Abstract
Characterizing the spatial arrangement of related individuals within populations can convey information about opportunities for the evolution of kin-selected social behaviors, the potential for inbreeding, and the geographic distribution of genetic variation. Prairie voles (Microtus ochrogaster) are socially monogamous rodents that sometimes breed cooperatively. Individuals of both sexes are highly philopatric, and among natal dispersers, the average dispersal distance is about 30 m. Such limited natal dispersal can result in the spatial clustering of kin and we used microsatellite data to estimate genetic relatedness among resident adult prairie voles in 2 natural populations to test the hypothesis that limited natal dispersal of male and female prairie voles results in the spatial clustering of kin. Spatial autocorrelation analyses of nest residency and microsatellite data indicated that proximate same-sex adult residents of both sexes were significantly more related than more spatially distant resident same-sex adults in Kansas. In Indiana, adult female voles residing less than 20 m apart were also significantly more related than more spatially distant resident adult females but spatial clustering of kin was not detected among resident adult males. The spatial clustering of kin indicates that opportunities for kin-selected behaviors exist in both populations, especially among females. Differences in the patterns of spatial genetic structure among resident males between the Kansas and Indiana populations may be due to population differences in factors such as demography and mating system, as well as in the extent of natal philopatry.
Key words: kin clusters, Microtus ochrogaster, philopatry, relatedness, spatial genetic autocorrelation
At least 2 factors, a high level of philopatry and short natal dispersal distances, can result in the formation of spatial clusters of kin (Greenwood 1980; Emlen 1982). In some polygynous mammals, dispersal is sex biased, with females more philopatric than males (Greenwood 1980; Waser 1985; Pusey 1987; Handley and Perrin 2007), whereas in some monogamous mammalian species, there is no evidence of sex-biased dispersal (Dobson 1982). In addition, offspring dispersing from the natal area should go no further than necessary before settling down as a resident at a new site (Murray 1967; Waser 1985).
The spatial distribution of related individuals within animal populations can be an important factor influencing social and mating behavior. The spatial proximity of kin is a critical initial step for facilitating kin-mediated social interactions that can lead to the evolution and maintenance of cooperation and sociality via kin selection (West-Eberhard 1975; Wilson 1975; Emlen 1982; Clutton-Brock 2002). In many social species, groups often contain adults of at least one sex that are closely related (Bourke 1997; Emlen 1997). Spatially clustered kin can also create opportunities for matings between related individuals, leading to the evolution of inbreeding avoidance mechanisms (e.g., kin recognition) to diminish the deleterious consequences of inbreeding (Pusey and Wolf 1996). Documenting the spatial arrangement of related individuals in natural populations can impart information about the opportunities for kin selection, the potential for inbreeding, as well as, the geographic distribution of genetic variation within populations.
Prairie voles (Microtus ochrogaster) are socially monogamous rodents that typically form pair bonds (Carter and Getz 1993; Getz et al. 1993; Young and Wang 2004) and sometimes breed cooperatively (Solomon and Getz 1997). Since social monogamy is so rare among mammals (< than 5% of species—Kleiman 1977), the social organization and behavior of prairie voles is among the most well studied of any nondomesticated species of small mammal. Getz et al. (1993) described 3 different types of social units found in natural populations of prairie voles: male–female pairs with or without offspring, single females with or without offspring, and groups containing at least 2 adults of the same sex with or without offspring. Most adults live as residents at a single nest site in one of these types of social units. However, some adult males and females, called wanderers, do not settle at a permanent nest site and continue to move among multiple nest sites for a period of time (Getz et al. 1994). During the breeding season, up to 45% of the adult males and 25% of the adult females may be classified as wanderers at any given time (Getz et al. 1993). Almost 75% of male and female juvenile prairie voles never disperse from their natal nest, and the formation and composition of communal groups is strongly influenced by the high levels of natal philopatry (McGuire et al. 1993). However, the percentage of voles that disperse is influenced by environmental factors. For example, a greater proportion of voles disperse at low (< 100/ha) population density than at high density (> 100/ha—McGuire et al. 1993). In a restored tallgrass prairie, a habitat with lower quality food, more than 92% juveniles remained philopatric (Getz 1997). Among the juveniles that do disperse, the average natal dispersal distance of both sexes is similar (~30 m—McGuire et al. 1993).
While there are extensive data on demography, social organization, and dispersal from natural populations of prairie voles (e.g., Getz et al. 1993; Getz et al. 2003; Mabry et al. 2011), the spatial arrangement of kin within populations has received relatively little attention considering the potential impact it can have on sociosexual behavior. Given that both male and female prairie voles exhibit a high degree of natal philopatry and that dispersing individuals typically move short distances, we expect there to be some degree of clustering of closely related prairie voles within populations. However, no study has used genetic data to examine the spatial pattern of relatedness among adults in a natural population of prairie voles. Therefore, the objective of this study was to use polymorphic microsatellite loci to estimate genetic relatedness among resident adult prairie voles to examine the spatial distribution of kin within 2 natural populations. We hypothesize that limited natal dispersal of males and females will result in the spatial clustering of kin among adult voles of both sexes. We predicted that, for both male and female voles, the relatedness between same-sex resident adults would be negatively correlated with increasing distance between the nests at which they reside.
Materials and Methods
Study sites.
We collected data from 2 natural populations of prairie voles. One study site was located at the Indiana University Bayles Road Preserve (Bloomington, Indiana, 39°13′00″N, 86°32′27″W), and the other at the Nelson Environmental Study Area (12 km northeast of Lawrence, Kansas, 39°03′07″N, 95°11′27″W) at the University of Kansas. Each site was located in an old field dominated by grasses and forbs, and both fields were mowed periodically to prevent the invasion of woody plants via ecological succession. We conducted fieldwork during 3 years at each of the sites with fieldwork occurring May–June 2005, 2006, and 2008 in Kansas and July−August 2006–2008 in Indiana (see Streatfeild et al. 2011; Chesh et al. 2012 for details). Fieldwork in Kansas began earlier in the year because voles in the Kansas population experience a hiatus in reproduction during midsummer (Rose and Gaines 1978) and also because the breeding season begins approximately 1 month earlier in Kansas than in Indiana (Myers and Krebs 1971; Rose and Gaines 1978).
Field methods.
We conducted 4 consecutive weeks of live-trapping per field season at each site. During either the first week (2005–2007) or first 2 weeks (2008) of each field season, trapping was conducted on a grid with 10 m spacing between live-traps. The size of the area that was live-trapped varied between sites and among years (range: 1–2.2 ha; Table 1). During grid trapping, a single Ugglan multiple capture live-trap (Grahnab, Hillerstorp, Sweden) was placed in a vole runway within 1 m of each grid marker. We set traps in the late afternoon and checked in the evening and the next morning from Sunday afternoon through Friday morning for a total of 10 trap checks per week (see Streatfeild et al. 2011; Chesh et al. 2012 for details). Traps were baited with cracked corn and covered with an aluminum shield or a wooden board overlaid with vegetation to protect any trapped animal from extreme heat or rain. When we were not trapping, traps were left in place unset.
Table 1.
Number of resident and unclassified adult male and female prairie voles (Microtus ochrogaster), study area size, and minimum number known alive estimates of population density (adult voles/ha ± SE) at each study site during each year. An adult prairie vole was classified as a resident at a nest if it was captured at least once per week during each of the first 2 weeks of nest trapping, and ≥ 75% of all captures during these 2 weeks were at a single nest site.
| Population | Resident | Unclassified | Resident | Unclassified | Study area | Density |
|---|---|---|---|---|---|---|
| Males | Males | Females | Females | Size (ha) | ||
| Indiana 2006 | 27 | 49 | 24 | 33 | 1.5 | 40.0±5.4 |
| Indiana 2007 | 34 | 64 | 52 | 109 | 2.2 | 84.2±9.0 |
| Indiana 2008 | 13 | 65 | 29 | 143 | 1.5 | 90.0±13.6 |
| Kansas 2005 | 6 | 28 | 11 | 26 | 1.0 | 44.3±5.3 |
| Kansas 2006 | 3 | 9 | 6 | 10 | 1.0 | 27.0±2.7 |
| Kansas 2008 | 17 | 35 | 18 | 56 | 1.0 | 58.1±5.6 |
For each adult female captured during grid trapping weeks, we attempted to locate her nest site using either radiotracking or fluorescent-powder tracking (see Lucia et al. 2008 for a complete description of methods for nest location). After the nest of an adult female was located, we recorded the nest coordinates with a hand-held global positioning unit (eTrex Legend, Garmin, Olathe, Kansas) and placed 4 Ugglan multiple capture live-traps within 30cm of the entrance(s) of the nest. At all nest sites that we located, we set live-traps for either 3 (2005–2007) or 2 (2008) consecutive weeks immediately following the initial grid trapping. During nest trapping weeks, we checked traps in the mornings and evenings from Sunday evening until Tuesday evening, and again from Wednesday evening until Friday evening, for a total of 10 trap checks per week. Traps were baited and covered as described for grid trapping.
When first captured, we marked all prairie voles with a unique toe-clip for identification. Toes were stored without a preservative in 1.5ml microcentrifuge tubes at −20°C for 2−4 months for subsequent genetic analysis of relatedness. At every capture, we recorded an individual’s ID number, capture location, sex, body mass (g), age class, and reproductive condition. We used body mass to assign individuals to age classes: juvenile (< 21g), subadult (21–29g), or adult (> 30 g—Gaines et al. 1979; Getz et al. 1993). None of the animals marked in 1 year was captured in the succeeding year in either population.
All procedures we used involving the trapping, marking, and handling of prairie voles were approved by the animal care and use committees of Miami University, University of Kansas, and Indiana University and were consistent with the guidelines published by the American Society of Mammalogists (Sikes et al. 2011) for the use of wild animals in research.
Nest residency.
We only used data from nest trapping to determine which adult males and females were residing at a particular nest site. An adult prairie vole was classified as a resident at a nest if it was captured at least once per week during each of the first 2 weeks of nest trapping, and ≥ 75% of all captures during these 2 weeks were at a single nest site (Cochran and Solomon 2000). Adults trapped at least once a week during the first 2 nest trapping weeks but less than 75% of the time at any one nest or that were not caught during each of the first 2 nest trapping weeks were not classified as a resident (see also Chesh et al. 2012). The unclassified individuals were wanderers (nonterritorial voles—Getz et al. 1993), dispersers, or residents at nests located off the study grid.
Population density.
We estimated prairie vole density on our study sites using the minimum number known alive (MNKA), which has previously been shown to be highly correlated with estimates of prairie vole population density using other methods (Slade and Blair 2000). The MNKA is equal to the number of animals captured at time t plus those individuals not captured at time t but present before and after time t. The mean adult density (± SE) at each site for each year was estimated by dividing the average MNKA estimate of adults during each of the 4 trapping weeks by the effective trapping area. The effective area sampled was considered to be the size of the trapping grid for each population plus a surrounding boundary strip with a width equal to 5 m, which is half the distance between adjacent grid trap locations.
Genetic relatedness.
To estimate genetic relatedness among individuals, we genotyped all prairie voles from which we collected tissue samples at 6 microsatellite loci known to be polymorphic in prairie voles (Keane et al. 2007). Genomic DNA was extracted from tissue samples using either standard phenol/chloroform extraction techniques (Sambrook et al. 1989) or DNeasy extraction kits (Qiagen, Valencia, California) and used to conduct polymerase chain reactions (PCR) to amplify alleles at the 6 microsatellite loci (for details on PCR conditions, see Keane et al. 2007; Solomon et al. 2009). PCR products were diluted, combined with an internal size standard (Liz 500; Applied Biosystems, Foster City, California) and detected using an ABI 3130xl or 3730 DNA sequencer (Applied Biosystems). Base pair lengths of the fluorescently labeled DNA fragments were determined with GeneMapper 3.7 software (Applied Biosystems) and alleles were binned into discrete size classes using FlexiBin (Amos et al. 2006). We have previously estimated the genotyping error rate at these loci in prairie voles due to mutation and mis-scoring to be approximately 0.02 (Solomon et al. 2004).
We calculated expected and observed heterozygosities, probabilities of identity among full siblings, and probabilities of null alleles using CERVUS 3.0 (Kalinowski et al. 2007). Departures from Hardy–Weinberg equilibrium were tested for each locus separately for each population and year using CERVUS 3.0 with significance levels set at α < 0.05 with a Bonferroni correction for multiple tests. Pairwise relatedness between resident adult voles within a population for each year was estimated using RELATEDNESS 5.0 (Queller and Goodnight 1989), which uses the frequency of alleles in a population to calculate the probability that 2 individuals share alleles identical by descent. Relatedness values between 2 individuals may range from −1 to +1, where a positive value indicates that 2 individuals share more alleles that are identical by descent than expected by chance (i.e., more related), whereas a negative value indicates they share fewer alleles identical by descent than expected by chance (i.e., less related). If a population is in Hardy−Weinberg equilibrium, 1st-degree relatives (e.g., parent–offspring or full siblings) should have relatedness values of 0.5, whereas pairs of unrelated individuals should have relatedness values of 0.
Relationship between relatedness and geographic distance.
We compared the mean pairwise geographic distances separating related (r ≥ 0.25) versus nonrelated (r < 0.25) voles separately for each sex within each population using permutation tests for 2 means (JMP 11.2 with the simple permutation test add-in). We ran 1,000 permutations for each analysis. Means ± SE were calculated by combining the 3 years of data collected within each population. Test were considered significant if P < 0.05.
We investigated relationships between relatedness and the geographic distance separating individuals by conducting spatial autocorrelation analyses using GENALEX 6.5 (Peakall and Smouse 2006). GENALEX 6.5 uses pairwise geographic and genetic distance matrices to provide a measure of genetic similarity between pairs of individuals whose geographic separation falls within a specified distance class. We constructed total genetic distance matrices utilizing genotypic data across all 6 microsatellite loci. Within a population, the 3 years of genetic and geographic data were pooled and we conducted separate analyses for males and females. We present results using 20-m distance intervals since this is approximately the average home range diameter of adult prairie voles in the 2 populations (Streatfeild et al. 2011). Results were qualitatively similar when using distance intervals or 10 and 40 m. We plotted the autocorrelation coefficients (r) calculated by GENALEX 6.5 for each distance interval to produce spatial genetic autocorrelograms. We ran 999 permutations to estimate the 95% confidence intervals (CI) around the null distribution for r when there is no genetic structure (r = 0). We also ran 1,000 bootstrapping simulations to calculate the 95% CI about the estimate of r for each distance class. Following the recommendations of Peakall et al. (2003), we rejected the null hypothesis of no spatial autocorrelation at a specific distance class when both of the following criteria were met: a) r exceeded the 95% CI around the null hypothesis of r = 0 and b) the 95% CI about r (derived from bootstrapping) did not contain 0.
Results
Residency and population density.
The number of male and female prairie voles classified as residents based on nest trapping data and MNKA estimates of population density were highly variable between study sites and among years (Table 1). The total number of residents in Indiana in each of the 3 years of study was greater than that in every year in Kansas. Densities tended to be higher in Indiana than in Kansas, but in every year, population densities at both sites were considered “low” (< 100 voles/ha) according to the criteria established by Getz et al. (1993).
Microsatellite loci analyses.
For the microsatellite loci used to estimate pairwise relatedness, the proportion of loci typed ranged from 0.94 to 0.98 per year per population, and the probability of identity between 2 randomly chosen full siblings varied from 2×10−3 to 3×10−3. The number of alleles per locus ranged from 9 to 23 in the Kansas population (Table 2) and 4 to 21 in the Indiana population (Table 3). In the Kansas population, observed heterozygosity ranged from 0.40 to 0.97 while expected heterozygosity ranged from 0.42 to 0.94 (Table 2). In Indiana, the observed and expected heterozygosity ranged from 0.08 to 0.89 and 0.09 to 0.90, respectively (Table 3). After Bonferroni correction, only a single locus in 1 population in 1 year (Indiana 2008; Table 3) deviated significantly from Hardy−Weinberg expectations, likely due to a null allele. Since we used a small number of loci to assess relatedness in this study, the influence of a single locus on estimates of spatial genetic structure could potentially be substantial. Therefore, we did not include the genotypic data from the locus (MSMM3) that was not in Hardy−Weinberg equilibrium in the Indiana population in 2008, in our spatial autocorrelation analyses to examine the relationship between genetic and geographic distance among same-sex voles in Indiana.
Table 2.
Number of prairie voles (Microtus ochrogaster) genotyped (n), number of alleles per locus, and observed (H o) and expected (H e) heterozygosities for the 2005, 2006, and 2008 field seasons in Kansas.
| Locus | n | Number of alleles | H o | H e | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2005 | 2006 | 2008 | 2005 | 2006 | 2008 | 2005 | 2006 | 2008 | 2005 | 2006 | 2008 | |
| AV13 | 57 | 31 | 143 | 17 | 16 | 14 | 0.74 | 0.81 | 0.83 | 0.91 | 0.92 | 0.87 |
| MOE2 | 55 | 29 | 143 | 15 | 15 | 16 | 0.91 | 0.93 | 0.87 | 0.88 | 0.92 | 0.89 |
| MSMM2 | 54 | 29 | 136 | 17 | 17 | 17 | 0.96 | 0.97 | 0.90 | 0.92 | 0.94 | 0.92 |
| MSMM3 | 56 | 30 | 140 | 13 | 16 | 15 | 0.88 | 0.93 | 0.79 | 0.89 | 0.89 | 0.87 |
| MSMM5 | 57 | 31 | 138 | 20 | 19 | 23 | 0.77 | 0.61 | 0.87 | 0.93 | 0.90 | 0.93 |
| MSMM6 | 57 | 30 | 144 | 9 | 10 | 10 | 0.46 | 0.40 | 0.46 | 0.46 | 0.50 | 0.42 |
Table 3.
Number of prairie voles (Microtus ochrogaster) genotyped (n), number of alleles per locus, and observed (H o) and expected (H e) heterozygosities for the 2006, 2007, and 2008 field seasons in Indiana.
| Locus | n | Number of alleles | H o | H e | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2006 | 2007 | 2008 | 2006 | 2007 | 2008 | 2006 | 2007 | 2008 | 2006 | 2007 | 2008 | |
| AV13 | 109 | 283 | 326 | 14 | 14 | 13 | 0.88 | 0.85 | 0.85 | 0.87 | 0.86 | 0.87 |
| MOE2 | 119 | 282 | 328 | 16 | 17 | 15 | 0.76 | 0.82 | 0.85 | 0.86 | 0.85 | 0.84 |
| MSMM2 | 120 | 276 | 328 | 16 | 14 | 14 | 0.88 | 0.85 | 0.89 | 0.88 | 0.87 | 0.89 |
| MSMM3 | 117 | 259 | 299 | 11 | 12 | 10 | 0.87 | 0.82 | 0.81 | 0.85 | 0.86 | 0.85a |
| MSMM5 | 102 | 272 | 327 | 21 | 20 | 21 | 0.82 | 0.82 | 0.86 | 0.90 | 0.90 | 0.89 |
| MSMM6 | 120 | 285 | 328 | 8 | 10 | 4 | 0.14 | 0.08 | 0.09 | 0.17 | 0.10 | 0.09 |
a Locus not in Hardy–Weinberg equilibrium.
Relationship between relatedness and geographic distance.
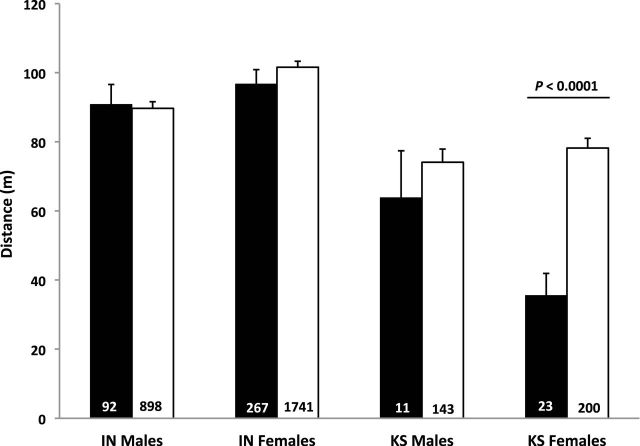
Estimates of pairwise relatedness among males and females in Kansas ranged from −0.4 to 0.47 and −0.36 to 1.0, respectively. In Indiana, pairwise relatedness ranged from −0.45 to 0.71 among males and −0.75 to 1.0 among females. The mean distance separating unrelated females in Kansas (78.2 m ± 6.4) was more than twice the mean distance separating related females (35.5 m ± 6.4; P < 0.0001; Fig. 1). No significant differences were detected between the mean distances separating unrelated and related males in Kansas (P = 0. 483), or voles of either sex in Indiana (females: P = 0.256; males: P = 0.852).
Fig. 1.
The mean distance (± SE) separating related (r ≥ 0.25, ■) versus unrelated (r < 0.25, □) resident adult voles (Microtus ochrogaster) for males and females compared separately within the Kansas (KS) and Indiana (IN) populations. Means were determined by combining the 3 years of data collected for each sex within each population. Sample sizes listed on bars.
The spatial autocorrelation coefficient r was significantly greater than expected by chance for both females (P = 0.001; Fig. 2a) and males (P = 0.013; Fig. 2b) at 20 m in the Kansas population, indicating that prairie voles separated by less than 20 m were significantly more related to same-sex conspecifics than expected by chance. None of the values of r were significant at distances greater than 20 m for males, but females showed significant negative spatial autocorrelation at distances of 80 m (P = 0.002) and 100 m (P = 0.002). In the Indiana population, we detected a significant positive spatial autocorrelation among females at distances less than 20 m (P = 0.033; Fig. 3a) but females appeared to be randomly distributed with respect to relatedness at all greater distances. Among males in the Indiana population, spatial autocorrelation analysis revealed a significant negative spatial autocorrelation at 80 m (P = 0.008; Fig. 3b), and a significant positive spatial autocorrelation at 220 m (P = 0.031; Fig. 3b).
Fig. 2.
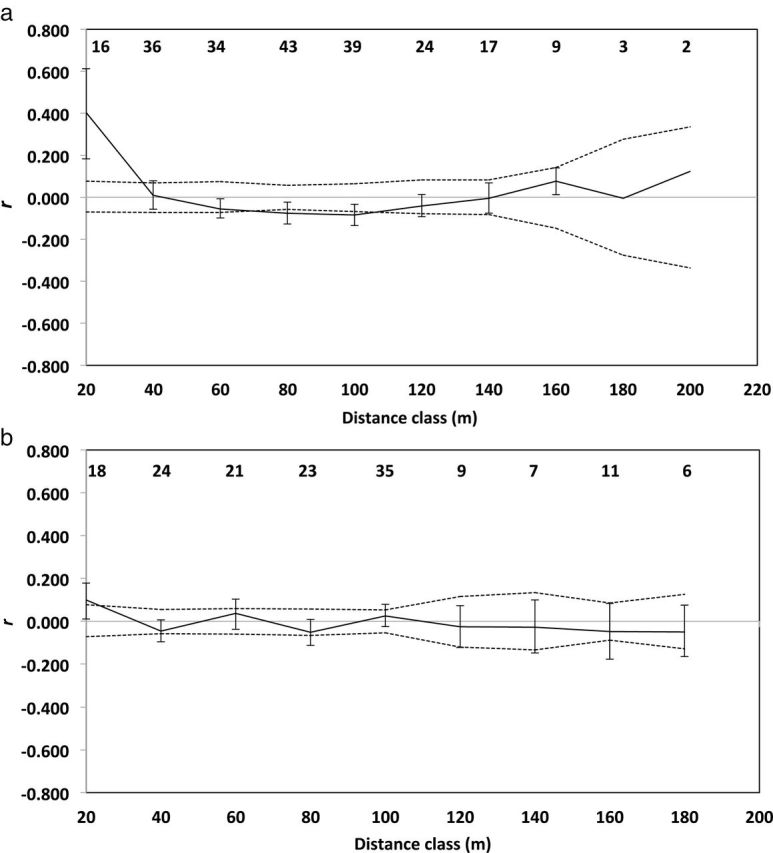
Correlograms illustrating the genetic correlation coefficient (r) as a function of geographical distance for resident adult a) female and b) male prairie voles (Microtus ochrogaster) in Kansas. Upper and lower permuted 95% CI (dashed lines) about the null hypothesis of no genetic structure (r = 0), and bootstrapped 95% CI error bars about r are shown. Sample sizes for each distance category are indicated at top of correlograms.
Fig. 3.
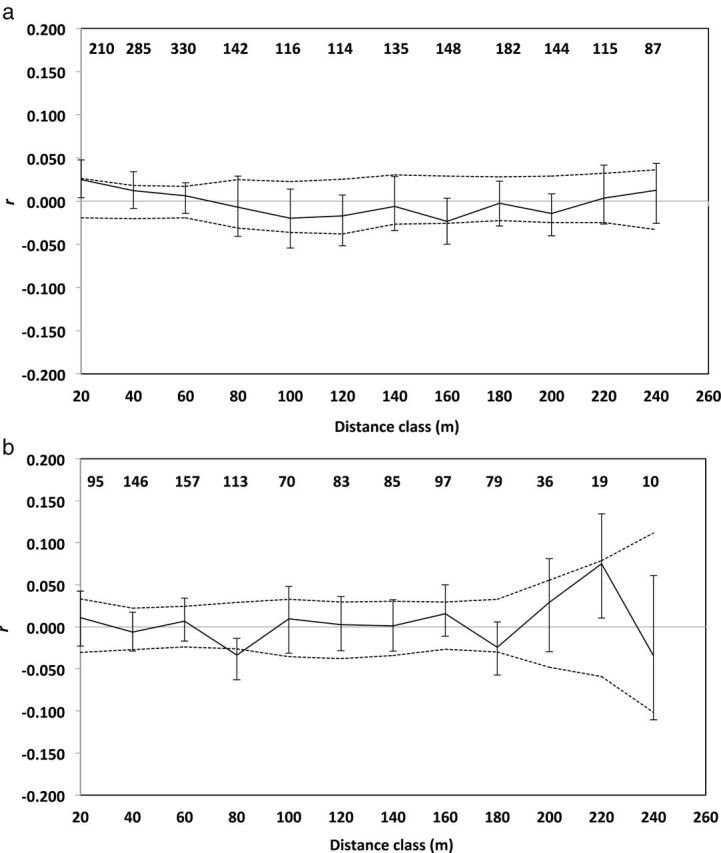
Correlograms illustrating the genetic correlation coefficient (r) as a function of geographical distance for resident adult a) female and b) male prairie voles (Microtus ochrogaster) in Indiana. Upper and lower permuted 95% CI (dashed lines) about the null hypothesis of no genetic structure (r = 0), and bootstrapped 95% CI error bars about r are shown. Sample sizes for each distance category are indicated at top of correlograms.
Discussion
High levels of natal philopatry are correlated with the spatial clustering of kin in a number of species of rodents (e.g., Australian bush rats—Peakall et al. 2003; dusky-footed woodrats—McEachern et al. 2007; banner tailed kangaroo rats—Busch et al. 2009; woodchucks—Maher 2009) and the previously documented high levels of natal philopatry in male and female prairie voles led us to expect that we would find spatial clustering among kin of both sexes within the 2 populations we studied. Our spatial autocorrelation analyses of nest residency and relatedness indicated significant positive genetic autocorrelations among adult females in both populations and males in the Kansas population residing at nests separated by distances up to 20 m, which is approximately the diameter of adult male and female home ranges in these populations (Streatfeild et al. 2011). These results are consistent with the hypothesis that limited natal dispersal results in the spatial clustering of kin among adult prairie voles of both sexes. However, contrary to our expectations, analyses of voles in the Indiana population showed no evidence that closely related males were spatially clustered. In general, the genetic similarity between resident adult male voles in Indiana was not significantly different from that expected for nonrelatives regardless of the geographic distance separating the nests at which individuals resided for distances up to about 250 m.
Whether neighboring prairie voles of either sex tend to be related or not has implications for the evolution of social behavior in this species. The development and maintenance of behaviors by kin selection requires relatively stable spatial associations of closely related individuals (Hamilton 1964). Our data indicate that opportunities for kin-selected social behaviors exist among adult females in both of the populations we studied. A study by Sera and Gaines (1994) indicated that female prairie voles display greater home range overlap with familiar relatives compared to nonrelatives. Although female survival and reproductive success was not correlated with the relatedness of neighboring individuals during the 7-week study of voles maintained in enclosed populations by Sera and Gaines (1994), the spatial clustering of female kin may have beneficial consequences over a longer time scale in natural populations. In Townsend’s voles, the 2-week survival of pups was significantly greater in a field population in which kin structure was manipulated to increase relatedness among neighboring resident female voles compared to a field population where the average relatedness among neighboring females was decreased through the removal of related individuals (Lambin and Yoccoz 1998).
One interpretation of our results for male voles from the Indiana population is that limited natal dispersal does not, in and of itself, result in the spatial clustering of kin. Fine-scale spatial genetic patterns within populations are almost certainly the consequence of factors (e.g., life history traits, mating system) other than natal dispersal and these factors may sometimes act to decrease the likelihood that close relatives reside in close proximity even when natal dispersal is limited. The average life expectancy for both male and female prairie voles is approximately 2−3 months, although estimates vary extensively due to factors such as population density, season of birth, and the type of social unit within which individuals are living (Getz et al. 1997). Male and female prairie voles become sexually mature at about 1 month of age (Solomon 1991), so, on average, a female will rear 1 litter of 3−4 pups during her lifetime. Therefore, there are unlikely to be more than a few siblings coexisting at the same time in most populations. In addition, we have found extra-pair paternity, which reduces relatedness among littermates, in both the Indiana and Kansas populations, with the frequency of genetic monogamy in the Kansas population (72%) about twice as great as in the Indiana population (39%—Streatfeild et al. 2011). Furthermore, in a population in Illinois, about a quarter of resident adult male and females prairie voles engage in additional dispersal events following natal dispersal (McGuire et al. 2013). Non-natal dispersal may take individuals farther from their natal nest and from related conspecifics. Thus, differences between the Kansas and Indiana populations in factors such as demography and mating system may result in different patterns of spatial genetic structure even when natal philopatry is widespread.
Furthermore, our expectation that we would detect evidence of the spatial clustering of kin in the 2 prairie vole populations we studied was based on the assumption that limited natal dispersal by both sexes is a relatively invariant feature of prairie vole populations. However, the reality for many species is that dispersal tends to vary with environmental conditions (Waser and Jones 1983; Matthysen 2005) and natal dispersal rates and distances in prairie voles are influenced by population density (Getz et al. 1993; Lucia et al. 2008), adult sex ratio (Sanders and Gaines 1991), and habitat quality (Lin et al. 2006). The average distance separating related resident adults of the same sex in Kansas was less than in Indiana for both males (~25 m less) and females (~60 m less) and in Kansas, the average distance separating related females (~36 m) was about half that of related males (~64 m). One explanation of these results is that voles in Kansas were more philopatric than in Indiana with Kansas voles also displaying female biased philopatry. If natal dispersal rates and distances differ between the 2 populations, these differences in philopatry may be attributable to population differences in ecological conditions that can differentially shape spatial genetic structure through effects on dispersal (Berthier et al. 2006; Busch et al. 2009). Although vole densities at both sites were considered low in each year of our study according to the criteria established by Getz et al. (1993), densities tended to be higher in Indiana. However, the Indiana site displayed no evidence of the spatial clustering of male kin despite higher densities.
The results of our study provide further evidence that a greater understanding of social behavior can be achieved by combining behavioral data from long-term field studies with genetic data. While the behavioral data on dispersal would lead one to expect kin clustering to be common in prairie vole populations due to high levels of natal philopatry in both sexes, and opportunities for kin-selected social behaviors to be high, the genetic data from the current study demonstrates that this is not necessarily the case. Our results indicate that spatial genetic structure may vary among prairie vole populations, possibly due to population-specific factors such as demography, mating behavior, and habitat quality, and that extrapolating findings from one population to others may be questionable. How typical the spatial clustering of kin is in prairie vole populations can only be addressed through further studies of populations where ecological conditions (e.g., density, mating system) vary temporally or spatially.
Acknowledgments
We thank G. Pittman and D. Kettle for logistical support at the Kansas University field site and K. Clay for logistical support at the Indiana University field site. Thanks to A. Kiss and C. Wood for their help at the Miami University Center for Bioinformatics and Functional Genomics. We also thank the numerous undergraduate students who assisted with the field and laboratory research. K. Anderson, T. Scheetz, and M. Eiswerth assisted with the construction of data matrices. Funding was provided by the National Science Foundation (IOS-0614015 to BK and NGS) and the National Institutes of Health (NIGMS GM 06409-01 to NGS).
Literature Cited
- Amos W., Hoffman J. I., Frodsham A., Zhang L., Best S., Hill A. V. S. 2006. Automated binning of microsatellite alleles: problems and solutions. Molecular Ecology Notes 7:10−14. [Google Scholar]
- Berthier K., Charbonnel N., Galan M., Chaval Y., Cosson J. F. 2006. Migration and recovery of the genetic diversity during the increasing density phase in cyclic vole populations. Molecular Ecology 15:2665−2676. [DOI] [PubMed] [Google Scholar]
- Bourke A. F. G. 1997. Sociality and kin selection in Insects. Pp. 203−227 in Behavioural ecology: an evolutionary approach ( J. R. Krebs and N. B. Davies , eds.) Blackwell Science, Cambridge, Massachusetts. [Google Scholar]
- Busch J. D., Waser P. M., DeWoody J. A. 2009. The influence of density and sex on patterns of fine-scale genetic structure. Evolution 63:2302−2314. [DOI] [PubMed] [Google Scholar]
- Carter C. S., Getz L. L. 1993. Monogamy and the prairie vole. Scientific American 268:100−106. [DOI] [PubMed] [Google Scholar]
- Chesh A. S., Mabry K. E., Noe D. A., Keane B., Solomon N. G. 2012. Influence of body mass and parasites on social partnerships and mating in prairie voles (Microtus ochrogaster). Journal of Mammalogy 93:229−238. [Google Scholar]
- Clutton-Brock T. 2002. Breeding together: kin selection and mutualism in cooperative vertebrates. Science 296:69−72. [DOI] [PubMed] [Google Scholar]
- Cochran G. R., Solomon N. G. 2000. Effects of food supplementation on the social organization of prairie voles (Microtus ochrogaster). Journal of Mammalogy 3:746−757. [Google Scholar]
- Dobson S. F. 1982. Competition for mates and predominant juvenile dispersal in mammals. Animal Behaviour 30:1183−1192. [Google Scholar]
- Emlen S. T. 1982. The evolution of helping: an ecological constraints model. American Naturalist 119:29–39. [Google Scholar]
- Emlen S. T. 1997. Predicting family dynamics in social vertebrates. Pp. 228−253 in Behavioural ecology: an evolutionary approach ( J. R. Krebs and N. B. Davies , eds.) Blackwell Science, Cambridge, Massachusetts. [Google Scholar]
- Gaines M. S., Vivas A. M., Baker C. L. 1979. An experimental analysis of dispersal in fluctuating vole populations: demographic parameters. Ecology 60:814−828. [Google Scholar]
- Getz L. L. 1997. Natal philopatry in the prairie vole, Microtus ochrogaster, in a low food habitat. American Midland Naturalist 138:412–413. [Google Scholar]
- Getz L. L., McGuire B., Carter C. S. 2003. Social behavior, reproduction and demography of the prairie vole Microtus ochrogaster . Ethology, Ecology and Evolution 15:105−118. [Google Scholar]
- Getz L. L., McGuire B., Hofmann J. E., Pizzuto T., Frase B. 1994. Natal dispersal and philopatry in prairie voles Microtus ochrogaster: settlement, survival, and potential reproductive success. Ethology, Ecology and Evolution 5:267−284. [Google Scholar]
- Getz L. L., McGuire B., Pizzuto T., Hofmann J. E., Frase B. 1993. Social organization of the prairie vole (Microtus ochrogaster). Journal of Mammalogy 74:44−58. [Google Scholar]
- Getz L. L., Simms L. E., McGuire B., Snarski M. E. 1997. Factors affecting life expectancy of the prairie vole, Microtus ochrogaster . Oikos 80:362−370. [Google Scholar]
- Greenwood P. J. 1980. Mating systems, philopatry and dispersal in birds and mammals. Animal Behaviour 28:1140−1162. [Google Scholar]
- Hamilton W. D. 1964. The genetical evolution of social behavior. Journal of Theoretical Biology 7:1−52. [DOI] [PubMed] [Google Scholar]
- Handley L. J. L., Perrin N. 2007. Advances in our understanding of mammalian sex-biased dispersal. Molecular Ecology 16:1559−1578. [DOI] [PubMed] [Google Scholar]
- Kalinowski S. T., Taper M. L., Marshall T. C. 2007. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Molecular Ecology 16:1099−1106. [DOI] [PubMed] [Google Scholar]
- Keane B., et al. 2007. No effect of body condition at weaning on survival and reproduction in prairie voles. Canadian Journal of Zoology 85:718−727. [Google Scholar]
- Kleiman D. 1977. Monogamy in mammals. Quarterly Review of Biology 52:39−69. [DOI] [PubMed] [Google Scholar]
- Lambin X., Yoccoz N. G. 1998. The impact of population kin-structure on nestling survival in Townsend’s voles, Microtus townsendii . Journal of Animal Ecology 67:1–16. [Google Scholar]
- Lin Y. K., Keane B., Isenhour A., Solomon N. G. 2006. Effects of patch quality on dispersal and social organization of prairie voles: an experimental approach. Journal of Mammalogy 87:446–453. [Google Scholar]
- Lucia K. E., Keane B., Hayes L. D., Kirk Lin Y., Schaefer R. L., Solomon N. G. 2008. Philopatry in prairie voles: an evaluation of the habitat saturation hypothesis. Behavioral Ecology 19:774−783. [Google Scholar]
- Mabry K. E. Streatfeild C. A. Keane B. and Solomon N. G.. 2011. avpr1a length polymorphism is not associated with either social or genetic monogamy in free-living prairie voles (Microtus ochrogaster). Animal Behaviour 81:11−18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher C. R. 2009. Genetic relatedness and space use in a behaviorally flexible species of marmot, the woodchuck (Marmota monax). Behavioral Ecology and Sociobiology 63:857−868. [Google Scholar]
- Matthysen E. 2005. Density-dependent dispersal in birds and mammals. Ecography 28:403−416. [Google Scholar]
- McEachern M. B., Eadie J. M., Van Vuren D. H. 2007. Local genetic structure and relatedness in a solitary mammal, Neotoma fuscipes . Behavioral Ecology and Sociobiology 61:1459–1469. [Google Scholar]
- McGuire B., Getz L. L., Bemis W. E., Oli M. K. 2013. Social dynamics and dispersal in free-living prairie voles (Microtus ochrogaster). Journal of Mammalogy 94:40−49. [Google Scholar]
- McGuire B., Getz L. L., Hofmann J. E., Pizzuto T., Frase B. 1993. Natal dispersal in prairie voles, Microtus ochrogaster, in relation to population density, season, and natal social environment. Behavioral Ecology and Sociobiology 32:293−302. [Google Scholar]
- Murray B. G. 1967. Dispersal in vertebrates. Ecology 48:975−978. [Google Scholar]
- Myers J. H., Krebs C. J. 1971. Genetic, behavioral, and reproductive attributes of dispersing field voles Microtus pennsylvanicus and Microtus ochrogaster . Ecological Monographs 41:53−78. [Google Scholar]
- Peakall R., Ruibal M., Lindenmayer D. B. 2003. Spatial autocorrelation analysis offers new insights into gene flow in the Australian bush rat, Rattus fuscipes . Evolution 57:1182−1195. [DOI] [PubMed] [Google Scholar]
- Peakall R., Smouse P. E. 2006. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes 6:288−295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pusey A. E. 1987. Sex-biased dispersal and inbreeding avoidance in birds and mammals. Trends in Ecology and Evolution 2:295−299. [DOI] [PubMed] [Google Scholar]
- Pusey A., Wolf M. 1996. Inbreeding avoidance in animals. Trends in Ecology and Evolution 11:201−206. [DOI] [PubMed] [Google Scholar]
- Queller D. C., Goodnight K. F. 1989. Estimating relatedness using genetic markers. Evolution 43:258−275. [DOI] [PubMed] [Google Scholar]
- Rose R. H., Gaines M. S. 1978. The reproductive cycle of Microtus ochrogaster in eastern Kansas. Ecological Monographs 48:21−42. [Google Scholar]
- Sambrook J., Fritch E. F., Maniatus T. 1989. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor Press, Cold Spring Harbor, New York. [Google Scholar]
- Sanders A. K., Gaines M. S. 1991. The influence of sex ratio on dispersal in the prairie vole, Microtus ochrogaster . Transactions of the Kansas Academy of Science 94:142–152. [Google Scholar]
- Sera W., Gaines M. S. 1994. The effect of relatedness on spacing behavior and fitness of female prairie voles. Ecology 75:1560–1566. [Google Scholar]
- Sikes R. S., W. L., Gannon, and the Animal Care and Use Committee of the American Society of Mammalogists. 2011. Guidelines of the American Society of Mammalogists for the use of wild mammals in research. Journal of Mammalogy 92:235−253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slade N. A., Blair S. M. 2000. An empirical test of counts of individuals captured as indices of population size. Journal of Mammalogy 81:1035−1045. [Google Scholar]
- Solomon N. G. 1991. Age of pairing affects reproduction in prairie voles. Laboratory Animal 25:232−235. [DOI] [PubMed] [Google Scholar]
- Solomon N. G., Getz L. L. 1997. Examination of alternative hypotheses for cooperative breeding in rodents. Pp. 199−230 in Cooperative breeding in mammals ( N. G. Solomon and J. A. French , eds.). Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- Solomon N. G., Keane B., Knoch L. R., Hogan P. J. 2004. Multiple paternity in socially monogamous prairie voles (Microtus ochrogaster). Canadian Journal of Zoology 82:1667−1671. [Google Scholar]
- Solomon N. G., et al. 2009. Polymorphism at the avpr1a locus in male prairie voles correlated with genetic but not social monogamy in field populations. Molecular Ecology 18:4680−4695. [DOI] [PubMed] [Google Scholar]
- Streatfeild C. A., Mabry K. E., Keane B., Crist T. O., Solomon N. G. 2011. Intraspecific variability in the social and genetic mating systems of prairie voles (Microtus ochrogaster). Animal Behaviour 82:1387−1398. [Google Scholar]
- Waser P. M. 1985. Does competition drive dispersal? Ecology 66:1170−1175. [Google Scholar]
- Waser P. M., Jones W. T. 1983. Natal philopatry among solitary mammals. Quarterly Review of Biology 58:355−390. [Google Scholar]
- West-Eberhard M. J. 1975. The evolution of social behavior by kin selection. Quarterly Review of Biology 50:1−33. [Google Scholar]
- Wilson E. O. 1975. Sociobiology: the new synthesis. Harvard University Press, Cambridge, Massachusetts. [Google Scholar]
- Young L. J., Wang Z. 2004. The neurobiology of pair-bonding. Nature Reviews Neuroscience 7:1048−1054. [DOI] [PubMed] [Google Scholar]