Abstract
In metastatic breast cancer, the acquisition of malignant traits has been associated with the increased rate of cell growth and division, mobility, resistance to chemotherapy, and invasiveness. While screening for the key regulators of cancer metastasis, we observed that neurotrophin receptor TrkB is frequently overexpressed in breast cancer patients and breast cancer cell lines. Additionally, we demonstrate that TrkB expression and clinical breast tumor pathological phenotypes show significant correlation. Moreover, TrkB expression was significantly upregulated in basal-like, claudin-low, and metaplastic breast cancers from a published microarray database and in patients with triple-negative breast cancer, which is associated with a higher risk of invasive recurrence. Interestingly, we identified a new TrkB-regulated functional network that is important for the tumorigenicity and metastasis of breast cancer. We demonstrated that TrkB plays a key role in regulation of the tumor suppressors Runx3 and Keap1. A markedly increased expression of Runx3 and Keap1 was observed upon knockdown of TrkB, treatment with a TrkB inhibitor, and in TrkB kinase dead mutants. Additionally, the inhibition of PI3K/AKT activation significantly induced Runx3 and Keap1 expression. Furthermore, we showed that TrkB enhances metastatic potential and induces proliferation. These observations suggest that TrkB plays a key role in tumorigenicity and metastasis of breast cancer cells through suppression of Runx3 or Keap1 and that it is a promising target for future intervention strategies for preventing tumor metastasis and cancer chemoprevention.
Keywords: breast cancer, keap1, metastasis, runx3, TrkB
INTRODUCTION
TrkB belongs to the neurotrophin receptor family, and its primary ligand is neurotrophin, brain-derived neurotrophic factor (BDNF). This receptor plays an important role in the context of neuronal proliferation, differentiation, and survival (Chao and Bothwell, 2002); recently, however, it has also been associated with a variety of human cancers, ranging from prostate cancer to lung cancer (Chao and Bothwell, 2002; Dionne et al., 1998; Eggert et al., 2001; Miknyoczki et al., 1999; Zhang et al., 2010). Several recent reports show that ectopic TrkB overexpression in rat and human kidney epithelial cells acts as a potent anoikis suppressor through AKT activation (Douma et al., 2004; Geiger and Peeper, 2005). Additionally, this ectopic overexpression promotes epithelial-to-mesenchymal transition (EMT) through induction of Zeb1, an E-cadherin repressor (Smit and Peeper, 2011). Recent studies have demonstrated that the TrkB gene is mutated within its kinase-encoding domain in colorectal and lung cancer (Bardelli et al., 2003; Ding et al., 2008; Marchetti et al., 2008). The effect of these mutations on TrkB kinase function remains to be investigated. Furthermore, TrkB enhances resistance to therapeutic agents (Yilmaz et al., 2010). These results indicate that TrkB activation/overexpression may play a crucial role in the initiation, progression, and metastasis of many tumors, and these observations suggest that TrkB remains an attractive therapeutic target for anti-metastatic therapies. Although TrkB may play an important role in cancer, TrkB signaling mechanisms that induce and then maintain tumorigenicity and metastatic potential of the breast cancer cells have remained poorly understood.
Other recent studies described that Runx3 and Keap1, as tumor suppressors, are frequently downregulated in a number of malignancies. Runx3 is frequently inactivated in breast cancer by hypermethylation of the Runx3 promoter, and it inhibits estrogen receptor α-dependent (ER-α) transactivation by reducing the stability of this receptor (Chen, 2012; Huang et al., 2012). In addition, hypermethylation of Keap1 promoter in breast and colorectal cancer suppresses its expression. Inactivation or somatic mutations of Keap1 are associated with poor survival of breast cancer patients (Hanada et al., 2012; Hartikainen et al., 2015). This raises the possibility that TrkB may play a role in the regulation of Runx3 and Keap1 during the process of tumorigenesis and metastasis, and may help in disseminating cancer cells.
Together, these diverse lines of evidence suggest a possible link between the loss of tumor suppression and TrkB-mediated tumor metastasis. In this report, we identify a signaling network present in metastatic cells that is regulated and coordinated by TrkB. Surprisingly, we found that TrkB is overexpressed in human breast cancers and that it acts as a key inhibitor of Runx3 and Keap1-mediated tumor suppression. Our study provides molecular insight into the tumor metastasis and has important implications in elucidating oncogenic processes.
MATERIALS AND METHODS
Cell culture and reagents
HMLEs (immortalized human mammary epithelial cells), human breast cancer (MCF10A, ZR-75-1, BT-549, SUM149, MDA-MB-231, MDA-MB-435, MDA-MB-468, and Hs578T), and canine kidney (MDCK) cell lines were maintained as previously described (Yang et al., 2004). The protein kinase inhibitor K252a and PI3K inhibitor LY294002 were purchased from Calbiochem.
Human breast tumor samples
RNA and proteins extracted from human breast normal and tumor samples were obtained from the Gangnam Severance Hospital after approval by the Institutional review board and the ethics committee of Gangnam Severance Hospital (IRB approval number: 3-2011-0191).
Plasmids
pLKO shAKT1 lentiviral vector were obtained from Sigma-Aldrich. shRNA that did not match any known human cDNA was used as a control.
Soft agar assay, anchorage-independent cell growth assay, wound healing assay, and matrigel invasion assay
All assays were performed as previously described (Jin et al., 2010; Lu et al., 2009).
RT-PCR
The primer sequences used to amplify the investigated genes are listed in the supplemental table (Supplementary Table S1). Total RNA was isolated using RNeasy Mini Kits (Qiagen) according to the manufacturer’s instructions and reverse transcription was done using a One-Step RT-PCR kit (Qiagen). The resulting PCR products were separated on 1% agarose gels and visualized.
Immunohistochemistry
A tissue microarray slide (IMX-364) was purchased from Super BioChips. Briefly, after deparaffinization and rehydration, 4-μm sections were subjected to heat-induced epitope retrieval in 0.01 mol/L citrate buffer (pH 6.0). Following this, the activity of endogenous peroxidase was blocked for 10 min in 3% hydrogen peroxide, after which non-specific binding was blocked with 5% goat serum for 1 h at room temperature. The slides were subsequently incubated with anti-TrkB antibody overnight at 4°C, and immunodetection was performed using the LSAB2 system (DakoCytomation). During immunodetection, the color was developed using 3-3′-diaminobenzidine and counterstaining was performed with hematoxylin.
In silico analysis of clinical microarray data
In silico analysis of the published clinical microarray data was performed using the NKI295 and TCGA datasets available at www.oncomine.org. TrkB gene expression signatures in the datasets from breast cancer patients were extracted and averaged. ANOVA was performed and graphs with gene expression and survival analysis were plotted using GraphPad Prism v 5.0 (GraphPad Software, Inc.).
Statistical analysis
All data are expressed as mean ± SEM. Statistical analyses of these data were conducted using Student’s t test (two-tailed).
RESULTS
Increased expression of TrkB correlates with breast cancer pathological phenotypes
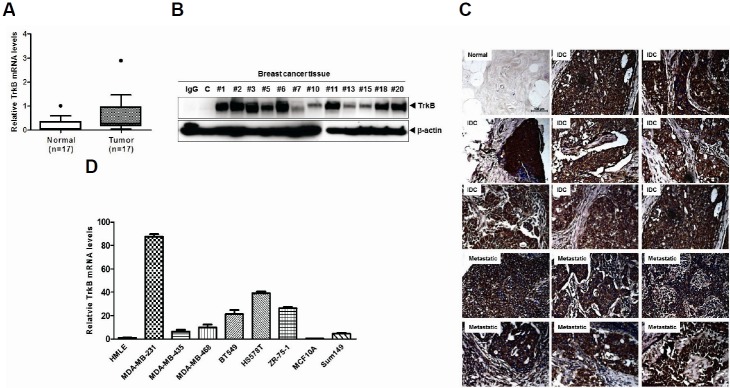
Although previous studies have already suggested that TrkB overexpression plays a crucial role in cancer progression, TrkB expression patterns have not been well characterized in human breast cancer tissues. To evaluate its potential involvement and role in breast cancer, we first examined human tumor specimens to determine whether TrkB expression correlates with a certain pathological phenotype. Strikingly, TrkB mRNA levels were significantly elevated in 14 out of 17 tumor tissues taken from breast cancer patients (80%) compared with patient-matched normal tissue samples. These samples also showed increased TrkB protein expression (Figs. 1A and 1B). Based on these observations, we evaluated TrkB expression in a series of 59 breast cancer samples using immunohistochemistry. We found significant correlation between TrkB overexpression and pathological phenotypes. Normal breast tissue samples demonstrated weak immuno-reactivity to anti-TrkB antibody; however, elevated TrkB levels were detected in infiltrating ductal carcinoma and metastatic breast cancer samples (Fig. 1C). We next examined TrkB expression in a panel of established metastatic and non-metastatic human cancer cell lines. TrkB was highly expressed in several invasive and metastatic human tumor cell lines, including MDA-MB-231, MDA-MB-435, MDA-MB-468, BT549, and Hs578T. In contrast, non-metastatic MCF10A cancer cells and human mammary epithelial cells (HMLEs) expressed low to undetectable TrkB levels (Fig. 1D).
Fig. 1.
Elevated TrkB expression in breast cancer cells and breast cancer patients. (A) The relative expression of TrkB mRNA in 17 human invasive breast carcinoma samples, relative to that of healthy tissues, as determined by quantitative RT-PCR. Expression levels were normalized to 18S mRNA levels. (B) Western blot analysis of TrkB expression in 12 invasive breast carcinoma samples. The β-actin was used as a loading control. (C) Representative immunohistochemical TrkB staining images of normal human breast tissue, infiltrating duct carcinoma, and metastatic carcinoma in the lymph nodes (magnification: 200×). (D) Relative TrkB mRNA expression levels in a panel of eight human nonmetastatic and metastatic cell lines, compared with HMLE cells, as determined by quantitative RT-PCR. The 18S mRNA expression level was used to normalize the TrkB mRNA expression levels.
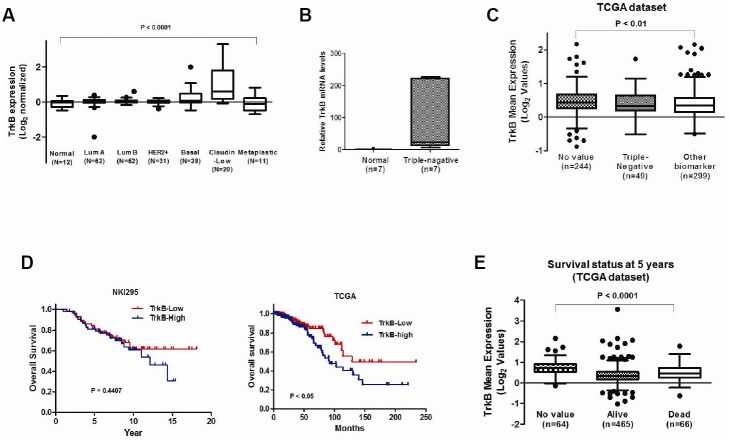
Additionally, TrkB expression analysis by mining of a published microarray database (NKI295 dataset) revealed that increased TrkB expression strongly correlates with the gene expression signatures derived from different breast cancer subtypes (Hennessy et al., 2009; van de Vijver et al., 2002). TrkB expression was significantly upregulated in basal, claudin low and metaplastic breast cancers than in other breast cancer subtypes (Fig. 2A). Based on these observations, we evaluated TrkB expression in normal tissues and triple-negative breast cancer tissues. Indeed, TrkB expression was significantly elevated in tissues of patients with triple-negative breast cancer compared with normal tissue samples (Fig. 2B). Furthermore, analysis of TrkB expression using publicly available RNA sequences and clinical data from 893 patients found in TCGA (Cancer Genome Atlas, 2012), showed that TrkB levels were increased in tumor tissues of triple-negative breast cancer patients compared with other biomarkers (Fig. 2C). These results suggest that high TrkB levels in breast cancer cells correlate with a higher recurrence risk of invasive cancer. We performed Kaplan-Meier survival analysis of the breast cancer patients to investigate whether patients with tumors that show increased TrkB expression exhibit poorer survival outcomes than those with tumors expressing low TrkB levels. This analysis revealed that patients with higher TrkB levels have worse prognosis than those that have low TrkB levels, which corresponded to the survival status in the TCGA dataset (Figs. 2D and 2E) (Cancer Genome Atlas, 2012). Our observations indicated that TrkB expression coincides with certain breast cancer subtypes, and may play a causal role in enabling metastatic dissemination.
Fig. 2.
Expression of TrkB correlates with pathological phenotypes of breast tissues. (A) The gene expression data were plotted as Box-and-whisker (Tukey) plots of mean TrkB expression levels in breast cancer patients. The TrkB level was extracted from the dataset and averaged for each tumor. Points below and above the whiskers are drawn as individual dots. A P < 0.0001 was considered to be significant in ANOVA. (B) The relative expression of the TrkB mRNA in normal or triple-negative breast carcinoma samples as determined by quantitative RT-PCR. Normalization was done using 18S mRNA levels. (C) The mean TrkB expression in other subtypes or triple-negative subtypes of human breast cancer patients. The TrkB gene expression level was extracted from the dataset and averaged for each tumor. Points below and above the whiskers are drawn as individual dots. A P < 0.0001 was considered statistically significant in ANOVA. (D) Patients were divided into those who expressed high and low TrkB levels, and their survival rates from two publicly available data sets (NKI295 and TCGA dataset) were compared. The P value was calculated by a log-rank test. (E) The mean TrkB expression in surviving or deceased breast cancer patients. The TrkB gene expression level was extracted from the dataset and averaged for each tumor. Points below and above the whiskers are drawn as individual dots. A P < 0.0001 was considered significant in ANOVA.
TrkB regulates invasion-metastasis cascade through the inhibition of tumor suppressors Runx3 and Keap1
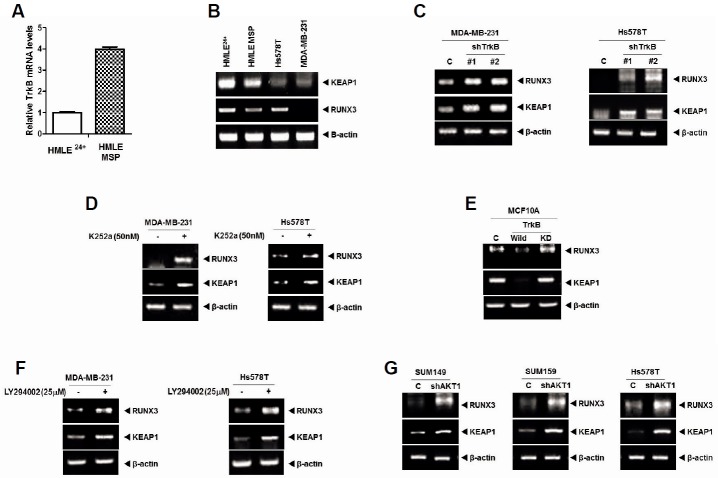
Previous studies demonstrated that activated Ras downregulates Runx3, and that Ras/MEK/ERK pathway promotes ROS detoxification and tumorigenesis through the induction of Nrf2 expression, and induces Keap1 mutations in solid tumors (DeNicola et al., 2011; Lee et al., 2013). In addition, inhibition of the PI3K/AKT pathway induces Keap1-dependent synthetic lethality (Dai et al., 2013). We recently demonstrated that c-Src activation by TrkB induces activation of the PI3K/AKT and Ras/MAPK or cyclin D1 expression (Kim et al., 2015). These diverse lines of evidence suggested a possible link between TrkB overexpression-induced loss of tumor suppression and acquisition of tumorigenic and metastatic potential in cancer cells. However, the signaling mechanism by which TrkB suppresses Keap1 and Runx3 has not been elucidated. To investigate the relationship between loss of Runx3 or Keap1 and TrkB-induced tumorigenesis, we first examined whether the expression of TrkB influences Runx3 and Keap1 expression, using two distinct human mammary epithelial cell models. Both cell lines are derived from human mammary epithelial cells (HMLE). HMLE 24+ cells are a subgroup of the naturally occurring CD24+/CD44− HMLE cells while HMLE MSP cells are a spontaneously arising mesenchymal subpopulation (MSP) of these cells. HMLE MSP cells express several mesenchymal markers and EMT transcription factors, and display a CD24low/CD44high cell-surface marker expression profile, which indicates cells with cancer stem cells (CSCs) properties (Scheel et al., 2011). TrkB expression was significantly increased in HMLE MSP cells in comparison with HMLE 24+ cells (Fig. 3A). In addition, compared with the levels in HMLE MSP cells, HMLE 24+ cells exhibited markedly increased Runx3 and Keap1 expression. Moreover, RT-PCR analyses revealed that the mRNA expression levels of Runx3 and Keap1 were greatly reduced in highly metastatic Hs578T and MDA-MB-231 cells, which have high TrkB expression compared with HMLE 24+ cells (Fig. 3B).
Fig. 3.
TrkB represses Runx3 and Keap1 expression through AKT activation. (A) The relative expression of mRNA encoding TrkB in HMLE24+ and HMLE MSP cells as determined by quantitative RT-PCR. Normalization was done using 18S mRNA expression levels. (B) RT-PCR analysis of Keap1 and Runx3 expression in HMLE24+, HMLE MSP, Hs578T, and MDA-MB-231 cells. β-actin was used as a loading control. (C) RT-PCR analysis of Keap1 and Runx3 expression in MDA-MB-231 and Hs578T control-shRNA or TrkB-shRNA cells. The β-actin was used as a loading control. (D) RT-PCR analysis of Keap1 and Runx3 expression in MDA-MB-231 and Hs578T with or without K252a (50 nM) treatment. The β-actin was used as a loading control. (E) RT-PCR analysis of Keap1 and Runx3 expression in MCF10A, MCF10A-TrkB, and MCF10A-TrkB kinase dead (K571N) cells. The β-actin was used as a loading control. (F) RT-PCR analysis of Keap1 and Runx3 expression in MDA-MB-231 and Hs578T with or without LY294002 (PI3K inhibitor) treatment. The β-actin was used as a loading control. (G) RT-PCR analysis of Keap1 and Runx3 expression in SUM149, SUM159, and Hs578T control-shRNA or AKT1-shRNA cells. β-actin was used as a loading control.
We performed RT-PCR experiments to confirm that TrkB overexpression induces downregulation of Runx3 and Keap1 expression. Upon TrkB silencing, Runx3 and Keap1 expression were significantly reduced in Hs578T and MDA-MB-231 cells relative to control-shRNA cells (Fig. 3C). We next assessed the effects of TrkB inhibitor on Runx3 or Keap1 expression. The inhibition of TrkB activation in Hs578T and MDA-MB-231 cells by K252a, a TrkB inhibitor as acting selectively blocks the effects of brain-derived neurotrophic factor (BDMF) (Knusel and Hefti, 1992; Koizumi et al., 1988), significantly increased Runx3 and Keap1 expression (Fig. 3D). These results indicate that the inhibition of tyrosine kinase activity of TrkB correlated with increased expression of Runx3 and Keap1. To avoid off-target effects of K252a, such as inhibition of TrkA or TrkC, we used a TrkB tyrosine kinase dead mutant (TrkB K571N mutant). As shown in Fig. 3E, Runx3 and Keap1 expression levels were significantly reduced in MCF10A-TrkB compared with MCF10A control or MCF10A-TrkB K571N cells. These findings suggest that the tyrosine kinase activity of TrkB is required for repression of Runx3 and Keap1.
Runx3 suppresses lung and gastric tumorigenesis through induction of p21WAF1/Cip1 (p21) (Chi et al., 2005; Lee et al., 2013), so we investigated TrkB regulation of p21 expression. p21 expression was significantly reduced in MCF10A-TrkB compared with MCF10A control or MCF10A-TrkB kinase dead (K571N) cells (Supplementary Fig. S1A). Further experiments showed that inhibition of TrkB by K252a in MDA-MB-231 cells significantly increased p21 expression (Supplementary Fig. S1B). Moreover, p21 expression was significantly increased in MDAMB-231 TrkB-shRNA cells relative to control-shRNA cells (Supplementary Fig. S1C). These findings suggest that the tyrosine kinase activity of TrkB promotes tumorigenicity of breast cancer cells, through suppression of Runx3-mediated induction of p21.
A recent study has demonstrated that Src phosphorylates Runx3 at multiple tyrosine residues through formation of a Src-Runx3 complex, and induces cytoplasmic mislocalization of Runx3 (Goh et al., 2010). Another study reported that Runx3 can induce AKT1 repression, which leads to inhibition of cell proliferation through inhibition of AKT1/β-catenin/cyclin D1 signaling pathway (Lin et al., 2012). In addition, Keap1 knockout showed increased Nrf2 expression level and AKT phosphorylation (Ke et al., 2013). It was demonstrated that AKT activation induces GSK3β inactivation and binds to Nrf2-Keap1, which strengthens cell defense and survival mechanisms through activation of Nrf2/Keap1 pathway (Jaramillo and Zhang, 2013; Rizvi et al., 2014). Nevertheless, the signaling network that suppresses Runx3 and Keap1 expression in cancer cells has remained poorly understood. The previously described observations led us to speculate that TrkB-mediated AKT activation might contribute to reduction in Runx3 and Keap1 expression. To test this, we assessed the effects of AKT inhibitor on Runx3 and Keap1 expression in the presence of TrkB. Interestingly, the inhibition of AKT activation by LY294002 in MDA-MB-231 and Hs578T cells significantly increased Runx3 and Keap1 expression (Fig. 3F). We further inhibited AKT activation to elucidate whether the inhibition of Runx3 and Keap1 expression in MDA-MB-231 and Hs578T cells was due to transcriptional repression by AKT. Indeed, expression of Runx3 and Keap1 was significantly higher in SUM149, SUM159, or Hs578T AKT1-shRNA cells relative to control-shRNA cells (Fig. 3G). In addition, the inhibition of AKT activity by LY294002 and AKT1-shRNA in MDA-MB-231 and Hs578T cells markedly increased Runx3-induced p21 expression (Supplementary Figs. S2A and S2B). These results indicate that the major mechanism underlying the reduction in Runx3 and Keap1 expression levels is direct transcriptional repression by TrkB through AKT activation.
TrkB enhances the metastatic potential of breast cancer cells
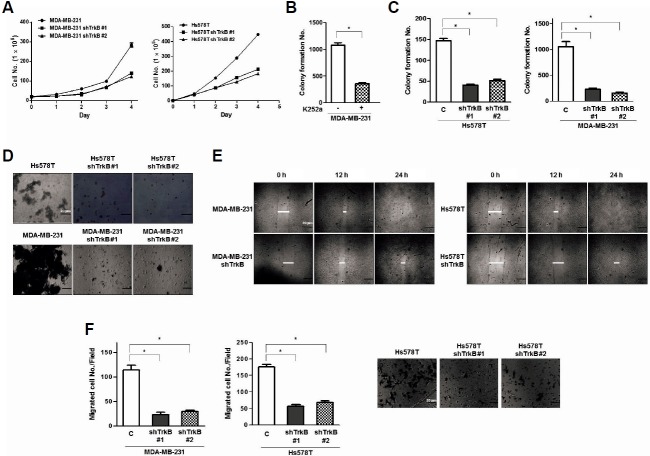
Based on the previous experimental results, we speculated that TrkB might contribute to tumorigenicity and metastatic potential of breast cancer cells. To test this possibility, we examined whether loss of TrkB affected the proliferation of MDA-MB-231 and Hs578T cells in vitro. We found that MDA-MB-231 and Hs578T TrkB-shRNA cells grew slower compared with control-shRNA cells (Fig. 4A). In addition, we examined how pharmacological inhibition of TrkB with K252a influences anchorage-independent growth of MDA-MB-231 cells. After growing in soft agar for 15 days, MDA-MB-231 cells treated with K252a produced a significantly lower number of colonies in comparison with the control MDA-MB-231 cells (Fig. 4B). Moreover, we found that MDA-MB-231 and Hs578T TrkB-shRNA cells in anchorage-independent growth formed a significantly lower number of colonies than control-shRNA cells (Fig. 4C).
Fig. 4.
Suppression of TrkB expression with stable TrkB-shRNA reduces cell proliferation, anoikis, cell migration, and colony formation. (A) Population doubling of MDA-MB-231 and Hs578T control-shRNA or TrkB-shRNA cells. Each data point represents the mean number of cells counted in three dishes. (B) Colony-forming assay of MDA-MB-231 cells treated with K252a (50 nM) (n = 3). *P < 0.001, t-test. (C) Colony-forming assay of MDA-MB-231 and Hs578T control-shRNA or TrkB-shRNA cells (n = 3). *P < 0.001, t-test. (D) Phase-contrast images of spheroid colonies of MDA-MB-231 and Hs578T control-shRNA or TrkB-shRNA cells, which were grown during a seven-day period, in ULC plates. Spheroid colonies in suspension were then photographed at 200× magnification. (E) Wound healing assay of MDA-MB-231 and Hs578T control-shRNA or TrkB-shRNA cells. Wound closures were photographed at 0 and 24 h after wounding. (F) Migration assay of MDA-MB-231 and Hs578T control-shRNA or TrkB-shRNA cells. The cells that migrated to the bottom of the chamber were counted in five fields under 100× magnification (n = 3). *P < 0.001, t-test.
The loss of interaction with the extracellular matrix causes normal epithelial cells to undergo programmed cell death, termed anoikis (Douma et al., 2004). Therefore, we examined whether TrkB knockdown could influence the ability of MAD-MB-231 and Hs578T cells to survive and proliferate. We used cell culture dishes to which MDA-MB-231 and Hs578T cells were unable to attach. Here, MDA-MB-231 and Hs578T control-shRNA cells proliferated as large spheroid aggregates in suspension, whereas MDA-MB-231 and Hs578T TrkB-shRNA cells demonstrated a significantly lower survival rate (Fig. 4D). Furthermore, investigations of migration and wound healing, functional hallmarks of cancer, demonstrated that MDA-MB-231 and Hs578T control-shRNA cells had increased cell motility, whereas TrkB-shRNA cells had low motility. This suggests that the increased cell motility was largely due to TrkB expression (Figs. 4E and 4G). These results suggest that TrkB expression increases tumorigenicity and metastatic potential of cancer cells.
DISCUSSION
TrkB expression patterns have not been extensively characterized in human breast cancers. In this study, we primarily focused on TrkB expression in breast cancer. We found that TrkB is frequently overexpressed in highly metastatic breast cancer cell lines and clinical breast cancer samples, and that it is associated with a high-grade duct carcinoma phenotype. Furthermore, TrkB expression was significantly upregulated in basal, claudin-low, and metaplastic breast cancer samples in comparison with other breast cancer subtypes. Elevated TrkB expression was also detected in the luminal and HER2+ breast cancer subtypes. This observation is common in breast cancer progression because gene expression alterations conferring the potential for invasive growth are already present in the preinvasive stages (Ma et al., 2003). Additionally, our results are consistent with other published results demonstrating that Slug, HOXB13, HER2/neu, and Goosecoid, which promote the metastatic behavior and poor prognosis for aggressive breast cancers, are already overexpressed in clinical specimens and breast cancer cells before the appearance of the malignant tumor phenotype (Hartwell et al., 2006).
In order to acquire tumorigenic and metastatic abilities, cancer cells are required to have following malignant traits: sustained proliferative signaling, evasion of growth suppressors, resistance to cell death, enabled replicative immortality, induction of angiogenesis, and activation of invasion and metastasis (Hanahan and Weinberg, 2011). Here, we demonstrated that TrkB plays a critical role in tumorigenesis and metastasis of breast cancer cells, by induction of proliferation and suppression of anoikis. In addition, TrkB increased cell migration and wound healing, both of which are considered functional hallmarks of cancer. Our findings suggest that TrkB activation leads to the acquisition of metastatic ability of cancer cells, and that this can contribute to the invasion and metastasis of these cells.
Taken together, these results elucidate the role of TrkB in tumorigenesis and metastasis of breast cancer. A number of studies suggest that the activation or expression of tumor suppressors in cancer cells needs to be inhibited in order for a cancer cell to acquire the ability to initiate the invasion-metastasis cascade. Runx3 and Keap1 proteins are both well-known tumor suppressors. Inactivation or loss of Runx3, caused by promoter hypermethylation and protein mislocalization, has been found in gastric, lung, bladder, pancreas, liver, colon, and breast cancers (Huang et al., 2012; Lee et al., 2013; Lin et al., 2012). Additionally, hypermethylation of Keap1 promoter, which is a negative regulator of NRF2, suppresses its expression in gliomas and colon, prostate, lung, and breast cancers (Eades et al., 2011; Hanada et al., 2012). Moreover, loss of Runx3 or Keap1 expression results in increased metastasis, reduced apoptosis, and increased resistance to chemotherapeutic drugs (Hanada et al., 2012; Shibata et al., 2008). Our results suggest that TrkB is sufficient for tumorigenesis and metastasis of breast cancer cells, through inhibition of Runx3 and Keap1 expression. Indeed, TrkB knockdown and inactivation of its tyrosine kinase activity markedly induced Runx3 and Keap1 expressions. These findings shed a new light on the possibility that molecular components of the TrkB signaling cascade can be potential candidates for induction of inactivation or loss of Runx3 and Keap1.
In normal epithelial cells, Runx3 and Keap1 transcriptionally repress AKT1 expression, which leads to the inhibition of proliferation (Ke et al., 2013; Lin et al., 2012). Also, Src activation induces mislocalization of Runx3, but not its loss (Goh et al., 2010). The mechanisms of the inhibition of tumor suppressors by TrkB in tumor progression have not been clearly determined.
Although increasing evidence implies that TrkB promotes tumor formation and metastasis, the loss of Runx3 and Keap1 expression by TrkB-mediated PI3K/AKT modulation in breast cancer have remained unknown, and none of the findings reported to date hinted at a link between these two phenomena. In this study, we show that TrkB inhibits Runx3 and Keap1 expression. In addition, TrkB-induced AKT activation leads to the loss of Runx3 and Keap1 expression. Therefore, the results presented here fill the apparent gap in our understanding of the inhibition of cellular defenses during the initial stage of tumor progression. Moreover, we demonstrated that TrkB may be a new potential target for improving the treatment efficacy of breast cancer.
Supplemental Figure Legends
Acknowledgments
This work was supported by grants from National Research Foundation of Korea (NRF-2010-0002525, NRF-2012R1A2A 2A01002728 to J.W. and 2015R1D1A1A01059406 to MS, K) and the Gachon University Gil Medical Center (2014-14 to J. W.).
Footnotes
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Bardelli A., Parsons D.W., Silliman N., Ptak J., Szabo S., Saha S., Markowitz S., Willson J.K., Parmigiani G., Kinzler K.W., et al. Mutational analysis of the tyrosine kinome in colorectal cancers. Science. 2003;300:949. doi: 10.1126/science.1082596. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao M.V., Bothwell M. Neurotrophins: to cleave or not to cleave. Neuron. 2002;33:9–12. doi: 10.1016/s0896-6273(01)00573-6. [DOI] [PubMed] [Google Scholar]
- Chen L.F. Tumor suppressor function of RUNX3 in breast cancer. J. Cell Biochem. 2012;113:1470–1477. doi: 10.1002/jcb.24074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X.Z., Yang J.O., Lee K.Y., Ito K., Sakakura C., Li Q.L., Kim H.R., Cha E.J., Lee Y.H., Kaneda A., et al. RUNX3 suppresses gastric epithelial cell growth by inducing p21(WAF1/Cip1) expression in cooperation with transforming growth factor (Bierie and Moses)-activated SMAD. Mol. Cell Biol. 2005;25:8097–8107. doi: 10.1128/MCB.25.18.8097-8107.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai B., Yoo S.Y., Bartholomeusz G., Graham R.A., Majidi M., Yan S., Meng J., Ji L., Coombes K., Minna J.D., et al. KEAP1-dependent synthetic lethality induced by AKT and TXNRD1 inhibitors in lung cancer. Cancer Res. 2013;73:5532–5543. doi: 10.1158/0008-5472.CAN-13-0712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeNicola G.M., Karreth F.A., Humpton T.J., Gopinathan A., Wei C., Frese K., Mangal D., Yu K.H., Yeo C.J., Calhoun E.S., et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475:106–109. doi: 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L., Getz G., Wheeler D.A., Mardis E.R., McLellan M.D., Cibulskis K., Sougnez C., Greulich H., Muzny D.M., Morgan M.B., et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionne C.A., Camoratto A.M., Jani J.P., Emerson E., Neff N., Vaught J.L., Murakata C., Djakiew D., Lamb J., Bova S., et al. Cell cycle-independent death of prostate adenocarcinoma is induced by the trk tyrosine kinase inhibitor CEP-751 (KT6587) Clin. Cancer Res. 1998;4:1887–1898. [PubMed] [Google Scholar]
- Douma S., Van Laar T., Zevenhoven J., Meuwissen R., Van Garderen E., Peeper D. S. Suppression of anoikis and induction of metastasis by the neurotrophic receptor TrkB. Nature. 2004;430:1034–1039. doi: 10.1038/nature02765. [DOI] [PubMed] [Google Scholar]
- Eades G., Yang M., Yao Y., Zhang Y., Zhou Q. miR-200a regulates Nrf2 activation by targeting Keap1 mRNA in breast cancer cells. J. Biol. Chem. 2011;286:40725–40733. doi: 10.1074/jbc.M111.275495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggert A., Grotzer M.A., Ikegaki N., Zhao H., Cnaan A., Brodeur G. M., Evans A. E. Expression of the neurotrophin receptor TrkB is associated with unfavorable outcome in Wilms' tumor. J. Clin. Oncol. 2001;19:689–696. doi: 10.1200/JCO.2001.19.3.689. [DOI] [PubMed] [Google Scholar]
- Geiger T.R., Peeper D.S. The neurotrophic receptor TrkB in anoikis resistance and metastasis: a perspective. Cancer Res. 2005;65:7033–7036. doi: 10.1158/0008-5472.CAN-05-0709. [DOI] [PubMed] [Google Scholar]
- Goh Y.M., Cinghu S., Hong E.T., Lee Y.S., Kim J.H., Jang J.W., Li Y.H., Chi X.Z., Lee K.S., Wee H., et al. Src kinase phosphorylates RUNX3 at tyrosine residues and localizes the protein in the cytoplasm. J. Biol. Chem. 2010;285:10122–10129. doi: 10.1074/jbc.M109.071381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada N., Takahata T., Zhou Q., Ye X., Sun R., Itoh J., Ishiguro A., Kijima H., Mimura J., Itoh K., et al. Methylation of the KEAP1 gene promoter region in human colorectal cancer. BMC Cancer. 2012;12:66. doi: 10.1186/1471-2407-12-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hartikainen J.M., Tengstrom M., Winqvist R., Jukkola-Vuorinen A., Pylkas K., Kosma V.M., Soini Y., Mannermaa A. KEAP1 genetic polymorphisms associate with breast cancer risk and survival outcomes. Clin. Cancer Res. 2015;21:1591–1601. doi: 10.1158/1078-0432.CCR-14-1887. [DOI] [PubMed] [Google Scholar]
- Hartwell K.A., Muir B., Reinhardt F., Carpenter A.E., Sgroi D.C., Weinberg R.A. The Spemann organizer gene, Goosecoid, promotes tumor metastasis. Proc. Natl. Acad. Sci. USA. 2006;103:18969–18974. doi: 10.1073/pnas.0608636103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennessy B.T., Gonzalez-Angulo A.M., Stemke-Hale K., Gilcrease M. Z., Krishnamurthy S., Lee J.S., Fridlyand J., Sahin A., Agarwal R., Joy C., et al. Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res. 2009;69:4116–4124. doi: 10.1158/0008-5472.CAN-08-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B., Qu Z., Ong C. W., Tsang Y. H., Xiao G., Shapiro D., Salto-Tellez M., Ito K., Ito Y., Chen L.F. RUNX3 acts as a tumor suppressor in breast cancer by targeting estrogen receptor alpha. Oncogene. 2012;31:527–534. doi: 10.1038/onc.2011.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaramillo M.C., Zhang D.D. The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev. 2013;27:2179–2191. doi: 10.1101/gad.225680.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W., Kim G.M., Kim M.S., Lim M.H., Yun C., Jeong J., Nam J.S., Kim S.J. TrkC plays an essential role in breast tumor growth and metastasis. Carcinogenesis. 2010;31:1939–1947. doi: 10.1093/carcin/bgq180. [DOI] [PubMed] [Google Scholar]
- Ke B., Shen X.D., Zhang Y., Ji H., Gao F., Yue S., Kamo N., Zhai Y., Yamamoto M., Busuttil R.W., et al. KEAP1-NRF2 complex in ischemia-induced hepatocellular damage of mouse liver transplants. J. Hepatol. 2013;59:1200–1207. doi: 10.1016/j.jhep.2013.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M. S., Lee W. S., Jeong J., Kim S. J., Jin W. Induction of metastatic potential by TrkB via activation of IL6/JAK2/STAT3 and PI3K/AKT signaling in breast cancer. Oncotarget. 2015;6:40158–40171. doi: 10.18632/oncotarget.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knusel B., Hefti F. K-252 compounds: modulators of neurotrophin signal transduction. J. Neurochem. 1992;59:1987–1996. doi: 10.1111/j.1471-4159.1992.tb10085.x. [DOI] [PubMed] [Google Scholar]
- Koizumi S., Contreras M. L., Matsuda Y., Hama T., Lazarovici P., Guroff G. K-252a: a specific inhibitor of the action of nerve growth factor on PC 12 cells. J. Neurosci. 1988;8:715–721. doi: 10.1523/JNEUROSCI.08-02-00715.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.S., Lee J.W., Jang J.W., Chi X.Z., Kim J.H., Li Y.H., Kim M.K., Kim D.M., Choi B.S., Kim E.G., et al. Runx3 inactivation is a crucial early event in the development of lung adenocarcinoma. Cancer Cell. 2013;24:603–616. doi: 10.1016/j.ccr.2013.10.003. [DOI] [PubMed] [Google Scholar]
- Lin F.C., Liu Y.P., Lai C.H., Shan Y.S., Cheng H.C., Hsu P.I., Lee C.H., Lee Y.C., Wang H.Y., Wang C.H., et al. RUNX3-mediated transcriptional inhibition of Akt suppresses tumorigenesis of human gastric cancer cells. Oncogene. 2012;31:4302–4316. doi: 10.1038/onc.2011.596. [DOI] [PubMed] [Google Scholar]
- Lu J., Guo H., Treekitkarnmongkol W., Li P., Zhang J., Shi B., Ling C., Zhou X., Chen T., Chiao P.J., et al. 14-3-3zeta Cooperates with ErbB2 to promote ductal carcinoma in situ progression to invasive breast cancer by inducing epithelial-mesenchymal transition. Cancer Cell. 2009;16:195–207. doi: 10.1016/j.ccr.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X.J., Salunga R., Tuggle J.T., Gaudet J., Enright E., McQuary P., Payette T., Pistone M., Stecker K., Zhang B.M., et al. Gene expression profiles of human breast cancer progression. Proc. Natl. Acad. Sci. USA. 2003;100:5974–5979. doi: 10.1073/pnas.0931261100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetti A., Felicioni L., Pelosi G., Del Grammastro M., Fumagalli C., Sciarrotta M., Malatesta S., Chella A., Barassi F., Mucilli F., et al. Frequent mutations in the neurotrophic tyrosine receptor kinase gene family in large cell neuroendocrine carcinoma of the lung. Hum. Mutat. 2008;29:609–616. doi: 10.1002/humu.20707. [DOI] [PubMed] [Google Scholar]
- Miknyoczki S.J., Dionne C.A., Klein-Szanto A.J., Ruggeri B.A. The novel Trk receptor tyrosine kinase inhibitor CEP-701 (KT-5555) exhibits antitumor efficacy against human pancreatic carcinoma (Panc1) xenograft growth and in vivo invasiveness. Ann. N Y Acad. Sci. 1999;880:252–262. doi: 10.1111/j.1749-6632.1999.tb09530.x. [DOI] [PubMed] [Google Scholar]
- Rizvi F., Shukla S., Kakkar P. Essential role of PH domain and leucine-rich repeat protein phosphatase 2 in Nrf2 suppression via modulation of Akt/GSK3beta/Fyn kinase axis during oxidative hepatocellular toxicity. Cell Death Dis. 2014;5:e1153. doi: 10.1038/cddis.2014.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheel C., Eaton E.N., Li S.H., Chaffer C.L., Reinhardt F., Kah K.J., Bell G., Guo W., Rubin J., Richardson A.L., et al. Paracrine and autocrine signals induce and maintain mesenchymal and stem cell states in the breast. Cell. 2011;145:926–940. doi: 10.1016/j.cell.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata T., Kokubu A., Gotoh M., Ojima H., Ohta T., Yamamoto M., Hirohashi S. Genetic alteration of Keap1 confers constitutive Nrf2 activation and resistance to chemotherapy in gallbladder cancer. Gastroenterology. 2008;135:1358–1368. doi: 10.1053/j.gastro.2008.06.082. 1368 e1351–1354. [DOI] [PubMed] [Google Scholar]
- Smit M.A., Peeper D.S. Zeb1 is required for TrkB-induced epithelial-mesenchymal transition, anoikis resistance and metastasis. Oncogene. 2011;30:3735–3744. doi: 10.1038/onc.2011.96. [DOI] [PubMed] [Google Scholar]
- van de Vijver M.J., He Y.D., van't Veer L.J., Dai H., Hart A.A., Voskuil D.W., Schreiber G.J., Peterse J.L., Roberts C., Marton M. J., et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl. J. Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- Yang J., Mani S.A., Donaher J.L., Ramaswamy S., Itzykson R.A., Come C., Savagner P., Gitelman I., Richardson A., Weinberg R.A. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Yilmaz T., Jiffar T., de la Garza G., Lin H., Milas Z., Takahashi Y., Hanna E., MacIntyre T., Brown J.L., Myers J.N., et al. Theraputic targeting of Trk supresses tumor proliferation and enhances cisplatin activity in HNSCC. Cancer Biol. Ther. 2010;10:644–653. doi: 10.4161/cbt.10.6.12782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Guo D., Luo W., Zhang Q., Zhang Y., Li C., Lu Y., Cui Z., Qiu X. TrkB is highly expressed in NSCLC and mediates BDNF-induced the activation of Pyk2 signaling and the invasion of A549 cells. BMC Cancer. 2010;10:43. doi: 10.1186/1471-2407-10-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.