Abstract
Specimens and associated data in natural history collections (NHCs) foster substantial scientific progress. In this paper, we explore recent contributions of NHCs to the study of systematics and biogeography, genomics, morphology, stable isotope ecology, and parasites and pathogens of mammals. To begin to assess the magnitude and scope of these contributions, we analyzed publications in the Journal of Mammalogy over the last decade, as well as recent research supported by a single university mammal collection (Museum of Southwestern Biology, Division of Mammals). Using these datasets, we also identify weak links that may be hindering the development of crucial NHC infrastructure. Maintaining the vitality and growth of this foundation of mammalogy depends on broader engagement and support from across the scientific community and is both an ethical and scientific imperative given the rapidly changing environmental conditions on our planet.
Key words: data integration, ethics, GenBank, IACUC, natural history collections, specimens, voucher collection
Natural history collections (NHCs) are a cornerstone of mammalogy. As in most taxon-specific disciplines, specimens form the basis for fundamental insights into mammalian systematics, biogeography, ecology, and evolution. Indeed, specimen-based inquiry has a long tradition with origins before the time of Darwin and Wallace. That tradition has been championed by C. Hart Merriam, E. Raymond Hall, James Patton, Robert Baker, and many other mammalogists in North America over the past century and a half. The result is a tangible record of mammal specimens spanning broad spatial and deep temporal scales, which today is leveraged not only for new insights in evolution and ecology, but also to address unanticipated questions about public health, toxicology, invasive species, wildlife forensics, and food security (Suarez and Tsutsui 2004; Pyke and Ehrlich 2010). These investigations are made possible through technological and methodological advances (e.g., genomic, isotopic, pathologic) that increase our ability to extract, track, and synthesize data from specimens. Such novel uses of NHCs, and their utility for addressing issues of global change, reiterate that collections remain fundamental components of scientific infrastructure and that their expansion should be a global priority.
The importance of collections (Winker 2004; Edwards 2005; Pyke and Ehrlich 2010; Funk 2014; Rocha et al. 2014; Kemp 2015) and of scientific collecting in particular (Winker 1996; Patterson 2002; Edwards 2005; Rocha et al. 2014) has been voiced broadly in recent literature. Still, shifting social and scientific priorities continue to negatively impact NHCs (Dalton 2003; Gropp 2013; Stokstad 2015), and the essential act of specimen collecting is subject to increased regulation (Sikes et al. 2012; Sikes and Paul 2013). Conversely, there have been relatively few papers emphasizing who may be responsible for continued collections growth, or exactly how NHCs can continue to expand their interface with the biological sciences. Finally, few papers have focused specifically on mammals. Because of the strong history of collections-based research in mammalogy, it is important to periodically assess how this research is evolving and also identify ways that a diverse body of mammal biologists (academics, managers, museum professionals) might continue to foster a culture of collecting and collections, to the general benefit of mammalogical science.
In this paper, we use 2 independent quantitative approaches and review recent literature to assess how NHCs are contributing to current mammalogy. The 1st quantitative approach (specimen usage across a decade of Journal of Mammalogy [JM] articles) provides perspective on the magnitude of NHC contributions across a broad array of mammalogical research. The 2nd approach (loan and publication data from the Museum of Southwestern Biology, Division of Mammals [MSB DOM]) provides an example of the breadth of collections-based research that can be supported by a single NHC. In more detail, we also review 5 major research agendas that NHCs support (systematics and biogeography, genomics, morphometrics, stable isotope ecology, and parasites and pathogens), utilizing examples from a broad sample of the recent literature (including JM).
We also use the JM data to identify ways that mammalogists can ensure continued growth and utility of NHCs. Central to this goal is consistent and standardized deposition of research materials in accredited collections, provision of financial resources for proper curation, improved collaboration between researchers and resource managers, and more integrated data management and accessibility. Opportunities for achieving these goals currently exist across many stages of the scientific process but are too often missed. Individuals at many different levels have important roles to play, including funding program officers, Institutional Animal Care and Use Committees, resource managers, regulatory managers, peer reviewers and journal editors, and NHC staff. We believe vigorous discussion of these issues is critical and can help revitalize archival practices, meet ethical guidelines, and spur renewed growth and diversification of NHCs. Just as many research questions permitted by NHCs were unanticipated by original collectors, we cannot fully anticipate the utility of today’s specimens for tomorrow’s questions.
Modern Contributions of Collections
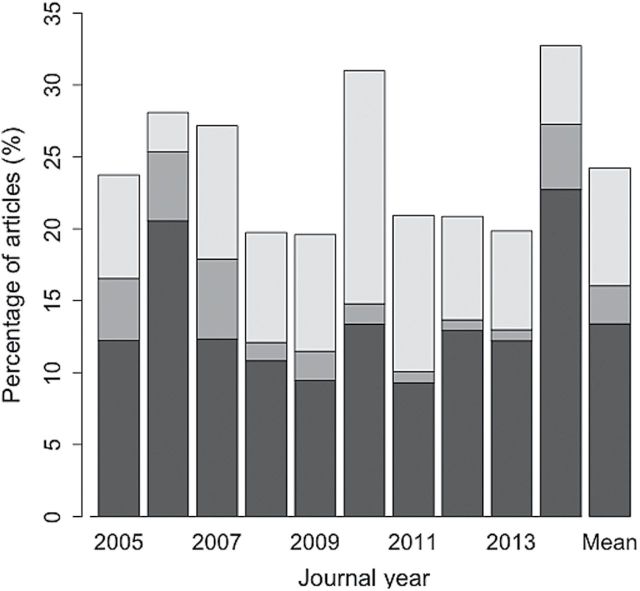
The importance of NHCs in mammalogy: quantitative perspectives.—To begin to quantify the contributions of NHCs to recent research in mammalogy, we assessed (Supporting Information S1) specimen usage in articles published in JM over the last decade (2005–2014, 1,403 articles). We chose JM because it broadly covers the field of mammalogy (e.g., taxonomy, evolution, ecology, conservation), thereby potentially offering insights into larger-scale trends in NHC usage. From the articles assessed, we found that approximately 25% (340) from 2005 to 2014 utilized museum material (Fig. 1; Supporting Information S2). We further distinguished among these studies if they: 1) used only existing specimens in NHCs, 2) collected new material that was eventually deposited in NHCs, or 3) used both existing and new material. We found that studies using only existing specimens are the largest of these categories (pairwise t-tests, P < 0.01, annual values as the unit of replication). Moreover, significantly more studies utilized historical material in combination with newly collected specimens than studies only generating new specimens (P = 0.001). Together, these results demonstrate that NHCs are critical infrastructure supporting substantial numbers of research publications annually. They also reveal that use of historic specimens in addition to ongoing voucher collection remains an integral approach to many research questions in mammalogy.
Fig. 1.
Percentage of articles published in Journal of Mammalogy for the period 2005–2014 utilizing natural history collections. Illustrated within each bar are the articles that used specimens already contained in natural history collections (dark gray), those that used new collected material that was subsequently deposited in natural history collections (medium gray), and those that used both (light gray). The rightmost bar illustrates decadal mean values.
The JM data provide one perspective on the magnitude of museum contributions in mammalogy. However, the scope of research supported by NHCs will typically be published across numerous journals in various scientific disciplines, and thus our JM analysis may only reveal part of the story. Therefore, we also classified loan and publication data from a single university-based NHC (MSB DOM) for a 5-year period (2010–2014; Supporting Information S1 and S2). The MSB data are useful in that they demonstrate the range and magnitude of research that a single NHC may support; this metric is therefore complimentary to the JM data.
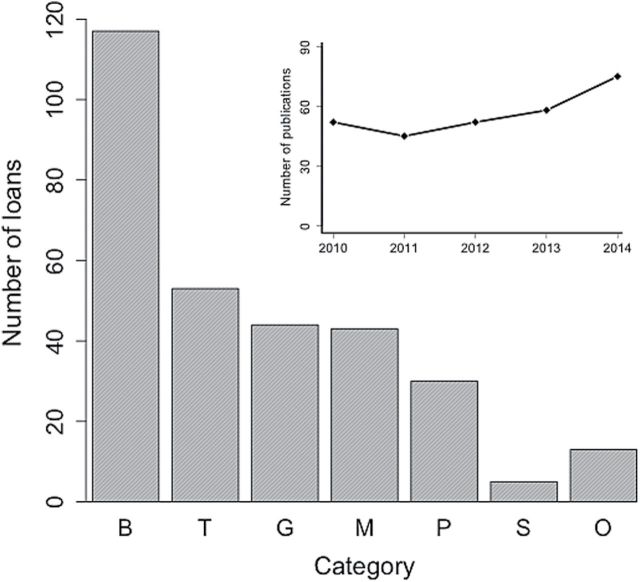
The number of research loans and publications supported by MSB during this period exceed the number of JM publications utilizing NHCs over the same 5 years (Fig. 2; 5 year totals = 252 loans for research/282 publications, 162 JM articles). This suggests that the scope of NHC contributions can be significantly underestimated using analysis of a single journal. Nearly 50% of MSB DOM loans (Fig. 2) are classified under the umbrella of “Systematics and Biogeography,” with remaining loans distributed among 4 additional research categories. This is consistent with our qualitative observations from the JM dataset and demonstrates that traditional roles of NHCs in systematics and biogeography are significant and persistent. An additional 17% of loans (53 of 305 total loans) were made for nonresearch (educational or exhibition) purposes. Together, these data show that single collections can have widespread impacts in biology, with individual specimens forming a nexus between otherwise disparate research programs. Aggregating specimen-based publications across many collections will be an important next step for demonstrating the true magnitude of NHC contributions.
Fig. 2.
Research and teaching in mammalogy supported by the Division of Mammals and Division of Genomic Resources at the Museum of Southwestern Biology. Main graph shows the number of loans in 7 categories (B = systematics/biogeography, T = teaching, G = genomics, M = morphometrics, P = parasitology and pathogen research, S = stable isotope analysis, O = other). Inset shows the total number of publications supported by the Divisions over the same time period.
Systematics and biogeography.
The traditional role of NHCs has been primarily to provide material for the fields of systematics and biogeography, collectively the exploration of the extent, evolution and distribution of Earth’s biodiversity. Because new mammals continue to be described at high rates (Patterson 2000; Reeder et al. 2007), the role of NHCs as primary infrastructure for systematics (mammalian and otherwise) demands continued support, particularly in biologically megadiverse nations (Paknia et al. 2015). Beyond providing samples and data for circumscribing the taxonomic, genetic, and ecological diversity of mammals, NHCs also support more integrative research agendas from phylogeographic and demographic signatures of past climate change (Lessa et al. 2003) to assessment of modern ecosystem services provided by mammals (Ceballos and Ehrlich 2009).
As primary archives of biogeographical data, NHCs permit rigorous analysis of changing mammalian distributions through both space and time in response to climatic and other environmental perturbations. Resurvey projects have used museum data to assess temporal change in distributions and community structure. For example, Moritz et al. (2008) and Rowe et al. (2014) resurveyed elevational transects originally established by Joseph Grinnell and colleagues in California and documented altitudinal shifts in several mammal species since the early 20th century. Further, Rubidge et al. (2012) explored population genetics of historic and contemporary specimens along one of these transects to demonstrate a loss in genetic diversity for alpine chipmunks (Tamias alpinus) in Yosemite National Park. Resurvey projects anchored by museum specimens now contribute to our understanding of climate-driven range shifts in a variety of organisms (Chen et al. 2011), and specimens coupled with climatic and ecological data are increasingly necessary for parsing the idiosyncratic morphological, ecological, and phenological responses of species to rapid global changes (Butler 2013; Calinger et al. 2013; Rowe et al. 2014).
Ongoing digitization of NHC specimens, growth of multi-institutional specimen databases, and increased web connectivity have become important facilitators of biodiversity research (Hanken 2013). For example, georeferenced specimens permit refined species distribution models (SDMs) to explore environmental and ecological factors that limit mammal distributions (e.g., Gutiérrez et al. 2014). Other applications of SDMs include forecasting distributional changes, extinction events, and biological invasions (Peterson and Vieglais 2001; Hope et al. 2013), and also advancing our understanding of fundamental concepts such as niche conservation through time (Peterson et al. 2000). Specimen-based SDMs also can be applied to issues as varied as locating elusive or poorly understood species and addressing food security for humans (Sanchez-Cordero and Martinez-Meyer 2000). Relating SDMs to specimen-derived genetic data (i.e., sequences in GenBank linked with vouchered NHC specimens) offers an even more powerful way to interpret genetic variation across spatial and temporal gradients.
Genomics.
Genomics is a key axis along which NHC-based research is evolving. Application of next-generation sequencing (NGS) to the vast store of mammalian diversity in NHCs is augmenting their roles in taxonomy, phylogenetics, phylogeography, and population genetics. NGS vastly increases sequencing coverage and is less inhibited by degraded DNA, unlocking historic and prehistoric tissues for genetic analysis such as fossil material (Paijmans et al. 2013) and desiccated skin and muscle, bone, and feces (Kimura et al. 2011; Mason et al. 2011), including from extinct species (Miller et al. 2009). In this way, NHCs are uniquely poised to contribute to eventual generic- and species-level phylogenies of Mammalia (Helgen 2011). Improving technologies and decreasing costs (van Dijk et al. 2014) will extend many different kinds of genomic investigations beyond a few model organisms (McCormack and Faircloth 2013).
NHCs are providing the crucial specimens for resolving longstanding problems in mammalian evolution that span a range of timescales. For example, genomes of Neanderthal (Green et al. 2010), Denisovan (Meyer et al. 2012), and possible ancestral (Meyer et al. 2014) specimens are illuminating the convoluted relationships and biogeography of early hominids and the selective forces operating in the lineage leading to modern Homo sapiens. The phylogeny and timescale of ursid evolution (Yu et al. 2007), timing of equid evolution (Orlando et al. 2013), and potential paraphyly of echolocation in chiropterans (Tsagkogeorga et al. 2013) were all clarified using specimen-derived genomic data. Further, such data are refining our understanding of the root of placental mammal phylogeny (Teeling and Hedges 2013) and the evolutionary origins of mammalian and reptilian traits of the duck-billed platypus (Ornithorhynchus anatinus—Warren et al. 2008).
Transcriptomic approaches applied to cryogenic museum collections can provide unique insights into molecular adaptation by illuminating functional genetic differences among cell types, developmental conditions, disease states, environmental conditions, populations, or species. Direct transcriptome sequencing (RNA-Seq) has enabled investigations of adaptation in numerous wild species (Alvarez et al. 2015), such as the function of lipid metabolism as a thermogenesis strategy in hypoxic Peromyscus (Cheviron et al. 2012). Because transcriptome research requires properly preserved samples of multiple tissues, NHCs will likely foster the quick extension of this approach to wild species via cryogenic collections (Lessa et al. 2014).
Conservation will continue to be an important application of specimen-derived genomic data. Recently, Mason et al. (2011) examined mitogenomic diversity in Sunda colugos (Galeopterus variegatus) using dried tissue from museum skins, revealing deep divergence among landmasses in these enigmatic, threatened, and geographically restricted mammals. Bi et al. (2013) used exome capture methods on museum skins to resurvey genetic diversity in alpine chipmunks (T. alpinus), identifying greater genetic subdivision among modern populations due to recent range contraction. NHCs can provide much of the taxonomic sampling required by these approaches, allowing conservation priorities and management plans to be refined. Still, ongoing field sampling will be a crucial part of comprehensive conservation efforts (Rocha et al. 2014).
Morphometrics.
NHCs achieve some of their broadest scientific contributions through studies of morphology and development. Recent specimen-based insights are as disparate as the evolution of bite force in polar bears (Ursus maritimus—Slater et al. 2010), feeding ecology of metatherian and eutherian saber-toothed cats (Wroe et al. 2013), and rates of behavioral and morphological adaptation in ancient proboscideans (Lister 2013). While traditional morphometric techniques remain widely utilized, much of this progress has been fueled by theoretical, methodological, and software advances in geometric morphometrics, which permit richer and more rigorous analysis of shape (e.g., Adams et al. 2013).
NHCs continue to provide substantial morphological data in support of taxonomy and systematics of fossil and modern taxa, and these remain crucial for elucidating relationships of taxa known either from few specimens or specimens unsuitable for genetic analysis (Wiens 2004). However, when morphological data can be combined with molecular phylogenies, powerful tests of complex evolutionary hypotheses are possible such as integration and modularity among morphological traits (Martín-Serra et al. 2015), ontogenetic and evolutionary allometry (Klingenberg and Marugán-Lobón 2013), morphological convergence (Mahler et al. 2013), and trends in morphological evolution during radiations (Monteiro and Nogueira 2011; Zelditch et al. 2015). Temporal morphological change due to environmental and ecological forces has also been illuminated using historical specimens, such as the post-Pleistocene body size decrease in several mammals (e.g., ground squirrels [Blois et al. 2007]; red deer [Rosvold et al. 2014]), as well as century-scale changes in body size and shape that are attributed to climatic or environmental changes (shrews [Yom-Tov and Yom-Tov 2005]; ground squirrels [Eastman et al. 2012]) or anthropogenic alteration of environments (carnivores [Yom-Tov 2003]; rodents, [Pergams and Lawler 2009]; bats [Tomassini et al. 2014]).
NHCs also provide the basis for studies of human evolution and animal domestication, topics long important in biology (Darwin 1859; Wright 1920). Chronologically comprehensive collections permitted robust analysis of hominoid speciation and phylogeny (Lockwood et al. 2004) and facilitated more precise characterization of morphological and taxonomic diversity in early Homo (Lordkipanidze et al. 2013). Qualitative and quantitative morphological data have also illuminated the evolution and biogeography of a range of mammalian domestication events (dogs [Drake and Klingenberg 2010]; pigs [Ottoni et al. 2013]). There will continue to be unprecedented opportunities for integrating phenotypic and genotypic data from nonmodel species using NHCs, as well as establishing stronger links between morphologies and their adaptive significance and studying embryological and developmental aspects of integration, heterochrony, and genetic bases of specific morphological traits.
Stable isotope ecology.
Stable isotope analyses (SIA) have become routine across a variety of biological subdisciplines over the past 2 decades (Martinez del Rio et al. 2009) and are now common for inference of diet, migration, resource partitioning, and other trophic dynamics in mammalogy (Ben-David and Flaherty 2012). NHCs facilitate a wide variety of stable isotope research due primarily to the temporal depth and spatial breadth that specimens embody. Currently, increasing numbers of isotope systems, knowledge of fractionation across different tissues (Koch 2007), compound-specific analyses (Evershed et al. 2007), refinement of analytical methods (Passey and Cerling 2006), and mixing models are all expanding the power of SIA.
Yeakel et al. (2009) used isotopic signatures of hair (reflecting days to weeks) and bone/tooth collagen (reflecting years) to reconstruct dietary trends of > 100-year-old specimens of man-eating lions from Tsavo, Kenya. Results of this work were consistent with the narrative of increased predation on humans from historical and popular accounts. Yeakel et al. (2009) used specimens from mammalogical and archaeological collections, and their study aptly illustrates the power of utilizing multiple types of collections as well as the depth of information that can be gained via assays of multiple tissue types (with different rates of isotopic turnover).
SIA applied to specimens can also provide insight into organismal biology, ontogeny, and phenology that is difficult to obtain in the field. Cryan et al. (2004) used hydrogen isotopes in hair to reconstruct long-distance migratory behavior of hoary bats (Lasiurus cinereus), while others explored migration dynamics, population connectivity, and poorly known breeding and/or wintering ranges of a variety of taxa (Wassenar and Hobson 1998; Rubenstein et al. 2002; Sullivan et al. 2012). Moreover, isotopic data derived from NHC specimens can permit a better understanding of connectivity and pathogen transmission among populations and species (e.g., Britzke et al. 2012).
Another major feature of SIA is that they can powerfully link neontological and paleontological collections to address environmental and ecological processes operating over extended (geologic) timescales. Chamberlain et al. (2005) analyzed Pleistocene megafaunal specimens (pinnipeds and ungulates) alongside modern samples to demonstrate recent shifts in the diets of California condors away from marine mammals. MacFadden et al. (1999) used SIA of fossil equid teeth to reconstruct latitudinal gradients in New World vegetation over the Pleistocene, and Badgley et al. (2008) demonstrated that Miocene climate change drove large-scale shifts in both vegetation structure and mammalian herbivore community structure in southern Asia.
Parasites and pathogens.
NHCs are a primary resource for discovery, description, and mitigation of mammalian parasites and pathogens (hereafter, “pathogens” includes parasites) and are crucial for rigorous integration of the fields of mammalogy and parasitology. Specimens not only form a strong basis for traditional research on mammal parasites such as evolutionary and ecological descriptions (Reed et al. 2007), but they can also be used to reveal coevolutionary histories with hosts (Hafner et al. 2003), first emergences in a host species or population (Hartigan et al. 2010), and even host evolutionary history (Galbreath and Hoberg 2011). Recent studies utilizing NHCs have also investigated parasite range shifts and host-switching events in light of past and contemporary climate change (Kutz et al. 2005; Hoberg et al. 2008) and the evolutionary and ecological correlates of parasitism across entire host faunas (Patterson et al. 2008). Spatially extensive parasite collections that are directly tied to host collections are an essential part of all these inquiries (Hoberg et al. 2003; Cook et al. 2005).
Host specimens can also yield insights into pathogen biology. Researchers have used NHCs to screen for external fungal infections and internal helminthes in fluid-preserved amphibians and for ectoparasites on bird and mammal study skins (Hellenthal and Price 1991; Clayton and Walther 1997; Johnson et al. 2003; Bodinof et al. 2011), as well as viral strains from both dry and fluid-preserved specimens and tissue subsamples (Yates et al. 2002; Ávila-Arcos et al. 2013). Beyond analysis of pathogen presence and diversity, host examinations can provide information on infection rates and magnitude of host exposure (Pinto et al. 2010; Deardorff et al. 2013). For example, retrospective genetic analysis of century-old museum specimens successfully detected Lyme bacillus (Borrelia burgdorferi—Persing et al. 1990; Marshall et al. 1994). That finding was inconsistent with a previous hypothesis of recent introduction (Steere et al. 1977), instead revealing that the first outbreak in 1975 was likely precipitated by suburban reforestation and domestic landscaping that increased human exposure to tick bites (Marshall et al. 1994). Even deeper in time, analysis of dental pulp from European mass graves enabled implication of Yersinia pestis as the etiologic agent of the Black Death (i.e., bubonic plague), which is strong proof-of-concept that specimens of many ages have potential in modern pathogen research (Haensch et al. 2010).
Discovery and description of Sin Nombre Virus in the American Southwest provides one of the best-documented examples of NHC-based pathogen research (Yates et al. 2002). Screening frozen tissue collections demonstrated that the previously unknown hantavirus had been circulating in North American deer mouse (Peromyscus maniculatus) populations well before this outbreak. Yet, equally remarkable are the descriptions of > 40 previously unknown hantaviruses worldwide since 1993—many from museum specimens—demonstrating that these viruses have a deep coevolutionary relationship with mammals and infect a number of nonrodent hosts such as shrews (Arai et al. 2008), moles (Kang et al. 2009), and bats (Gu et al. 2014). These descriptions are fueling a richer understanding of hantavirus evolution and diversity (Yanagihara et al. 2014). The ability to probe mammalian diversity in NHCs for viruses and other pathogens demonstrates their potential to establish host viability and also inform our understanding of temporal and spatial dynamics of infections, allowing more effective response to pathogen emergence in humans and other mammals.
Fostering Continued Growth and Expansion of Collections
There has never been a more pressing need to support NHCs in their traditional roles of surveying and describing global biodiversity. Yet specimens also enable an array of exciting (and evolving) research questions that extends beyond what we include here, such as studies of agricultural pests and productivity, environmental toxicology, chemical contamination of food and water supplies, and bioterrorism (Suarez and Tsutsui 2004; Winker 2004). Ensuring NHCs continue to expand and further interface with the biological sciences is crucial for all these roles, but how can mammalogists help facilitate this?
Voucher collection.
The materials contained in NHCs are typically referred to as voucher specimens, or simply vouchers. These terms capture the essential role of museum specimens, serving as references for the taxonomy, morphology, evolution, and ecology of species by allowing these attributes to be physically verified by researchers. However, vouchers embody far more. Because each specimen is sampled from a particular time and place on our planet, it represents a potential storehouse of information on 1) the multitude of organisms hosted by that voucher (from RNA viruses and bacteria to helminth worms and ectoparasites), 2) the substances consumed throughout its life (their biological and chemical composition and/or toxicity), and 3) a genetic and morphological record of the population and species, and potentially the environments they inhabit (via epigenetic interactions). Furthermore, vouchers allow previous investigations to be replicated, a critical step in the scientific process that otherwise becomes difficult, if not impossible.
Ongoing voucher collection (and appropriate preservation) will be paramount for biodiversity research in the 21st century, particularly because it will establish rigorous baselines for investigating species- and community-level response to global change. These collections will be most powerfully leveraged when they are built systematically, over pertinent temporal and spatial scales, and across taxonomic diversity. Future collections should also be unbiased with regards to sampled species. For example, nonnative and invasive species, often overlooked by museums in the past (e.g., Rattus—Lack et al. 2012), can have important ecological, evolutionary, and public health effects on other species and surrounding ecosystems.
Sentiments for collection of wild mammals are often mixed, but it is important to note that voucher collection can be accomplished ethically in regard to the welfare of individual organisms as well as the stability of populations and species. Indeed, the ethics of animal-based research is an important topic in the biological sciences, one that the American Society of Mammalogists has actively addressed (Sikes et al. 2011, 2012). Communicating this to the lay public, permitting agencies, Institutional Animal Care and Use Committees, and scientists should be a top priority of American Society of Mammalogists and other taxon-specific societies. In situations where specimen collections are curtailed by permitting bodies (e.g., federal or state/provincial wildlife agencies), effectively communicating how voucher collections achieve a shared goal of biodiversity conservation is imperative. Specifically, collections provide a critical basis for objective, evidence-based management directives (Cook and MacDonald 2013).
Formal education avenues also play a vital—but underutilized—role in conveying the importance of NHCs, voucher collection, and ongoing documentation of biodiversity (Schmidly 2005). Educators, museum professionals, and even many scientists who regularly utilize NHC specimens can contribute to education and public outreach in this regard. One new and dynamic way to transform biological education is through use of large-scale digitization of NHC specimens and associated data in the classroom (Cook et al. 2014).
Specimen deposition and curation.
Many permitted research projects involve surveying, sampling, or collecting wild organisms. When possible, those projects should give serious consideration to specimen deposition in accredited NHCs. Archiving research specimens (or specimen parts) serves to maximize the information gained from sacrificed individuals by ensuring their availability to other researchers for future projects (Edwards 2005). Maximizing the scientific potential of wild organisms that are handled, euthanized, incidentally killed, salvaged, or otherwise sampled is also an ethical imperative.
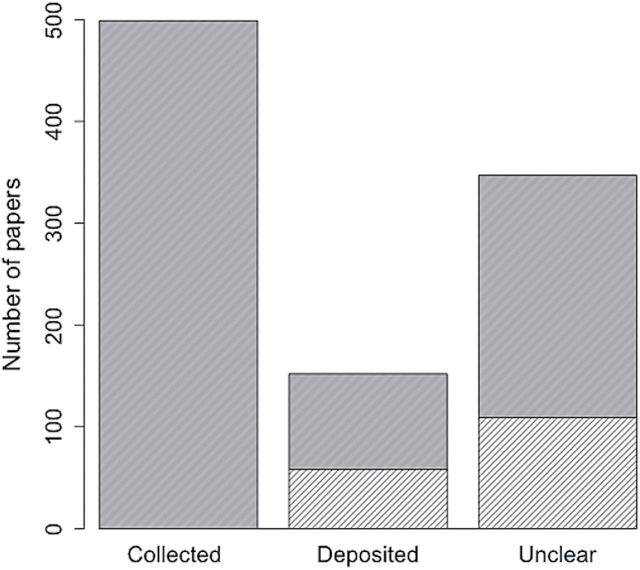
Using the JM dataset, we were able to assess trends in specimen deposition over the past decade. We recorded data from articles where animal material was collected in the field or lab, employing a broad definition of “animal material” that encompasses traditional voucher specimens, samples of tissue from field-sacrificed mammals, and other specimens or tissues from passive monitoring or salvage (Supporting Information S1). We then assessed whether collected materials were deposited in a referenced collection. Finally, we recorded whether projects were supported in some measure by United States federal funding; this was used as a coarse metric of extramural support. The data reveal a large discrepancy in the fate of mammal materials obtained for research (Fig. 3). In greater than two-thirds of papers that collected such material, it was unclear if materials were deposited in a NHC (347 papers), whereas less than one third reported completing this important step (151 papers). We note that many of these materials would qualify as voucher specimens, if properly preserved and associated with collection data (Kageyama et al. 2007). The materials used in JM articles during this time period represent diverse taxa from localities across the globe and, if not placed in a permanent repository, constitute a significant loss of potential information on mammalian biology whose temporal representation cannot be replaced.
Fig. 3.
Number of articles published in Journal of Mammalogy for the period 2005–2014 in which animal material was collected (left). Also shown is the number of those studies in which animal material was definitely deposited in natural history collections (middle) and the number of studies in which it was unclear if material was deposited (right). Lighter hatched areas represent the number within each category funded at least partly by federal (United States) agencies.
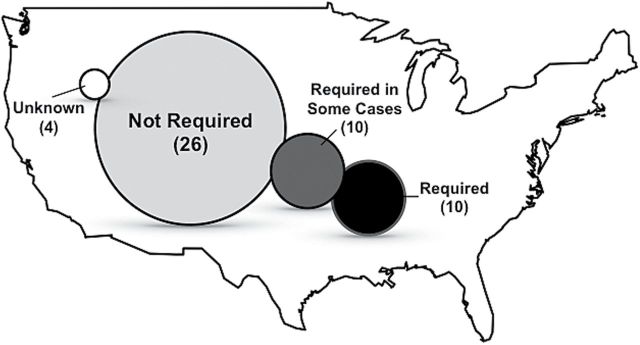
Unfortunately, deposition of research specimens is still an inconsistent requirement of permitting and funding agencies or journal editors, even for mammals and other vertebrates whose handling, collection, and transport are otherwise intensely regulated (e.g., Paul and Sikes 2013). For example, in the JM metadata, we found a negligible relationship between federal funding and the depositional fate of animal material (Fig. 3); federally funded projects comprised 38% and 31% of studies in “Deposited” and “Unclear if Deposited” categories, respectively. To further broaden this line of inquiry, we compiled a separate dataset of specimen deposition requirements of United States wildlife management agencies (n = 50, Alaska and Hawaii included) by polling via email or phone. Specifically, we determined whether specimen deposition is a requirement to obtain a state scientific collection permit. We found that a majority of states (n = 26) have no requirements for deposition of sacrificed mammals, while 10 additional states require voucher deposition for select taxa or studies only (Fig. 4).
Fig. 4.
Requirements of United States wildlife agencies for deposition of collected wildlife specimens in museum collections. “Required” signifies deposition is required, “Not Required” signifies it is not. “Required in some cases” signifies that deposition requirements are dependent on the taxon in question or other circumstances or the proposed collection. “Unknown” signifies uncertainty in requirements or inability to successfully contact agency personnel. Alaska and Hawaii were included.
Voucher deposition is a reasonable requirement for many studies by federal, state, and institutional (IACUCs) permitting and funding agencies and would leverage the long-term impact of public funds and ensure associated data remain available. Preservation of tissue subsamples from a small percentage of game mammals and skeletal carcasses collected by commercial trappers represents yet another cost-effective way to build archives. Currently, such series are largely unavailable for hunted or trapped species, either throughout their ranges or for most years, despite large annual harvests of these species (e.g., Patterson 2002). Another potential source of specimens are disease outbreak investigations, which typically involve reactive field collections (often mammals), but too often do not result in specimen preservation. Specimens of wild host species (Frey et al. 1992) should be preserved to provide a stronger basis for understanding pathogen natural history, zoonotic potential, and host-parasite coevolution through space and time.
Subsamples of tissues and other animal materials (“secondary vouchers” sensu Kageyama et al. 2007) retain scientific potential far beyond original research objectives, and technological advances such as genomic sequencing further augment their roles as voucher specimens. There is therefore a pressing need to discuss all of the above sources of museum material among researchers, managers, and NHCs. This discussion should focus on best practices for secondary voucher preservation and data collection as well as those financial provisions that are necessary for specimen curation (Bradley et al. 2014).
Data management, reporting, and integration.
A growing number of funding agencies and peer-reviewed journals now require data management plans and permanent data archiving, respectively (e.g., Holdren 2013; Dryad Digital Repository 2015). Increased data accessibility stimulates repeatability, extension, and quality control (Whitlock et al. 2010). Because access to datasets from peer-reviewed publications decreases significantly over time (Wolkovich et al. 2012; Vines et al. 2014), practices that standardize and promote access to data are critical in an era of rapid global change (Wolkovich et al. 2012). In the same vein, NHCs are themselves permanent archives and some have built web-accessible databases with strong links to derivative data, research publications, and other specimen-associated projects (e.g., Dunnum and Cook 2012).
GenBank, which is often cited as an exemplary public data repository (Strasser 2008; Nature Editorial 2014), provides a relevant example of why maintaining such specimen associations is critical. While it has become a key genetic and genomic database that spans the Tree of Life (see GenBank 2015), a relatively small proportion of sequence data derived from NHC materials is clearly linked to voucher specimens. Nevertheless, this capability exists, and standardizing and promoting these links within GenBank is essential due to occasionally high contamination rates (roughly one quarter—Longo et al. 2011) and biases in quality control and analysis (e.g., chromatogram interpretation—Fietz et al. 2013). Investigating and rectifying such issues is facilitated if sequences are tied to voucher specimens. Valkiūnas et al. (2008) also note that many sequence submissions from parasites are from inconclusively identified samples or are unassociated with traditional morphospecies or vouchered specimens (also see Salazar-Bravo et al. 2006 for virus hosts). Explicit links between NHC specimens and genetic data derived from those specimens also facilitates reexamination when taxonomic changes occur.
The emergence of large, web-distributed databases poses unique challenges, but also opportunities, for NHCs. In this arena, specimens are best perceived as a nexus to link genetic (GenBank), morphological (MorphBank), virological (ViPr), isotopic (Pauli et al. 2015), and other derivative data. Proper cross-linking of specimens with these data and with publications is a reasonable requirement of reviewers, editors, publishers, curators of data repositories, and NHCs and would meet federal requirements for permanent data management plans. The American Society of Mammalogists, other taxon-based societies, and NHC curators and staff have important roles to play in facilitating discussions of best practices in data management and integration.
Substantial scientific progress is made possible by the specimens and associated data collectively housed in NHCs. These contributions stand in contrast to inconsistent public, political, and institutional support that threaten NHCs and ultimately undermine our ability to address major environmental and ecological questions, both now and into the future. However, the vitality and growth of NHCs also hinges on the community of scientists, managers, and educators who benefit from availability of archived material. While utilizing specimens in novel ways, we must also implement standards that maximize opportunities for growth and expansion of NHCs, and their integration with burgeoning amounts of specimen-derived data. If current trends are an indication, specimens that are archived today will provide many answers to the crucial questions of tomorrow. But only if building collections remains a collective priority.
Supporting Information
The Supporting Information documents are linked to this manuscript and are available at Journal of Mammalogy online (jmammal.oxfordjournals.org). The materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supporting data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Supporting Information S1.—Description of methods used (data-gathering, scoring criteria, and statistical analysis).
Supporting Information S2.—Summary data for Journal of Mammalogy publication data compiled and analyzed in this paper.
Supporting Information S3.—Summary data for loans and research publications supported by Museum of Southwestern Biology, Division of Mammals.
Acknowledgments
We thank E. J. Heske for important insights on an earlier draft of this manuscript, as well as B. Patterson, 1 anonymous reviewer, and the editor for thoughtful and constructive comments that helped us to hone our message. Development of these ideas was supported in part by the National Science Foundation (grants DEB 0956129 and DEB 1258010), with support from the USGS Molecular Ecology Laboratory in Anchorage, Alaska. FS-M was supported by an NIH Maximizing Access to Research Careers grant (T34GM008751-15). The authors declare no conflict of interest.
Literature Cited
- Adams D. C., Rohlf F. J., Slice D. E. 2013. A field comes of age: geometric morphometrics in the 21st century. Hystrix 24:7–14. [Google Scholar]
- Alvarez M., Schrey A. W., Richards C. L. 2015. Ten years of transcriptomics in wild populations: what have we learned about their ecology and evolution? Molecular Ecology 24:710–725. [DOI] [PubMed] [Google Scholar]
- Arai S., et al. 2008. Phylogenetically distinct hantaviruses in the masked shrew (Sorex cinereus) and montane shrew (Sorex monticolus) in the United States. American Journal of Tropical Medicine and Hygiene 78:348–351. [PMC free article] [PubMed] [Google Scholar]
- Ávila-Arcos M. C., et al. 2013. One hundred twenty years of koala retrovirus evolution determined from museum skins. Molecular Biology and Evolution 30:299–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badgley C., et al. 2008. Ecological changes in Miocene mammalian record show impact of prolonged climatic forcing. Proceedings of the National Academy of Sciences of the United States of America 105:12145–12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-David M., Flaherty E. A. 2012. Stable isotopes in mammalian research: a beginner’s guide. Journal of Mammalogy 93:312–328. [Google Scholar]
- Bi K., Linderoth T., Vanderpool D., Good J. M., Nielsen R., Moritz C. 2013. Unlocking the vault: next-generation museum population genomics. Molecular Ecology 22:6018–6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blois J. L., Feranec R. S., Hadly E. A. 2007. Environmental influences on spatial and temporal patterns of body-size variation in California ground squirrels (Spermophilus beecheyi). Journal of Biogeography 35:602–613. [Google Scholar]
- Bodinof C. M., Briggler J. T., Duncan M. C., Beringer J., Millspaugh J. J. 2011. Historic occurrence of the amphibian chytrid fungus Batrachochytrium dendrobatidis in hellbender Cryptobranchus alleganiensis populations from Missouri. Diseases of Aquatic Organisms 96:1–7. [DOI] [PubMed] [Google Scholar]
- Bradley R. D., Bradley L. C., Garner H. J., Baker R. J. 2014. Assessing the value of natural history collections and addressing issues regarding long-term growth and care. BioScience 64:1150–1158. [Google Scholar]
- Britzke E. R., Loeb S. C., Romanek C. S., Hobson K. A., Vonhof M. J. 2012. Variation in catchment areas of Indiana bat (Myotis sodalis) hibernacula inferred from stable hydrogen (δ2H) isotope analysis. Canadian Journal of Zoology 90:1243–1250. [Google Scholar]
- Butler L. K. 2013. The grass is always greener: do monsoon rains matter for molt of the Vermillion Flycatcher (Pyrocephalus rubinus)? The Auk 130:297–307. [Google Scholar]
- Calinger K. M., Queenborough S., Curtis P. S. 2013. Herbarium specimens reveal the footprint of climate change on flowering trends across north-central North America. Ecology Letters 16:1037–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceballos G., Ehrlich P. R. 2009. Discoveries of new mammal species and their implications for conservation and ecosystem services. Proceedings of the National Academy of Sciences of the United States of America 106:3841–3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain C. P., et al. 2005. Pleistocene to recent dietary shifts in California condors. Proceedings of the National Academy of Sciences of the United States of America 102:16707–16711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I.-C., Hill J. K., Ohlemüller R., Roy D. B., Thomas C. D. 2011. Rapid range shifts of species associated with high levels of climate warming. Science 333:1024–1026. [DOI] [PubMed] [Google Scholar]
- Cheviron Z. A., Bachman G. C., Connaty A. D., McClelland G. B., Storz J. F. 2012. Regulatory changes contribute to the adaptive enhancement of thermogenic capacity in high-altitude deer mice. Proceedings of the National Academy of Sciences of the United States of America 109:8635–8640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton D. H. and Walther B. A.. 1997. Appendix C, Collection and quantification of arthropod parasites of birds. Pp. 419–440 in Host-parasite evolution: general principles and avian models ( D. H. Clayton and B. A. Walther , eds.). Oxford University Press, Oxford, United Kingdom. [Google Scholar]
- Cook J. A., et al. 2014. Natural history collections as emerging resources for innovative education. BioScience 64:725–734. [Google Scholar]
- Cook, J. A., et al. 2005. Beringia: intercontinental exchange and diversification of high latitude mammals and their parasites during the Pliocene and Quaternary. Mammal Study 30:33–44. [Google Scholar]
- Cook J. A., MacDonald S. O. 2013. Island life: coming to grips with the insular nature of North Pacific Coastal Forests. Pp. 19–42 in Conservation of North Pacific Coastal Forests ( G. H. Orians and J. W. Schoen , eds). University of Washington Press, Seattle, Washington. [Google Scholar]
- Cryan P. M., Bogan M. A., Rye R. O., Landis G. P., Kester C. L. 2004. Stable hydrogen isotope analysis of bat hair as evidence for seasonal molt and long-distance migration. Journal of Mammalogy 85:995–1001. [Google Scholar]
- Dalton R. 2003. Natural history collections in crisis as funding is slashed. Nature 423:575. [DOI] [PubMed] [Google Scholar]
- Darwin C. 1859. On the origin of species by means of natural selection, or the preservation of favoured races in the struggle for life. 1st ed. John Murray, London, United Kingdom. [PMC free article] [PubMed] [Google Scholar]
- Deardorff E. R., et al. 2013. Serological evidence of Powassan Virus in mammals from Russia, Alaska and New Mexico, 2004–2007. Emerging and Infectious Diseases 19:2012–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake A. G., Klingenberg C. P. 2010. Large-scale diversification of skull shape in domestic dogs: disparity and modularity. The American Naturalist 175:289–301. [DOI] [PubMed] [Google Scholar]
- Dryad Digital Repository. . 2015. Joint Data Archiving Policy (JDAP) http://datadryad.org/pages/jdap Accessed 15 October 2015.
- Dunnum J. L., Cook J. A. 2012. Gerrit Smith Miller: his influence on the enduring legacy of natural history collections. Mammalia 76:365–373. [Google Scholar]
- Eastman L. M., Morelli T. L., Rowe K. C., Conroy C. J., Moritz C. 2012. Size increase in high elevation ground squirrels over the last century. Global Change Biology 18:1499–1508. [Google Scholar]
- Edwards S. V. 2005. Bird collections: development and use of a scientific resource. The Auk 122:966–971. [Google Scholar]
- Evershed R. P., et al. 2007. Compound-specific stable isotope analysis in ecology and paleoecology. Pp. 480–540 in Stable isotopes in ecology and environmental science ( R. Michener and K. Lajtha , eds.). 2nd ed. Blackwell Publishing, Malden, Massachusetts. [Google Scholar]
- Fietz K., Graves J. A., Olsen M. T. 2013. Control control control: a reassessment and comparison of GenBank and chromatogram mtDNA sequence variation in Baltic grey seals (Halichoerus grypus). PLos ONE 8:e72853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey J. K., Yates T. L., Duszynski D. W., Gannon W. L., Gardner S. L. 1992. Designation and curatorial management of type host specimens (symbiotypes) for new parasite species. The Journal of Parasitology 78:930–932. [Google Scholar]
- Funk V. A. 2014. The erosion of collections-based science: alarming trend or coincidence? Plant Press 17. [Google Scholar]
- Galbreath K. E., Hoberg E. P. 2011. Return to Beringia: parasites reveal cryptic biogeographic history of North American pikas. Proceedings of the Royal Society B: Biological Sciences 279:371–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GenBank 2015. Growth of GenBank and WGS. National Center for Biotechnology Information, National Institutes of Health http://www.ncbi.nlm.nih.gov/genbank/statistics Accessed 15 October 2015.
- Green R. E., et al. 2010 A draft sequence of the Neandertal genome. Science 328:710–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gropp R. E. 2013. Are university natural science collections going extinct? Bioscience 53:550. [Google Scholar]
- Gu S. H., et al. 2014. Molecular phylogeny of hantaviruses harbored by insectivorous bats in Côte d’Ivoire and Vietnam. Viruses 6:1897–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez E. E. Boria R. A. and Anderson R. P.. 2014. Can biotic interactions cause allopatry? Niche models, competition, and distributions of South American mouse opossums. Ecography 37:741–753. [Google Scholar]
- Haensch S., et al. 2010. Distinct clones of Yersinia pestis caused the black death. PLoS Pathogens 6:e1001134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M. S., Demastes J. W., Spradling T. A., Reed D. L. 2003. Cophylogeny between pocket gophers and chewing lice. Pp. 195–220 in Tangled trees: phylogeny, cospeciation, and coevolution ( R. D. M. Page , ed.). University of Chicago Press, Chicago, Illinois. [Google Scholar]
- Hanken J. 2013. Biodiversity online: toward a network integrated biocollections alliance. BioScience 63:789–790. [Google Scholar]
- Hartigan A., Phalen D. N., Šlapeta J. 2010. Museum material reveals a frog parasite emergence after the invasion of the cane toad in Australia. Parasites & Vectors 3:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgen K. M. 2011. The mammal family tree. Science 334:458–459. [DOI] [PubMed] [Google Scholar]
- Hellenthal R. A., Price R. D. 1991. Biosystematics of the chewing lice of pocket gophers. Annual Review of Entomology 36:185–203. [DOI] [PubMed] [Google Scholar]
- Hoberg E. P., Kutz S. J., Galbreath K. E., Cook J. A. 2003. Arctic biodiversity: from discovery to faunal baselines – revealing the history of a dynamic system. Journal of Parasitology 89:S84–S95. [Google Scholar]
- Hoberg E. P., et al. 2008. Integrated approaches and empirical models for investigation of parasitic diseases in northern wildlife. Emerging Infectious Diseases 10:10–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdren J. P. 2013. Increasing access to the results of federally funded scientific research. Memorandum for the heads of executive departments and agencies. Executive Office of the President, Office of Science and Technology Policy, Washington, D.C. [Google Scholar]
- Hope A. G., Waltari E., Payer D. C., Cook J. A., Talbot S. L. 2013. Future distribution of tundra refugia in northern Alaska. Nature Climate Change 3:931–938. [Google Scholar]
- Johnson P. T. J., Lunde K. B., Zelmer D. A., Werner J. K. 2003. Limb deformities as an emerging parasitic disease in amphibians: evidence from museum specimens and resurvey data. Conservation Biology 17:1724–1737. [Google Scholar]
- Kageyama M., Monk R. R., Bradley R. D., Edson G. F., Baker R. J. 2007. The changing significance and definition of the biological voucher. Pp. 257–264 in Museum studies: perspectives and innovations ( S. L. Williams and C. A. Hawks , eds.). Society for the Preservation of Natural History Collections, Washington, D.C. [Google Scholar]
- Kang H. J., et al. 2009. Evolutionary insights from a genetically divergent hantavirus harbored by the European common mole (Talpa europaea). PLoS ONE 4:e6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp C. 2015. The endangered dead. Nature 518:292–294. [DOI] [PubMed] [Google Scholar]
- Kimura B., et al. 2011. Ancient DNA from Nubian and Somali wild ass provides insights into donkey ancestry and domestication. Proceedings of the Royal Society B: Biological Sciences 278:50–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingenberg C. P., Marugán-Lobón J. 2013. Evolutionary covariation in geometric morphometric data: analyzing integration, modularity, and allometry in a phylogenetic context. Systematic Biology 62:591–610. [DOI] [PubMed] [Google Scholar]
- Koch P. L. 2007. Isotopic study of the biology of modern and fossil vertebrates. Pp. 99–154 in Stable isotopes in ecology and environmental science ( R. Michener and K. Lajtha , eds.). 2nd edition. Blackwell Publishing, Malden, Massachusetts. [Google Scholar]
- Kutz S. J., Hoberg E. P., Polley L., Jenkins E. J. 2005. Global warming is changing the dynamics of Arctic host–parasite systems. Proceedings of the Royal Society B: Biological Sciences 272:2571–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lack J. B., et al. 2012. Invasion facilitates hybridization with introgression in the Rattus rattus species complex. Molecular Ecology 21:3545–3561. [DOI] [PubMed] [Google Scholar]
- Lessa E. P., Cook J. A., D’Elia G., Opazo J. C. 2014. Rodent diversity in South America: transitioning into the genomics era. Frontiers in Ecology and Evolution 2:1–7. [Google Scholar]
- Lessa E. P., Cook J. A., Patton J. L. 2003. Genetic footprints of demographic expansion in North America, but not Amazonia, during the Late Quaternary. Proceedings of the National Academy of Sciences of the United States of America 100:10331–10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister A. M. 2013. The role of behaviour in adaptive morphological evolution of African proboscideans. Nature 500:331–334. [DOI] [PubMed] [Google Scholar]
- Lockwood C. A., Kimbel W. H., Lynch J. M., Pilbeam D. 2004. Morphometrics and hominoid phylogeny: support for a chimpanzee-human clade and differentiation among great ape subspecies. Proceedings of the National Academy of Sciences of the United States of America 101:4356–4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo M. S., O’Neill M. J., O’Neill R. J. 2011. Abundant human DNA contamination identified in non-primate genome databases. PLoS ONE 6:e16410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lordkipanidze D., et al. 2013. A complete skull from Dmanisi, Georgia, and the evolutionary biology of early Homo . Science 342:326–331. [DOI] [PubMed] [Google Scholar]
- MacFadden B. J., Cerling T. E., Harris J. M., Prado J. 1999. Ancient latitudinal gradients of C3/C4 grasses interpreted from stable isotopes of New World Pleistocene horse (Equus) teeth. Global Ecology and Biogeography 8:137–149. [Google Scholar]
- Mahler D. L., Ingram T., Revell L. J., Losos J. B. 2013. Exceptional convergence on the macroevolutionary landscape in island lizard radiations. Science 341:292–295. [DOI] [PubMed] [Google Scholar]
- Marshall W. F., III, et al. 1994. Detection of Borrelia burgdorferi DNA in museum specimens of Peromyscus leucopus . The Journal of Infectious Diseases 170:1027–1032. [DOI] [PubMed] [Google Scholar]
- Martín-Serra A., Figueirido B., Pérez-Claros J. A., Palmqvist P. 2015. Patterns of morphological integration in the appendicular skeleton of mammalian carnivores. Evolution 69:321–340. [DOI] [PubMed] [Google Scholar]
- Martinez del Rio C., Wolf N., Carleton S. A., Gannes L. Z. 2009. Isotopic ecology ten years after a call for more laboratory experiments. Biological Reviews 84:91–111. [DOI] [PubMed] [Google Scholar]
- Mason V. C., Li G., Helgen K. M., Murphy W. J. 2011. Efficient cross-species capture hybridization and next-generation sequencing of mitochondrial genomes from noninvasively sampled museum specimens. Genome Research 21:1695–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack J. E., Faircloth B. C. 2013. Next-generation phylogenetics takes root. Molecular Ecology 22:19–21. [DOI] [PubMed] [Google Scholar]
- Meyer M., et al. 2012. A high coverage genome sequence from an archaic Denisovan individual. Science 338:222–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M., et al. 2014. A mitochondrial genome sequence of a hominin from Sima de los Huesos. Nature 505:403–406. [DOI] [PubMed] [Google Scholar]
- Miller W., et al. 2009. The mitochondrial genome sequence of the Tasmanian tiger (Thylacinus cynocephalus). Genome Research 19:213–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro L. R., Nogueira M. R. 2011. Evolutionary patterns and processes in the radiation of phyllostomid bats. BMC Evolutionary Biology 11:137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz C., Patton J. L., Conroy C. J., Parra J. L., White G. C., Beissinger S. R. 2008. Impact of a century of climate change on small-mammal communities in Yosemite National Park, USA. Science 322:261–264. [DOI] [PubMed] [Google Scholar]
- Nature Editorial. . 2014. Share alike: research communities need to agree on standard etiquette for data-sharing. Nature 507:140.24627917 [Google Scholar]
- Orlando L., et al. 2013. Recalibrating Equus evolution using the genome sequence of an early Middle Pleistocene horse. Nature 499:74–78. [DOI] [PubMed] [Google Scholar]
- Ottoni C., et al. 2013. Pig domestication and human-mediated dispersal in western Eurasia revealed through ancient DNA and geometric morphometrics. Molecular Biology and Evolution 30:824–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paijmans J. L. A., Gilbert M. T. P., Hofreiter M. 2013. Mitogenomic analyses from ancient DNA. Molecular Phylogenetics and Evolution 69:404–416. [DOI] [PubMed] [Google Scholar]
- Paknia, O., R. S. Hossein, and A. Koch. 2015. Lack of well-maintained natural history collections and taxonomists in megadiverse developing countries hampers global biodiversity exploration. Organisms Diversity & Evolution 15:619–629. [Google Scholar]
- Passey B., Cerling T. 2006 In situ stable isotope analysis (δ13C, δ18O) of very small teeth using laser ablation GC/IRMS. Chemical Geology 235:238–249. [Google Scholar]
- Patterson B. D. 2000. Patterns and trends in the discovery of new Neotropical mammals. Diversity and Distributions 6:145–151. [Google Scholar]
- Patterson B. D. 2002. On the continuing need for scientific collecting of mammals. Mastozoologia Neotropical 9:253–262. [Google Scholar]
- Patterson B. D., Dick C. W., Dittmar K. 2008. Parasitism by bat flies (Diptera: Streblidae) on Neotropical bats: effects of host body size, distribution and abundance. Parasitology Research 103:1091–1100. [DOI] [PubMed] [Google Scholar]
- Paul E., Sikes R. S. 2013. Wildlife researchers running the permit maze. ILAR Journal 54:14–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli J. N., Steffan S. A., Newsome S. D. 2015. It is time for Isobank. BioScience 65:229–230. [Google Scholar]
- Pergams O. R. W., Lawler J. J. 2009 Recent and widespread rapid morphological change in rodents. PLoS ONE 4:e6452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persing D. H., et al. 1990. Detection of Borrelia burgdorferi DNA in museum specimens of Ixodes dammini ticks. Science 249:1420–1423. [DOI] [PubMed] [Google Scholar]
- Peterson A. T., Soberon J., Sanchez-Cordero V. 2000. Conservatism of ecological niches in evolutionary time. Science 285:1265–1267. [DOI] [PubMed] [Google Scholar]
- Peterson, A. T., and D. A. Vieglais. 2001. Predicting species invasions using ecological niche modeling: new approaches from bioinformatics attack a pressing problem. Bioscience 51:363–372. [Google Scholar]
- Pinto C. M., et al. 2010. Using museum collections to detect pathogens. Emerging Infectious Diseases 16:356–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke G. H., Ehrlich P. R. 2010. Biological collections and ecological/environmental research: a review, some observations and a look to the future. Biological Reviews 85:247–266. [DOI] [PubMed] [Google Scholar]
- Reed D. L., Light J. E., Allen J. M., Kirchman J. J. 2007. Pair of lice lost or parasites regained: the evolutionary history of anthropoid primate lice. BMC Biology 5:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder D. M., Helgen K. M., Wilson D. E. 2007. Global trends and biases in new mammal species discoveries. Occasional Papers of the Museum of Texas Tech University 269:1–35. [Google Scholar]
- Rocha L. A., et al. 2014. Specimen collection: an essential tool. Science 344:814–815. [DOI] [PubMed] [Google Scholar]
- Rosvold J., Herfindal I., Andersen R., Hufthammer A. K. 2014. Long-term morphological changes in the skeleton of red deer (Artiodactyla, Cervidae) at its northern periphery. Journal of Mammalogy 95:626–637. [Google Scholar]
- Rowe K. C., et al. 2014. Spatially heterogeneous impact of climate change on small mammals of montane California. Proceedings of the Royal Society B: Biological Sciences 282:20141857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubenstein D. R., et al. 2002. Linking breeding and wintering ranges of a migratory songbird using stable isotopes. Science 295:1062–1065. [DOI] [PubMed] [Google Scholar]
- Rubidge E. M., Patton J. L., Lim M., Burton A. C., Brashares J. S., Moritz C. 2012. Climate-induced range contraction drives genetic erosion in an alpine mammal. Nature Climate Change 2:258–288. [Google Scholar]
- Salazar-Bravo J., Phillips C. J., Bradley R. D., Baker R. J., Yates T. L., Ruedas L. A. 2006. Voucher specimens for SARS-linked bats. Science 311:1099–1100. [DOI] [PubMed] [Google Scholar]
- Sanchez-Cordero V., Martinez-Meyer E. 2000. Museum specimen data predict crop damage by tropical rodents. Proceedings of the National Academy of Sciences of the United States of America 97:7074–7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidly D. J. 2005. What it means to be a naturalist and the future of natural history at American universities. Journal of Mammalogy 86:449–456. [Google Scholar]
- Sikes R. Gannon W., and the Animal Care and Use Committee of the American Society of Mammalogists. 2011. Guidelines of the American Society of Mammalogists for the use of wild mammals in research. Journal of Mammalogy 92:235–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikes R. S., Paul E. 2013. Fundamental differences between wildlife and biomedical research. ILAR Journal 54:5–13. [DOI] [PubMed] [Google Scholar]
- Sikes R. S., Paul E., Beaupre S. J. 2012 Standards for wildlife research: taxon-specific guidelines versus US public health service policy. BioScience 62:830–834. [Google Scholar]
- Slater G. J., Figueirido B., Louis L., Yang P., Van Valkenburgh B. 2010. Biomechanical consequences of rapid evolution in the polar bear lineage. PLoS ONE 5:e13870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steere A. C., et al. 1977. An epidemic of oligoarticular arthritis in children and adults in three Connecticut communities. Arthritis & Rheumatology 20:7–17. [DOI] [PubMed] [Google Scholar]
- Stokstad E. 2015. Research at Kew overhauled for leaner times. Science 347:936. [DOI] [PubMed] [Google Scholar]
- Strasser B. J. 2008. GenBank - natural history collections in the 21st century? Science 322:537–538. [DOI] [PubMed] [Google Scholar]
- Suarez A. V., Tsutsui N. D. 2004. The value of museum collections for research and society. BioScience 54:66–74. [Google Scholar]
- Sullivan A. R., Bump J. K., Kruger L. A., Peterson R. O. 2012. Bat-cave catchment areas: using stable isotopes (δD) to determine the probable origins of hibernating bats. Ecological Applications 22:1428–1434. [DOI] [PubMed] [Google Scholar]
- Teeling E. C., Hedges S. B. 2013. Making the impossible possible: rooting the tree of placental mammals. Molecular Biology and Evolution 30:1999–2000. [DOI] [PubMed] [Google Scholar]
- Tomassini A., Colangelo P., Agnelli P., Jones G., Russo D. 2014. Cranial size has increased over 133 years in a common bat, Pipistrellus kuhlii: a response to changing climate or urbanization? Journal of Biogeography 41:944–953. [Google Scholar]
- Tsagkogeorga G., Parker J., Stupka E., Cotton J. A., Rossiter S. J. 2013. Phylogenomic analyses elucidate the evolutionary relationships of bats. Current Biology 23:2262–2267. [DOI] [PubMed] [Google Scholar]
- Valkiūnas G., Atkinson C. T., Bensch S., Sehgal R. N. M., Ricklefs R. E. 2008. Parasite misidentifications in GenBank: how to minimize their number? Trends in Parasitology 24:247–248. [DOI] [PubMed] [Google Scholar]
- van Dijk E. L., Auger H., Jaszczyszyn Y., Thermes C. 2014. Ten years of next-generation sequencing technology. Trends in Genetics 30:418–426. [DOI] [PubMed] [Google Scholar]
- Vines T. H., et al. 2014. The availability of research data declines rapidly with article age. Current Biology 24:94–97. [DOI] [PubMed] [Google Scholar]
- Warren W. C., et al. 2008. Genome analysis of the platypus reveals unique signatures of evolution. Nature 453:175–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenar L. I., Hobson K. A. 1998. Natal origins of migratory monarch butterflies at wintering colonies in Mexico: new isotopic evidence. Proceedings of the National Academy of Sciences of the United States of America 95:15436–15439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock M. C., McPeek M. A., Rausher M. D., Rieseberg L., Moore A. J. 2010. Data archiving. The American Naturalist 175:145–146. [DOI] [PubMed] [Google Scholar]
- Wiens J. 2004. The role of morphological data in phylogeny reconstruction. Systematic Biology 53:653–661. [DOI] [PubMed] [Google Scholar]
- Winker K. 1996. The crumbling infrastructure of biodiversity: the avian example. Conservation Biology 10:703–707. [Google Scholar]
- Winker K. 2004. Natural history museums in a postbiodiversity era. BioScience 54:455–459. [Google Scholar]
- Wolkovich E. M., Regetz J., O’Connor M. I. 2012. Advances in global change research require open science by individual researchers. Global Change Biology 18:2102–2110. [Google Scholar]
- Wright S. 1920. The relative importance of heredity and environment in determining the piebald pattern of guinea-pigs. Proceedings of the National Academy of Sciences of the United States of America 6:320–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wroe S., Chamoli U., Parr W. C. H., Clausen P., Ridgely R., Witmer L. 2013. Comparative biomechanical modeling of metatherian and placental saber-tooths: a different kind of bite for an extreme pouched predator. PLoS ONE 8:e66888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagihara R., Gu S. H., Arai S., Kang H. J., Song J.-W. 2014. Hantaviruses: rediscovery and new beginnings. Virus Research 187:6–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates T. L., et al. 2002. The ecology and evolutionary history of an emergent disease: Hantavirus Pulmonary Syndrome. BioScience 52:989–998. [Google Scholar]
- Yeakel J. D., et al. 2009. Cooperation and individuality among man-eating lions. Proceedings of the National Academy of Sciences of the United States of America 106:19040–19043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yom-Tov Y. 2003. Body sizes of carnivores commensal with humans have increased over the past 50 years. Functional Ecology 17:323–327. [Google Scholar]
- Yom-Tov Y., Yom-Tov J. 2005. Global warming, Bergmann’s rule and body size in the masked shrew Sorex cinereus Kerr in Alaska. Journal of Animal Ecology 74:803–808. [Google Scholar]
- Yu L., Li Y.-W., Ryder O. A., Zhang Y.-P. 2007. Analysis of complete mitochondrial genome sequences increases phylogenetic resolution of bears (Ursidae), a mammalian family that experienced rapid speciation. BMC Evolutionary Biology 7:198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelditch M. L., Li J., Tran L. A. P., Swiderski D. L. 2015. Relationships of diversity, disparity and their evolutionary rates in squirrels. Evolution 69:1284–2000. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.