Abstract
Osteoarthritis (OA) is a common chronic degenerative joint disease, with complicated pathogenic factors and undefined pathogenesis. Various signaling pathways play important roles in OA pathogenesis, including genetic expression, matrix synthesis and degradation, cell proliferation, differentiation, apoptosis, and so on. MicroRNA (miRNA) is a class of non-coding RNA in Eukaryon, regulating genetic expression on the post-transcriptional level. A great number of miRNAs are involved in the development of OA, and are closely associated with different signaling pathways. This article reviews the roles of miRNAs and signaling pathways in OA, looking toward having a better understanding of its pathogenesis mechanisms and providing new therapeutic targets for its treatment.
Keywords: MicroRNA, Signaling pathway, Osteoarthritis, Pathogenesis
1. Introduction
Osteoarthritis (OA) is a common chronic degenerative joint disease, which is characterized by degeneration of the articular cartilage, synovial inflammation, and changes in the periarticular subchondral bone (Pelletier et al., 1983; 1992; Gu et al., 2014). Although there are a lot of predispositions contributing to OA, including joint injury, heredity, obesity, aging, mechanics, and inflammation (Goldring and Goldring, 2007; Miyaki and Asahara, 2012), its pathogenesis is complicated and not fully understood. The current clinical treatment for OA is unsatisfactory. Drugs such as non-steroidal antiinflammatory drugs (NSAIDs), selective cyclooxygenase 2 (COX-2) inhibitors, steroids, hyaluronic acid have limited effectiveness in alleviating its symptoms, and fail to reverse the loss of articular cartilage (Shamoon and Hochberg, 2000). Total joint arthroplasty (TJA) is an effective treatment for end stage OA. However, we have to accept its associated risks such as infection, peri-prosthetic fracture, deep vein thrombosis (DVT), and joint dislocation. So it is important to continue in-depth studies on OA pathogenesis, which may help to find new therapeutic targets and methods for treating this disease.
In the developing stages of OA, various signaling pathways play important roles, including nuclear factor-κB (NF-κB) pathways, bone morphogenetic protein (BMP) pathways, transforming growth factor β (TGF-β) pathways, SRY-related protein 9 (SOX9) pathways, insulin-like growth factor (IGF) pathways, and so on. These signaling pathways are involved in chondrocyte metabolisms, cell proliferation, differentiation, apoptosis, synthesis and degradation of the extracellular matrix (ECM), pro-inflammation, and anti-inflammation. If these signaling pathways were able to be interrupted, then the advancement of OA may be either hindered or even inverted.
MicroRNAs (miRNAs) are a class of naturally occurring, small non-coding RNA molecules about 20–22 nucleotides long, in Eukaryon, regulating the genetic expression on the level of post-transcription by interacting with the 3' untranslated regions (UTRs) (Cheng and Jin, 2012; Lu et al., 2014). As more and more miRNAs were discovered, their functions in biological processes are being given greater attention. A large number of current studies have reported that various miRNAs play different roles in OA pathogenesis. It is therefore necessary to have a systematic understanding of miRNA in OA, which will help to provide new therapeutic targets.
With the purpose of developing a foundation for providing a better understanding of OA pathogenesis and new therapeutic targets, we reviewed the miRNAs and signaling pathways which are involved in OA pathogenesis, clarifying their functional mechanisms and showing how they interact with each other.
2. miRNA
Currently, the biochemical progress and mechanisms of miRNA have been identified (Wang and Luo, 2015; Yuan et al., 2014): miRNA genes are transcribed to form primary (pri)-miRNAs, which are subjected to cleavage by a miRNA processor (a protein complex composed of Drosh associated with DGCR8) to form a shorter precursor miRNA called a pre-miRNA (Lee et al., 2003). A pre-miRNA is transported from the nucleus to the cytoplasm by exportin-5 (Lund et al., 2004), and then is sliced by another RNase III, called a Dicer, to form a mature miRNA (Bernstein et al., 2001). miRNA is then combined together with Agonature proteins (Agos), the core unit of the RNA-induced silencing complex (RISC) (Bartel, 2004; Farh et al., 2005; Calin and Croce, 2006). The miRNA-RISC complex binds the targeted mRNA and mediates the translational repression or degradation of the mRNA (Bartel, 2004).
Although the information about miRNA expression and function in the musculoskeletal system is not fully understood, its importance in cartilage and chondrocyte studies has been established. Loss of a Dicer in the chondrocytes results in a reduction in the number of proliferation chondrocytes by decreased proliferation or accelerated differentiation into post-mitotic hypertrophic chondrocytes (Cobb et al., 2005; Kanellopoulou et al., 2005). Limb or cartilage specific Dicer deficiency may lead to a severe phenotype with reduced limb size but normal patterning (Harfe et al., 2005; Kobayashi et al., 2008). As a Dicer plays a key role in miRNA synthesis, the importance of miRNA in biological processes of the musculoskeletal system is self-evident.
Iliopoulos et al. (2008) tested the expressions of 365 miRNAs in articular cartilage obtained from patients with OA and total knee arthroplasty (TKA), and from normal individuals without a history of joint disease, finding that 16 miRNAs were differentially expressed in osteoarthritic versus normal cartilage. Hundreds of miRNAs take part in gene expression, cell cycle regulation, ECM metabolism, inflammation process, and so on (Table 1). In the meantime, various inflammation cytokines play important roles in OA by regulating the different miRNAs. For example, interleukin (IL)-1β can increase the expression of miR-491-3p and decrease the expressions of 42 miRNAs, including miR-23-3p, miR-610, and miR-27b (Yasuda, 2011). The degradation of ECM is a feature of articular cartilage degeneration, while collagen II and proteoglycan are important compositions of ECM (Goldring and Goldring, 2010). Matrix metalloproteinases (MMPs) and a disintegrin and metalloproteinase with thrombospondin motifs (ADAMTS) are vital ECM-degrading enzymes, participating in the degradation of collagen II and proteoglycan (Tortorella et al., 2009; Li and Wu, 2010). MMP13 can degrade a large number of ECM components, including collagen. Because of its powerful degradation ability, MMP13 is a key factor in the biology studies of articular cartilage and OA pathogenesis (Fosang et al., 1996; Knauper et al., 1996; Billinghurst et al., 1997). ADAMTS4 and ADAMTS5 are major enzymes in the degradation of proteoglycan, and are important targets for treatment of OA (Wittwer et al., 2007; Gilbert et al., 2008). A great number of miRNAs are involved in the regulation of MMPs, ADAMTSs, and other related factors by using different pathways to control the progress of OA.
Table 1.
miRNAs and their targets
| miRNA | Target | Species | Effect | Function | Reference |
| 140 | MMP13 | Homo sapiens | ↓ | Matrix-degrading enzyme | Liang et al., 2012 |
| ADAMTs | H. sapiens | ↓ | Matrix-degrading enzyme | Miyaki et al., 2009 | |
| ADAMTs | Mus musculus | ↓ | Matrix-degrading enzyme | Miyaki et al., 2010 | |
| ACAN | H. sapines | ↑ | ECM component | Miyaki et al., 2009 | |
| CXCL12 | M. musculus | ↓ | Signaling | Jones et al., 2009 | |
| SMAD3 | M. musculus | ↓ | Signaling | Bazzoni et al., 2009 | |
| DNPEP | M. musculus | ↓ | Signaling | Ohgawara et al., 2009 | |
| HDAC4 | M. musculus | ↓ | Transcription | Tuddenham et al., 2006 | |
| PDGFRA | Danio rerio | ↓ | Skeletogenesis | Eberhart et al., 2008 | |
| IGFBP5 | H. sapiens | ↓ | Signaling | Tardif et al., 2009 | |
| SP1 | H. sapiens | ↓ | Cell cycle regulation | Martinez-Sanchez et al., 2012 | |
| RALA | H. sapiens | ↓ | Regulate SOX9 | Karlsen et al., 2014 | |
| 146 | IRAK1/TRFA6 | H. sapiens | ↓ | Signaling | Taganov et al., 2006 |
| TNFα (IL-1-induced) | H. sapiens | ↓ | Inflammation mediators | Jones et al., 2009 | |
| 9 | TNFα | H. sapiens | ↓ | Inflammation mediators | Bazzoni et al., 2009 |
| TIR | H. sapiens | ↓ | Signaling | Bazzoni et al., 2009 | |
| MMP13 (secretion) | H. sapiens | ↓ | Matrix-degrading enzyme | Jones et al., 2009 | |
| 18A | CCN2 | H. sapiens | ↓ | Signaling | Ohgawara et al., 2009 |
| 21 | MMP-1/2/3/9 | H. sapiens | ↓ | Matrix-degrading enzyme | Zhang et al., 2014 |
| GDF5 | H. sapiens | ↓ | Signaling | Zhang et al., 2014 | |
| 22 | BMP-7 | H. sapiens | ↓ | Signaling | Iliopoulos et al., 2008 |
| PPRA | H. sapiens | ↓ | Signaling | Iliopoulos et al., 2008 | |
| 27 | IGFBP5 | H. sapiens | ↓ | Signaling | Tardif et al., 2009 |
| MMP13 | H. sapiens | ↓ | Matrix-degrading enzyme | Akhtar et al., 2010 | |
| 34 | COL2A1 | H. sapiens | ↓ | ECM component | Abouheif et al., 2010 |
| iNOS2 | H. sapiens | ↓ | Signaling | Abouheif et al., 2010 | |
| 98 | TNFα | H. sapiens | ↓ | Inflammation mediators | Tardif et al., 2009 |
| 125B | ADAMTS4 | H. sapiens | ↓ | Matrix-degrading enzyme | Matsukawa et al., 2013 |
| 127-5P | MMP13 | H. sapiens | ↓ | Matrix-degrading enzyme | Park et al., 2013a |
| 145 | SOX9 | H. sapiens | ↓ | Transcription | Martinez-Sanchez et al., 2012 |
| SOX9 | M. musculus | ↓ | Transcription | Yang et al., 2011 | |
| 365 | HDAC4 | Gallus gallus | ↓ | Transcription | Guan et al., 2011 |
| 455-3P | ACVR2B | H. sapines | ↓ | Signaling | Swingler et al., 2012 |
| SMAD2 | |||||
| CHRD1 | |||||
| 558 | COX2 (IL-1β-induced) | H. sapines | ↓ | Inflammation mediators | Park et al., 2013b |
| 675 | COLA1 | H. sapines | ↑ | ECM component | Dudek et al., 2010 |
3. miRNAs and signaling pathways
3.1. NF-κB signaling pathway
NF-κB proteins constitute a family of ubiquitously expressed transcription factors involved inimmunity, stress responses, inflammatory diseases, cell proliferation, and cell death (Oeckinghaus and Ghosh, 2009). NF-κB can be stimulated by pro-inflammatory cytokines, chemokines, stress-related factors, and ECM degradation products (Yasuda, 2011; Rigoglou and Papavassiliou, 2013). The activation of an NF-κB signaling pathway can trigger the expressions for various amounts of immunomodulatory proteins, cytokines, chemokines, proteases, angiogenic factors, and proliferation- or apoptosis-related molecules (Niederberger and Geisslinger, 2008). There are two distinct pathways for activating the NF-κB signaling cascades. The first one, called canonical or classical pathway, is mediated by a tumor necrosis factor, Toll-like or T-cell receptor (TNF-R, TL-R, and TC-R, respectively), and induces the activation of the inhibitor of nuclear factor κB kinase α (IKKα)/IKKβ/IKKγ-NEMO (NF-κB essential modulator) complex, which results in degradation of the Iκ proteins. The other one, called non-canonical or alternative pathway, involves stimulation of the B-cell activating factor, CD40 or lymphotoxin β receptors (BAFF-R, CD40-R, LTβ-R), and relies on NF-κB-inducing kinase (NIK) that activates the IKKα kinase (Rigoglou and Papavassiliou, 2013).
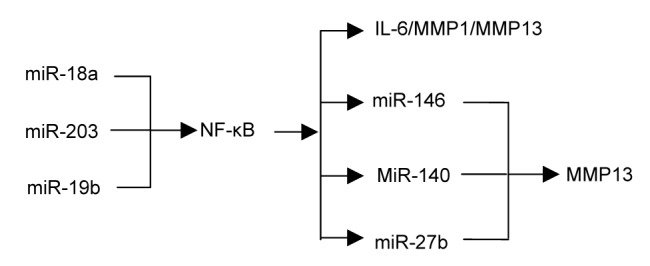
A great number of miRNAs were found to be involved in the NF-κB signaling pathway. Akhtar et al. (2010) reported that the NF-κB signaling pathway could suppress miR-27b, which regulated the expression of MMP13. miR-146a/b has the function of decreasing the expressions of TNF receptor-associated factor 6 (TRAF6) and IL-1 receptor-associated kinase 1 (IRK1) at a post-transcriptional level (Taganov et al., 2006). TRAF6 and IRK1 play important roles in triggering the activation of Iκ kinase and JNK, and then the downstream NF-κB and AP-1 transcription factors which result in the up-regulation of the immune-responsive gene (Taganov et al., 2006). In IL-1β-stimulated C28/I2 cells, expressions of miRNA-140 and MMP13 were elevated. However, their expressions decreased when the IL-1β-stimulated C28/I2 cells were treated with DHMEQ, an NF-κB inhibitor. So the expressions of miRNA-140 and MMP13 were shown to be NF-κB-dependent. miRNA-140 down-regulates the expression of MMP13, which will be up-regulated when transfecting C28/I2 with anti-miR-140 (Liang et al., 2012).
In rheumatoid arthritis synovial fibroblasts (RASFs), TNFα induced the expression of miR-17-92 in an NF-κB-dependent manner. miR-17-92-derived miR-18a contributes to cartilage destruction and chronic inflammation in the joints through a positive feedback loop in the NF-κB signaling (Trenkmann et al., 2013). Gantier et al. (2012) found that miR-19b controlled NF-κB signaling by suppressing its regulon of negative regulators (including A20/Tnfainp3, Rnf11, Fbx11/Kdm2a, and Zbtb16). What’s more, miR-203 up-regulated the expressions of MMP13 and IL-6 through the NF-κB signaling pathway (Stanczyk et al., 2011). As miRNA is relatively conservative, the roles that these miRNAs play in rheumatoid arthritis (RA) may also be played in OA (Fig. 1).
Fig. 1.
miRNAs and NF-κB signaling pathway
3.2. BMP signaling pathway and TGF-β signaling pathway
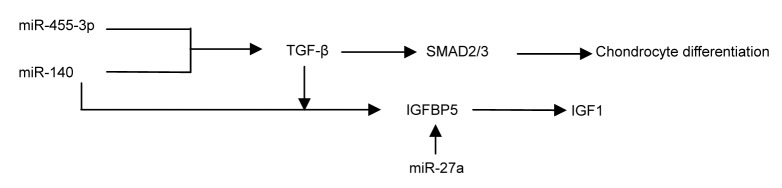
BMP/TGF-β-mediated signaling pathways involve the development of OA and are potent regulatory systems in chondrocytic cell types (Ballock et al., 1993; Serra et al., 1999; Grimsrud et al., 2001). In the growth plate, BMP signaling promotes chondrocyte terminal differentiation through SMAD1/5/8; conversely, TGF-β signaling blocks this process through SMAD2/3 (van der Kraan et al., 2012).
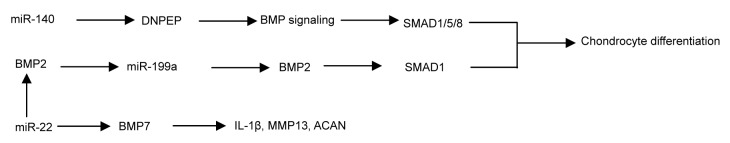
By comparing the profiles of RNA associated with Argonature 2 (Ago2) between the wild-type and miR-140−/− chondrocytes, it was found that aspartyl aminopeptidase (DNPEP) was identified as a miR-140 target gene. In miR-140−/− chondrocytes, the increased expression of DNPEP showed a mild antagonistic effect on BMP signaling at a position downstream of the SMAD activation and the lower-than-normal basal BMP signaling in miR-140−/− chondrocytes was reversed by applying a DNPEP knockdown. miR-140 was essential for normal endochondral bone development and the reduced BMP signaling caused by DNPEP up-regulation played a causal role in skeletal defects of miR-140−/− mice (Nakamura et al., 2011). The knockdown of miR-140 in limb bud micromass cultures resulted in the arrest of chondrogenic proliferation by regulating SP1, acting downstream from the BMP signaling (Yang et al., 2011). miR-140 has plenty of targets conserved between human and chicken and validated BMP2 as a direct target gene (Nicolas et al., 2011). miR-140 targets the CXC group of chemokine ligand 12 (CXCL12) and SMAD3 (Nicolas et al., 2008; Pais et al., 2010), both of which are involved in chondrocyte differentiation. Through repressing SMAD3, miR-140 suppresses the TGF-β pathway (Tuddenham et al., 2006, Araldi and Schipani, 2010). In conclusion, miR-140 promotes chondrocyte terminal differentiation by enhancing the BMP pathway and suppressing the TGF-β pathway.
During BMP2-induced chondrogenesis, miR-199a expression is decreased, indicating that it may function as a suppressor during the early stages in the chondrogenic program (Lin et al., 2009). Enforced miR-199a expression in Murine C3H10T1/2 stem cells or in the prechondrogenic cell line ATDC5 suppresses multiple markers of early chondrogenesis, including the type II collagen and cartilage oligomeric matrix protein (COMP), while anti-miR-199a has an opposite, stimulatory effect (Lin et al., 2009). SMAD1, a positive downstream mediator of BMP2 signaling, was shown to be a direct target of miR-199a (Lin et al., 2009). So the post-transcriptional repression of SMAD1 mediated by miR-199a will be prevented by BMP2-mediated repression of miR-199a.
Functional experiments on selected miR-gene pairs verified the presence of miR-22-regulated BMP7 and peroxisome proliferator-activated receptor α (PPARA) at the RNA and protein levels, respectively (Iliopoulos et al., 2008). The up-regulation of miR-22 or the down-regulation of BMP7 and PPARA can result in increases in the IL-1β and MMP13 protein levels (Iliopoulos et al., 2008). miR-455-3p appears to regulate TGF-β signaling by suppressing the SMAD2/3 pathway (Swingler et al., 2012). In other words, various miRNAs play important roles in chondrocyte differentiation, the regulation of inflammatory factors and ECM-degrading enzymes through the BMP/TGF-β signaling pathway (Figs. 2 and 3).
Fig. 2.
miRNAs and BMP signaling pathway
Fig. 3.
miRNAs and TGF-β signaling pathway
3.3. SOX9-related signaling pathway
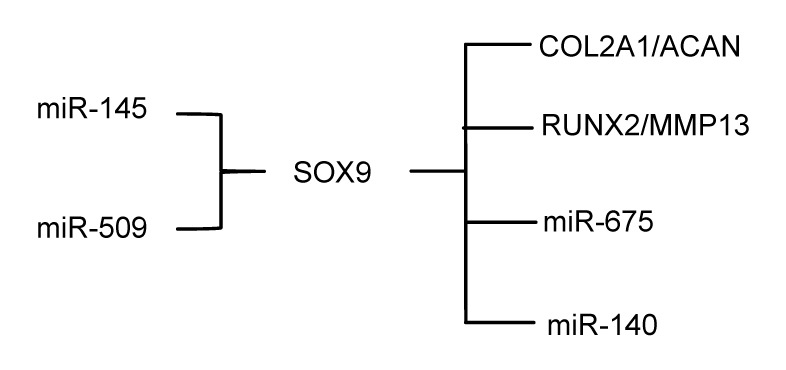
SOX9 is an essential transcription factor regulating the expression of many ECM genes, such as ACAN (Bi et al., 1999) and COL2A1 (Bell et al., 1997), and is essential for converting medenchymal stem cells (MSCs) into chondrocytes (Kronenberg, 2003). The CAMP-PKA-CREB pathway synergized with SOX9 at the nuclear and cytoplasmic levels to promote BMP2-induced osteochondrogenic differentiation (Zhao et al., 2009). TGF-β is shown to stimulate the expression of SOX9 mRNA (Roman-Blas et al., 2007; Kim et al., 2014). In addition, SMAD3 acts in cooperation with p300 and SOX9 to control gene expression during chondrogenesis (Furumatsu et al., 2009).
It was confirmed that miR-140 was directly induced by SOX9 and that the suppression of miR-140 is partially due to the inhibition of SOX9 by Wnt/catenin signaling in the micro mass cultures and the ATDC5 cell line (Yang et al., 2011). It was reported that RALA, a small GTPase not previously known to be involved in chondrogenesis, acted as a new direct target of miR-140-5p and showed that a knockdown of RALA during early chondrogenesis led to a significant up-regulation of SOX9 protein expression (Kartsen et al., 2014). SOX9 itself is directly targeted by miR-145 during the early stages of chondrogenic differentiation (Yang et al., 2011; Martinez-Sanchez et al., 2012). Through regulating SOX9, increasing miR-145 leads to down-regulation of the critical cartilage ECM genes (COL2A1 and ACAN) and tissue-specific miRNAs (miR-675 and miR-140), and up-regulation of RUNX2 and MMP13 (Martinez-Sanchez et al., 2012). OA cartilage revealed several miRNA-gene target pairs potentially involved in cartilage homeostasis and structure, including miR-509-SOX9 (Iliopoulos et al., 2008).
Multiple signaling pathways and miRNAs result in various bio-effects in articular cartilage through SOX9, which is a key factor in the progress of OA. So controlling the expression of SOX9 may help us to intervene these signaling pathways and miRNAs, providing new treatments for OA (Fig. 4).
Fig. 4.
miRNAs and SOX9 signaling pathway
3.4. IGF signaling pathway
IGF-1, a main anabolic mediator in articular cartilage, enhances cell proliferation and the synthesis of ECM proteins, and inhibits apoptosis through PI3K and ERK (Ashraf et al., 2015). Insulin-like growth factor binding proteins (IGFBPs), whose expression is very low in human OA chondrocytes (Tardif et al., 2009), are known to play an important role for IGF1 in joint treatment (Jones et al., 1993). Increasing the IGFBP5 concentration results in the increase of IGF-1, which is associated with a reduction of cartilage destruction in a dog OA model (Clemmons et al., 2002).
Transfection with pre-miR-140 significantly decreased IGFBP5 expression, while transfection with anti-miR-140 had the opposite effect, suggesting that IGFBP5 is a direct target of miR-140 (Ashraf et al., 2015). When human OA chondrocytes were treated with TGF-β, the expression of IGFBP5 was increased and the expression of miR-140 was decreased, indicating that both of them are regulated by TGF-β (Ashraf et al., 2015). It was found that miR-27a down-regulated the levels of MMP13 and IGFBP5 indirectly, and both of them were up-regulated when transfected with anti-miR-27a (Tardif et al., 2009) (Fig. 3).
4. Conclusions
In summary, through different signaling pathways, various miRNAs and their targets play important roles in the OA process, including genetic expression, matrix synthesis and degradation, cell proliferation, differentiation, apoptosis, and so on. Although a larger amount of work has already been done, OA pathogenesis requires future studies to have a better understanding of how it works and the implications for ongoing treatment. The knowledge about the functions of signaling pathways and miRNAs in OA will provide the potential means for diagnosis and treatment of OA.
Footnotes
Project supported by the Ministry of Health of the People’s Republic of China (No. 201302007)
Compliance with ethics guidelines: Bin XU, Yao-yao LI, Jun MA, and Fu-xing PEI declare that they have no conflicts of interest.
The article does not contain any studies with human or animal subjects performed by any of the authors.
References
- 1.Abouheif MM, Nakasa T, Shibuya H, et al. Silencing microRNA-34a inhibits chondrocyte apoptosis in a rat osteoarthritis model in vitro. Rheumatology (Oxford) 2010;49(11):2054–2060. doi: 10.1093/rheumatology/keq247. [DOI] [PubMed] [Google Scholar]
- 2.Akhtar N, Rasheed Z, Ramamurthy S, et al. MicroRNA-27b regulates the expression of matrix metalloproteinase 13 in human osteoarthritis chondrocytes. Arthritis Rheum. 2010;62(5):1361–1371. doi: 10.1002/art.27329. (Available from: http://dx.doi.org/10.1002/art.27329) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Araldi E, Schipani E. MicroRNA-140 and the silencing of osteoarthritis. Genes Dev. 2010;24(11):1075–1080. doi: 10.1101/gad.1939310. (Available from: http://dx.doi.org/10.1101/gad.1939310) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashraf S, Cha BH, Kim JS, et al. Regulation of senescence associated signaling mechanisms in chondrocytes for cartilage tissue regeneration. Osteoarthr Cartilage. 2015;24(2):196–205. doi: 10.1016/j.joca.2015.07.008. (Available from: http://dx.doi.org/10.1016/j.joca.2015.07.008) [DOI] [PubMed] [Google Scholar]
- 5.Ballock RT, Heydemann A, Wakefield LM, et al. TGF-β 1 prevents hypertrophy of epiphyseal chondrocytes: regulation of gene expression for cartilage matrix proteins and metalloproteases. Dev Biol. 1993;158(2):414–429. doi: 10.1006/dbio.1993.1200. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. (Available from: http://dx.doi.org/10.1016/S0092-8674(04)00045-5) [DOI] [PubMed] [Google Scholar]
- 7.Bazzoni F, Rossato M, Fabbri M, et al. Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to proinflammatory signals. PNAS. 2009;106(13):5282–5287. doi: 10.1073/pnas.0810909106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bell DM, Leung KK, Wheatley SC, et al. SOX9 directly regulates the type-II collagen gene. Nat Genet. 1997;16(2):174–178. doi: 10.1038/ng0697-174. (Available from: http://dx.doi.org/10.1038/ng0697-174) [DOI] [PubMed] [Google Scholar]
- 9.Bernstein E, Caudy AA, Hammond SM, et al. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409(6818):363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 10.Bi WM, Deng JM, Zhang ZP, et al. Sox9 is required for cartilage formation. Nat Genet. 1999;22(1):85–89. doi: 10.1038/8792. [DOI] [PubMed] [Google Scholar]
- 11.Billinghurst RC, Dahlberg L, Ionescu M, et al. Enhanced cleavage of type II collagen by collagenases in osteoarthritic articular cartilage. J Clin Invest. 1997;99(7):1534–1545. doi: 10.1172/JCI119316. (Available from: http://dx.doi.org/10.1172/JCI119316) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 13.Cheng G, Jin Y. MicroRNAs: potentially important regulators for schistosome development and therapeutic targets against schistosomiasis. Parasitology. 2012;139(5):669–679. doi: 10.1017/S0031182011001855. (Available from: http://dx.doi.org/10.1017/S0031182011001855) [DOI] [PubMed] [Google Scholar]
- 14.Clemmons DR, Busby WJ, Garmong A, et al. Inhibition of insulin-like growth factor binding protein 5 proteolysis in articular cartilage and joint fluid results in enhanced concentrations of insulin-like growth factor 1 and is associated with improved osteoarthritis. Arthritis Rheum. 2002;46(3):694–703. doi: 10.1002/art.10222. (Available from: http://dx.doi.org/10.1002/art.10222) [DOI] [PubMed] [Google Scholar]
- 15.Cobb BS, Nesterova TB, Thompson E, et al. T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer. J Exp Med. 2005;201(9):1367–1373. doi: 10.1084/jem.20050572. (Available from: http://dx.doi.org/10.1084/jem.20050572) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dudek KA, Lafont JE, Martinez-Sanchez A, et al. Type II collagen expression is regulated by tissue-specific miR-675 in human articular chondrocytes. J Biol Chem. 2010;285(32):24381–24387. doi: 10.1074/jbc.M110.111328. (Available from: http://dx.doi.org/10.1074/jbc.M110.111328) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eberhart JK, He X, Swartz ME, et al. MicroRNA Mirn140 modulates Pdgf signaling during palatogenesis. Nat Genet. 2008;40(3):290–298. doi: 10.1038/ng.82. (Available from: http://dx.doi.org/10.1038/ng.82) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farh KK, Grimson A, Jan C, et al. The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science. 2005;310(5755):1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 19.Fosang AJ, Last K, Knauper V, et al. Degradation of cartilage aggrecan by collagenase-3 (MMP-13) FEBS Lett. 1996;380(1-2):17–20. doi: 10.1016/0014-5793(95)01539-6. (Available from: http://dx.doi.org/10.1016/0014-5793(95)01539-6) [DOI] [PubMed] [Google Scholar]
- 20.Furumatsu T, Ozaki T, Asahara H. Smad3 activates the Sox9-dependent transcription on chromatin. Int J Biochem Cell Biol. 2009;41(5):1198–1204. doi: 10.1016/j.biocel.2008.10.032. (Available from: http://dx.doi.org/10.1016/j.biocel.2008.10.032) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gantier MP, Stunden HJ, McCoy CE, et al. A miR-19 regulon that controls NF-κB signaling. Nucleic Acids Res. 2012;40(16):8048–8058. doi: 10.1093/nar/gks521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gilbert AM, Bursavich MG, Lombardi S, et al. N-((8-Hydroxy-5-substituted-quinolin-7-yl)(phenyl)methyl)-2-phenyloxy/amino-acetamide inhibitors of ADAMTS-5 (Aggrecanase-2) Bioorg Med Chem Lett. 2008;18(24):6454–6457. doi: 10.1016/j.bmcl.2008.10.065. (Available from: http://dx.doi.org/10.1016/j.bmcl.2008.10.065) [DOI] [PubMed] [Google Scholar]
- 23.Goldring MB, Goldring SR. Osteoarthritis. J Cell Physiol. 2007;213(3):626–634. doi: 10.1002/jcp.21258. (Available from: http://dx.doi.org/10.1002/jcp.21258) [DOI] [PubMed] [Google Scholar]
- 24.Goldring MB, Goldring SR. Articular cartilage and subchondral bone in the pathogenesis of osteoarthritis. Ann N Y Acad Sci. 2010;1192(1):230–237. doi: 10.1111/j.1749-6632.2009.05240.x. (Available from: http://dx.doi.org/10.1111/j.1749-6632.2009.05240.x) [DOI] [PubMed] [Google Scholar]
- 25.Grimsrud CD, Romano PR, D'Souza M, et al. BMP signaling stimulates chondrocyte maturation and the expression of Indian hedgehog. J Orthop Res. 2001;19(1):18–25. doi: 10.1016/S0736-0266(00)00017-6. (Available from: http://dx.doi.org/10.1016/S0736-0266(00)00017-6) [DOI] [PubMed] [Google Scholar]
- 26.Gu YJ, Ge P, Mu Y, et al. Clinical and laboratory characteristics of patients having amyloidogenic transhyretin deposition in osteoarthritic knee joints. J Zhejiang Univ-Sci B (Biomed & Biotechnol) 2014;15(1):92–99. doi: 10.1631/jzus.B1300046. (Available from: http://dx.doi.org/10.1631/jzus.B1300046) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guan YJ, Yang X, Wei L, et al. MiR-365: a mechanosensitive microRNA stimulates chondrocyte differentiation through targeting histone deacetylase 4. FASEB J. 2011;25(12):4457–4466. doi: 10.1096/fj.11-185132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harfe BD, McManus MT, Mansfield JH, et al. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. PNAS. 2005;102(31):10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iliopoulos D, Malizos KN, Oikonomou P, et al. Integrative microRNA and proteomic approaches identify novel osteoarthritis genes and their collaborative metabolic and inflammatory networks. PLoS ONE. 2008;3(11):e3740. doi: 10.1371/journal.pone.0003740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones JI, Gockerman A, Busby WJ, et al. Extracellular matrix contains insulin-like growth factor binding protein-5: potentiation of the effects of IGF-I. J Cell Biol. 1993;121(3):679–687. doi: 10.1083/jcb.121.3.679. (Available from: http://dx.doi.org/10.1083/jcb.121.3.679) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones SW, Watkins G, Le Good N, et al. The identification of differentially expressed microRNA in osteoarthritic tissue that modulate the production of TNF-α and MMP13. Osteoarthr Cartilage. 2009;17(4):464–472. doi: 10.1016/j.joca.2008.09.012. (Available from: http://dx.doi.org/10.1016/j.joca.2008.09.012) [DOI] [PubMed] [Google Scholar]
- 32.Kanellopoulou C, Muljo SA, Kung AL, et al. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19(4):489–501. doi: 10.1101/gad.1248505. (Available from: http://dx.doi.org/10.1101/gad.1248505) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karlsen TA, Jakobsen RB, Mikkelsen TS, et al. microRNA-140 targets RALA and regulates chondrogenic differentiation of human mesenchymal stem cells by translational enhancement of SOX9 and ACAN . Stem Cells Dev. 2014;23(3):290–304. doi: 10.1089/scd.2013.0209. (Available from: http://dx.doi.org/10.1089/scd.2013.0209) [DOI] [PubMed] [Google Scholar]
- 34.Kim YI, Ryu JS, Yeo JE, et al. Overexpression of TGF-β1 enhances chondrogenic differentiation and proliferation of human synovium-derived stem cells. Biochem Biophys Res Commun. 2014;450(4):1593–1599. doi: 10.1016/j.bbrc.2014.07.045. (Available from: http://dx.doi.org/10.1016/j.bbrc.2014.07.045) [DOI] [PubMed] [Google Scholar]
- 35.Knauper V, Lopez-Otin C, Smith B, et al. Biochemical characterization of human collagenase-3. J Biol Chem. 1996;271(3):1544–1550. doi: 10.1074/jbc.271.3.1544. (Available from: http://dx.doi.org/10.1074/jbc.271.3.1544) [DOI] [PubMed] [Google Scholar]
- 36.Kobayashi T, Lu J, Cobb BS, et al. Dicer-dependent pathways regulate chondrocyte proliferation and differentiation. PNAS. 2008;105(6):1949–1954. doi: 10.1073/pnas.0707900105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kronenberg HM. Developmental regulation of the growth plate. Nature. 2003;423(6937):332–336. doi: 10.1038/nature01657. [DOI] [PubMed] [Google Scholar]
- 38.Lee Y, Ahn C, Han J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425(6956):415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 39.Li X, Wu JF. Recent developments in patent anti-cancer agents targeting the matrix metalloproteinases (MMPs) Recent Pat Anticancer Drug Discov. 2010;5(2):109–141. doi: 10.2174/157489210790936234. (Available from: http://dx.doi.org/10.2174/157489110791233540) [DOI] [PubMed] [Google Scholar]
- 40.Liang ZJ, Zhuang H, Wang GX, et al. MiRNA-140 is a negative feedback regulator of MMP-13 in IL-1β-stimulated human articular chondrocyte C28/I2 cells. Inflamm Res. 2012;61(5):503–509. doi: 10.1007/s00011-012-0438-6. [DOI] [PubMed] [Google Scholar]
- 41.Lin EA, Kong L, Bai XH, et al. miR-199a*, a bone morphogenic protein 2-responsive microRNA, regulates chondrogenesis via direct targeting to Smad1. J Biol Chem. 2009;284(17):11326–11335. doi: 10.1074/jbc.M807709200. (Available from: http://dx.doi.org/10.1074/jbc.M807709200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu YL, Jing W, Feng LS, et al. Effects of hypoxic exercise training on microRNA expression and lipid metabolism in obese rat livers. J Zhejiang Univ-Sci B (Biomed & Biotechnol) 2014;15(9):820–829. doi: 10.1631/jzus.B1400052. (Available from: http://dx.doi.org/10.1631/jzus.B1400052) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lund E, Guttinger S, Calado A, et al. Nuclear export of microRNA precursors. Science. 2004;303(5654):95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 44.Martinez-Sanchez A, Dudek KA, Murphy CL. Regulation of human chondrocyte function through direct inhibition of cartilage master regulator SOX9 by microRNA-145 (miRNA-145) J Biol Chem. 2012;287(2):916–924. doi: 10.1074/jbc.M111.302430. (Available from: http://dx.doi.org/10.1074/jbc.M111.302430) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matsukawa T, Sakai T, Yonezawa T, et al. MicroRNA-125b regulates the expression of aggrecanase-1 (ADAMTS-4) in human osteoarthritic chondrocytes. Arthritis Res Ther. 2013;15(1):R28. doi: 10.1186/ar4164. (Available from: http://dx.doi.org/10.1186/ar4164) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miyaki S, Asahara H. Macro view of microRNA function in osteoarthritis. Nat Rev Rheumatol. 2012;8(9):543–552. doi: 10.1038/nrrheum.2012.128. (Available from: http://dx.doi.org/10.1038/nrrheum.2012.128) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miyaki S, Nakasa T, Otsuki S, et al. MicroRNA-140 is expressed in differentiated human articular chondrocytes and modulates interleukin-1 responses. Arthritis Rheum. 2009;60(9):2723–2730. doi: 10.1002/art.24745. (Available from: http://dx.doi.org/10.1002/art.24745) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miyaki S, Sato T, Inoue A, et al. MicroRNA-140 plays dual roles in both cartilage development and homeostasis. Genes Dev. 2010;24(11):1173–1185. doi: 10.1101/gad.1915510. (Available from: http://dx.doi.org/10.1101/gad.1915510) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakamura Y, Inloes JB, Katagiri T, et al. Chondrocyte-specific microRNA-140 regulates endochondral bone development and targets Dnpep to modulate bone morphogenetic protein signaling. Mol Cell Biol. 2011;31(14):3019–3028. doi: 10.1128/MCB.05178-11. (Available from: http://dx.doi.org/10.1128/MCB.05178-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nicolas FE, Pais H, Schwach F, et al. Experimental identification of microRNA-140 targets by silencing and overexpressing miR-140. RNA. 2008;14(12):2513–2520. doi: 10.1261/rna.1221108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nicolas FE, Pais H, Schwach F, et al. mRNA expression profiling reveals conserved and non-conserved miR-140 targets. RNA Biol. 2011;8(4):607–615. doi: 10.4161/rna.8.4.15390. (Available from: http://dx.doi.org/10.4161/rna.8.4.15390) [DOI] [PubMed] [Google Scholar]
- 52.Niederberger E, Geisslinger G. The IKK-NF-κB pathway: a source for novel molecular drug targets in pain therapy? . FASEB J. 2008;22(10):3432–3442. doi: 10.1096/fj.08-109355. (Available from: http://dx.doi.org/10.1096/fj.08-109355) [DOI] [PubMed] [Google Scholar]
- 53.Oeckinghaus A, Ghosh S. The NF-κB family of transcription factors and its regulation. Cold Spring Harb Perspect Biol. 2009;1(4):a000034. doi: 10.1101/cshperspect.a000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ohgawara T, Kubota S, Kawaki H, et al. Regulation of chondrocytic phenotype by micro RNA 18a: involvement of Ccn2/Ctgf as a major target gene. FEBS Lett. 2009;583(6):1006–1010. doi: 10.1016/j.febslet.2009.02.025. [DOI] [PubMed] [Google Scholar]
- 55.Pais H, Nicolas FE, Soond SM, et al. Analyzing mRNA expression identifies Smad3 as a microRNA-140 target regulated only at protein level. RNA. 2010;16(3):489–494. doi: 10.1261/rna.1701210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Park SJ, Cheon EJ, Lee MH, et al. MicroRNA-127-5p regulates matrix metalloproteinase 13 expression and interleukin-1β-induced catabolic effects in human chondrocytes. Arthritis Rheum. 2013;65(12):3141–3152. doi: 10.1002/art.38188. (Available from: http://dx.doi.org/10.1002/art.38188) [DOI] [PubMed] [Google Scholar]
- 57.Park SJ, Cheon EJ, Kim HA. MicroRNA-558 regulates the expression of cyclooxygenase-2 and IL-1β-induced catabolic effects in human articular chondrocytes. Osteoarthr Cartilage. 2013;21(7):981–989. doi: 10.1016/j.joca.2013.04.012. (Available from: http://dx.doi.org/10.1016/j.joca.2013.04.012) [DOI] [PubMed] [Google Scholar]
- 58.Pelletier JP, Martel-Pelletier J, Altman RD, et al. Collagenolytic activity and collagen matrix breakdown of the articular cartilage in the Pond-Nuki dog model of osteoarthritis. Arthritis Rheum. 1983;26(7):866–874. doi: 10.1002/art.1780260708. (Available from: http://dx.doi.org/10.1002/art.1780260708) [DOI] [PubMed] [Google Scholar]
- 59.Pelletier JP, Martel-Pelletier J, Mehraban F, et al. Immunological analysis of proteoglycan structural changes in the early stage of experimental osteoarthritic canine cartilage lesions. J Orthop Res. 1992;10(4):511–523. doi: 10.1002/jor.1100100406. (Available from: http://dx.doi.org/10.1002/jor.1100100406) [DOI] [PubMed] [Google Scholar]
- 60.Rigoglou S, Papavassiliou AG. The NF-κB signalling pathway in osteoarthritis. Int J Biochem Cell Biol. 2013;45(11):2580–2584. doi: 10.1016/j.biocel.2013.08.018. (Available from: http://dx.doi.org/10.1016/j.biocel.2013.08.018) [DOI] [PubMed] [Google Scholar]
- 61.Roman-Blas JA, Stokes DG, Jimenez SA. Modulation of TGF-β signaling by proinflammatory cytokines in articular chondrocytes. Osteoarthr Cartilage. 2007;15(12):1367–1377. doi: 10.1016/j.joca.2007.04.011. (Available from: http://dx.doi.org/10.1016/j.joca.2007.04.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Serra R, Karaplis A, Sohn P. Parathyroid hormone-related peptide (PTHrP)-dependent and -independent effects of transforming growth factor β (TGF-β) on endochondral bone formation. J Cell Biol. 1999;145(4):783–794. doi: 10.1083/jcb.145.4.783. (Available from: http://dx.doi.org/10.1083/jcb.145.4.783) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shamoon M, Hochberg MC. Treatment of osteoarthritis with acetaminophen: efficacy, safety, and comparison with nonsteroidal anti-inflammatory drugs. Curr Rheumatol Rep. 2000;2(6):454–458. doi: 10.1007/s11926-000-0020-z. (Available from: http://dx.doi.org/10.1007/s11926-000-0020-z) [DOI] [PubMed] [Google Scholar]
- 64.Stanczyk J, Ospelt C, Karouzakis E, et al. Altered expression of microRNA-203 in rheumatoid arthritis synovial fibroblasts and its role in fibroblast activation. Arthritis Rheum. 2011;63(2):373–381. doi: 10.1002/art.30115. (Available from: http://dx.doi.org/10.1002/art.30115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Swingler TE, Wheeler G, Carmont V, et al. The expression and function of microRNAs in chondrogenesis and osteoarthritis. Arthritis Rheum. 2012;64(6):1909–1919. doi: 10.1002/art.34314. (Available from: http://dx.doi.org/10.1002/art.34314) [DOI] [PubMed] [Google Scholar]
- 66.Taganov KD, Boldin MP, Chang KJ, et al. NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. PNAS. 2006;103(33):12481–12486. doi: 10.1073/pnas.0605298103. (Available from: http://dx.doi.org/10.1073/pnas.0605298103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tardif G, Hum D, Pelletier JP, et al. Regulation of the IGFBP-5 and MMP-13 genes by the microRNAs miR-140 and miR-27a in human osteoarthritic chondrocytes. BMC Musculoskelet Disord. 2009;10(1):148. doi: 10.1186/1471-2474-10-148. (Available from: http://dx.doi.org/10.1186/1471-2474-10-148) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tortorella MD, Tomasselli AG, Mathis KJ, et al. Structural and inhibition analysis reveals the mechanism of selectivity of a series of aggrecanase inhibitors. J Biol Chem. 2009;284(36):24185–24191. doi: 10.1074/jbc.M109.029116. (Available from: http://dx.doi.org/10.1074/jbc.M109.029116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Trenkmann M, Brock M, Gay RE, et al. Tumor necrosis factor α-induced microRNA-18a activates rheumatoid arthritis synovial fibroblasts through a feedback loop in NF-κB signaling. Arthritis Rheum. 2013;65(4):916–927. doi: 10.1002/art.37834. (Available from: http://dx.doi.org/10.1002/art.37834) [DOI] [PubMed] [Google Scholar]
- 70.Tuddenham L, Wheeler G, Ntounia-Fousara S, et al. The cartilage specific microRNA-140 targets histone deacetylase 4 in mouse cells. FEBS Lett. 2006;580(17):4214–4217. doi: 10.1016/j.febslet.2006.06.080. [DOI] [PubMed] [Google Scholar]
- 71.van der Kraan PM, Goumans MJ, Blaney DE, et al. Age-dependent alteration of TGF-β signalling in osteoarthritis. Cell Tissue Res. 2012;347(1):257–265. doi: 10.1007/s00441-011-1194-6. (Available from: http://dx.doi.org/10.1007/s00441-011-1194-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang W, Luo YP. MicroRNAs in breast cancer: oncogene and tumor suppressors with clinical potential. J Zhejiang Univ-Sci B (Biomed & Biotechnol) 2015;16(1):18–31. doi: 10.1631/jzus.B1400184. (Available from: http://dx.doi.org/10.1631/jzus.B1400184) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wittwer AJ, Hills RL, Keith RH, et al. Substrate-dependent inhibition kinetics of an active site-directed inhibitor of ADAMTS-4 (Aggrecanase 1) Biochemistry. 2007;46(21):6393–6401. doi: 10.1021/bi7000642. [DOI] [PubMed] [Google Scholar]
- 74.Yang B, Guo H, Zhang Y, et al. MicroRNA-145 regulates chondrogenic differentiation of mesenchymal stem cells by targeting Sox9. PLoS ONE. 2011;6(7):e21679. doi: 10.1371/journal.pone.0021679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang J, Qin S, Yi C, et al. MiR-140 is co-expressed with Wwp2-C transcript and activated by Sox9 to target Sp1 in maintaining the chondrocyte proliferation. FEBS Lett. 2011;585(19):2992–2997. doi: 10.1016/j.febslet.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 76.Yasuda T. Activation of Akt leading to NF-κB up-regulation in chondrocytes stimulated with fibronectin fragment. Biomed Res. 2011;32(3):209–215. doi: 10.2220/biomedres.32.209. (Available from: http://dx.doi.org/10.2220/biomedres.32.209) [DOI] [PubMed] [Google Scholar]
- 77.Yuan ZM, Yang ZL, Zheng Q. Deregulation of microRNA expression in thyroid tumors. J Zhejiang Univ-Sci B (Biomed & Biotechnol) 2014;15(3):212–224. doi: 10.1631/jzus.B1300192. (Available from: http://dx.doi.org/10.1631/jzus.B1300192) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang Y, Jia J, Yang S, et al. MicroRNA-21 controls the development of osteoarthritis by targeting GDF-5 in chondrocytes. Exp Mol Med. 2014;46(2):e79. doi: 10.1038/emm.2013.152. (Available from: http://dx.doi.org/10.1038/emm.2013.152) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhao L, Li G, Zhou GQ. SOX9 directly binds CREB as a novel synergism with the PKA pathway in BMP-2-induced osteochondrogenic differentiation. J Bone Miner Res. 2009;24(5):826–836. doi: 10.1359/jbmr.081236. (Available from: http://dx.doi.org/10.1359/jbmr.081236) [DOI] [PubMed] [Google Scholar]