Abstract
Insult-provoked transformation of neuronal networks into epileptic ones involves multiple mechanisms. Intervention studies have identified both dysregulated inflammatory pathways and NRSF-mediated repression of crucial neuronal genes as contributors to epileptogenesis. However, it remains unclear how epilepsy-provoking insults (e.g., prolonged seizures) induce both inflammation and NRSF, and whether common mechanisms exist. We examined miR-124 as a candidate dual regulator of NRSF- and inflammatory-pathways. Status epilepticus (SE) led to reduced miR-124 expression via SIRT1, and in turn MiR-124 repression, via C/EBPα, upregulated NRSF. We tested whether augmenting miR-124 after SE would abort epileptogenesis by preventing inflammation and NRSF upregulation. SE-sustaining animals developed epilepsy but supplementing miR-124 did not modify epileptogenesis. Examining this result further, we found that synthetic miR-124 effectively blocked NRSF upregulation and rescued NRSF target genes, but also augmented microglia activation and inflammatory cytokines. Thus, miR-124 attenuates epileptogenesis via NRSF while promoting epilepsy via inflammation.
Introduction
The transformative mechanisms converting neuronal networks into epileptic ones (Vezzani et al., 2011; Engel et al., 2013a; Simonato et al., 2014) are driven at least partly by altered expression and function of crucial neuronal genes (Brooks-Kayal et al., 2009; Cacheaux et al., 2009; Goldberg and Coulter, 2013; Rossignol et al., 2014), in parallel with activation of inflammatory cells (Pernot et al., 2011; Vezzani et al., 2011). Recently, interfering with the repression of numerous target genes by the transcriptional repressor Neuron Restrictive Silencer Factor (NRSF) was found to attenuate the development of epilepsy in the short-term (McClelland et al., 2011), supporting a mechanistic role in epileptogenesis (Gao et al., 2011; Baldelli and Meldolesi 2015). In addition, anti-inflammatory strategies have modified the course of this disease, supporting the contribution of inflammation to insult-provoked epilepsy (Maroso et al., 2011; Li et al., 2013). It is unclear how epilepsy-provoking insults such as status epilepticus (SE) initiate both NRSF dysregulation and pro-epileptogenic inflammation, and if these concurrent processes are co-orchestrated. Identification of regulatory molecules which govern multiple epileptogenic pathways may allow for the development of effective preventative therapies.
MicroRNAs are potent and pleiotropic molecules with numerous gene targets and conserved across evolution (Lall et al., 2006; O'Carroll and Schaefer, 2013; Izaurralde, 2015). Their targets include transcription factors and inflammatory cascades (Quinn and O'Neill, 2011; Neo et al., 2014; Su et al., 2015). Recent studies have found aberrant expression of miRNAs during epileptogenesis and consequent gene dysregulation, including miR-134 (Jimenez-Mateos et al., 2012), miR-128 (Tan et al., 2013), miR-146 (Aronica et al., 2010) and others (Risbud and Porter, 2013; Gorter et al., 2014). Here we focused on miR-124 because it is strongly expressed in hippocampus (Smirnova et al., 2005), has been linked to NRSF expression in developing neurons (Conaco et al., 2006; Visvanathan et al., 2007; Yoo et al., 2009), and has recently been reported to be required for microglial quiescence (Ponomarev et al., 2011). Using a SE model of epilepsy generation, we demonstrate that miR-124 plays dual and opposing roles in epileptogenesis. We find that preventing the SE-provoked reduction of miR-124 levels is anti-epileptogenic because it prevents increased NRSF expression and activity. However, preventing reduction in miR-124 levels surprisingly also exacerbates inflammation, with a pro-epileptogenic effect.
Results
Both inflammatory and transcriptional pathways are activated by an epilepsy-promoting insult
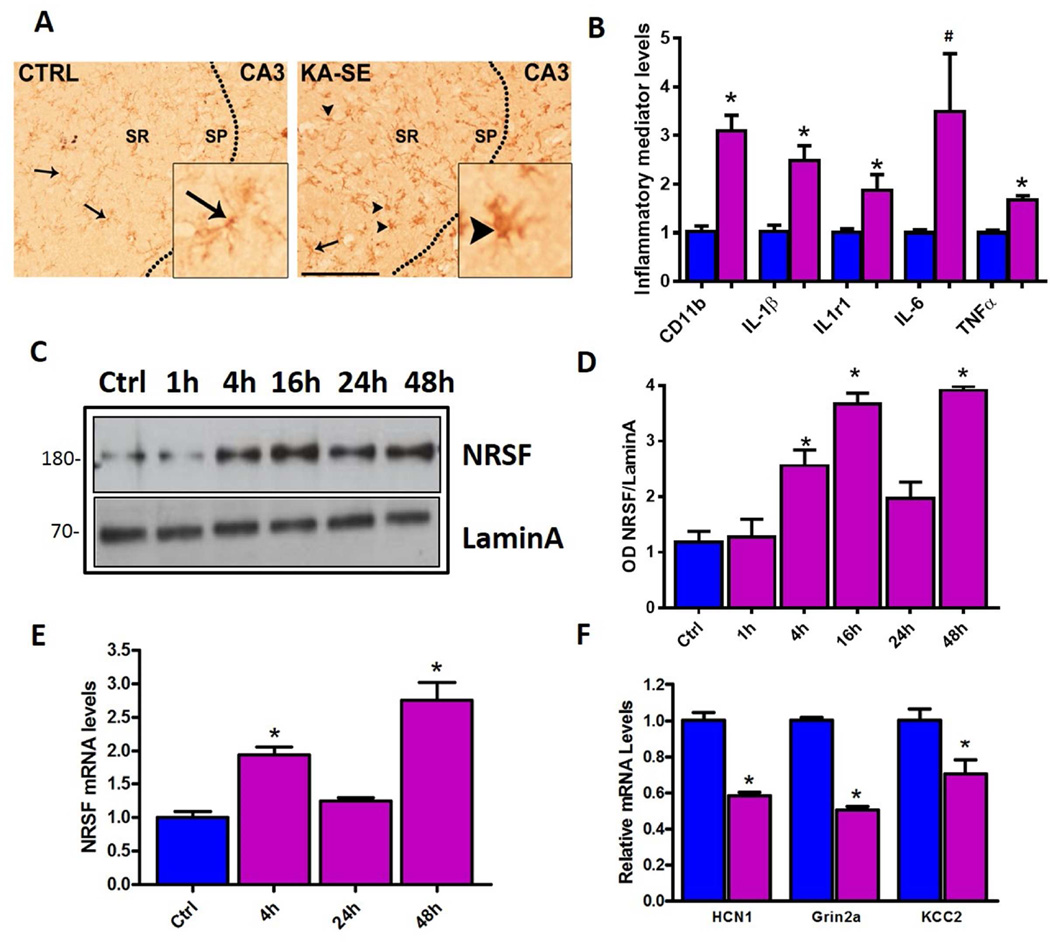
Within hours of SE provoked by the systemic administration of the glutamate receptor agonist kainic acid (KA) (KA-SE), many gene networks were activated. First, pro-inflammatory cascades were induced in the hippocampus evident by microglial activation, as apparent from the altered microglia morphology from ramified to globular (Figure 1A), augmented CD11b expression (Figure 1B), and robust induction of pro-inflammatory cytokines. Levels of the cytokines IL-1β, IL-6, and TNF-α, and of IL-1β receptor, were significantly enhanced compared with control hippocampus (Figure 1B). Concurrently, as reported, the transcriptional repressor NRSF was upregulated at both mRNA and protein level already within 4h of KA-SE, and remained elevated up to 48h (Palm et al., 1998; McClelland et al., 2011; McClelland et al., 2014) (Figures 1C–1E). Furthermore, NRSF activation by SE led to repression of NRSF-regulated target genes including HCN1, GRIN2A and KCC2 (Figure 1F).
Figure 1. Concurrent induction of inflammatory mediators and NRSF expression by epilepsy-provoking status epilepticus.
Microglia activation in KA-SE rat hippocampus (A). Compared with control hippocampus, where the microglial marker IBA1 is found in wispy ramified cells, (inset) microglia in KA-SE hippocampus appear globular (arrowheads). Sp, sr, strata pyramidale and radiatum. Bar= 100µm. (B). KA-SE induced mRNA expression of inflammatory markers CD11b, and pro-inflammatory cytokines in hippocampus as detected by qPCR n=4/group. *= p<0.05, #p=0.07. (C) Representative W. blot showing NRSF protein levels from hippocampal nuclear fractions. NRSF levels increased at 4h following SE and remained elevated at 48h. One hippocampus per lane, n=5 animals/time-point. (D) Densitometric analysis of NRSF protein levels after KA-SE, normalized and compared to control. (E) qPCR analysis of NRSF mRNA expression in control hippocampus and following KA-SE shows upregulation of NRSF compared to control, n=9/group. (F) Repression of NRSF target genes including HCN1, GRIN2A and KCC2 at 48h post SE, n=4/group.
Hippocampal miR-124 levels decline rapidly following epilepsy-provoking seizures
As both inflammatory cells and NRSF are induced by SE and might contribute to epileptogenesis we sought to identify a potential common upstream regulatory mechanism. However in-silico approaches failed to identify canonical transcriptional controls common to both. Therefore, we considered non-canonical mechanisms of regulation including non-coding RNAs, and focused on microRNAs (miRNA) because they are expressed and function in all cell types including neurons and microglia (Ponomarev et al., 2011; Jovicic et al., 2013; Malmevik et al., 2015). In-silico analysis identified a group of miRNAs predicted to regulate NRSF (Figure S1A), those reported to regulate neuro-inflammation (Soreq and Wolf, 2011) and those, such as miR-132, 134, 128, 9 and 124 that have been implicated in epilepsy (Jimenez-Mateos et al., 2012; Kan et al., 2012; Risbud and Porter, 2013). Because miR-9 and miR-124 also interact with NRSF during neuronal development (Conaco et al., 2006; Visvanathan et al., 2007; Yoo et al., 2009) we focused on them..
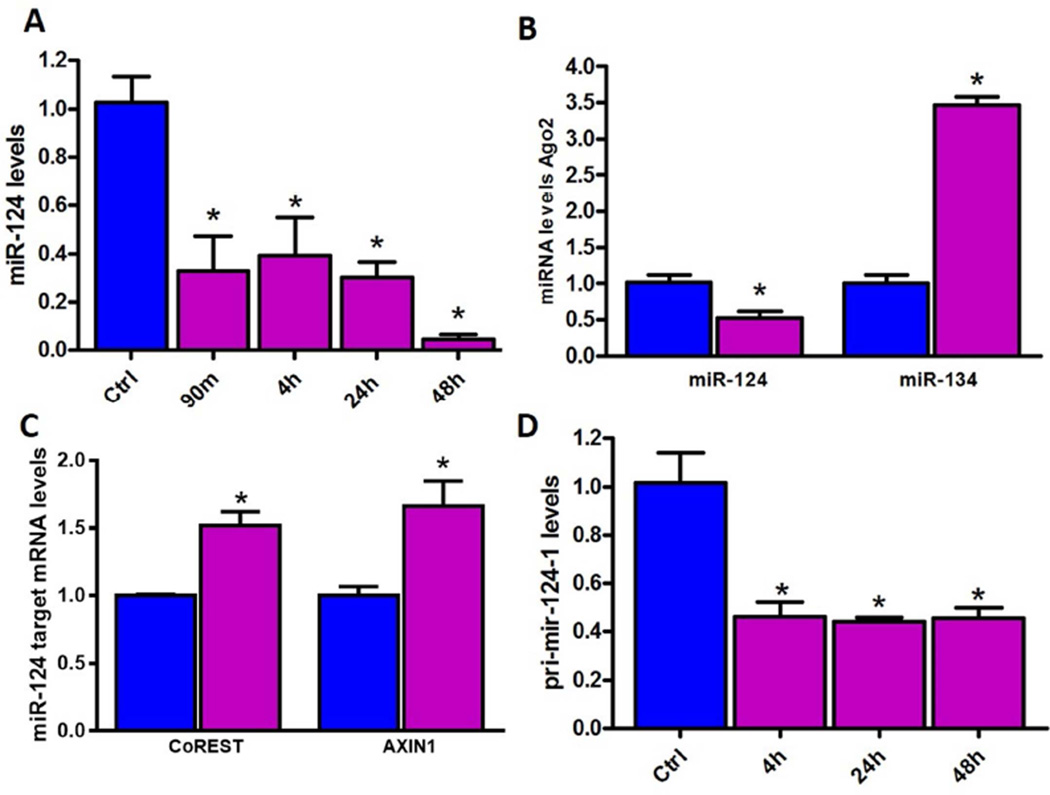
We found no change in hippocampal miR-9 levels following KA-SE (Figure S1C). In contrast, miR-124 levels were rapidly and significantly reduced by 90min following KA-SE and remained depressed for up to 48h (Figure 2A). We then measured the levels of functional miR-124 by measuring levels bound to Argonaute 2 (Ago2) within RISC. Levels of miR-124 within RISC were lower in KA-SE compared to control hippocampi (Figure 2B). This reduction was selective because miR-134 RISC levels were increased, as reported (Jimenez-Mateos et al., 2012); Figure 2B). Decreased miR-124 expression was biologically important because levels of target mRNAs such as CoREST and AXIN1 were increased (de-repressed) in hippocampus 48h after KA-SE (Figure 2C).To test if reduced miR-124 in the RISC complex stemmed from diminished total miR-124 levels or from potential SE induced perturbation of miRNA uptake by RISC we measured pri-mir-124-1, 2 and 3, precursor molecules expressed from the three genes which code for the same mature miR-124. Primir-124-1 expression was significantly attenuated after SE (Figure 2D), whereas pri-mir-124-2 and 3 were unaffected (Figure S1D), indicating that reduced mature miR-124 levels derived from reduced output of the miR-124-1 gene. Together, the findings indicated that the expression, activity and function of miR-124 were repressed rapidly following SE, preceding the augmentation of inflammatory cytokines and of NRSF levels.
Figure 2. Reduction of levels and function of miR-124 in hippocampus following KA-SE.
(A) qPCR analysis of mature miR-124 levels in control hippocampus and after KA-SE. miR-124 levels declined as soon as 90min post SE and this persisted at 48h, n=9/time-point. (B) Ago2-bound (active) miR-124 levels significantly reduced following KA-SE compared with controls, whereas 134 levels were elevated. n=5/group. (C) qPCR analysis of miR-124 target genes CoREST and AXIN1. Both miR-124 targets were de-repressed following KA-SE compared to controls; n=6/group. (D) Pri-mir-124-1 expression in hippocampus was significantly reduced following KA-SE as compared to controls, n=6/timepoint.
The miR-124-1 gene is repressed via histone deacetylation that is mediated by SIRT1
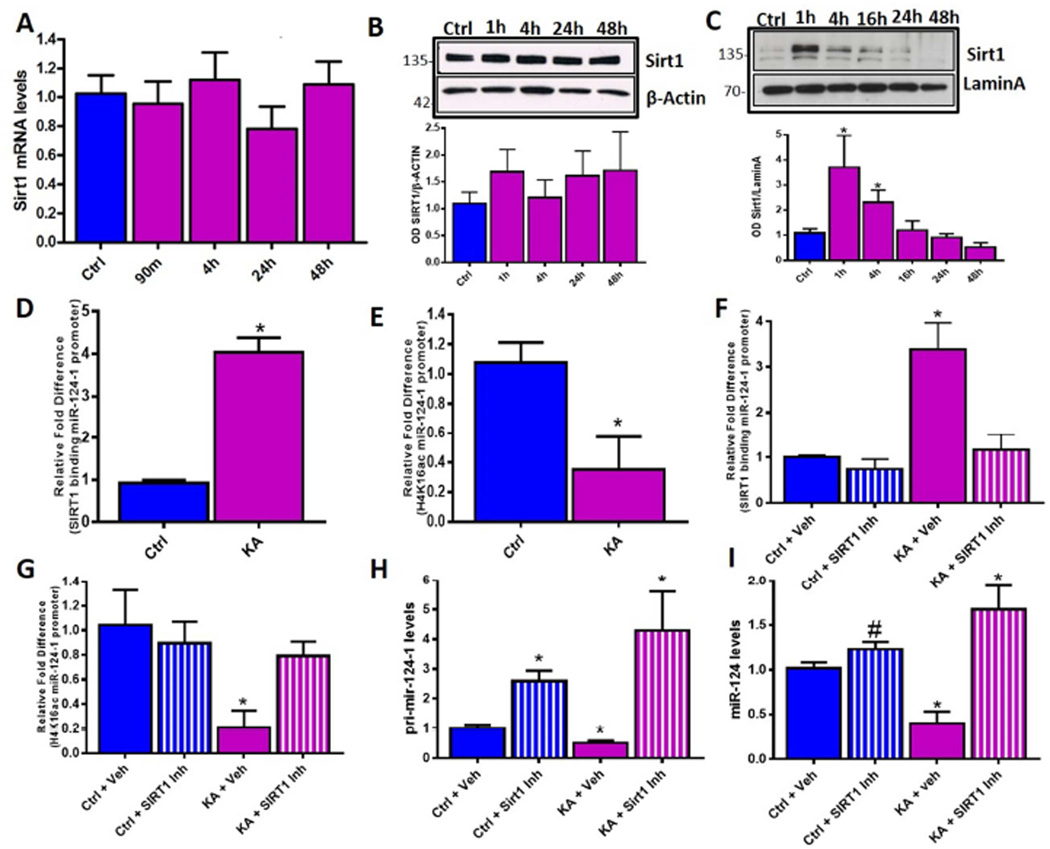
The above data demonstrated that KA-SE rapidly reduced miR-124-1 gene expression, prompting a search for the underlying mechanisms. Histone deacetylation can take place within minutes as a result of neuronal activity during memory processes (Roth et al., 2010; White and Wood, 2014). Therefore, we probed the deacetylation of chromatin surrounding miR-124-1 gene locus as a candidate mechanism for miR-124 repression, and found we found enhanced de-acetylation of H4K16 (Figure 3E) de-acetylation of which typically denotes gene repression (Ferguson et al., 2015). Looking for the responsible deacetylating enzyme, we reasoned that SE represents an extremely metabolically demanding process (Duffy et al., 1975; Fujikawa et al., 1988; Carmody and Brennan, 2010; Jupp et al., 2012; Tantama et al., 2013; Choy et al., 2014). Therefore we tested for the involvement of members of the lysine deacetylase family, Sirtuins, which are activated by changes in cellular energy levels (Blander and Guarente, 2004; Herskovits and Guarente, 2014). KA-SE did not alter mRNA or total cellular protein levels of SIRT1 (Figure 3A, 3B) however the levels of SIRT1 within the nuclear fraction of hippocampal tissue were significantly augmented already at 1h post KA-SE (Figure 3C). (Cytoplasmic levels of SIRT1 were low and did not change appreciably; Figure S2A). Augmented nuclear SIRT1 was associated with increased binding to the miR-124-1 gene promoter (Figure 3D) whereas binding to miR-124-2 and 3 genes did not change (Figure S2B and S2C), and reduced H4K16 acetylation (Figure 3E). Whereas the above data supported that SIRT1 binding caused the repression of the miR-124-1 gene, we tested this directly by inhibiting SIRT1 using the specific inhibitor Ex-527. Using organotypic hippocampal slice cultures we replicated our in vivo findings from above by treating with kainic acid to induce seizure-like events (Figure S3A S3B). Following 3h of seizure-like activity we applied the SIRT1 inhibitor and found that this reduced SIRT1 binding to the chromatin surrounding the miR-124-1 promoter region (Figure 3F) and prevented deacetylation of the H4K16 locus (Figure 3G). Inhibition of SIRT1 rescued the expression of pri-mir-124-1 (Figure 3H), of mature miR-124 (Figure 3I), and of another established target of SIRT1, miR-134; (Gao et al., 2010) (Figure S2D). Together these data indicated that SIRT1 directly repressed miR-124 by binding to the miR-124-1 gene and deacetylating histone lysine residues, promoting the formation of heterochromatin and consequent gene silencing.
Figure 3. SIRT1 mediated repression of miR-124 following KA-SE.
(A) SIRT1 mRNA expression in hippocampus remains stable after KA-SE, n=8/time-point. (B) SIRT1 protein levels, analysed using W. blot, did not change significantly in whole cell extracts obtained from hippocampus from control and KA-SE rats over a 48h time-course. n=3 per/time-point. (C) In contrast, analysis of nuclear fractions from control and KA-SE hippocampal tissue revealed significant increase of SIRT1 by 1h following SE n=4/group. (D) SIRT1 binding to the miR-124-1 gene, analysed using ChIP-qPCR, was enhanced significantly 60min following SE. n=6/group. (E) Acetylation levels of H4K16ac, an established SIRT1 target, were determined by ChIP-qPCR surrounding the miR-124-1 gene promoter. H4K16ac levels were reduced in hippocampus of KA-SE rats compared to controls. n=4/group. (F) Enhanced SIRT1 binding to miR-124-1 gene promoter after SE was attenuated by SIRT1 inhibition. n=4/group. (G) The SIRT1 inhibitor also abolished SE-induced H4K16 deacetylation at the miR-124-1 promoter. ChIP-qPCR quantification of H4K16ac levels in control and KA-SE groups with vehicle or SIRT1 inhibitor treatment. n=4/group. (H) And (I) qPCR analysis of pri-mir-124-1 (H) and mature miR-124 (I) expression in control or KA-SE hippocampal slices, after vehicle or SIRT1 inhibition. SIRT1 inhibition prevented repression of pri-mir-124-1 and restored levels of mature miR-124, #p=0.06, n=4/group.
Reduced miR-124 expression is required for increased NRSF expression
MiR-124 has been shown to repress microglia activation and the ensuing inflammatory molecular cascades (Ponomarev et al., 2011). The relationship between repression of miR-124 and the expression of NRSF has not been studied in the context of epileptogenesis. Therefore, having established the link between epilepsy-promoting metabolic insults such as KA-SE and repression of miR-124, we asked if depleted miR-124 was a cause of NRSF upregulation during epileptogenesis (McClelland et al., 2011; McClelland et al., 2014).
To test this possibility, we induced seizure-like events in organotypic hippocampal slice cultures and prevented cellular depletion of miR-124 levels by providing synthetic microRNA (agomir) to a subgroup of the cultures after termination of seizure-like activity. Restitution of miR-124 levels abrogated the upregulation of NRSF expression for up to 96h (Figure S3C) and prevented repression of NRSF target genes including GRIN2A (Figure S3d) (McClelland et al., 2014). The specificity of the effects of miR-124 was examined by supplementing the medium with miR-134 agomirs which did not prevent seizure-induced upregulation of NRSF (Figure S3E).
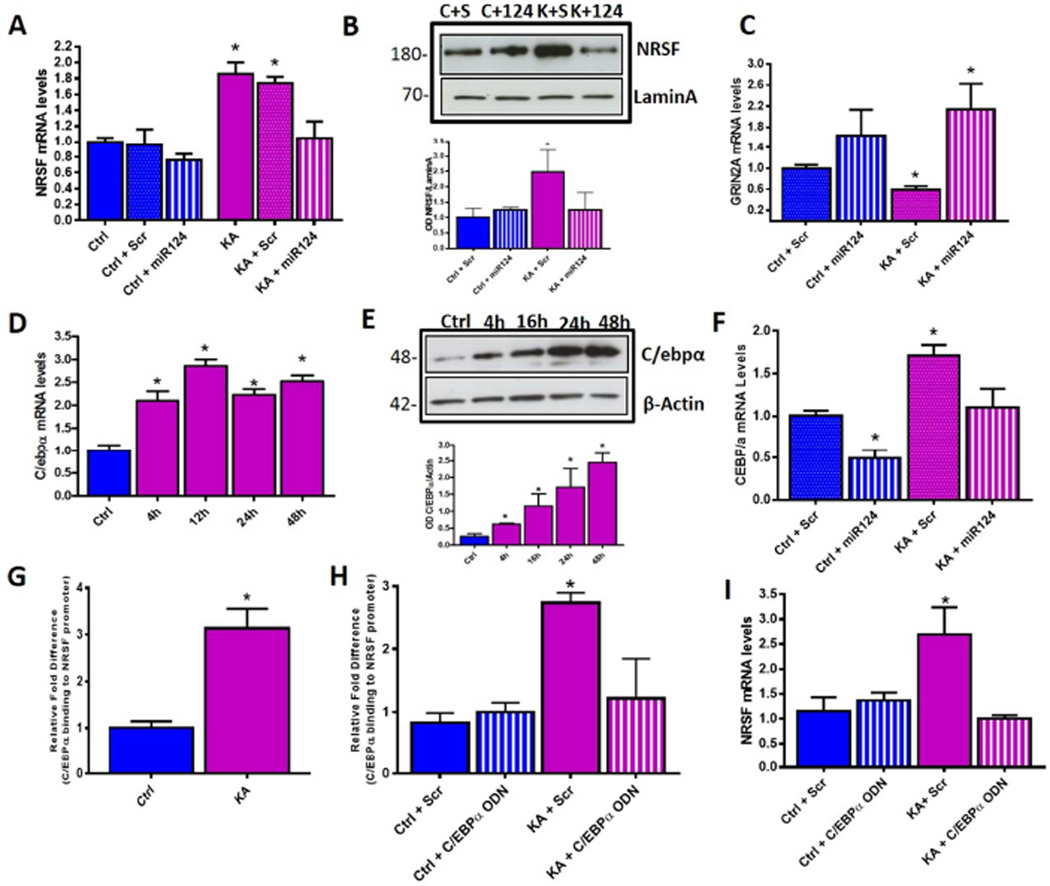
We then tested the role of miR-124 in regulating NRSF in vivo. We infused miR-124 agomirs into the lateral cerebral ventricles (ICV) of rats following KA-SE and measured mRNA and protein levels of hippocampal NRSF using qPCR and Western blots respectively. MiR-124 agomirs prevented seizure-induced upregulation of NRSF at both mRNA (Figure 4A) and protein level (Figure 4B) and prevented NRSF mediated repression of GRIN2A (Figure 4C). The combined in vitro and in vivo approaches demonstrated that the rapid repression of miR-124 levels after prolonged seizures caused NRSF upregulation and consequent repression of critical neuronal genes.
Figure 4. MiR-124 repression is required for increased NRSF expression and this regulation is mediated by miR-124 target C/EBPα.
(A) Hippocampal NRSF mRNA levels in controls vs KA-SE rats given either miR-124 agomir or Scr infusion. Hippocampal NRSF mRNA levels increased 48h following KA-SE+Scr and this increase was blocked in rats given miR-124 agomirs immediately following SE (KA+miR-124); n=6/group. (B) NRSF protein levels in nuclear fraction of hippocampus extracts in control and KA animals receiving either Scr or miR-124 infusion. NRSF protein levels were higher in SE+Scr hippocampus 48h following insult compared to SE+miR-124, where levels were comparable to controls. n=6/group, 1 hippocampus/lane. (C) MiR-124 agomir, but not Scr, treatment prevented seizure-induced NRSF-mediated repression of GRIN2A; n=6/group. (D) Hippocampal C/EBPα mRNA levels were enhanced significantly 4h following KA-SE and remained elevated up to 48h, n=8/time-point; qPCR. (E) C/EBPα protein levels increased 4h post SE and remain elevated for at least 48h. Representative W. blot and quantification. n=5/group. (F) MiR-124 agomir treatment following KA-SE prevented seizure-induced increase of C/EBPα expression compared to KA-SE which received Scr agomirs; n=6/group. (G) C/EBPα occupancy at NRSF gene increased significantly following KA-SE as compared to controls, n = 6/group. See Figure S4 for examined binding sites using ChIP-qPCR. (H) C/EBPα binding to the NRSF gene after KA-seizures was attenuated in hippocampi treated with ordered ODNs, compared to Scr ODN, n=3/group. (I) Inhibition of C/EBPα binding prevented seizure-induced NRSF upregulation. n=3/group.
MiR-124 regulates NRSF levels via the transcription factor C/EBPα
In silico analysis revealed no canonical binding site for miR-124 on the 3’UTR of the NRSF mRNA, suggesting the existence of intermediate miR-124 targets which, in turn, control NRSF expression. We initially tested a potential role of the miR-124 target PTBP1, (Visvanathan et al., 2007) however, the presence of increased REST4 splicing (Figure S4B) and no changes in PTBP1 levels (Figure S4A) suggested alternative regulatory mechanisms. The robust increase in NRSF mRNA levels provoked by SE suggested that transcriptional mechanisms were involved. In silico analysis of miR-124 targets combined with analysis of the NRSF gene, identified transcription factors that were predicted to both be targets of miR-124 and bind the NRSF gene. Among these, the CCAAAT enhancer binding protein (C/EBPα) has been described as a target of miR-124 that exerted actions related to microglia regulation (Ponomarev et al., 2011), and potential C/EBPα binding sites exist within the NRSF gene promoter (Figure S4C). C/EBPα mRNA and protein levels increased significantly in hippocampus of rats sustaining KA-SE (Figures 4D, 4E) and this increase was abrogated by administration of miR-124 agomirs immediately following KA-SE (Figure 4F). Chromatin immunoprecipitation (ChIP) demonstrated that C/EBPα binding to the NRSF promoter region, but not to the C/EBPα binding site within the first intron of the NRSF gene (Figure S4D), was significantly higher in hippocampus of KA-SE animals compared to control (Figure 4G). To test if C/EBPα binding was required for increased NRSF expression, we blocked C/EBPα binding by using decoy oligodeoxynucleotides (ODNs) composed of the C/EBPα binding recognition site (McClelland et al., 2011). These ODNs, but not scrambled (Scr) ODNs, decreased C/EBPα binding to the NRSF gene promoter (Figure 4H). Importantly, blocking C/EBPα binding abrogated upregulation of NRSF following seizure-like activity (Figure 4I). Together, these data identified C/EBPα as a seizure- and miR-124 dependent regulator of NRSF expression in hippocampus. Specifically, seizure-induced repression of miR-124 levels led to increased levels of C/EBPα, which, in turn, augmented NRSF expression by binding to the gene promoter and enhancing transcription.
Effects of post-insult miR-124 replenishment on the development of epilepsy
The data above suggested that rapid, KA-SE-induced repression of miR-124 levels in hippocampus promotes the development of epilepsy by two mechanisms: first, miR-124 represses inflammation, therefore, its depletion should promote increased cytokine expression (Figure S1B and Figure 5). Second, miR-124 depletion augments NRSF levels, implicated in short-term epileptogenesis (McClelland et al., 2011). Because these combined data poise miR-124 depletion as an important upstream mediator of epileptogenesis, we tested if miR-124 restitution would prevent epileptogenesis by both preventing upregulation of NRSF and by maintaining quiescent microglia and preventing inflammation.
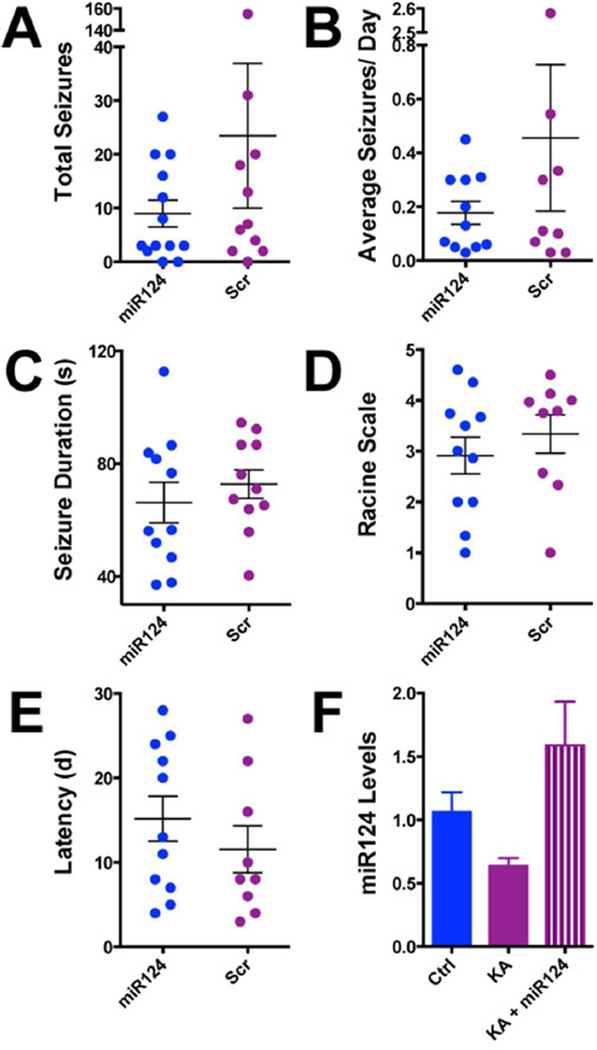
Figure 5. MiR-124 restitution following KA-SE in rats does not prevent the development of epilepsy.
(A) The vast majority (21/24) of KA-SE rats developed epilepsy regardless of treatment. Blue circles represent individual KA-SE sustaining rats given ICV miR-124 after the SE, Fuchsia circles represent the Scr agomirs. Total seizure numbers were similar in both groups. (B) Average number of seizures per day was not different between rats who received Scr agomirs and rats who received miR-124 agomirs. (C) EEG seizure duration was not significantly different between the two groups. (D) Seizure severity did not differ significantly between epileptic rats who received Scr agomir and those who received miR-124 agomirs. (E) The latency to onset of the 1st seizure, shown as days from the beginning of recording (day 2 post SE) was comparable in both groups. (F) Hippocampal miR-124 levels in animals who received miR-124 agomirs following KA-SE, measured using qPCR were similar to those in naïve controls.
We infused miR-124 agomirs or scrambled sequences directly into brain (ICV) of adult male rats immediately following the termination of KA-SE. The experimental groups consisted of rats ‘destined’ to become epileptic (KA-SE with Scr agomirs; n=12), rats sustaining KA-SE and treated with miR-124 agomirs (n=12), and control rats treated with either Scr agomir or ordered miR-124 agomir (n=5/group). All rats underwent continuous two month video-EEG monitoring (Hellier and Dudek, 2005). We evaluated the presence or absence of spontaneous seizures (epilepsy), the latency to onset of the first seizure, seizure duration, severity and the number of seizures per rat and per group. Mir-124 did not influence basal EEG including rhythms and amplitude in either controls or KA-SE rats and control rats did not develop seizures. The number of rats developing epilepsy was similar in KA-SE miR-124 agomir and Scr groups (11/12 and 10/12, respectively) (Figure 5A). Detailed analyses of video-EEG data revealed that the number of seizures (Figure 5A), average seizure number per day (Figure 5B), seizure duration (Figure 5C), seizure severity on the Racine scale (Figure 5D), and latency to the onset of the first seizure (Figure 5E) did not distinguish between groups. Together, these data indicated that administration of miR-124 agomirs after an epilepsy-provoking insult does not prevent or modify significantly the disease process.
Unexpected and complex actions and expression patterns of miR-124 in the brain
The lack of effect of miR-124 infusion on epileptogenesis was not a result of insufficient agomir levels. We employed a chemically-protected agomir (see methods), and detected levels in vivo in ranges comparable to endogenous miR-124 in naïve hippocampus (Figure 5F).Additionally, infused agomirs prevented the SE-induced upregulation of NRSF levels and activity (Figures 4A–C). We therefore examined if restitution of the SE-depleted miR-124 levels exerted the expected suppressive actions on SE-induced inflammatory cascades.
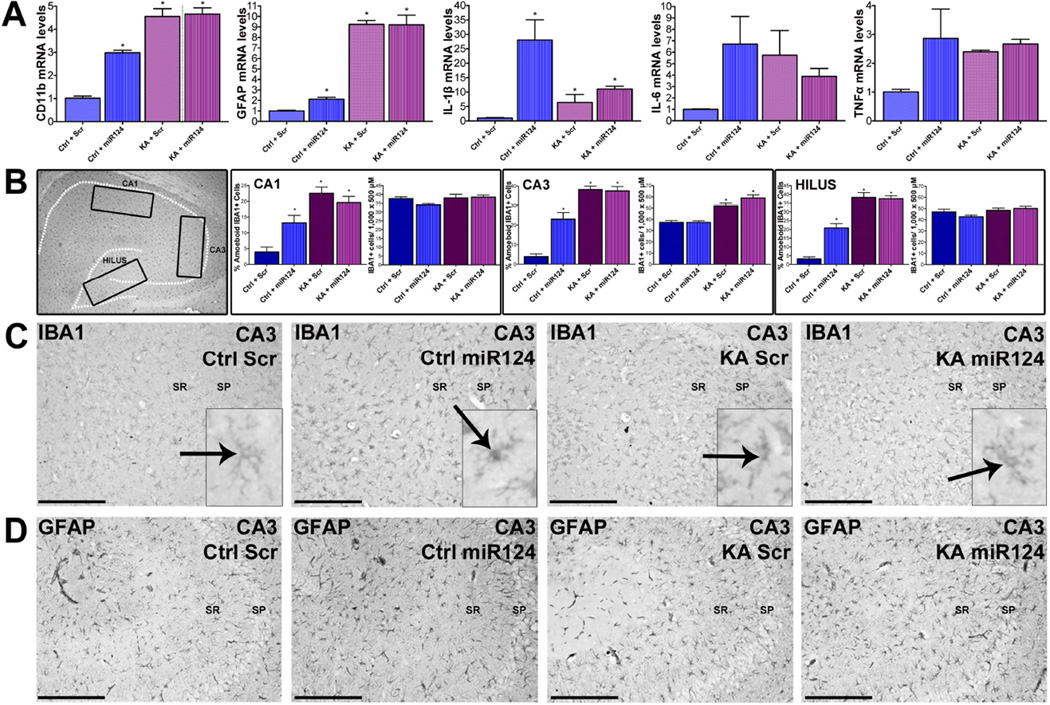
We first measured expression of inflammatory markers and cytokines and found that, as expected, they were elevated in hippocampi of KA-SE rats compared with controls (Figure 6A) (Jarvela et al., 2011; Pernot et al., 2011; Vezzani et al., 2011; Jiang et al., 2013). Unexpectedly, in hippocampi of rats receiving miR-124 agomirs, cytokine levels were not repressed but increased (Figure 6A) including IL-1β, TNF-α and IL-6. Robust increase in CD11b and modest upregulation of astrocytic GFAP were also observed after miR-124 infusion (Figure 6A). These striking effects were not detected in animals which received scrambled agomirs, suggesting specific and surprising pro-inflammatory actions of miR-124.
Figure 6. MiR-124-induced inflammatory cascades and microglia activation.
(A) Hippocampal cytokine mRNA levels, including IL-1β, TNF-α, IL-6 and IL-1 receptor and CD11b, were dramatically increased by miR-124 but not scr treatment in control brain. KA-SE increased inflammatory markers (including the astrocytic marker GFAP). (B,C) ICC and quantification of total and activated microglia. Insets denote the regions assessed in C. In control hippocampus, miR124 increased the activation of microglia, assessed as a transformation of IBA1-cells from thin and ramified into globular. (Ctrl-miR124; inset). No change in total IBA1-expressing cells was noted in areas CA1, CA3 and the DG hilus. Microglia activation was not found in control rats given a scr agomir (Ctrl+Scr, inset). Moderate microglia activation was observed in KA-SE rats. (D) In contrast to microglia, astrocytes did not seem to be affected by miR-124 agomir infusion. As expected, astrocytes were activated by KA-SE. n=5/group.
We then examined the cell types influenced by miR-124. We assessed the four groups described for evidence of inflammatory cell activation using ICC. We measured the numbers of cells expressing the microglial IBA1, and quantified the proportion of these cells that were activated (Figure 6B–C inset). While moderate microglia activation was observed in KA-SE rats (Figures 6B–C), in rats that received miR-124 agomirs, there was a dramatic and unexpected activation of microglia not only after KA-SE but also in control rats given miR-124 (Figures 6B–C). The same activation was not found in control rats given Scr miRNA. Astrocytes did not seem to be affected by miR-124 agomir infusion, though they were activated by KA-SE (Figure 6D).
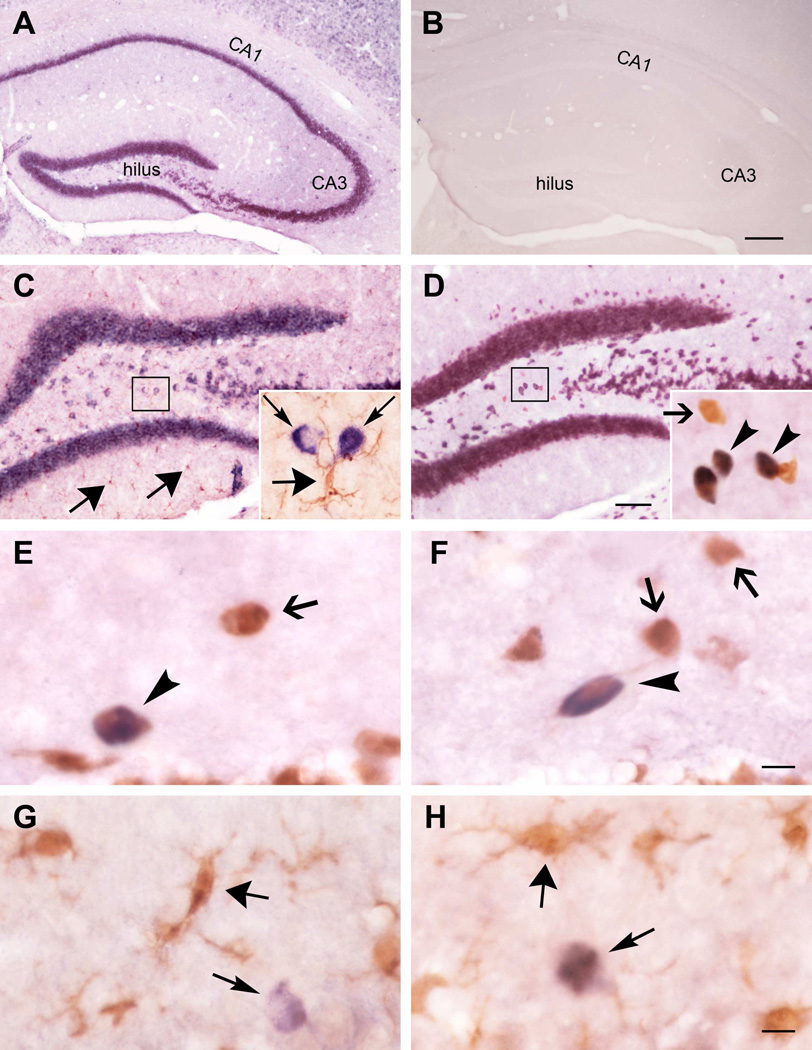
These surprising results suggested that miR-124 and/or miR-124 agomirs activated rather than repressed microglia, promoting inflammation. MiR-124 was described in microglia where it was considered to have suppressive effects (Ponomarev et al., 2011), and the findings described above might arise from endogenous or synthetic microglial miR-124. Alternatively, microglial activation might derive from augmented levels of endogenous and/or synthetic miR-124 in neurons. To resolve this question, we examined the cellular localization of both endogenous and synthetic miR-124 agomir in hippocampus (Figure 7). In naïve hippocampus, non-radioactive in situ hybridization (ISH) suggested a neuronal distribution of the miRNA (Figure 7A). A sense miR-124 probe did not detect any miRNA expression (Figure 7B). Dual ISH and ICC demonstrated an exclusive location of the miR-124 in cells expressing the neuronal marker NeuN, and lack of co-localization of miR-124 with cells with the typical features of microglia and expressing the microglial marker IBA1 (Figures 7C, 7D). We employed mouse hippocampus in these studies, to obviate potential species differences with published reports. Then, we examined the cellular distribution of endogenous and synthetic miR-124 in the same rats used in the current studies. Neuronal expression of miR-124 was found in controls administered synthetic miRNA (Figures 7E, 7G), as well as in rats infused with the miR-124 agomir following KA-SE (Figure 7F, 7H). In both cases, the neuronal nature of cells expressing miR-124 was identified using NeuN. We did not detect endogenous or synthetic miR-124 in cells co-expressing the microglial marker IBA1.
Figure 7. MiR-124 resides exclusively in neurons in mature mouse and rat hippocampus.
(A) Typical neuronal distribution of miR-124 in naïve adult mouse hippocampus visualized using ISH. (B). A sense miR-124 probe did not detect any miRNA expression. (C) Dual ISH and ICC for IBA1 demonstrated no cellular co-expression of miR-124 signal (thin arrows) and microglia (thick arrows). (D) ISH/ICC miR-124/NeuN demonstrated dually-labelled neurons (arrowheads) as well as neurons lacking miR-124 (arrow). (E–H) Distribution of miR-124 in the same rats used in the current studies. Neuronal (E) but no microglia (G) expression of miR-124 was found in controls administered synthetic miRNA, as well as in rats infused with the miR-124 agomir following KA-SE (F, H). Arrowheads indicate co-expression of miR-124 and NeuN. Thick arrows delineate delicate ramified microglia in controls (G), and globular/amoeboid microglia in KA-SE hippocampus. n=3/group.
Taken together, the data demonstrate that rather than reducing brain inflammation, miR-124 led to a profound activation of microglia with dramatic increase in the production of several inflammatory cytokines. These effects of miR-124 were robust enough to negate the mitigating actions of the miRNA against aberrant upregulation of NRSF. Thus, the combinatorial effect of administration of miR-124 after the epilepsy-inducing insult was a minimal change of the disease process. However, the mechanisms discovered by the series of experiments described here highlight important actions of miR-124 on neurons and on neuron-glia interactions (Figure S5).
Discussion
The studies described here uncover several aspects of the mechanisms by which the brain becomes epileptic following an insult. First, they provide direct molecular links between the cellular effects of the insult and the disruption of inflammatory and gene-expression regulatory programs. Second, they highlight the complex and opposing roles of miR-124 in this disease process, which likely apply to a number of brain disorders. Finally, the studies inform us about cell-specific actions of miR-124 on neuronal and microglia components of the normal brain.
It has become increasingly clear the acute epileptogenesis, the process by which an insult leads to the development of spontaneous seizures, is complex. Roles have been described for inflammation (Vezzani et al., 2011) as well to a variety of molecular and cellular changes (Brooks-Kayal et al., 2009; Cacheaux et al., 2009). However, interventions targeting one of these mechanisms have not been successful in preventing the disease (Maroso et al., 2011; Jiang et al., 2013; McClelland et al., 2011). Therefore, here we sought mechanisms that might regulate several epilepsy-mediating cellular pathways. We focused on inflammation as well as on the NRSF pathway. Inflammation accompanies and exacerbates epileptogenesis (Dube et al., 2010; Vezzani et al., 2011). For NRSF, the repressor’s levels and function are increase soon after epilepsy-provoking insults. In addition, a one-week blockade of NRSF actions reduced significantly the development of spontaneous seizures for up to two weeks, suggesting that NRSF contributes to the acute epileptogenic phase. Here we identified microRNAs and specifically miR-124 as a ‘master-regulator’ of both inflammation and NRSF, and established a direct molecular link between the epileptogenic trigger (SE) and miR-124.
Epilepsy promoting insults activate metabolic sensors that can modify gene expression
Epilepsy-promoting insults including SE, TB and febrile-SE provoke cellular metabolic stress because they are either metabolically demanding or constrain nutrient and oxygen supplies (Duffy et al., 1975; Fujikawa et al., 1988; Kahles and Brandes, 2012; Sada et al., 2015). Thus, common cellular pathways for these insults include metabolic derangements including altered NAD+/NADH ratio. Here we find the rapid activation of the NAD+ sensitive deacetylase SIRT1 following SE. The activation of this potent histone deacetylase (Imai et al., 2000) causes repression of the miR-124-1 gene expression via H4K16 deacetylation. It is likely that SE-induced SIRT1 activity causes repression of other genes and thus represents a direct mechanism by which epilepsy-promoting insults rapidly induce large-scale changes in the gene networks. Notably, SIRT1 inhibition is currently in clinical trials as a treatment for Huntington disease (Smith et al., 2014; Sussmuth et al., 2015), where repressed levels of miR-124 and NRSF dysregulation have been reported (Johnson and Buckley, 2009). These observations support a commonality in the pathogenesis of brain disorders including Huntington disease and epilepsy, based on metabolic dysregulation, SIRT1 activation and miR-124 repression.
MiR-124: a prevalent miRNA with unexpected actions on microglia
Aberrant miRNA expression and function influences numerous and diverse molecular and cellular cascades (Lallet al., 2006;, Makeyev et al., 2007). Here, we provide the evidence for the role of rapid depletion of miR-124 in the aberrant upregulation of NRSF, reported for a number of brain insults (Calderone et al., 2003; Zuccato et al., 2007; Doeppner et al., 2013). This is mediated by the miR-124 target transcription factor C/EBPα. Whereas this transcription factor has yet to be studied in in the context of epilepsy, other members of the C/CAAT enhancer protein are activated by SE (Engel et al., 2013; Brennan et al., 2015).
MiR-124, in addition to modulating neuronal phenotype via protean changes in gene expression, is implicated in microglial function. Specifically, reduced miR-124 levels in microglia caused their activation (Ponomarev et al., 2011). Thus, in view of the crucial roles for CNS glial cells and their production of inflammatory molecules in insult-provoked epilepsy, we considered a second, microglial-mediated anti-epileptogenic function of miR-124. Surprisingly, prevention of miR-124 depletion in hippocampus provoked robust microglial activation and striking augmentation of pro-inflammatory cytokine expression including IL-1β (Maroso et al., 2011).
Basis of the surprising effects of miR-124 on microglia
These unexpected findings were not a result of non-specific or toxic actions of the synthetic miR-124. We employed a chemically-protected agomir to provide stability in vivo, so it was conceivable that the modification might induce an inflammatory response. However, the same modification was present in the random/ Scr control-RNA which did not induce inflammation. The miR-124 dose used in this study did not lead to overt toxicity and no animals died or appeared ill for the 2 month duration of the study. Importantly, the effects of the miR-124 agomir were selective to microglia: NeuN-stained neurons appeared healthy, and astrocytes were unaffected. These data suggested that the synthetic miR-124 was not toxic but rather recapitulated the actions of endogenous microRNA.
Then how might endogenous and synthetic miR-124 promote the activation of microglia? The most straightforward mechanisms involve the intrinsic function of microRNAs in cells where they are produced. Therefore, we examined the cell types harboring endogenous miR-124 as well as the synthetic agomir. We detected miR-124 in NeuN-expressing neurons in adult mouse and rat hippocampi but failed to detect miR-124 in IBA1-expressing microglia. This was the case in both naïve hippocampus as well as following KA-SE. These findings strongly suggested that in rodent hippocampus, miR-124 was expressed exclusively in neurons. In addition, in hippocampi of rats given synthetic miR-124, we were unable to detect any miR-124 (endogenous or agomir) in microglia. It is conceivable that the DIG-labelled in situ hybridization probes did not bind the modified miR-124 agomirs due to steric hindrance, and miR-124 agomirs were taken up by microglia. This possibility does not influence the conclusion that endogenous miR-124 is located in, and therefore acts within, neurons. Notably, our results, using in situ hybridization combined with ICC are in close accord with a recently published study (Akerblom et al., 2012), utilizing a transgenic approach. This study employed a mouse expressing GFP modified to contain miR-124 binding sites in its mRNA. If miR-124 and GFP were expressed in the same cell, then miR-124 should inhibit GFP expression. Akerblom et al., failed to detect microglia lacking GFP expression whereas all neurons lacked GFP-expression, indicating that miR-124 was confined to neurons. Whereas both our study and Akerblom et al., employed rigorous single-cell resolution in situ approaches, microglial miR-124 was previously reported based on FACS sorting, using cellular profiles (Ponomarev et al., 2011). Potentially, technical challenges in obtaining pure microglial cell populations might account for the apparent miR-124 expression in microglia using FACS.
Its neuronal localization suggests that both endogenous and synthetic miR-124 activate microglia via neuronal-microglia signalling. A plausible mechanism involves repressing target mRNAs such as Co-REST (Baudet et al., 2011), important for the regulation of NF-kB (Saijo et al., 2009). Whereas both neuronal and glial mechanisms will require future study, the fundamental discoveries here are that miR-124 is neuronally expressed and exerts proinflammatory functions in epileptogenesis. Indeed, reduced miR-124 expression following SE may be an adaptive response, aiming to curb excessive inflammation in the brain and limit its potential adverse effects on neuronal survival (Maroso et al., 2011; Vezzani et al., 2011). In the context of the generation of neurological diseases such as epilepsy (and potentially others), the activation of microglia and inflammation by miR-124 counterbalanced disease-preventing effects through NRSF inhibition. Thus, miR-124 plays a pivotal role in epileptogenesis albeit via dual and opposing mechanisms.
Experimental Procedures
For more details regarding the materials and methods used in this study please see the supplemental experimental procedures.
All experiments were approved by the University of California-Irvine Institutional Animal Care and Use Committee and conformed to NIH guidelines.
Animals
Adult male Sprague-Dawley rats were housed under a 12h light-dark cycle, with ad libitum access to food and water.
Surgery, induction of status epilepticus and miR-124 agomir treatment in vivo
Rats (Ctrl+Scr n=5; Ctrl+miR124 agomir n=5, KA+Scr n=12; KA+miR124 agomir n=12) were implanted with cannula and bilateral hippocampal electrodes as reported (Dube et al., 2010), and as described in the supplemental methods.
Chromatin Immunoprecipitation
ChIP was performed as previously described (McClelland et al., 2011) and delineated in the supplemental methods.
Argonaute2 (Ago2) Immunoprecipitation (IP)
Ago2 IP was performed as described by Schratt et al., 2006. Briefly, hippocampi were solubilized in immunoprecipitation buffer (300mM NaCl, 5mM MgCl2, 0.1% NP-40, 50mM Tris-HCl) and centrifuged. 400µl supernatant was incubated overnight with 5µg of either Argonaute2 antibody (C34C6) or rabbit IgG. Protein A agarose beads were added at 4°C on a rotator, samples were then centrifuged and the supernatant was removed. Beads were washed and processed for miRNA analysis using reverse transcription and qPCR. Ago bound miRNAs were normalized to RNU6B.
In situ hybridization (ISH) and combined ISH and immunocytochemistry (ICC)
ISH and combined ISH/ICC were performed as reported (Noam et al., 2012), and detailed in the supplemental methods. To detect miR-124, Digoxigenin (DIG)-3’-conjugated miR124 sense and antisense oligonucleotide probes were generated. Specificity of hybridization was verified by substituting labelled sense for the antisense probe and by omitting the antisense probe or alkaline phosphatase-conjugated antibody. For combined ISH/ICC, free-floating sections were first processed for ISH then processed for ICC.
Statistical Analyses
All analyses were performed without knowledge of treatment group. Standard t-test was used to compare two groups; for analyses involving >two groups, one way or two way ANOVA were used. Statistical significance was set at p<0.05. Experiments were repeated at least twice.
Supplementary Material
Highlights.
Epilepsy-provoking insults (SE) repress miR-124 via SIRT1 activation
miR-124 repression controls both inflammation and NRSF
miR-124 replenishment does not prevent epileptogenesis
miR-124 activates microglia in control and SE hippocampus
Acknowledgments
Support: NIH R37 NS35439, T32 NS45540, R01 NS78279. GPB support: The George Hewitt Foundation for Biomedical research. Pam See for technical support, and Dr Black for the PTBP1 antibody.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contribution
Conceptualization: GPB, TZB; Methodology and Investigation: GPB, DD, YC, KPP, EM, AH, CD, YTM; Writing: Original draft: GPB, TZB; Review & Editing: GPB, TZB; Funding Acquisition: GPB, TZB; Resources: TZB; Supervision: TZB.
References
- Akerblom M, Sachdeva R, Barde I, Verp S, Gentner B, Trono D, Jakobsson J. MicroRNA-124 is a subventricular zone neuronal fate determinant. J. Neurosci. 2012;32:8879–8889. doi: 10.1523/JNEUROSCI.0558-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronica E, Fluiter K, Iyer A, Zurolo E, Vreijling J, van Vliet EA, Baayen JC, Gorter JA. Expression pattern of miR-146a, an inflammation-associated microRNA, in experimental and human temporal lobe epilepsy. Eur. J. Neurosci. 2010;31:1100–1107. doi: 10.1111/j.1460-9568.2010.07122.x. [DOI] [PubMed] [Google Scholar]
- Baldelli P, Meldolesi J. The Transcription Repressor REST in Adult Neurons: Physiology, Pathology, and Diseases. 2015 doi: 10.1523/ENEURO.0010-15.2015. eNeuro. 10.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudet ML, Zivraj KH, Abreu-Goodger C, Muldal A, Armisen J, Blenkiron C, Goldstein LD, Miska EA, Holt CE. miR-124 acts through CoREST to control onset of Sema3A sensitivity in navigating retinal growth cones. Nat. Neurosci. 2011;15:29–38. doi: 10.1038/nn.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu Rev Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- Brennan GP, Jimenez-Mateos EM, Sanz-Rodriguez A, Mooney CM, Tzivion G, Henshall DC, Engel T. Overexpression of 14-3-3zeta Increases Brain Levels of C/EBP Homologous Protein CHOP. J. Mol. Neurosci. 2015;56:255–262. doi: 10.1007/s12031-015-0510-0. [DOI] [PubMed] [Google Scholar]
- Brooks-Kayal AR, Raol YH, Russek SJ. Alteration of epileptogenesis genes. Neurotherapeutics. 2009;6:312–318. doi: 10.1016/j.nurt.2009.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacheaux LP, Ivens S, David Y, Lakhter AJ, Bar-Klein G, Shapira M, Heinemann U, Friedman A, Kaufer D. Transcriptome profiling reveals TGF-beta signaling involvement in epileptogenesis. J. Neurosci. 2009;29:8927–8935. doi: 10.1523/JNEUROSCI.0430-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderone A, Jover T, Noh KM, Tanaka H, Yokota H, Lin Y, Grooms SY, Regis R, Bennett MV, Zukin RS. Ischemic insults derepress the gene silencer REST in neurons destined to die. J. Neurosci. 2003;23:2112–2121. doi: 10.1523/JNEUROSCI.23-06-02112.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmody S, Brennan L. Effects of pentylenetetrazole-induced seizures on metabolomic profiles of rat brain. Neurochem. Int. 2010;56:340–344. doi: 10.1016/j.neuint.2009.11.004. [DOI] [PubMed] [Google Scholar]
- Choy M, Dube CM, Patterson K, Barnes SR, Maras P, Blood AB, Hasso AN, Obenaus A, Baram TZ. A novel, noninvasive, predictive epilepsy biomarker with clinical potential. J. Neurosci. 2014;34:8672–8684. doi: 10.1523/JNEUROSCI.4806-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaco C, Otto S, Han JJ, Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc. Natl. Acad. Sci. USA. 2006;103:2422–2427. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doeppner TR, Doehring M, Bretschneider E, Zechariah A, Kaltwasser B, Muller B, Koch JC, Bahr M, Hermann DM, Michel U. MicroRNA-124 protects against focal cerebral ischemia via mechanisms involving Usp14-dependent REST degradation. Acta Neuropathol. 2013;126:251–265. doi: 10.1007/s00401-013-1142-5. [DOI] [PubMed] [Google Scholar]
- Dube CM, Ravizza T, Hamamura M, Zha Q, Keebaugh A, Fok K, Andres AL, Nalcioglu O, Obenaus A, Vezzani A, Baram TZ. Epileptogenesis provoked by prolonged experimental febrile seizures: mechanisms and biomarkers. J. Neurosci. 2010;30:7484–7494. doi: 10.1523/JNEUROSCI.0551-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy TE, Howse DC, Plum F. Cerebral energy metabolism during experimental status epilepticus. J. Neurochem. 1975;24:925–934. doi: 10.1111/j.1471-4159.1975.tb03657.x. [DOI] [PubMed] [Google Scholar]
- Engel J, Jr, Thompson PM, Stern JM, Staba RJ, Bragin A, Mody I. Connectomics and epilepsy. Curr. Opin. Neurol. 2013a;26:186–194. doi: 10.1097/WCO.0b013e32835ee5b8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel T, Sanz-Rodgriguez A, Jimenez-Mateos EM, Concannon CG, Jimenez-Pacheco A, Moran C, Mesuret G, Petit E, Delanty N, Farrell MA, O’Brien DF, Prehn JH, Lucas JJ, Henshall DC. CHOP regulates the p53-MDM2 axis and is required for neuronal survival after seizures. Brain. 2013b;136:577–592. doi: 10.1093/brain/aws337. [DOI] [PubMed] [Google Scholar]
- Fujikawa DG, Vannucci RC, Dwyer BE, Wasterlain CG. Generalized seizures deplete brain energy reserves in normoxemic newborn monkeys. Brain Res. 1988;454:51–59. doi: 10.1016/0006-8993(88)90802-5. [DOI] [PubMed] [Google Scholar]
- Gao J, Wang WY, Mao YW, Graff J, Guan JS, Pan L, Mak G, Kim D, Su SC, Tsai LH. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature. 2010;466:1105–1109. doi: 10.1038/nature09271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Ure K, Ding P, Nashaat M, Yuan L, Ma J, Hammer RE, Hsieh J. The master negative regulator REST/NRSF controls adult neurogenesis by restraining the neurogenic program in quiescent stem cells. J. Neurosci. 2011;31:9772–9786. doi: 10.1523/JNEUROSCI.1604-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg EM, Coulter DA. Mechanisms of epileptogenesis: a convergence on neural circuit dysfunction. Nat. Rev. Neuro. 2013;14:337–349. doi: 10.1038/nrn3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorter JA, Iyer A, White I, Colzi A, van Vliet EA, Sisodiya S, Aronica E. Hippocampal subregion-specific microRNA expression during epileptogenesis in experimental temporal lobe epilepsy. Neurobiol. Dis. 2014;62:508–520. doi: 10.1016/j.nbd.2013.10.026. [DOI] [PubMed] [Google Scholar]
- Hellier JL, Dudek FE. Chemoconvulsant model of chronic spontaneous seizures. Curr. Protoc. Neurosci. 2005;9 doi: 10.1002/0471142301.ns0919s31. [DOI] [PubMed] [Google Scholar]
- Herskovits AZ, Guarente L. SIRT1 in neurodevelopment and brain senescence. Neuron. 2014;81:471–483. doi: 10.1016/j.neuron.2014.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Izaurralde E. Breakers and blockers—miRNAs at work. Science. 2015;349:380–382. doi: 10.1126/science.1260969. [DOI] [PubMed] [Google Scholar]
- Jarvela JT, Lopez-Picon FR, Plysjuk A, Ruohonen S, Holopainen IE. Temporal profiles of age-dependent changes in cytokine mRNA expression and glial cell activation after status epilepticus in postnatal rat hippocampus. J Neuroinflammation. 2011;8:1742–2094. doi: 10.1186/1742-2094-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Quan Y, Ganesh T, Pouliot WA, Dudek FE, Dingledine R. Inhibition of the prostaglandin receptor EP2 following status epilepticus reduces delayed mortality and brain inflammation. Proc. Natl. Acad. Sci. USA. 2013;110:3591–3596. doi: 10.1073/pnas.1218498110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Mateos EM, Engel T, Merino-Serrais P, McKiernan RC, Tanaka K, Mouri G, Sano T, O'Tuathaigh C, Waddington JL, Prenter S, Delanty N, Farrell MA, O’Brien DF, Conroy RM, Stallings RL, deFelipe J, Henshall DC. Silencing microRNA-134 produces neuroprotective and prolonged seizure-suppressive effects. Nat. Med. 2012;18:1087–1094. doi: 10.1038/nm.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R, Buckley NJ. Gene dysregulation in Huntington's disease: REST, microRNAs and beyond. Neuromolecular Med. 2009;11:183–199. doi: 10.1007/s12017-009-8063-4. [DOI] [PubMed] [Google Scholar]
- Jovicic A, Roshan R, Moisoi N, Pradervand S, Moser R, Pillai B, Luthi-Carter R. Comprehensive expression analyses of neural cell-type-specific miRNAs identify new determinants of the specification and maintenance of neuronal phenotypes. J. Neurosci. 2013;33:5127–5137. doi: 10.1523/JNEUROSCI.0600-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jupp B, Williams J, Binns D, Hicks RJ, Cardamone L, Jones N, Rees S, O'Brien TJ. Hypometabolism precedes limbic atrophy and spontaneous recurrent seizures in a rat model of TLE. Epilepsia. 2012;53:1233–1244. doi: 10.1111/j.1528-1167.2012.03525.x. [DOI] [PubMed] [Google Scholar]
- Kahles T, Brandes RP. NADPH oxidases as therapeutic targets in ischemic stroke. Cell Mol. Life Sci. 2012;69:2345–2363. doi: 10.1007/s00018-012-1011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan AA, van Erp S, Derijck AA, de Wit M, Hessel EV, O'Duibhir E, de Jager W, Van Rijen PC, Gosselaar PH, de Graan PN, Pasterkamp RJ. Genome-wide microRNA profiling of human temporal lobe epilepsy identifies modulators of the immune response. Cell Mol. Life Sci. 2012;69:3127–3145. doi: 10.1007/s00018-012-0992-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lall S, Grün D, Krek A, Chen K, Wang YL, Dewey CN, Sood P, Colombo T, Bray N, Macmenamin P, Kao HL, Gunsalus KC, Pachter L, Piano F, Rajewsky N. A genome-wide map of conserved microRNA targets in C. elegans. Curr. Biol. 2006;16:460–471. doi: 10.1016/j.cub.2006.01.050. [DOI] [PubMed] [Google Scholar]
- Li Z, Li B, Zhu X, Yin P, Liu J, Huang S, Sun R. Neuroprotective effects of anti-high-mobility group box 1 antibody in juvenile rat hippocampus after kainic acid-induced status epilepticus. Neuroreport. 2013;24:785–790. doi: 10.1097/WNR.0b013e328363fed3. [DOI] [PubMed] [Google Scholar]
- Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmevik J, Petri R, Klussendorf T, Knauff P, Akerblom M, Johansson J, Soneji S, Jakobsson J. Identification of the miRNA targetome in hippocampal neurons using RIP-seq. Sci. Rep. 2015;5 doi: 10.1038/srep12609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroso M, Balosso S, Ravizza T, Liu J, Bianchi ME, Vezzani A. Interleukin-1 type 1 receptor/Toll-like receptor signalling in epilepsy: the importance of IL-1beta and high-mobility group box 1. J. Int. Med. 2011;270:319–326. doi: 10.1111/j.1365-2796.2011.02431.x. [DOI] [PubMed] [Google Scholar]
- McClelland S, Brennan GP, Dube C, Rajpara S, Iyer S, Richichi C, Bernard C, Baram TZ. The transcription factor NRSF contributes to epileptogenesis by selective repression of a subset of target genes. eLife. 2014;3:e01267. doi: 10.7554/eLife.01267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland S, Flynn C, Dube C, Richichi C, Zha Q, Ghestem A, Esclapez M, Bernard C, Baram TZ. Neuron-restrictive silencer factor-mediated hyperpolarization-activated cyclic nucleotide gated channelopathy in experimental temporal lobe epilepsy. Ann Neurol. 2011;70:454–464. doi: 10.1002/ana.22479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neo WH, Yap K, Lee SH, Looi LS, Khandelia P, Neo SX, Makeyev EV, Su IH. MicroRNA miR-124 controls the choice between neuronal and astrocyte differentiation by fine-tuning Ezh2 expression. J. Biol. Chem. 2014;289:20788–20801. doi: 10.1074/jbc.M113.525493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Carroll D, Schaefer A. General Principals of miRNA Biogenesis and Regulation in the Brain. Neuropsychopharmacology. 2013;38:39–54. doi: 10.1038/npp.2012.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm K, Belluardo N, Metsis M, Timmusk T. Neuronal expression of zinc finger transcription factor REST/NRSF/XBR gene. J. Neurosci. 1998;18:1280–1296. doi: 10.1523/JNEUROSCI.18-04-01280.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G, Watson C, Pennisi M, Topple A. Bregma, lambda and the interaural midpoint in stereotaxic surgery with rats of different sex, strain and weight. J. Neurosci. Meth. 1985;13:139–143. doi: 10.1016/0165-0270(85)90026-3. [DOI] [PubMed] [Google Scholar]
- Pernot F, Heinrich C, Barbier L, Peinnequin A, Carpentier P, Dhote F, Baille V, Beaup C, Depaulis A, Dorandeu F. Inflammatory changes during epileptogenesis and spontaneous seizures in a mouse model of mesiotemporal lobe epilepsy. Epilepsia. 2011;52:2315–2325. doi: 10.1111/j.1528-1167.2011.03273.x. [DOI] [PubMed] [Google Scholar]
- Ponomarev ED, Veremeyko T, Barteneva N, Krichevsky AM, Weiner HL. MicroRNA-124 promotes microglia quiescence and suppresses EAE by deactivating macrophages via the C/EBP-alpha-PU.1 pathway. Nat. Med. 2011;17:64–70. doi: 10.1038/nm.2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn SR, O'Neill LA. A trio of microRNAs that control Toll-like receptor signalling. Int. Immunol. 2011;23:421–425. doi: 10.1093/intimm/dxr034. [DOI] [PubMed] [Google Scholar]
- Racine RJ. Modification of seizure activity by electrical stimulation. I. After-discharge threshold. Electroencephalography and clinical neurophysiology. 1972;32:269–279. doi: 10.1016/0013-4694(72)90176-9. [DOI] [PubMed] [Google Scholar]
- Risbud RM, Porter BE. Changes in microRNA expression in the whole hippocampus and hippocampal synaptoneurosome fraction following pilocarpine induced status epilepticus. PloS one. 2013;8:7. doi: 10.1371/journal.pone.0053464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossignol E, Kobow K, Simonato M, Loeb JA, Grisar T, Gilby KL, Vinet J, Kadam SD, Becker AJ. WONOEP appraisal: new genetic approaches to study epilepsy. Epilepsia. 2014;55:1170–1186. doi: 10.1111/epi.12692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth TL, Roth ED, Sweatt JD. Epigenetic regulation of genes in learning and memory. Essays Biochem. 2010;48:263–274. doi: 10.1042/bse0480263. [DOI] [PubMed] [Google Scholar]
- Routbort MJ, Bausch SB, McNamara JO. Seizures, cell death, and mossy fiber sprouting in kainic acid-treated organotypic hippocampal cultures. Neuroscience. 1999;94:755–765. doi: 10.1016/s0306-4522(99)00358-9. [DOI] [PubMed] [Google Scholar]
- Sada N, Lee S, Katsu T, Otsuki T, Inoue T. Targeting LDH enzymes with a stiripentol analog to treat epilepsy. Science. 2015;347:1362–1367. doi: 10.1126/science.aaa1299. [DOI] [PubMed] [Google Scholar]
- Saijo K, Winner B, Carson CT, Collier JG, Boyer L, Rosenfeld MG, Gage FH, Glass CK. A Nurr1/CoREST pathway in microglia and astrocytes protects dopaminergic neurons from inflammation-induced death. Cell. 2009;137:47–59. doi: 10.1016/j.cell.2009.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- Simonato M, Brooks-Kayal AR, Engel J, Jr, Galanopoulou AS, Jensen FE, Moshe SL, O'Brien TJ, Pitkanen A, Wilcox KS, French JA. The challenge and promise of anti-epileptic therapy development in animal models. Lancet Neurol. 2014;13:949–960. doi: 10.1016/S1474-4422(14)70076-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova L, Grafe A, Seiler A, Schumacher S, Nitsch R, Wulczyn FG. Regulation of miRNA expression during neural cell specification. Eur. J. Neurosci. 2005;21:1469–1477. doi: 10.1111/j.1460-9568.2005.03978.x. [DOI] [PubMed] [Google Scholar]
- Smith MR, Syed A, Lukacsovich T, Purcell J, Barbaro BA, Worthge SA, Wei SR, Pollio G, Magnoni L, Scali C, Massai L, Franceschini D, Camarri M, Gianfriddo M, Diodato E, Thomas R, Gokce O, Tabrizi SJ, Caricasole A, Landwehrmeyer B, Menalled L, Murphy C, Ramboz S, Luthi-Carter R, Westerberg G, Marsh JL. A potent and selective Sirtuin 1 inhibitor alleviates pathology in multiple animal and cell models of Huntington's disease. Hum. Mol. Gen. 2014;23:2995–3007. doi: 10.1093/hmg/ddu010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soreq H, Wolf Y. NeurimmiRs: microRNAs in the neuroimmune interface. Trends Mol. Med. 2011;17:548–555. doi: 10.1016/j.molmed.2011.06.009. [DOI] [PubMed] [Google Scholar]
- Su W, Aloi MS, Garden GA. MicroRNAs mediating CNS inflammation: Small regulators with powerful potential. Brain Behav. Immun. 2015;3:00237–00238. doi: 10.1016/j.bbi.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussmuth SD, Haider S, Landwehrmeyer GB, Farmer R, Frost C, Tripepi G, Andersen CA, Di Bacco M, Lamanna C, Diodato E, et al. An exploratory double-blind, randomized clinical trial with selisistat, a SirT1 inhibitor, in patients with Huntington's disease. Br. J. Clin. Pharmacol. 2015;79:465–476. doi: 10.1111/bcp.12512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan CL, Plotkin JL, Veno MT, von Schimmelmann M, Feinberg P, Mann S, Handler A, Kjems J, Surmeier DJ, O'Carroll D, Greengard P, Schaefer A. MicroRNA-128 governs neuronal excitability and motor behavior in mice. Science. 2013;342:1254–1258. doi: 10.1126/science.1244193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantama M, Martinez-Francois JR, Mongeon R, Yellen G. Imaging energy status in live cells with a fluorescent biosensor of the intracellular ATP-to-ADP ratio. Nat. Comm. 2013;4 doi: 10.1038/ncomms3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vezzani A, French J, Bartfai T, Baram TZ. The role of inflammation in epilepsy. Nat. Rev. Neurol. 2011;7:31–40. doi: 10.1038/nrneurol.2010.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvanathan J, Lee S, Lee B, Lee JW, Lee SK. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 2007;21:744–749. doi: 10.1101/gad.1519107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White AO, Wood MA. Does stress remove the HDAC brakes for the formation and persistence of long-term memory? Neurobiol. Learn. Mem. 2014;112:61–67. doi: 10.1016/j.nlm.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo AS, Staahl BT, Chen L, Crabtree GR. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature. 2009;460:642–646. doi: 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccato C, Belyaev N, Conforti P, Ooi L, Tartari M, Papadimou E, MacDonald M, Fossale E, Zeitlin S, Buckley N, Cattaneo E. Widespread disruption of repressor element-1 silencing transcription factor/neuron-restrictive silencer factor occupancy at its target genes in Huntington's disease. J. Neurosci. 2007;27:6972–6983. doi: 10.1523/JNEUROSCI.4278-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.