 2 other features, which were significant in one or at most 2 time groups but still had significant p-values. The 16 features in Table 1 showed a pattern of decreased metabolite level versus control level at every time point examined. Two of these features are plotted as a function of each time period in Supplementary Figure S4 (A, B). The last 2 features in Table 1 had elevated abundances compared to control levels in at least one time group (Supplementary Figure S4 (C, D). Supplementary Figure S4 (E-H) also shows results for 4 features from our data set, which have been reported to be changed after acute IR exposure (11). These included creatinine (Supplementary Figure S4 E) and uric acid (Supplementary Figure S4 G).
Table 1.
Features having high classification scores between 0 and 5 Gy in Random Forest Analysis
| ESI mode | RT (min) | m/z | Rank score* | Highest Peak Intensity | Smallest p-value |
|---|---|---|---|---|---|
| - | 4.6196 | 429.1951† | 12 | 13.8916 | 1.3E-10 |
| - | 4.8581 | 363.2147 | 12 | 10.8167 | 1.74E-11 |
| - | 4.599 | 525.2718 | 12 | 5.2848 | 1.81E-10 |
| - | 5.56 | 603.3193 | 12 | 3.6994 | 8.08E-08 |
| - | 4.1257 | 379.2128 | 12 | 3.4885 | 1.07E-10 |
| - | 5.6552 | 605.3356 | 12 | 3.4373 | 1.2E-07 |
| - | 4.5149 | 521.2396 | 12 | 1.0468 | 9.26E-12 |
| - | 4.6265 | 497.184 | 12 | 0.9409 | 9.62E-07 |
| + | 4.5885 | 549.2666 | 12 | 2.412 | 1.29E-07 |
| - | 4.4558 | 365.2326 | 11 | 2.0792 | 1.07E-06 |
| - | 3.7191 | 541.2681 | 11 | 0.7425 | 6.04E-07 |
| - | 4.5962 | 794.4357 | 11 | 0.3345 | 3.62E-05 |
| + | 4.6 | 297.2174 | 11 | 3.1995 | 3.79E-08 |
| + | 4.5858 | 544.313 | 11 | 0.9968 | 1.01E-06 |
| - | 5.6148 | 607.3496 | 10 | 3.3545 | 6.73E-06 |
| - | 4.9593 | 509.2739 | 10 | 1.4376 | 2.51E-09 |
| - | 5.5729 | 335.0591 | --- | 29.852 | 1.72E-09 |
| - | 2.5719 | 366.1552 | --- | 110.83 | 4.08E-05 |
Rank Score = The number of times the feature found in the 12 comparisons between 0 and 5.4 Gy including IRH and IRS. Feature used for random forest are about 250.
Putatively identified as taurine conjugated steroid acid
Discussion
Early detection of cancer can be an important determinant of successful treatment (4). While it has been known for many years that IR can cause cancer, there have been few studies directed toward identifying metabolites that would predict its emergence or presence. With the threat of IR exposure to large populations of people, particularly non-lethal exposures, the ability to identify individuals early before cancer appears would be advantageous. Likewise, cancer patients who have completed successful radiation treatment of their tumor have an increased risk of developing a second malignancy (22, 23). An early assay for cancer would also benefit this population.
The findings of the present study suggest that UHPLC-QTOFMS-based metabolomics coupled with sophisticated longitudinal data analysis can identify distinct patterns of urinary features that emerge from IR-treated mice that are predictive of cancer induction at a later time. When using unsupervised clustering (PCA) of control versus IR treated mice, we were unsuccessful in clearly separating these groups at time period. However, using supervised PLS-DR such separation was achievable. In particular, Figure 1E, clusters 1 and 2 clearly show different profiles for IR treated mice compared to controls. These patterns emerged well before the animal exhibited any observable health effects from the IR exposure or was diagnosed with a neoplasm. The feature profiles in cluster 1 further suggest that IR produces long-term changes in metabolism. Conducting a long-term study where fate of each animal with regard to cancer induction was documented enabled supervised clustering which associated the feature patterns with animals with known outcomes post-IR. This example emphasizes the need to use a variety of data analyses to extract meaningful biological outcomes. Determination of the chemical structures of these metabolites would be desirable in that knowing these might aid in elucidating the complex biochemical mechanisms associated with cancer induction, progression and/or accelerated aging. Given the large number of features studied and limited sample volumes available, only a few of the features in these clusters were identified by tandem MS as shown in Supplementary Figure S3. Three of the four metabolites identified decreased over the 1 year time period following IR exposure, while indoxyl sulfate showed no change. Changes in urine metabolites in rodents as a result of TBI over the dose range of 1-15 Gy with metabolite determinations to a maximum of 30 days post-TBI have been reported (7-11). Indole-3-carboxylic acid levels were reported to decrease following TBI measured 2 days post-TBI (11). A feature with similar m/z ratio and retention time to this metabolite was also decreased by TBI in the present study (Supplementary Figure S4D) indicating that IR-induced suppression of this metabolite could be detected within days following TBI and maintained at low levels out to 1 year post-IR (current study). Changes in dihydroxyquinoline suggest changes in bacterial metabolism as DHQ has been shown to be strongly associated with changes in the gut microbiota (24). Whether DHQ reflects a change in the gut microbiota composition or a change in gut microbiota metabolism remains to be determined. Decreased uric acid levels in the IR treated mice (Supplementary Figure S4G) were also observed. Uric acid is thought to be an antioxidant and this could contribute increased susceptibility to cancer induction/progression due to increased oxidative stress (25). Future studies will be focused on determining chemical structures of metabolites that either increase or decrease as a result of IR exposure.
Feature profile patterns that predicted different types of neoplasms was an unexpected finding. Figure 2 clearly indicates that there are different cluster patterns of features among the three neoplasm types; however, at this time it is not possible to assign a single specific metabolite with a particular neoplasm. Instead a linear combination of various features will be required to uniquely identify each neoplasm. Another question posed was whether there would be a difference in feature profiles between solid neoplasms induced by IR with spontaneously occurring solid neoplasms in unirradiated mice. Although the number of spontaneously occurring neoplasms in control mice was low, the feature profile was similar (Figure 3).
IR exposure can cause premature aging leading to a shortened lifespan. In the past the shortened lifespan was attributed primarily to the induction of neoplasms. A number of laboratories explored other contributions to IR-induced life shortening in the 1960s. For example, it had shown that young mice were better able to withstand cold-induced stress than older animals (26). Nine month old mice receiving TBI exhibited the same level of cold-induced stress as unirradiated 16 month old control mice (27), suggesting that TBI accelerated aging with respect to altered metabolic and biochemical response to cold stress. It is now clear that IR exposure is also associated with other age-related disorders including cardiovascular disease, stem cell depletion, and inflammation (28). A recent study comparing the feature profiles of young versus 2 year old mice showed distinct changes in glucose and fatty acid metabolism and redox homeostasis with aging (29). In the present study feature profiles of control mice were followed for the entire lifespan (Figure 4). Clearly feature profiles changed dramatically as the animals aged. As can be seen in Figure 5 comparing feature profiles for mice at 6 months of age varied considerably from mice at 18 months of age. Interestingly, the feature profile of mice between 4.5 and 6 months of age that received TBI closely resembled the profile of 18 month control mice. This profile was evident before the mice were diagnosed with neoplasms, suggesting that from a metabolic perspective, TBI accelerated aging. The reason for this change might be increased inflammatory processes, oxidative stress, or redox perturbations that can be associated with aging animals. More research will be required to fully support this notion as well as a concentrated effort to determine the chemical structures of metabolites altered.
What is clear from the data in the current study is that clusters of features, not necessarily single metabolites, predict for IR-induced neoplasms and neoplasm types. This conclusion was based on PLS-DA, ANOVA (heat maps), and template matching (Supplementary Methods). RF analysis provided a different perspective toward identifying groups of features important in predicting IR-induced neoplasm formation. RF allowed for a rapid classification of the various treatment groups with the ability to rank metabolites. Of the features shown in Table 1, four were found in cluster 1, Figure 1E. This cluster clearly shows a difference in feature profile among IR-treated mice and perhaps important biomarkers for future studies.
Supplementary Material
Kaplan-Meier survival plots for mice receiving 0 Gy or 5.4 Gy TBI. Curve labeled IR Ave is the average survival for mice with all three neoplasms. Survival curves are included for subsets of mice determined to have benign (IRB), hematopoietic (IRH), or solid (IRS) neoplasms. The vertical markings indicate two of the time periods within one year where there was one death among all the groups (139-182 days) or the time period of 302-365 days where more death had occurred.
Comparison of metabolite profiles of 5.4 Gy (IR) to 0 Gy (Control) using 525 samples (Control = 141 from 17 mice, IR = 384 from 51 mice). Scores plots of the first three components of OPLS-DA using \m=~\7100 features that passed intensity filter.
Temporal profiles of a few selected metabolites. A-D: Four metabolites previously reported in literature. A, creatinine, B, phenylacetyl glycine, C, uric acid, and D, xanthurenic acid. These were identified by m/z ratios and retention times given in previous reports. E-H: Four features identified by random forest classification.
Temporal profiles of 4 metabolic features identified by random forest analysis of control versus IR treated mice (taken from Table 1). A, B) metabolites that were reduced in the IR treated mice compared to control mice. C, D) features which were elevated in at least one time point post IR. E, F) time profiles of features having similar m/z ratios and retention times in this study with metabolites identified in the literature which were associated with IR-treated mice.
Six patterns of relative IRB, IRH and IRS levels considered for template matching. The features correlated to these templates at coefficient \x=gt\ 0.4 and having VIP\x=gt\1 in PLS-DA in each time group are shown in heat maps in Figure 2.
Acknowledgments
This research was supported by the NIAID Medical Countermeasures against Radiological and Nuclear Threats Program and the Intramural Research Program of the Center of Cancer Research, National Cancer Institute, National Institutes of Health and in part with federal funds from the NCI, NIH, under Contract No. HHSN261200800001E. The authors declare that there are no potential conflicts of interest.
References
- 1.Rios CI, Cassatt DR, Dicarlo AL, et al. Building the strategic national stockpile through the NIAID Radiation Nuclear Countermeasures Program. Drug Dev Res. 2014;75:23–8. doi: 10.1002/ddr.21163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pierce DA, Shimizu Y, Preston DL, Vaeth M, Mabuchi K. Studies of the mortality of atomic bomb survivors. Report 12, Part I. Cancer: 1950-1990. Radiat Res. 1996;146:1–27. [PubMed] [Google Scholar]
- 3.Thompson DE, Mabuchi K, Ron E, et al. Cancer incidence in atomic bomb survivors. Part II: Solid tumors, 1958-1987. Radiat Res. 1994;137:S17–67. [PubMed] [Google Scholar]
- 4.Etzioni R, Urban N, Ramsey S, et al. The case for early detection. Nat Rev Cancer. 2003;3:243–52. doi: 10.1038/nrc1041. [DOI] [PubMed] [Google Scholar]
- 5.Mathe EA, Patterson AD, Haznadar M, et al. Noninvasive urinary metabolomic profiling identifies diagnostic and prognostic markers in lung cancer. Cancer Res. 2014;74:3259–70. doi: 10.1158/0008-5472.CAN-14-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patterson AD, Lanz C, Gonzalez FJ, Idle JR. The role of mass spectrometry-based metabolomics in medical countermeasures against radiation. Mass Spectrom Rev. 2010;29:503–21. doi: 10.1002/mas.20272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tyburski JB, Patterson AD, Krausz KW, et al. Radiation metabolomics. 1. Identification of minimally invasive urine biomarkers for gamma-radiation exposure in mice. Radiat Res. 2008;170:1–14. doi: 10.1667/RR1265.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lanz C, Patterson AD, Slavik J, et al. Radiation metabolomics. 3. Biomarker discovery in the urine of gamma-irradiated rats using a simplified metabolomics protocol of gas chromatography-mass spectrometry combined with random forests machine learning algorithm. Radiat Res. 2009;172:198–212. doi: 10.1667/RR1796.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson CH, Patterson AD, Krausz KW, et al. Radiation metabolomics. 4. UPLC-ESI-QTOFMS-Based metabolomics for urinary biomarker discovery in gamma-irradiated rats. Radiat Res. 2011;175:473–84. doi: 10.1667/RR2437.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laiakis EC, Hyduke DR, Fornace AJ. Comparison of mouse urinary metabolic profiles after exposure to the inflammatory stressors gamma radiation and lipopolysaccharide. Radiat Res. 2012;177:187–99. doi: 10.1667/rr2771.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goudarzi M, Mak TD, Chen C, Smilenov LB, Brenner DJ, Fornace AJ. The effect of low dose rate on metabolomic response to radiation in mice. Radiat Environ Biophys. 2014;53:645–57. doi: 10.1007/s00411-014-0558-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Warrack BM, Hnatyshyn S, Ott KH, et al. Normalization strategies for metabonomic analysis of urine samples. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877:547–52. doi: 10.1016/j.jchromb.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 13.Barr DB, Wilder LC, Caudill SP, Gonzalez AJ, Needham LL, Pirkle JL. Urinary creatinine concentrations in the U.S. population: implications for urinary biologic monitoring measurements. Environ Health Perspect. 2005;113:192–200. doi: 10.1289/ehp.7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manna SK, Patterson AD, Yang Q, et al. UPLC-MS-based urine metabolomics reveals indole-3-lactic acid and phenyllactic acid as conserved biomarkers for alcohol-induced liver disease in the Ppara-null mouse model. J Proteome Res. 2011;10:4120–33. doi: 10.1021/pr200310s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Breiman L. Random forests. Machine Learning. 2001;45:5–32. [Google Scholar]
- 16.Patterson AD, Slanar O, Krausz KW, et al. Human urinary metabolomic profile of PPARalpha induced fatty acid beta-oxidation. J Proteome Res. 2009;8:4293–300. doi: 10.1021/pr9004103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith CA, O'Maille G, Want EJ, et al. METLIN: a metabolite mass spectral database. Ther Drug Monit. 2005;27:747–51. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- 18.Wishart DS, Knox C, Guo AC, et al. HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res. 2009;37:D603–10. doi: 10.1093/nar/gkn810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitchell JB, Anver MR, Sowers AL, et al. The antioxidant tempol reduces carcinogenesis and enhances survival in mice when administered after nonlethal total body radiation. Cancer Res. 2012;72:4846–55. doi: 10.1158/0008-5472.CAN-12-1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.LINDOP PJ, ROTBLAT J. Shortening of life and causes of death in mice exposed to a single whole-body dose of radiation. Nature. 1961;189:645–8. doi: 10.1038/189645a0. [DOI] [PubMed] [Google Scholar]
- 21.Metalli P. Life span shortening. Int J Radiat Oncol Biol Phys. 1979;5:1123–30. doi: 10.1016/0360-3016(79)90629-1. [DOI] [PubMed] [Google Scholar]
- 22.Ng AK, Kenney LB, Gilbert ES, Travis LB. Secondary malignancies across the age spectrum. Semin Radiat Oncol. 2010;20:67–78. doi: 10.1016/j.semradonc.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berrington de Gonzalez A, Gilbert E, Curtis R, et al. Second solid cancers after radiation therapy: a systematic review of the epidemiologic studies of the radiation dose-response relationship. Int J Radiat Oncol Biol Phys. 2013;86:224–33. doi: 10.1016/j.ijrobp.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li F, Jiang C, Krausz KW, et al. Microbiome remodelling leads to inhibition of intestinal farnesoid X receptor signalling and decreased obesity. Nat Commun. 2013;4:2384. doi: 10.1038/ncomms3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roszkowski K, Gackowski D, Rozalski R, et al. Small field radiotherapy of head and neck cancer patients is responsible for oxidatively damaged DNA/oxidative stress on the level of a whole organism. Int J Cancer. 2008;123:1964–7. doi: 10.1002/ijc.23700. [DOI] [PubMed] [Google Scholar]
- 26.GRAD B, KRAL VA. The effect of senescence on resistance to stress. I. Response of young and old mice to cold. J Gerontol. 1957;12:172–81. doi: 10.1093/geronj/12.2.172. [DOI] [PubMed] [Google Scholar]
- 27.TRUJILLO TT, SPALDING JF, LANGHAM WH. A study of radiation-induced aging: response of irradiated and nonirradiated mice to cold stress. Radiat Res. 1962;16:144–50. [PubMed] [Google Scholar]
- 28.Richardson RB. Ionizing radiation and aging: rejuvenating an old idea. Aging (Albany NY) 2009;1:887–902. doi: 10.18632/aging.100081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Houtkooper RH, Argmann C, Houten SM, et al. The metabolic footprint of aging in mice. Sci Rep. 2011;1:134. doi: 10.1038/srep00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Kaplan-Meier survival plots for mice receiving 0 Gy or 5.4 Gy TBI. Curve labeled IR Ave is the average survival for mice with all three neoplasms. Survival curves are included for subsets of mice determined to have benign (IRB), hematopoietic (IRH), or solid (IRS) neoplasms. The vertical markings indicate two of the time periods within one year where there was one death among all the groups (139-182 days) or the time period of 302-365 days where more death had occurred.
Comparison of metabolite profiles of 5.4 Gy (IR) to 0 Gy (Control) using 525 samples (Control = 141 from 17 mice, IR = 384 from 51 mice). Scores plots of the first three components of OPLS-DA using \m=~\7100 features that passed intensity filter.
Temporal profiles of a few selected metabolites. A-D: Four metabolites previously reported in literature. A, creatinine, B, phenylacetyl glycine, C, uric acid, and D, xanthurenic acid. These were identified by m/z ratios and retention times given in previous reports. E-H: Four features identified by random forest classification.
Temporal profiles of 4 metabolic features identified by random forest analysis of control versus IR treated mice (taken from Table 1). A, B) metabolites that were reduced in the IR treated mice compared to control mice. C, D) features which were elevated in at least one time point post IR. E, F) time profiles of features having similar m/z ratios and retention times in this study with metabolites identified in the literature which were associated with IR-treated mice.
Six patterns of relative IRB, IRH and IRS levels considered for template matching. The features correlated to these templates at coefficient \x=gt\ 0.4 and having VIP\x=gt\1 in PLS-DA in each time group are shown in heat maps in Figure 2.