Abstract
CYP107W1 from Streptomyces avermitilis is a cytochrome P450 enzyme involved in the biosynthesis of macrolide oligomycin A. A previous study reported that CYP107W1 regioselectively hydroxylated C12 of oligomycin C to produce oligomycin A, and the crystal structure of ligand free CYP107W1 was determined. Here, we analyzed the structural properties of the CYP107W1-oligomycin A complex and characterized the functional role of the Trp178 residue in CYP107W1. The crystal structure of the CYP107W1 complex with oligomycin A was determined at a resolution of 2.6 Å. Oligomycin A is bound in the substrate access channel on the upper side of the prosthetic heme mainly by hydrophobic interactions. In particular, the Trp178 residue in the active site intercalates into the large macrolide ring, thereby guiding the substrate into the correct binding orientation for a productive P450 reaction. A Trp178 to Gly mutation resulted in the distortion of binding titration spectra with oligomycin A, whereas binding spectra with azoles were not affected. The Gly178 mutant’s catalytic turnover number for the 12-hydroxylation reaction of oligomycin C was highly reduced. These results indicate that Trp178, located in the open pocket of the active site, may be a critical residue for the productive binding conformation of large macrolide substrates.
Keywords: CYP, CYP107W1, oligomycin, P450, Streptomyces avermitilis, X-ray crystal structure
INTRODUCTION
Streptomyces avermitilis produces a variety of human and veterinary medicines including avermectins and its genome contains 33 Cytochrome P450 enzyme (CYP, P450) genes (Burg et al., 1979; Dyson, 2011; Kelly et al., 2005). So far, four P450 enzymes have been characterized as participating in the biosynthesis of macrolide metabolites. CYP171A1 catalyzes the formation of the furan ring in the avermectins, and CYP105P1 and CYP105D6 participate in hydroxylation during filipin biosynthesis (Ikeda et al., 1999; Lamb et al., 2011; Xu et al., 2010). Our previous study characterized the biochemical properties of CYP107W1 from S. avermitilis (Han et al., 2015). This enzyme efficiently catalyzes oligomycin C 12-hydroxylation to produce oligomycin A, and the interaction of the substrate oligomycin C with purified CYP107W1 shows a typical substrate binding titration (Han et al., 2015). The initial structural analysis of CYP107W1 without a ligand displayed a large substrate access pocket with a conserved P450 folding (Han et al., 2015). However, the underlying mechanism for the macrolide substrate recognition by P450 and the productive orientation of this enzyme is unknown, due to the limited structural information from CYP107W1 without a ligand. P450 enzymes are involved in various metabolic pathways and drug metabolisms (Durairaj et al., 2015; Min et al., 2015). The detail structural insight of CYP107W1’s substrate recognition will be helpful for the better understanding of P450 enzymes.
In this study, we determined the structure of the CYP107W1-oligomycin A bound complex and characterized the role of the Trp178 residue. Structural and enzymatic analyses indicated that hydrophobic interactions and a Trp178 residue in the active site played a key role to assist in the proper binding of the large macrolide substrate for regioselective hydroxylation.
MATERIALS AND METHODS
Chemicals
Oligomycin A, econazole, miconazole, spinach ferredoxin, ferredoxin reductase, and NADPH were purchased from SigmaAldrich (USA). Oligomycin C was purchased from Santa Cruz Biotechnology (USA). Crystal screening kits were obtained from Hampton Research (USA) and Emerald Biosystems (USA). All chemicals were of the highest grade that is commercially available.
Protein crystallization and data collection
Crystallization of the CYP107W1-oligomycin A bound complex was carried out as previously described with some modifications (Han et al., 2015). Briefly, the sitting-drop vapor-diffusion method was employed for the initial crystallization at 14°C, using a Hydra II e-Drop automated pipetting system (Matrix) and the Index, Crystal Screen, Crystal Screen Cryo, Crystal Screen Lite, PEGRx, SaltRx, PEG/Ion, Wizard, and Wizard precipitant synergy screening kits. To determine the structure of the oligomycin A complex, the native CYP107W1 enzyme was co-crystallized with oligomycin A in a 1:10 molecular ratio. Crystals of the oligomycin A complex appeared in Wizard precipitant synergy D2 conditions, with 3.35% (v/v) isopropanol and 1.34 M ammonium citrate/citric acid (pH 6.5).
The initial crystals were reproduced in the same conditions by the hanging-drop method, in which drops consisted of 1.0 μl protein solution mixed with 1.0 μl reservoir solution. X-ray diffraction data were collected from the cryoprotected crystal (at 100 K) with 1° rotation at a crystal-to-detector distance of 250 mm using an ADSC Q270 detector at beamline 7A-SBI of the Pohang Light Source (PLS, Pohang, Korea). Diffraction data were integrated and scaled using the HKL-2000 program package (Otwinowski and Minor, 1997).
Structure determination
The structure of CYP107W1 complexed with oligomycin A (CYP107W1-OliA) was solved by molecular replacement with Molrep (Vagin and Teplyakov, 1997), using the native CYP107W1 ligand-free structure (PDB ID: 4WPZ). One monomer of CYP107W1-OliA was present in the asymmetric unit. After refinement of the protein model, the resulting map was examined for the bound ligand density. The oligomycin A ligand model from the PRODRG2 server (Schuttelkopf and van Aalten, 2004) was fitted into the electron-density map with Coot (Emsley and Cowtan, 2004). The overall structure and volume of the substrate binding pocket of CYP107W1-OliA were calculated and generated in PyMol (Schrodinger, 2010). Data collection statistics are provided in Table 1.
Table 1.
Data collection and refinement statistics
| CYP107W1-OliA | |
|---|---|
| Data collection | |
| Beamline | PAL-7A (Korea) |
| Wavelength (Å) | 0.97935 |
| Resolution range (Å) | 50.00−2.60 (2.94−2.60) |
| Space group | P43212 |
| Unit-cell parameters (Å) | a = 127.3, b = 127.3, c = 75.5 α = γ = β = 90.0 |
| Total No. of reflections | 173,672 |
| No. of unique reflections | 19,627 |
| Completeness (%) | 99.6 (99.8) |
| Molecules per asymmetric unit | 1 |
| Solvent content (%) | 64.74 |
| Average I/σ (I) | 26.2 (2.2) |
| Rsym † (%) | 9.8 (58.3) |
| Multiplicity | 8.8 (7.4) |
| Refinement | |
| Resolution range (Å) | 38.62−2.60 |
| Rwork/Rfree | 20.8/28.2 |
| Wilson B factor | 67.3 |
| Protein | 67.0 |
| Heme | 50.4 |
| OliA (occupancy: 0.89) | 96.1 |
| Waters | 60.6 |
| R.M.S.D | |
| Bond | 0.018 |
| Angle | 2.248 |
Rsym = ΣhΣi|I(h)i–<I(h)>|/ΣhΣiI(h)i, where I(h) is the intensity of reflection h, Σh is the sum over all reflections, and Σi is the sum zover i measurements of reflection h.
Values in parentheses are for highest-resolution shell.
Site-directed mutagenesis to construct W178G mutant
Site-directed mutagenesis was performed to construct the CYP107W1 W178G mutant using the QuikChange mutagenesis kit (Stratagene, USA) with the following primers: 5′-TGCTCGGGGACGGGCAGCAGGTGGT-3′, 5′-ACCACCTGC TGCCCGTCCCCGAGCA-3′. The constructed mutant pET vector was confirmed by nucleotide sequencing analysis.
Spectral analysis of binding titrations
Spectroscopic characterization and spectral binding titration were carried out as previously described (Han et al., 2015). Purified CYP107W1 W178G mutant enzyme was diluted to 3 μM in 100 mM potassium phosphate buffer (pH 7.4) and divided between two glass cuvettes. Spectra between 350 and 500 nm were recorded using a CARY Varian spectrophotometer (Choi et al., 2013; Lee et al., 2014), while adding various ligands (oligomycin A and azole agents). The difference in absorbance between wavelength maximum and minimum was plotted against ligand concentration to calculate the binding constants (Schenkman et al., 1967).
Enzymatic activity
Oligomycin C hydroxylation by the CYP107W1 W178G mutant was analyzed as previously described (Han et al., 2015). Briefly, the enzyme reaction included 200 pmol purified P450 enzyme (W178G mutant), 40 μg/ml spinach ferredoxin, and 0.04 U/ml spinach ferredoxin reductase, in 0.50 ml of 100 mM potassium phosphate buffer (pH 7.4), along with various concentrations of oligomycin C. Reactions were initiated by adding 50 μl of an NADPH-generating system and were terminated after 30 min of incubation at 37°C by adding 1 ml of CH2Cl2, followed by vortex mixing and centrifugation. The reaction products were recovered from the organic phase after drying under N2. For the analysis of oxidized products, LC-mass spectrometry was performed as previously described (Han et al., 2015), using a Shimadzu LCMS-2010 EV system (Shimadzu, Japan).
RESULTS
Overall structure of the CYP107W1-oligomycin A complex
A crystal structure of the CYP107W1-oligomycin A complex (CYP107W1-OliA) was determined at 2.6 Å resolution by molecular replacement using the previously reported CYP107W1 ligand-free (CYP107W1-LF) structure (Han et al., 2015) (Table 1). The overall P450-folding was well conserved in the CYP107W1-OliA complex structure (Fig. 1A). In the structure of the CYP107W1-OliA complex, the F and G helices and the loop between them appeared to move inward to hold oligomycin A. The bulky and circular oligomycin A was bound to the upper side of the substrate-binding cavity with the occupancy and B factor of 0.89 and 96.1, respectively (Fig. 1B and Supplementary Fig. 1). The heme was bound with a full occupancy and B factor of 50.4 (Supplementary Fig. 2).
Fig. 1.
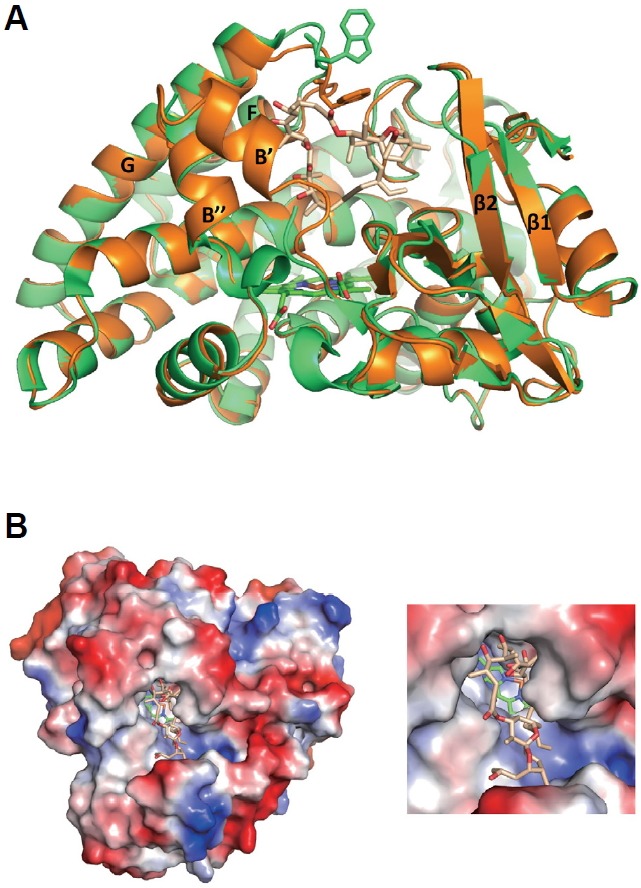
Structures of the CYP107W1-oligomycin A complex (CYP107W1-OliA). (A) Superimposed structures of the CYP107W1-OliA (orange) and CYP107W1-LF (ligand free CYP107W1, green). The bound OliA is represented as salmon. Heme is represented as green. (B) Surface structures of CYP107W1-OliA involving macrolide oligomycin A. Coordinates and structure factors of CYP107W1-OliA have been deposited in the Protein Data Bank with accession number 4WQ0.
Oligomycin A binding to the CYP107W1 enzyme
In the substrate binding cavity of CYP107W1, the bulky dioxaspiro[5.5]undecane group of oligomycin A was bound in the space between the B′-helix and the β2 strand, while the remaining circular ring slid into the central heme, mainly through hydrophobic interactions (Figs. 1A and 2A). Met69 and Met76 in the B′-helix and Met85 in the B″-helix showed van der Waals interactions with oligomycin A within a distance of 4 Å (Fig. 2A). The three methionine residues, Met69, Met76, and Met85, as well as Ile292 were positioned on one side of the ligand, while on the other side, Pro12 and Ile393, also directly interacted with oligomycin A by van der Waals interactions (Fig. 2A). Only Lys184 had an H-bond with the carbonyl oxygen atom at C7 of oligomycin A. Specifically, Trp178 sandwiched the circular ring of oligomycin A from the top, with the B′-helix at the bottom (Fig. 2A). Relative to the CYP107W1-LF structure, Trp178 in the CYP107W1-OliA complex structure moved in toward oligomycin A, to a distance of 3.6 Å from the Cα position and 8.4 Å from the side-chain indole ring (Figs. 1A and 2A). With this movement, Trp178 seemed to intercalate into the macrolide ring of oligomycin to accommodate the correct orientation of the bound substrate in the large pocket of the active site.
Fig. 2.
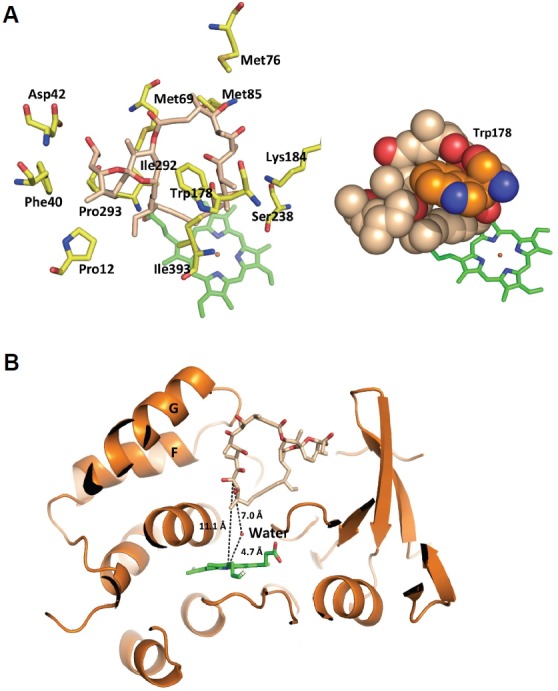
Oligomycin A recognition in CYP107W1-OliA. (A) Oligomycin A was bound to Met69, Met76, Met85, and Ile292 on one side and Pro12, Ile393, and Trp178 on the other side via hydrophobic interactions. (B) Interactions between CYP107W1 and oligomycin A. In the CYP107W1-OliA structure, the distance between the iron atom of heme and C12 of oligomycin A is 11.1 Å.
The relative position of oligomycin A and heme
In the CYP107W1-OliA complex structure, oligomycin A was bound in the heme plane via C12 (the catalytic position) of its macrolide ring, extended toward the iron atom of the heme prosthetic group (Fig. 2B). However, the distance between the iron atom of heme and C12 was not close (11.1 Å) enough to allow for catalysis of the P450 reaction. A water molecule was found at the active site, which was coordinated with an H-bond with Gly290 and observed with 4.7 Å distance at the tilted upper position of the heme iron atom in this structure (Fig. 2B). A similar orientation of the macrolide ligand in the active site of the P450 enzyme was previously reported in MycG (Li et al., 2012). In the structure of MycG, reaction sites for the substrate are also away from the heme iron (8.9–10.0 Å) than distances that would allow for the interactions with activated oxygen species (Li et al., 2012).
Catalytic alteration of the CYP107W1 W178G mutant
In the structure of the CYP107W1-OliA complex, Trp178 seemed to be a key residue to orient the ring structure of oligomycin A for productive binding conformation. To explore the role of Trp178 in substrate and product binding, a Trp178 to Gly (W178G) mutant was constructed and purified (Supplementary Fig. 3). The spectral analysis of binding to the purified W178G mutant enzyme displayed a distorted, shifted type I binding of oligomycin A, indicating an incomplete binding mode (Fig. 3A). These spectral changes were very different from those of wild type CYP107W1 (Han et al., 2015). In contrast, the type II spectra of W178G binding to azole agents were very similar to those of wild type CYP107W1 (Fig. 3B). Steady-state kinetic analysis of the W178G mutant displayed a kcat value of 0.061 ± 0.001 min−1 and a Km value of 3.4 ± 0.4 μM (Fig. 4). This result indicated a substantial decrease (∼ 30 %) in the turnover number relative to that of the CYP107W1 wild type, and suggested that Trp178 is important for the productive binding conformation of this large macrolide substrate.
Fig. 3.
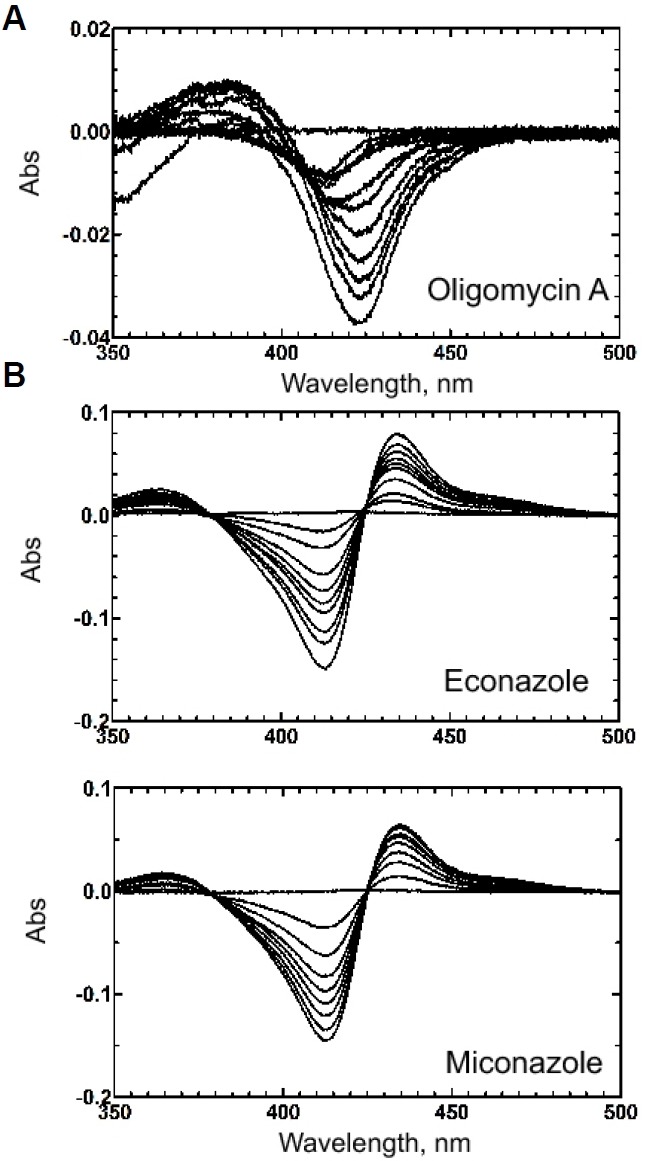
Binding spectra of the purified CYP107W1 W178G mutant. (A) Oligomycin A titration of the CYP107W1 W178G mutant. The calculated Kd value was 4.3 ± 0.7 μM. (B) Azole binding to the CYP107W1 W178G mutant. The binding affinities of azoles were calculated; the calculated Kd values are econazole: 5.3 ± 0.4 μM, miconazole: 3.1 ± 0.3 μM, respectively.
Fig. 4.
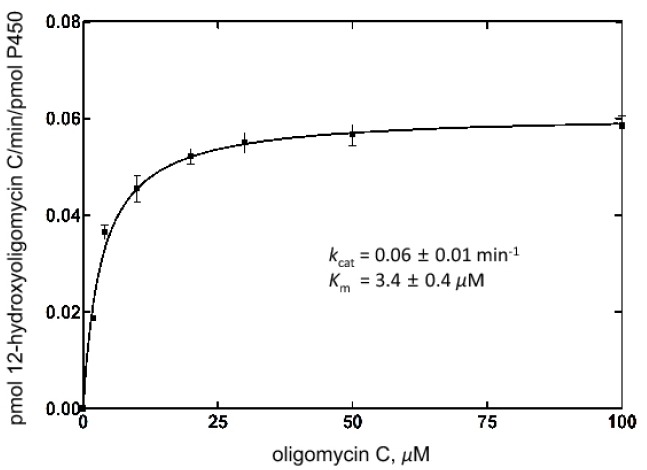
Steady-state kinetic analysis of oligomycin C hydroxylation activity by the CYP107W1 W178G mutant. The kinetic parameters for the CYP107W1 W178G mutant were estimated to be a kcat value of 0.06 ± 0.01 min−1 and a Km value of 3.4 ± 0.4 μM. (Results are presented as the mean ± SD (range) of duplicate assays)
DISCUSSION
In this study, we identified a large, open substrate-binding pocket and a special structural architecture that accommodates large substrates in CYP107W1. In the structure of the CYP107W1-OliA complex, oligomycin A was bound in the substrate access channel on the upper side of the prosthetic heme, mainly by hydrophobic interactions, and some part of the large substrate moiety was exposed to solvent. The Trp178 residue of CYP107W1 moved and intercalated into the large macrolide ring to facilitate the correct orientation for substrate binding in the P450 reaction.
The CYP107W1-oligomycin A structure could represent an intermediate step between the hydroxylation reaction and the release of the product, oligomycin A. Since the structures of oligomycin A and oligomycin C are similar except for the C12 position, we speculated that the structure of the CYP107W1 in complex with substrate oligomycin C would be similar to the product-bound structure of CYP107W1-oligomycin A. For oligomycin C to access the catalytic center of the heme group, a conformational change in CYP107W1 or oligomycin itself is expected. Hydrophobic macrolide ring of oligomycin might slide into the catalytic center through the smooth hydrophobic surface of substrate access channel. For example, oligomycin C might be folded into a rod-shaped structure similar to filipin in the CYP105P1-filipin complex structure via maximizing its internal hydrophobic interactions among the carbon atoms in the ring (Xu et al., 2009; 2010), of which conformation can allow oligomycin C12 atom to reach deeper into the heme structure for the P450 oxidation reaction. However, further studies are necessary to elucidate the detailed catalytic mechanism.
Oligomycin A was mainly bound by the B′ and B″ helices and the β1 and β2 strands, and was also covered by the Trp178 residue (Fig. 1A). The loop containing Trp178 was available for conformational changes that allowed oligomycin A to be recognized. The folded B′ and B″ helices generated a larger substrate-binding pocket on the upper side of heme, and an extra bulb space for the bulky dioxaspiro[5.5]undecane group of oligomycin A appeared between the B−-helix and the β2 strand. Compared with the linearly folded shape of filipin in CYP105P1, oligomycin A showed a more circular shape, similar to the structural configuration observed in the ATPases bound complex (Symersky et al., 2012)(Fig. 2A). Interestingly, almost all interactions between oligomycin A and the substrate-binding site residues of CYP107W1 were van der Waals interactions, except for one H-bond with Lys184. Even Lys184 is flexible, due to its long side chain, which could enable the sliding movement of oligomycin into the substrate-binding pocket. Trp178, which was located in a central position of the oligomycin A ring, appeared to be more flexible without oligomycin A bound. The W178G mutant showed a dramatically altered binding conformation and reduced catalytic activity for oligomycin C, indicating the substrate-recognizing role of Trp178 via van der Waals interactions.
We compared three structures of macrolide synthesis-related P450 enzymes in the regions of the F/G and B′/B″ helices, as well as the rest of the proteins (Supplementary Fig. 4). The root-mean-square deviations (RMSDs) of the B′/B″ and F/G helices of the CYP107W1-oligomycin A structure from those of the CYP105P1 and the MycG structure were 1.716 Å for the 283 Cα atoms and 2.203 Å for the 259 Cα atoms, respectively. The RMSDs of the rest of the CYP107W1-oligomycin A structure from those of the CYP105P1 and the MycG structure were 1.040 Å for the 1602 Cα atoms and 0.971 Å for the 1655 Cα atoms, respectively. This shows that the conformations of the F/G and B′/B″ helices are important for substrate specificity of the P450s, particularly if they have big molecules as substrates. In many of P450 structures, the region of the B′ and B″ helices showed diverse secondary structures and flexible conformations (Li et al., 2012; Xu et al., 2009) and sometimes even disappeared due to the flexibility (Lim et al., 2012; Xu et al., 2010) that allowed the enzyme to easily change conformations to accommodate substrates. In CYP107W1, the presumed flexible region was folded into a more stable secondary structure of two α helices, changing the general substrate-binding pocket geometry of this P450 enzyme and enabling substrate binding in a different conformation.
The Km value of W178G mutant was lower than that of CYP107W1 wild type (∼18 μM, Fig. 4). Generally, the Km value indicates the affinities to the substrate. However, this is not the case of CYP107W1 since the lower binding affinity was observed from the calculated Kd value of W178G mutant to oligomycin A (4.3 ± 0.7 μM, Fig 3A). Our postulation is that the catalytic activity of CYP107W1 may be mainly dependent on the kcat value because the series of biosynthesis process are performed in the limited place where the biosynthesis cluster gene products are concentrated. Therefore, the Km value (or the substrate affinity) may not affect the catalysis of this enzyme significantly. In previous study, it was also reported that the Km value of the P450 4A11 enzyme product is much lower than that of the substrate (Kim et al., 2014).
The structural analysis of the CYP107W1-oligomycin A complex suggested a way to recognize and bind a large macrolide substrate in a substrate-binding pocket. Our results will be useful for elucidating novel substrate recognition mechanism of P450s, while extending the existing knowledge of these enzymes and their function in macrolide synthesis.
Supplemental Data
Acknowledgments
This work was supported by a National Research Foundation of Korea (NRF) grant funded by the Korea government (NRF-2013R1A1A2061502) and by Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ01127901) funded by Rural Development Administration, Korea.
Footnotes
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Burg R.W., Miller B.M., Baker E.E., Birnbaum J., Currie S.A., Hartman R., Kong Y.L., Monaghan R.L., Olson G., Putter I., et al. Avermectins, new family of potent anthelmintic agents: producing organism and fermentation. Antimicrob. Agents Chemother. 1979;15:361–367. doi: 10.1128/aac.15.3.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S., Han S., Lee H., Chun Y.J., Kim D. Evaluation of Luminescent P450 analysis for directed evolution of human CYP4A11. Biomol. Ther. 2013;21:487–492. doi: 10.4062/biomolther.2013.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durairaj P., Malla S., Nadarajan S.P., Lee P.G., Jung E., Park H.H., Kim B.G., Yun H. Fungal cytochrome P450 monooxygenases of Fusarium oxysporum for the synthesis of omega-hydroxy fatty acids in engineered Saccharomyces cerevisiae. Microb Cell Fact. 2015;14:45. doi: 10.1186/s12934-015-0228-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson P. Streptomyces: Molecular Biology and Biotechnology. Norfolk, UK: Caister Academic Press; 2011. [Google Scholar]
- Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Han S., Pham T.V., Kim J.H., Lim Y.R., Park H.G., Cha G.S., Yun C.H., Chun Y.J., Kang L.W., Kim D. Functional characterization of CYP107W1 from Streptomyces avermitilis and biosynthesis of macrolide oligomycin A. Arch. Biochem. Biophys. 2015;575:1–7. doi: 10.1016/j.abb.2015.03.025. [DOI] [PubMed] [Google Scholar]
- Ikeda H., Nonomiya T., Usami M., Ohta T., Omura S. Organization of the biosynthetic gene cluster for the polyketide anthelmintic macrolide avermectin in Streptomyces avermitilis. Proc. Natl. Acad. Sci. USA. 1999;96:9509–9514. doi: 10.1073/pnas.96.17.9509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly S.L., Kelly D.E., Jackson C.J., Warrilow A.G.S., Lamb D.C. The diversity and importance of microbial cytochrome P450. In: Ortiz de Montellano P.R., editor. In cytochrome P450: structure, mechanism, and biochemistry. New York: Plenum Press; 2005. pp. 585–617. [Google Scholar]
- Kim D., Cha G.S., Nagy L.D., Yun C.H., Guengerich F.P. Kinetic analysis of lauric acid hydroxylation by human cytochrome P450 4A11. Biochemistry. 2014;53:6161–6172. doi: 10.1021/bi500710e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb D.C., Zhao B., Guengerich F.P., Kelly S.L., Waterman M.R. Genomics of Streptomyces cytochrome P450. In: Dyson P., editor. In streptomyces molecular biology and biotechnology. Norfolk, UK: Caister Academic Press; 2011. pp. 233–253. [Google Scholar]
- Lee H., Kim J.H., Han S., Lim Y.R., Park H.G., Chun Y.J., Park S.W., Kim D. Directed-evolution analysis of human cytochrome P450 2A6 for enhanced enzymatic catalysis. J. Toxicol. Environ. Health A. 2014;77:1409–1418. doi: 10.1080/15287394.2014.951757. [DOI] [PubMed] [Google Scholar]
- Li S., Tietz D.R., Rutaganira F.U., Kells P.M., Anzai Y., Kato F., Pochapsky T.C., Sherman D.H., Podust L.M. Substrate recognition by the multifunctional cytochrome P450 MycG in mycinamicin hydroxylation and epoxidation reactions. J. Biol. Chem. 2012;287:37880–37890. doi: 10.1074/jbc.M112.410340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim Y.R., Hong M.K., Kim J.K., Doan T.T., Kim D.H., Yun C.H., Chun Y.J., Kang L.W., Kim D. Crystal structure of cytochrome P450 CYP105N1 from Streptomyces coelicolor, an oxidase in the coelibactin siderophore biosynthetic pathway. Arch. Biochem. Biophys. 2012;528:111–117. doi: 10.1016/j.abb.2012.09.001. [DOI] [PubMed] [Google Scholar]
- Min H., Kawasaki I., Gong J., Shim Y.H. Caffeine induces high expression of cyp-35A family genes and inhibits the early larval development in Caenorhabditis elegans. Mol. Cells. 2015;38:236–242. doi: 10.14348/molcells.2015.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Schenkman J.B., Remmer H., Estabrook R.W. Spectral studies of drug interaction with hepatic microsomal cytochrome P-450. Mol. Pharmacol. 1967;3:113–123. [PubMed] [Google Scholar]
- Schrodinger L. The PyMOL Molecular Graphics System, Version 1.3r1. 2010. [Google Scholar]
- Schuttelkopf A.W., van Aalten D.M. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D. Biol. Crystallogr. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- Symersky J., Osowski D., Walters D.E., Mueller D.M. Oligomycin frames a common drug-binding site in the ATP synthase. Proc. Natl. Acad. Sci. USA. 2012;109:13961–13965. doi: 10.1073/pnas.1207912109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin A., Teplyakov A. MOLREP: an Automated Program for Molecular Replacement. J. Appl. Cryst. 1997;30:1022–1025. [Google Scholar]
- Xu L.H., Fushinobu S., Ikeda H., Wakagi T., Shoun H. Crystal structures of cytochrome P450 105P1 from Streptomyces avermitilis: conformational flexibility and histidine ligation state. J. Bacteriol. 2009;191:1211–1219. doi: 10.1128/JB.01276-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L.H., Fushinobu S., Takamatsu S., Wakagi T., Ikeda H., Shoun H. Regio- and stereospecificity of filipin hydroxylation sites revealed by crystal structures of cytochrome P450 105P1 and 105D6 from Streptomyces avermitilis. J. Biol. Chem. 2010;285:16844–16853. doi: 10.1074/jbc.M109.092460. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


