Abstract
The ability of cancer cells to evade apoptosis is dictated by a shift in the balance between pro- and anti-apoptotic gene expression programs. Monocyte chemotactic protein induced protein 1 (MCPIP1) is a zinc finger RNA binding protein with important roles in mediating inflammatory responses. Overexpression of MCPIP1 in different cancer cell types has been implicated in eliciting an antitumor response, but a direct role of MCPIP1 in apoptosis has not been established. In this study, we demonstrate that MCPIP1 functions as a potent tumor suppressor that induces apoptosis of breast tumor cells by selectively enhancing mRNA decay of anti-apoptotic gene transcripts including Bcl2L1, Bcl2A1, RelB, Birc3, and Bcl3. Mechanistically, MCPIP1 physically interacted with a stem-loop structure in the 3'UTR of these transcripts through its PIN domain, causing mRNA destabilization. Furthermore, we found that MCPIP1 expression was repressed in breast tumor cells, and overexpression of MCPIP1 induced apoptosis, whereas its depletion enhanced cancer cell proliferation. Moreover, MCPIP1 induction in vivo resulted in complete regression of established tumors and a significant reduction in metastatic disease. Notably, low MCPIP1 expression in tumor samples from breast cancer patients was strongly associated with poor survival over 13 years of follow up. Collectively, our results highlight MCPIP1 is a new tumor suppressor in breast cancer that induces cell death by tipping the balance in favor of pro-apoptotic gene expression.
Keywords: Breast cancer, RNA-binding protein, Tumor suppressor, mRNA decay, Apoptosis
Introduction
A hallmark of cancer is that cancer cells can evade apoptosis and proliferate uncontrollably (1,2), which also plays a significant role in their resistance to conventional therapeutic regimens (3,4). While recent progress has broadened our understanding of the apoptosis-evasion mechanisms by cancer cells, how apoptosis resistance develops in cancer cells through posttranscriptional mechanisms, especially by the newly discovered monocyte chemotactic protein induced protein 1 (MCPIP1), remains unknown.
MCPIP1, a Cys-Cys-Cys-His-type zinc finger protein encoded by the zc3h12a gene, was initially discovered as the most highly induced mRNA by monocyte chemotactic protein-1 (MCP-1) in human peripheral blood monocytes (5). MCPIP1 is rapidly induced in macrophages upon stimulation with proinflammatory molecules, such as TNF, IL-1β, and LPS (6-8). MCPIP1 has RNase activity and inhibits the expression of proinflammatory cytokines (IL-1, IL-6, and IL-12) by binding to their 3’UTRs for mRNA degradation. MCPIP1 is also named as Regnase-1 based on the RNase activity (8). In addition, MCPIP1 can act like a brake for T cell activation (9). Therefore, MCPIP1 is believed to be a key negative regulator involved in the control of inflammation and maintenance of homeostasis. Mice deficient of MCPIP1 develop a complex phenotype, including autoimmune disorders, anemia, and a severe inflammatory response (8,10). It is recently reported that MCPIP1 degrades viral RNA and thus acts as a host defense against virus infection (11-13). MCPIP1 also involves in controlling cytokines-induced endothelial inflammation (14) and inducing endothelial dysfunction (15). However, it remains unknown whether MCPIP1 plays a role in cancer progression and apoptosis evasion.
Apoptosis plays an important role in many diseases, including cancer (16,17). Though the mechanisms of apoptosis are complex and involve many pathways, the ratio of pro-apoptotic to anti-apoptotic genes determines whether cancer cells undergo apoptosis or survival (18). Most tumor cells evade apoptosis by either increasing the expression of anti-apoptotic genes or decreasing the expression of pro-apoptotic genes. Overexpression of anti-apoptotic proteins in the BCL2 family is associated with a poor cancer prognosis (19,20). Therefore, current efforts are ongoing to interfere with BCL2 and its fellow pro-survival family members to help restore the sensitivity of cancer cells to pro-apoptotic signals.
We have previously shown that overexpression of MCPIP1 sensitizes mouse macrophages for apoptosis in response to stress signals (21). Treatment of HeLa and HepG2 cells with proteasome inhibitor MG-132 reduces cell viability along with MCPIP1 expression (22). In human neuroblastoma cells MCPIP1 overexpression decreases cell viability and proliferation (23). MCPIP1 also stabilizes RGS2 protein through its deubiquitinase activity to suppress breast cancer cell growth (24). In this study, we identify that MCPIP1 is a potent tumor suppressor by inducing tumor apoptosis through selectively suppressing the expression of anti-apoptotic gene transcripts, including Bcl2L1, Bcl2A1, RelB, Birc3, and Bcl3. The expression of MCPIP1 is repressed in breast tumor cells that may help them evade apoptosis. Ectopic expression of MCPIP1 triggered apoptosis of breast tumor cells, whereas further suppression of MCPIP1 expression with shRNA enhanced tumor cell proliferation. In agreement with these observations, MCPIP1 expression in vivo abolished existing tumors and significantly reduced metastases. By surveying a gene array dataset derived from the excised breast tumors of 251 patients (25), we found that low MCPIP1 levels correlated strongly with poor survival of breast cancer patients over 13 years of follow up. These results suggest that MCPIP1 is a potent tumor suppressor involved in regulating apoptotic pathway through suppression of anti-apoptotic gene expression.
Materials and Methods
Mice
6~8 week old female Balb/c mice and NSG mice were obtained from The Jackson Laboratories and respectively housed in cages with filter tops in a laminar flow hood, fed food and acid water ad libitum and in pathogen-free condition. All experimental procedures were performed with the approval of the IACUC at Saint Louis University.
Cells and Plasmids
MDA-MB-231, MDA-MB-453, MCF-10A, MCF-12A, 4T1, Ts/A, and HEK293 cells were obtained from ATCC and maintained in DMEM with 10% FBS. Mouse mammary gland epithelial cells FSK4 and CommD were kindly provided by Janet S Butel and cultured as described (26). Isogenic tumorigenic line 67NR, 168FARN, 4TO7, 66cl4 and 4T1 were kindly provided by Yibin Kang and cultured as described (27). Retro-X Tet-On 3G inducible MCPIP1 cell lines were established in 4T1 and MDA-MB-231 cells according to the manufacturer's instruction (Clontech). A set of luciferase-expressing 3’UTR reporter plasmids (BCL2L13’UTR, BCL2A13’UTR, BIRC33’UTR, RELB3’UTR, BCL33’UTR, and β-ACTIN3’UTR) were cloned by inserting their 3’UTRs into the pGL3 control vector (Promega) between XbaI and FseI sites. For stem-loop deletion constructs, mutated and truncated BCL2L1 3’UTR (1-1559 bp) and BIRC3 3’UTR (1-544 bp) were amplified, sequenced, and inserted into pGL3 control vector as described above.
qPCR and PCR Array
The TaqMan® Human Apoptosis PCR Array (Life Technologies) was used to analyze apoptosis-related gene expression. Total RNA was extracted with Trizol from MCPIP1-expressed and -unexpressed MDA-MB-231 cells, purified using the RNA Cleanup Kit (Fisher Scientific), reverse-transcribed to cDNA, and added to each well of the PCR Array plates combined with TaqMan Master Mix according to the manufacturer's instructions. Data analysis was based on the ΔΔCt method, with normalization to three different housekeeping genes. The genes in the array and primer sequences used are described in Supplemental Table 1 and 2.
RNA Immunoprecipitation
GFP/MCPIP1 fusion protein was induced by Dox for 30 h in MDA-MB-231/Tet-On cells. Then cells were lysed and whole cell lysates pre-cleared with IgG, followed by incubation with anti-GFP antibody at 40C for 4 h. The RNA-protein complexes were pulled-down by protein A/G agarose beads and RNA extracted with Trizol, followed by detecting apoptosis-related genes with RT-PCR.
mRNA Stability
MCPIP1 was induced by Dox in MDA-MB-231/Tet-On cells for 36 h. Then 5 μg/ml of ActD and 5 μg/ml of DRB were added to block de novo RNA synthesis. Total RNA was harvested at indicated time points and mRNA expression analyzed by qRT-PCR. The mRNA levels were normalized to GAPDH mRNA and half-life of the mRNA determined by comparing to the levels of mRNA before adding ActD and DRB.
shRNA Lentivirus
Three lentiviral shRNAs targeting human MCPIP1 mRNA and the lentiviral shRNAs targeting the mRNAs of BCL2, RELB, BCL3, FAS and DEDD2 were obtained from Sigma (St. Louis, MO). The #1 MCPIP1 shRNA targets CDS (NM_025079.1-1777s1c1), #2 targets 3’UTR (NM_025079.1-2532s1c1), and #3 targets CDS (NM_025079.1-260s1c1). A scramble non-targeting shRNA was used as control. Lentiviral particles were packaged in HEK293T cells by co-transfecting shRNA-pLKO.1, pCMV-dR8.2, and pMD2.G constructs. 48 h later, supernatants were collected and centrifuged to discard cell debris. For infection, virus supernatants with 1 μg/ml Polybrene were added to MDA-MB-231 and MDA-MB-453 cells for overnight. After two rounds infection, the infected cells were selected with puromycin (2.5 μg/ml) for 2 weeks, followed by experimental analysis.
Immunofluorescence Staining
Cells in cover slips were fixed with 4% paraformaldehyde and permeabilized with 0.5% Triton X-100 in PBS. After blocking, cells were probed with primary antibody overnight and then incubated with Alexa Fluor 488- or 594-conjugated secondary antibodies. Nuclei were counterstained with DAPI. The images were taken by confocal fluorescence microscopy.
Immunohistochemistry and TUNEL Staining
Tumor and lung tissues were stained with hematoxylin and eosin (H&E). Images were taken and analyzed using Zeiss Imaging System. For frozen section, tumor tissues were embedded with Tissue-Tek OCT compound (VWR) and slowly submerged into liquid nitrogen. The frozen sections were placed on polylysine-treated glass slides and used for GFP, MCPIP1 immunofluorescent staining, and TUNEL staining (Roche).
Luciferase Reporter Assays
Luciferase assay was performed as described previously (28). All transfections were performed in triplicate and repeated at least three times. pGL3 luciferase reporter plasmids containing 3’UTRs of different genes were transfected into HEK293 cells along with Flag-tagged MCPIP1, Flag-ΔZF (zinc finger), Flag-ΔPIN (D141N) and Flag-control plasmids. The luciferase activity was measured 36 h after transfection using a dual-luciferase reporter assay system (Promega).
6-Thioguanine Lung Metastasis Single Clone Selection
Lungs of tumor-bearing mice were minced and digested with 100 U/ml Collagenase IV (Sigma). Digested lung tissues were passed through a 70-μm strainer. Filtered cell suspension was serially diluted in a 1:10 ratio and plated into 15 cm plates. 60 μM of 6-thioguanine (Sigma) was added for selection of drug-resistant 4T1 tumor cells for 2 weeks. After counted the number of colonies, Dox was added to induce GFP/MCPIP1 expression.
Statistical Analysis
All data are presented as mean plus SD. Unless indicated otherwise, statistical analysis was performed with the Student's t test. Statistical significance was defined as a p value of <0.05.
Results
MCPIP1 expression is reduced in breast tumors and breast tumor cell lines
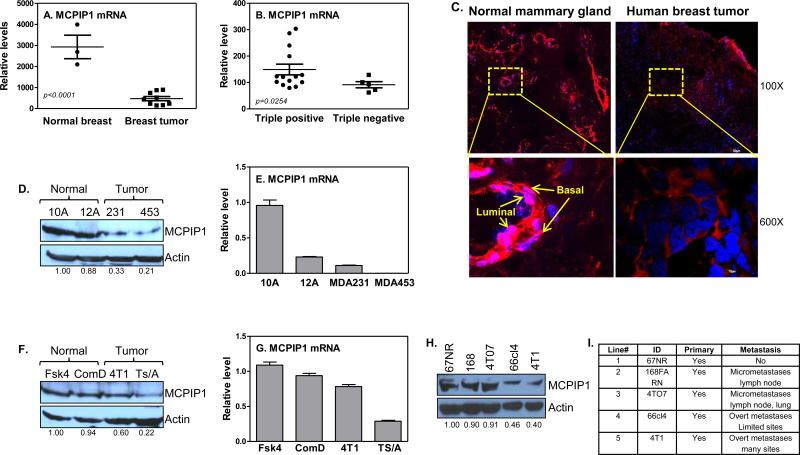
To determine the levels of MCPIP1 in breast tumors, we obtained fresh tumor tissues and the surrounding ‘healthy’ tissues from breast cancer patients undergoing surgery and measured MCPIP1 mRNA expression. MCPIP1 mRNA expression was significantly reduced in tumors compared with ‘healthy’ tissues (Figure 1A). Since triple negative breast tumors have the worst outcome, we further compared MCPIP1 levels between triple negative (ER−/PR−/HER2−) and positive (ER+/PR+/HER2+) tumors. MCPIP1 expression was significantly lower in triple negative tumors than it in triple positive tumors (Figure 1B). Immunofluorescence staining of MCPIP1 protein indicated a lower MCPIP1 levels in breast tumors compared to normal mammary gland tissues, with expression of the MCPIP1 in both luminal and basal/myoepithelial cells of the normal mammary gland tissues (Figure 1C). MCPIP1 levels were also significantly reduced in breast tumors of nude mice induced with MDA-MB-231 cells compared to normal mammary glands (Supplemental Figure 1A). To determine if breast tumor cells had MCPIP1 expression repressed, we examined MCPIP1 in human MDA-MB-231 and MDA-MB-453 as well as mouse 4T1 and TS/A breast tumor cell lines and compared them to normal mammary gland epithelial cells. MCPIP1 protein (Figure 1D) and mRNA (Figure 1E) were significantly reduced in human breast tumor cells compared to it in normal mammary gland epithelial cells. A similar reduction of MCPIP1 protein (Figure 1F) and mRNA (Figure 1G) were also displayed in mouse breast tumor cells. Immunofluorescence staining of MCPIP1 and detection of MCPIP1 by Western blot and qRT-PCR also indicated a lower MCPIP1 levels in MCF7 (Supplemental Figures 1C-E) and 4T1 (Supplemental Figure 1B) breast tumor cells compared to MCF-12A and CommD cells, respectively, suggesting that the impaired MCPIP1 expression in tumor mass may be due to MCPIP1 reduction in tumor cells. To determine whether MCPIP1 levels were associated with tumor aggressive feature, we measured MCPIP1 in five isogenic tumorigenic lines (67NR, 168FARN, 4TO7, 66cl4, and 4T1), with each line having unique tumorigenic feature and increased metastatic capability in order (27). The most tumorigenic and metastatic 4T1 cells showed the least levels of MCPIP1 compared to the other metastatic cell lines (Figures 1H & I), indicating a correlation of MCPIP1 level with tumor progression.
Figure 1.
MCPIP1 expression is impaired in breast tumors and breast tumor cell lines. (A) MCPIP1 expression was measured by qRT-PCR in human breast tumor specimens (n=9) compared to surrounding ‘normal’ breast tissue (n=3). The tumors have mixture molecular subtypes, including three luminal A, two luminal B and four triple negative tumors. (B) MCPIP1 mRNA levels were measured in triple negative breast tumors (n=5) and compared to triple positive breast tumors (n=14). (C) Immunofluorescence staining of MCPIP1 with anti-MCPIP1 antibody (red color) in human breast tumor tissues and normal mammary gland tissues. MCPIP1 protein (D) and mRNA (E) levels were measured respectively by western blot and qRT-PCR in human breast tumor cell lines (MDA-MB-231 and MDA-MB-453) and human normal mammary gland epithelial cell lines (MCF-10A and MCF-12A). MCPIP1 protein (F) and mRNA (G) expression were measured respectively by western blot and qRT-PCR in mouse breast tumor cell lines (4T1 and Ts/A) and mouse normal mammary gland epithelial cell lines (Fsk4 and CommD). Immunoblotting analysis of MCPIP1 protein with anti-MCPIP1 antibody with whole cell lysates extracted from mouse breast tumor cell lines with different metastatic potential (H) and their characteristics (I).
MCPIP1 induces apoptosis of breast tumor cells
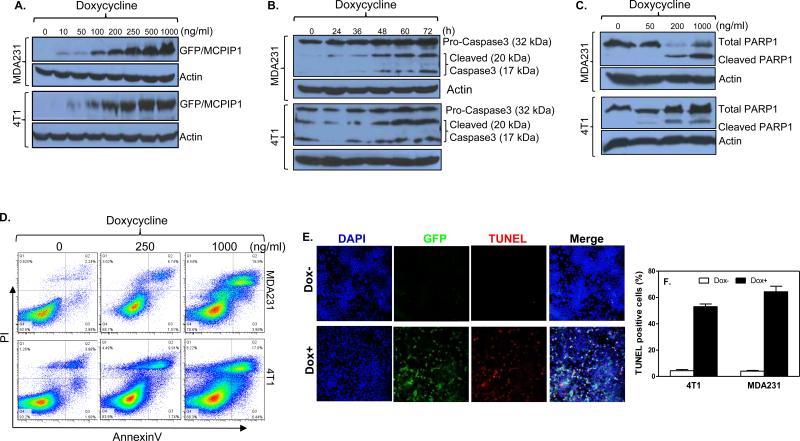
To study MCPIP1 function we generated inducible expression of MCPIP1 in human MDA-MB-231 and mouse 4T1 breast tumor cells with the Tet-on system. Addition of Doxycycline (Dox) dose-dependently induced GFP/MCPIP1 fusion protein in both cells (Figure 2A and Supplemental Figure 2A). When MCPIP1 was induced, we noticed a loss of both tumor cells (Supplemental Figures 2B&C). To determine whether MCPIP1 induced cell death by apoptosis, we measured the expression of two apoptosis markers, Caspase3 and PARP1, in cells expressing MCPIP1. MCPIP1, in a time-dependent and dose-dependent manner, caused cleavages of Caspase3 (Figure 2B) and PARP1 (Figure 2C) in MDA-MB-231 cells (upper panel) and 4T1 cells (lower panel). In addition, MCPIP1 caused an increase in PI+ and Annexin V+ apoptotic MDA-MB-231 and 4T1 tumor cells in a dose-dependent manner (Figure 2D). By TUNEL staining, we found more TUNEL+ apoptotic MDA-MB-231 cells (Figures 2E&F) and 4T1 cells (Supplemental Figure 2D) after MCPIP1 induction by Dox. These data clearly demonstrate that MCPIP1 induces apoptosis of breast tumor cells.
Figure 2.
MCPIP1 induces apoptosis of breast tumor cells. (A) MDA-MB-231 or 4T1 breast tumor cells were generated to express GFP/MCPIP1 fusion protein with a Tet-on inducible system. GFP/MCPIP1 fusion protein was measured by Immunoblotting with anti-GFP antibody after addition of different amounts of Doxycycline. (B) Caspase 3 and its cleaved products were measured by Immunoblotting with anti-caspase3 antibody at different times after adding 500 ng/ml of Dox to the above two Tet-On cells. (C) Poly (ADP-ribose) polymerase (PARP)-1 and its cleaved product were measured by Immunoblotting with anti-PARP1 antibody after addition of different amounts of Dox to the above cells. (D) Apoptosis was measured by flow cytometry in the above cells at 48 hours after adding different amounts of Dox. TUNEL staining was performed in MDA-MB-231/Tet-On cells at 48 hours after adding 500 ng/ml of Dox (Dox+). The ratio of TUNEL-positive cells were calculated and plotted on the histogram (F).
MCPIP1 targets anti-apoptotic genes for apoptosis
We next identified the genes in apoptosis pathway affected by MCPIP1 using the TaqMan® Human Apoptosis PCR Array kit (Life Technologies). In MCPIP1-expressing MDA-MB-231 cells, we identified 31 transcripts affected by MCPIP1 expression (Figure 3A), with 6 anti-apoptotic genes down-regulated and 25 pro-apoptotic genes up-regulated (Supplemental Table 1). To validate the array results, we measured the mRNAs of five down-regulated anti-apoptotic genes (Bcl2L1, Bcl2A1, Birc3, RelB, and Bcl3) and four up-regulated pro-apoptotic genes (Bad, Ripk2, Fas, and Dedd2) by real-time PCR. The expressions of five anti-apoptotic gene transcripts were reduced in a time-dependent manner in cells expressed MCPIP1 (Figures 3B-E). In contrast, no changes in pro-apoptotic BAD (Figure 3F) and RIPK2 (Figure 3G) transcripts and minor increase in FAS (Figure 3H) and DEDD2 (Figure 3I) transcripts were observed upon MCPIP1 induction. To determine whether MCPIP1 bound to these anti-apoptotic transcripts, we first induced GFP/MCPIP1 fusion protein in MDA-MB-231/Tet-On cells with Dox, and then pull-downed the MCPIP1-binding complex with anti-GFP antibody from cytoplasmic extracts (29), followed by detecting the bound mRNAs by RT-PCR. All five anti-apoptotic gene mRNAs were amplified by PCR but not the GAPDH mRNA (Figure 3J) and the pro-apoptotic mRNAs (Figure 3K). IL-6 was served as positive control (Supplemental Figure 3B). These data indicate that MCPIP1 preferentially inhibits the mRNA expression of anti-apoptotic genes for inducing apoptosis in breast tumor cells.
Figure 3.
MCPIP1 selectively inhibits the mRNA expression of anti-apoptotic genes. (A) Clustergram of the apoptosis related genes affected differentially in MDA-MB-231/Tet-On cells at 32 hours after adding Dox (500 ng/ml). MCPIP1: cells with Dox; Control: cells without Dox. MDA-MB-231/Tet-On cells were induced to express MCPIP1 by Dox (500 ng/ml) for different times. Then total RNA was collected to detect mRNA levels of selected anti-apoptotic genes by qRT-PCR, including bcl2l1 (B), bric3 (C), bcl3 (D), and relb (E). The mRNA levels of pro-apoptotic genes in the above cells were measured by qRT-PCR, including bad (F), ripk2 (G), fas (H), and dedd2 (I). MDA-MB-231/Tet-On cells were induced to express GFP/MCPIP1 fusion protein by Dox (500 ng/ml) for 36 h, followed by lysate extraction, immunoprecipitation with anti-GFP antibody or IgG, and RNA extraction. RT-PCR was performed to detect anti-apoptotic gene transcripts (J) and pro-apoptotic gene transcripts (K). IL-6 transcript was used as positive control and GAPDH served as negative control.
MCPIP1 targets the mRNA of anti-apoptotic genes for degradation via the PIN domain
Since MCPIP1 inhibits the mRNA expression of anti-apoptotic genes, we speculated that MCPIP1 might trigger apoptosis through destabilizing the mRNAs of these anti-apoptotic genes. We first induced MCPIP1 expression and then blocked de novo synthesis of mRNA with Actinomycin D and DRB, followed by measuring the remaining mRNAs at different time points. The half-lives of anti-apoptotic mRNAs, including Bcl2L1 (Figure 4A), Bcl3 (Figure 4B), BIRC3 (Figure 4C), RelB (Figure 4D), and Bcl2A1 (Supplemental Figure 4A), were shorten about two folds in MCPIP1-expressing cells (Dox+) compared to cells without MCPIP1 induction (Dox−), whereas MCPIP1 expression had little effects on half-lives of the mRNAs encoding pro-apoptotic DEDD2, BAD, FAS, LTA, and RIPK2 (Supplemental Figures 4B-F), demonstrating that MCPIP1 induces apoptosis through directly targeting mRNAs of the anti-apoptosis genes but not pro-apoptosis gene for degradation.
Figure 4.
MCPIP1 destabilizes the mRNAs of anti-apoptotic genes via the PIN domain. MDA-MB-231/Tet-On cells were treated with 500 ng/ml of Dox for 36 hours to induce MCPIP1 expression and then ActD and DRB added for different times. The remained anti-apoptotic gene transcripts, bcl2l1 (A), bcl3 (B), birc3 (C), and relb (D), were measured by qRT-PCR. HEK293 cells were co-transfected with luciferase reporter constructs containing full-length 3’UTRs of the anti-apoptotic genes and either MCPIP1 or control vector. Luciferase activities were measured in cell lysates after 36 h. β-actin 3’UTR was used as negative control. Results shown represent the mean ± SD of four independent experiments (*: p< 0.05). (F) Schematic representation of the RNase and zinc finger domains in MCPIP1, and their mutant, D141N and ΔZF. (G) HEK293 cells were co-transfected with luciferase reporters containing 3’UTRs of the anti-apoptotic genes and WT MCPIP1 as well as the two mutants. After 36 h, luciferase activities were measured in cell lysates and compared to empty vector transfected cells. (H) MDA-MB-231 cells were transiently transfected with MCPIP1 and two mutants, respectively. 36 h later, total RNA was extracted to measure the mRNA expression of different genes as indicated by qRT-PCR. (I) MDA-MB-231 cells were transiently transfected with MCPIP1 and two mutants. 48 h later, floating dead cells were counted in each group. n=3. (J) MDA-MB-231 cells were transiently transfected with MCPIP1 and two mutants. 48 h later, whole cell lysates were collected to measure total PARP1 and cleaved PARP1 by western blot.
To determine if the mRNA decay was mediated through 3’UTR, we cloned the 3’UTRs of Bcl2L1, RelB, BIRC3 and Bcl3 downstream of the luciferase gene as previously described (28), and then co-transfected the 3’UTR reporter constructs with MCPIP1 expression plasmid into HEK293 cells, followed by measuring luciferase activity. As shown in Figure 4E, MCPIP1 suppressed luciferase activities of all four 3’UTR reporter constructs compared to cells transfected with control vector. Since β-actin mRNA is quite stable and not affected by its 3’UTR, MCPIP1 did not affect its luciferase activity. Luciferase mRNA linked to the 3’UTRs of Bcl2L1 and BRIC3 was also pulled down by GFP antibody (Supplemental Figure 4G). These results indicate that MCPIP1 acts through the 3’UTR to decay anti-apoptotic gene transcripts.
MCPIP1 protein contains a PIN-like domain which is reported to have an RNase activity and a zinc finger domain thought to interact with AU-rich elements (ARE) located in the 3’UTR of mRNAs (8,10). To determine which domains in MCPIP1 are responsible for mRNA decay, we mutated the PIN domain (D141N) and the ZF domain (ΔZF) as illustrated in Figure 4F, and respectively co-transfected the mutants as well as wild type MCPIP1 with different 3’UTR reporter constructs into HEk293 cells. Wild type and ZF mutant MCPIP1 suppressed the luciferase activities of all four 3’UTR reporter constructs (Figure 4G) and their mRNA expression (Figure 4H), while D141N mutant MCPIP1 completely loosed its suppressive effects. In addition, MDA-MB-231 cells transfected with the D141N mutant MCPIP1 did not cause cell death (Figure 4I) and failed to cleave the PARP1 (Figure 4J). These results demonstrate that the PIN domain in MCPIP1 is responsible for mRNA decay of the anti-apoptotic genes and for apoptosis induction.
MCPIP1 depletion stabilizes anti-apoptotic gene transcripts and promotes tumor cell proliferation
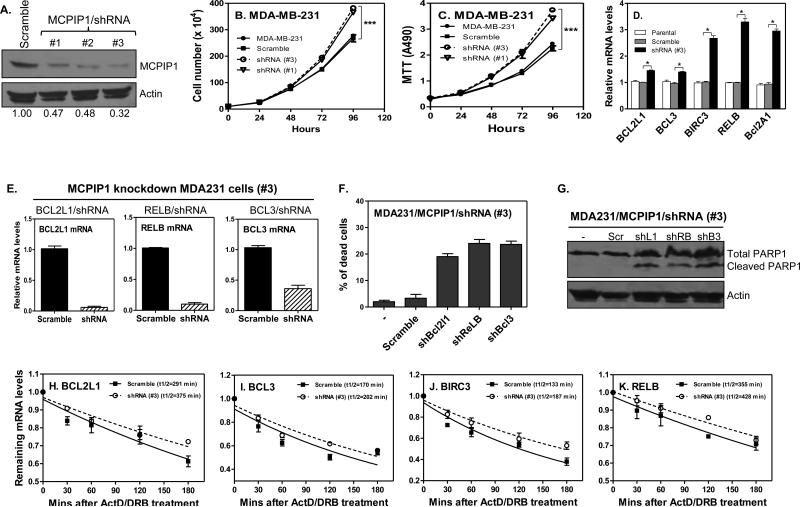
To confirm the inductive effects of MCPIP1 on tumor apoptosis, we further reduced MCPIP1 expression with shRNA in MDA-MB-231 cells. The MCPIP1 protein was respectively knocked-down about 53%-68% by three shRNAs (Figure 5A). Therefore, we used the #1 and #3 shRNA/MCPIP1 for subsequent study. Though MCPIP1 expression is repressed in tumor cells, further knocking-down MCPIP1 by shRNA significantly enhanced tumor cell proliferation as indicated by cell counting (Figure 5B) and by MTT assay (Figure 5C). In agreement with the overexpression results, knocking-down MCPIP1 expression further increased the mRNA expression of anti-apoptotic genes, including bcl2l1, bcl3, birc3, relb and bcl2a1 (Figure 5D), while the mRNA of pro-apoptotic genes (bad, ripk2, fas and dedd2) did not change between MCPIP1 knocked-down and scramble cells (data not shown). To confirm whether the pro-apoptotic genes are involved in MCPIP1-induced apoptosis, we knocked down FAS and DEDD2 by shRNA lentivirus in MDA-MB-231/Tet-on cells. After confirmed their knockdown efficiency (Supplemental Figure 5A), we checked their effects on apoptosis by measuring cell survival and PARP1 cleavage. Knocking-down FAS and DEDD2 had no effects on MCPIP1-induced cell death (Supplemental Figure 5B) and on PARP-1 cleavage (Supplemental Figure 5C), which is consistent with the above data that MCPIP1 induces apoptosis through directly affecting anti-apoptotic genes. To define further the roles of the anti-apoptotic genes in MCPIP1-induced apoptosis, we respectively knocked down BCL2L1, RELB and BCL3 with shRNA in MCPIP1 knocked-down MDA-MB-231 cells, and then checked cell survival and PARP1 cleavage. These shRNAs effectively knocked down the expression of Bcl2l1, Relb and Bcl3 mRNA, respectively (Figure 5E). As a consequence of Bcl2l1, Relb and Bcl3 knockdown, the floating dead cells were increased after MCPIP1 induction (Figure 5F). In line with these findings, knockdown these anti-apoptotic genes with their respect shRNAs led to PARP1 cleavage (Figure 5G), further confirming that MCPIP1 induces tumor cell apoptosis through suppressing expression of the anti-apoptotic genes.
Figure 5.
knocking-down MCPIP1 enhances anti-apoptotic gene transcript stability and increases cell proliferation. Cell lystaes were isolated from the MDA-MB-231 cells infected with scramble-shRNA/lentivirus and three MCPIP1-shRNAs/lentivirus, followed by western blot analysis with MCPIP1 and β-actin antibodies (A). (B) Total numbers of parental MDA-MB-231 cells and cells infected with scramble/lentivirus or MCPIP1-shRNA/lentivirus were counted at the indicated time (n = 3). ***: p< 0.0001. (C) MTT was performed in the above cells to measure cell proliferation (n = 3). ***: p< 0.0001. (D) Anti-apoptotic gene mRNAs were measured in the parental MDA-MB-231 cells and cells infected with scramble/lentivirus or MCPIP1-shRNA#3/lentivirus by qRT-PCR. *: p< 0.05. MDA-MB-231/MCPIP1 knockdown cells (#3) were infected with scramble/lentivirus or shRNA-lentivirus targeting BCL2L1, RELB, and BCL3, respectively, and then total RNA extracted to measure mRNAs of BCL2L1, RELB, and BCL3 (E). The number of floating cells (dead cells) were counted from the above cells 72 hours after infection (F). Cell lystaes were isolated from the above cells to detect PARP1 and its fragment by western blot (G). (H-K) MDA-MB-231 cells were infected with scramble/lentivirus or MCPIP1-shRNA#3/lentivirus and then treated with ActD and DRB, followed by collecting RNA at the indicated times to detect the remaining mRNAs of anti-apoptotic genes by qRT-PCR.
Since MCPIP1 overexpression enhanced mRNA degradation of the anti-apoptosis genes, we wondered whether deletion of MCPIP1 could stabilize the mRNAs of the anti-apoptotic genes. Indeed, the half-lives of four MCPIP1 targets were increased in MDA-MB-231 cells after knocking-down MCPIP1 by shRNA, with the half-life of Bcl2l1 mRNA being increased from 291 min to 375 min (Figure 5H), Bcl3 from 170 min to 202 min (Figure 5I), BIRC3 from 133 min to 187 min (Figure 5J), and RELB from 355 min to 428 min (Figure 5K), respectively. Furthermore, we also observed a similar pattern of increase in cell proliferation of MDA-MB-453 cells after knocking-down MCPIP1 (data not shown). To determine the effects of MCPIP1 knockdown on cell survival in response to apoptotic insult, we used different concentrations of TNF-α to treat the MDA-MB-231 cells with or without MCPIP1 knockdown, and then checked its effects on PARP-1 cleavage and cell survival. The cleavage of PARP1 was decreased in MCPIP1 knockdown cells compared to cells without MCPIP knockdown (Supplemental Figure 5D). In addition, two independent MCPIP1 knockdown cell lines (#1 and #3) showed increased number of survival cells in response to TNF-α treatment compared to control cells (Supplemental Figure 5E), suggesting that MCPIP1 knockdown results in resistance to apoptotic insult. These results confirm that MCPIP1 repression indeed promotes breast cancer cell proliferation and induces tumor cell apoptosis-resistance through stabilizing anti-apoptotic transcripts.
MCPIP1 binds to a stem-loop structure in the 3’UTR of anti-apoptotic mRNAs for degradation
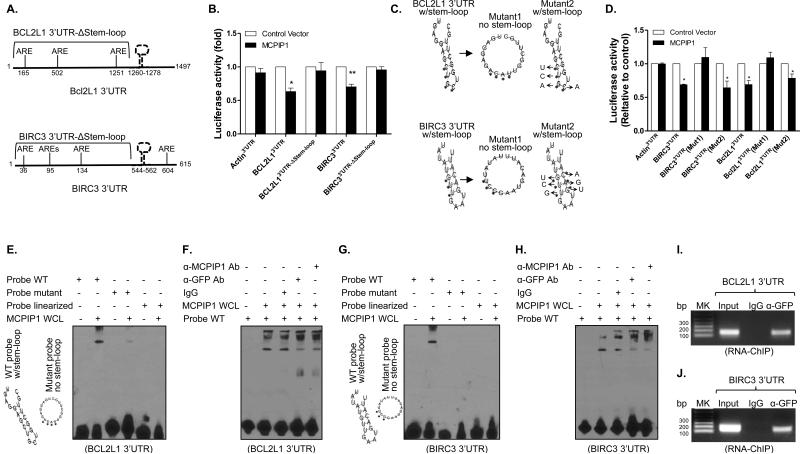
MCPIP1 is known to decay Il6 and Il2 mRNAs through stem-loop structure (30,31). We compared the 3’UTR sequences of five anti-apoptotic genes among different species and identified a conserved sequence. Interestingly, a stem-loop structure could be formed within the conserved sequences when analyzed the sequences with RNAfold (32) (Supplemental Figures 6A-E). To define the role of stem-loop structure in MCPIP1-mediated mRNA decay, we generated deletion constructs by deleting the sequences containing the stem-loop in the 3’UTRs of BCL2L1 and BIRC3, and then co-transfected wild type and deletion reporters with MCPIP1 into HEK293 cells, followed by luciferase measurement. MCPIP1 suppressed the luciferase activities in cells transfected with full-length BCL2L1 and BIRC3 3’UTR constructs, but had no effects on cells transfected with the deletion constructs (Figure 6B), suggesting a requirement for stem-loop structure but not ARE for mRNA decay. To define whether the stem-loop structure or absolute nucleotide is required for mRNA decay, we generated two 3’UTR mutation constructs of BIRC3 and BCL2L1, mutant1 with the step-loop being disrupted by substitution of four nucleotides and mutant2 still kept the stem-loop though substitution of four different nucleotides in stem and loop region as illustrated in Figure 6C, and then co-transfected the two mutants with MCPIP1 for measuring luciferase activity. Disruption of the stem-loop structure in the 3’UTRs of BIRC3 and Bcl2L1 (mutant1) made them completely resistant to MCPIP1 inhibition, whereas the mutant kept the intact step-loop structure (mutant2) were still susceptible to MCPIP1 suppression (Figure 6D), further confirming that MCPIP1 destabilizes the mRNAs of anti-apoptotic genes through the stem-loop structure in their 3’UTRs independent of the absolute nucleotide sequence.
Figure 6.
MCPIP1 binds to stem-loop structure in the 3’UTR of anti-apoptotic genes for mRNA degradation. (A) Schematic representation of the luciferase reporter constructs of bcl2l1 and birc3 containing truncated 3’UTRs without the stem-loop (Δstem-loop). (B) HEK293 cells were co-transfected with luciferase reporters containing full-length 3’UTRs or truncated 3’UTRs (Δstem-loop) of bcl2l1 and birc3 in the presence of MCPIP1 or control vector. 48 hour later, luciferase activity was measured in cell lysates and shown as relative levels compared to cells transfected with empty vector. β-actin 3’UTR was used as negative control. Results shown represent the mean ± SD of four independent experiments. *: p< 0.05, **: p<0.01 between two groups. (C) The predicted stem-loop structures of Bcl2l1 (upper) and Birc3 (lower) in their 3’UTRs and mutation strategy (asterisks indicate base substitution). Mutant1 becomes unable to form stem-loop structure (middle) and Mutant2 still forms a stem-loop structure (left). (D) HEK293 cells were co-transfected with luciferase reporters containing full-length 3’UTRs or mutant 3’UTRs of bcl2l1 and birc3 in the presence of either MCPIP1 or control vector. Luciferase activity was measured in cell lysates and shown as relative levels compared to cells transfected with empty vector. Results shown represent the mean ± SD of four independent experiments (p< 0.05). (E) RNA-EMSA was performed with biotin-labeled probes corresponding to the BCL2L1 3’UTR, including WT probe, mutant probe and linearized probe, in the presence of whole cell lysates (WCL) extracted from MDA-MB-231/Tet-on cells after adding Dox. (F) WCL of MCPIP1-expressed MDA-MB-231/Tet-on cells were incubated with 1 ug of anti-MCPIP1 and anti-GFP antibody and IgG for 30 min, followed by performing RNA-EMSA assay as described above. (G & H) RNA-EMSA and supershift EMSA were respectively performed with biotin-labeled probes corresponding to the BIRC3 3’UTR, including WT probe, mutant probe and linearized probe, in the presence of whole cell lysates (WCL) extracted from MDA-MB-231/Tet-on cells after adding Dox. (I & J) RNA-ChIP was performed with genome fragments from MDA-MB-231/Tet-On cells 24 h after GFP/MCPIP1 induction. The pull-down 3’UTR sequences were amplified by PCR with primers flanking the putative stem-loop sequences.
To determine if MCPIP1 physically bound to the stem-loop in the 3’UTRs of BCL2L1 and BIRC3, we performed RNA-EMSA with wild type RNA probes harboring the step-loop structure, and RNA probes with the step-loop structure either disrupted by nucleotide substitution (mutant probe) or eliminated by physical force (linearized probe) according to manufacturer's manual (Fisher). There was a unique RNA-cytoplasmic protein binding complex formed only to the WT probe but not to the mutant and linearized probes of BCL2L1 (Figure 6E) and BIRC3 (Figure 6G). To confirm the MCPIP1 binding, we added anti-GFP and anti-MCPIP1 antibodies to the cytoplasmic proteins and performed supershift RNA-EMSA. The binding intensity was significantly reduced by adding either anti-GFP antibody or anti-MCPIP1 antibody but not control IgG (Figures 6F & H), indicating that MCPIP1 indeed physically binds to the stem-loop structures in the 3’UTRs of BCL2L1 and BIRC3. Furthermore, we performed a modified RNA immunoprecipitation–chromatin immunoprecipitation assay (RIP-ChIP) (29) to determine whether MCPIP1 bound to the 3’UTRs of anti-apoptotic genes in vivo. After induced MCPIP1/GFP fusion protein in MDA-MB-231 cells, we precipitated the complex with anti-GFP antibody and fragmented the binding mRNAs, followed by amplifying the stem-loop sequence by PCR. The stem-loop sequences in the 3’UTRs of BCL2L1 (Figure 6I) and BIRC3 (Figure 6J) were amplified in the groups pulled down with anti-GFP antibody but not the IgG, indicating the binding of MCPIP1 to the 3’UTRs of these anti-apoptotic mRNAs inside breast tumor cells. Taken together, these data demonstrate that MCPIP1 recognizes the stem-loop structure in the 3’UTRs of anti-apoptotic gene transcripts for degradation and apoptosis induction.
MCPIP1 suppresses breast tumor growth and metastasis
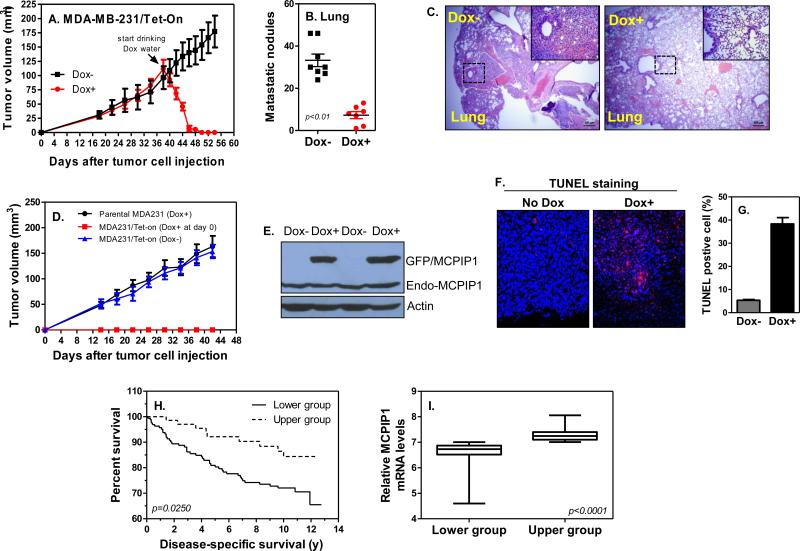
To determine if MCPIP1 suppressed breast tumor progression, we inoculated the MDA-MB-231/Tet-On tumor cells into mammary glands of immune-compromised NSG mice. When tumor mass reaching 5 mm in diameter, we induced MCPIP1 by feeding the tumor-bearing mice with Dox-containing water (Dox+) (Supplemental Figure 7A). One day after drinking Dox water, tumors started to shrink and then rapidly disappeared within six days, while tumors in mice fed with normal water (Dox−) kept growing (Figure 7A and Supplemental Figure 7B). 54 days after tumor cell inoculation, we euthanized the mice, counted metastatic lung nodules, and compared it between mice with and without Dox water. There was a significant reduction in lung metastases in mice drinking Dox water compared to mice fed with normal water (Figure 7B and Supplemental Figure 7C). Histological analysis also showed much less micro-metastatic sites in the lungs of Dox-feeding mice compared to mice without Dox water (Figure 7C). When Dox water was fed at the same time as tumor cell inoculation (day0), no tumors were formed; again tumors kept growth in mice fed with normal water (Dox−) (Figure 7D). Meanwhile, Dox-containing water had no effects on tumor growth in mice inoculated with parental tumor cells (Figure 7D), indicating specific anti-tumor effects of MCPIP1. Indeed, MCPIP1 was highly induced in tumors of mice fed with dox-water (Figure 7E). To determine whether tumor suppression was due to apoptosis, we performed TUNEL staining and results showed that apoptotic cells were significantly increased in tumors of mice drinking dox-water compared with mice fed with normal water (Figures 7F&G). To further confirm broader anti-tumor effects of MCPIP1 in vivo, we inoculated the 4T1/Tet-on tumor cells into mammary glands of immunocompromised NSG mice (Supplemental Figures 8A-D) and into mammary glands of immunocompetent Balb/c mice (Supplemental Figures 9A-G). Similar to MDA-MB-231 cells, a strong suppression on tumor growth and metastases by MCPIP1 were observed in both models (Supplemental Figures 8 & 9). Taken together, these results demonstrate that MCPIP1 is a potent tumor suppressor that strongly suppresses breast tumor growth and metastasis by activation of apoptosis.
Figure 7.
MCPIP1 suppresses breast tumor growth and metastasis and inversely correlate with survival of breast cancer patients. (A) Tumor growth curves in NSG mice received 2 × 106 MDA-MB-231/Tet-On cells (n = 8/group). The day of mice fed with Dox-containing water (1 μg/ml) to induce MCPIP1 was indicated with arrow. (B) Quantification of the number of nodules on lungs of each mouse 12 days after fed with Dox water. (C) H&E staining of lung sections of tumor-bearing mice fed with Dox water or normal water. Lung tumor colonization was shown with dot lines and the metastatic lesions enlarged in upper-right box. Scale bar, 200 μm and 50 μm, respectively. (D) Tumor growth curves in NSG mice received parental MDA-MB-231 and MDA-MB-231/Tet-On cells (n=3/group). Dox-water was provided at the same time as tumor cell inoculation (day0). (E) Total protein was extracted from tumors of mice 4 days after fed with dox-water and used to detect exogenous GFP/MCPIP1 expression by immunoblotting with anti-MCPIP1 antibody. (F) TUNEL staining on tumor tissues of mice 4 days after fed with dox-water or normal water. (G) Quantification of the percentage of TUNEL-positive cells per slide. (H) Kaplan-Meier survival curve of breast tumor patients with low and high tumor MCPIP1 transcripts (n=251). (I) All patients were grouped into MCPIP1 lower (n=164) and higher (n=72) groups based on the levels of MCPIP1 transcript in tumors.
To determine if MCPIP1 was associated with tumor aggressiveness in breast cancer patients, we surveyed MCPIP1 expression across a gene array dataset derived from the excised breast tumors of 251 patients (25). Using this cohort, we analyzed breast cancer-specific survival of these 251 patients between high and low MCPIP1 expression groups. As shown in Figure 7H, patients with low tumor MCPIP1 mRNA levels were more likely to die from recurrent breast cancer following tumorectomy than patients whose tumors expressed high levels of MCPIP1 at excision. The difference in MCPIP1 expression between two groups was statistically significant (Figure 7I). To determine whether MCPIP1 is predictive in triple negative, HER2+, and/or ER+ subsets, we analyzed this dataset and found that the levels of MCPIP1 in tumors are associated with patient survival in PR+ subset but not in ER+ subset (data not shown). Because this dataset does not provide the HER2 staining result, we analyzed another dataset (GSE21653) and found there is no association of MCPIP1 levels in tumors with patient survival in either HER2+ or triple negative (TN) subset, though there is a trend that higher levels of MCPIP1 in tumors have better survival in ER+, HER2+, and TN subsets (data not shown). More study is needed to explore further its role as a predictive marker in long-term survival of breast patients with different subsets. Collectively, these results indicate that MCPIP1 is a strong tumor suppressor and increasing MCPIP1 expression in tumors is a potentially new therapy for breast tumor treatment.
Discussion
RNA-binding protein (RBP) is a group of proteins able to bind to RNA, including the MCPIP1 and tristetraprolin (TTP). In difference to TTP which controls mRNA stability through deadenylation (33-35), MCPIP1 can directly digest the targeted mRNAs (8,9). MCPIP1 is a conserved zinc finger RBP with more than 85% homology between human and mouse. MCPIP1 has been shown to play an important role in suppressing chronic inflammation by promoting mRNA decay of proinflammatory cytokines. Using five isogenic tumorigenic lines that originated from one spontaneous tumor in the BALB/cfC3H mouse, we found the most tumorigenic and metastatic 4T1 cells expressed the lowest levels of MCPIP1 (Figures 1H&I). This inverse correlation between MCPIP1 expression and metastatic potential suggests that MCPIP1 is a tumor suppressor. Unlike the well-known tumor suppressor p53, MCPIP1 is a zinc finger protein mainly localized in cytoplasm (Supplemental Figure 1C). This unique location makes MCPIP1 a second layer to prevent tumorigenesis and tumor progression in addition of the nuclear p53.
When we tried to express MCPIP1 in breast cancer cells, we encountered a difficulty in establishing a standard stable cell line due to massive death of tumor cells after MCPIP1 expression (Supplemental Figures 2B&C). The relationship between MCPIP1 and apoptosis was initially suggested in cardiomyocytes (5). We later found that MCPIP1 itself does not induce apoptosis in macrophages but increases the sensitivity of macrophages to stress signal (21). To our knowledge, so far, there is no direct evidence in inducing apoptosis by MCPIP1 in breast tumor cells. Using different methods we demonstrate that MCPIP1 can strongly induce apoptosis in breast cancer cells (Figures 2A-F). Apoptosis is triggered upon the balance between pro-apoptotic and anti-apoptotic gene being broken. MCPIP1 inhibited the expression of anti-apoptotic gene transcripts (Figures 3B-E) while has minor or no effects on pro-apoptotic gene transcripts (Figures 3F-I). We chose these 4 pro-apoptotic genes based on two reasons: (1) they are among the most affected genes by MCPIP1 induction; and (2) the changes of these gene expressions are further confirmed by independent experiments after MCPIP1 induction. Indeed, among the apoptotic genes affected by MCPIP1, only FAS and DEDD2 fit the above two criteria. The binding of MCPIP1 to anti-apoptotic mRNAs but not to pro-apoptotic mRNAs further confirms a select targeting of MCPIP1 in apoptosis pathway, causing imbalance between pro-apoptotic and anti-apoptotic genes. We have also observed that MCPIP1 induces apoptosis of other types of cells, including HEK293 cells, Jurkat cells and CEM cells (data not shown), indicating that MCPIP1 may play a broader role in regulation of apoptosis pathway. Collectively, our results indicate that MCPIP1 suppresses breast tumor progression through induction of cell apoptosis.
Steady-state mRNA level is determined by both transcription and mRNA stability. Our results indicate that MCPIP1 suppresses the expression of anti-apoptotic genes at the post-transcriptional level through enhancing mRNA degradation. Among the MCPIP1-targeted genes, Bcl2L1 (also called Bcl-xl) (36) and Bcl2A1 (37) are mainly involved in intrinsic mitochondria anti-apoptosis signaling pathway, while BIRC3 (38), RelB (39), and Bcl3 (40) are involved in activation of NF-kB pathway. The broad targets of MCPIP1 in apoptosis pathway suggest that MCPIP1 induces apoptosis through multi molecular events in breast tumor cells. It has been known that the elements regulating mRNA decay are mainly located in the 3’UTR, such as ARE and GRE (41,42). Indeed, MCPIP1 inhibits luciferase activity through the 3’UTRs of anti-apoptotic genes (Figure 4E). In contrast to TTP (43) and Roquin (44) which recruit other decadenylases to the 3’UTR for mRNA degradation, MCPIP1 is known to decay its target mRNAs by itself. It has been reported that MCPIP1 possesses a PIN-like domain at its N-terminal with endonuclease activity (45). This PIN-domain is essential for MCPIP1's mRNA degradation and anti-viral effects. By mutating PIN and ZF domain, we demonstrate that the PIN-domain is also required for apoptosis induction.
MCPIP1 destabilizes mRNA stability of IL-6 and IL-2 by targeting stem-loop but ARE in the 3’UTR. Our results demonstrate that it is the stem-loop structure but not the absolute nucleotide sequence required for MCPIP1-mediated mRNA decay. Roquin has been shown to recognize a constitutive decay element (CDE) in the 3’UTR of TNF-α mRNAs and this CDE can fold into a stem-loop structure (44). We found no consensus stem-loop sequences among the five anti-apoptotic genes, which support our hypothesis that MCPIP1 mainly recognizes the secondary hairpin structure but not linear sequence in the 3’UTR. MCPIP1 could cleave and process pre-miRNAs via the specific loop structure (46). These results demonstrate that MCPIP1 seems to modulate not only biogenesis of the miRNAs, but also translation of the mRNAs important for tumor growth and apoptosis. As a RBP localized in cytoplasm, MCPIP1 readily controls the fate of tumor cells by tipping the balance towards apoptosis.
Though the primary tumors are completely eliminated in mice fed with Dox water, there are still a few metastatic nodules and cells presented in the lungs. To figure out the reason, we isolated the metastatic tumor cells from lung and cultured them in vitro. We found that the remained lung metastatic tumor cells can still express MCPIP1 after adding Dox into culture medium and die within a few days (Supplemental Figures 10B-D), indicating that MCPIP1 was not induced in those cells previously. A possible reason for it might be due to a low concentration of Dox in the lung, which failed to reach sufficient level for inducing MCPIP1 in lung tumor cells. It also suggests that increasing the concentration of Dox in metastatic organs, including lung, should be able to eliminate metastatic cells entirely. Therefore, drugs or small molecules able to induce MCPIP1 expression can be used therapeutically for treatment of metastatic breast cancer. It has been showed that MCPIP is cleaved by Malt1 and inhibition of Malt1 protease activity could increase MCPIP1 levels in T cells (9). We speculate that Malt1 inhibitor such as MI-2 (47) could be used to prevent MCPIP1 cleavage and therefore increase MCPIP1 levels in tumor cells. MCPIP1-expressing adenovirus can also be used to introduce MCPIP1 into tumor cells as gene therapy. We are currently investigating the effectiveness of these methods on MCPIP1 induction and tumor progression. The strong association between levels of MCPIP1 in tumors and survival of patients with breast tumor over 13 years follow-up further demonstrate the biological relevance of inducing MCPIP1 for breast tumor treatment.
Based on our study, we propose a model to illustrate the potent role for MCPIP1 in suppression of breast tumor growth and metastasis (Supplemental Figure 11). MCPIP1 selectively targets the mRNAs of anti-apoptotic genes for degradation through the PIN domain via recognizing and binding to a stem-loop structure in the 3’UTRs. As an RNA-binding protein localized in the cytoplasm, MCPIP1 can quickly control cell fate by tipping the balance towards apoptosis in tumor cells. Our findings also add a new regulator in the apoptosis pathway for apoptosis.
Supplementary Material
Acknowledgments
We thank Janet S Butel (Baylor College of Medicine) for mouse mammary gland epithelial FSK4 and CommD cells, Yibin Kang (Princeton University) for isogenic tumorigenic line 67NR, 168FARN, 4TO7, 66cl4 and 4T1 cells. We also thank Tianyi Wang (SRI International) for assistance in cloning the Tet-On tumor cells. We also thank Nadim Alkharouf (Towson University) for assistance in analyzing the microarray dataset.
Grant Support
This work was supported by the National Cancer Institute of the National Institutes of Health under Award Number R01CA163808 (J Liu) and AI103618 (M Fu). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
DISCLOSURES
All authors have no conflict of interest in this study.
Supplemental Information
Supplemental information, including additional methods, 11 figures, and two tables, can be found with this article online.
Disclosure of Potential Conflicts of Interest
All authors have no conflict of interest in this study.
Authors’ Contributions
Concept and design: J. Liu
Development of methodology: J. Liu, W. Lu, H. Ning, L. Gu
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): W. Lu, H. Ning, L. Gu, D. Hoft, Q. Wang, H. Peng, J. Liu
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): J. Liu, W. Lu, H. Ning
Writing, review, and/or revision of the manuscript: J. Liu, W. Lu, H. Ning, D. Hoft, R. Hou
Administrative, technical, or material support (i.e., reporting or organizing data, construction database): M. Fu
Study supervision: J. Liu
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Lowe SW, Lin AW. Apoptosis in cancer. Carcinogenesis. 2000;21(3):485–95. doi: 10.1093/carcin/21.3.485. [DOI] [PubMed] [Google Scholar]
- 3.Igney FH, Krammer PH. Death and anti-death: tumour resistance to apoptosis. Nat Rev Cancer. 2002;2(4):277–88. doi: 10.1038/nrc776. [DOI] [PubMed] [Google Scholar]
- 4.Kang MH, Reynolds CP. Bcl-2 inhibitors: targeting mitochondrial apoptotic pathways in cancer therapy. Clin Cancer Res. 2009;15(4):1126–32. doi: 10.1158/1078-0432.CCR-08-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou L, Azfer A, Niu J, Graham S, Choudhury M, Adamski FM, et al. Monocyte chemoattractant protein-1 induces a novel transcription factor that causes cardiac myocyte apoptosis and ventricular dysfunction. Circ Res. 2006;98(9):1177–85. doi: 10.1161/01.RES.0000220106.64661.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liang J, Wang J, Azfer A, Song W, Tromp G, Kolattukudy PE, et al. A novel CCCH-zinc finger protein family regulates proinflammatory activation of macrophages. J Biol Chem. 2008;283(10):6337–46. doi: 10.1074/jbc.M707861200. [DOI] [PubMed] [Google Scholar]
- 7.Huang S, Miao R, Zhou Z, Wang T, Liu J, Liu G, et al. MCPIP1 negatively regulates toll-like receptor 4 signaling and protects mice from LPS-induced septic shock. Cell Signal. 2013;25(5):1228–34. doi: 10.1016/j.cellsig.2013.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matsushita K, Takeuchi O, Standley DM, Kumagai Y, Kawagoe T, Miyake T, et al. Zc3h12a is an RNase essential for controlling immune responses by regulating mRNA decay. Nature. 2009;458(7242):1185–90. doi: 10.1038/nature07924. [DOI] [PubMed] [Google Scholar]
- 9.Uehata T, Iwasaki H, Vandenbon A, Matsushita K, Hernandez-Cuellar E, Kuniyoshi K, et al. Malt1-induced cleavage of regnase-1 in CD4(+) helper T cells regulates immune activation. Cell. 2013;153(5):1036–49. doi: 10.1016/j.cell.2013.04.034. [DOI] [PubMed] [Google Scholar]
- 10.Liang J, Saad Y, Lei T, Wang J, Qi D, Yang Q, et al. MCP-induced protein 1 deubiquitinates TRAF proteins and negatively regulates JNK and NF-kappaB signaling. J Exp Med. 2010;207(13):2959–73. doi: 10.1084/jem.20092641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin RJ, Chien HL, Lin SY, Chang BL, Yu HP, Tang WC, et al. MCPIP1 ribonuclease exhibits broad-spectrum antiviral effects through viral RNA binding and degradation. Nucleic Acids Res. 2013;41(5):3314–26. doi: 10.1093/nar/gkt019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu S, Qiu C, Miao R, Zhou J, Lee A, Liu B, et al. MCPIP1 restricts HIV infection and is rapidly degraded in activated CD4+ T cells. Proc Natl Acad Sci U S A. 2013;110(47):19083–8. doi: 10.1073/pnas.1316208110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin RJ, Chu JS, Chien HL, Tseng CH, Ko PC, Mei YY, et al. MCPIP1 suppresses hepatitis C virus replication and negatively regulates virus-induced proinflammatory cytokine responses. J Immunol. 2014;193(8):4159–68. doi: 10.4049/jimmunol.1400337. [DOI] [PubMed] [Google Scholar]
- 14.Qi Y, Liang J, She ZG, Cai Y, Wang J, Lei T, et al. MCP-induced protein 1 suppresses TNFalpha-induced VCAM-1 expression in human endothelial cells. FEBS Lett. 2010;584(14):3065–72. doi: 10.1016/j.febslet.2010.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He M, Liang X, He L, Wen W, Zhao S, Wen L, et al. Endothelial dysfunction in rheumatoid arthritis: the role of monocyte chemotactic protein-1-induced protein. Arterioscler Thromb Vasc Biol. 2013;33(6):1384–91. doi: 10.1161/ATVBAHA.113.301490. [DOI] [PubMed] [Google Scholar]
- 16.Steller H. Mechanisms and genes of cellular suicide. Science. 1995;267(5203):1445–9. doi: 10.1126/science.7878463. [DOI] [PubMed] [Google Scholar]
- 17.Thompson CB. Apoptosis in the pathogenesis and treatment of disease. Science. 1995;267(5203):1456–62. doi: 10.1126/science.7878464. [DOI] [PubMed] [Google Scholar]
- 18.Kumar R, Vadlamudi RK, Adam L. Apoptosis in mammary gland and cancer. Endocr Relat Cancer. 2000;7(4):257–69. doi: 10.1677/erc.0.0070257. [DOI] [PubMed] [Google Scholar]
- 19.Yip KW, Reed JC. Bcl-2 family proteins and cancer. Oncogene. 2008;27(50):6398–406. doi: 10.1038/onc.2008.307. [DOI] [PubMed] [Google Scholar]
- 20.Placzek WJ, Wei J, Kitada S, Zhai D, Reed JC, Pellecchia M. A survey of the anti-apoptotic Bcl-2 subfamily expression in cancer types provides a platform to predict the efficacy of Bcl-2 antagonists in cancer therapy. Cell Death Dis. 2010;1:e40. doi: 10.1038/cddis.2010.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qi D, Huang S, Miao R, She ZG, Quinn T, Chang Y, et al. Monocyte chemotactic protein-induced protein 1 (MCPIP1) suppresses stress granule formation and determines apoptosis under stress. J Biol Chem. 2011;286(48):41692–700. doi: 10.1074/jbc.M111.276006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skalniak L, Koj A, Jura J. Proteasome inhibitor MG-132 induces MCPIP1 expression. FEBS J. 2013;280(11):2665–74. doi: 10.1111/febs.12264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skalniak A, Boratyn E, Tyrkalska SD, Horwacik I, Durbas M, Lastowska M, et al. Expression of the monocyte chemotactic protein-1-induced protein 1 decreases human neuroblastoma cell survival. Oncol Rep. 2014;31(5):2385–92. doi: 10.3892/or.2014.3076. [DOI] [PubMed] [Google Scholar]
- 24.Lyu JH, Park DW, Huang B, Kang SH, Lee SJ, Lee C, et al. RGS2 suppresses breast cancer cell growth via a MCPIP1-dependent pathway. J Cell Biochem. 2014 doi: 10.1002/jcb.24964. [DOI] [PubMed] [Google Scholar]
- 25.Miller LD, Smeds J, George J, Vega VB, Vergara L, Ploner A, et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci U S A. 2005;102(38):13550–5. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hollmann CA, Kittrell FS, Medina D, Butel JS. Wnt-1 and int-2 mammary oncogene effects on the beta-catenin pathway in immortalized mouse mammary epithelial cells are not sufficient for tumorigenesis. Oncogene. 2001;20(52):7645–57. doi: 10.1038/sj.onc.1204980. [DOI] [PubMed] [Google Scholar]
- 27.Lu X, Bennet B, Mu E, Rabinowitz J, Kang Y. Metabolomic changes accompanying transformation and acquisition of metastatic potential in a syngeneic mouse mammary tumor model. J Biol Chem. 2010;285(13):9317–21. doi: 10.1074/jbc.C110.104448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qian X, Ning H, Zhang J, Hoft DF, Stumpo DJ, Blackshear PJ, et al. Posttranscriptional regulation of IL-23 expression by IFN-gamma through tristetraprolin. J Immunol. 2011;186(11):6454–64. doi: 10.4049/jimmunol.1002672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keene JD, Komisarow JM, Friedersdorf MB. RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat Protoc. 2006;1(1):302–7. doi: 10.1038/nprot.2006.47. [DOI] [PubMed] [Google Scholar]
- 30.Iwasaki H, Takeuchi O, Teraguchi S, Matsushita K, Uehata T, Kuniyoshi K, et al. The IkappaB kinase complex regulates the stability of cytokine-encoding mRNA induced by TLR-IL-1R by controlling degradation of regnase-1. Nat Immunol. 2011;12(12):1167–75. doi: 10.1038/ni.2137. [DOI] [PubMed] [Google Scholar]
- 31.Li M, Cao W, Liu H, Zhang W, Liu X, Cai Z, et al. MCPIP1 down-regulates IL-2 expression through an ARE-independent pathway. PLoS One. 2012;7(11):e49841. doi: 10.1371/journal.pone.0049841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31(13):3429–31. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanduja S, Blanco FF, Young LE, Kaza V, Dixon DA. The role of tristetraprolin in cancer and inflammation. Front Biosci (Landmark Ed) 2012;17:174–88. doi: 10.2741/3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ross CR, Brennan-Laun SE, Wilson GM. Tristetraprolin: roles in cancer and senescence. Ageing Res Rev. 2012;11(4):473–84. doi: 10.1016/j.arr.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qian X, Gu L, Ning H, Zhang Y, Hsueh EC, Fu M, et al. Increased Th17 cells in the tumor microenvironment is mediated by IL-23 via tumor-secreted prostaglandin E2. J Immunol. 2013;190(11):5894–902. doi: 10.4049/jimmunol.1203141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eichhorn JM, Sakurikar N, Alford SE, Chu R, Chambers TC. Critical role of anti-apoptotic Bcl-2 protein phosphorylation in mitotic death. Cell Death Dis. 2013;4:e834. doi: 10.1038/cddis.2013.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ottina E, Grespi F, Tischner D, Soratroi C, Geley S, Ploner A, et al. Targeting antiapoptotic A1/Bfl-1 by in vivo RNAi reveals multiple roles in leukocyte development in mice. Blood. 2012;119(25):6032–42. doi: 10.1182/blood-2011-12-399089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zarnegar BJ, Wang Y, Mahoney DJ, Dempsey PW, Cheung HH, He J, et al. Noncanonical NF-kappaB activation requires coordinated assembly of a regulatory complex of the adaptors cIAP1, cIAP2, TRAF2 and TRAF3 and the kinase NIK. Nat Immunol. 2008;9(12):1371–8. doi: 10.1038/ni.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tando T, Ishizaka A, Watanabe H, Ito T, Iida S, Haraguchi T, et al. Requiem protein links RelB/p52 and the Brm-type SWI/SNF complex in a noncanonical NF-kappaB pathway. J Biol Chem. 2010;285(29):21951–60. doi: 10.1074/jbc.M109.087783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maldonado V, Melendez-Zajgla J. Role of Bcl-3 in solid tumors. Mol Cancer. 2011;10:152. doi: 10.1186/1476-4598-10-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vlasova-St Louis I, Bohjanen PR. Post-transcriptional regulation of cytokine signaling by AU-rich and GU-rich elements. J Interferon Cytokine Res. 2014;34(4):233–41. doi: 10.1089/jir.2013.0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ivanov P, Anderson P. Post-transcriptional regulatory networks in immunity. Immunol Rev. 2013;253(1):253–72. doi: 10.1111/imr.12051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sandler H, Kreth J, Timmers HT, Stoecklin G. Not1 mediates recruitment of the deadenylase Caf1 to mRNAs targeted for degradation by tristetraprolin. Nucleic Acids Res. 2011;39(10):4373–86. doi: 10.1093/nar/gkr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leppek K, Schott J, Reitter S, Poetz F, Hammond MC, Stoecklin G. Roquin promotes constitutive mRNA decay via a conserved class of stem-loop recognition motifs. Cell. 2013;153(4):869–81. doi: 10.1016/j.cell.2013.04.016. [DOI] [PubMed] [Google Scholar]
- 45.Xu J, Peng W, Sun Y, Wang X, Xu Y, Li X, et al. Structural study of MCPIP1 N-terminal conserved domain reveals a PIN-like RNase. Nucleic Acids Res. 2012;40(14):6957–65. doi: 10.1093/nar/gks359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suzuki HI, Arase M, Matsuyama H, Choi YL, Ueno T, Mano H, et al. MCPIP1 ribonuclease antagonizes dicer and terminates microRNA biogenesis through precursor microRNA degradation. Mol Cell. 2011;44(3):424–36. doi: 10.1016/j.molcel.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 47.Fontan L, Yang C, Kabaleeswaran V, Volpon L, Osborne MJ, Beltran E, et al. MALT1 small molecule inhibitors specifically suppress ABC-DLBCL in vitro and in vivo. Cancer cell. 2012;22(6):812–24. doi: 10.1016/j.ccr.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.