Abstract
Autophagy is a highly conserved mechanism that is activated during cellular stress. We hypothesized that autophagy may be induced by acid reflux, which causes injury and inflammation, and therefore contributes to the pathogenesis of Barrett’s esophagus (BE) and esophageal adenocarcinoma (EAC). Currently, the role of autophagy in BE and EAC is poorly studied. We quantitatively define autophagy levels in human BE cell lines, a transgenic mouse model of BE, and human BE and EAC biopsies. Human non-dysplastic BE had the highest basal number of autophagic vesicles (AVs), while AVs were reduced in normal squamous cells and dysplastic BE cells, and nearly absent in EAC. To demonstrate a functional role for autophagy in BE pathogenesis, normal squamous (STR), non-dysplastic BE (CPA), dysplastic BE (CPD), and esophageal adenocarcinoma (OE19) cell lines were exposed to an acid pulse (pH3.5) followed by incubation in the presence or absence of chloroquine, an autophagy inhibitor. Acid exposure increased reactive oxygen species (ROS) levels in STR and CPA cells. Chloroquine alone had a small impact on intracellular ROS or cell survival. However, combination of chloroquine with the acid pulse resulted in a significant increase in ROS levels at 6 hours in STR and CPA cells, and increased cell death in all cell lines. These findings establish increased numbers of AVs in human BE compared to normal squamous or EAC, and suggest that autophagy functions to improve cell survival after acid reflux injury. Autophagy may thus play a critical role in BE pathogenesis and progression.
Keywords: Barrett’s esophagus, autophagy, oxidative stress, esophageal adenocarcinoma, chloroquine
INTRODUCTION
Esophageal adenocarcinoma (EAC) is associated with approximately 15,000 deaths in the U.S. annually [1]. Over the past 30 years, EAC incidence has risen at an alarming rate, making it an increasingly important cause of morbidity and mortality [2]. EAC does not arise in normal squamous esophageal mucosa but develops in an altered, premalignant mucosa known as Barrett’s esophagus (BE) [3,4]. BE arises in the setting of chronic gastric acid and bile reflux that results in injury and inflammation. It is characterized by the replacement of normal squamous epithelium with a metaplasia consisting of columnar cells with a morphology and gene expression pattern more typical of intestinal epithelium[3,5]. The prevalence of Barrett’s esophagus is estimated at 15% of patients with gastroesophageal reflux disease (GERD)[6]. Given the high prevalence of BE, its association with EAC, and the increasing incidence of EAC, understanding BE pathogenesis is an important unmet clinical and research objective. However, little is known about the molecular mechanisms giving rise to BE.
The current paradigm for the induction of BE requires gastric acid and bile reflux to provoke tissue injury and the release of pro-inflammatory eicosanoids and cytokines[7]. One aspect of immune system activation with important implications for the induction of metaplasia and cancer is the increased production of highly reactive free-radical species such as reactive oxygen species (ROS). These agents contribute to cellular oxidative stress [8]. Interestingly, bile salts, a common component of GERD refluxate, can increase ROS [9,10]. Several genes function to protect cells from ROS, including superoxide dismutase, glutathione peroxidase and glutathione-S-transferase [11]. However, these important detoxifying pathways are diminished in BE [12].
Macroautophagy (referred to hereafter as “autophagy”) is a highly conserved cellular mechanism by which organelles and proteins are sequestered in autophagic vesicles (AV) and degraded after fusion with lysosomes [13]. Autophagy is induced by a variety of processes including normal embryonic development. More typically autophagy is induced as an adaptive response to cellular stress, either from nutrient and growth factor limitation, infection, hypoxia, oxidative stress, or accumulation of protein aggregates and endoplasmic reticulum stress [14,15]. The latter three pathways are all likely involved when autophagy is induced by an injury and inflammatory response [15]. Lastly, while autophagy’s role in carcinogenesis as a tumor promoter and/or suppressor remains unsettled, it clearly has a role in established cancers enhancing tumor cell survival in response to chemo- and radiation therapies [14,16,17].
The molecular mechanisms giving rise to BE from normal esophageal mucosa are poorly understood. Although GERD is reported to induce inflammation and oxidative stress in the esophageal epithelium, and this stress is thought to contribute to the pathogenesis of BE [10,18–22], there is only a single study on autophagy in BE, and it is focused on the Beclin gene [23]. Thus this important pathway is essentially unexplored in BE and EAC pathogenesis. Using multiple complimentary techniques, we have quantified autophagy levels across the BE disease spectrum, from normal squamous, to esophagitis, non-dysplastic BE and dysplastic BE, and EAC, and explored the functional contributions of the autophagic response upon cellular oxidative stress and cell survival in an in vitro model of acid reflux.
MATERIALS AND METHODS
Cell Culture
Immortalized human primary esophageal epithelial cells STR (EPC-hTERT) were developed and maintained as previously described [24–26]. CPA and CPD cells were kindly provided by Peter Rabinovich, University of Washington [27]. Cells were adapted to serum-free conditions in keratinocyte serum-free medium (KSFM, Invitrogen). OE19 and OE33 cells were purchased from Sigma Aldrich and maintained in RPMI 1640 with 2mM Glutamine and 10% Fetal Calf Serum. Stable transduction of esophageal cells with retroviral vectors has been described previously [26,28]. Lentiviral vector GFP-LC3 was obtained from Dr Craig Thompson (Memorial Sloan Kettering) and transfected into 293T cells along with the packaging plasmids using Lipofectamine 2000 (Invitrogen) reagent following the manufacturer’s instructions. Then STR, CPA, CPD, OE19, and OE33 cells were exposed to virus in the presence of 8 mg/mL polybrene for 16–18h respectively. GFP-LC3 expressing cell populations were isolated by sorted for GFP using flow cytometry. LC3-GFP expression was confirmed by examination of the cells by confocal fluorescent microscopy on a Nikon Eclipse Ti-U microscope. LC3-GFP+ vesicles were identified and quantified using the spot finder application in the Volocity image analysis software package (Perkin Elmer).
To acid-stress the cells as a mimic for GERD, cell culture media was acidified with hydrochloric acid to pH 3.5. Cells were incubated in the acidified media for the specified lengths of time, then rinsed briefly with PBS before returning to normal cell-culture media. Some cells were studied at 6 hours post pulse for ROS and AV levels by FACS analysis approaches. The remainder were continued on for 24 hours, then stained using the Live/Dead Assay kit (Life Technologies). To quantify percentage survival, cells were imaged by epifluorescence microscopy and the living (green) cells and dead (red) cells determined in 3 different visual fields. At least 300 cells were counted per well. Percent survival was calculated as the number of living cells over the total number of cells (living + dead).
Human BE and EAC tissue biopsies
All human normal esophagus, BE, and EAC biopsy tissues used in this study were collected under a study protocol (IRB# 813841/UPCC# 12211) that was reviewed and approved by the Institutional Review Board for human research at the Hospital of the University of Pennsylvania. All study participants were recruited at the University of Pennsylvania, Philadelphia, PA. Study participants were recruited from among patients having a scheduled esophagogastroduodenoscopy (EGD) evaluation as part of the patient’s routine clinical care. During the endoscopy, in addition to the clinically indicated biopsies, the subject had additional research biopsies taken from the squamous esophagus, the metaplastic Barrett’s esophagus, and/or from the esophageal adenocarcinoma.
Inclusion Criteria--Patients who have GERD (with and without Barrett’s esophagus) or esophageal adenocarcinoma and who were scheduled for elective endoscopic examination of the upper gastrointestinal tract for clinical purposes were eligible for this study. GERD was defined by a history of heartburn at least once a week when patients were not taking antisecretory medications. Barrett’s esophagus was defined as biopsy-verified specialized intestinal metaplasia extending >2 cm proximal to the gastroesophageal junction (defined as the most proximal extent of the gastric folds with the stomach partially deflated).
Exclusion Criteria--Patients were excluded if they were unwilling or unable to provide informed consent, had squamous esophageal cancer, esophageal varices, were taking warfarin, clopidogrel, or newer anticoagulation agents or had a coagulopathy that precluded safe biopsy of the esophagus, or had a comorbidity that precluded safe participation in the study.
Transmission Electron Microscopy (TEM) analysis
For TEM, cells and tissues were fixed and analyzed as described previously [29]. Briefly, tissues were fixed in 2.5% (w/v) glutaraldehyde and 2.0% paraformaldehyde in 0.1 M cacodylate buffer (pH 7.4) at 4°C overnight. After buffer wash, the samples were post-fixed in 2% osmium tetroxide for 1 h, washed again in buffer, and dehydrated in a graded ethanol series. Samples were treated with several changes of hexamethyldisilazane (HMDS) and then allowed to air dry prior to mounting and sputter-coating with gold. TEM examinations were made, and TEM photos were taken with a JEOL 1010 electron microscope fitted with a Hamamatsu digital camera and AMT Advantage imaging software. AVs were identified based on published ultrastructural features [30], most significantly the presence of double or multiple membranes as well as vesicles which contain cytoplasmic material. Counts were performed by two independent investigators and averaged for each cell. Cells counts were then averaged and statistical testing performed as described.
IL-1β mouse model for Barrett’s Esophagus
All studies with the mouse models were fully approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Pennsylvania. The generation and genotyping of L2-IL-1β [31] transgenic mice has been previously described. Mice were placed on drinking water containing deoxycholic acid (0.2% DCA, pH 7.0) at age of 8 weeks. After 12-months of treatment, mice were sacrificed and tissues isolated for further analysis.
Flow cytometry analysis
For human biopsies, a single cell suspension was obtained by mashing tissues through 70µm mesh filters (BD Biosciences, San Jose, CA). For murine experiments, esophagi were dissected then incubated in 1U/ml dispase (Sigma-Aldrich) for at 37°C for 10 minutes. Esophagi were then opened longitudinally and epithelium was peeled from submucosa. Minced epithelium was incubated at 37°C with 1000RPM shaking in Dulbecco’s modified Eagle medium (DMEM, Invitrogen) containing 1mg/ml collagenase I (Sigma-Aldrich) for 15 minutes then in 0.5% trypsin-EDTA (Invitrogen) for 5 minutes. Cell suspension in trypsin was passed through a 70µm mesh filter and trypsin was quenched by addition of DMEM+10% FBS. Following centrifugation, cell pellets were washed in 1:1 mix of KSFM and DPBS then stained with Cyto-ID (Enzo Life Sciences) for 30 minutes at 37°C. Cyto-ID was used at 1:100 for human and murine studies and at 1:1000 for cell line studies.
For human studies, cells were concurrently stained with APC-anti-E-cadherin (1:20, BD Biosciences). ROS were determined by flow cytometry with 2’, 7’-dichlorodihydrofluorescein diacetate (DCF) dye (Life Technologies) as described previously [32]. In brief, cells were incubated with 10µM DCF at 37°C for 30 min and further cultured for up to 3 hours prior to analysis. Following staining, cells were washed then analyzed in DPBS containing 1% BSA using a FACS Calibur (BD Biosciences). FlowJo software (Tree Star, Ashland, OR) was used for data analysis. Similarly, cell death measures at 24 hours were carried out using staining with 7AAD (7-amino-actinomycin D). 7AAD has a high DNA binding constant and is efficiently excluded by intact cells. It is useful for DNA analysis and dead cell discrimination during flow cytometric analysis. After cells were isolated from the culture plate, washed and centrifuged, they were resuspended in 0.5 mL of FACS Buffer and 5 µl of 7-AAD (Biolegend) and incubate for 5–10 minutes in the dark before analysis and quantitation by flow cytometry.
Protein Extraction and Western Blot Analysis
Whole cell lysates were prepared as described previously [33]. Protein concentration of samples and bovine serum albumin standard was determined using the Bradford protein assay (Bio-Rad Laboratories, Hercules, CA, USA). 50 µg of denatured protein was fractionated on a NuPAGE Bis-Tris 4–12% gel (Life Technologies). Following electrotransfer, Immobilon-P membranes (Millipore) were blocked with PBST containing 5% milk, followed by overnight incubation with the following primary antibodies: rabbit anti-LC3B (1:1000, Cell Signaling Technology), rabbit anti-Beclin1 antibody (1:1000, Cell Signaling Technology; 1), and mouse anti-actin (1:10000, Sigma-Aldrich) at 4 °C. The secondary antibodies used were all from Sigma-Aldrich and used at 1:10000. Oxyblot (anti-carbonyl antibody; EMD Millipore) was used according to the manufacturer’s guidelines. Targeted proteins were visualized using a chemiluminescence detection system (Amersham ECL or ECL Prime; GE Healthcare Life Sciences) and exposed to Blue Lite Autorad film (ISC-BioExpress).
Immunostaining
All specimens were isolated, rinsed in ice-cold PBS, fixed, and analyzed histologically by staining sections with hematoxylin and eosin (H&E) or immunohistochemically using standard methods as described [34]. 5 mm paraffin-embedded sections were pretreated with xylene and then placed in a pressure cooker in 10 mmol/L citric acid buffer (pH 6.0) for 2 hours. Endogenous peroxidases were quenched using hydrogen peroxide before sections were incubated in avidin D blocking reagent and biotin blocking reagent. Antibodies used include rabbit anti-cleaved LC3 (1:250, Abgent). Sections were incubated with primary and biotinylated secondary antibodies and an avidin-horseradish peroxidase conjugate (Vectastain Elite ABC kit; Vector Laboratories, Burlingame, CA) following the manufacturer’s protocol. The signal was developed using the 3,39-diaminobenzidine substrate kit (Vector Laboratories). Sections were counterstained with hematoxylin.
Statistical analysis
GraphPad Prism version 3.04 was used for all statistical analyses (GraphPad, San Diego, CA, USA). Data from multiple cells or multiple treatments were analyzed using One-way ANOVA and Tukey’s multiple comparison test. All data are represented as mean and standard deviation unless otherwise stated.
RESULTS
Cell lines representing the disease spectrum from normal esophagus to EAC demonstrate different levels of basal autophagic vesicle (AV) content
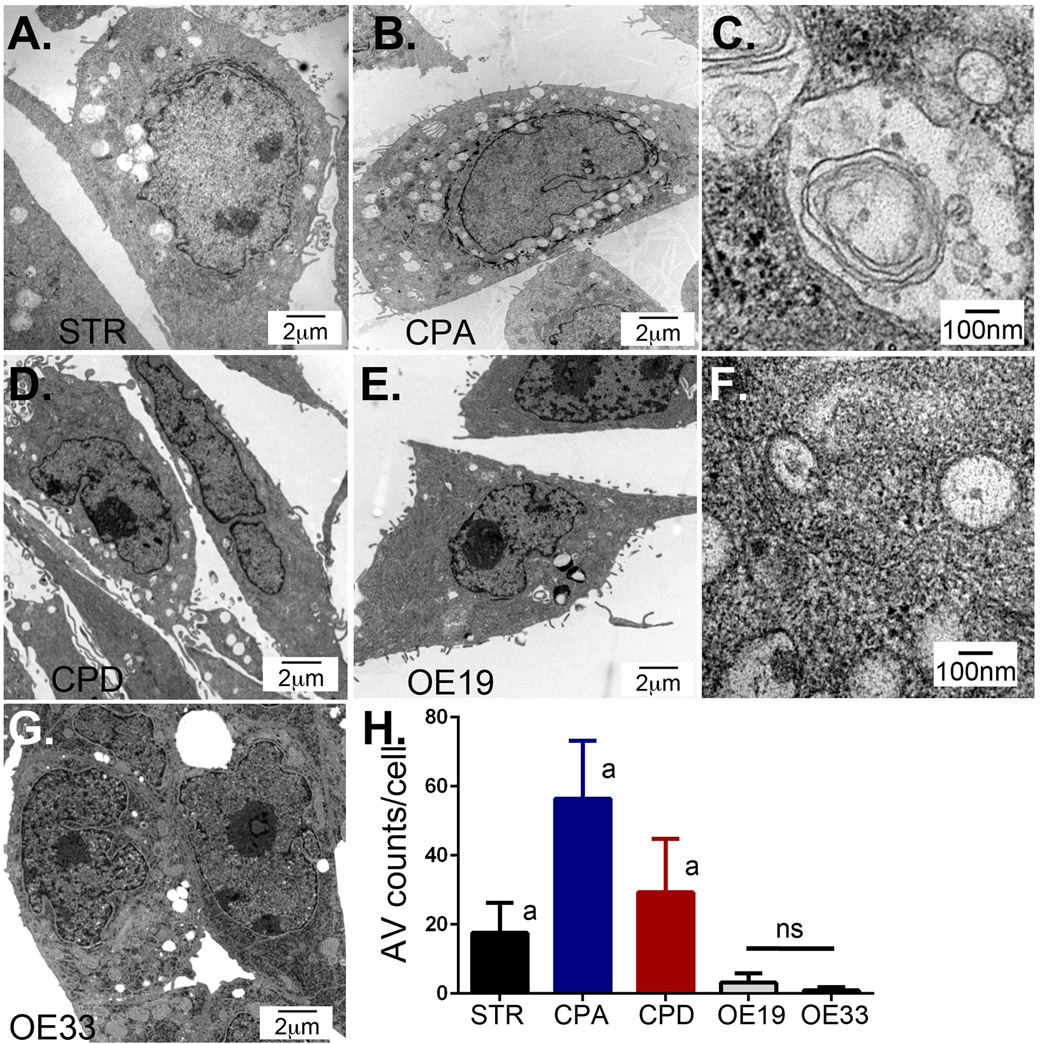
Autophagy is a highly-conserved pathway to degrade and recycle organelles, proteins, and cell membranes when they are damaged or when the cell is nutritionally stressed. Transmission electron microscopy (TEM) remains the gold standard for identifying and quantifying autophagic vesicles [30]. STR (aka EPC2-hTERT) is a normal human esophageal cell line that is immortalized by telomerase and which maintains wild-type p16 and p53 [25]. CPA and CPD are human BE cell lines isolated from non-dysplastic (CPA) and dysplastic (CPD) BE, both are also immortalized with telomerase and have allelic losses and mutations [27]. CPA cells have mutant p16 but wild-type p53, and CPD cells are mutant in both genes. OE19 and OE33 cells were isolated from human EAC and have mutations in their p53 genes [35]. Under fully replete nutritional nonconfluent growth conditions, we evaluated these cells for the presence of AVs by transmission electron microscopy (TEM) (Figure 1). By definition, AVs are double or multiple membranes vesicles that contain cytoplasmic material [30] (Figure 1C, 1F). We found that CPA cells had the highest baseline level of AVs (56.4 AV/cell±16.8; n=74), while AVs were rare in EAC cell lines OE19 (3.1 AV/cell±2.7; n=85) and OE33 (0.9 AV/cell±0.9; n=69), with an intermediate AV level in STR (17.5 AV/cell±8.7; n=103) and dysplastic CPD cells (29.3 AV/cell±15.5; n=134) (Figure 1).
Figure 1. Transmission electron microscopy (TEM) of human esophageal cell lines to quantify autophagy vesicle presence.
All cell lines were maintained in basal growth media. Cells were imaged by TEM to visualize AVs. A. Normal human esophageal squamous keratinocyte STR cells. B. Non-dysplastic Barrett’s Esophagus CPA cells. C. Higher power examination of CPA cell AV demonstrating double-membrane and enclosed material. D. Dysplastic Barrett’s esophagus CPD cells. E. Barrett’s-associated esophageal adenocarcinoma OE19 cells. F. Higher power examination of OE19 cell AV demonstrating double-membrane and enclosed material. G. Barrett’s-associated Esophageal adenocarcinoma OE33 cells. H. AV counts per cell. Counts were made by two investigators on 70 to 130 separate cells for each cell line imaged by TEM at 10,000× enhancement. a=significantly differs from all other cell lines by 1 way ANOVA testing and Tukey’s Multiple Comparison Testing p<0.05. ns= not significantly different from each other.
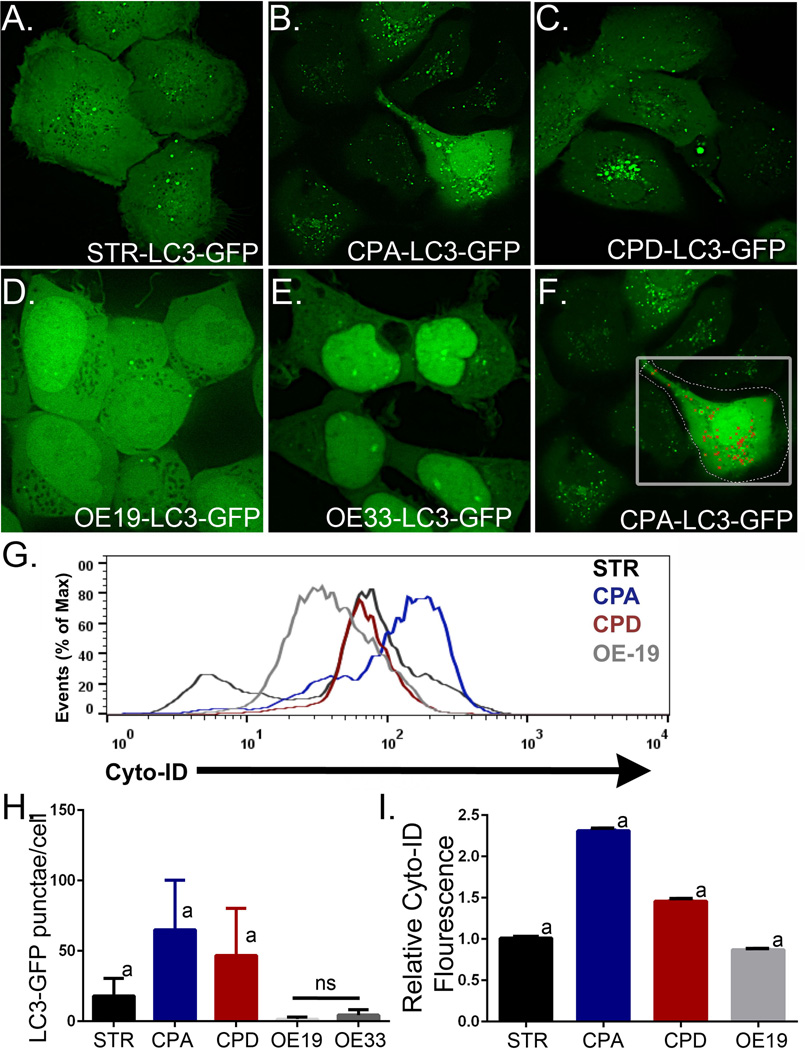
In order to further validate our ability to quantitate AV levels reliably, we utilized two additional techniques to quantify relative AV levels in these cell lines. We first transduced these same cell lines with a retrovirus to express LC3-GFP, a fluorescently marked protein that is lipidated during normal autophagy and localized into AVs [36]. An examination by confocal microscopy and quantitation of the LC3-GFP+ puncta using Volocity image analysis software (PerkinElmer) found a similar pattern with regard to AV content as the TEM studies (Figure 2A–2G). Using this approach we found that CPA cells had 65.1 AV/cell±35.2; (n=36), while AVs were rare in EAC cell lines OE19 (1.6 AV/cell±1.5; n=55) and OE33 (4.4 AV/cell±3.8; n=55), and an intermediate AV level in STR (17.9 AV/cell±12.5; n=36) and dysplastic CPD cells (46.8 AV/cell±33.4; n=42). Of importance, the AV counts by TEM and LC3-GFP puncta quantitation were in close agreement (Figures 1H and 2H), substantiating the utility of this approach to quantify AV numbers.
Figure 2. LC3-GFP fluorescence-mediated identification of autophagic vacuoles in human Barrett’s esophagus cell lines.
STR, CPA, CPD, OE19, and OE33 cell lines were retrovirally transduced to express LC3B-GFP. All cell lines were maintained in basal growth media. Cells were imaged by epifluorescence microscopy to visualize fluorescent AV puncta, indicating autophagic vesicles [36]. A. Normal human esophageal squamous keratinocyte STR cells. B. Non-dysplastic Barrett’s Esophagus CPA cells. C. Dysplastic Barrett’s Esophagus CPD cells. D. Barrett’s-associated Esophageal adenocarcinoma OE19 cells. E. Barrett’s-associated esophageal adenocarcinoma OE33 cells. F. Demonstration of Volocity (Perkin Elmer)-medicated identification and counting of fluorescent AV puncta. G. Cyto-ID Autophagy fluorescence histogram of four cell lines STR (Black), CPA (Blue), CPD (Red), and OE19 (Grey) after gating on E-cadherin+ cells. H. Fluorescent AV puncta counts per cell. Counts were made on 36 to 55 separate cells for each cell line imaged. a=significantly differs from all other cell lines by 1 way ANOVA testing and Tukey’s Multiple Comparison Testing p<0.05. ns= not significantly different from each other. I. Summary of FACs quantitation of Cyto-ID Autophagy fluorescence in cell lines maintained under growth conditions. n=3 for each cell line. a=significantly differs from all other cell lines by 1 way ANOVA testing and Tukey’s Multiple Comparison Testing p<0.05.
Finally, a FACS-based detection of AVs was explored and compared to our validated TEM and LC3-GFP approaches. Cyto-ID is a cationic amphiphilic tracer dye that rapidly partitions into cells and labels vacuoles associated with the autophagy pathway (Enzo Lifesciences). Quantitation of Cyto-ID Autophagy fluorescence by FACS analysis revealed a pattern similar to that observed by the other two validated approaches (Figure 2G). CPA cells yielded the greatest level of baseline Cyto-ID fluorescence, nearly 2.5-fold greater than STR (n=3), and the esophageal adenocarcinoma OE19 cells the least (about 80% of STR fluorescence, n=3), with STR and CPD cells (nearly 1.5-fold greater than STR, n=3) falling within these limits (Figure 2I). Together, these three techniques suggest autophagy vesicles are increased in Barrett’s esophagus when compared to normal esophagus, with levels progressively diminishing as the cells become increasingly dysplastic, the lowest being observed in the neoplastic cell lines. This pattern has been observed in other human cancers [37,38]. Moreover, these findings validate our use of each these techniques to quantify autophagy levels in human and animal tissues as well cell lines in response to stressors including acid reflux.
Autophagy in human normal squamous, BE, and EAC biopsies
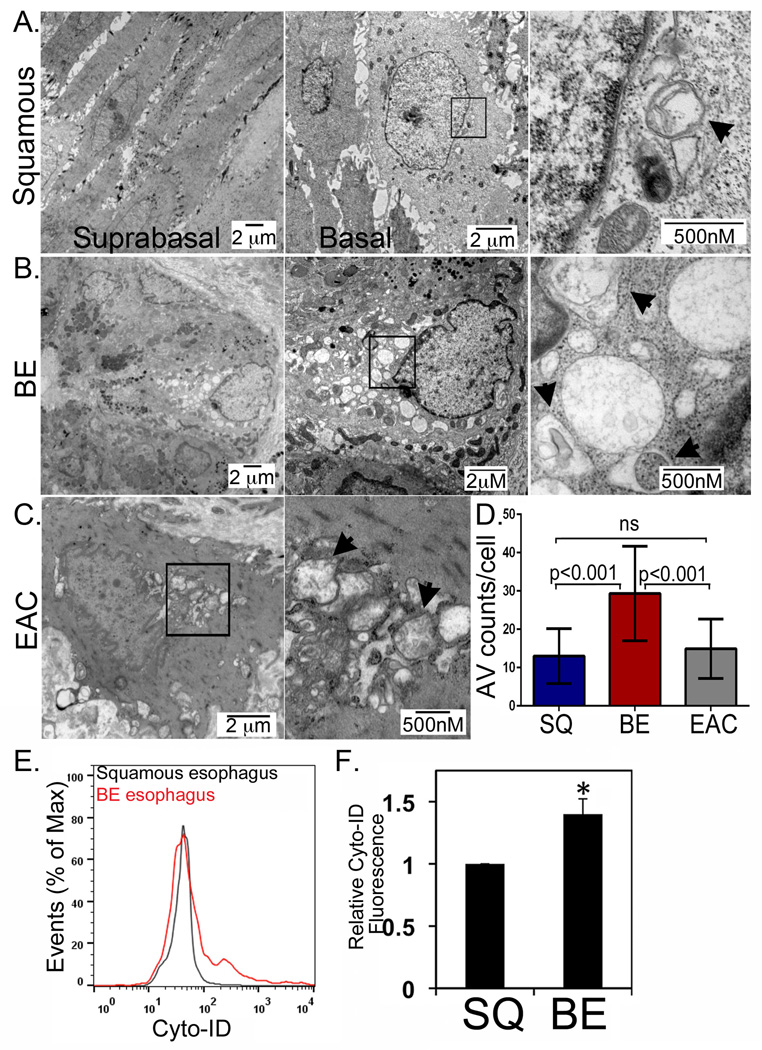
Inflammation and oxidative stress are two of several known triggers of cellular autophagy [37,39,40]. While both inflammation and oxidative stress have been documented in GERD and BE, the presence of autophagic vesicles in human BE tissues has not been adequately explored. We have obtained endoscopic biopsies from 10 patients, one with a normal squamous esophagus, six with BE, and three with EAC and subjected them to TEM analysis. From four of the BE patients and all three of the EAC patients, we obtained matching biopsies from normal esophagus proximal to the metaplasia or cancer. We observed on average 12.9 ±1.5 AVs/cell (n=198) in the normal squamous epithelium, with more AVs in basal cells and fewer AVs in the suprabasal compartment (Figure 3A). In the BE biopsies, there was much greater heterogeneity, with most cells having a few dozen AVs, and, less frequently, cells with 40 or more AVs but averaged 29.6±12.3 AVs/cell (n=112)(Figure 3B). In the EAC, AV levels were reduced compared to BE to 14.1±7.8 AVs/cell (n=74), similar to levels seen in the normal esophagus (Figure 3C and 3D).
Figure 3. Steady-state levels of AV in normal human squamous epithelium, BE, and EAC.
A. Human squamous esophagus examined by TEM under increasing magnification. Black box-region extremely magnified in frame to the left. B. Barrett’s esophagus TEM by increasing magnification. Black box-region extremely magnified in frame to the left. C. Human EAC imaged by TEM under increasing magnification. Black box-region extremely magnified in frame to the left. D. Counts of AVs in 74 or more cells per patient sample were obtained averaged across all patient samples, with the average number of AVs per cells graphed. SQ: squamous; BE: Barrett’s esophagus: EAC: esophageal adenocarcinoma. One-way ANOVA and Tukey Rank mean with q< 0.005, n=9 subjects/198 cells, 5 subjects/112 cells, and 3 subjects/74 cells for squamous, BE, and EAC samples, respectively. E. Autophagic vesicles quantified by Cyto-ID stain and flow cytometry in the Barrett’s (Red) and squamous epithelium (Black) from a single subject. F. Averaged relative Cyto-ID fluorescence from three subjects for squamous esophagus and BE tissue. *, p<0.05.
As an additional quantitation of autophagy levels in normal human squamous esophagus and human Barrett’s esophagus, we obtained additional biopsies from the BE tissue and from the proximal normal squamous epithelium during a clinically indicated endoscopy in three BE patients. The biopsies were processed by mechanical disruption and then all cells labeled with Cyto-ID Autophagy and an antibody to the epithelial marker E-cadherin. After gating on the E-cadherin+ cells, relative autophagy levels were determined based on Cyto ID Autophagy fluorescence. Overall, BE cells have on average 1.5-fold (n=3, p<0.05) greater levels of autophagy than squamous cells (Figure 3E and 3F). Moreover, in BE cells there appears to be a subpopulation with increased levels of autophagic vesicles (Figure 3E). Both of these findings agree with our TEM observations. In summary, our findings confirm increased numbers of autophagy vesicles in human BE compared to normal squamous or EAC.
L2-IL-1β transgenic mice, a model for human BE and EAC, demonstrate oxidative stress, and increased autophagy preceding the onset of metaplasia and dysplasia
Mechanistically in the setting of GERD, autophagy may be induced after chemical damage from acidic reflux or the actions of bile acids. Indeed, Beclin-1 protein and LC3-GFP+ autophagy vesicles were reported to be increased in CPA cells after treatment with the bile acid deoxycholic acid [23]. However, other mechanisms for autophagy induction were not explored. The role of an acidic pH stress alone was not examined, nor was the role for inflammation, a potent inducer of autophagic responses through a combined effect on hypoxia, oxidative stress, and the accumulation of protein aggregates and endoplasmic reticulum stress [15].
Recently, a physiologically relevant transgenic mouse model for BE-like metaplasia was reported [31]. It utilized an Epstein-Barr virus L2 promoter to over-express human IL-1β in the oral cavity, esophagus, and squamous forestomach of mice [43]. These L2-IL-1β mice develop a chronic[43] inflammatory esophagitis by 3 months (Figure 1A) that is followed subsequently by the development of a columnar metaplasia with intestinal features that later progresses to dysplasia and cancer. The strength of this transgenic mouse model is that in many ways it strongly phenocopies the pathogenesis of the human Barrett’s esophagus as it is presently believed to occur [44,45]. This metaplasia is intestinalized, as confirmed by the presence of intestinal mucins established by Alcian blue staining and immunohistochemistry for Muc2, an intestinal mucin [31]. Most importantly, this disease model absolutely requires inflammation. When these L2-IL-1β mice were crossed with IL-6 knockout mice, the metaplasia and cancer were completely abrogated [31]. Moreover, while the bile acid deoxycholic acid (DCA) in the drinking water accelerates disease progression, DCA treatment alone is insufficient to induce the metaplasia or cancer phenotype [31].
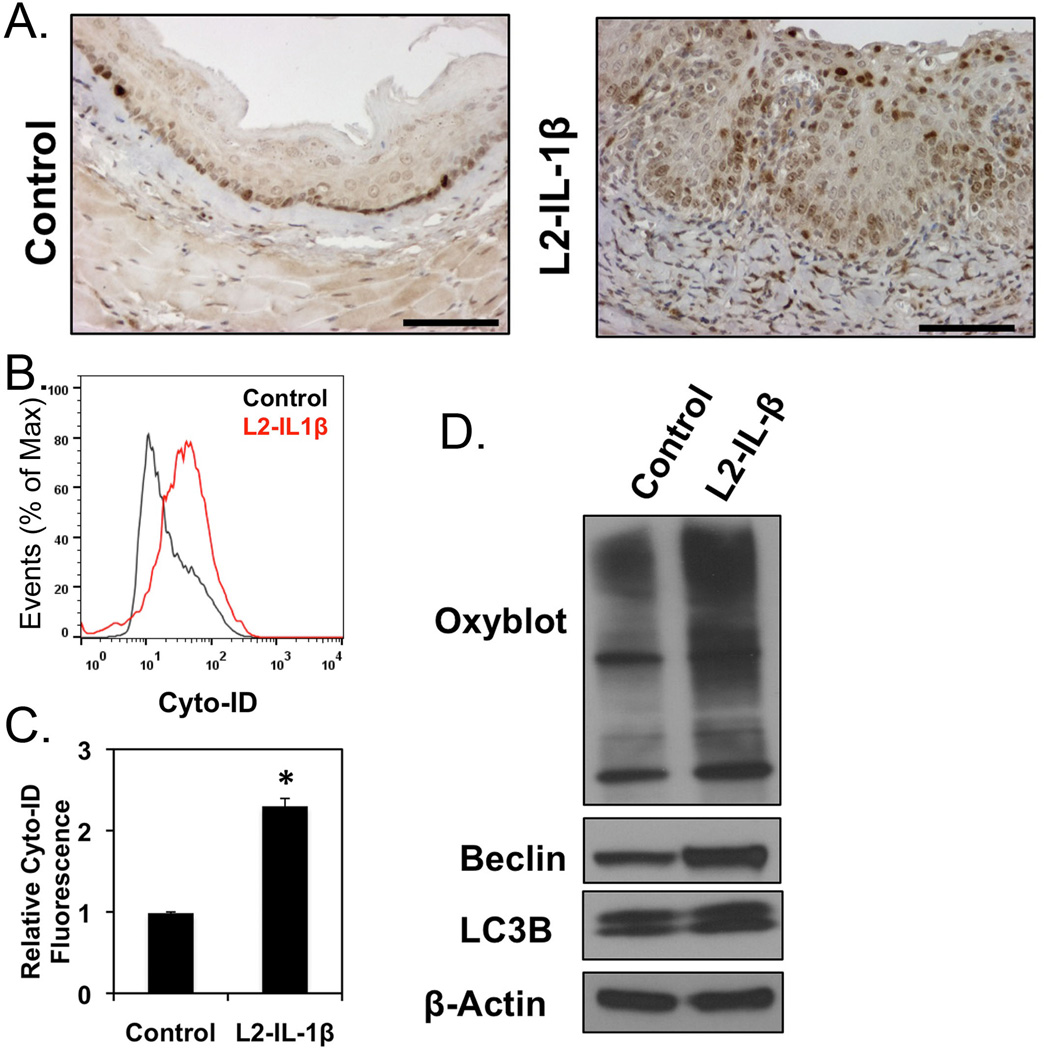
We therefore utilized this mouse model to determine if an inflammatory microenvironment alone, in the absence of a reflux injury, could induce oxidative stress and autophagy. We thus isolated esophageal tissue and epithelial cells from several 3 month-old L2-IL-1β mice and control mice. This is well before the onset of metaplasia in the L2-IL-1β mice [31]. IHC staining for the autophagy-activated cleaved form of LC3 was increased in the esophageal epithelium of L2-IL-1β mice compared to controls (Figure 4A). This is supported by flow cytometric studies with Cyto-ID Autophagy. Esophageal cells from L2-IL-1β mice demonstrated relative 2-fold increase in fluorescent intensity (Figure 4B and 4C), compared to esophageal cells derived from littermate controls. Mechanistically, this increase in autophagy correlates with an increase in oxidative stress, as evidenced by Western blot studies for carbonyl groups that are byproducts of ROS interactions with proteins (Oxyblot, Millipore). The Oxyblot analysis revealed a significant increase in carbonyl adducts in L2-IL-1β mice compared to controls, suggesting elevated ROS and oxidative stress in these cells. In addition, simultaneously with this elevated oxidative stress there was an increase in the autophagy-associated proteins LC3B and Beclin1 in esophageal epithelial cells from L2-IL-1β mice compared to wild-type controls, suggesting that oxidative stress and autophagic activity are increased together (Figure 4D). We conclude that squamous esophageal epithelial cells in L2-IL-1β mice with esophagitis experience significant oxidative stress and activation of autophagy preceding the onset of the intestinal metaplasia.
Figure 4. Oxidative stress and autophagy in the L2-IL-1β transgenic mouse model of BE.
A. IHC staining for active cleaved form of LC3 in normal mouse esophagus or transgenic L2-IL-1β mice. B. Cyto-ID autophagy profile of wild- (Black) and L2-IL-1β esophageal epithelium (Red) by flow cytometry. C. Averaged relative Cyto-ID fluorescence, n=3. *, p<0.05. D. Western blot for oxidized proteins (Oxiblot, Millipore) and autophagy proteins (LC3B and Beclin1) in the squamous epithelium from 3 month old L2-IL-1β mice. One of three blots is shown.
Normal human esophageal squamous STR cells are much more sensitive to an acid exposure that mimics GERD compared to BE and EAC cell lines
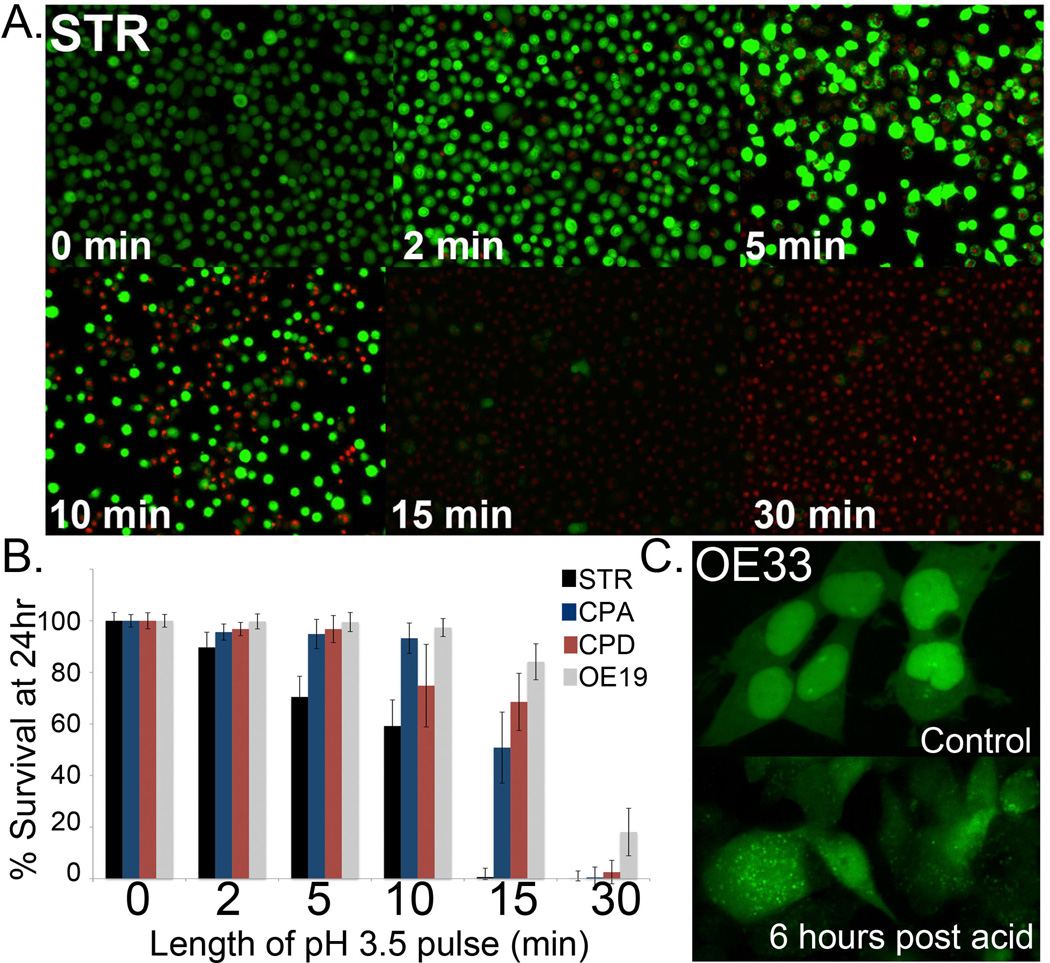
In previous studies, an acidic injury to human esophageal Seg-1 and Het-1A cells or Barrett’s cells NES and BAR-T lead to the induction of reactive oxygen species within 20 minutes of the exposure [21,46,47]. In order to explore a potential role for autophagy in responding to a GERD mediated cell injury and oxidative stress, we characterized the human BE cell lines in our study for their ability to tolerate an acid pulse (pH 3.5) for increasing lengths of time. We used cell viability (Live/Dead) quantitation at 24 hours as our measure of sensitivity. In terms of viability after acid exposure, the cancer cell line OE19 was the most tolerant and the normal human esophageal cells (STR) the least, with the following pattern emerging with regard to the panel: OE19> CPD> CPA> STR (Figure 5A and 5B). Using conditions yielding 20–40% cell death at 24 hours, we repeated the study using LC3-GFP expressing cells. We exposed the cells to acid pulses (pH3.5) for 10 minutes (STR), 15 minutes (CPA and CPD) or 20 minutes (OE19). The increase in autophagic vesicles, as indicated by increased fluorescent punctae, was most evident in the OE19 and OE33 cell lines given their low initial levels (Figure 5C and data not shown). Generally, the increase in AV punctae was evident by 6 hours post acid exposure (5C and data not shown). In summary, we have established that STR esophageal keratinocytes are more sensitive to acid exposure conditions than BE or EAC cell lines, and that this exposure induces an increase in AV numbers by 6 hours post exposure.
Figure 5. Modeling GERD in vitro by pH 3.5 media pulse.
A. Live/dead imaging of STR cells 24 hours post pH 3.5 acid pulse. Time length of pulse is indicated in minutes. B. Quantitation of Live/dead fluorescence for STR (Black bar), CPA (Blue bar), CPD (Red bar), and OE19 (Grey bar) cells, respectively. n=3. Length of pulse is in minutes. C. OE33 cells were transduced with a retrovirus to express LC3B-GFP. These cells were imaged at T=0, then exposed to acidified culture media (pH=3.5) for 20 minutes then returned to normal growth media and reimaged at 6 hours. Increased AV steady state levels are visible as increase in GFP+ small vacuoles.
Pharmacologic inhibition of autophagy following acidic stress increases ROS production and diminishes cell viability
To assess the role of autophagy in cell response to acid stress, we exposed each of the cell lines to the acid pulse followed by immediate treatment with an inhibitor of autophagy (chloroquine, 50 µM) [48,49] or vehicle control. Chloroquine inhibits autophagy by blocking fusion of the AV with lysosomes, leading to an inhibition in AV flux and an accumulation of blocked AVs. We focused on six hours post acid stress as a timepoint in which to determine ROS levels and autophagic responses. Six hours post exposure cells were stained with 2, 7’-dichlorofluorescein diacetate (DCF) or Cyto-ID autophagy. DFC is lipophilic and non-fluorescent compound that is oxidized to fluorescent DCF by ROS, and is widely used to evaluate cellular oxidative stress. DCF and Cyto-ID autophagy fluorescence were quantified by flow cytometry and confirmed by confocal microscopy. Unstained cells in additional plates were maintained until 24 hours post acid exposure, at which time an assay for cell viability was performed.
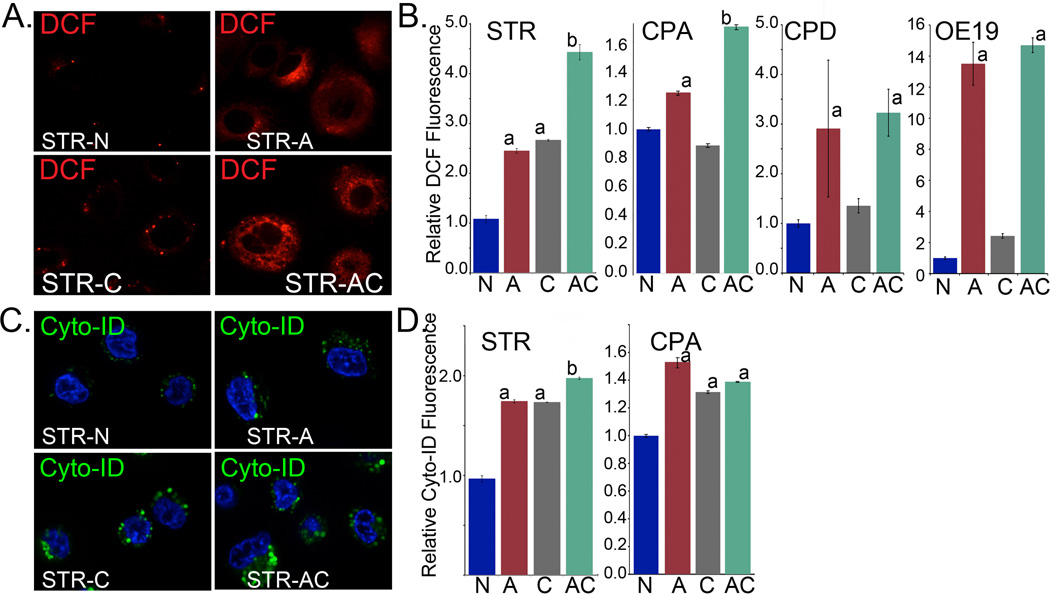
In both STR and CPA cells, acid treatment significantly increased ROS levels at 6 hours post exposure (Figure 6A and 6B). Chloroquine alone had a mixed impact on cellular ROS levels-increasing them in STR but not CPA cells. However, when CQ was combined with acid stress, there was an additional significant increase in ROS levels experienced by both STR and CPA cells compared to acid only exposed cells (Figure 6A and 6B). This significant increase is consistent with autophagy acting to reduce intracellular ROS stress after an injury. Autophagy levels were similarly responsive to these treatments. Cyto-ID Autophagy levels were increased by all three conditions, acid exposure, CQ, as well as the combined treatments. The increased observed with CQ is due to the accumulation of blocked AVs (Figure 6C and 6D).
Figure 6. Effects of autophagy inhibition on intracellular oxidative stress and AV formation after GERD-like acid exposure.
A. Relative intracellular ROS levels as determined by reporter DCF fluorescence in STR cells at 6 hours post treatment; imaged by confocal microscopy. N=control nonacidic media; A=acid [pH 3.5] pulsed; C=chloroquine treated; AC=acid pulsed followed by chloroquine treatment. B. Summary of DCF fluorescence quantified by flow cytometry in representative normal (STR) and BE (non-dysplastic/CPA) cell lines treated as before and measured at 6 hours post-treatment; n=6 experiments for each. Significance testing was by 1 way ANOVA followed by Tukey’s multiple comparison test; p values adjusted for multiple comparisons are reported. a = significantly differs from control and chloroquine treated cells; adjusted p≤0.0073. b= significantly differs from acid and chloroquine treated cells by 1 way ANOVA and Tukey’s multiple comparison test; p<.05. C. Relative autophagy induction 6 hours post treatment with acid or/and chloroquine as determined by reporter Cyto-ID fluorescence in STR cells and imaged by confocal microscopy. D. Summary of Cyto-ID fluorescence and relative autophagy induction quantified by flow cytometry in representative normal (STR), BE (non-dysplastic/CPA and dysplastic/CPD) and EAC (OE19) cell lines treated as before and measured at 6 hours post-treatment; n=6 experiments for each. a= significantly differs from control treated cells by 1 way ANOVA and Tukey’s multiple comparison test; p>.001. b= significantly differs from acid and chloroquine treated cells by 1 way ANOVA and Tukey’s multiple comparison test; p<.05.
In both CPD and OE19 cells, the responses were different. As with the non-dysplastic cells, the acid treatment led to a significantly increased ROS levels at 6 hours post exposure in these cell lines (Figure 6B). Chloroquine alone had no significant effect on cellular ROS levels. However, when CQ was combined with acid stress, there was no additional increase in ROS levels experienced by either the CPD or OE19 cells as compared to acid only exposed cells (Figure 6B). This suggests these dysplastic cell lines do not utilized autophagy to manage the ROS stress induced by an acidic environment.
Similarly we were surprised by our findings using Cyto-ID to quantify the autophagic response of these cells to an acidic stress. In CPD cells, we found no change in Cyto-ID fluorescent signal with any treatment (Supplemental Figure S1A), including after chloroquine treatment alone. In OE19 cells, not only was there was no increase in relative Cyto-ID fluorescence after chloroquine treatment, treatment with acid significantly reduced this relative fluorescence (Figure S1B). As these observations ran contrary to our expectations, especially with respect to chloroquine treatment, we were concerned that there was a problem with the Cyto-ID fluorescence as a measure of autophagy responses. We therefore examined the Cyto-ID fluorescent by confocal microscopy in OE19 cells after these same treatments and in parallel treated LC3-GFP labeled OE19 cells as a second measure of the autophagic response. Unexpectedly, there was a significant difference between the two methodologies. In untreated OE19 cells, Cyto-ID fluoresces brightly in cytoplasmic vesicles, while LC3-GFP does not similarly collect (Supplemental Figure S1C and D). Moreover, while LC3-GFP vesicles were increased weakly by acid treatment and strongly after chloroquine treatment, Cyto-ID fluorescence pattern recapitulated that measured by flow cytometry, with diminished signal in acid-treated cells (Supplemental Figure S1C and D). Based on these conflicting observations, we conclude that the autophagic response in CPD and OE19 cells is biologically different from that functioning in the non-dysplastic STR and CPA cells.
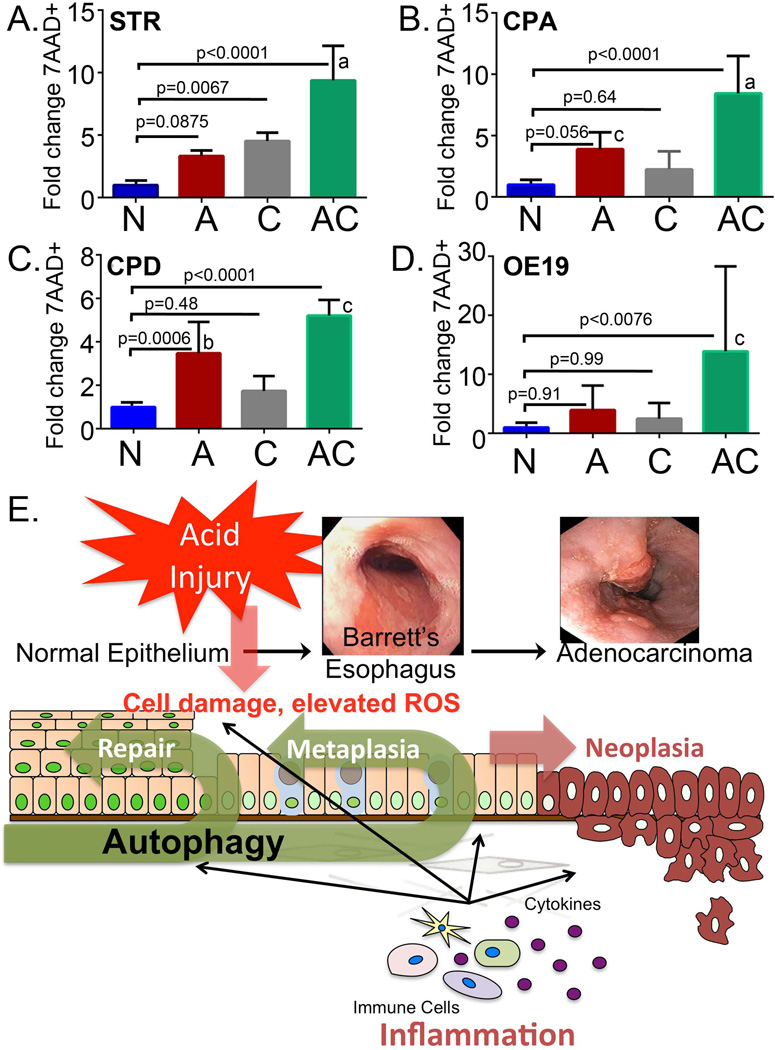
In a final study, we were interested to establish whether the autophagic responses we were observing at baseline and in response to acid treatment provided any functional benefit to the cells. Most important would be demonstration of a survival advantage provided by an autophagic response. We adopted a new, a highly quantitative flow cytometry approach to measure cell death; staining with 7AAD (7-amino-actinomycin D). 7AAD is fluorescent after binding DNA and is normally excluded by living, intact cells. It is therefore frequently used for dead cell discrimination during flow cytometry studies. We quantified 7AAD+ dead cells 24 hours after treatment with acid, chloroquine, or the combination of acid and chloroquine in all four cell lines representative of the BE disease spectrum. Cell death after acid treatment was increased in three of four cell lines (STR, CPA, and CPD), but this was most significant for CPD cells (Figure 7A, B, and C). STR cells were uniquely sensitive to chloroquine, with a nearly 5-fold increase in 7AAD+ dead cells after 24 hours of treatment. Most important, for all cell lines examined, the combination of acidic stress and chloroquine treatment led to a very significant increase in 7AAD+ dead cells at 24 hours (Figure 7A, B, C, and D). In STR, CPA, and OE19 cells, the combination of acidic stress and inhibition of autophagy synergized and led to greater level of cell death than the sum of the individual treatments (Figure 7A, B, and D). Together, these findings suggest that autophagy functions to reduce ROS levels in non-dysplastic cells and improve cell survival after acid exposure in all cell lines. Moreover, these observations suggest that the drugs which inhibit autophagy may worsen GERD related esophageal injury.
Figure 7. Inhibition of autophagy increases cell death after GERD-like acid exposure.
Flow cytometric quantitation of 7-amino-actinomycin D+ (7AAD+) cell staining at 24 hours in A) STR cells, B) CPA cells, C) CPD cells, and D) OE19 cells. 7AAD+ cells are expressed as fold-increased over non-acid treated control cells. N=control nonacidic media; A=acid pulsed [pH3.5]; C=chloroquine treated; AC=acid pulsed followed by chloroquine treatment; n=6 experiments for each. Significance testing was by 1 way ANOVA followed by Tukey’s multiple comparison test; p values adjusted for multiple comparisons are reported. a= significantly differs from acid and chloroquine treatments, p≤0.0018. b= significantly differs from chloroquine treatments, p≤0.015. c= significantly differs from acid and chloroquine treatments, p≤0.028. E) Model for role of autophagy in the esophagus in the response to acid reflux injury. Gastric acid and bile reflux into the esophagus causes tissue injury and cellular damage, as well as an inflammatory response. With the inflammation there is production of pro-inflammatory eicosanoids and cytokines. Together, the acid injury and inflammatory response contribute to an increase in intracellular reactive oxygen species (ROS). Autophagy is induced to help the cell remove damaged proteins and organelles and reduce ROS levels.
DISCUSSION
The role of autophagy in human disease processes is complex and likely depends on both the environmental and genetic context in which it occurs [50]. In addition to the role of autophagy in response to cellular stress, autophagy clearly has a role in promoting tumor cell survival. Larger tumors typically have hypoxic and nutritionally starved centers; tumor cells located within the centers typically utilize autophagy to enhance tumor cell survival. More importantly, the adaptive role for autophagy in responding to cellular stress has important therapeutic implications. Tumor cells frequently utilize autophagy responses to enhance survival after chemotherapy and radiotherapy treatments [14,16,17]. For these reasons, pharmacological autophagy inhibitors including chloroquine (CQ) and its derivatives like Lys05 [51] have been effective in enhancing the cytotoxic effects of radiation or chemotherapy and improving survival in several small trials [52–55]. Based on these successes, larger randomized trials of CQ versus placebo with anticancer treatments are underway.
Less clear is whether autophagy plays a paradoxical role as a tumor suppressor in premalignant diseases. Haploinsufficiency of Beclin-1 promotes tumorigenesis in mouse models, as Beclin-1 deficient mice display a broad increase in spontaneous tumor formation [56]. In addition, genetically engineered mouse models of oncogene driven, lung and pancreas cancers with autophagy defects demonstrated a pattern of accelerated transformation from premalignant lesions to malignant lesions [57–60]. In general, however, once tumors are formed autophagy defects limited the progression of established cancers, supporting the hypothesis that autophagy suppresses tumorigenesis but once a cell is transformed it can promote cancer cell growth and survival
One mechanism by which autophagy can function as tumor suppressor is by facilitating the maintenance of genetic stability and energy homeostasis [50]. As an example, loss of Autophagy-related protein 7 (Atg7), which is required for fusion of peroxisomal and vacuolar membranes and is critical for normal autophagy responses, leads to increased oxidative stress and elevated ROS in cells [61,62]. Associated with this increased ROS is a noticeable genomic instability that has culminated in neoplastic transformation [61].
By a related mechanism, autophagy may function in BE to help reduce ROS levels and resolve the ROS-mediated organelle damage that occurs in response to GERD. Previous studies have established that after exposure to an acidic pulse, intracellular ROS levels are significantly elevated [21,46,47]. Several studies have established that excessive ROS can damage DNA leading to mutations and epigenetic changes in gene expression, fostering neoplastic transformation [63–65]. Taking into account all these observations, recent models of BE pathogenesis have incorporated elevations in ROS in response to GERD as a key process driving disease onset and progression to cancer (Figure7) [66,67].
Given this well-established relationship between elevated ROS and an increase in autophagy steady-state levels, it is surprising that this pathway has not been better studied in GERD and BE. To our knowledge, there is a single published paper on this subject, and the focus of this work is on Beclin-1 (BECN1) and how its loss correlates with progression to esophageal adenocarcinoma [67]. This published study demonstrated that Becin-1 protein and mRNA levels were increased in BE and progressively diminished with progression to dysplasia and cancer. Moreover, they demonstrated that the bile acid deoxycholic acid induced Beclin-1 mRNA and protein expression in several human BE-related cell lines.
Our work here is a more comprehensive study with regards to the autophagy pathway in BE. We utilized multiple techniques to document steady-state autophagy levels in normal esophagus, BE and EAC, as well as a mouse model of BE. That these varied techniques were in close agreement with each other greatly enhanced the significance of our findings, and established the usefulness of these techniques for future studies. Despite the apparent ease of Beclin-1 immunohistochemistry, it is not an established surrogate biomarker for autophagy [30]. Utilizing these well-validated techniques, we established a clear pattern for autophagy steady-state levels in the normal esophagus and as it transitions to esophagitis, BE, and EAC. This pattern is similar to that reported for other pre-neoplastic to cancer transitions [37,38]. In the normal esophagus, autophagic vacuoles can be observed, and the levels of vacuoles are increased in esophagitis and in BE cells. However, these steady-state levels are decreased as the cells become progressively neoplastic. While this pattern is widely observed in carcinogenesis, the reasons for it are uncertain. However, a number of possible explanations have been raised. Several studies implicate the oncogene activation and the loss of tumor suppressor function which occurs with cellular transformation. Several classic oncoproteins when activated can inhibit autophagic responses [68]. For example, BECN1 is sequestered and its function inhibited by members of the Bcl-2 protein family [68]. Similarly, hyperactivation of receptor tyrosine kinases (RTKs) either by activating mutations or gene amplifications, can inhibit autophagy through their downstream effects on mTOR1 and Akt signaling [68]. It may also be associated with the change in cellular metabolism that occurs, including the adoption of aerobic glycolysis known as the Warberg effect [69], or the activation of mTOR, a negative regulator of autophagy that is frequently induced in cancer. Understanding how the transforming events critical for the progression of BE to EAC impact autophagic responses is an important area for future research efforts.
Equally important to describing the patterns of autophagy in each of these conditions, we have established a role for autophagy responses in GERD and BE. The injury sustained by cells after an acidic pulse includes inflammation as well as elevation of intracellular ROS. Using the L2-IL-1β transgenic mouse model for BE-like dysplasia, we establish that inflammation alone in the esophagus is sufficient to induce oxidative stress and autophagy (Figure 4). Moreover, we establish that an acid injury alone, in the absence of bile acids, is sufficient to induce autophagy in several cell lines. Most significantly, we observed that the inhibition of autophagy after an acidic insult leads to significantly greater cell death at 24 hours in all cell lines tested. This suggests two important role for autophagy in the response to an acid insult: the modulation of the oxidative stress and enhancing cell survival.
How might autophagy function to reduce intracellular ROS after GERD or an acidic insult? It is unclear at the moment, and a question we are currently exploring. One speculation is that autophagy may be removing damaged mitochondria. Mitochondria are well known to be an important source of ROS for cell signaling [70]. However excessive ROS can damage mitochondria as well as other organelles. Damaged mitochondria leak ROS, worsening oxidative stress within a cell. Typically damaged mitochondria undergo mitochondria-targeted autophagy, termed mitophagy [40]. Autophagy can alleviate oxidative stress through other mechanisms, including the upregulation of nuclear factor erythroid 2-related factor 2 (NRF2), a transcription factor regulating the expression of a number of antioxidant proteins including Thioredoxin reductase and Glutathione-s-transferase, among others [71]. NRF2 is sequesterd in the cytoplasm by kelch-like ECH-associated protein 1 (KEAP1). However, p62, an autophagy pathway component and cargo adapter, can bind and target KEAP1 to the autophagosomes, leading to release of NRF2 and its translocation to the nucleus and antioxidant gene expression [71]. Therefore, by inhibiting autophagy we may prevent cells from recycling these damaged, leaky mitochondria and increasing the expression of antioxidant defense factors, thereby worsening cellular oxidative stress and diminishing cell survival.
One unexpected finding from our study is that in dysplastic CPD and OE19 cells, Cyto-ID and LC3-GFP appear to mark different vesicle subsets. This is the only explanation available to explain the disconnect between these two well-established approaches to quantifying autophagy vesicle content of cells. We tend to favor the LC3-GFP results as being more representative of the autophagy rsponse, given that control OE19 cells by TEM had vesicles but these were not double-walled and did not contain cellular debris. Thus the Cyto-ID dye identifies these vesicles based on a shared biochemistry with autophagosomes. However, we cannot explain why, only in OE19 cells, the Cyto-ID signal was lost after acid treatment. Together these findings do suggest autophagosome biochemistry may be different in the dysplastic cell line. The mechanism for this alteration, and the physiologic role it may play, cannot be determined by the present studies and is an area of focus for future work.
One important reason for our interest in the role of autophagy in the pathogenesis of BE and EAC is that this is a pathway with immediate translational potential; there are already several drugs approved for human use that are known to modulate autophagic responses, with many others under investigation [48,49]. In addition to the autophagy inhibiting drugs, (Chloroquine, and Hydroxychloroquine), there are a number of other drugs available to induce the autophagy pathway, including mTOR inhibitors (Rapamycin, Everolimus, Temsirolimus), Tyrosine kinase inhibitors (Imatinib, Dasatinib), Akt inhibitors (Perifosine, Triciribine), and AMPK activators (Metformin). Therefore, it is conceivable that therapies can be directed to fine-tune autophagy in patients to achieve therapeutic goals using these established drugs. In future studies we plan to further explore the effect of manipulating autophagy responses on BE disease onset and progression using cell culture and the mouse models.
In summary, autophagy is a common cell stress response mechanism that is induced by GERD-like acidic stress and inflammation where it functions to reduce intracellular oxidative stress and improve cell survival. In addition, autophagy may be a novel therapeutic target in Barrett’s esophagus that deserves to be explored. Our work exploring the activity of autophagy in Barrett’s esophagus and esophageal adenocarcinoma is thus important both for these mechanistic insights as well as the potential application of these novel therapeutic agents to intervene in BE and EAC onset and progression.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank the University of Pennsylvania’s Morphology and Pathology Imaging Core, Electron Microscopy Resource Laboratory, and Flow Cytometry and Cell Sorting facility for technical expertise and assistance. This work is supported as part of the NCI BETRNet program (CA163004 JPL), the Integrated Microphysiological Systems program and the (TR 000536), an NIH career development award to JPL (OD 012097), a K26 RR 032714 (HN) and University of Pennsylvania Transdisciplinary Research on Energetics and Cancer (TREC) Survivor Center (HN, GWF and JPL), and K01DK103953 (KAW). This work was also supported by the Morphology, Cell Culture, and Molecular Biology Core Facilities of the Center for Molecular Studies in Digestive and Liver Disease at the University of Pennsylvania (P30-DK050306 and PO1 CA098101).
REFERENCES
- 1.Society AC. Cancer Facts and Figures 2010. Atlanta, Ga: American Cancer Society; 2010. [Google Scholar]
- 2.Shaheen NJ. What is behind the remarkable increase in esophageal adenocarcinoma? Am J Gastroenterol. 2014;109(3):345–347. doi: 10.1038/ajg.2014.35. [DOI] [PubMed] [Google Scholar]
- 3.Morales CP, Souza RF, Spechler SJ. Hallmarks of cancer progression in Barrett's oesophagus. Lancet. 2002;360(9345):1587–1589. doi: 10.1016/S0140-6736(02)11569-8. [DOI] [PubMed] [Google Scholar]
- 4.Paulson TG, Reid BJ. Focus on Barrett's esophagus and esophageal adenocarcinoma. Cancer Cell. 2004;6(1):11–16. doi: 10.1016/j.ccr.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 5.Kong J, Stairs DB, Lynch JP. Modeling Barrett’s Esophagus. Biochemical Society Transactions. 2010;38 doi: 10.1042/BST0380321. In Press. [DOI] [PubMed] [Google Scholar]
- 6.Sharma P. Clinical practice. Barrett's esophagus. N Engl J Med. 2009;361(26):2548–2556. doi: 10.1056/NEJMcp0902173. [DOI] [PubMed] [Google Scholar]
- 7.Moons LM, Kusters JG, Bultman E, et al. Barrett's oesophagus is characterized by a predominantly humoral inflammatory response. J Pathol. 2005;207(3):269–276. doi: 10.1002/path.1847. [DOI] [PubMed] [Google Scholar]
- 8.Federico A, Morgillo F, Tuccillo C, Ciardiello F, Loguercio C. Chronic inflammation and oxidative stress in human carcinogenesis. Int J Cancer. 2007;121(11):2381–2386. doi: 10.1002/ijc.23192. [DOI] [PubMed] [Google Scholar]
- 9.Chen X, Ding YW, Yang G, Bondoc F, Lee MJ, Yang CS. Oxidative damage in an esophageal adenocarcinoma model with rats. Carcinogenesis. 2000;21(2):257–263. doi: 10.1093/carcin/21.2.257. [DOI] [PubMed] [Google Scholar]
- 10.Inayama M, Hashimoto N, Tokoro T, Shiozaki H. Involvement of oxidative stress in experimentally induced reflux esophagitis and esophageal cancer. Hepatogastroenterology. 2007;54(75):761–765. [PubMed] [Google Scholar]
- 11.Valko M, Leibfritz D, Moncol J, Cronin MT, Mazur M, Telser J. Free radicals and antioxidants in normal physiological functions and human disease. Int J Biochem Cell Biol. 2007;39(1):44–84. doi: 10.1016/j.biocel.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 12.Clements DM, Oleesky DA, Smith SC, et al. A study to determine plasma antioxidant concentrations in patients with Barrett's oesophagus. J Clin Pathol. 2005;58(5):490–492. doi: 10.1136/jcp.2004.023721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lum JJ, DeBerardinis RJ, Thompson CB. Autophagy in metazoans: cell survival in the land of plenty. Nat Rev Mol Cell Biol. 2005;6(6):439–448. doi: 10.1038/nrm1660. [DOI] [PubMed] [Google Scholar]
- 14.Amaravadi RK, Thompson CB. The roles of therapy-induced autophagy and necrosis in cancer treatment. Clin Cancer Res. 2007;13(24):7271–7279. doi: 10.1158/1078-0432.CCR-07-1595. [DOI] [PubMed] [Google Scholar]
- 15.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amaravadi RK. Autophagy-induced tumor dormancy in ovarian cancer. J Clin Invest. 2008;118(12):3837–3840. doi: 10.1172/JCI37667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amaravadi RK, Yu D, Lum JJ, et al. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest. 2007;117(2):326–336. doi: 10.1172/JCI28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee JS, Oh TY, Ahn BO, et al. Involvement of oxidative stress in experimentally induced reflux esophagitis and Barrett's esophagus: clue for the chemoprevention of esophageal carcinoma by antioxidants. Mutat Res. 2001;480–481:189–200. doi: 10.1016/s0027-5107(01)00199-3. [DOI] [PubMed] [Google Scholar]
- 19.Farhadi A, Fields J, Banan A, Keshavarzian A. Reactive oxygen species: are they involved in the pathogenesis of GERD, Barrett's esophagus, and the latter's progression toward esophageal cancer? Am J Gastroenterol. 2002;97(1):22–26. doi: 10.1111/j.1572-0241.2002.05444.x. [DOI] [PubMed] [Google Scholar]
- 20.Sihvo EI, Salminen JT, Rantanen TK, et al. Oxidative stress has a role in malignant transformation in Barrett's oesophagus. Int J Cancer. 2002;102(6):551–555. doi: 10.1002/ijc.10755. [DOI] [PubMed] [Google Scholar]
- 21.Dvorak K, Payne CM, Chavarria M, et al. Bile acids in combination with low pH induce oxidative stress and oxidative DNA damage: relevance to the pathogenesis of Barrett's oesophagus. Gut. 2007;56(6):763–771. doi: 10.1136/gut.2006.103697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McQuaid KR, Laine L, Fennerty MB, Souza R, Spechler SJ. Systematic review: the role of bile acids in the pathogenesis of gastro-oesophageal reflux disease and related neoplasia. Aliment Pharmacol Ther. 2011 doi: 10.1111/j.1365-2036.2011.04709.x. [DOI] [PubMed] [Google Scholar]
- 23.Roesly HB, Khan MR, Chen HD, et al. The decreased expression of Beclin-1 correlates with progression to esophageal adenocarcinoma: the role of deoxycholic acid. Am J Physiol Gastrointest Liver Physiol. 2012;302(8):G864–G872. doi: 10.1152/ajpgi.00340.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harada H, Nakagawa H, Oyama K, et al. Telomerase induces immortalization of human esophageal keratinocytes without p16INK4a inactivation. Mol Cancer Res. 2003;1(10):729–738. [PubMed] [Google Scholar]
- 25.Okawa T, Michaylira CZ, Kalabis J, et al. The functional interplay between EGFR overexpression, hTERT activation, and p53 mutation in esophageal epithelial cells with activation of stromal fibroblasts induces tumor development, invasion, and differentiation. Genes Dev. 2007;21(21):2788–2803. doi: 10.1101/gad.1544507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kong J, Nakagawa H, Isariyawongse B, et al. Induction of Intestinalization in Human Esophageal Keratinocytes is a Multi-step Process. Carcinogenesis. 2009;30(1):122–130. doi: 10.1093/carcin/bgn227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palanca-Wessels MC, Klingelhutz A, Reid BJ, et al. Extended lifespan of Barrett's esophagus epithelium transduced with the human telomerase catalytic subunit: a useful in vitro model. Carcinogenesis. 2003;24(7):1183–1190. doi: 10.1093/carcin/bgg076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stairs DB, Nakagawa H, Klein-Szanto A, et al. Cdx1 and c-Myc foster the initiation of transdifferentiation of the normal esophageal squamous epithelium toward Barrett's esophagus. PLoS ONE. 2008;3(10):e3534. doi: 10.1371/journal.pone.0003534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keller MS, Ezaki T, Guo RJ, Lynch JP. Cdx1 or Cdx2 Expression Activates E-Cadherin-mediated Cell-cell Adhesion and Compaction in Human Colo 205 cells. Am J Physiol Gastrointest Liver Physiol. 2004;287(1):G104–G114. doi: 10.1152/ajpgi.00484.2003. [DOI] [PubMed] [Google Scholar]
- 30.Klionsky DJ, Abdalla FC, Abeliovich H, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8(4):445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quante M, Bhagat G, Abrams JA, et al. Bile Acid and inflammation activate gastric cardia stem cells in a mouse model of barrett-like metaplasia. Cancer Cell. 2012;21(1):36–51. doi: 10.1016/j.ccr.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Natsuizaka M, Kinugasa H, Kagawa S, et al. IGFBP3 promotes esophageal cancer growth by suppressing oxidative stress in hypoxic tumor microenvironment. Am J Cancer Res. 2014;4(1):29–41. [PMC free article] [PubMed] [Google Scholar]
- 33.Kagawa S, Natsuizaka M, Whelan KA, et al. Cellular senescence checkpoint function determines differential Notch1-dependent oncogenic and tumor-suppressor activities. Oncogene. 2014;0 doi: 10.1038/onc.2014.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kong J, Crissey MA, Funakoshi S, Kreindler JL, Lynch JP. Ectopic Cdx2 Expression in Murine Esophagus Models an Intermediate Stage in the Emergence of Barrett's Esophagus. PLoS ONE. 2011;6(4):e18280. doi: 10.1371/journal.pone.0018280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rockett JC, Larkin K, Darnton SJ, Morris AG, Matthews HR. Five newly established oesophageal carcinoma cell lines: phenotypic and immunological characterization. Br J Cancer. 1997;75(2):258–263. doi: 10.1038/bjc.1997.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang L, Yu J, Pan H, et al. Small molecule regulators of autophagy identified by an image-based high-throughput screen. Proc Natl Acad Sci U S A. 2007;104(48):19023–19028. doi: 10.1073/pnas.0709695104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo JY, Xia B, White E. Autophagy-mediated tumor promotion. Cell. 2013;155(6):1216–1219. doi: 10.1016/j.cell.2013.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lorin S, Hamai A, Mehrpour M, Codogno P. Autophagy regulation and its role in cancer. Semin Cancer Biol. 2013;23(5):361–379. doi: 10.1016/j.semcancer.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 39.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451(7182):1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12(1):9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hartman KG, Bortner JD, Jr, Falk GW, et al. Modeling human gastrointestinal inflammatory diseases using microphysiological culture systems. Exp Biol Med (Maywood) 2014;239(9):1108–1123. doi: 10.1177/1535370214529388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hartman KG, Bortner JD, Falk GW, et al. Modeling inflammation and oxidative stress in gastrointestinal disease development using novel organotypic culture systems. Stem Cell Res Ther. 2013;4(Suppl 1):S5. doi: 10.1186/scrt366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakagawa H, Wang TC, Zukerberg L, et al. The targeting of the cyclin D1 oncogene by an Epstein-Barr virus promoter in transgenic mice causes dysplasia in the tongue, esophagus and forestomach. Oncogene. 1997;14(10):1185–1190. doi: 10.1038/sj.onc.1200937. [DOI] [PubMed] [Google Scholar]
- 44.Nakagawa H, Whelan K, Lynch JP. Mechanisms of Barrett's oesophagus: Intestinal differentiation, stem cells, and tissue models. Best Pract Res Clin Gastroenterol. 2015;29(1):3–16. doi: 10.1016/j.bpg.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spechler SJ, Fitzgerald RC, Prasad GA, Wang KK. History, molecular mechanisms, and endoscopic treatment of Barrett's esophagus. Gastroenterology. 2010;138(3):854–869. doi: 10.1053/j.gastro.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dvorak K, Fass R, Dekel R, et al. Esophageal acid exposure at pH< or = 2 is more common in Barrett's esophagus patients and is associated with oxidative stress. Dis Esophagus. 2006;19(5):366–372. doi: 10.1111/j.1442-2050.2006.00596.x. [DOI] [PubMed] [Google Scholar]
- 47.Feagins LA, Zhang HY, Zhang X, et al. Mechanisms of oxidant production in esophageal squamous cell and Barrett's cell lines. Am J Physiol Gastrointest Liver Physiol. 2008;294(2):G411–G417. doi: 10.1152/ajpgi.00373.2007. [DOI] [PubMed] [Google Scholar]
- 48.Li X, Xu HL, Liu YX, An N, Zhao S, Bao JK. Autophagy modulation as a target for anticancer drug discovery. Acta Pharmacol Sin. 2013;34(5):612–624. doi: 10.1038/aps.2013.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Janku F, McConkey DJ, Hong DS, Kurzrock R. Autophagy as a target for anticancer therapy. Nat Rev Clin Oncol. 2011;8(9):528–539. doi: 10.1038/nrclinonc.2011.71. [DOI] [PubMed] [Google Scholar]
- 50.Kimmelman AC. The dynamic nature of autophagy in cancer. Genes Dev. 2011;25(19):1999–2010. doi: 10.1101/gad.17558811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McAfee Q, Zhang Z, Samanta A, et al. Autophagy inhibitor Lys05 has single-agent antitumor activity and reproduces the phenotype of a genetic autophagy deficiency. Proc Natl Acad Sci U S A. 2012;109(21):8253–8258. doi: 10.1073/pnas.1118193109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sotelo J, Briceno E, Lopez-Gonzalez MA. Adding chloroquine to conventional treatment for glioblastoma multiforme: a randomized, double-blind, placebo-controlled trial. Ann Intern Med. 2006;144(5):337–343. doi: 10.7326/0003-4819-144-5-200603070-00008. [DOI] [PubMed] [Google Scholar]
- 53.Rangwala R, Chang YC, Hu J, et al. Combined MTOR and autophagy inhibition: phase I trial of hydroxychloroquine and temsirolimus in patients with advanced solid tumors and melanoma. Autophagy. 2014;10(8):1391–1402. doi: 10.4161/auto.29119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rangwala R, Leone R, Chang YC, et al. Phase I trial of hydroxychloroquine with dose-intense temozolomide in patients with advanced solid tumors and melanoma. Autophagy. 2014;10(8):1369–1379. doi: 10.4161/auto.29118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rosenfeld MR, Ye X, Supko JG, et al. A phase I/II trial of hydroxychloroquine in conjunction with radiation therapy and concurrent and adjuvant temozolomide in patients with newly diagnosed glioblastoma multiforme. Autophagy. 2014;10(8):1359–1368. doi: 10.4161/auto.28984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100(25):15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang A, Rajeshkumar NV, Wang X, et al. Autophagy is critical for pancreatic tumor growth and progression in tumors with p53 alterations. Cancer Discov. 2014;4(8):905–913. doi: 10.1158/2159-8290.CD-14-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guo JY, Karsli-Uzunbas G, Mathew R, et al. Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes Dev. 2013;27(13):1447–1461. doi: 10.1101/gad.219642.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guo JY, White E. Autophagy is required for mitochondrial function, lipid metabolism, growth, and fate of KRAS(G12D)-driven lung tumors. Autophagy. 2013;9(10):1636–1638. doi: 10.4161/auto.26123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Strohecker AM, White E. Autophagy promotes BrafV600E-driven lung tumorigenesis by preserving mitochondrial metabolism. Autophagy. 2014;10(2):384–385. doi: 10.4161/auto.27320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takamura A, Komatsu M, Hara T, et al. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25(8):795–800. doi: 10.1101/gad.2016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Y, Goldman S, Baerga R, Zhao Y, Komatsu M, Jin S. Adipose-specific deletion of autophagy-related gene 7 (atg7) in mice reveals a role in adipogenesis. Proc Natl Acad Sci U S A. 2009;106(47):19860–19865. doi: 10.1073/pnas.0906048106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Suzuki N, Yasui M, Geacintov NE, Shafirovich V, Shibutani S. Miscoding events during DNA synthesis past the nitration-damaged base 8-nitroguanine. Biochemistry. 2005;44(25):9238–9245. doi: 10.1021/bi050276p. [DOI] [PubMed] [Google Scholar]
- 64.Yermilov V, Rubio J, Becchi M, Friesen MD, Pignatelli B, Ohshima H. Formation of 8-nitroguanine by the reaction of guanine with peroxynitrite in vitro. Carcinogenesis. 1995;16(9):2045–2050. doi: 10.1093/carcin/16.9.2045. [DOI] [PubMed] [Google Scholar]
- 65.O'Hagan HM, Wang W, Sen S, et al. Oxidative damage targets complexes containing DNA methyltransferases, SIRT1, and polycomb members to promoter CpG Islands. Cancer Cell. 2011;20(5):606–619. doi: 10.1016/j.ccr.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Poehlmann A, Kuester D, Malfertheiner P, Guenther T, Roessner A. Inflammation and Barrett's carcinogenesis. Pathol Res Pract. 2012;208(5):269–280. doi: 10.1016/j.prp.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 67.Hartman KG, Bortner JD, Jr, Falk GW, et al. Modeling human gastrointestinal inflammatory diseases using microphysiological culture systems. Exp Biol Med (Maywood) 2014 doi: 10.1177/1535370214529388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Galluzzi L, Pietrocola F, Bravo-San Pedro JM, et al. Autophagy in malignant transformation and cancer progression. EMBO J. 2015 Apr 1;34(7):856–880. doi: 10.15252/embj.201490784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Martinez-Outschoorn UE, Pavlides S, Howell A, et al. Stromal-epithelial metabolic coupling in cancer: integrating autophagy and metabolism in the tumor microenvironment. Int J Biochem Cell Biol. 2011;43(7):1045–1051. doi: 10.1016/j.biocel.2011.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lambert AJ, Brand MD. Reactive oxygen species production by mitochondria. Methods Mol Biol. 2009;554:165–181. doi: 10.1007/978-1-59745-521-3_11. [DOI] [PubMed] [Google Scholar]
- 71.Poillet-Perez L, Despouy G, Delage-Mourroux R, Boyer-Guittaut M. Interplay between ROS and autophagy in cancer cells, from tumor initiation to cancer therapy. Redox Biol. 2015;4:184–192. doi: 10.1016/j.redox.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.