Abstract
A balance between production and degradation of reactive oxygen species (ROS) is critical for maintaining cellular homeostasis. Increased levels of ROS during oxidative stress are associated with disease conditions. Antioxidant enzymes, such as extracellular superoxide dismutase (EC-SOD), in the extracellular matrix (ECM) neutralize the toxicity of superoxide. Recent studies have emphasized the importance of EC-SOD in protecting the brain, lungs, and other tissues from oxidative stress. Therefore, EC-SOD would be an excellent therapeutic drug for treatment of diseases caused by oxidative stress. We cloned both the full length (residues 1–240) and truncated (residues 19–240) forms of human EC-SOD (hEC-SOD) into the donor plasmid pFastBacHTb. After transposition, the bacmid was transfected into the Sf9-baculovirus expression system and the expressed hEC-SOD purified using FLAG-tag. Western blot analysis revealed that hEC-SOD is present both as a monomer (33 kDa) and a dimer (66 kDa), as detected by the FLAG antibody. A water-soluble tetrazolium (WST-1) assay showed that both full length and truncated hEC-SOD proteins were enzymatically active. We showed that a potent superoxide dismutase inhibitor, diethyldithiocarbamate (DDC), inhibits hEC-SOD activity.
Keywords: extracellular matrix (ECM), human extracellular superoxide (hEC-SOD), LC-MS/MS, reactive oxygen species (ROS), sodiumdiethyldithiocarbamate
INTRODUCTION
The body produces reactive oxygen species (ROS) as a result of aerobic respiration and substrate oxidation. These ROS play several important roles in cell signaling, defense against microbial infections, maintaining cellular homeostasis, and lipid oxidation. However, high levels of ROS are observed during oxidative stress and, if unchecked, can lead to cardiovascular diseases (e.g., heart failure, hypercholesterolemia, arthrosclerosis, hypertension, and diabetes). Enzymes, such as superoxide dismutase (SOD), help to neutralize the harmful effect of these ROS. SOD converts free radicals (ROS) into less harmful molecules, viz., hydrogen peroxide (H2O2) and oxygen (O2), which are further metabolized by relevant cellular pathways.
Based on its distribution, SOD is mainly categorized into three forms: SOD1 (Cu/Zn-SOD, a homodimer) is localized in the cytosol, SOD2 (Mn-SOD, a homodimer/tetramer) is found in the mitochondria, and SOD3 (EC-SOD, a tetramer) is localized in the extracellular space (Beyer et al., 1991; Zelko et al., 2002). Extracellular superoxide dismutase (EC-SOD)/SOD3 catalyzes dis-mutation of the superoxide (O2−) radical into either ordinary molecular oxygen (O2) or hydrogen peroxide (H2O2). Blood vessel walls, kidneys, the uterus, and lungs usually contain high SOD3 levels (Nozik-Grayck et al., 2005; Petersen et al., 2005). SOD3 is synthesized by vascular smooth muscle cells, which subsequently binds to the heparan sulfate on the endothelial cell surface, from where it is internalized by the endothelial cells (Ohta et al., 1994). SOD3 prevents destruction of the NO• from superoxide anions in the extracellular space (Carlsson et al., 1995), which is necessary to stimulate smooth muscle relaxation (Oury et al., 1996a). Furthermore, SOD3 is the only antioxidant enzyme in the extracellular matrix (ECM) that neutralizes superoxide toxicity. It has been proposed that EC-SOD protects the brain, lungs, and other tissues from oxidative stress (Oury et al., 1996b). For example, SOD3 plays an important role in hypoxic lung injury, bleomycin toxicity, viral pneumonia, and asbestosis (Bowler et al., 2002; 2004; Folz et al., 1999; Mamo et al., 2004; Oury et al., 2002; Tan et al., 2004). In both the brain and kidney, SOD3 has protective effects related to ischemia/reperfusion injury (Demchenko et al., 2002), while hypoxic activation of erythropoietin in the kidney also requires SOD3 (Suliman et al., 2004).
The potential of SOD3 as a therapeutic drug for various diseases has led to several attempts at expression and purification of EC-SOD in Escherichia coli (He et al., 2002; Ryu et al., 2008), insect cells (He et al., 2002), Pichia pastoris (Chen et al., 2006), CHO cells (Tibell et al., 1987), Lactobacillus casei (Chen et al., 2013), and transgenic chicken (Byun el al., 2013). However, expression of EC-SOD has been limited by factors such as inclusion bodies, insufficient yield, inhomogeneity, and lack of activity.
Here, we engineered FLAG-tagged full length (residues 1–240; hEC-SODf) and truncated (residues 19–240; hEC-SODtr) forms of human extracellular superoxide dismutase (SOD3/hEC-SOD) and expressed these in the Spodoptera frugiperda (Sf9) insect cell system. We then purified the recombinant hEC-SOD, using anti-DYKDDDK G1 affinity resin as a soluble and uniformly active form of the enzyme. Furthermore, diethyldithiocarbamate (DDC), a potent superoxide dismutase inhibitor was also tested which was able to inhibit hEC-SOD activity. Our study forms the basis for developing active and homogenous hEC-SOD as a therapeutic drug using the Sf9 expression system.
MATERIALS AND METHODS
Plasmid construction of hEC-SOD
Nucleotide sequences encoding the full length (residues 1–240) and truncated/mature form (residues 19–240) of hEC-SOD were first amplified by PCR, using primers encoding a FLAG-tag (DYKDDDDK) and PreScission cleavage enzyme recognition sequence (LEVLFQG) at the N-termini, using EcoRI and XhoI restriction enzyme sites. The amplified DNA fragments were inserted into the pFastBacHTb™ vector (Invitrogen, USA) to encode a (His)6-FLAG-tagged-PreScission cleavage site fusion of the hEC-SOD protein. After cloning, sequences of both constructs were confirmed by sequencing the plasmids (Macrogen Co.).
Insect cell transfection and expression
Cloned plasmids were transformed into DH10Bac™ Escherichia coli. Transposition into the bacmid was performed as described in the protocol of the Bac-to-Bac® Baculovirus Expression System. 1000ug of recombinant bacmid DNA containing hEC-SODf and hEC-SODtr were transfected into Sf9 cells (0.5 × 106 cells/ml) using 8 μl of Cellfectin® II Reagent (Invitrogen™ by Life Technologies) and 100 μl CCM3 media (HyClone™), after which cells were incubated for 3 days at 27°C. Virus stock amplification was performed using the Bac-to-Bac Baculovirus Expression method as per the manufacturer’s instructions. Sf9 cells (2 × 106 cells/ml) were infected with P3 virus at an optimal multiplicity of infection (MOI: number of virus particles/number of cells) of 3 in Insectagro® media (Corning) and incubated at 27°C for 72 h before harvesting (2500 rpm at 4°C).
Purification of recombinant hEC-SOD
Frozen cell pellets were thawed and resuspended at 4°C for 30 min using lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 10 mM β-mercaptoethanol, an EDTA-free protease inhibitor cocktail (Roche), 0.5% (v/v) Nonidet®P-40, and 2 mM phenylmethylsulfonylfluoride [PMSF]). The lysate was subjected to ultracentrifugation at 15,000 rpm for 30 min at 4°C and the supernatant was loaded onto the GenScript Anti-DYKDDDK G1 Affinity resin (Cat. No. L00432). FLAG-tagged hEC-SOD was then purified following the protocol provided by GenScript. Purified hEC-SOD was simultaneously concentrated to 0.3 mM (9 mg/ml). The final buffer was exchanged to phosphate-buffered saline (PBS) using 10 kDa Amicon Ultra centrifugal filter tube (Millipore).
Western blot analysis
Whole cell expressed in Sf9 cells, the soluble fraction obtained after lysis, and purified hEC-SODf and hEC-SODtr were mixed with 5× sample buffer and resolved by 12% SDS-PAGE at 120 V for 2 h. Western blot analysis was performed as described by Yun et al. (2005). Monoclonal anti-FLAG M2 alkaline phosphatase antibody (Sigma), and SIGMAFAST BCIP/NBT (Sigma) were used for visualization. A mouse monoclonal primary antibody was used at a dilution of 1:2000 to detect the FLAG-tagged recombinant hEC-SOD.
Anti-Superoxide Dismutase 3 antibody (abcam® - ab21974) was used to detect native form of hEC-SOD before and after DTT titration. A secondary anti-rabbit antibody was used at a dilution of 1:5000 to detect the recombinant hEC-SOD. The 12% Native PAGE was run at 90V for 3 h at 4°C to avoid denaturing the protein.
Circular dichroism (CD) and fluorescence spectroscopy
CD spectra of both the hEC-SODf and hEC-SODtr were measured in PBS buffer at pH 7.4 on a JASCO J-815 spectropolarimeter (Jasco, Japan), calibrated with ammonium D-10-camphorsulfonate at 290 nm and equipped with a thermostatically controlled cell holder attached to a water bath with an accuracy of ± 0.1°C. The CD spectra were measured in the presence and absence of 50 μM Cu/Zn buffer of pH 7.4. Inbuilt secondary structure prediction (Yang’s and Reed’s Reference) was used to calculate the tendency for taking on a helical to β-sheet secondary structure upon Cu/Zn addition.
Fluorescence assay was performed at 298 K (25°C) using an LS55 spectrofluorophotometer (Perkin Elmer, USA). Both hEC-SOD forms (10 μM) and an inhibitor, Na-DDC (1 mM), were prepared in PBS (pH 7.4). The fluorescence of hEC-SOD (Ex. 280 nm) was collected from 270 nm to 450 nm (data shown from 300 nm to 450 nm) upon titrating with 5-μM increments of Na-DDC (0 μM to 100 μM), with a 1-min incubation time for each increment.
DTT titration of purified hEC-SOD
Purified hEC-SODf and hEC-SODtr were incubated for 60 min with and without 10 mM DTT in PBS buffer, at 4°C. Protein samples were centrifuged at 14,000 rpm at 4°C to remove any aggregates. hEC-SOD was also analyzed by size exclusion chromatography(SEC) using a HiLOAD™ 10/300 Superdex™ 200 (GE Healthcare) after injection of a 500-μl sample. The protein was eluted at 4°C, at a flow rate of 0.5 ml/min, using PBS (pH 7.4) as the mobile phase, with and without 10 mM DTT.
Activity assay of hEC-SOD
The activity of the purified hEC-SODf and hEC-SODtr was assayed in vitro using the SOD Assay Kit WST-1 (Dojindo Laboratories) (Tan and Berridge, 2000). This assay is based on the detection of a water-soluble formazan dye that is formed upon reduction of water-soluble tetrazolium salt, WST-1, by the superoxide anion. Purified EC-SOD (25 μM) was incubated with the reaction mixture, including a final concentration of 0.05 M sodium carbonate (pH 9.0), 1.0 U catalase, 10−4 M xanthine, 5.6 × 10−5 M WST-1, and 3.7 mU xanthine oxidase, at 37°C in the cell compartment of the spectrophotometer, and the sample was spectrophotometrically monitored at 450 nm at various time points, up to 30 min.
SOD Activity calculation
One unit of SOD is defined as the amount of the enzyme in 20 μl of sample solution that inhibits the reduction reaction of WST-1 with superoxide anion by 50%.
In-gel tryptic digestion
1DE band of hEC-SOD were excised from the preparative gel, and the spots were transferred into each 1.5 ml tube. The band was washed with 100 μl of distilled water; then, 100 μl of 50 mM NH4HCO3 (pH 7.8) and acetonitrile (6:4) were added to the band and shook for in 10 min. This process was repeated at least three times until the Coomassie brilliant blue G250 dye disappeared. The supernatant was decanted, and the band was dried in speed vacuum concentrator (LaBoGeneAps, Denmark) for 10 min. Reduction of the sample was performed in 10 mM dithiothreitol/25 mM NH4HCO3 (pH 8.0) for over 30 min at 56°C. The solution was decanted and the band was stored at room temperature in 55 mM iodoacetamide in the dark for 20 min. The solution was decanted and then digested with sequence-grade modified trypsin (Promega Co., USA) (enzyme to substrate ratio = 1:50) at 37°C with shaking for 16 h.
Deglycosylation
After Trypsin digestion, 1 ul of NH4HCO3 buffer (10 mM, pH 8.0) with 0.5 U PNGase F enzymes was added and shook at 37°C for 6 h for glycopeptide deglycosylation. The N-linked site was identified on the basis of the difference in mass from the native sequence associated with conversion of Asn to Asp (0.98 Da) after PNGase F treatment.
LC-MS/MS peptide analysis and database searching
Nano LC-MS/MS analysis of the purified hEC-SODf and hEC-SODtr was performed with an Easy n-LC (Thermo Fisher, USA). The capillary column used for LC-MS/MS analysis (150 mm × 0.075 mm) was obtained from Proxeon (Denmark) and the slurry packed in-house with a 5-μm, 100-A pore size Magic C18 stationary phase resin (Michrom BioResources, USA). LTQ-Orbitrap mass spectrometry (Thermo Fisher, USA) was used for peptide identification. Mass spectra were acquired using data-dependent acquisition with a full mass scan (350–1200 m/z) followed by five MS/MS scans. For MS1 full scans, the Orbitrap resolution was 30,000 and the AGC was 2 × 105. For MS/MS in the LTQ, the AGC was 1 × 104. The mascot algorithm (Matrix-science, USA) was used to identify peptide sequences present in a protein sequence database. Database search criteria were: taxonomy; target protein (hEC-SODf and hEC-SODtr), fixed modification; carboxyamidomethylated at cysteine residues; variable modification; oxidized at methionine residues, deamidated at asparagine residue, maximum allowed missed cleavage of 2, MS tolerance of 10 ppm, MS/MS tolerance of 0.8 Da. The peptides were filtered with a significance threshold of P < 0.05.
RESULTS
Construction and bacmid preparation of the expression plasmid
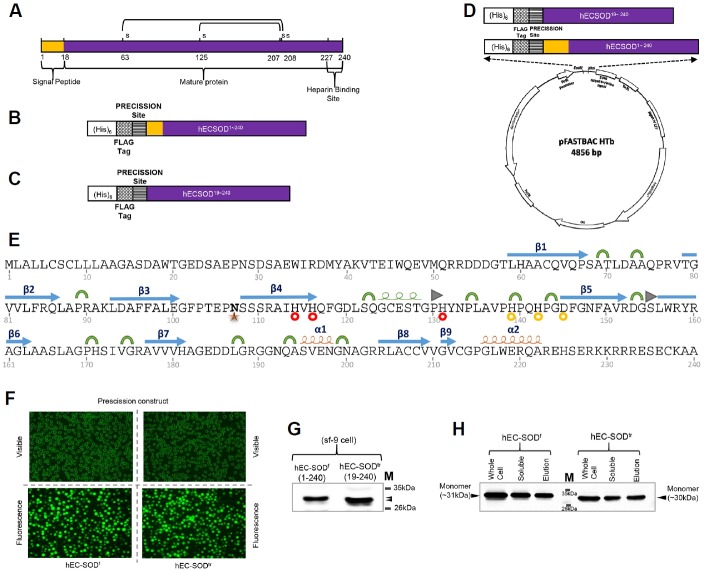
The coding sequence of hEC-SOD was amplified by PCR from cDNA using primers that included both the N-terminal signal peptide and the C-terminal heparin-/heparin sulfate-binding domain, and cloned into the pFastBacHTb™ vector to construct an expression plasmid, as described in materials and methods section (Fig. 1). The cloned plasmids of both hEC-SODf and hEC-SODtr contained an N-terminal Flag-tag, and a PreScission cleavage site, followed by the hEC-SOD sequence, as confirmed by sequencing (Figs. 1A–1D). The cDNA was transpositioned into the bacmid in E. coli using a blue/white colony selection method (Invitrogen™ by Life Technologies). Transposition occurred by transforming the donor plasmid into the E. coli DH10Bac, which contained the Bacmid and helper phage. Recombinant bacmid DNA was isolated by ethanol precipitation and the concentration determined by nanodrop. Primary sequence with secondary structures of hSOD, metail ion binding and glycosylation site are shown in Fig. 1E.
Fig. 1.
Construct design, expression, and purification of hEC-SOD protein from Sf9 insect cells. (A) Schematic illustration of hEC-SOD. The protein consists of an N-terminal signal peptide, mature protein, and a C-terminal heparin-binding site. The design of the construct of (B) hEC-SODf and (C) hEC-SODtr, with an N-terminal (His)6, a FLAG-tag and Precession enzyme site. (D) Map of the hEC-SOD expression vector, pFastBac HTB, containing the pH promoter, a pBR322 origin, and Ampicillin and Gentamycin resistance genes. A full length (residues 1–240) and truncated/mature form (residues 19–240) of hEC-SOD were first amplified by PCR, using primers encoding a FLAG-tag (DYKDDDDK) and PreScission cleavage enzyme recognition sequence (LEVLFQG) at the N-terminus, as well as EcoRI and XhoI restriction enzyme sites. (E) Secondary structure showing the glycosylation site and Cu/Zn binding site of hEC-SOD1–240 based on the crystal structure. PDB code: 2JLP. [
 α-helix,
α-helix,
 310-helix,
310-helix,
 β-strand,
β-strand,
 Turn,
Turn,
 β-bridge,
β-bridge,
 N107 Glycosylation Site,
N107 Glycosylation Site,
 Cu binding site,
Cu binding site,
 Zn binding site]. (F) Transfected recombinant hEC-SOD expressed in Sf9 cells observed at the EGFP-excitation wavelength. (G) Western blot analysis of hEC-SOD expressed in Sf9 cells that had been incubated for 3 days at 27°C, detected using an anti-FLAG antibody. (H) Western analysis of purified hEC-SOD resolved by 12% SDS–PAGE and detected using an anti-FLAG antibody. The size of the expressed hEC-SODf and hEC-SODtr was about 31 and 30 kDa respectively. hEC-SODf: Lane 1, whole cell lysate; lane 2, soluble fraction; and lane 3, purified protein. hEC-SODtr: Lane 5, whole cell lysate; lane 6, soluble fraction; and lane 7, purified protein. Lane 4, molecular marker.
Zn binding site]. (F) Transfected recombinant hEC-SOD expressed in Sf9 cells observed at the EGFP-excitation wavelength. (G) Western blot analysis of hEC-SOD expressed in Sf9 cells that had been incubated for 3 days at 27°C, detected using an anti-FLAG antibody. (H) Western analysis of purified hEC-SOD resolved by 12% SDS–PAGE and detected using an anti-FLAG antibody. The size of the expressed hEC-SODf and hEC-SODtr was about 31 and 30 kDa respectively. hEC-SODf: Lane 1, whole cell lysate; lane 2, soluble fraction; and lane 3, purified protein. hEC-SODtr: Lane 5, whole cell lysate; lane 6, soluble fraction; and lane 7, purified protein. Lane 4, molecular marker.
Expression and purification of hEC-SOD in Sf-9 cells
Transposition of the recombinant vector to the baculovirus DNA in E. coli was followed by preparation of high-titer virus stocks. This recombinant baculovirus DNA was infected into Sf9 cells to amplify the virus stock from a P1 to P3 virus titer. For hEC-SOD expression, 300 ml Sf9 cells were infected with P3 virus at an MOI of 3.0 and harvested 72 h post-infection. The infection was confirmed by monitoring YFP (Yellow Fluorescence Protein/Green filter; Fig. 1F). hEC-SOD cDNA is expressed under the control of the polyhedrine promoter after recombinant baculovirus infection of Sf9 cells.
The cell pellet harvested after 72 h was resuspended in lysis buffer and purified using anti-DYKDDDK G1 Affinity Resin (section 2.3). Western blot analysis confirmed that the molecular weights of hEC-SODf and hEC-SODtr expressed in Sf9 cells were around 31 kDa and 30 kDa, respectively, as expected (Figs. 1G and 1H).
Oligomerization of hEC-SOD
To obtain the native form of hEC-SOD, we purified both hEC-SODf and hEC-SODtr without using reducing agents, such as DTT or β-mercaptoethanol. Purified proteins were then loaded onto HiLOAD™ 10/300 Superdex™ 200 (GE Healthcare), previously calibrated with standard protein markers and equilibrated with PBS buffer, which resulted in elution of almost all injected proteins in the void volume, including some in the monomeric form (Fig. 2A). This confirmed that both hEC-SODf and hEC-SODtr form octamer. To assess the possible effect of DTT on the oligomerization of hEC-SOD, 10 mM DTT was added to the previously purified hEC-SODf and hEC-SODtr, as described in materials and methods section. SEC of the DTT-treated hEC-SOD showed that the majority of the oligomeric hEC-SOD (both full length and truncated forms) shifted towards the monomeric fraction (Fig. 2A.), this was reconfirmed by running native page detected using anti-Superoxide Dismutase 3 antibody (Fig. 2B), a result of protein being reduced by excess DTT, as previously published (Bae et al., 2013).
Fig. 2.
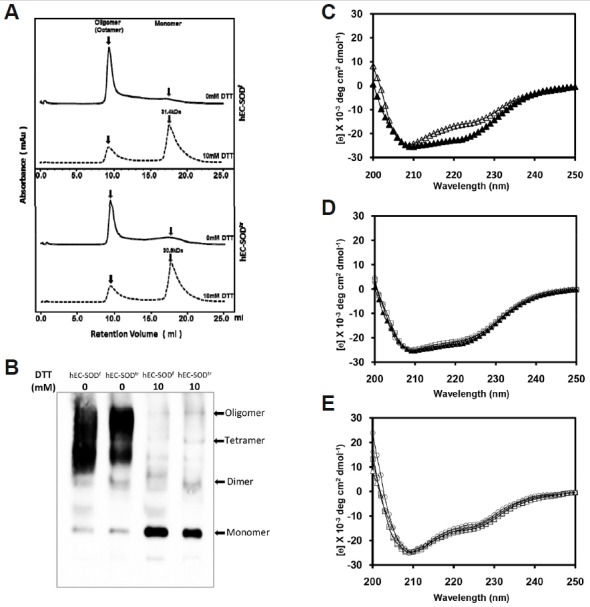
Biophysical and biochemical characterization of purified hEC-SOD expressed in Sf9 cells. (A) Analytic size exclusion chromatography profile of the recombinant hEC-SOD (0 mM DTT and 10 mM DTT), using a Superdex 200 column calibrated with standard protein markers. (B) Effect of DTT (0 to 10 mM) on the oligomerization of hEC-SOD analyzed by Western blot under native condition. (C) Far-UV circular dichroism of hEC-SODf and hEC-SODtr in the presence and absence of 50 µM Cu/Zn. Comparison of CD spectra of hEC-SODf (▲) and hEC-SODtr (Δ) in the presence of 50 μM Cu/Zn. (D) hEC-SODtr: free Δ, 50 μM Zn □, 50 μM Cu ⋄, and 50 μM Cu/Zn ○. (E) hEC-SODtr: free Δ, 50 μM Zn □, 50 μM Cu ⋄, and 50 μM Cu/Zn ○.
Secondary structure determination by circular dichroism (CD)
Circular dichroism experiments have been performed on both hEC-SODf and hEC-SODtr to compare their secondary structure, using 20 μM of protein in PBS (pH 7.4) at 25°C (Fig. 2C). Superimposing the CD spectra of the full length and truncated form showed slight differences in the secondary structure. This may be because hEC-SODf contains an extra N-terminal sequence that contribute to the difference in secondary structure. We assessed the effect of Zn2+; Cu2+, and Zn2+/Cu2+ (50 μM) on the secondary structure of hEC-SOD, and found no significant perturbation in the presence of metal ions (Figs. 2D and 2E). Prediction of the average secondary structure using Yang’s prediction showed that hEC-SOD consisted of 36.4% β-sheet and 17.9% α-helix, whereas hEC-SODtr consisted of 38.8% β-sheet and 16.9% α-helix. The crystal structure of hEC-SOD from E. coli has been reported to be mainly composed of β-sheet, where the N- and C-termini were not visible due to high flexibility (Antonyuk et al., 2009). Thus, we conclude that the hEC-SOD purified from Sf9 insect cells is mainly composed of β-sheet structures.
Enzyme activity of hEC-SOD
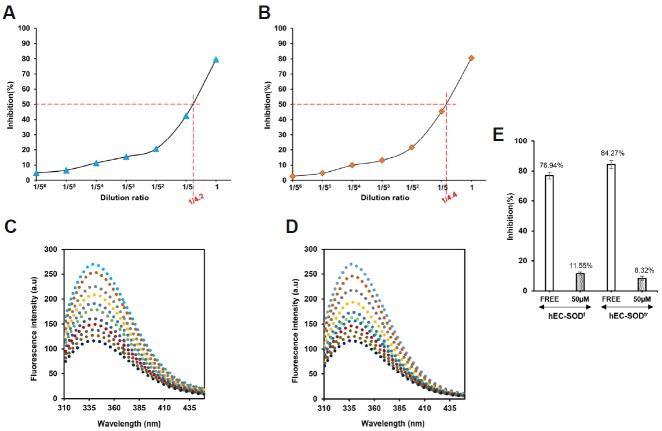
The enzymatic activity of hEC-SODf and hEC-SODtr was measured in the presence of Zn2+/Cu2+ and 0.1% BSA (Tan and Berridge, 2000) as described in materials and methods section. The activities of purified hEC-SODf and hEC-SODtr from Sf9 cells were calculated to be 210 U/ml and 220 U/ml, respectively (Figs. 3A and 3B). This implies that both the full length and truncated form of hEC-SOD have enzymatic activity. DDC, a thiol-containing molecule known as a potent metal ion-chelating agent (Arnelle et al., 1997; Iqbal and Whitney, 1991), is an inhibitor of Cu-Zn SOD. DDC inhibits the detoxification of ROS and increases superoxide concentrations (Didion et al., 2001; Li et al., 2002). Since Cu-Zn SOD and hEC-SOD are both forms of SOD, we tested whether sodium-diethyldithiocarbamate trihydrate, Na-DDC (Sigma life science) also inhibits hEC-SOD. Data from a fluorescence assay showed that Na-DDC has a higher affinity for hEC-SODf than for hEC-SODtr. Dissociation constants (Kd) were determined as 7.14 × 10−6 M for hEC-SODf and 34.17 × 10−6 M for hEC-SODtr (Figs. 3C and 3D). Inhibition of both hEC-SODf and hEC-SODtr in the presence of 50 μM Na-DDC is shown in Fig. 3E.
Fig. 3.
Enzyme activity and binding affinity of an inhibitor (DDC) of hEC-SODf and hEC-SODtr in the presence of 0.1% BSA & 50 μM Cu/Zn. (A) Activity assay of hEC-SODf in the presence of 0.1% BSA, and 50 μM Cu/Zn ions. (B) Activity assay of hEC-SODtr in the presence of 0.1% BSA, and 50 μM Cu/Zn ions. (C) Fluorescence assay of hEC-SODf with inhibitor (Na-DDC) in the presence of 0.1% BSA; 50 μM Cu/Zn. (D) Fluorescence assay of hEC-SODtr with inhibitor (Na-DDC) in the presence of 0.1% BSA; 50 μM Zn/Cu. Protein samples were excited at 280 nm and emission spectra were recorded from 300 to 450 nm. All the assays were performed in phosphate-buffered saline (PBS; pH 7.4) using 20 μM hEC-SOD with an increasing concentration of inhibitor (Na-DDC). [
 0 μM;
0 μM;
 10 μM;
10 μM;
 20 μM;
20 μM;
 30 μM;
30 μM;
 40 μM;
40 μM;
 50 μM;
50 μM;
 60 μM;
60 μM;
 70 μM;
70 μM;
 80 μM;
80 μM;
 90 μM and
90 μM and
 100 μM]. (E) Effect of Na-DDC (inhibitor) on hEC-SOD activity. Enzyme activity of hEC-SOD was determined using the SOD Assay Kit WST-1 (Dojindo Laboratories) in PBS buffer (pH 7.4) in the absence and presence of 50 μM of inhibitor (Na-DDC).
100 μM]. (E) Effect of Na-DDC (inhibitor) on hEC-SOD activity. Enzyme activity of hEC-SOD was determined using the SOD Assay Kit WST-1 (Dojindo Laboratories) in PBS buffer (pH 7.4) in the absence and presence of 50 μM of inhibitor (Na-DDC).
LC-MS analysis of hEC-SOD
The details of hEC-SODf and hEC-SODtr analyzed using LC-MS are shown in Table 1. Sequence coverage for hEC-SODf and hEC-SODtr were 60% and 62%, respectively. For hEC-SODf, several tryptic peptides were obtained, of which peptide 1 (R.AKLDAFFALEGFPTEPNSSSR.A) and peptide 2 (K.LDAFFALEGFPTEPNSSSR.A) showed a molecular weight difference of 0.984 Da after deamidation (Figs. 4A and 4B). Similarly, for hEC-SODtr, peptide 1 (R.AKLDAFFALEGFPTEPNSSSR.A) and peptide 2 (K.LDAFFALEGFPTEPNSSSR.A) also showed a molecular weight difference of 0.984 Da after deamidation (Figs. 4C and 4D). Table 2 summarizes the tryptic peptide of interest subjected to deamidation for molecular weight calculation. Taken together, this data confirmed that both hEC-SODf and hEC-SODtr were glycosylated at Asn107.
Table 1.
Identification of target protein based on LC-MS analysis
| Details | hEC-SOD | hEC-SODtr |
|---|---|---|
| Nominal mass (Mr) | 31835 (Da) | 30927 (Da) |
| Sequence coverage | 60% | 62% |
| Calculated pI value | 5.71 | 5.72 |
Fig. 4.
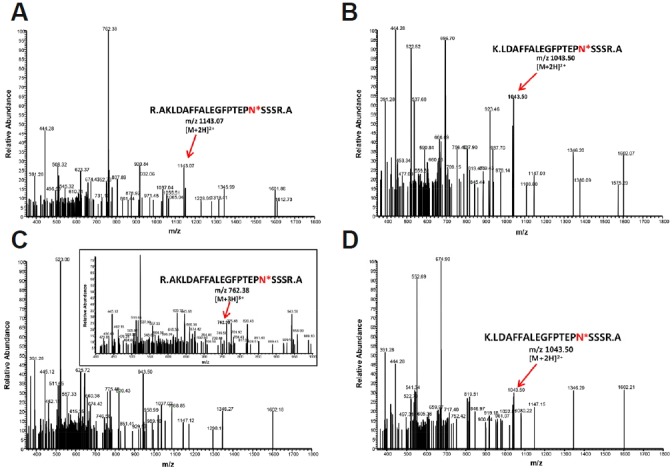
LC-MS/MS analysis of hEC-SODf and hEC-SODtr purified from Sf9 cells. (A,B) Map of hEC-SOD, peptide 1, and peptide 2 after trypsin cleavage and deamidation. (A) Fragmentation of the low-abundance ion at m/z 1143.07 resulted in identification of the fully tryptic peptide R.AKLDAFFALEGFPTEPN*SSSR.A. (B) This peptide was identified from the fragmentation of the low-abundance ion at m/z 1043.50 as the fully tryptic peptide K.LDAFFALEGFPTEPN*SSSR.A. (C, D) Map of hEC-SODtr, peptide 1, and peptide 2 after trypsin cleavage and deamidation. (C) Fragmentation of the low-abundance ion at m/z 762.38 resulted in identification of the fully tryptic peptide R.AKLDAFFALEGFPTEPN*SSSR.A. (D) This peptide was identified from the fragmentation of the low-abundance ion at m/z 1043.50 as the fully tryptic peptide K.LDAFFALEGFPTEPN*SSSR.A. (Asterisk indicates the N-glycosylation site in each peptide).
Table 2.
Tryptic peptide molecular weights after deamidation
| Tryptic peptide | hEC-SOD | Mr (calc) | Ions Score | hEC-SODtr | Mr (calc) | Ions score |
|---|---|---|---|---|---|---|
| 1 | R.AKLDAFFALEGFPTEPNSSSR.A | 2283.1171 | 104 | R.AKLDAFFALEGFPTEPNSSSR.A | 2283.1171 | 89 |
| R.AKLDAFFALEGFPTEPNSSSR.A (deamidated) | 2284.1012 | 91 | R.AKLDAFFALEGFPTEPNSSSR.A (deamidated) | 2284.1012 | 40 | |
| 2 | K.LDAFFALEGFPTEPNSSSR.A | 2083.9851 | 66 | K.LDAFFALEGFPTEPNSSSR.A | 2083.9851 | 50 |
| K.LDAFFALEGFPTEPNSSSR.A (deamidated) | 2084.9691 | 72 | K.LDAFFALEGFPTEPNSSSR.A (deamidated) | 2084.9691 | 83 |
DISCUSSION
The potential of SOD3 as an important therapeutic drug for treating various diseases has been evaluated; however, various limitations have hindered the achievement of a homogenous, active form of hEC-SOD. Here, hEC-SOD was cloned into a pFastBacHTb™ vector to obtain a (His)6-FLAG tagged-PreScission cleavage site-fusion of the hEC-SOD protein. hEC-SOD protein purified from human aorta has been reported to form stable tetramer as well as octamer (Due et al., 2006). However, it has been reported that hEC-SOD purified from E. coli (Bae et. al., 2013) forms octamer in the absence of reducing agent. Addition of reducing agent such as DTT drives the formation of tetramer, dimer and monomer. Our result from Sf9 cells also shows hEC-SOD forms octamer in the absence of reducing agent, whereas addition of DTT disrupts oligomer formation, driving to monomer formation. This was confirmed by the analytical size exclusion chromatography (Fig. 2A) and native gel (Fig. 2B). hEC-SOD expressed in eukaryotic Sf9 insect cells might have similar behavior with that purified from E. coli.
We have found that hEC-SOD expressed in Sf9 insect cells is glycosylated at Asn107, although the role of this modification is not clear (Liu et al., 2005). The glycosylated form of hEC-SOD is soluble, whereas the unglycosylated form is not, but it can be solubilized by increasing the pH and ionic strength of the solution (Edlund et al., 1992). LC-MS data revealed that both hEC-SODf and hEC-SODtr expressed in Sf9 cells are uniformly glycosylated. DDC, a potent metal ion-chelating agent (Arnelle et al., 1997; Iqbal and Whitney, 1991), also inhibited the hEC-SOD protein purified from Sf9 insect cells (Fig. 3E). We showed that DDC, an inhibitor of superoxide dismutase(Cu-ZnSOD and MnSOD), inhibits the activity of hEC-SOD. Possibly, by similar mechanism as in Cu-ZnSOD, hSOD becomes inactive after the removal of copper from Cu-ZnSOD in the presence of DDC (Arnelle et al., 1997; Iqbal and Whitney, 1991). However, the detailed binding mode of DDC to superoxide dismutase has unrevealed. We are working on solving crystal structure of hEC-SOD/DDC complex.
Taken together, we found that both full length and truncated hEC-SOD expressed in Sf9 insect cell possess the native oligomeric conformation, with enzymatic activity, which is essential for developing potent therapeutic hEC-SOD.
Acknowledgments
This research was supported by the Bio & Medical Technology Development Program of the National Research Foundation (NRF) funded by the Ministry of Science, ICT & Future Planning (NRF-2013M3A9A9050577). This work was partially supported by the Brain Korea plus (BK+) graduate student scholarship program.
REFERENCES
- Antonyuk S.V., Strange R.W., Marklund S.L., Hasnain S.S. The structure of human extracellular copper-zinc super-oxide dismutase at 1.7 Å resolution: insights into heparin and collagen binding. J. Mol. Biol. 2009;388:310–326. doi: 10.1016/j.jmb.2009.03.026. [DOI] [PubMed] [Google Scholar]
- Arnelle D.R., Day B.J., Stamler J.S. Diethyl dithiocarbamate-induced decomposition of S-nitrosothiols. Nitric Oxide. 1997;1:56–64. doi: 10.1006/niox.1996.0107. [DOI] [PubMed] [Google Scholar]
- Bae J.Y., Koo B.K., Ryu H.B., Song J.A., Nguyen M.T., Vu T.T., Son Y.J., Lee H.K., Choe H. Cu/Zn incorporation during purification of soluble human EC-SOD from E. coli stabilizes proper disulfide bond formation. Appl. Biochem. Biotechnol. 2013;169:1633–1647. doi: 10.1007/s12010-012-0025-x. [DOI] [PubMed] [Google Scholar]
- Beyer W., Imlay J., Fridovich I. Superoxide dismutases. Prog. Nucl. Acid Res. Mol. Biol. 1991;40:221–253. doi: 10.1016/s0079-6603(08)60843-0. [DOI] [PubMed] [Google Scholar]
- Bowler R.P., Nicks M., Warnick K., Crapo J.D. Role of extracellular superoxide dismutase in bleomycin-induced pulmonary fibrosis. Am. J. Physiol. Lung Cell Mol. Physiol. 2002;282:L719–726. doi: 10.1152/ajplung.00058.2001. [DOI] [PubMed] [Google Scholar]
- Bowler R.P., Nicks M., Tran K., Tanner G., Chang L.Y., Young S.K., Worthen G.S. Extracellular superoxide dismutase attenuates lipopolysaccharide-induced neutrophilic inflammation. Am. J. Respir. Cell. Mol. Biol. 2004;31:432–439. doi: 10.1165/rcmb.2004-0057OC. [DOI] [PubMed] [Google Scholar]
- Byun S.J., Ji M.R., Jang Y.J., Hwang A.I., Chung H.K., Kim J.S., Kim K.W., Yoo J.G., Kim T.Y. Human extracellular superoxide dismutase (EC-SOD) expression in transgenic chicken. BMB Rep. 2013;46:404–409. doi: 10.5483/BMBRep.2013.46.8.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson L.M., Jonsson J., Edlund T., Marklund S.L. Mice lacking extracellular superoxide dismutase are more sensitive to hyperoxia. Proc. Natl. Acad. Sci. USA. 1995;92:6264–6268. doi: 10.1073/pnas.92.14.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.L., Yen C.C., Tsai T.C., Yu C.H., Liou Y.J., Lai Y.W. Production and characterization of human extracellular superoxide dismutase in the methylotrophic yeast Pichia pastoris. J. Agric. Food Chem. 2006;54:8041–8047. doi: 10.1021/jf061379x. [DOI] [PubMed] [Google Scholar]
- Chen C.M., Lai Z.L., Yen C.C., Wang M.L., Chen H.L. Cloning and expression of the human extracellular superoxide dismutase (EC-SOD) gene in probiotic lactobacillus casei. Int. J. Biosci. Biochem. Bioinforma. 2013;3:557–561. [Google Scholar]
- Demchenko I.T., Oury T.D., Crapo J.D., Piantadosi C.A. Regulation of the brain's vascular responses to oxygen. Circ. Res. 2002;91:1031–1037. doi: 10.1161/01.res.0000043500.03647.81. [DOI] [PubMed] [Google Scholar]
- Didion S.P., Hathaway C.A., Faraci F.M. Superoxide levels and function of cerebral blood vessels after inhibition of CuZn-SOD. Am. J. Physiol. Heart Circ. Physiol. 2001;281:H1697–H1703. doi: 10.1152/ajpheart.2001.281.4.H1697. [DOI] [PubMed] [Google Scholar]
- Due A.V., Petersen S.V., Valnickova Z., Ostergaard L., Oury T.D., Crapo J.D. Extracellular superoxide dismutase exists as an octamer. FEBS Lett. 2006;580:1485–1489. doi: 10.1016/j.febslet.2006.01.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlund A., Edlund T., Hjalmarsson K., Marklund S.L., Sandstrom J., Stromqvist M., Tibell L. A non-glycosylated extra-cellular superoxide dismutase variant. Biochem. J. 1992;288:451–456. doi: 10.1042/bj2880451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folz R.J., Abushamaa A.M., Suliman H.B. Extracellular superoxide dismutase in the airways of transgenic mice reduces inflammation and attenuates lung toxicity following hyperoxia. J. Clin. Invest. 1999;103:1055–1066. doi: 10.1172/JCI3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H.J., Yuan Q.S., Yang G. Z., Wu X.F. High-level expression of human extracellular superoxide dismutase in Escherichia coli and insect cells. Protein Expr. Purif. 2002;24:13–17. doi: 10.1006/prep.2001.1529. [DOI] [PubMed] [Google Scholar]
- Iqbal J., Whitney P. Use of cyanide and diethyldithiocarbamate in the assay of superoxide dismutases. Free Radic. Biol. Med. 1991;10:69–77. doi: 10.1016/0891-5849(91)90023-v. [DOI] [PubMed] [Google Scholar]
- Li J., Li P.F., Dietz R., von Harsdorf R. Intracellular superoxide induces apoptosis in VSMCs: role of mitochondrial membrane potential, cytochrome C and caspases. Apoptosis. 2002;7:511–517. doi: 10.1023/a:1020639025669. [DOI] [PubMed] [Google Scholar]
- Liu T., Qian W.J., Gritsenko M.A., Camp D.G., 2nd, Monroe M.E., Moore R.J., Smith R.D. Human plasma N-glycoproteome analysis by immunoaffinity subtraction, hydrazide chemistry, and mass spectrometry. J. Proteome Res. 2005;4:2070–2080. doi: 10.1021/pr0502065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamo L.B., Suliman H.B., Giles B.L., Auten R.L., Piantadosi C.A., Nozik-Grayck E. Discordant extracellular superoxide dismutase expression and activity in neonatal hyperoxic lung. Am. J. Respir. Crit. Care Med. 2004;170:313–318. doi: 10.1164/rccm.200309-1282OC. [DOI] [PubMed] [Google Scholar]
- Nozik-Grayck E., Suliman H.B., Piantadosi C.A. Extracellular superoxide dismutase. Int. J. Biochem. Cell Biol. 2005;37:2466–2471. doi: 10.1016/j.biocel.2005.06.012. [DOI] [PubMed] [Google Scholar]
- Ohta H., Adachi T., Hirano K. Internalization of human extracellular- superoxide dismutase by bovine aortic endothelial cells. Free Rad. Biol. Med. 1994;16:501–507. doi: 10.1016/0891-5849(94)90128-7. [DOI] [PubMed] [Google Scholar]
- Oury T.D., Day B.J., Crapo J.D. Extracellular superoxide dismutase: a regulator of nitric oxide bioavailability. Lab. Invest. 1996a;75:617–636. [PubMed] [Google Scholar]
- Oury T.D., Day B.J., Crapo J.D. Extracellular superoxide dismutase in vessels and airways of humans and baboons. Free Radic. Biol. Med. 1996b;20:957–965. doi: 10.1016/0891-5849(95)02222-8. [DOI] [PubMed] [Google Scholar]
- Oury T.D., Schaefer L.M., Fattman C.L., Choi A., Weck K.E., Watkins S.C. Depletion of pulmonary EC-SOD after exposure to hyperoxia. Am J. Physiol. Lung Cell Mol. Physiol. 2002;283:L777–784. doi: 10.1152/ajplung.00011.2002. [DOI] [PubMed] [Google Scholar]
- Petersen S.V., Olsen D.A., Kenney J.M., Oury T.D., Valnickova Z., Thogersen I.B., Crapo J.D., Enghild J.J. The high concentration of Arg213 → Gly extracellular superoxide dismutase (EC-SOD) in plasma is caused by a reduction of both heparin and collagen affinities. Biochem. J. 2005;385:427–432. doi: 10.1042/BJ20041218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu K., Kim Y.H., Lee J.S., Jeon B., Kim T.Y. Increased yield of high-purity and active tetrameric recombinant human EC-SOD by solid phase refolding. J. Microbiol. Biotechnol. 2008;18:1648–1654. [PubMed] [Google Scholar]
- Suliman H.B., Ali M., Piantadosi C.A. Superoxide dismutase-3 promotes full expression of the EPO response to hypoxia. Blood. 2004;104:43–50. doi: 10.1182/blood-2003-07-2240. [DOI] [PubMed] [Google Scholar]
- Tan A.S., Berridge M.V. Superoxide produced by activated neutrophils efficiently reduces the tetrazolium salt, WST-1 to produce a soluble formazan: a simple colorimetric assay for measuring respiratory burst activation and for screening anti-inflammatory agents. J. Immunol. Methods. 2000;238:59–68. doi: 10.1016/s0022-1759(00)00156-3. [DOI] [PubMed] [Google Scholar]
- Tan R.J., Fattman C.L., Watkins S.C., Oury T.D. Redistribution of pulmonary EC-SOD after exposure to asbestos. J. Appl. Physiol. 2004;97:2006–2013. doi: 10.1152/japplphysiol.00480.2004. [DOI] [PubMed] [Google Scholar]
- Tibell L., Hjalmarsson K., Edlund T., Skogman G., Engstrom V., Marklund S.L. Expression of human extracellular superoxide dismutase in Chinese hamster ovary cells and characterization of the product. Proc. Natl. Acad. Sci. USA. 1987;84:6634–6638. doi: 10.1073/pnas.84.19.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun J.H., Kim K., Jung Y., Park J.H., Cho H.S., Lee W. Co-expression of human agouti-related protein enhances expression and stability of human melanocortin-4 receptor. Biochem. Biophys. Res. Commun. 2015;456:116–121. doi: 10.1016/j.bbrc.2014.11.044. [DOI] [PubMed] [Google Scholar]
- Zelko I.N., Mariani T.J., Folz R.J. Superoxide dismutase multigene family: a comparison of the CuZn-SOD (SOD1), Mn-SOD (SOD2), and EC-SOD (SOD3) gene structures, evolution, and expression. Free Radical. Biol. Med. 2002;33:337–349. doi: 10.1016/s0891-5849(02)00905-x. [DOI] [PubMed] [Google Scholar]