Abstract
Abiotic stresses such as drought and low temperature critically restrict plant growth, reproduction, and productivity. Higher plants have developed various defense strategies against these unfavorable conditions. CaPUB1 (Capsicum annuum Putative U-box protein 1) is a hot pepper U-box E3 Ub ligase. Transgenic Arabidopsis plants that constitutively expressed CaPUB1 exhibited drought-sensitive phenotypes, suggesting that it functions as a negative regulator of the drought stress response. In this study, CaPUB1 was over-expressed in rice (Oryza sativa L.), and the phenotypic properties of transgenic rice plants were examined in terms of their drought and cold stress tolerance. Ubi:CaPUB1 T3 transgenic rice plants displayed phenotypes hypersensitive to dehydration, suggesting that its role in the negative regulation of drought stress response is conserved in dicot Arabidopsis and monocot rice plants. In contrast, Ubi:CaPUB1 progeny exhibited phenotypes markedly tolerant to prolonged low temperature (4°C) treatment, compared to those of wild-type plants, as determined by survival rates, electrolyte leakage, and total chlorophyll content. Cold stress-induced marker genes, including DREB1A, DREB1B, DREB1C, and Cytochrome P450, were more up-regulated by cold treatment in Ubi:CaPUB1 plants than in wild-type plants. These results suggest that CaPUB1 serves as both a negative regulator of the drought stress response and a positive regulator of the cold stress response in transgenic rice plants. This raises the possibility that CaPUB1 participates in the cross-talk between drought and low-temperature signaling pathways.
Keywords: cold stress, cross-talk in stress response, drought stress, transgenic rice (Oryza sativa) plants, u-box E3 Ub ligase
INTRODUCTION
Rice is a staple crop worldwide and a typical monocot model plant. Like other plant species, rice is faced with a variety of environmental stresses throughout its life cycle. Drought is a major abiotic stress responsible for the reduction of rice productivity worldwide. In addition, rice is often exposed to unfavorable temperatures below 20°C in the sub-tropical and high-elevation zones, leading to chilling injury. These stresses cause stunted growth, weakened photosynthetic ability, reduced tillering, delayed heading, and greatly reduced agricultural production (Benjamin and Nielsen, 2006; Kim and Tai, 2011; Mackill and Lei, 1997; Praba et al., 2009).
Higher plants have developed molecular and physiological response mechanisms to tolerate such detrimental conditions. Under stress conditions, a series of signaling pathways regulate gene expression to produce various kinds of defensive proteins and molecules. Diverse sets of genes are induced under both drought and cold stress, but a subset of genes respond either only to dehydration or only to low temperature (Shinozaki et al., 2000). The presence of these suggests that there is cross-talk between drought and cold stress signaling pathways.
The ubiquitin (Ub)-mediated post-translational modification of proteins regulates numerous cellular processes, including cell cycle progression, environmental stress responses, and hormone signaling, (Lee and Kim, 2011; Lyzenga and Stone, 2012; Viersta, 2009; Yee and Goring, 2009). Recent studies show that U-box E3 Ub ligases are integrally involved in responses to biotic and abiotic stresses. In Arabidopsis, AtACRE276 (AtPUB17) was determined to be a U-box E3 ligase, which acts as a positive regulator of the hypersensitive response (HR) to pathogenic infection (Yang et al., 2006). AtPUB18/AtPUB19 and AtPUB22/AtPUB23 function as ABA-dependent and ABA-independent negative regulators in drought responses, respectively (Bergler and Hoth, 2011; Cho et al., 2008; Liu et al., 2011; Seo et al., 2012). In addition, three homologous AtPUB22, AtPUB23, and AtPUB24 E3 ligases were proposed as negative regulators of plant defense responses to pathogen infection (Stegmann et al., 2012; Trujillo et al., 2008). Rice OsSPL11 is a negative regulator in the pathogenic defense reaction and HR-associated cell death (Zeng et al., 2004). OsPUB15 plays a role in reducing cellular oxidative stress and cell death during seedling establishment in rice (Park et al., 2011).
CaPUB1 (Capsicum annuum Putative U-box protein 1) is a hot pepper U-box E3 Ub ligase (Cho et al., 2006). The CaPUB1 gene activity was enhanced by environmental stresses, including drought and cold, in hot pepper seedlings. Transgenic Arabidopsis plants that constitutively expressing CaPUB1 exhibited drought-sensitive phenotypes, suggesting that its function is to act as a negative regulator of the drought stress response.
In this report, CaPUB1 was over-expressed in rice (Oryza sativa L.) and the phenotypic properties of transgenic rice plants were examined in terms of their drought and cold stress tolerance. Ubi:CaPUB1 transgenic rice plants exhibited phenotypes hypersensitive to water deficit, implying that its function in drought stress responses is conserved in rice and Arabidopsis. In contrast, CaPUB1-overexpressing rice plants displayed phenotypes markedly more tolerant to prolonged low temperature (4°C) treatment than those of wild-type plants. These results suggest that CaPUB1 plays an opposite role in the response of rice to drought and cold stress. These introduced the possibility that CaPUB1 is involved in the cross-talk between drought and cold stress responses.
MATERIALS AND METHODS
Plant materials and growth conditions
Dry rice (Oryza sativa L. japonica variety ‘Dong-jin’) seeds were sequentially washed with 70% ethanol and distilled water. Rice seeds were sterilized with 0.4% NaClO solution for 30 min, rinsed with sterilized water, and germinated on half-strength Murashige and Skoog (MS) medium containing vitamins (Duchefa Biochemie, The Netherlands), 3% sucrose, and 0.7% phytoagar. Rice seedlings were grown for 10 days at 28°C under long-day conditions (16 h light/8 h dark), and then transplanted to soil in a growth chamber or greenhouse.
Generation of Ubi:CaPUB1 transgenic rice plants
To generate CaPUB1-overexpressing transgenic rice plants, the full-length CaPUB1 was introduced into the pGA2897 vector. The resulting binary vector was transformed into Agrobacterium tumefaciens strain LBA4404 via electroporation, as described by Shaw (1995), with some modifications. Wild-type rice embryonic calli were co-cultivated with Agrobacterium cells containing the Ubi:CaPUB1 construct via the standard transformation method, as recently described by Zeng et al. (2014). The transformed callus tissues were selected based on their hygromycin resistance. Transgenic T0 plants were regenerated from selected calli as described by Byun et al. (2015), and T3 plants were used for the phenotypic analysis.
Genomic DNA extraction and genomic Southern blot analysis
To identify independent lines of Ubi:CaPUB1 transgenic plants, a genomic Southern blot analysis was performed. Total genomic DNA was extracted from leaves using CTAB solution (2% CTAB, 2% PVP-40, 1.4 M NaCl, 100 mM Tris-HCl, pH 8.0, and 20 mM EDTA). Total DNAs from wild-type and transgenic plants were digested with EcoRI restriction enzyme (Roche, USA), precipitated, and separated by electrophoresis on 0.8% agarose gels. The gels were serially soaked in depurination, denaturation, and neutralization buffer. DNA on the gel was transferred to a nylon membrane (GE healthcare, Sweden) via the capillary transfer method. The DNA gel blot was hybridized with 32P-labeled hygromycin B phosphotransferase (HPT) probe under high-stringency hybridization and washing conditions (65°C). The autoradiography signals were visualized using the Bio-Imaging Analyzer (BAS2500; Fuji Film, Japan).
RT-PCR and quantitative RT-PCR analysis
Total RNA was extracted from various tissues from 2-week-old and 2-month-old mature wild-type and Ubi:CaPUB1 transgenic rice plants using Easy Spin Plant Total RNA Extraction kits (iN-tRON Biotechnology, Korea) according to the manufacturer’s protocol. Single-strand cDNA was synthesized from 2 μg of total RNA with TOPscript Reverse Transcriptase (Enzynomics, Daejeon, Korea) and oligo (dT) primers as described by Han et al. (2014). For RT-PCR analysis, CaPUB1 and OsUBQ10 gene were amplified from synthesized cDNA product using a gene-specific primer set. Real-time qRT-PCR was performed using an IQ5 light cycler (Bio-Rad, USA) with SYBR Green Premix Ex TaqII (Takara, Japan), as described previously (Shen et al., 2014). The OsACTIN was used as an internal control to normalize the data. The primer sets used in this study are listed in Table 1.
Table 1.
PCR primer sequences used for this article
| Primer Name | Sequence (5′-3′) | |
|---|---|---|
| RT-PCR | OsUBQ10 RT FW | 5′-ATGCAGATCTTTGTGAAGAC-3′ |
| OsUBQ10 RT RV | 5′-TTACTGACCACCACGGAGGC-3′ | |
| CaPUB1 RT FW | 5′-CAATCCCTTACGAGCATAGAG-3′ | |
| CaPUB1 RT RV | 5′-TTGGTAGTGTGGATGCTTCT-3′ | |
| Genomic Southern blot | HPT probe FW | 5′-GAGCCTGACCTATTGCATCTC-3′ |
| HPT probe RV | 5′-AGTACTTCTACACAGCCATCGG-3′ | |
| Real-time qRT-PCR | OsDREB1A FW | 5′-AGCCGTCCTCGTGTAGAAACAG-3′ |
| OsDREB1A RV | 5′-TCGTCACTGTAGTTCTCGTGCAG-3′ | |
| OsDREB1B FW | 5′-AGAGAGTCATCCATGGAGGTGGAG-3′ | |
| OsDREB1B RV | 5′-TCGTCTCCCTGAACTTGGTCCTTC-3′ | |
| OsDREB1C FW | 5′-AGCTACTGATGATCGCGAGTTGG-3′ | |
| OsDREB1C RV | 5′-AGGAGGAGCAAAGCTGGTTGAG-3′ | |
| OsCytochromeP450 FW | 5′-CCCACAACTGAGCATGGATGAG-3′ | |
| OsCytochromeP450 RV | 5′-GCAAGTGTGATGTGGTGTCATGC-3′ | |
| OsACTIN FW | 5′-CGTATGAGCAAGGAGATCAC-3′ | |
| OsACTIN RV | 5′-CACATCTGTTGGAAGGTGCT-3′ |
Stress treatments of rice plants
Wild-type and Ubi:CaPUB1 transgenic rice plants were grown for 6 weeks under normal growth conditions and subjected to drought or cold stress. To examine the water stress responses, 10-day-old wild-type and Ubi:CaPUB1 lines were transplanted into the same pot and grown in the growth chamber. Light-grown, 6-week-old plants were further grown without irrigation for 9 days, and recovered by re-watering. To investigate the effect of CaPUB1 on cold stress tolerance, 6-week-old plants were transferred to a growth chamber maintained at 4°C for 6 days, after which plants were recovered at 28°C.
Analysis of drought and cold stress phenotypes of wild-type and Ubi:CaPUB1 transgenic plants
For water loss rate measurement of wild-type and Ubi:CaPUB1 transgenic lines, the cut aerial parts of 5-week-old plants were incubated at 25°C under dim light and their weights were estimated at various time points. The water loss rates were expressed as a percentage of initial fresh weights as described previously (Cho et al., 2011).
Total chlorophyll (chlorophyll a + chlorophyll b) was extracted from drought and cold stress-treated wild-type and Ubi:CaPUB1 transgenic rice leaves as previously described (Lichtenthaler, 1987) with slight modifications. Chlorophyll content was estimated with a spectrophotometer (model DU800; Beckman Coulter). Chlorophyll a and chlorophyll b were measured at 664.2 nm and 648.6 nm, respectively. Total chlorophyll content was calculated using the following formula: chlorophyll a + chlorophyll b (mg/g DW) = 8.1 × (5.24 A648.6 + 22.24 A664.2)/DW.
An electrolyte leakage assay was conducted with the leaves of 5-week-old wild-type and Ubi:CaPUB1 rice plants after 7 days of 4°C treatment. The conductivity of each sample was estimated before and after autoclaving with a conductivity meter (Orion Star A212, Thermo Scientific, USA).
RESULTS
Generation of Ubi:CaPUB1 transgenic rice plants
To investigate the role of CaPUB1 in the environmental stress responses of crop plants, CaPUB1 was constitutively expressed in rice under the control of the maize ubiquitin promoter (Ubi) via the Agrobacterium-mediated transformation method (Fig. 1A). The T0 transgenic plants regenerated from callus tissue were isolated according to their hygromycin resistance (Fig. 1A). Based on the semi-quantitative RT-PCR analysis, the mRNA level of CaPUB1 was markedly enhanced in transgenic (lines #1 and #2) rice plants (Fig. 1B). The genomic Southern blot analysis indicated that these CaPUB1-overexpressing transgenic plants were independent lines and both possessed a single copy of the transgene (Fig. 1C). Next, we examined the developmental phenotypes of T3 Ubi:CaPUB1 transgenic lines and compared them to those of wild-type plants. There were no detectable phenotypic differences between the transgenic and wild-type rice plants under normal growth conditions (Fig. 1D). These T3 CaPUB1-overexpressing transgenic plants were used for the subsequent phenotypic analysis.
Fig. 1.
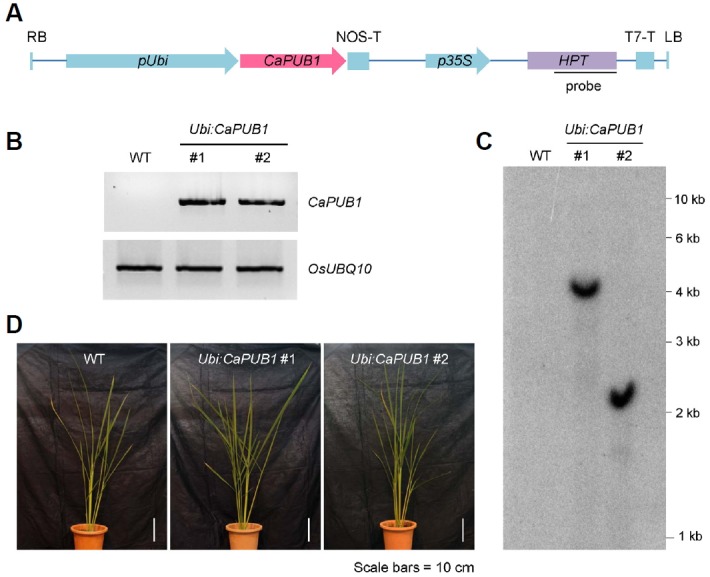
Construction of CaPUB1-overexpressing transgenic rice plants. (A) Schematic representation of the CaPUB1 overexpression binary vector construct. RB, right border; pUbi, maize ubiquitin promoter; NOS-T, NOS terminator; p35S, 35S CaMV promoter; Hpt, hygromycin phosphotransferase; T7-T, T7 terminator; LB, left border. A solid line indicates a DNA probe for genomic Southern blot analysis. (B) RT-PCR analysis of wild-type and Ubi:CaPUB1 T3 transgenic (lines #1 and #2) rice plants. OsUBQ10 was used as a loading control. (C) Identification of two independent transgenic lines by DNA gel blot analysis. Genomic DNA was isolated from mature leaf tissues and digested with EcoRI restriction enzyme. The DNA gel blot was hybridized with 32P-labeled HPT probe. (D) Morphology of 3-month-old wild-type and Ubi:CaPUB1 T3 transgenic rice plants under normal growth conditions. Scale bars = 10 cm.
Over-expression of CaPUB1 in rice decreases tolerance to drought stress
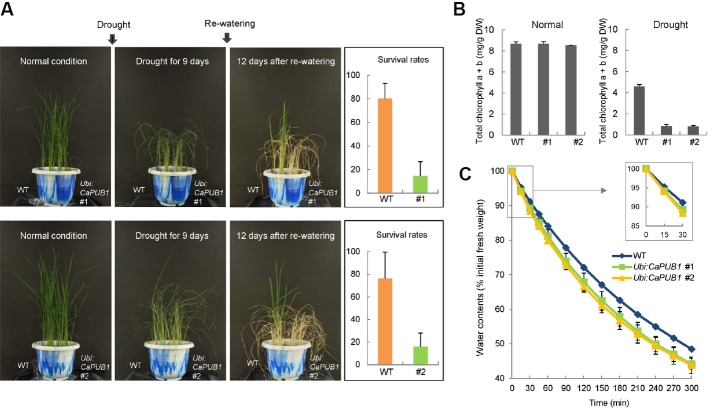
To elucidate the effect of CaPUB1 overexpression on abiotic stress responses in rice, we assessed drought stress tolerance. Light-grown, 6-week-old wild-type and transgenic (lines #1 and #2) plants were sown in the same pot and grown for 9 days without irrigation. These water-stressed plants were then irrigated normally. As shown in Fig. 2A, wild-type plants exhibited 76.3–80.4% survival rates after re-watering. However, only 14.6% of Ubi:CaPUB1 line #1 and 15.9% of Ub:CaPUB1 line #2 survived.
Fig. 2.
The Ubi:CaPUB1 transgenic rice plants displayed decreased tolerance to drought stress. (A) Light-grown, 6-week-old wild-type and T3 Ubi:CaPUB1 transgenic rice plants were subjected to drought stress (left panel). Survival rates of wild-type and CaPUB1-overexpressing transgenic lines were determined after drought stress. Data represent means ± SD (n = 38 plants) from four biological replicates for line #1 and five biological replicates for line #2 (right panel). (B) Total chlorophyll content of wild-type and CaPUB1-overexpressing plants before and after drought stress. Wild-type and Ubi:CaPUB1 transgenic plants were grown for 6 weeks and further grown with or without irrigation. Total amounts of chlorophyll a + chlorophyll b in leaf discs were measured. Data indicate means ± SD (n = 20 plants) from two independent experiments. (C) CaPUB1-overexpressing rice plants lost water more rapidly than wild-type plants during drought stress. Aerial parts of 5-week-old wild-type and CaPUB1-overexpressing T3 transgenic plants were detached and their fresh weights were measured at indicated time points. Data represent means ± SD from three independent experiments.
Consistent with survival rates, after drought stress the total chlorophyll content of wild-type leaves was much higher than that of CaPUIB1-overexpressing progeny. Wild-type and Ubi:CaPUB1 leaves contained very similar amounts of chlorophyll a + chlorophyll b (8.5 ± 0.2 mg/g DW) under normal growth conditions (Fig. 2B). The chlorophyll content of detached wild-type leaves was reduced to 4.6 ± 0.2 mg/g DW after drought stress. However, Ubi:CaPUB1 transgenic lines #1 and #2 exhibited pale green and bleached leaves, and their total chlorophyll contents were only 0.9 ± 0.1 mg/g DW and 0.8 ± 0.1 mg/g DW, respectively, after drought stress (Fig. 2B).
In addition, during dehydration CaPUB1-overexpressing rice plants lost water more rapidly than wild-type plants. The aerial parts of 5-week-old wild-type and transgenic rice plants were detached, incubated for 0–5 h at room temperature, and their water loss rates were monitored. After 1 h incubation, the aboveground parts of the wild-type and Ubi:CaPUB1 (lines #1 and #2) plants retained approximately 84% and 80–81% of their fresh weights, respectively (Fig. 2C). After 4 h incubation, wild-type leaves lost approximately 45% of their starting weights, while those of transgenic lines lost almost 50% of their fresh weights (Fig. 2C). These results indicate that more rapid water loss occurred in Ubi:CaPUB1 transgenic lines than in wild-type rice plants. Taken together, the over-expression of CaPUB1 decreased drought-stress tolerance in rice, suggesting that negative role played by U-box E3 ligase CaPUB1 in response to drought stress is conserved in Arabidopsis (Cho et al., 2006) and rice.
Ubi:CaPUB1 transgenic rice plants are more tolerant to cold stress than wild-type plants
Because the CaPUB1 transcript levels were elevated by cold temperature (4°C) as well as dehydration (Cho et al., 2006), we next examined whether the constitutive expression of CaPUB1 alters response to cold stress in rice. Both wild-type and CaPUB1-overexpressors were grown at 28°C for 6-weeks and subjected to cold stress (4°C) for 6 days. Then, they were transferred to a 28°C growth chamber and their growth was observed. After the cold treatment, all wild-type plants examined wilted, lost their chlorophyll, were unable to grow, and died, with survival rates of 0–2.8%. In contrast, most of the Ubi:CaPUB1 transgenic rice plants displayed a healthy morphology and resumed growth. The survival rates of Ubi:CaPUB1 transgenic lines #1 and #2 were 60.9% and 64.5%, respectively (Fig. 3A).
Fig. 3.
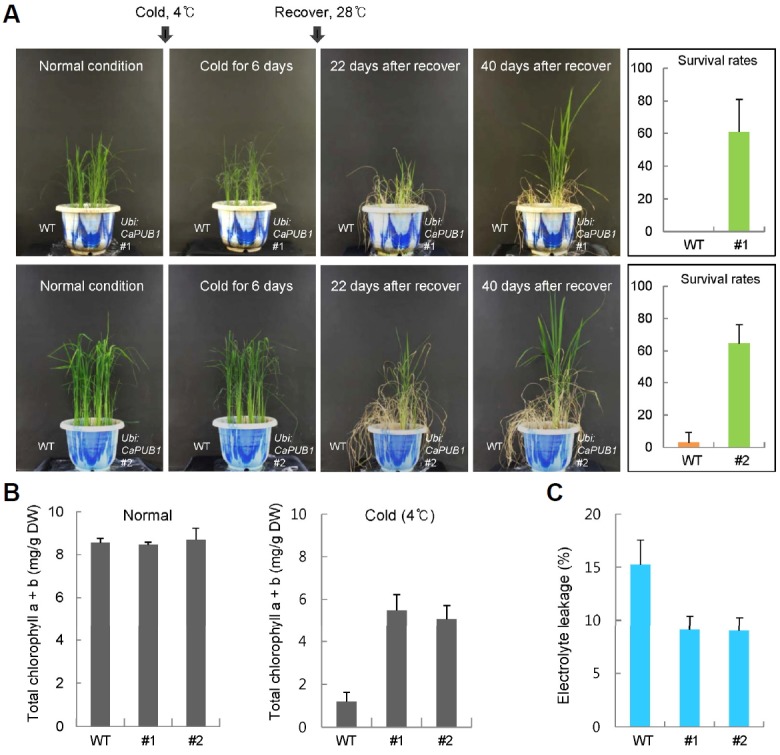
Ubi:CaPUB1 transgenic rice plants were tolerant to cold stress. (A) Overexpression of CaPUB1 conferred enhanced tolerance to cold stress on transgenic rice plants. Wild-type and T3 transgenic plants were grown for 6 weeks under normal growth conditions. After another 6 days of exposure to 4°C, plants were transferred to 28°C for recovery (left panel). The cold stress survival rates of wild-type and Ubi:CaPUB1 transgenic rice plants were monitored. Data represent means ± SD (n > 36 plants) from five biological replicates (right panel). (B) Total chlorophyll content of wild-type and CaPUB1-overexpressing plants before and after cold stress. Light-grown, 6-week-old wild-type and Ubi:CaPUB1 transgenic rice plants were grown for another 6 days under normal or low temperature conditions. Amount of total chlorophyll a + chlorophyll b in leaves was estimated. Data indicate the means ± SD (n = 30 plants) from three independent experiments. (C) Electrolyte leakage assay. Electrolyte leakage values were determined after 4°C treatment. Data represent the means ± SD (n = 30 plants) from three independent experiments.
These results were further validated by measuring the total chlorophyll content. Both cold stress-treated wild-type and transgenic plants were allowed to recover for 7 days, after which their leaf chlorophyll contents were determined. Most wild-type leaves lost their green color after stress treatment and contained 1.2 ± 0.4 mg/g DW of total chlorophyll. However, Ubi:CaPUB1 leaves retained 5.5 ± 0.7 mg/g DW (line #1) and 5.1 ± 0.6 mg/g DW (line #2) of chlorophyll a + chlorophyll b, respectively after low temperature treatment (Fig. 3B).
Subsequently, the mature leaves of 5-week-old wild-type and CaPUB1-overexpressing plants were exposed to 4°C for 7 days, after which their electrolyte leakage was determined. The results revealed that after cold treatment, the Ubi:CaPUB1 leaves displayed lower conductivity means (8.8% for lines #1 and #2) than wild-type leaves (14.7%) (Fig. 3C). These results suggest that Ubi:CaPUB1 transgenic rice plants were more tolerant of cold stress (4°C) than wild-type plants.
Cold stress-induced marker genes were more enhanced in Ubi:CaPUB1 transgenic plants than in wild-type plants under low temperature conditions
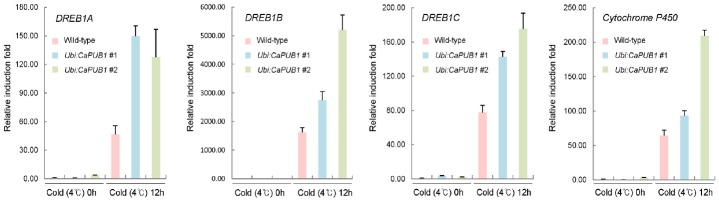
To investigate whether CaPUB1 affects the expression profiles of cold-stress related genes in rice, we conducted real-time quantitative RT-PCR (qRT-PCR). Both wild-type and transgenic lines were subjected to cold stress (4°C) for 0 or 24 h, after which total RNA was isolated from the shoot tissue. The expression patterns of cold stress-induced marker genes, including DREB1A, DREB1B, DREB1C, and Cytochrome P450, were observed by qRT-PCR using gene-specific primer sets (Table 1). The results in Figure 4 demonstrate that there was no significant difference between the transcripts levels of these four marker genes in wild-type and transgenic lines under normal growth conditions. However, after cold-stress treatment, all four marker genes displayed higher induction level in CaPUB1-overexpressors than in wild-type plants. These results suggest that the cold stress-tolerant phenotypes of the Ubi:CaPUB1 plants were correlated with increased levels of cold stress-induced gene expression.
Fig. 4.
Real-time qRT-PCR analysis of cold stress-inducible genes in wild-type and Ubi:CaPUB1 transgenic plants. Total RNA was extracted from wild-type and two CaPUB1-overexpressing independent lines before and after cold treatment (4°C). Total RNA was analyzed by qRT-PCR using the gene-specific primer sets listed in Table 1. The graphs indicate the relative expression levels of OsDREB1A, OsDREB1B, OsDREB1C, and OsCytochromeP450 in wild-type and Ubi:CaPUB1 plants. The mean values of the three technical replicates were normalized to the level of OsACTIN mRNA as an internal control.
DISCUSSION
CaPUB1 was previously identified as an abiotic stress-induced gene that encodes a U-box E3 Ub ligase in hot pepper (Capsicum annuum c.v. pukang) (Cho et al., 2006). Ectopic expression of CaPUB1 in the dicot model plant Arabidopsis results in phenotypes that exhibit hyper-sensitive responses to dehydration, suggesting that it plays a negative role in the drought stress response (Cho et al., 2006). In this study, CaPUB1 was constitutively overexpressed in rice, a monocot model plant, and CaPUB1-overexpressing transgenic plants (Ubi:CaPUB1) were characterized in terms of their drought and cold stress responses. The results reveal that Ubi:CaPUB1 lines are more sensitive to water deficit than wild-type rice plants, as determined by survival rates, total leaf chlorophyll content, and the rate of water loss from above-ground parts (Fig. 2). These results are consistent with the drought-sensitive phenotypes of CaPUB1-overexpressing Arabidopsis plants, implying that the role of the molecule as a negative regulator of the response to drought is conserved in both dicot Arabidopsis and monocot rice plants.
In contrast to drought stress responses, the Ubi:CaPUB1 transgenic plants exhibited phenotypes highly tolerant to low temperature stress. After 6 days of exposure to 4°C, the CaPUB1-overexpressing lines displayed healthy green leaves with larger amounts of total chlorophyll and lower rates of electrolyte leakage, along with over 60% survival rates, as compared to wild type plants (0–2.8% survival rates) (Fig. 3). Thus, during the extended low temperature treatment Ubi:CaPUB1 plants appeared to retain a higher level of photosynthetic ability and experience less membrane disruption than wild-type plants. Based on these results, we concluded that the overexpression of CaPUB1 conferred tolerance to cold stress on transgenic rice plants.
qRT-PCR analysis showed that after cold treatment, the expression levels of OsDREB1A/CBF3, OsDREB1B/CBF1, and OsDREB1C/CBF2 were much higher in Ubi:CaPUB1 transgenic lines than in wild-type plants (Fig. 4). These transcription factor genes are induced during an early stage of the cold response, in order to activate diverse sets of cold-responsive genes in rice (Dubouzet et al., 2003; Ito et al., 2006; Su et al., 2010). A number of studies proposed that DREBs serve as a major regulatory network acting in response to low temperature conditions (Agarwal et al., 2013; Thomashow, 2010; Zhang et al., 2013). The over-expression of DREBs increased tolerance to cold stress in transgenic rice plants, in terms of growth, ROS scavenging, and hormone metabolism (Ito et al., 2006; Wang et al., 2008; Zhang et al., 2013). However, the constitutive expression of DREBs often resulted in dwarf phenotypes even under normal growth conditions (Dubouzet et al., 2003; Gilmour et al., 2000; Xu et al., 2011). It is worth noting that Ubi:CaPUB1 transgenic rice plants did not exhibit detectable growth retardation (Fig. 1D). This could be due to that, in contrast to constitutive expression, the basal levels of OsDREB1A/CBF3, OsDREB1B/CBF1, and OsDREB1C/CBF2 were very low under normal conditions, but were more up-regulated by the cold treatment in Ubi:CaPUB1 plants than in wild-type plants.
In addition, Ubi:CaPUB1 showed the enhanced expression level of Cytochrome P450 (CYP709C9) after 4°C treatment. Although the roles of Cytochrome P450 (CYP709C9) have not been clearly understood, it was previously reported that it could function as a potential marker gene for oxidative stress tolerance in rice (Liu et al., 2010). Consistent with this notion, Cytochrome P450 (CYP709C9) was induced by exposure to low temperatures, methyl viologen, and arsenate treatments (Su et al., 2010; Tripathi et al., 2012). Furthermore, Cytochrome P450 (CYP709C9) was down-regulated in Ubi:OsSPX1-antisense transgenic rice plants that displayed phenotypes hypersensitive to cold stress (Wang et al., 2013).
Thus, we speculate that CaPUB1 increases the expression of DREB cold-responsive transcription factors and cytochrome P450 by an as-yet unidentified mechanism, which, in turn, confers tolerance to cold stress on transgenic rice plants. Because CaPUB1 is a U-box E3 Ub ligase and possesses neither a DNA binding domain nor a transcription activator domain, we ruled out the possibility that CaPUB1 directly binds to the upstream regions of OsDREBs and Cytochrome P450 and induces them. Alternatively, we hypothesized that CaPUB1 ubiquitinates and triggers the proteasome-dependent degradation of a negative factor that inhibits the expression of cold-responsive genes.
Ubi:CaPUB1 transgenic rice plants displayed similar morphological phenotypes as compared to wild-type rice under normal growth conditions (Fig. 1D). It was previously reported that over-expression of CaPUB1 in Arabidopsis caused enhanced vegetative growth and early bolting (Cho et al., 2006). These results suggest that ectopic expression of CaPUB1 may exert different effects on growths of monocot rice and dicot Arabidopsis plants.
A database search revealed three potential CaPUB1 orthologs, OsPUB31, OsPUB32, and OsPUB33 with 41%, 43%, and 44% amino-acid identities, respectively, in rice (Supplementary Fig. S1). Cellular functions of these three rice U-box E3 homologs are not currently unraveled. Elucidation of the roles of rice U-box E3 Ub ligases will provide more detailed responses to drought and cold stresses in crop plants.
The over-expression of CaPUB1 decreased tolerance to water deficit, but enhanced tolerance to cold stress in rice (Figs. 2 and 3). This raises the possibility that CaPUB1 is involved in the cross-talk of the signaling pathways of two different environmental stresses: drought and cold. Although a number of studies proposed that diverse E3 Ub ligases play regulatory roles in response to abiotic stresses, understanding their roles in the cross-talk between different stress responses is still rudimentary. In Arabidopsis, the suppression of AtATL78, a RING-type E3 Ub ligase, resulted in increased cold tolerance, but decreased drought tolerance, suggesting that it plays a role in the crosstalk between the drought and cold stress responses (Kim and Kim, 2013). Thus, additional experiments are required to elucidate the detailed mechanisms, by which CaPUB1 plays an opposite role in the drought stress and cold stress responses of transgenic rice plants.
In conclusion, our data suggest that the constitutive expression of a U-box E3 ubiquitin ligase CaPUB1 increases tolerance to cold stress and decreases tolerance to drought stress in rice, a monocot model crop plant. Our study also suggests that CaPUB1 could be utilized as a genetic material to investigate the novel regulatory module for two environmental stresses, drought and low temperature. In addition, enhanced tolerance to cold stress of CaPUB1-overexpressing rice plants may have an advantage for preventing decrease in rice production caused by chilling injury during their vegetative stage in temperate regions.
Acknowledgments
This work was supported by grants from the National Center for GM Crops (Project No.PJ01113801 funded by the Rural Development Administration, Republic of Korea) and the National Research Foundation (Project No. 2014R1A2A2A01003891) to W.T.K., and from the Korea Institute of Science and Technology Information to B.G.K.
Footnotes
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Agarwal P.K., Agarwal P., Reddy M., Sopory S.K. Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep. 2006;25:1263–1274. doi: 10.1007/s00299-006-0204-8. [DOI] [PubMed] [Google Scholar]
- Benjamin J.G., Nielsen D.C. Water deficit effects on root distribution of soybean, field pea and chickpea. Field Crops Res. 2006;97:248–253. [Google Scholar]
- Bergler J., Hoth S. Plant U-box armadillo repeat proteins AtPUB18 and AtPUB19 are involved in salt inhibition of germination in Arabidopsis. Plant Biol. (Stuttg) 2011;13:725–730. doi: 10.1111/j.1438-8677.2010.00431.x. [DOI] [PubMed] [Google Scholar]
- Byun M.Y., Lee J., Cui L.H., Kang Y., Oh T.K., P H., Lee H., Kim W.T. Constitutive expression of DaCBF7, an Antarctic vascular plant, Deschampsia antarctica CBF homolog, resulted in improved cold tolerance in transgenic rice plants. Plant Science. 2015;236:61–74. doi: 10.1016/j.plantsci.2015.03.020. [DOI] [PubMed] [Google Scholar]
- Cho S.K., Chung H.S., Ryu M.Y., Park M.J., Lee M.M., Bahk Y.Y., Kim J., Pai H.S., Kim W.T. Heterologous expression and molecular and cellular characterization of CaPUB1 encoding a hot pepper U-Box E3 ubiquitin ligase homolog. Plant Physiol. 2006;142:1664–1682. doi: 10.1104/pp.106.087965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S.K., Ryu M.Y., Song C., Kwak J.M., Kim W.T. Arabidopsis PUB22 and PUB23 are homologous U-Box E3 ubiquitin ligases that play combinatory roles in response to drought stress. Plant Cell. 2008;20:1899–1914. doi: 10.1105/tpc.108.060699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S.K., Ryu M.Y., Seo D.H., Kang B.G., Kim W.T. The Arabidopsis RING E3 ubiquitin ligase AtAIRP2 plays combinatory roles with AtAIRP1 in abscisic acid-mediated drought stress responses. Plant physiol. 2011;157:2240–2257. doi: 10.1104/pp.111.185595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubouzet J.G., Sakuma Y., Ito Y., Kasuga M., Dubouzet E.G., Miura S., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J. 2003;33:751–763. doi: 10.1046/j.1365-313x.2003.01661.x. [DOI] [PubMed] [Google Scholar]
- Gilmour S.J., Sebolt A.M., Salazar M.P., Everard J.D., Thomashow M.F. Over-expression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol. 2000;124:1854–1865. doi: 10.1104/pp.124.4.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M., Kim C.-Y., Lee J., Lee S.-K., Jeon J.-S. OsWRKY42 represses OsMT1d and induces reactive oxygen species and leaf senescence in rice. Mol. Cells. 2014;37:532–539. doi: 10.14348/molcells.2014.0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y., Katsura K., Maruyama K., Taji T., Kobayashi M., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol. 2006;47:141–153. doi: 10.1093/pcp/pci230. [DOI] [PubMed] [Google Scholar]
- Kim S.I., Tai T.H. Evaluation of seedling cold tolerance in rice cultivars: a comparison of visual ratings and quantitative indicators of physiological changes. Euphytica. 2011;178:437–447. [Google Scholar]
- Kim S.J., Kim W.T. Suppression of Arabidopsis RING E3 ubiquitin ligase AtATL78 increases tolerance to cold stress and decreases tolerance to drought stress. FEBS Lett. 2013;587:2584–2590. doi: 10.1016/j.febslet.2013.06.038. [DOI] [PubMed] [Google Scholar]
- Lee J-H., Kim W.T. Regulation of abiotic stress signal transduction by E3 ubiquitin ligases in Arabidopsis. Mol. Cells. 2011;31:201–208. doi: 10.1007/s10059-011-0031-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler H.K. Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. Methods Enzymol. 1987;148:350–382. [Google Scholar]
- Liu F., Xu W., Wei Q., Zhang Z., Xing Z., Tan L., Di C., Yao D., Wang C., Tan Y., et al. Gene expression profiles deciphering rice phenotypic variation between Nipponbare (Japonica) and 93-11 (Indica) during oxidative stress. PLoS One. 2010;5:e8632. doi: 10.1371/journal.pone.0008632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.C., Wu Y.R., Huang X.H., Sun J., Xie Q. AtPUB19, a U-box E3 ubiquitin ligase, negatively regulates abscisic acid and drought responses in Arabidopsis thaliana. Mol. Plant. 2011;4:938–946. doi: 10.1093/mp/ssr030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyzenga W.J., Stone S.L. Abiotic stress tolerance mediated by protein ubiquitination. J. Exp. Bot. 2012;63:599–616. doi: 10.1093/jxb/err310. [DOI] [PubMed] [Google Scholar]
- Mackill D.J., Lei X. Genetic variation for traits related to temperature adaptation of rice cultivars. Crop Sci. 1997;37:1340–1346. [Google Scholar]
- Park J.J., Yi J., Yoon J., Cho L.H., Ping J., Jeong H.J., Cho S.K., Kim W.T., An G. OsPUB15, an E3 ubiquitin ligase, functions to reduce cellular oxidative stress during seedling establishment. Plant J. 2011;65:194–205. doi: 10.1111/j.1365-313X.2010.04416.x. [DOI] [PubMed] [Google Scholar]
- Praba M.L., Cairns J.E., Babu R.C., Lafitte H.R. Identification of physiological traits underlying cultivar differences in drought tolerance in rice and wheat. J. Agron. Crop Sci. 2009;195:30–46. [Google Scholar]
- Seo D.H., Ryu M.Y., Jammes F., Hwang J.H., Turek M., Kang B.G., Kwak J.M., Kim W.T. Roles of four Arabidopsis U-box E3 ubiquitin ligases in negative regulation of abscisic acid-mediated drought stress responses. Plant Physiol. 2012;160:556–568. doi: 10.1104/pp.112.202143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw C.H. In Plant gene transfer and expression protocols. Vol. 49. Springer; New York: 1995. Introduction of cloning plasmids into Agrobacterium tumefaciens; pp. 33–37. [DOI] [PubMed] [Google Scholar]
- Shen C., Li D., He R., Fang Z., Xia Y., Gao J., Shen H., Cao M. Comparative transcriptome analysis of RNA-seq data for cold-tolerant and cold-sensitive rice genotypes under cold stress. J. Plant Biol. 2014;56:337–348. [Google Scholar]
- Shinozaki K., Yamaguchi-Shinozaki K. Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 2000;3:217–223. [PubMed] [Google Scholar]
- Stegmann M., Anderson R.G., Ichimura K., Pecenkova T., Reuter P., Žárský V., McDowell J.M., Shirasu K., Trujillo M. The ubiquitin ligase PUB22 targets a subunit of the exocyst complex required for PAMP-triggered responses in Arabidopsis. Plant Cell. 2012;24:4703–4716. doi: 10.1105/tpc.112.104463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su C.F., Wang Y.C., Hsieh T.H., Tseng T.H., Lu C.A., Tseng T.H., Yu S.M. A novel MYBS3-dependent pathway confers cold tolerance in rice. Plant Physiol. 2010;153:145–158. doi: 10.1104/pp.110.153015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow M.F. Molecular basis of plant cold acclimation: insights gained from studying the CBF cold response pathway. Plant Physiol. 2010;154:571–577. doi: 10.1104/pp.110.161794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi R.D., Tripathi P., Dwivedi S., Dubey S., Chatterjee S., Chakrabarty D., Trivedi P.K. Arsenomics: omics of arsenic metabolism in plants. Front. Physiol. 2012;3:275. doi: 10.3389/fphys.2012.00275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo M., Ichimura K., Casais C., Shirasu K. Negative regulation of PAMP-triggered immunity by an E3 ubiquitin ligase triplet in Arabidopsis. Curr. Biol. 2008;18:1396–1401. doi: 10.1016/j.cub.2008.07.085. [DOI] [PubMed] [Google Scholar]
- Vierstra R.D. The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell. Biol. 2009;110:385–397. doi: 10.1038/nrm2688. [DOI] [PubMed] [Google Scholar]
- Wang Q., Guan Y., Wu Y., Chen H., Chen F., Chu C. Overexpression of a rice OsDREB1F gene increases salt, drought, and low temperature tolerance in both Arabidopsis and rice. Plant Mol. Biol. 2008;67:589–602. doi: 10.1007/s11103-008-9340-6. [DOI] [PubMed] [Google Scholar]
- Wang C., Wei Q., Zhang K., Wang L., Liu F., Zhao L., Tan Y., Di C., Yan H., Yu J., et al. Down-regulation of OsSPX1 causes high sensitivity to cold and oxidative stresses in rice seedlings. PLoS One. 2013;8:e81849. doi: 10.1371/journal.pone.0081849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M., Li L., Fan Y., Wan J., Wang L. ZmCBF3 over-expression improves tolerance to abiotic stress in transgenic rice (Oryza sativa) without yield penalty. Plant Cell Rep. 2011;30:1949–1957. doi: 10.1007/s00299-011-1103-1. [DOI] [PubMed] [Google Scholar]
- Yang C.W., González-Lamothe R., Ewan R.A., Rowland O., Yoshioka H., Shenton M., Ye H., O'Donnell E., Jones J.D., Sadanandom A. The E3 ubiquitin ligase activity of Arabidopsis PLANT U-BOX17 and its functional tobacco homolog ACRE276 are required for cell death and defense. Plant Cell. 2006;18:1084–1098. doi: 10.1105/tpc.105.039198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee D., Goring D.R. The diversity of plant U-box E3 ubiquitin ligases: from upstream activators to downstream target substrates. J. Exp. Bot. 2009;60:1109–1121. doi: 10.1093/jxb/ern369. [DOI] [PubMed] [Google Scholar]
- Zeng L.R., Qu S., Bordeos A., Yang C., Baraoidan M., Yan H., Xie Q., Nahm B.H., Leung H., Wang G.L. Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell. 2004;16:2795–2808. doi: 10.1105/tpc.104.025171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng D-E., Hou P., Xiao F., Liu Y. Overexpressing a novel RING-H2 finger protein gene, OsRHP1, enhances drought and salt tolerance in rice (Oryza sativa L.) J. Plant Biol. 2014;57:357–365. [Google Scholar]
- Zhang Q., Jiang N., Wang G.L., Hong Y., Wang Z. Advances in understanding cold sensing and the cold-responsive network in rice. Adv. Crop Sci. Tech. 2013;1:104. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.