Heritable single nucleotide polymorphisms (SNPs) in six genomic loci have been associated with childhood ALL risk in genome-wide association studies (GWAS) of European-ancestry populations. Four of these heritable ALL risk loci are near genes crucial for hematopoietic differentiation, including CEBPE1,2. CEBPE encodes one of a family of six known CCAAT-targeting bZIP transcription factors which critically regulate myelopoiesis. To identify causal polymorphisms underlying the childhood ALL association peak near CEBPE, we performed SNP genotyping, imputation-based fine-mapping, and functional validation analyses of the chromosome 14q11.2 locus in a multi-ethnic case-control population of Hispanic children from the California Childhood Leukemia Study (CCLS) and European-ancestry children from the Children’s Oncology Group (COG), totaling 1277 cases and 3078 controls.
Supplementary Figure 1 and Supplementary Table 1 summarize the study design and study populations (full methods in online supplement). Briefly, 297 Hispanic B-cell ALL (B-ALL) cases and 454 Hispanic controls underwent genotyping and targeted fine-mapping. Additional GWAS data was used to perform imputation-based fine-mapping in 980 European-ancestry COG ALL subjects and 2624 Wellcome Trust Case Control Consortium controls (WTCCC)3. An additional 271 cases and 436 controls from the CCLS underwent targeted Sequenom genotyping. Association tests were performed using an allelic additive model, adjusted for ancestry-informative principal components.
Using all available genome-wide SNP data, we imputed all the common SNPs in the CEBPE region using a multi-ethnic reference panel from the 1000 Genomes Project. Three SNPs (rs2239635, rs8007237, rs56031127) were more significantly associated than either of the previously reported GWAS hits (rs2239633 and rs4982731) (Supplementary Table 2)1,4. The most statistically significant association detected in analysis of the combined CCLS and COG data was at rs2239635 (Pmeta=5.1×10−11, O.R.=1.45, 95% C.I.=1.30-1.62) (Figure 1a). Imputed genotype and directly assayed genotype at rs2239635 were 97.9% concordant, validating imputation accuracy.
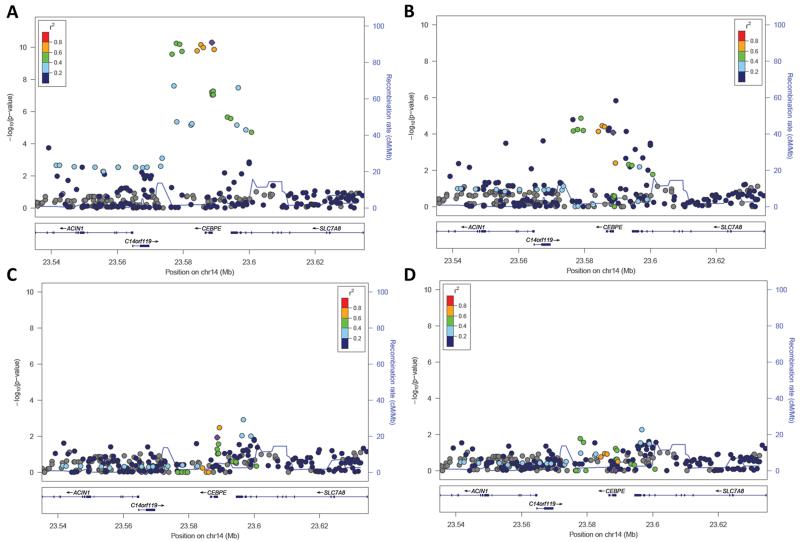
Figure 1. Association of SNPs in the CEBPE locus with ALL risk, with adjustment for previous GWAS lead SNPs and novel lead SNP rs2339635.
Lead SNP rs2339635 appears in purple, with other SNPs colored to indicate their degree of linkage with rs2339635. A) Association of SNPs in the CEBPE locus with ALL risk. B) Association of SNPs in the CEBPE locus with ALL risk, adjusted for rs2239633, the top hit from two previous GWAS1,15. C) Association of SNPs in the CEBPE locus with ALL risk, adjusted for rs4982731, the top hit from a previous GWAS4. D) Association of SNPs in the CEBPE locus with ALL risk, adjusted for rs2239635, the top hit from our fine-mapping analyses.
In addition to being more significantly associated than the previously reported GWAS hits, rs2239635 remained significantly associated with ALL risk in conditional analyses adjusted for rs2239633 and for rs4982731 (P= 8.4×10−5 and 0.011, respectively) (Figure 1b-c). Furthermore, in conditional analyses adjusted for this new top hit, the previous ALL GWAS hits at rs2239633 and rs4982731 no longer reached statistical significance (P=0.076 and 0.11, respectively) (Figure 1d), suggesting that rs2239635, or a variant in strong LD, underlies the CEBPE association signal first detected by GWAS.
In the 100kb CEBPE imputation region, Haploview analysis identified 15 distinct haplotype blocks among CCLS Hispanics compared with only 8 in the COG European-ancestry population (Supplementary Figure 2). Taking advantage of this reduced LD, we performed stratified association analyses in CCLS Hispanics to further refine the CEBPE association signal. Among 297 Hispanic B-ALL cases and 454 Hispanic controls with GWAS data available in the CCLS, rs2239635 was again the most significantly associated SNP by nearly two orders of magnitude (Supplementary Table 2). The minor allele was associated with a 1.87-fold increase in the odds of B-ALL in Hispanics (95% CI=1.51-2.30; P=6.2×10−9).
SNPs in the CEBPE region generally showed stronger associations with the high-hyperdiploid B-ALL subgroup (Supplementary Table 2 and Supplementary Figure 3). The minor allele of rs2239635 conferred 2.94-fold increased odds of high-hyperdiploid B-cell ALL (95% CI = 2.12-4.06; P = 8.2×10−11), but only 1.51-fold increased odds of non-hyperdiploid B-ALL (95% CI = 1.11-2.04; P = 8.8×10−3). In case-case comparisons, rs2239635 was associated with a 1.66-fold increase in the odds of high-hyperdiploid B-ALL relative to non-hyperdiploid B-ALL (P=0.0034), demonstrating a robust difference in the magnitude of effect across subtypes.
Top associated SNPs were functionally annotated using ENCODE2 data5,6, and assessed if they were cis eQTLs for CEBPE7. Functional annotations indicated that the top-ranked SNP, rs2239635, impacts CEBPE expression (P=0.01) and is among the most functionally-relevant variants in the region (Supplementary Table 2). These annotations also suggest a potential mechanism by which rs2239635 affects expression, as the SNP is located within an Ikaros transcription factor binding site and is predicted to affect TF binding8. Ikaros is encoded by the IKZF1 gene, a critical mediator of hematopoietic differentiation which has also been associated with ALL risk in GWAS1,2.
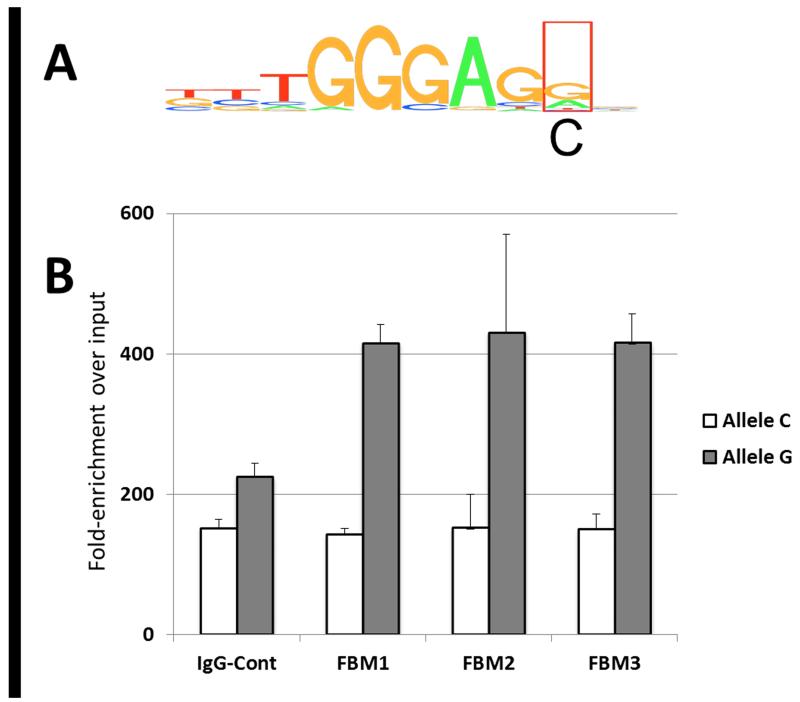
To test the predicted disruption of the Ikaros binding site, we identified three normal fetal bone marrow samples heterozygotes at rs2239635. Such heterozygotes contain a natural “test allele” along with a “control allele” for comparison. In ChIP experiments, the wild-type (WT) allele, which maintains the canonical Ikaros binding motif (TTTGGGAGG, Figure 2a), immunoprecipitated significantly more efficiently than the risk allele (P<0.05) (Figure 2b). This indicates that the risk allele “C” disrupts Ikaros binding near CEBPE in its native physiological state. However, it is unlikely that the “C” allele completely abolishes binding due to its location at the periphery of the Ikaros binding motif and it is noted that the IgG control also displayed a slight assay readout bias towards the “C” allele.
Figure 2. Chromatin immunoprecipitation of CEBPE promoter region.
(A) The region of the promoter of CEBPE showing the perfect match for IKZF1 binding (Regulome database) and the position of rs2339635 (boxed). The identity of the risk allele is indicated below the box. (B) Bone marrow from 3 independent subjects heterozygous for rs2239635 was fixed, sonicated, and precipitated with an antibody for IKZF1. Assays for each allele of CEBPE pulled down only the “G” variant of rs2339635, without pulling down the “C” variant any more than a nonspecific IgG control antibody (P<0.05, t-test) using a ddPCR assay. ddPCR analysis of the input (prior to ChIP with either IKZF1 or isotype control) is displayed on the left side of the figure.
Using a model expression vector, we tested whether the genomic region including rs2239635 affects transcription of CEBPE. Since Ikaros is a known transcriptional repressor we cloned the segment into a vector with a known strong transcriptional activation sequence, the promoter of PTPRG9. Plasmids containing the CEBPE promoter segment were significantly repressive compared to the PTPRG promoter alone (P < 0.001), confirming the repressive properties of the CEBPE promoter segment. Gene repression was not significantly different for the “C” allele than the “G” allele possibly due to the comparatively low expression of Ikaros in 293 cells.
Because a previous GWAS identified an ALL risk locus at rs4132601 in IKZF1 that is associated with reduced gene expression1, and because our experiments suggest biologic interaction between IKZF1 and CEBPE, we tested for statistical interaction between SNPs. In case-control analyses of high-hyperdiploid B-ALL, rs2239635 and rs4132601 displayed significant multiplicative interaction in CCLS Hispanics (P=0.021; N=147 cases, 714 controls). A weaker interaction was detected in analysis of all CCLS B-ALL samples with genotyping data for both SNPs (P=0.061; N=449 cases, 714 controls). The direction of the interaction indicates that the combined effect of rs2239635 (CEBPE) and rs4132601 (IKZF1) risk alleles is greater than would be expected if they operated independently. Although rs2239635 showed a stronger effect in high-hyperdiploid leukemia, effects were similar in cases with IKZF1 deletions as in those without (OR=2.17 and 2.23, respectively).
CEBPE and IKZF1 expression levels were compared to investigate changes in expression during B-cell development. CEBPE expression was highest in multipotent progenitor cells (S1) and declined sharply as cells progressed to B-cell-committed progenitors, including pre-B-I cells (S2), pre-B-II cells (S3) and immature B-cells (S4) (Supplementary Figure 4a). CEBPE expression dropped again when cells transitioned to mature B-cells. This pattern was tightly and inversely coordinated with expression of IKZF1 (Supplementary Figure 4b). As expected, expression of both CEBPE and IKZF1 in patient leukemic B-cells was most similar to that in early stage B-cells (Supplementary Figure 4), believed to be the B-ALL cell-of-origin.
We identify an allele in the CEBPE gene promoter that impacts risk of childhood ALL by disrupting binding of the Ikaros transcriptional repressor. Additionally, we note a cytogenetic subtype-specific association between the functional SNP of CEBPE and high-hyperdiploid B-ALL. Because high-hyperdiploid leukemias typically carry an extra chromosome 14, leading to 3 copies of CEBPE, overexpression of CEBPE may be critical for development of high-hyperdiploid B-ALL.
CEBPE is a critical modulator of myelopoiesis. Disruption of C/EBP proteins, including C/EBPε, alters myelopoiesis10. C/EBPε is not required for B-cell maturation or function, opening a question as to why a polymorphism affecting a subtype of pre-B-ALL may be located proximal to the gene. As progenitor cells choose their lineage fate, transcription factors that are required for other fates must be shut off permanently11. Also, maturation towards a pre-B cell from a common lymphoid progenitor is associated with strong expression of Ikaros, a transcription factor essential for B-cell development with both activating and repressive properties12. Our results suggest that lineage commitment for pre-B cells involves the suppression of CEBPE by Ikaros, and a polymorphism that disrupts this repression promotes B-ALL risk. Incomplete suppression of CEBPE by Ikaros may lead to lineage confusion, a common feature of leukemogenesis13.
That overexpression of a non-B-cell specific transcription factor would increase risk of a B-cell malignancy is not without precedent. A heritable polymorphism in the T-cell-specific transcription factor GATA3 is associated with higher GATA3 expression and also with increased B-ALL risk14. Together, these observations suggest that disruption of the precise control of lineage choice and commitment may be a more common feature of leukemogenesis than represented by the rare diagnosis of mixed-lineage leukemias (typically associated with MLL1 or BCR-ABL1 gene translocations). Common heritable genetic modifiers are likely to contribute much more subtle and specific alterations in hematopoietic cell development than the broad epigenetic changes induced by acquired gene fusions, but may add significantly to the burden of disease due to their high prevalence. Additional work is needed to conclusively determine the role of CEBPE control in lymphoid neoplasia; however, its connection with Ikaros function is supported by our ChIP experiments and a demonstrated statistical interaction between the two SNPs in conferring leukemia risk.
Supplementary Material
ACKNOWLEDGEMENTS
This work was financially supported by NCI R01CA155461 (J.L.W., K.M.W.), NCI R25CA112355 (K.M.W.), NIEHS and EPA P01ES018172 (C.M, J.L.W), NIEHS R01ES09137 (C.M., J.L.W), and the National Institute of Diabetes and Digestive and Kidney Diseases at the National Institutes of Health P01DK088760 (M.O.M. and M.E.F.). A full list of acknowledgements can be found in the online supplement.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
JLW and KMW designed experiments, analyzed the data, and wrote the manuscript. AJdS and JQ designed and performed experiments and helped write the manuscript. S-TL helped analyze the data. MOM and MEF supplied key reagents and helped perform experiments. MZ, and HH performed experiments. AT and CM secured clinical and population samples. All authors read and finalized the manuscript.
Supplementary Information accompanies this paper on the Leukemia website (http://www.nature.com/leu)
REFERENCES
- 1.Papaemmanuil E, Hosking FJ, Vijayakrishnan J, Price A, Olver B, Sheridan E, et al. Loci on 7p12.2, 10q21.2 and 14q11.2 are associated with risk of childhood acute lymphoblastic leukemia. Nature genetics. 2009 Sep;41(9):1006–1010. doi: 10.1038/ng.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Trevino LR, Yang W, French D, Hunger SP, Carroll WL, Devidas M, et al. Germline genomic variants associated with childhood acute lymphoblastic leukemia. Nature genetics. 2009 Sep;41(9):1001–1005. doi: 10.1038/ng.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wellcome Trust Case Control C Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007 Jun 7;447(7145):661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu H, Yang W, Perez-Andreu V, Devidas M, Fan Y, Cheng C, et al. Novel susceptibility variants at 10p12.31-12.2 for childhood acute lymphoblastic leukemia in ethnically diverse populations. Journal of the National Cancer Institute. 2013 May 15;105(10):733–742. doi: 10.1093/jnci/djt042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012 Jan;40:D930–934. doi: 10.1093/nar/gkr917. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012 Sep;22(9):1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Consortium GT The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013 Jun;45(6):580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pique-Regi R, Degner JF, Pai AA, Gaffney DJ, Gilad Y, Pritchard JK. Accurate inference of transcription factor binding from DNA sequence and chromatin accessibility data. Genome Res. 2011 Mar;21(3):447–455. doi: 10.1101/gr.112623.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiao J, Lee ST, Xiao Y, Ma X, Andres Houseman E, Hsu LI, et al. PTPRG inhibition by DNA methylation and cooperation with RAS gene activation in childhood acute lymphoblastic leukemia. Int J Cancer. 135(5):1101–9. doi: 10.1002/ijc.28759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamanaka R, Barlow C, Lekstrom-Himes J, Castilla LH, Liu PP, Eckhaus M, et al. Impaired granulopoiesis, myelodysplasia, and early lethality in CCAAT/enhancer binding protein epsilon-deficient mice. Proc Natl Acad Sci U S A. 1997 Nov 25;94(24):13187–13192. doi: 10.1073/pnas.94.24.13187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Starnes LM, Sorrentino A. Regulatory circuitries coordinated by transcription factors and microRNAs at the cornerstone of hematopoietic stem cell self-renewal and differentiation. Curr Stem Cell Res Ther. 2011 Jun;6(2):142–161. doi: 10.2174/157488811795495431. [DOI] [PubMed] [Google Scholar]
- 12.Schwickert TA, Tagoh H, Gultekin S, Dakic A, Axelsson E, Minnich M, et al. Stage-specific control of early B cell development by the transcription factor Ikaros. Nat Immunol. 2014 Mar;15(3):283–293. doi: 10.1038/ni.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmidt CA, Przybylski GK. What can we learn from leukemia as for the process of lineage commitment in hematopoiesis? Int Rev Immunol. 2001 Feb;20(1):107–115. doi: 10.3109/08830180109056725. [DOI] [PubMed] [Google Scholar]
- 14.Perez-Andreu V, Roberts KG, Harvey RC, Yang W, Cheng C, Pei D, et al. Inherited GATA3 variants are associated with Ph-like childhood acute lymphoblastic leukemia and risk of relapse. Nature genetics. 2013 Dec;45(12):1494–1498. doi: 10.1038/ng.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Migliorini G, Fiege B, Hosking FJ, Ma Y, Kumar R, Sherborne AL, et al. Variation at 10p12.2 and 10p14 influences risk of childhood B-cell acute lymphoblastic leukemia and phenotype. Blood. 2013 Nov 7;122(19):3298–3307. doi: 10.1182/blood-2013-03-491316. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.