Abstract
Background
A variety of biological functions of estrogens, including regulation of energy metabolism, are mediated by neurons expressing estrogen receptor-α (ERα) in the brain. However, complex intracellular processes in these ERα-expressing neurons are difficult to unravel, due to the lack of strategy to visualize ERα-expressing neurons, especially in unfixed brain tissues.
Results and Conclusions
Here we generated a novel ERα-ZsGreen reporter mouse line in which expression of a green fluorescent reporter protein, ZsGreen, is driven by a 241 kb ERα gene promoter. We validated that ZsGreen is highly colocalized with endogenous ERα in the brain. Native ZsGreen signals were visualized in unfixed brain tissue, and were used to assist single cell collection and electrophysiological recordings. Finally, we demonstrated that this ERα-ZsGreen mouse allele can be used in combination with other genetic reporter alleles to allow experiments in highly selective neural populations.
Keywords: transgenic mouse, brain, estrogen receptor-α, electrophysiology
Introduction
Estrogen receptor-α (ERα) plays important roles in many functions throughout the central nervous system (CNS). For example, during early development, signals mediated by brain ERα, as well as other ER isoforms (e.g. ERβ), are essential to establish sex differences, promote neurite growth, and regulate synaptic patterning [1]. In addition, ERα in adult brains plays important roles in a variety of physiological processes, including female fertility [2, 3], sexual behaviors [4], cardiovascular activities [5] and cognition [6].
In particular, accumulating evidence indicates that ERα-expressing neurons in the brain are critically involved in the regulation of energy homeostasis [7–9]. This notion was first supported by early observations that microinjections of ERα agonists into various brain regions change animal’s feeding behavior and body weight [10, 11]. Confirming a critical role of brain ERα in body weight control, we showed that mutant mice lacking ERα selectively in the brain develop obesity despite higher circulating levels of 17β-estradiol [12]. Recent efforts in the field have led to an appreciation that estrogenic actions on different aspects of energy balance are mediated by distinct ERα neural populations in the brain. For example, the inhibitory effects of 17β-estradiol on food intake are mediated by ERα expressed in the arcuate hypothalamic nucleus (ARH) [12], the dorsal nucleus raphe (DR) [13] and the nucleus of the solitary tract (NTS) [14]; ERα in the ventrolateral subdivision of the ventromedial hypothalamic nucleus (vlVMH), however, mediates estrogenic actions to enhance energy expenditure but does not affect feeding [12, 15–17]; ERα in the medial amygdalar nucleus (MeA) is functionally required to maintain normal physical activity [18].
With these advances in understanding the critical brain ERα populations in energy homeostasis, increasing efforts have been spent on unravelling the complex cellular behaviors and the underlying mechanisms of these ERα neurons [15, 19, 20]. However, characterizing the phenotype and understanding the functions of ERα-expressing neurons in the brain has been challenging, mainly because of the lack of tools to allow direct visualization of ERα-expressing cells, which are often located in brain regions comprised of heterogeneous neurons with various neurochemical identities. For example, ERα expressed by pro-opiomelanocortin (POMC) neurons in the ARH has been implicated in the regulation of food intake [12]. However, the effects of ERα signals on POMC neuron functions are difficult to assess, because only a portion (about 30%) of POMC neurons express ERα while many other POMC neurons do not [12]. Adding to the complexity, within the ARH, there are abundant ERα-expressing neurons that do not express POMC [21]. Thus, in order to rigorously study a specific ERα population (e.g. ERα (+) POMC neurons), one would need a tool to reliably identify ERα-expressing cells of interest.
In this study, we used the bacterial artificial chromosome (BAC) engineering to generate a new transgenic mouse line, namely ERα-ZsGreen, in which expression of a green fluorescent reporter protein (ZsGreen) was driven by a long (241 kb) ERα gene promoter. We used several different approaches to validate that the ZsGreen reporter can be used, in either fixed or unfixed brain tissue, to identify ERα-expressing cells. We further tested effects of a selective ERα agonist on the firing activities of ZsGreen (+) cells in the ARH. Finally, we demonstrated that this ERα-ZsGreen mouse allele can be used in combination with other genetic reporter alleles to allow experiments in double-selected neural populations.
Methods
Generation of ERα-ZsGreen transgenic mice
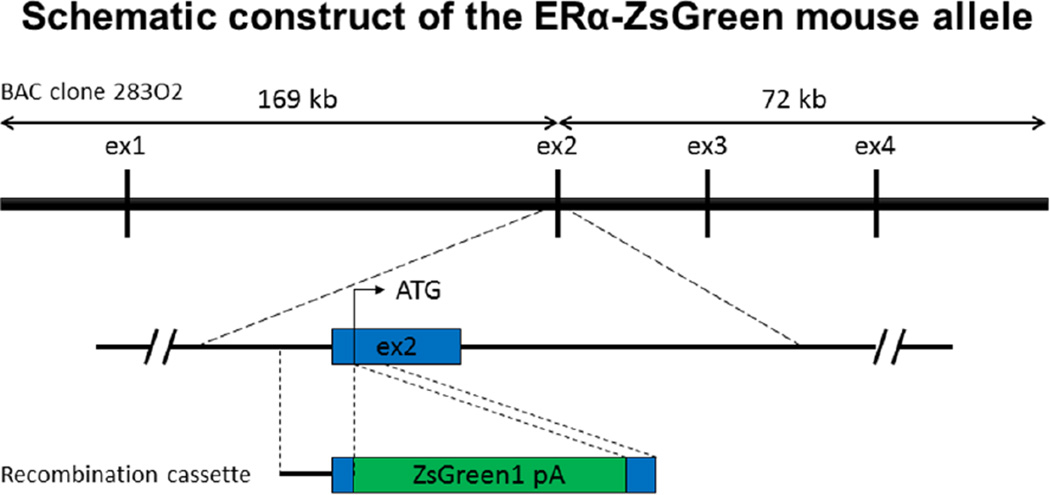
A BAC clone (#RP23-283O2) was obtained from BAC PAC Resource Center at Children’s Hospital Oakland Research Institute (CHORI). This BAC clone was derived from the C57BL/6J mouse strain and covers exon 1 to 4 of ERα gene. ZsGreen1 cDNA was prepared from pIRES-ZsGreen1 plasmid (Clontech, Mountain View, CA). The engineering of BAC transgenic vector was conducted in E. coli DH10B cells, using the Counter-Selection BAC Modification Kit (Gene Bridges, Heidelberg, Germany) following the manufacturer’s manual. As illustrated in Figure 1, the ZsGreen cDNA was introduced into translation start site in exon 2 of the ERα gene in the BAC clone.
Figure 1. Schematic construction of the ERα-ZsGreen transgenic mouse allele.
The BAC clone 283O2 covers 241 kb around Esr1 genomic sequence (5’, 169 kb; 3’, 72 kb), which contains exon1–4 out of the Esr1 gene. For BAC recombination cassette, 393-bp of 5’ flanking region and 148-bp of 3’ flanking region were used as homology arms. By homologous recombination in bacteria, the ZsGreen cDNA was introduced into translation start site of the Esr1 gene in the BAC clone.
Large scale of the recombinant ERα-ZsGreen BAC DNA was prepared by NucleoBond Xtra BAC Kit (Macherey-Nagel, Germany), which were linearized by PI-SceI digestion, purified with phenol-chloroform, precipitated with ethanol. Transgenic mice were obtained by injecting the linearized ERα-ZsGreen BAC DNA into pronuclei of C57BL/6 mice in the Genetically Engineered Mouse Core at Baylor College of Medicine. ERα-ZsGreen founders were maintained at a C57BL/6 background. Tail biopsies were used for the genotyping on PCR with primers Esr1ATG-393 F 5’-CCAGCAGCGAAGACCTGGAAAGT-3’ and ZsGr+236 R 5’ – ATGTCCTGGGGGTACTCGGTGAA-3’ producing the amplicon of 629 bp long.
Mice
ERα-ZsGreen mice were bred with C57Bl6 mice to produce ERα-ZsGreen mice for studies. In parallel, ERα-ZsGreen mice were bred with mice that carried both POMC-CreER [22] and Rosa26-tdTOMATO alleles [23] to produce ERα-ZsGreen/POMC-CreER/ Rosa26-tdTOMATO triple transgenic offspring. These ERα-ZsGreen/POMC-CreER/ Rosa26-tdTOMATO mice received i.p. injection of tamoxifen (0.2 mg/g) at 6 weeks of age to induce Cre recombinase activity.
Care of all animals and procedures were approved by Baylor College of Medicine Institutional Animal Care and Use Committee. Mice were housed in a temperature-controlled environment at 22°C–24°C using a 12 hr light/12 hr dark cycle. The mice were fed on standard chow with minimal phytoestrogens (4% fat, #2916, Harlan-Teklad) and water was provided ad libitum. All mice were on a pure C57Bl/6 background.
Tissue processing for histology
Both male and female ERα-ZsGreen mice (8–10 weeks of age) were anesthetized with isofluorane inhalation and were perfused intracardially with saline followed by 10% formalin. To avoid the influences of the estrous cycle on ERα expression, all female mice were perfused at the diestrus which was determined by vaginal cytology. The brains were removed, postfixed two hours in the same fixative, and then immersed in 20% sucrose in PBS at 4°C until sinking in the solution. 25-µm-thick coronal sections throughout whole brains were cut by freezing microtome, collected in cryoprotective solution (20% glycerol, 30% ethylene glycol in PBS) and stored at −20°C until use.
DAB-immunohistochemistry for ZsGreen
Whole immunostaining procedures were carried out with free-floating sections at room temperature. After thorough washing in PBS, to block endogenous peroxidase activity, sections were treated with 3% H2O2 in methanol for 30 min and washed in PBS. Sections were blocked for 1 hr with blocking solution (PBS containing 3% normal goat serum and 0.25% Triton-X), and then incubated overnight in blocking solution containing rabbit anti-ZsGreen antibody (1:1000, Clontech, Mountain View, CA). After washing in PBS, sections were incubated in blocking solution containing biotinylated goat anti-rabbit secondary antibody (1:1000, Jackson Immunoresearch Laboratories, Inc. West Grove, PA) for 2 hours, then washed again and incubated in avidin-biotin complex (Vectastain ABC Elite kit, Vector laboratories, Inc. Burlingame, CA). Immunolabeled ZsGreen was visualized with 0.1% diaminobenzidine/0.01% cobalt chloride/0.01% nickel sulfate/0.01% H2O2 in PBS. Coloring was stopped by rinsing sections thoroughly in PBS. Sections were then mounted on Poly-L-Lysine glass slides and air dried. Finally, they were dehydrated with ascending series of ethanol, cleared in xylene, and coverslipped with Cytoseal (Richard Allen Scientific, Kalamazoo, MI). Photographs were taken with Leica Microscope using MM AF software. Anatomical identification of brain structures was conducted with the aid of Allen Reference Atlas (mouse.brain-map.org).
Colocalization of ERα and ZsGreen
Brain sections were prepared as same as in immunoperoxidase labelling. After thorough washing in PBS, sections were incubated for 1 hr with blocking solution (PBS containing 3% normal goat serum and 0.25% Triton-X), and then incubated overnight in blocking solution containing rabbit anti-ERα antibody (1:1000, Millipore, Billerica, MA). After washing in PBS, sections were incubated in blocking solution containing biotinylated goat anti-rabbit secondary antibody (1:1000, Jackson Immunoresearch Laboratories, Inc) for 2 hours, then washed again and incubated in blocking solution containing Dylight 594 streptavidin (1:500, Jackson Immunoresearch Laboratories, Inc.). After washing in PBS, the sections were mounted on Poly-L-Lysine glass slides and air dried. Finally, they were coverslipped with Vectashield mounting medium with DAPI (Vector Laboratories). Under the Leica 5500 fluorescence microscope equipped with optigrid illumination, native ZsGreen signals were directly visualized with the green fluorescence filter and ERα immunoreactivity was visualized with the red fluorescence filter.
Single-cell RT-PCR analysis
Six to twelve-week old ERα-ZsGreen mice were deeply anesthetized with isoflurane inhalation in early morning. Mice were then decapitated, and the entire brain was removed and immediately submerged in ice-cold sucrose-based cutting solution (adjusted to pH 7.3) containing (in mM) 10 NaCl, 25 NaHCO3, 195 Sucrose, 5 Glucose, 2.5 KCl, 1.25 NaH2PO4, 2 Na pyruvate, 0.5 CaCl2, 7 MgCl2 bubbled continuously with 95% O2 and 5% CO2, as we did before [13]. The slices containing the ARH region (250 µm) were cut with a Microm HM 650V vibratome (Thermo Scientific). Slices were incubated for 15 min at room temperature in PIPES buffer (in mM): 120 NaCl, 5 KCl, 1 CaCl2, 1 MgCl2, 25 D-glucose, 20 PIPES, 100% O2 [24], and then for 60 min at 33°C in PIPES containing trypsin (Sigma-Aldrich, type XI; 0.5 mg/ml). After enzymatic treatment, slices were rinsed and maintained in PIPES buffer at room temperature for 60 min. Slices were transferred to DMEM buffer (Invitrogen, Life Technologies), and the ARH region was identified and excised under a fluorescence-equipped dissecting microscope. The tissue was triturated gently in DMEM buffer using a series of fire-polished Pasteur pipettes (600, 300, and 150 µm, inner diameter) and the DMEM/neuron suspension was placed in a recording chamber on a fixed-stage fluorescence microscope (Eclipse FN-1, Nikon).
16 Single ZsGreen(+) and 16 ZsGreen (−) cells were visualized and collected via micropipette, and put in a PCR tube containing reverse transcription reaction mixture. cDNA was synthesized using qScript cDNA SuperMix (Quanta Biosciences, MD), according to the manufacturer’s instruction. After the RT reaction, regular PCR was performed to detect ERα and β-actin (as an internal control) using two-step PCR protocol with REDExtract-N-Amp PCR Reaction Mix (Sigma-Aldrich, St. Louis, MO) and SsoAdvanced Universal SYBR Green Supermix (Bio-Rad, Hercules, CA). The first round multiplex PCR contained 3 µl of cDNA, 2 µl of primer sets, 10 µl of REDExtract-N-Amp PCR Reaction Mix, and 5 µl of water. The PCR condition was composed of an initial 4 min at 94°C, then 30 cycles of 15 sec at 94°C, 60 sec at 60°C, 90 sec at 72°C, and 10 min at 72°C. The first PCR was purified using NucleoSpin PCR Clean-up Kit (Macherey-Nagel, Germany) and eluted with 25 µl of the bundled Buffer NE. The second round PCR was performed individually for each gene using SsoAdvanced Universal SYBR Green Supermix with 5µl SYBR Green, 1.2 µl primer set, 3.8 µl purified first round PCR. The amplicons of the second round PCR were separated on 12% polyacrylamide gel electrophoresis for ERα or on 1.5% agarose gel electrophoresis for β-actin. Primer sequences and the size of amplicon are shown below: ERα (5’-TGATGATTGGTCTCGTCTG-3’ and 5’-TACCTTGATTCCTGTCCAG-3’, 87 bp), and β-actin (5’-ATGGAGGGGAATACAGCC-3’ and 5’-TTCTTTGCAGCTCCTTCGTT-3’, 148 bp). For the ERα primers, the forward and reverse primers hybridize to exon 6 and exon 6/7 junction, respectively, which are not covered by the BAC clone. As a positive control, total cDNA extracted from a mouse hypothalamus was used and subjected to regular PCR using the same ERα primers.
Electrophysiology
ERα-ZsGreen mice or ZsGreen/POMC-CreER/Rosa26-tdTOMATO mice (at 6–10 weeks of age) were used for electrophysiological recordings. Briefly, mice were deeply anesthetized with isoflurane and then decapitated, and the entire brain was removed and immediately submerged in ice-cold sucrose-based cutting solution. Using a Microm HM 650V vibratome (Thermo Scientific), the brains were cut into coronal slices (without trimming at other dimension) at the thickness of 250 µm each. Usually, three to four consecutive brain slices per mouse, ranging from Bregma −2.54 mm to −1.46 mm, contained the ARH and adjacent nuclei (the ventromedial hypothalamic nucleus, the dorsal medial hypothalamus and the lateral hypothalamus) were used for recordings. The slices were recovered for 1 h at 34°C and then maintained at room temperature in artificial cerebrospinal fluid (aCSF, adjusted to pH7.3) containing (in mM) 126 NaCl, 2.5 KCl, 2.4 CaCl2, 1.2 NaH2PO4, 1.2 MgCl2, 11.1 glucose, and 21.4 NaHCO3) saturated with 95% O2 and 5% CO2 before recording.
Slices were transferred to the recording chamber and allowed to equilibrate for at least 10 min before recording. The slices were superfused at 34°C in oxygenated aCSF at a flow rate of 1.8–2 ml/min. ZsGreen(+) neurons, ZsGreen(−) neurons, or ZsGreen(+)/TOMATO(+) neurons in the ARH were visualized using epifluorescence and IR-DIC imaging on an upright microscope (Eclipse FN-1, Nikon) equipped with a moveable stage (MP-285, Sutter Instrument). Patch pipettes with resistances of 3–5 MΩ were filled with intracellular solution (adjusted to pH 7.3) containing (in mM) 128 K gluconate, 10 KCl, 10 HEPES, 0.1 EGTA, 2 MgCl2, 0.3 Na-GTP and 3 Mg-ATP. Recordings were made using a MultiClamp 700B amplifier (Axon Instrument), sampled using Digidata 1440A and analyzed offline with pClamp 10.3 software (Axon Instrument). Series resistance was monitored during the recording, and the values were generally < 10 MΩ and were not compensated. The liquid junction potential was +12.5 mV, and was corrected after the experiment. Data were excluded if the series resistance increased more than 20% during the experiment or without overshoot for action potential. Currents were amplified, filtered at 1 kHz, and digitized at 20 kHz. Frequency and peak amplitude were measured using the Mini Analysis program (Synaptosoft, Inc.). Current clamp was engaged to test neural firing frequency at the baseline and after bath perfusion of 100 nM propyl pyrazole triol (PPT, a selective ERα agonist) [25] for 6 min. The concentration of PPT was chosen based on previous studies showing that effects of PPT at this dose were blocked by genetic deletion of ERα [13]. The values for resting membrane potential and firing frequency were averaged within 2-min bin at the baseline or after PPT perfusion. In some experiments, the aCSF solution also contained 1 µM tetrodotoxin (TTX) [26] and a cocktail of fast synaptic inhibitors, namely bicuculline (50 µM; a GABA receptor antagonist) [27], AP-5 (30 µM; an NMDA receptor antagonist) [28] and CNQX (30 µM; an AMPA receptor antagonist) [28] to block the majority of presynaptic inputs.
Statistics
The data for electrophysiology are presented individually for each recorded neuron. Statistical analyses were performed using GraphPad Prism. Comparisons between before and after PPT treatment were made by paired t-test. P<0.05 was considered to be statistically significant.
Results
Distribution of ZsGreen in ERα-ZsGreen mouse brains
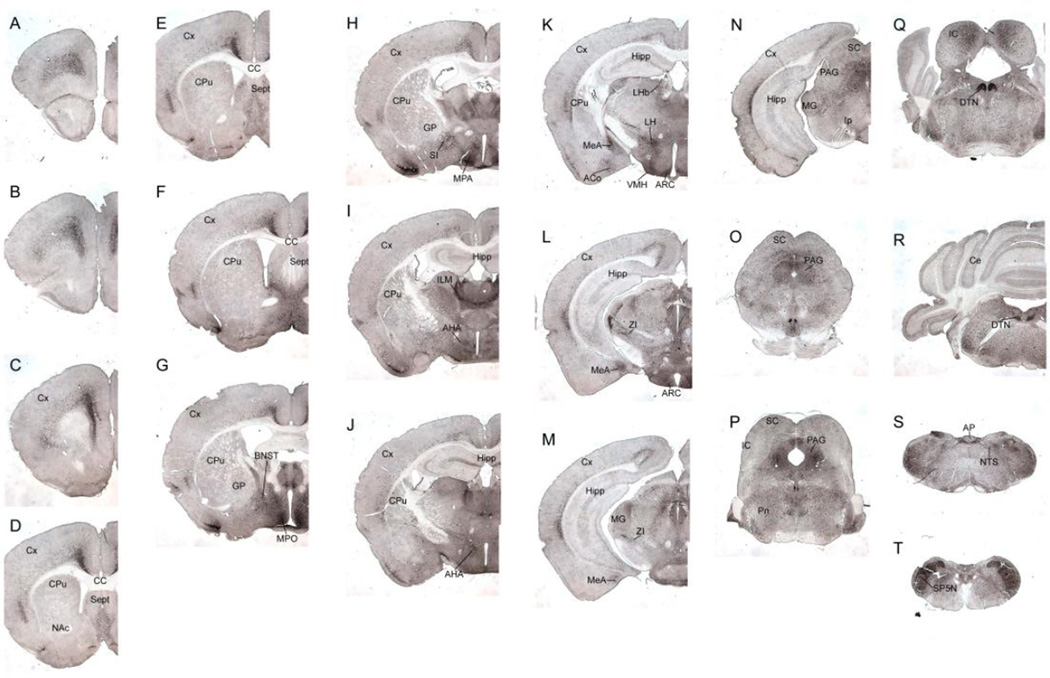
Three transgenic ERα-ZsGreen founder lines were obtained. We used an anti-ZsGreen antibody to detect ZsGreen immunoreactivity in the brains from each line. Line 1 did not express any ZsGreen in the brain; ZsGreen immunoreactivity was detectable in both line 2 and line 3, with stronger signals in line 2 than in line 3. We then systemically characterized the distribution of ZsGreen in fixed brain sections from line 2, using DAB-immunohistochemistry (Figure 2). As summarized in Table 1, the distribution pattern of ZsGreen in our ERα-ZsGreen mouse is largely consistent with distribution of endogenous ERα immunoreactivity in mouse brains as reported previously [29].
Figure 2. Distribution of ZsGreen (+) cells throughout the brain.
Images of coronal sections of the ERα-ZsGreen transgenic mouse brain are arranged from rostral to caudal separated by a distance of 400–600 µm. Images shown here were taken in same magnification. Scale bars=1 mm. AHA, anterior hypothalamic area; AON, anterior olfactory nucleus; AP, area postrema; ARH, arcuate hypothalamic nucleus; BNST, bed nucleus of the stria terminalis; CA, CA1–3; Cbx, cerebellum; CoA, cortical amygdalar nucleus; CPu, striatum; CT, central nucleus of the thalamus; Ctx, cerebral cortex; DG, dentate gyrus; DTN, dorsal tegmental nucleus; ENT, entorhinal cortex; IC, inferior colliculus; LG, lateral geniculate complex, thalamus; LHA, lateral hypothalamic area; LHb, lateral habenula; LPO, lateral preoptic area; LS, lateral septum; MeA, medial amygdalar nucleus; MPO, medial preoptic area; MS, medial septum; NAc, nucleus accumbens; NDB, diagonal band of Broca; NTS, nucleus of the solitary tract; PAG, periaqueductal grey; PIR, piriform cortex; PM, premammillary nucleus; PV, periventricular hypothalamic nucleus; PVT, paraventricular nucleus of the thalamus; SC, superior colliculus; SP5N, spinal trigeminal nucleus; VMH, ventromedial hypothalamic nucleus; ZI, zona incerta.
Table 1.
Distribution of ZsGreen expressing cells throughout the brain
| Anatomical structure | ZsGreen expression |
|---|---|
| Telencephalon | |
| Olfactory bulb | |
| Anterior olfactory nucleus | +/− |
| Isocortex | |
| Layers 2–3 | − |
| Layer 6 | ++ |
| Piriform cortex | − |
| Entorhinal cortex | + |
| Hippocampal formation | |
| CA1 – CA3 | + |
| Dentate gyrus | + |
| Septum | |
| Medial | ++ |
| Lateral | ++ |
| Diagonal band of Broca | + |
| Bed nucleus of the stria terminalis | ++ |
| Amygdala | |
| Basolateral nucleus | +/− |
| Basomedial nucleus | + |
| Central nucleus | +/− |
| Cortical nucleus | ++ |
| Medial nucleus | ++ |
| Nucleus accumbens | + |
| Striatum | + |
| Diencephalon | |
| Thalamus | |
| Anterior nuclei (paraventricular) | ++ |
| Central nuclei | ++ |
| Lateral geniculate nucleus | − |
| Zona incerta | ++ |
| Habenula | |
| Medial | − |
| Lateral | ++ |
| Hypothalamus | |
| Preoptic area | +/− |
| Periventricular | +/− |
| Anterior hypothalamic area | ++ |
| Periventricular nucleus | +/− |
| Paraventricular nucleus | +/− |
| Retrochiasmatic area | − |
| Suprachiasmatic nucleus | − |
| Supraoptic nucleus | − |
| Arcuate nucleus | ++ |
| Dorsomedial nucleus | +/− |
| Lateral hypothalamus | ++ |
| Ventromedial nucleus | |
| Dorsal | − |
| Ventral | ++ |
| Premammillary nuclei | + |
| Mesencephalon | |
| Interpeduncular nucleus | − |
| Periaqueductal gray | ++ |
| Raphe nuclei, dorsalis | +/− |
| Metencephalon | |
| Cerebellum (Purkinje cells) | +/− |
| Myelencephalon | |
| Area postrema | ++ |
| Nucleus of the solitary tract | + |
| Spinal nucleus of the trigeminal | ++ |
ZsGreen expression was evaluated based on number of intense ly-stained cells in each brain sites. Anatom ical structures were defined based on Allen Reference Atlas. −, not detected; +/−, low/faint; +, moderate; ++, high.
Colocalization of ZsGreen and endogenous ERα
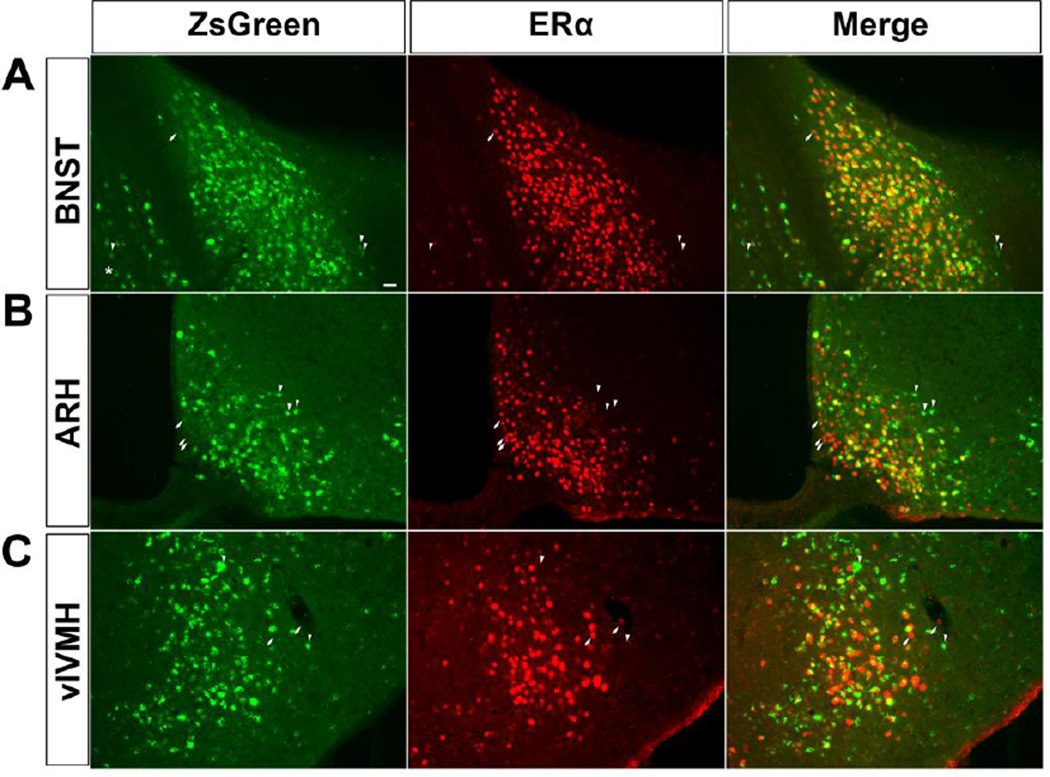
We then directly visualized native ZsGreen signals, without any immunohistochemical staining procedures, in fixed brain sections obtained from both male and female mice. ZsGreen signals were distributed broadly in the cells, including the nucleus and the cytosol (Figure 3). Interestingly, in some cells, ZsGreen signals appeared to punctuate in intracellular organelles (asterisk-pointed neurons in Figure 3). We also used immunofluorescence to detect the endogenous ERα in the same brain sections, and examined the colocalization of native ZsGreen and ERα immunoreactivity in several brain regions with abundant ERα, including the bed nucleus of the stria terminalis (BNST), the arcuate hypothalamic nucleus (ARH) and the ventrolateral subdivision of the ventromedial hypothalamic nucleus (vlVMH). In all these three regions, the majority of ZsGreen (+) cells co-expressed ERα; similarly, most of ERα (+) cells co-expressed ZsGreen (Figure 3). In particular, more than 90% of ZsGreen (+) neurons in female brains were confirmed to co-express ERα; this overlapping pattern was slightly lower in male brains (80–95%, Table 2). In addition, we found that about 70–87% ERα (+) neurons in male/female brains were labelled by native ZsGreen (Table 2).
Figure 3. Colocalization of ZsGreen and ERα.
Colocalization of native ZsGreen (left), ERα immunoreactivity (middle), and the merge (right) in the BNST (A), ARH (B) and vlVMH (C). Scale bar = 50 µm. Note that most cells co-express both ZsGreen and ERα; a few cells that only express ZsGreen are indicated by arrowheads; a few cells that only express ERα are indicated by arrows; the asterisk indicates a cell with punctuated ZsGreen signals.
Table 2.
Colocalization of native ZsGreen and ERα immunoreactivity
| Male | Female | |||
|---|---|---|---|---|
| Brain Regions |
Double/ZsGreen | Double/ERα | Double/ZsGreen | Double/ERα |
| BNST | 81.05±2.11% | 71.68±5.70% | 97.88±0.34% | 83.12±2.29% |
| ARH | 94.12±0.88% | 55.35±1.55% | 95.22±0.49% | 70.06±1.7% |
| vlVMH | 83.78±1.94% | 86.81±3.61% | 90.62±1.02% | 73.20±5.33% |
ZsGreen (+) single cells express ERα mRNA
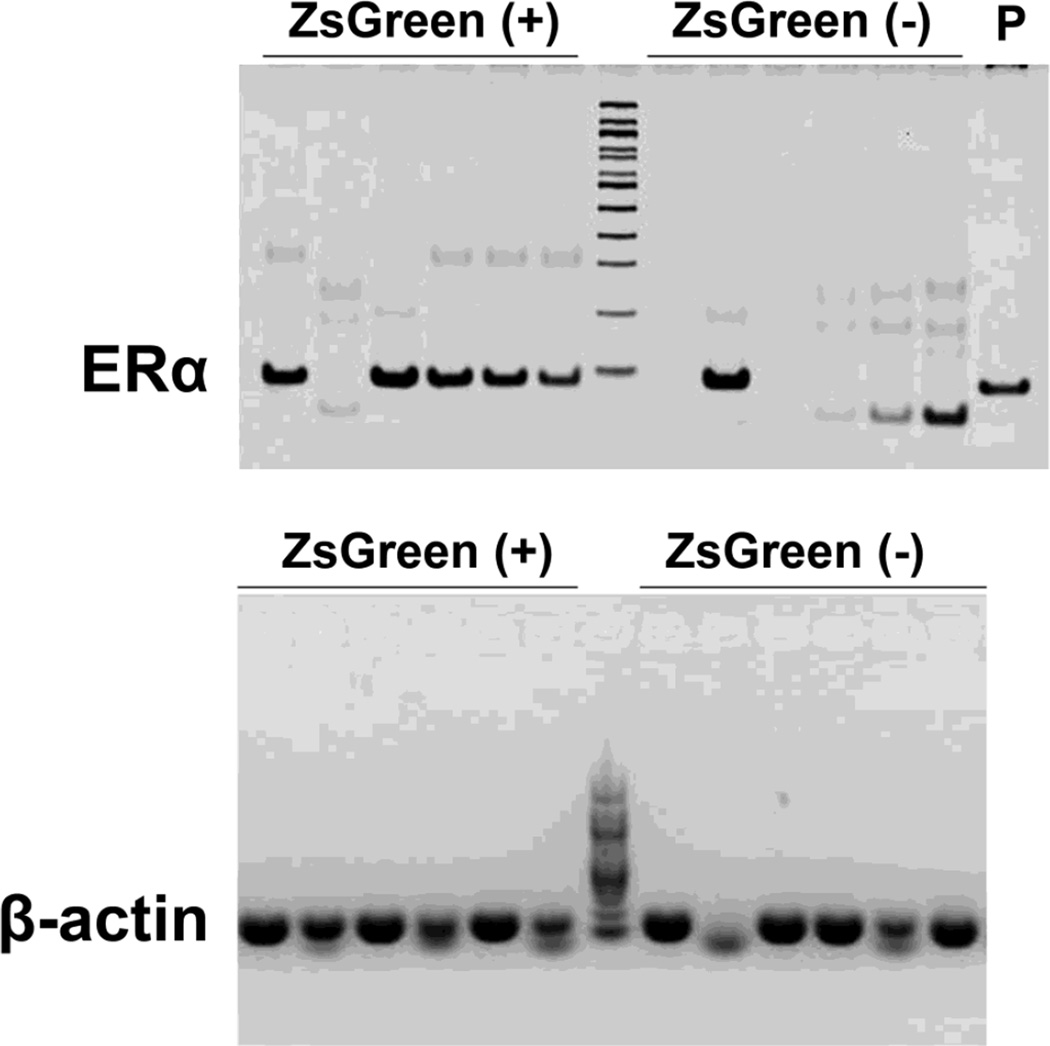
To further confirm that ZsGreen (+) cells do express ERα mRNA, we collected ZsGreen (+) single cells in the ARH from unfixed brain slices, and performed single cell RT-PCR to detect ERα mRNA. Remarkably, ERα mRNA was detected in 14 out of 16 ZsGreen (+) cells (see Figure 4 for representative data). As a negative control, we also isolated ZsGreen (−) single cells from the same brain slices, and only detected ERα mRNA in 4 out 16 cells (see Figure 4 for representative data). β-actin mRNA was detected in most ZsGreen (+) and ZsGreen (−) single cells, confirming good quality of mRNA samples (Figure 4).
Figure 4. ZsGreen(+) cells express ERα mRNA.
RT-PCR detecting ERα (upper panel) and β-actin (lower panel) mRNAs in single ZsGreen (+) or ZsGreen (−) cells isolated from the ARH of ERα-ZsGreen mice. P indicates a positive control, showing that the same ERα primers produced an amplicon from total cDNA extracted from a mouse hypothalamus.
An ERα agonist stimulates firing of ZsGreen (+) cells in the ARH
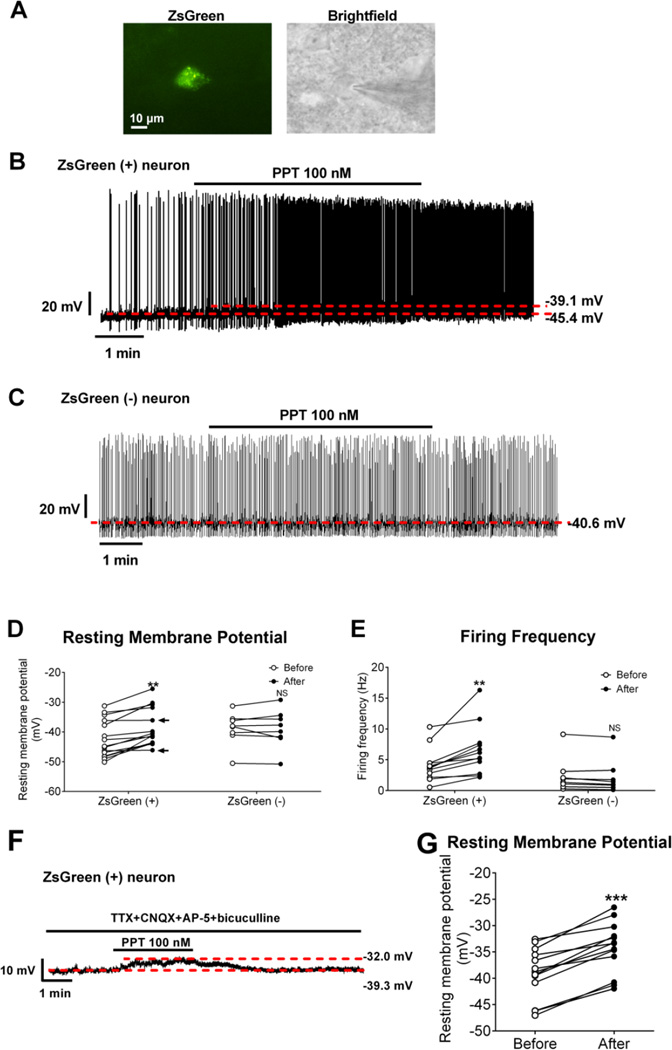
Using the whole-cell current clamp electrophysiology, we examined effects of a highly selective ERα agonist, propyl pyrazole triol (PPT) [25] on the firing activities of ZsGreen (+) cells in the ARH (Figure 5A). We showed that PPT (100 nM, bath for 6 min) depolarized 13/15 ZsGreen (+) cells tested (see Figure 5B for representative traces and Figure 5D for summary). We further analysed effects of PPT on firing frequency in 12 ZsGreen (+) cells with spontaneous firing activity and found that PPT significantly increased firing frequency in these cells (see Figure 5B for representative traces and Figure 5E for summary). We also tested effects of PPT on 8 ZsGreen (−) cells, and failed to detect any effects (see Figure 5C for representative traces and Figure 5D–5E for summary). In addition, we found that PPT depolarized 13/13 ZsGreen (+) cells in the presence of TTX and a cocktail of fast synaptic inhibitors (CNQX, AP-5 and bicuculline) (see Figure 5F for representative traces and Figure 5G for summary), which confirmed a direct action of PPT on these ZsGreen (+) cells.
Figure 5. Effects of a selective ERα agonist on neural activities of ZsGreen (+) and ZsGreen (−) cells in the ARH.
(A) Micrographic images showing a recorded ZsGreen (+) neuron in the ARH. Left panel: the neuron visualized under the green fluorescence microscope; right panel: the same neurons patch clamped by a micropipette visualized under the brightfield microscope. Scale bar=10 µm. (B–C) Representative responses to PPT (100 nM, bath) in ZsGreen (+) cells (B) and in ZsGreen (−) cells (C). (D–E) Summary data of resting membrane potential (D) and firing frequency (E) before and after PPT treatment. Data are presented for each cell. **, P<0.01 in paired t-tests. Note that the arrows in (D) point to the two ZsGreen (+) cells that were not depolarized by PPT. (F) Representative responses to PPT (100 nM, bath) in ZsGreen (+) cells in the presence of 1 µM TTX, 30 µM CNQX, 30 µM AP-5 and 50 µM bicuculline. (G) Summary data of resting membrane potential before and after PPT treatment in the presence of 1 µM TTX, 30 µM CNQX, 30 µM AP-5 and 50 µM bicuculline. Data are presented for each cell. ***, P<0.001 in paired t-tests.
Electrophysiological recordings from identified ERα (+) POMC neurons in the ARH
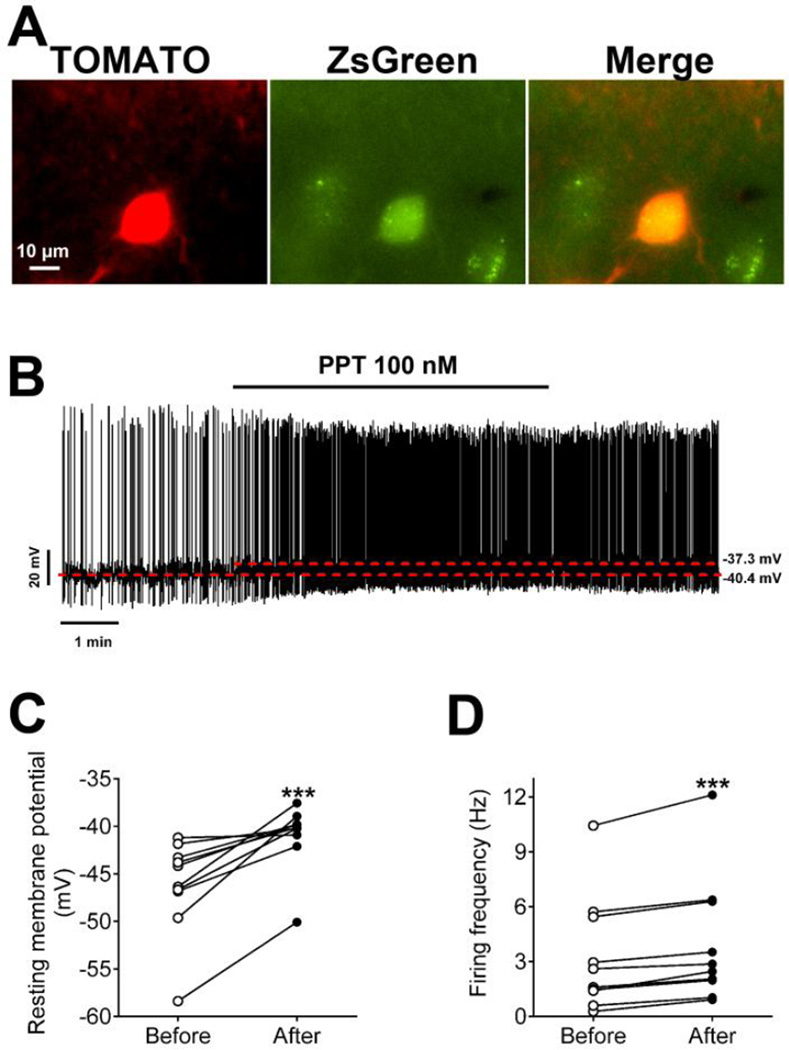
We then crossed both the POMC-CreER [22] and Rosa26-tdTOMATO alleles [23] onto our ERα-ZsGreen mice to produce ERα-ZsGreen/POMC-CreER/Rosa26-tdTOMATO mice. Tamoxifen was injected into these mice to induce Cre recombinase activity in POMC neurons, which led to expression of the red fluorescence protein, TOMATO, specifically in POMC neurons. Thus, cells labelled by both TOMATO and ZsGreen in the ARH were identified as ERα (+) POMC neurons (Figure 6A). Using the whole-cell current clamp electrophysiology, we examined effects of PPT on the firing activities of ERα (+) POMC neurons, and found that PPT (100 nM, bath for 6 min) significantly depolarized these neurons and increased their firing frequency (n = 10; see Figure 6B for representative traces and Figure 6C–6D for summary).
Figure 6. Effects of a selective ERα agonist on neural activities of ER (+) POMC neurons in the ARH.
(A) Micrographic images showing a recorded ERα (+) POMC neuron in the ARH. Left panel: TOMATO signal visualized under the red fluorescence microscope; middle panel: ZsGreen signal visualized under the green fluorescence microscope; right panel: merge. Scale bar=10 µm. (B) Representative responses to PPT (100 nM, bath). (C–D) Summary data of resting membrane potential (C) and firing frequency (D) before and after PPT treatment. Data are presented for each cell. ***, P<0.001 in paired t-tests.
Discussions
In the present study, we generated a new reporter mouse line, in which expression of a green fluorescence protein, namely ZsGreen, is driven by a long ERα promoter (about 241 kb). Several different approaches were used to validate this model. First, we showed that the distribution of ZsGreen is largely consistent with reported ERα expression pattern in mouse brains [29]. We further confirmed that the majority of neurons labelled by native ZsGreen signals co-express endogenous ERα. In addition, using native ZsGreen signals as a guide, we collected single cells from unfixed brain slices, and showed that most of these ZsGreen (+) cells express ERα mRNA. At the functional level, we showed that a selective ERα agonist exclusively stimulated ZsGreen (+) cells in the ARH, but not ZsGreen (−) cells in the same region. Collectively, these results validated that ZsGreen is faithfully expressed in ERα-expressing cells in the brain.
Prior to our studies, several ERα promoter-driven reporter lines have been described. However, the fidelity of reporter expression in these lines was less than ideal. For example, Hamada et al. generated a transgenic rat expressing eGFP under the control of a 2.1 kb rat ERα gene promoter [30]. In these rats, eGFP was barely detected in the ARH and vlVMH, regions where abundant ERα is expressed [29] and functional [12]. Further, in the BNST, only 70% of eGFP (+) neurons co-express ERα, and even a smaller portion of ERα (+) neurons are labelled by eGFP [12]. More recently, Matsuda et al. generated a transgenic mouse line in which expression of eGFP is driven by a 6 kb mouse ERα gene promoter [31]. Similar to the rat reporter, many ERα (+) neurons in the BNST, vlVMH and ARH do not express eGFP in this ERα-eGFP mouse line [31]. On the other hand, we demonstrated a greatly improved fidelity in our ERα-ZsGreen reporter line. We speculate that this improvement resulted from the much longer promoter (approximately 241 kb) we used to drive ZsGreen expression, which may have been more faithful to the endogenous ERα promoter activity.
Lee et al. developed and validated an Esr1Cre mouse line that expresses Cre recombinase in ERα-positive cells [32]. In combination with viral vectors expressing Cre-inducible optogenetic channels (e.g. channelrhodopsin and halorhodopsin), this Esr1Cre mouse line was used to achieve photostimulation or photoinhibition of ERα (+) neurons in the vlVMH [32]. Notably, one can use the similar strategy, injecting Cre-inducible reporter viral vectors, to label ERα (+) neurons in any region of mouse brains, although precise stereotaxic viral injections could be technically challenging. Alternatively, one can cross this Esr1Cre mouse line to a Cre-inducible reporter line (e.g. Rosa26-tdTOMATO) to generate offspring mice expressing the reporter selectively in ERα (+) cells. However, one caveat is that the reporter will be expressed not only in “current” ERα (+) cells, but also in cells with only transient ERα promoter activity. Given the dynamic changes in brain ERα expression during development [33] and depending on the estrogen milieu [34], these “transient” ERα (+) cells are expected to exist in large numbers and may substantially confound the experiments.
Our ERα-ZsGreen reporter mouse line offers a reliable and easy-to-use tool to visualize ERα (+) cells in mouse brains, which could benefit the field of estrogen research though various experimental designs. Obviously, the ZsGreen fluorescence reporter can replace the conventional staining procedures (e.g. immunohistochemistry or in situ hybridization) required to visualize ERα (+) cells; ZsGreen can also be easily combined with another staining protocol to determine if ERα (+) cells express other proteins or mRNAs. In addition to these histological applications in usually fixed tissues, one can also use ZsGreen as a marker to directly identify ERα (+) cells in unfixed tissues. For example, we took advantage of this feature to collect single ERα (+) cells. In addition, one can use flow cytometry to sort out highly purified ERα (+) cells; the similar approach has been applied to purify other neural populations which facilitated the discovery of novel factors that regulate functions of specific neural populations [35, 36].
In addition, as we did in this study, one can prepare fresh brain slices from ERα-ZsGreen mice, and perform electrophysiological recordings in identified ERα (+) cells. One step further is to cross the ERα-ZsGreen transgene onto mice that express another fluorescent reporter in selective neural population. For example, here we crossed the ERα-ZsGreen mice with POMC-CreER/Rosa26-tdTOMATO mice to generate mice carrying all these 3 transgenic alleles. In the brain slices obtained from the triple genetic offspring, we easily identified ERα (+) POMC neurons as double-labelled (yellow) neurons; electrophysiological recordings from these neurons demonstrated that the ERα agonist stimulated firing activities of these ERα (+) POMC neurons. The significance of these results is at least two folds. First, we provided direct evidence that activation of ERα can stimulate firing of hypothalamic POMC neurons, which could mediate the physiological functions of estrogenic actions in these neurons to prevent overeating and obesity [12]. In addition, the fact that all double-labelled neurons responded to the ERα agonist provided the proof of the principle that the ERα-ZsGreen mouse line could be used in combination with other genetically labelled reporter lines to allow experiments in highly selective neural populations. These will no doubt advance our understanding about molecular and cellular actions of ERα in various neural populations.
In summary, we generated and validated a new reporter mouse line which faithfully expresses a fluorescence protein in ERα (+) cells in the brain. Although the expression of this reporter in peripheral tissues warrants further characterizations in future, we propose that this tool can be used in a variety of experimental designs, with fixed or unfixed brains, to study estrogenic actions in specific neural populations.
Acknowledgments
This work was supported by grants from the NIH (R01DK093587 and R01DK101379 to YX; R01DK092605 to QT), American Heart Association postdoctoral fellowship (PX), American Heart Association National Scientist Development Grant (QT).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest. No potential conflicts of interest relevant to this article were reported.
References
- 1.McCarthy MM. Estradiol and the developing brain. Physiol Rev. 2008;88:91–124. doi: 10.1152/physrev.00010.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dubois SL, Acosta-Martinez M, DeJoseph MR, Wolfe A, Radovick S, Boehm U, et al. Positive, but not negative feedback actions of estradiol in adult female mice require estrogen receptor alpha in kisspeptin neurons. Endocrinology. 2015;156:1111–1120. doi: 10.1210/en.2014-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wintermantel TM, Campbell RE, Porteous R, Bock D, Grone HJ, Todman MG, et al. Definition of estrogen receptor pathway critical for estrogen positive feedback to gonadotropin-releasing hormone neurons and fertility. Neuron. 2006;52:271–280. doi: 10.1016/j.neuron.2006.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee H, Kim DW, Remedios R, Anthony TE, Chang A, Madisen L, et al. Scalable control of mounting and attack by Esr1 neurons in the ventromedial hypothalamus. Nature. 2014 doi: 10.1038/nature13169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spary EJ, Maqbool A, Batten TF. Oestrogen receptors in the central nervous system and evidence for their role in the control of cardiovascular function. J Chem Neuroanat. 2009;38:185–196. doi: 10.1016/j.jchemneu.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 6.Hara Y, Waters EM, McEwen BS, Morrison JH. Estrogen Effects on Cognitive and Synaptic Health Over the Lifecourse. Physiol Rev. 2015;95:785–807. doi: 10.1152/physrev.00036.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saito K, Cao X, He Y, Xu Y. Progress in the molecular understanding of central regulation of body weight by estrogens. Obesity (Silver Spring) 2015;23:919–926. doi: 10.1002/oby.21099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frank A, Brown LM, Clegg DJ. The role of hypothalamic estrogen receptors in metabolic regulation. Front Neuroendocrinol. 2014;35:550–557. doi: 10.1016/j.yfrne.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faulds MH, Zhao C, Dahlman-Wright K, Gustafsson JA. The diversity of sex steroid action: regulation of metabolism by estrogen signaling. J Endocrinol. 2012;212:3–12. doi: 10.1530/JOE-11-0044. [DOI] [PubMed] [Google Scholar]
- 10.Butera PC, Beikirch RJ. Central implants of diluted estradiol: independent effects on ingestive and reproductive behaviors of ovariectomized rats. Brain Res. 1989;491:266–273. doi: 10.1016/0006-8993(89)90062-0. [DOI] [PubMed] [Google Scholar]
- 11.Palmer K, Gray JM. Central vs. peripheral effects of estrogen on food intake and lipoprotein lipase activity in ovariectomized rats. Physiol Behav. 1986;37:187–189. doi: 10.1016/0031-9384(86)90404-x. [DOI] [PubMed] [Google Scholar]
- 12.Xu Y, Nedungadi TP, Zhu L, Sobhani N, Irani BG, Davis KE, et al. Distinct hypothalamic neurons mediate estrogenic effects on energy homeostasis and reproduction. Cell Metab. 2011;14:453–465. doi: 10.1016/j.cmet.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao X, Xu P, Oyola MG, Xia Y, Yan X, Saito K, et al. Estrogens stimulate serotonin neurons to inhibit binge-like eating in mice. J Clin Invest. 2014;124:4351–4362. doi: 10.1172/JCI74726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Asarian L, Geary N. Estradiol enhances cholecystokinin-dependent lipid-induced satiation and activates estrogen receptor-alpha-expressing cells in the nucleus tractus solitarius of ovariectomized rats. Endocrinology. 2007;148:5656–5666. doi: 10.1210/en.2007-0341. [DOI] [PubMed] [Google Scholar]
- 15.Martinez de Morentin PB, Gonzalez-Garcia I, Martins L, Lage R, Fernandez-Mallo D, Martinez-Sanchez N, et al. Estradiol Regulates Brown Adipose Tissue Thermogenesis via Hypothalamic AMPK. Cell Metab. 2014;20:41–53. doi: 10.1016/j.cmet.2014.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Musatov S, Chen W, Pfaff DW, Mobbs CV, Yang XJ, Clegg DJ, et al. Silencing of estrogen receptor alpha in the ventromedial nucleus of hypothalamus leads to metabolic syndrome. Proc Natl Acad Sci U S A. 2007;104:2501–2506. doi: 10.1073/pnas.0610787104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Correa SM, Newstrom DW, Warne JP, Flandin P, Cheung CC, Lin-Moore AT, et al. An estrogen-responsive module in the ventromedial hypothalamus selectively drives sex-specific activity in females. Cell Rep. 2015;10:62–74. doi: 10.1016/j.celrep.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu P, Cao X, He Y, Zhu L, Yang Y, Saito K, et al. Estrogen receptor-alpha in medial amygdala neurons regulates body weight. J Clin Invest. 2015;125:2861–2876. doi: 10.1172/JCI80941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park CJ, Zhao Z, Glidewell-Kenney C, Lazic M, Chambon P, Krust A, et al. Genetic rescue of nonclassical ERalpha signaling normalizes energy balance in obese Eralpha-null mutant mice. J Clin Invest. 2011;121:604–612. doi: 10.1172/JCI41702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu L, Zou F, Yang Y, Xu P, Saito K, Othrell Hinton A, Jr, et al. Estrogens prevent metabolic dysfunctions induced by circadian disruptions in female mice. Endocrinology. 2015;156:2114–2123. doi: 10.1210/en.2014-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frazao R, Cravo RM, Donato J, Jr, Ratra DV, Clegg DJ, Elmquist JK, et al. Shift in Kiss1 cell activity requires estrogen receptor alpha. J Neurosci. 2013;33:2807–2820. doi: 10.1523/JNEUROSCI.1610-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berglund ED, Liu C, Sohn JW, Liu T, Kim MH, Lee CE, et al. Serotonin 2C receptors in pro-opiomelanocortin neurons regulate energy and glucose homeostasis. J Clin Invest. 2013;123:5061–5070. doi: 10.1172/JCI70338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kay AR, Wong RK. Isolation of neurons suitable for patch-clamping from adult mammalian central nervous systems. J Neurosci Methods. 1986;16:227–238. doi: 10.1016/0165-0270(86)90040-3. [DOI] [PubMed] [Google Scholar]
- 25.Stauffer SR, Coletta CJ, Tedesco R, Nishiguchi G, Carlson K, Sun J, et al. Pyrazole ligands: structure-affinity/activity relationships and estrogen receptor-alpha-selective agonists. J Med Chem. 2000;43:4934–4947. doi: 10.1021/jm000170m. [DOI] [PubMed] [Google Scholar]
- 26.Sohn JW, Harris LE, Berglund ED, Liu T, Vong L, Lowell BB, et al. Melanocortin 4 receptors reciprocally regulate sympathetic and parasympathetic preganglionic neurons. Cell. 2013;152:612–619. doi: 10.1016/j.cell.2012.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Y, Liu H, Sauvey C, Yao L, Zarnowska ED, Zhang SC. Directed differentiation of forebrain GABA interneurons from human pluripotent stem cells. Nat Protoc. 2013;8:1670–1679. doi: 10.1038/nprot.2013.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu T, Kong D, Shah BP, Ye C, Koda S, Saunders A, et al. Fasting activation of AgRP neurons requires NMDA receptors and involves spinogenesis and increased excitatory tone. Neuron. 2012;73:511–522. doi: 10.1016/j.neuron.2011.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merchenthaler I, Lane MV, Numan S, Dellovade TL. Distribution of estrogen receptor alpha and beta in the mouse central nervous system: in vivo autoradiographic and immunocytochemical analyses. J Comp Neurol. 2004;473:270–291. doi: 10.1002/cne.20128. [DOI] [PubMed] [Google Scholar]
- 30.Hamada T, Wada-Kiyama Y, Sakuma Y. Visualizing forebrain-specific usage of an estrogen receptor alpha promoter for receptor downregulation in the rat. Brain Res Mol Brain Res. 2005;139:42–51. doi: 10.1016/j.molbrainres.2005.05.019. [DOI] [PubMed] [Google Scholar]
- 31.Matsuda KI, Yanagisawa M, Sano K, Ochiai I, Musatov S, Okoshi K, et al. Visualisation and characterisation of oestrogen receptor alpha-positive neurons expressing green fluorescent protein under the control of the oestrogen receptor alpha promoter. Eur J Neurosci. 2013;38:2242–2249. doi: 10.1111/ejn.12227. [DOI] [PubMed] [Google Scholar]
- 32.Lee H, Kim DW, Remedios R, Anthony TE, Chang A, Madisen L, et al. Scalable control of mounting and attack by Esr1+ neurons in the ventromedial hypothalamus. Nature. 2014;509:627–632. doi: 10.1038/nature13169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yokosuka M, Okamura H, Hayashi S. Postnatal development and sex difference in neurons containing estrogen receptor-alpha immunoreactivity in the preoptic brain, the diencephalon, and the amygdala in the rat. J Comp Neurol. 1997;389:81–93. doi: 10.1002/(sici)1096-9861(19971208)389:1<81::aid-cne6>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 34.Shughrue PJ, Bushnell CD, Dorsa DM. Estrogen receptor messenger ribonucleic acid in female rat brain during the estrous cycle: a comparison with ovariectomized females and intact males. Endocrinology. 1992;131:381–388. doi: 10.1210/endo.131.1.1612018. [DOI] [PubMed] [Google Scholar]
- 35.Ren H, Orozco IJ, Su Y, Suyama S, Gutierrez-Juarez R, Horvath TL, et al. FoxO1 Target Gpr17 Activates AgRP Neurons to Regulate Food Intake. Cell. 2012;149:1314–1326. doi: 10.1016/j.cell.2012.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Henry FE, Sugino K, Tozer A, Branco T, Sternson SM. Cell type-specific transcriptomics of hypothalamic energy-sensing neuron responses to weight-loss. Elife. 2015;4 doi: 10.7554/eLife.09800. [DOI] [PMC free article] [PubMed] [Google Scholar]