Abstract
Dysregulation of MYC expression is a hallmark of cancer, but the development of agents that target MYC has remained challenging. The oncogenic MUC1-C transmembrane protein is, like MYC, aberrantly expressed in diverse human cancers. The present studies demonstrate that MUC1-C induces MYC expression in KRAS mutant non-small cell lung cancer (NSCLC) cells, an effect that can be suppressed by targeting MUC1-C via shRNA silencing, CRISPR editing, or pharmacological inhibition with GO-203. MUC1-C activated the WNT/β-catenin (CTNNB1) pathway and promoted occupancy of MUC1-C/β-catenin/TCF4 complexes on the MYC promoter. MUC1-C also promoted the recruitment of the p300 histone acetylase (EP300) and, in turn, induced histone H3 acetylation and activation of MYC gene transcription. We also show that targeting MUC1-C decreased the expression of key MYC target genes essential for the growth and survival of NSCLC cells, such as TERT and CDK4. Based on these results, we found that the combination of GO-203 and the BET bromodomain inhibitor JQ1, which targets MYC transcription, synergistically suppressed MYC expression and cell survival in vitro as well as tumor xenograft growth. Furthermore, MUC1 expression significantly correlated with that of MYC and its target genes in human KRAS mutant NSCLC tumors. Taken together, these findings suggest a therapeutic approach for targeting MYC-dependent cancers and provide the framework for the ongoing clinical studies addressing the efficacy of MUC1-C inhibition in solid tumors.
Introduction
MYC functions as a DNA-binding transcription factor that activates a cellular program of genes contributing to the control of cell growth, metabolism, protein synthesis and survival (1, 2). Dysregulation of MYC expression occurs in diverse human cancers (3) and is sufficient to confer tumorigenesis in transgenic mouse models (4). MYC is also of importance for tumor maintenance as evidenced by tumor regressions in response to MYC downregulation (5) or treatment with an inhibitor of MYC heterodimerization (6). Moreover, targeting MYC expression with the bromodomain and extra-terminal (BET) bromodomain inhibitor JQ1 is active against models of multiple myeloma (7), leukemia (8) and NUT midline carcinoma (9). Other studies have provided evidence for the dependence of mutant KRAS tumors on MYC signaling (10). In this context, inducible expression of the dominant-negative MYC mutant, designated OmoMyc, eradicates KRAS-driven non-small cell lung cancer in mice (6, 11). Induction of a dominant-negative MYC allele in a KRAS-dependent mouse model of lung cancer has also demonstrated the effectiveness of inhibiting MYC function (12). In addition, BET bromodomain inhibition with JQ1 is effective against transgenic mouse models of NSCLC expressing mutant KRAS (13). These findings have provided support for the notion that MYC is an attractive target for the treatment of KRAS mutant NSCLC.
Mucin 1 (MUC1) is a transmembrane heterodimeric protein that is aberrantly expressed in over 80% of NSCLCs (14). Moreover, aberrant expression of MUC1 in NSCLC is associated with poor disease-free and overall survival (14–16). MUC1 consists of an extracellular N-terminal subunit (MUC1-N) that contains glycosylated tandem repeats, which are characteristic of the mucin family (17). MUC1-N forms a complex with the transmembrane MUC1 C-terminal subunit (MUC1-C) (17). MUC1-C includes an intrinsically disordered 72 amino acid cytoplasmic domain that is phosphorylated by diverse kinases and interacts with various effectors that have been linked to transformation (18). In this way, the MUC1-C cytoplasmic domain activates the WNT/β-catenin pathway by binding directly to β-catenin (19). In turn, MUC1-C stabilizes β-catenin and promotes the induction of certain WNT target genes, such as cyclin D1 (20, 21). The MUC1-C cytoplasmic domain contains a CQC motif that is necessary for MUC1-C homodimerization and function (22, 23). Notably, expression of MUC1-C with a CQC→AQA mutation inhibits anchorage-independent growth and tumorigenicity of cancer cells, indicating that the AQA mutant functions as a dominant-negative for transformation (22, 24). Accordingly, cell-penetrating peptides, such as GO-203, have been developed to target the MUC1-C CQC motif and block MUC1-C-mediated activation of growth and survival pathways in NSCLC cells (25). In addition, targeting MUC1-C in KRAS mutant NSCLC cells with GO-203 and other approaches, such as silencing, have shown that MUC1-C drives the epithelial-mesenchymal transition (EMT) and confers stemness (26).
The present studies demonstrate that MUC1-C activates MYC gene transcription in mutant KRAS NSCLC cells. Targeting MUC1-C thus suppresses MYC expression and the induction of key MYC target genes, such as hTERT and CDK4. We further show that (i) treatment of KRAS mutant NSCLC cells with GO-203 is synergistic with the BET bromodomain inhibitor JQ1 in targeting MYC activation and survival and (ii) the GO-203/JQ1 combination is significantly more active than GO-203 or JQ1 alone in inhibiting growth of tumor xenografts and downregulating MYC levels in vivo. In support of these results, analysis of datasets from KRAS mutant NSCLC tumors demonstrated a significant correlation between MUC1 and MYC expression.
Materials and Methods
Cell culture
Human A549/KRAS(G12S), H460/KRAS(Q61H), H358/KRAS(G12C), H441/KRAS(G12V), H2009/KRAS(G12A), H1975/EGFR(L858R/T790M), PC9GR/EGFR(delE746_A750) and H838s(mutant p53) NSCLC cells (ATCC) were grown in RPMI1640 medium supplemented with 10% heat-inactivated fetal bovine serum (HI-FBS), 100 μg/ml streptomycin, 100 units/ml penicillin and 2 mM L-glutamine. Cells were infected with lentiviral vectors expressing a scrambled control shRNA (CshRNA; Sigma) or a MUC1 shRNA (Sigma)(26). Cells stably expressing MUC1-C or MUC1-C(CQC→AQA) were generated as described (26). Cells were treated with the MUC1-C inhibitor peptide GO-203 or a control peptide CP-2 (25).
MUC1 silencing by CRISPR editing
The knockdown of MUC1 expression by CRISPR/cas9 was achieved as described (27). The sgRNAs targeting the first exon of the MUC1 gene were cloned into a lenti-CRISPR plasmid (Genome Engineering Production Group, Harvard Medical School). The viral vectors were produced in HEK293T cells as previously described (28). Cells were transduced with the vectors and cultured in the presence of puromycin. Single cell clones were selected by limiting dilution.
Immunoblot analysis
Whole cell lysates were prepared in NP-40 lysis buffer and analyzed by immunoblotting with anti-MUC1-C (LabVision), anti-MYC (Abcam), anti-β-actin (Sigma), anti-CDK4 (Cell Signaling Technology), anti-cyclin D1 (NeoMarkers), anti-phospho-Rb, and anti-Rb (BD Biosciences) as described (28). Immune complexes were detected with horseradish peroxidase secondary antibodies and enhanced chemiluminescence (GE Healthcare).
Quantitative RT-PCR
Whole cell RNA was isolated using the RNeasy mini kit (Qiagen). cDNAs were synthesized from 1 μg RNA using the High Capacity cDNA Reverse Transcription kit (Life Technologies). The cDNAs were amplified using the SYBR green qPCR assay kit with the ABI Prism 7000 Sequence Detector (Applied Biosystems). Primers used for detection of MYC, MUC1, CDK4 and hTERT mRNAs are listed in Supplemental Table S1.
Promoter-reporter assays
Cells growing in 6-well plates were transfected with 1.5 μg of TOPFlash (Addgene), pGL3 basic vector (Promega) or pGL3-MYC-Luc promoter vector (29) and SV-40-Renilla-Luc in the presence of Superfect (Qiagen). The pGL3-pMYC-Luc vector was also mutated using the QuickChange II kit (Agilent Technologies). At 48 h after transfection, the cells were lysed in passive lysis buffer. Lysates were analyzed with the Dual-Luciferase assay kit (Promega).
Chromatin immunoprecipitation (ChIP) assays
Soluble chromatin was prepared from 2–3 × 106 cells as described (30) and precipitated with anti-TCF4 (Santa Cruz) or a control nonimmune IgG. For re-ChIP assays, TCF4 complexes from the primary ChIP were released and reimmunoprecipitated with anti-MUC1-C (NeoMarkers), anti-β-catenin (Cell Signaling Technology), anti-p300 (Santa Cruz) and anti-acetyl histone H3 (Millipore) as described (21). The SYBR green qPCR kit was used for the qPCR analyses with the ABI Prism 7000 Sequence Detector (Applied Biosystems). Primers used for the MYC promoter and control GAPDH region are listed in Supplemental Table S2. Relative fold enrichment was calculated as described (21).
Colony formation assays
Cells were seeded in 6-well plates for 24 h and then treated with GO-203 each day and/or with JQ1 every 3 days. After 10 d, the cells were stained with 0.5% crystal violet in 25% methanol. Colonies >25 cells were counted in triplicate wells.
Determination of IC50 values and survival analysis
Cells were seeded on 96-well plates in 100 μl growth medium at a density of 1000–2000 cells per well. After 24 h, the cells were exposed to GO-203 and/or JQ1 for an additional 72 h. Cell viability was assessed using the CellTiter-Glo® One Solution (Promega). Triplicate wells for each treatment were analyzed and each experiment was performed three times. The IC50 was determined by nonlinear regression of the dose-response data using Prism 5.0 for Mac OSX (GraphPad Software). Cells were exposed to 1:1 ratios of the respective IC50 values for GO-203 and JQ1 at 1/4 × IC50, 1/2 × IC50, IC50, 2 × IC50 and 4 × IC50. Assessment of synergy was performed using CalcuSyn software (Biosoft). The combination index (CI) was calculated to assess synergism (CI<1) or antagonism (CI>1).
NSCLC tumor xenograft treatment studies
Six-week old female NCR nu/nu mice were injected with 3 × 106 H460 cells subcutaneously in the flank. When the tumors reached ~150 mm3, the mice were pair-matched into 4 groups of 5 mice each, and treated and with (i) control vehicle, (ii) 12 mg/kg GO-203 administered intraperitoneally (IP) each day, (iii) 50 mg/kg JQ1 administered IP each day, or (iv) both GO-203 and JQ1 administered IP each day. Tumor volumes were calculated using the formula V=L × W2/2, where L and W are the larger and smaller diameters, respectively.
Analysis of NSCLC databases
Clinical data sets of lung adenocarcinoma patients with corresponding KRAS and EGFR mutation status were downloaded from GEO under accession numbers GSE32867 and GSE29066. MYC-dependent genes were collected from the PCR-based MYC target gene array. Gene expression values were calculated relative to the median value of KRAS wild-type tumors. Multiple probe set IDs for a given gene were averaged for each patient sample after normalization to obtain a representative expression value for each gene. Heatmaps were created by hierarchical clustering via Ward’s method to display gene expression patterns. Statistical analysis and clustering were performed using JMP 9.0 (SAS Institute).
Results
Targeting MUC1 downregulates MYC expression
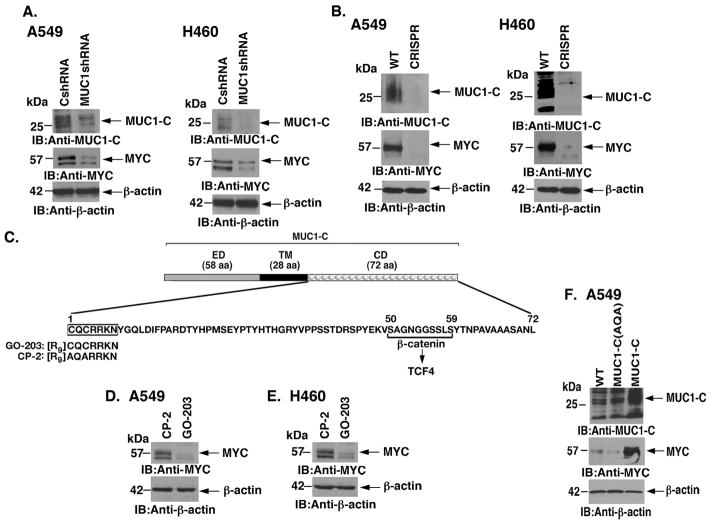
To determine whether MUC1-C contributes to MYC regulation, we first studied the effects of targeting MUC1-C in KRAS mutant A549 lung cancer cells (Fig. 1A). Partial suppression of MUC1-C with a MUC1shRNA was associated with downregulation of MYC levels (Fig. 1A, left). Similar results were obtained in KRAS mutant H460 lung cancer cells (Fig. 1A, right). To extend these studies, silencing MUC1-C with CRISPR genome editing also suppressed MYC expression in A549 and H460 cells (Figs. 1B, left and right). Moreover, silencing MUC1-C in KRAS mutant H441 (Supplemental Fig. S1A), H358 (Supplemental Fig. S1B) and H2009 (Supplemental Fig. S1C) cells was associated with suppression of MYC. Intriguingly, however, targeting MUC1-C in EGFR mutant H1975 and PC9 NSCLC cells, which also express both MUC1-C and MYC (Supplemental Fig. S2A), had no apparent effect on MYC levels (Supplemental Figs. S2B and S2C), indicating that MUC1-C selectively promotes MYC expression in KRAS mutant lung cancer cells. The MUC1-C cytoplasmic domain contains a CQC motif that is necessary for MUC1-C homodimerization and function (22, 23)(Fig. 1C). Accordingly, the cell penetrating peptide, designated GO-203, was developed to block the MUC1-C CQC motif in lung and other carcinoma cells (18, 25)(Fig. 1C). Notably, treatment of A549 and H460 cells with GO-203, but not the control peptide CP-2, was associated with downregulation of MYC (Figs. 1D and E). By contrast, targeting MUC1-C with GO-203 in H838s NSCLC cells (KRAS and EGFR wild-type; p53 mutant) had little if any effect on MYC expression (Supplemental Fig. S2D, left and right). A549 cells were also stably transduced to express MUC1-C or the MUC1-C(AQA) mutant (Fig. 1F). Overexpression of MUC1-C was associated with a marked increase in MYC levels (Fig. 1F). In addition, expression of the MUC1-C(AQA) mutant resulted in suppression of MYC as compared to that in wild-type cells (Fig. 1F). These findings collectively supported the premise that MUC1-C activates MYC expression.
Figure 1. Targeting MUC1-C downregulates MYC expression.
A. A549 (left) and H460 (right) lung cancer cells were transduced with lentiviral vectors to stably express a Control shRNA (CshRNA) or a MUC1 shRNA. Lysates from the indicated cells were immunoblotted with antibodies against MUC1-C, MYC and β-actin as a control. B. A549 (left) and H460 (right) cells were silenced for MUC1 using CRISPR/cas9. Lysates from wild-type (WT) and CRISPR cells were immunoblotted with the indicated antibodies. C. Schema of the MUC1-C subunit with a 58 amino acid (aa) extracellular domain (ED) and the 28 aa transmembrane domain (TM). The sequence of the 72 aa cytoplasmic domain (CD) is highlighted at the CQCRRKN motif, which is targeted by the cell penetrating GO-203 peptide and not the control CP-2 peptide. Also highlighted is the β-catenin binding site (SAGNGGSSLS). D and E. A549 (D) and H460 (E) cells were treated with 5 μM GO-203 or CP-2 for 48 h. Lysates were immunoblotted with the indicated antibodies. F. A549 cells were transduced to stably express MUC1-C or MUC1-C(AQA). Lysates from the indicated cells were immunoblotted with antibodies against MUC1-C, MYC and β-actin.
MUC1-C drives MYC transcription
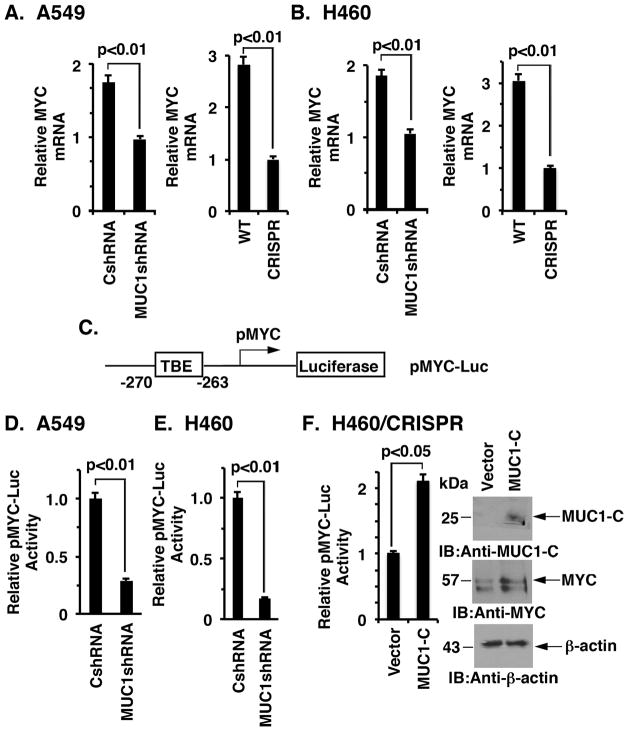
To define in part the mechanism by which MUC1-C contributes to MYC regulation, we assessed the effects of silencing MUC1-C on MYC mRNA levels. Downregulation of MUC1-C in A549 cells with shRNA was associated with suppression of MYC transcripts as determined by qRT-PCR (Fig. 2A, left). Similar results were obtained with A549 cells in which MUC1-C was silenced with CRISPR editing (Fig. 2A, right). Moreover, silencing MUC1-C in H460 cells with either approach resulted in downregulation of MYC mRNA levels (Fig. 2B, left and right). To determine whether MUC1-C activates the MYC promoter, we transfected cells to express a Del4 pMYC-Luc reporter that contains a TCF4 binding element (TBE) upstream of the MYC transcription start site (Fig. 2C). Using this approach, we found that targeting MUC1-C in A549 cells suppresses MYC promoter activation (Figs. 2D). Similar results were obtained when H460 cells were transfected to express pMYC-Luc (Fig. 2E). To confirm these results, we overexpressed MUC1-C in H460/CRISPR cells in a “rescue” experiment, which showed induction of pMYC-Luc activity and MYC expression (Fig. 2F, left and right). Similar results were obtained when A549/CRISPR cells were rescued with MUC1-C (Supplemental Fig. S3, left and right), indicating that MUC1-C induces MYC expression at least in part at the transcriptional level.
Figure 2. Targeting MUC1-C suppresses MYC transcription.
A and B. The indicated A549 (A) and H460 (B) cells were analyzed for MYC mRNA levels by qRT-PCR. The results (mean±SD of three determinations) are expressed as relative MYC mRNA levels as compared to that obtained for the MUC1-C silenced cells (assigned a value of 1). C. Schema of the Del4 pMYC-Luc reporter with positioning of the TCF4 binding element (TBE) at position −270 to −263 upstream to the transcription start site. D and E. The indicated A549 (D) and H460 (E) cells were transfected with the pMYC-Luc reporter for 48 h and then assayed for luciferase activity. The results are expressed as the relative luciferase activity (mean±SD of three determinations) compared with that obtained the cells expressing the control CshRNA (assigned a value of 1). F. H460/CRISPR cells were transiently transfected to express an empty vector or one expressing MUC1-C and the pMYC-Luc reporter for 48 h. The results are expressed as the relative luciferase activity (mean±SD of three determinations) compared with that obtained the cells expressing the control vector (assigned a value of 1)(left). Lysates from the indicated cells were immunoblotted with antibodies against MUC1-C, MYC and β-actin (right).
MUC1-C activates the MYC promoter through the TCF4 binding element
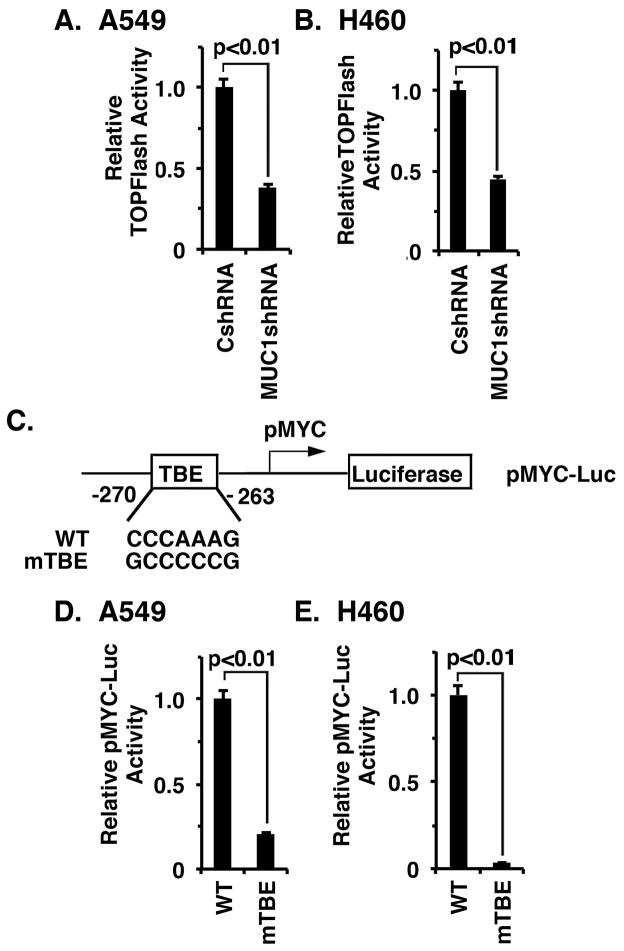
MUC1-C has been linked to activation of the WNT/β-catenin pathway (18). Moreover, MYC is a downstream effector of WNT/β-catenin signaling (1). Accordingly, we first asked if targeting MUC1-C in A549 cells affects activation of the TOPFlash reporter, which is driven by β-catenin/TCF4 complexes (31). Here, we found that silencing MUC1-C significantly decreases TOPFlash activity (Fig. 3A). Similar results were obtained in H460 cells silenced for MUC1-C (Fig. 3B), indicating that MUC1-C activates the WNT/β-catenin pathway in these NSCLC cells. To determine whether MUC1-C drives the MYC promoter through WNT/β-catenin signaling, we assessed the effects of mutating the TBE (Fig. 3C). Using this approach, we found that the TBE mutation markedly decreases pMYC-Luc activity in A549 (Fig. 3D) and H460 (Fig. 3E) cells, indicating that MUC1-C induces MYC gene expression through activation of the WNT/β-catenin/TCF4 pathway.
Figure 3. MUC1-C induces MYC transcription by the WNT/β-catenin/TCF4 pathway.
A and B. The indicated A549 (A) and H460 (B) cells were transfected with TOPFlash for 48 h and then assayed for luciferase activity. The results (mean±SD from 3 determinations) are expressed as relative TOPFlash activity as compared to that obtained in cells expressing the CshRNA (assigned a value of 1). C. Schema of the pMYC-Luc reporter highlighting the mutated TBE (mTBE) site. D and E. A549/CshRNA (D) and H460/CshRNA (E) cells were transfected with wild-type (WT) or TBE-mutated pMYC-Luc. The results (mean±SD of three determinations) are expressed as the relative pMYC-Luc activity compared to that for cells transfected with the WT pMYC-Luc (assigned a value of 1).
MUC1-C occupies the MYC promoter
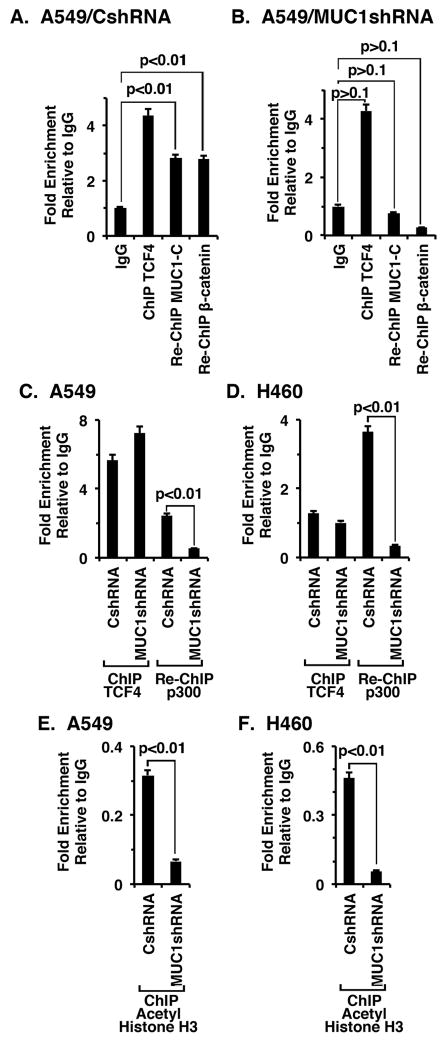
Previous work demonstrated that MUC1-C binds directly to β-catenin (19, 20) and forms a complex with β-catenin/TCF4 on the cyclin D promoter (21). In the present studies, ChIP analysis of the MYC promoter in A549 cells confirmed TCF4 occupancy (Fig. 4A). Moreover, the results of re-ChIP assays showed that TCF4 occupies the MYC promoter in complexes with MUC1-C and β-catenin (Fig. 4A). Silencing MUC1-C in A549 cells had little if any effect on TCF4 occupancy, but in re-ChIP assays was associated with significant decreases in both MUC1-C and β-catenin (Fig. 4B). In the absence of β-catenin, TCF4 recruits corepressors, such as HDAC1 (32, 33). By contrast, the interaction between TCF4 and β-catenin displaces corepressors and promotes recruitment of the p300 histone acetyltransferase and coactivator (34, 35). In concert those findings, silencing MUC1-C was associated with a significant decrease in p300 occupancy on the MYC promoter in A549 (Fig. 4C) and H460 (Fig. 4D) cells. Moreover, silencing MUC1-C was associated with a marked decrease in histone H3 acetylation of the MYC promoter (Figs. 4E and F).
Figure 4. MUC1-C occupies the MYC promoter with TCF4 and β-catenin and regulates histone H3 acetylation.
A and B. Soluble chromatin from the indicated A549/CshRNA (A), A549/MUC1shRNA (B) cells was precipitated with anti-TCF4 or a control IgG. In the re-ChIP experiments, TCF4 precipitates were released and reimmunoprecipitated with anti-MUC1-C or anti-β-catenin. The final DNA samples were amplified by qPCR with primers for the MYC promoter TBE binding region or as a control GAPDH. The results (mean±SD of three determinations) are expressed as the relative fold enrichment compared with that obtained with the IgG control. C and D. Soluble chromatin from the indicated A549 (C) and H460 (D) cells was precipitated with anti-TCF4 or a control IgG. In the re-ChIP experiments, TCF4 precipitates were released, reimmunoprecipitated with anti-p300 and then analyzed for MYC promoter sequences by qPCR. The results (mean±SD of three determinations) are expressed as the relative fold enrichment compared with that obtained with the IgG control. E and F. Soluble chromatin from the indicated A549 (E) and H460 (F) cells was precipitated with anti-acetylated histone H3 or a control IgG, and then analyzed for MYC promoter sequences by qPCR. The results (mean±SD of three determinations) are expressed as the relative fold enrichment compared with that obtained with the IgG control.
Silencing MUC1-C downregulates MYC-target genes
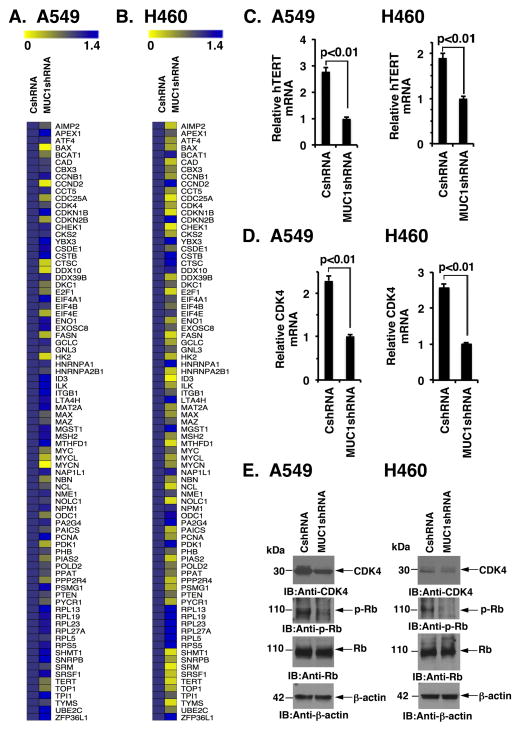
To assess the effects of silencing MUC1-C on MYC-induced transcription, we analyzed MYC target gene expression in cells using a commercially available PCR array. Other MYC family members (MYCN, MYCL) and MAX are also represented in this array. Interestingly and in addition to MYC, marked suppression of MYCN and MYCL mRNAs was observed in the response of A549 cells to MUC1-C silencing (Fig. 5A). Moreover, we found downregulation of MAX expression (Fig. 5A). Among others, suppression of the MYC target genes, hTERT (36) and CDK4 (37), was also detected in the A549/MUC1shRNA cells (Fig. 5A). Similar results were obtained when analyzing the effects of MUC1-C silencing in H460 cells (Fig. 5B). To confirm these findings, at least in part, we found that silencing MUC1-C is associated with suppression of hTERT (Fig. 5C, left and right) and CDK4 (Fig. 5D, left and right) mRNA levels. And, in concert with these results, we found that silencing MUC1-C results in downregulation of CDK4 protein and phosphorylation of its downstream target Rb (Fig. 5E, left and right).
Figure 5. Silencing MUC1-C downregulates MYC-target genes.
A and B. Expression of MYC family members and MYC target genes was analyzed in the indicated A549 (A) and H460 (B) cells using the commercially PCR array (Qiagen). The results obtained in cells without and with MUC1-C silencing are highlighted in the heat maps. C and D. The indicated A549 and H460 cells were analyzed for hTERT (C) and CDK4 (D) mRNA levels by qRT-PCR. The results (mean±SD of three determinations) are expressed as relative hTERT or CDK4 mRNA levels as compared to that obtained MUC1-C silenced cells (assigned a value of 1). E. Lysates from the indicated A549 and H460 cells were immunoblotted with antibodies against CDK4, phospho-Rb (p-Rb), Rb and β-actin.
Targeting MUC1-C is synergistic with JQ1 in the treatment of KRAS mutant NSCLC cells
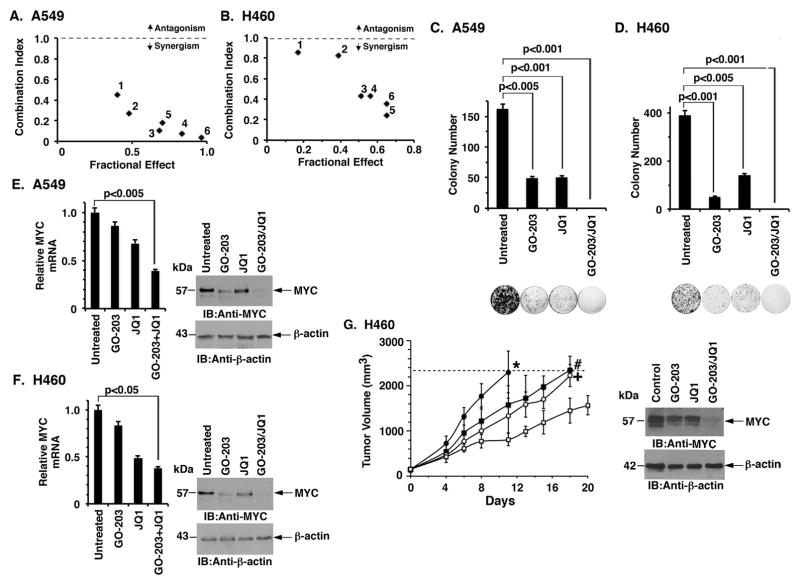
The JQ1 BET bromodomain inhibitor was the first agent that was found to suppress MYC transcription (7). Based on these and the present findings, we investigated whether combining GO-203 with JQ1 is effective against KRAS mutant NSCLC cells. Accordingly, we first defined the half-maximal inhibitory concentrations of these agents when used alone in the treatment of A549 cells and then performed an isobologram analysis. The results obtained demonstrated a synergistic interaction (Fig. 6A). Determination of the CI values at ED50, ED75 and ED90 demonstrated that the GO-203/JQ1 combination is synergistic (Supplemental Table S3A). Similar results were obtained when H460 cells were treated with GO-203 and JQ1 (Fig. 6B and Supplemental Table S3B); however, there was no evidence for synergy when H1975 (Supplemental Table S3C) or PC9GR (Supplemental Table S3D) cells were exposed to these agents. Treatment of A549 (Fig. 6C) and H460 (Fig. 6D) cells with synergistic combinations was also more effective in suppressing clonogenic survival than that obtained with GO-203 or JQ1 alone. We similarly found that treatment of A549 cells with a synergistic combination of GO-203/JQ1 results in more pronounced suppression of MYC mRNA (Fig. 6E, left) and protein (Fig. 6E, right) levels than that obtained with GO-203 or JQ1 alone. Moreover, in experiments with H460 cells, combining GO-203 and JQ1 was more effective in suppressing MYC expression (Fig. 6F, left and right) than either agent alone. To extend this analysis, treatment of established H460 tumor xenografts further showed that the GO-203/JQ1 combination is significantly more active than GO-203 or JQ1 alone in inhibiting tumor growth and downregulating MYC levels (Fig. 6G, left and right).
Figure 6. GO-203 is synergistic with JQ1 in the treatment of KRAS mutant NSCLC cells.
A and B. The indicated A549 (A) and H460 (B) were treated with (i) GO-203 alone each day for 72 h, (ii) JQ1 for 72 h, or (iii) GO-203 combined with JQ1 for 72 h. Numbers 1 to 6 in the graphs represent combinations listed in Supplemental Tables S3A and B. Mean cell survival was assessed in triplicate by CellTiter-Glo® One Solution assays. C and D. The indicated A549 (C) and H460 (D) cells were seeded at 1000 cells/well (6-well plate), left (i) untreated, (ii) treated with 2 μM of GO-203 alone every 24 h, (iii) 5 μM of JQ1 alone every 72 h, or (iv) the combination of GO-203 and JQ1. After 10 d, the cells were stained with crystal violet. Colony number (>25 cells) is expressed as the mean±SD of three replicates. E and F. A549 (E) and H460 (F) cells were treated with 2 μM of GO-203 or/and 5 μM of JQ1 for 72 h. Cells were analyzed for MYC mRNA levels by qRT-PCR (left). The results (mean±SD of three determinations) are expressed as relative MYC mRNA levels as compared to that obtained for the untreated cells (assigned a value of 1). Lysates from the indicated cells were immunoblotted with antibodies against MYC and β-actin (right). G. Mice bearing established H460 tumor xenografts (~150 mm3) were treated IP with daily administration of control vehicle (closed circles), 12 mg/kg GO-203 (closed squares), 50 mg/kg JQ1 (open circles) or the GO-203/JQ1 combination (open squares). The results are expressed as tumor volume (mean±SEM; 5 mice/group)(left). Statistical analysis (Student’s t-test) comparing the results from the GO-203/JQ1 combination-treated group with those from the (i) control group, p=0.011 (*); (ii) GO-203 group, p=0.033 (#); and (iii) JQ1 group, p=0.016 (+), where p<0.05 is significant. Tumor lysates from control (day 11) and the treated mice (day 18) were immunoblotted with the indicated antibodies (right).
MYC-dependent gene expression in human KRAS mutant lung cancers
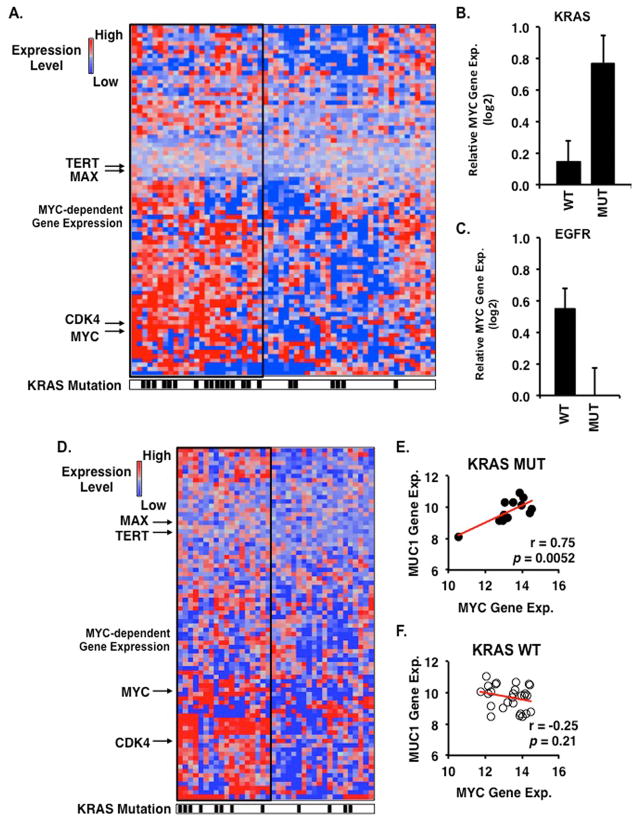
Our experimental results obtained from cell-based and mouse models lend support to a functional association of MUC1-C with MYC in KRAS mutant lung adenocarcinomas. However, there is no evidence that MUC1-C is linked to expression of MYC-dependent target genes in patients with lung cancer. We therefore investigated the expression of MYC target genes in clinical data sets of KRAS wild-type and mutant lung adenocarcinomas. Microarray analysis of 82 MYC-dependent genes obtained from the PCR-based MYC array identified ~40% of patients with overexpression of MYC target genes (Fig. 7A). We examined overexpression of MYC-dependent genes as a function of KRAS mutation status. The results demonstrate a significantly higher incidence of KRAS mutations in tumors with elevated levels of MYC target gene expression (64% vs. 18%, p=0.0008, 2-tailed Fisher’s exact test, Fig. 7A). In addition, tumors bearing KRAS mutations significantly overexpressed the MYC gene (p=0.0060, 2-tailed t-test) as compared to KRAS wild-type tumors (Fig. 7B). In contrast, EGFR mutant tumors demonstrated reduced levels of MYC expression as compared to EGFR wild-type tumors (Fig. 7C). In an independent clinical data set of lung adenocarcinomas, we confirmed an association between MYC-dependent target gene expression and KRAS mutations (Fig. 7D). Importantly, we found a strong correlation between MUC1 and MYC gene expression values in KRAS mutant tumors (Pearson correlation coefficient (r=0.75, p=0.0052), but not in KRAS wild-type tumors (r=−0.25, p=0.21). Taken together, these results demonstrate that KRAS mutant lung adenocarcinomas overexpress MYC and MYC-dependent genes and support a role for MUC1 in driving MYC expression in KRAS mutant tumors.
Figure 7. Activation of MYC-dependent gene expression in KRAS mutant lung adenocarcinomas.
A. Heatmap of 82 MYC-dependent genes in 58 lung adenocarcinomas (GSE32867) demonstrating increased gene expression in KRAS mutant tumors (n=22) (denoted by black hash marks below heatmap) as compared to KRAS wild-type tumors (n=36). Samples displaying overexpression of MYC target genes are enclosed within a thick black rectangle. B. Relative MYC gene expression in KRAS wild-type (WT) and mutant (MUT) tumors. C. Relative MYC gene expression in EGFR wild-type and mutant tumors. Graphs display mean±SEM of MYC gene expression normalized to the median value of wild-type tumors. D. Heatmap of 82 MYC-dependent genes in 38 lung adenocarcinomas (GSE29066) demonstrating increased gene expression in KRAS mutant tumors (n=12) as compared to KRAS wild-type tumors (n=26). Pearson correlation analysis of MUC1 and MYC gene expression in KRAS mutant (E) and wild-type (F) tumors.
Discussion
NSCLCs that harbor an oncogenic KRAS mutation are often resistant to conventional and targeted agents (38). In addition, inhibitors of activated KRAS have been clinically ineffective to date, supporting the need for identifying agents that block downstream KRAS signaling pathways. Previous work showed that targeting MUC1-C in KRAS mutant NSCLC cells is associated with decreases in self-renewal and tumorigenicity; however, the mechanism(s) responsible for these responses was unclear (26). The present studies provide new insights into the function of MUC1-C in KRAS mutant NSCLC cells by demonstrating that targeting MUC1-C is associated with downregulation of MYC expression. MYC is of functional importance in orchestrating transcriptional pathways that regulate cell cycle progression, metabolism, survival and stemness (1, 39, 40). Therefore, the effects of targeting MUC1-C on self-renewal of KRAS mutant NSCLC cells can be attributable, at least in part, to suppression of MYC signaling. Notably, we have used multiple approaches for targeting MUC1-C that include (i) silencing with MUC1 shRNA and with CRISPR editing, and (ii) treatment with the GO-203 inhibitor. Using these different experimental conditions, diverse KRAS mutant NSCLC cells responded with MYC downregulation, providing convincing support for a MUC1-C→MYC pathway. By contrast and interestingly, targeting MUC1-C in NSCLC cells harboring EGFR mutations had little if any effect on MYC expression, indicating that the link between MUC1-C and MYC in NSCLC may be limited to the KRAS mutant setting.
The MUC1-C cytoplasmic domain is largely an intrinsically disordered protein (41), which is often found in effectors at regulatory nodes that integrate posttranslational signaling mechanisms (42). In this regard, the MUC1-C cytoplasmic domain interacts with multiple kinases and transcription factors that have been linked to growth, survival and transformation (18). In the present studies, we found that MUC1-C drives MYC gene transcription by activation of the WNT/β-catenin pathway. The MUC1-C cytoplasmic domain contains a motif with homology to sequences found in E-cadherin and APC that function as β-catenin binding sites (19, 43). The MUC1-C cytoplasmic domain thereby binds directly to β-catenin Armadillo repeats and, in turn, blocks GSK3β-mediated phosphorylation and degradation of β-catenin (20). MUC1-C also interacts with the TCF4 transcription factor and promotes the formation of β-catenin/TCF4 complexes on WNT target genes (21). In concert with this model, we found that MUC1-C occupies the MYC promoter with TCF4 in NSCLC cells. In addition, MUC1-C increased occupancy of β-catenin on the MYC promoter and thereby contributed to the recruitment of the p300 histone acetylase coactivator with induction of MYC gene transcription. These findings thus support a model in which MUC1-C directly activates the MYC promoter in a complex with β-catenin/TCF4. MUC1 has been linked to activation of the WNT/β-catenin pathway in certain cancer types (18); however, to our knowledge, not previously in NSCLC cells. In addition, certain studies in breast cancer cells have shown that targeting MUC1 is associated with upregulation of MYC expression (44). Thus, the demonstration that MUC1-C drives MYC by a β-catenin-mediated mechanism in KRAS mutant NSCLC cells is of particular interest given their dependence on MYC for survival.
The MYC target gene network is estimated to comprise about 15% of all human genes that are subject to both MYC-induced activation and repression (45). To assess the effects of targeting MUC1-C on MYC target genes, we used a commercially available array that also included MYCN, MYCL and MAX. Surprisingly, we found that targeting MUC1-C is associated with downregulation of these genes, indicating that MUC1-C regulates all of the MYC family members, as well as MAX, the requisite binding partner for MYC-mediated transactivation (1). Using the same array, we found that targeting MUC1-C results in the upregulation and suppression of a number of different MYC target genes. For the present studies, we confirmed that targeting MUC1-C downregulates hTERT expression, based on the importance of this catalytic subunit of telomerase in maintaining telomere length and extending the life span of cells (46). Moreover, and concert with activated RAS and MYC, hTERT is an essential effector in models of transforming primary human cells (47). We also confirmed that targeting MUC1-C results in suppression of CDK4, a MYC target gene that is essential for cell cycle progression (39). In addition, CDK4 is of importance for the growth and survival of NSCLC cells expressing mutant KRAS (48). And, like hTERT, ectopic expression of CDK4 has been associated with immortalization of primary human cells (47). These findings and the significant correlation between MUC1 and MYC expression identified in datasets from KRAS mutant NSCLC tumors thus support the notion that MUC1-C drives MYC and thereby MYC target genes that are of importance for transformation.
Dysregulation of MYC expression is observed at high frequency in NSCLC and diverse other cancer types, supporting MYC as a highly attractive target (49). Blocking MYC/MAX heterodimerization with Omomyc in KRAS-driven lung cancer in mice has further validated MYC as a target with an acceptable therapeutic index (11). Moreover, treatment of a transgenic mouse KRAS mutant NSCLC model with JQ1 has shown that targeting MYC by BET bromodomain inhibition is a promising therapeutic strategy (13). The present studies demonstrate that targeting MUC1-C with GO-203 is another potential approach for suppressing MYC expression. Accordingly and based on the findings that GO-203 and JQ1 inhibit MYC transcription by distinct mechanisms, we investigated the potential for combining these agents. Indeed, our studies show that combining GO-203 and JQ1 is synergistic in inhibiting growth and that GO-203 potentiates the inhibitory effects of JQ1 on MYC expression. The GO-203/JQ1 combination was also more effective in inhibiting growth of H460 tumor xenografts than either agent alone. The BET bromodomain inhibitors GSK525762 and TEN-010 are being evaluated in Phase I clinical trials. Moreover, a Phase I trial of GO-203 has been completed in patients with refractory solid tumors and has been formulated in nanoparticles for assessing the effectiveness of this agent in treating NSCLC (50). Based on the present results, subsequent studies that assess the activity of GO-203 with BET bromodomain inhibitors could be potentially informative in the setting of KRAS mutant NSCLC and possibly other MYC-dependent cancers.
Supplementary Material
Acknowledgments
Financial Support: Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under award numbers CA166480 and CA97098, and the Lung Cancer Research Foundation.
Abbreviations
- MUC1
Mucin 1
- MUC1-C
MUC1 C-terminal transmembrane subunit
- NSCLC
non-small cell lung cancer
- CRISPR
clustered regularly interspaced short palindromic repeats
- BET
bromodomain and extra-terminal
- TCF
T cell factor
- TBE
TCF4 binding element
- ChIP
chromatin immunoprecipitation
Footnotes
Conflict of Interest: D.K. holds equity in Genus Oncology and is a consultant to the company. The other authors disclosed no potential conflicts of interest.
References
- 1.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li B, Simon MC. Molecular Pathways: Targeting MYC-induced metabolic reprogramming and oncogenic stress in cancer. Clin Cancer Res. 2013;19:5835–41. doi: 10.1158/1078-0432.CCR-12-3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leder A, Pattengale PK, Kuo A, Stewart TA, Leder P. Consequences of widespread deregulation of the c-myc gene in transgenic mice: multiple neoplasms and normal development. Cell. 1986;45:485–95. doi: 10.1016/0092-8674(86)90280-1. [DOI] [PubMed] [Google Scholar]
- 5.Arvanitis C, Felsher DW. Conditional transgenic models define how MYC initiates and maintains tumorigenesis. Semin Cancer Biol. 2006;16:313–7. doi: 10.1016/j.semcancer.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 6.Soucek L, Whitfield J, Martins CP, Finch AJ, Murphy DJ, Sodir NM, et al. Modelling Myc inhibition as a cancer therapy. Nature. 2008;455:679–83. doi: 10.1038/nature07260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–17. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–8. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–73. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yeh E, Cunningham M, Arnold H, Chasse D, Monteith T, Ivaldi G, et al. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat Cell Biol. 2004;6:308–18. doi: 10.1038/ncb1110. [DOI] [PubMed] [Google Scholar]
- 11.Soucek L, Whitfield JR, Sodir NM, Masso-Valles D, Serrano E, Karnezis AN, et al. Inhibition of Myc family proteins eradicates KRas-driven lung cancer in mice. Genes Dev. 2013;27:504–13. doi: 10.1101/gad.205542.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fukazawa T, Maeda Y, Matsuoka J, Yamatsuji T, Shigemitsu k, Morita I, et al. Inhibition of Myc effectively targets KRAS mutation-positive lung cancer expressing high levels of Myc. Anticancer Res. 2010;30:4193–200. [PubMed] [Google Scholar]
- 13.Shimamura T, Chen Z, Soucheray M, Carretero J, Kikuchi E, Tchaicha JH, et al. Efficacy of BET bromodomain inhibition in Kras-mutant non-small cell lung cancer. Clin Cancer Res. 2013;19:6183–92. doi: 10.1158/1078-0432.CCR-12-3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Situ D, Wang J, Ma Y, Zhu Z, Hu Y, Long H, et al. Expression and prognostic relevance of MUC1 in stage IB non-small cell lung cancer. Med Oncol. 2010;28:596–604. doi: 10.1007/s12032-010-9752-4. [DOI] [PubMed] [Google Scholar]
- 15.Khodarev N, Pitroda S, Beckett M, MacDermed D, Huang L, Kufe D, et al. MUC1-induced transcriptional programs associated with tumorigenesis predict outcome in breast and lung cancer. Cancer Res. 2009;69:2833–7. doi: 10.1158/0008-5472.CAN-08-4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.MacDermed DM, Khodarev NN, Pitroda SP, Edwards DC, Pelizzari CA, Huang L, et al. MUC1-associated proliferation signature predicts outcomes in lung adenocarcinoma patients. BMC Medical Genomics. 2010;3:16. doi: 10.1186/1755-8794-3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kufe D. Mucins in cancer: function, prognosis and therapy. Nature Reviews Cancer. 2009;9:874–85. doi: 10.1038/nrc2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kufe D. MUC1-C oncoprotein as a target in breast cancer: activation of signaling pathways and therapeutic approaches. Oncogene. 2013;32:1073–81. doi: 10.1038/onc.2012.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamamoto M, Bharti A, Li Y, Kufe D. Interaction of the DF3/MUC1 breast carcinoma-associated antigen and b-catenin in cell adhesion. J Biol Chem. 1997;272:12492–4. doi: 10.1074/jbc.272.19.12492. [DOI] [PubMed] [Google Scholar]
- 20.Huang L, Chen D, Liu D, Yin L, Kharbanda S, Kufe D. MUC1 oncoprotein blocks GSK3β-mediated phosphorylation and degradation of β-catenin. Cancer Res. 2005;65:10413–22. doi: 10.1158/0008-5472.CAN-05-2474. [DOI] [PubMed] [Google Scholar]
- 21.Rajabi H, Ahmad R, Jin C, Kosugi M, Alam M, Joshi M, et al. MUC1-C oncoprotein induces TCF7L2 activation and promotes cyclin D1 expression in human breast cancer cells. J Biol Chem. 2012;287:10703–13. doi: 10.1074/jbc.M111.323311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leng Y, Cao C, Ren J, Huang L, Chen D, Ito M, et al. Nuclear import of the MUC1-C oncoprotein is mediated by nucleoporin Nup62. J Biol Chem. 2007;282:19321–30. doi: 10.1074/jbc.M703222200. [DOI] [PubMed] [Google Scholar]
- 23.Raina D, Ahmad R, Rajabi H, Panchamoorthy G, Kharbanda S, Kufe D. Targeting cysteine-mediated dimerization of the MUC1-C oncoprotein in human cancer cells. Int J Oncol. 2012;40:1643–9. doi: 10.3892/ijo.2011.1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kufe D. Functional targeting of the MUC1 oncogene in human cancers. Cancer Biol Ther. 2009;8:1201–7. doi: 10.4161/cbt.8.13.8844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raina D, Kosugi M, Ahmad R, Panchamoorthy G, Rajabi H, Alam M, et al. Dependence on the MUC1-C oncoprotein in non-small cell lung cancer cells. Mol Cancer Therapeutics. 2011;10:806–16. doi: 10.1158/1535-7163.MCT-10-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kharbanda A, Rajabi H, Jin C, Alam M, Wong K, Kufe D. MUC1-C confers EMT and KRAS independence in mutant KRAS lung cancer cells. Oncotarget. 2014;5:8893–905. doi: 10.18632/oncotarget.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–23. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rajabi H, Alam M, Takahashi H, Kharbanda A, Guha M, Ahmad R, et al. MUC1-C oncoprotein activates the ZEB1/miR-200c regulatory loop and epithelial-mesenchymal transition. Oncogene. 2014;33:1680–9. doi: 10.1038/onc.2013.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, et al. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–12. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 30.Alam M, Ahmad R, Rajabi H, Kharbanda A, Kufe D. MUC1-C oncoprotein activates ERK→C/EBPβ-mediated induction of aldehyde dehydrogenase activity in breast cancer cells. J Biol Chem. 2013;288:30829–903. doi: 10.1074/jbc.M113.477158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, et al. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC−/− colon carcinoma. Science. 1997;275:1784–7. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 32.Brannon M, Brown JD, Bates R, Kimelman D, Moon RT. XCtBP is a XTcf-3 co-repressor with roles throughout Xenopus development. Development. 1999;126:3159–70. doi: 10.1242/dev.126.14.3159. [DOI] [PubMed] [Google Scholar]
- 33.Kioussi C, Briata P, Baek SH, Rose DW, Hamblet NS, Herman T, et al. Identification of a Wnt/Dvl/beta-Catenin --> Pitx2 pathway mediating cell-type-specific proliferation during development. Cell. 2002;111:673–85. doi: 10.1016/s0092-8674(02)01084-x. [DOI] [PubMed] [Google Scholar]
- 34.Daniels DL, Weis WI. Beta-catenin directly displaces Groucho/TLE repressors from Tcf/Lef in Wnt-mediated transcription activation. Nat Struct Mol Biol. 2005;12:364–71. doi: 10.1038/nsmb912. [DOI] [PubMed] [Google Scholar]
- 35.Evans PM, Chen X, Zhang W, Liu C. KLF4 interacts with beta-catenin/TCF4 and blocks p300/CBP recruitment by beta-catenin. Mol Cell Biol. 2010;30:372–81. doi: 10.1128/MCB.00063-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu KJ, Grandori C, Amacker M, Simon-Vermot N, Polack A, Lingner J, et al. Direct activation of TERT transcription by c-MYC. Nat Genet. 1999;21:220–4. doi: 10.1038/6010. [DOI] [PubMed] [Google Scholar]
- 37.Hermeking H, Rago C, Schuhmacher M, Li Q, Barrett JF, Obaya AJ, et al. Identification of CDK4 as a target of c-MYC. Proc Natl Acad Sci U S A. 2000;97:2229–34. doi: 10.1073/pnas.050586197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer. 2011;11:761–74. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bretones G, Delgado MD, Leon J. Myc and cell cycle control. Biochim Biophys Acta. 2015;1849:506–16. doi: 10.1016/j.bbagrm.2014.03.013. [DOI] [PubMed] [Google Scholar]
- 40.Conacci-Sorrell M, McFerrin L, Eisenman RN. An overview of MYC and its interactome. Cold Spring Harbor perspectives in medicine. 2014;4:a014357. doi: 10.1101/cshperspect.a014357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raina D, Agarwal P, Lee J, Bharti A, McKnight C, Sharma P, et al. Characterization of the MUC1-C cytoplasmic domain as a cancer target. PLoS One. 2015;10:e0135156. doi: 10.1371/journal.pone.0135156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gsponer J, Babu MM. The rules of disorder or why disorder rules. Progress in biophysics and molecular biology. 2009;99:94–103. doi: 10.1016/j.pbiomolbio.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 43.Huang L, Ren J, Chen D, Li Y, Kharbanda S, Kufe D. MUC1 cytoplasmic domain coactivates Wnt target gene transcription and confers transformation. Cancer Biol Ther. 2003;2:702–6. [PubMed] [Google Scholar]
- 44.Hattrup CL, Gendler SJ. MUC1 alters oncogenic events and transcription in human breast cancer cells. Breast Cancer Res. 2006;8:R37. doi: 10.1186/bcr1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dang CV, O’Donnell KA, Zeller KI, Nguyen T, Osthus RC, Li F. The c-Myc target gene network. Semin Cancer Biol. 2006;16:253–64. doi: 10.1016/j.semcancer.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 46.Wang J, Xie LY, Allan S, Beach D, Hannon GJ. Myc activates telomerase. Genes Dev. 1998;12:1769–74. doi: 10.1101/gad.12.12.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kendall SD, Adam SJ, Counter CM. Genetically engineered human cancer models utilizing mammalian transgene expression. Cell Cycle. 2006;5:1074–9. doi: 10.4161/cc.5.10.2734. [DOI] [PubMed] [Google Scholar]
- 48.Puyol M, Martin A, Dubus P, Mulero F, Pizcueta P, Khan G, et al. A synthetic lethal interaction between K-Ras oncogenes and Cdk4 unveils a therapeutic strategy for non-small cell lung carcinoma. Cancer Cell. 2010;18:63–73. doi: 10.1016/j.ccr.2010.05.025. [DOI] [PubMed] [Google Scholar]
- 49.McKeown MR, Bradner JE. Therapeutic strategies to inhibit MYC. Cold Spring Harbor perspectives in medicine. 2014;4:1–16. doi: 10.1101/cshperspect.a014266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hasegawa M, Sinha RK, Kumar M, Alam M, Yin L, Raina D, et al. Intracellular Targeting of the Oncogenic MUC1-C Protein with a Novel GO-203 Nanoparticle Formulation. Clin Cancer Res. 2015;21:2338–47. doi: 10.1158/1078-0432.CCR-14-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.