ABSTRACT
The positive-sense RNA genome of Sweet potato feathery mottle virus (SPFMV) (genus Potyvirus, family Potyviridae) contains a large open reading frame (ORF) of 3,494 codons translatable as a polyprotein and two embedded shorter ORFs in the −1 frame: PISPO, of 230 codons, and PIPO, of 66 codons, located in the P1 and P3 regions, respectively. PISPO is specific to some sweet potato-infecting potyviruses, while PIPO is present in all potyvirids. In SPFMV these two extra ORFs are preceded by conserved G2A6 motifs. We have shown recently that a polymerase slippage mechanism at these sites could produce transcripts bringing these ORFs in frame with the upstream polyprotein, thus leading to P1N-PISPO and P3N-PIPO products (B. Rodamilans, A. Valli, A. Mingot, D. San Leon, D. B. Baulcombe, J. J. Lopez-Moya, and J.A. Garcia, J Virol 89:6965–6967, 2015, doi:10.1128/JVI.00337-15). Here, we demonstrate by liquid chromatography coupled to mass spectrometry that both P1 and P1N-PISPO are produced during viral infection and coexist in SPFMV-infected Ipomoea batatas plants. Interestingly, transient expression of SPFMV gene products coagroinfiltrated with a reporter gene in Nicotiana benthamiana revealed that P1N-PISPO acts as an RNA silencing suppressor, a role normally associated with HCPro in other potyviruses. Moreover, mutation of WG/GW motifs present in P1N-PISPO abolished its silencing suppression activity, suggesting that the function might require interaction with Argonaute components of the silencing machinery, as was shown for other viral suppressors. Altogether, our results reveal a further layer of complexity of the RNA silencing suppression activity within the Potyviridae family.
IMPORTANCE Gene products of potyviruses include P1, HCPro, P3, 6K1, CI, 6K2, VPg/NIaPro, NIb, and CP, all derived from the proteolytic processing of a large polyprotein, and an additional P3N-PIPO product, with the PIPO segment encoded in a different frame within the P3 cistron. In sweet potato feathery mottle virus (SPFMV), another out-of-frame element (PISPO) was predicted within the P1 region. We have shown recently that a polymerase slippage mechanism can generate the transcript variants with extra nucleotides that could be translated into P1N-PISPO and P3N-PIPO. Now, we demonstrate by mass spectrometry analysis that P1N-PISPO is indeed produced in SPFMV-infected plants, in addition to P1. Interestingly, while in other potyviruses the suppressor of RNA silencing is HCPro, we show here that P1N-PISPO exhibited this activity in SPFMV, revealing how the complexity of the gene content could contribute to supply this essential function in members of the Potyviridae family.
INTRODUCTION
Potyviruses (family Potyviridae) are important viral pathogens with positive-sense, single-stranded RNA genomes that are able to infect a wide range of plant species. The genomic RNA of potyviruses, around 10 kb in size with a viral protein (VPg) at its 5′ end and polyadenylated at the 3′ end, contains a large open reading frame (ORF) that encodes a polyprotein comprising the following gene products from the N to the C terminus: P1, HCPro, P3, 6K1, CI, 6K2, VPg/NIaPro, NIb, and CP (1, 2). Despite the abundant sequence information available on potyviruses, it was not until 2008 that the presence of a well-conserved second short ORF of around 60 codons, termed PIPO (Pretty Interesting Potyviral ORF) by its discoverers, was predicted as an overlapping product within the P3 region in all members of the family. Thus, this ORF yields a fusion product with the upstream portion of P3 after frameshift (P3N-PIPO), as found in plants infected with turnip mosaic virus (TuMV) (3). More recently, another short ORF termed PISPO (Pretty Interesting Sweet potato Potyviral ORF) was predicted by bioinformatics analysis within the P1-coding sequence of a few members of the Potyvirus genus, all related to sweet potato feathery mottle virus (SPFMV), including sweet potato virus G (SPVG) and sweet potato virus 2 (SPV2), with the notable exception of sweet potato latent virus, a potyvirus described as latent (4, 44). In the reference genome of SPFMV (NC_001841.1) (5), the PISPO sequence is nested in the −1 frame (relative to the polyprotein ORF) within the P1-coding region (positions 118 to 2109), which corresponds to the first N-terminal gene product of the large polyprotein (ORF from position 118 to 10599). The PISPO sequence begins at position 1382 and spans 690 nucleotides from the GGAAAAAA (G2A6) motif. This motif is identical to the conserved consensus sequence for the PIPO frameshifting, which in SPFMV gives rise to a shorter coding sequence, also in the −1 frame.
Separation of P1 from the other viral products occurs by autoproteolysis (6), and except for the conserved C-terminal protease region, this product is the most variable of the potyvirus proteins (7). SPFMV P1 is the largest among all potyvirus P1 proteins, with 664 to 724 amino acids (aa), resulting in a protein of 74.1 to 80 kDa. Similarity between the N-terminal parts of the P1 proteins of SPFMV and the ipomovirus sweet potato mild mottle virus (SPMMV) has been reported earlier (7). Interestingly, the resemblance between these two proteins ends near the predicted frameshift point of PISPO.
Since the PIPO coding sequence was first described in potyviruses, the new gene product P3N-PIPO has attracted much interest, leading to the identification of several important associated functions (8–12). However, the mechanism by which P3N-PIPO is produced remained unclear until recently, when we and another team independently found evidence that PIPO is expressed by a polymerase slippage mechanism (13, 14). Our previous results also support the idea that PISPO, if it was expressed, would be synthesized by an equivalent polymerase slippage event (14).
One of the key functions of RNA silencing in plants is as a defense barrier against viral infections (15). Viruses, in turn, use diverse strategies to escape from RNA silencing, such as the expression of viral proteins with RNA silencing suppression activity (16). For example, in the Potyviridae family, all members of the genus Potyvirus described so far express HCPro to counteract RNA silencing. RNA silencing suppression in plant viruses was recently reviewed (17).
By using liquid chromatography coupled to mass spectrometry (LC-MS), we show here that P1N-PISPO is produced in sweet potato plants infected with SPFMV, demonstrating that the product derived from this trans-frame viral ORF is indeed expressed during virus infection. Next, we describe that P1N-PISPO exhibits RNA silencing suppression activity, which is associated with the presence of conserved WG/GW motifs, suggesting a mode of action similar to that of other RNA silencing suppressors (RSSs) (18, 19).
MATERIALS AND METHODS
Plant and virus materials.
Commercial sweet potato (Ipomea batatas) roots were acquired in a local market in Barcelona and were planted on soil to produce fresh aerial tissue (stems and leaves) that was further propagated through cuttings at Center for Research in Agricultural Genomics facilities. The presence of SPFMV in the vegetatively propagated plants was confirmed by reverse transcription-PCR (RT-PCR) of total RNA extracted with TRIzol reagent following the provider's instructions, using primers FMCPFdeg and MFRdeg (see Table S1 in the supplemental material for details on all primer sequences), which were designed to amplify a 389-nucleotide (nt) fragment within the CP-coding region. A robust plant, denoted AM-MB2, with occasional viral symptoms (weak mosaic, leaf spots, and distortions) that tested positive for SPFMV presence was selected for cutting propagation. The complete P1-coding region was also amplified by RT-PCR with FMP1F and 1PMFR specific primers and was sequenced to confirm the presence of the embedded PISPO sequence. The same infected AM-MB2 plant material described here was used for previously published experiments (13).
To boost accumulation of SPFMV in sweet potato AM-MB2 plants, coinfection with isolate Can181-9 of the crinivirus sweet potato chlorotic stunt virus (SPCSV) strain West African (SPCSV-WA), kindly provided by Jesús Navas-Castillo (IHSM-UMA-CSIC La Mayora, Málaga, Spain), was achieved by inoculation with Bemisia tabaci whiteflies. Briefly, groups of around 50 viruliferous whiteflies that had acquired SPCSV during 48 h in an infected Ipomea setosa plant were caged on fully expanded leaves of AM-MB2 cuttings for a 48-h inoculation period. After insect removal and insecticide treatment, plants were sampled and tested for detection of both SPFMV and SPCSV, in the later case with oligonucleotides SPCSVCP and PCVSCPS, which amplify a 790-nt fragment from the CP-coding sequence of SPCSV.
Isolates of two potyviruses, sweet potato virus C (SPVC) and SPV2, as well as one begomovirus and one badnavirus (both unclassified), were found to be coinfecting the sweet potato AM-MB2 plant after deep-sequencing analysis (see below).
Extraction of total RNA, construction and sequencing of RNA sequencing (RNA-seq) libraries, and preliminary data filtering.
Samples of symptomatic tissue of infected plants were collected for RNA extraction with TRIzol reagent. Contaminating DNA was removed from RNA samples with Turbo DNase (Ambion), and RNA was further purified using the RNeasy minikit (Qiagen). RNA libraries were constructed with the ScripSeq complete kit (plant leaf) (Epicenter, Illumina), including barcoding elements to identify the different samples, according to the provider's protocols. Libraries were submitted to BGI (Hong Kong) for Illumina sequencing on a HiSeq 2000 platform, and 100-bp (average) paired-end reads were generated. Sequences with an average quality below 20, as well as sequences with more than 10 nt with quality below 15, were removed using the FASTX toolkit (available from http://hannonlab.cshl.edu/fastx_toolkit/index.html). The quality process was driven with FASTQC (Babraham Bioinformatics) (available at http://www.bioinformatics.babraham.ac.uk/projects/fastqc/).
Alignments and indel analysis.
Filtered sequences were mapped versus the references with Bowtie2 (20), allowing a maximum of 3 mismatches, insertions, or deletions (indels) per read. To reduce the number of sequencing errors, only the central part of reads (80 nt) was used, and only paired alignments were considered (21). Alignments were analyzed with SAMtools to create a list of variants in which only indels were included. Reads that presented fewer than 3 nonredundant sequences and reads in which the frameshift caused by the indel was cancelled out by another indel in the same sequence were discarded. To reduce false positives caused by sequencing errors or random errors, the expected indel error was modeled as a Poisson distribution, which was calculated from Illumina indel calling-error rate, PCR error rate, and sample indel frequency. Indels with a false-discovery rate (FDR) higher than 0.05 were removed. Analysis was performed with in-house R scripts.
Phylogenetic and recombination analysis.
Initial phylogenetic analysis of the SPFMV, SPVC, and SPV2 isolates found in AM-MB2 was performed with data sets created with the full genome sequences corresponding to 9 SPFMV, 7 SPVC, and 6 SPV2 isolates found in the NCBI nucleotide database, with no filter by identity redundancy. Also, sequences of complete CP regions of all other SPFMV, SPVC, and SPV2 isolates available in the NCBI nucleotide database were considered, after exclusion of partial CP sequences and application of a filter at 97% of identity to reduce redundancy, leading to data sets of 63 SPFMV, 46 SPVC, and 6 SPV2 annotated sequences. Phylogenetic trees were created using the maximum-likelihood algorithm implemented in MEGA6 software (22). The bootstrapping test was driven with 1,000 replicates. CP sequence-based trees were used to estimate putative recombination events (see below).
The assembly of SPFMV used the reference genome already described (13), and was driven with Quasirecomb (23). The same procedure was applied to SPVC and SPV2. The reconstruction of sequences used coverage of >10.000 to reduce errors, thereby excluding both extremes, and the option to reduce the false positives was activated. The sequences obtained were used to search for putative recombination events with the software RDP (24) version 3.45, using a threshold of a P value equal or less than 0.05 with the methods RDP (25), BootScan (26), Geneconv (26), and MaxChi (27). The recombination points were identified using the SBP and GARD algorithms (28) implemented in the Datamonkey webserver (29).
Constructs of viral gene products for in vitro translation and for transient expression of viral proteins by agroinfiltration.
Cloning and plasmid production were performed using Escherichia coli strain DH5α and standard procedures. Viral products were RT-PCR amplified with appropriate primers, and the products were cloned directionally into pENTR/D-TOPO (Life Technologies). Primers are listed in Table S1 in the supplemental material, which includes information about their use for the modifications required to direct and/or force or avoid the expression of certain gene products in the different frames. Recombinant products with mutations introduced by PCR with appropriate primers were generated and were confirmed by sequencing the pENTR clones. The multiple mutant with the 4 WG/GWs motifs altered was synthesized as follows. A fragment flanked by NcoI and AscI sites engineered upstream and downstream from position 1 to 1964 (Met to Stop) in the P1N-PISPO sequence, with replacement of W with A residues in the WG/GW motifs located at positions 73, 1465, 1795, and 1918 (relative to the initial Met codon), was ordered from GeneScript (USA) and inserted using the unique restriction sites into the pENTR plasmid, linearized with the same enzymes, and ligated with T4 DNA ligase. Clones containing inserts were selected and sequenced to confirm the presence of the mutated motifs. The inserts in pENTR were mobilized using Gateway into the different destination vectors with LR Clonase II enzyme mix (Life Technologies). Reactions were performed, and two clones of the different plasmids were verified. For expression in plants, the pGWB7XX destination plasmid series was used (30, 31), which placed the construct under control of appropriate elements.
In vitro translation.
pENTR-P1 was used as the template for PCR amplifications of all constructs used for in vitro translation, including unmodified wild-type P1 and variants with P1 interrupted by a stop codon after the slippage site (P1Δ) and P1 with the PISPO sequence interrupted (P1N-PISPOΔ) generated by site-directed mutagenesis with the primers FMP1ΔF and Δ1PMFR for P1Δ and FMPISPOΔF and ΔOPSIPMFR for P1N-PISPOΔ (see Table S1 in the supplemental material). T7 promoter, 5′ untranslated region (5′UTR), and 3′UTR fragments were prepared as described previously (32). mRNAs were produced by in vitro transcription using the T7-Scribe Standard RNA IVT kit (Cellscript) and purified by organic extraction and ammonium acetate precipitation, and their quality and amount were assessed by NanoDrop (Thermo Fisher Scientific) and gel electrophoresis. The final concentration was adjusted to 1 g/liter, and in vitro translation was carried out using wheat germ extract (Promega) according to the manufacturer's instructions. Labeling of the synthesized proteins was done by including in the reaction mixture l-[35S]methionine and l-[35S]cysteine (PerkinElmer). Samples were resolved by 12% SDS-PAGE and detected with a PhosphorImager.
Agrobacterium tumefaciens infiltration of Nicotiana benthamiana leaves.
N. benthamiana leaves (fully expanded, from 3- to 4-week-old plants grown in a greenhouse) were agroinfiltrated as described previously (33), using A. tumefaciens cultures of strain C58C1 or EHA105 transformed with the relevant plasmids. Growth of bacteria was monitored by assessing the optical density at 600 nm (OD600) until it reached 0.5 units. Cultures were induced by acetosyringone and infiltrated with a needle-less syringe. For sampling material to be analyzed by LC-tandem MS (LC-MS/MS) (see below), the agroinfiltration was performed with mixed cultures incorporating a construct expressing P1b of CVYV (33) to increase expression of SPFMV proteins. For silencing suppression experiments (see below), negative (empty vector, delta) and positive (CVYV P1b and SPMMV P1) controls were included, following previously described procedures (34, 35).
SDS-PAGE and fractionation of protein products.
Plant samples were collected, deep-frozen, and homogenized in extraction buffer. After boiling for 5 min, aliquots were separated by 10% SDS-PAGE. Following staining with Coomassie blue G-250 (SimplyBlue safe stain; Life Technologies), the portion above the RuBisCO was excised and processed.
MS and protein identification by LC-MS/MS analysis.
Gel-excised fragments containing protein samples were washed with 25 mM NH4HCO3 and acetonitrile (ACN), reduced for 60 min at 60°C with 20 mM dithiothreitol (DTT), and alkylated for 30 min at 30°C with 55 mM iodoacetamide in the dark before being digested for 16 h at 37°C with 0.9 μg trypsin (porcine sequence-grade modified trypsin; Promega). Peptides were extracted from the gel matrix with 10% formic acid and ACN, dyed, and desalted with a C18 Top-tip (PolyLC), following the procedure of the provider. Dried-down tryptic peptides mixtures resuspended in 1% formic acid were injected for chromatographic separation in a nanoAcquity liquid chromatograph (Waters) coupled to an LTQ-Orbitrap Velos (Thermo Scientific) mass spectrometer. Peptides were trapped on a Symmetry C18 trap column (5 μm by 180 μm by 20 mm; Waters) and separated with a C18 reverse-phase capillary column (75 μm, 10 cm nanoAcquity; 1.7-μm BEH column; Waters). The gradient for elution was prepared with 0.1% formic acid in ACN and consisted of 1 to 30% for 60 min, 35 to 45% for 10 min, and 45 to 85% for 5 min, with a flow rate of 250 nl/min. Eluted peptides were subjected to electrospray ionization in an emitter needle (PicoTip; New Objective) with 2,000 V applied. Peptide masses (m/z 350 to 1700) were analyzed in full-scan MS data-dependent mode in the Orbitrap with 60,000 full width at half maximum (FWHM) resolution at 400 m/z. Up to the 10 most abundant peptides (minimum intensity of 500 counts) were selected for each MS scan and fragmented using collision-induced dissociation in a linear ion trap with helium as the collision gas and 38% normalized collision energy. The targeted mode was used to analyze the presence of predicted peptides in the ORF that contains PISPO (the seven peptides underlined in the sequence LVWEKTGRTIGHKERDQKRSQSKMEVGTLQTSQEDQEGQPKTAPTEAHGEGTAIIDGYATSSSDGHLHCWGSIGESGNDSNSEWEDFLHAFHEEEENFKISQINTRENSRAHAGSSENCVQEKDEHRIGGQEVHKRAVQEISRSKLFVPSFKTYGRLKRVSGFKNSHNNSKPRTSSCQGWSMEKTCKDNNVVQRFKWHGAESRQTVGPKRSCTTRNARGAWGFTRSAIRRTNETW) or in the C-terminal part of the P1 (the three peptides underlined in the sequence KLDEQLATRNEIRKGLKVKWRWGLYRLVKKTRKDNQRQRRQRRMEKEQQLLMAMPPQVLTGISIAGGPSASLEMTPTPNGKIFCTPSMKKKKTLKSPKLTQEKIHELTQAVLKIACRKRMSIELVGKKSTKGQYRKFQGANYLFLHLKHMEGLRESVDLRIHTTTQNLVLQAAKVGAWKRPVKTTMLSKGSSGMVLNPDKLLGPRGHAPHGMLVVRGALRGVLYDARMKLGRSVLPYIIQY). Generated raw data were collected with Thermo Xcalibur (v.2.2). For database searching, custom databases were created merging all UniProt entries (as of December 2014) for I. batatas plus the predicted list of mature protein sequences from SPFMV plus a list of predicted mature gene products deriving from viruses present in the AM-MB2 sample or for N. benthamiana and A. tumefaciens plus P1 and P1N-PISPO. Entries from common laboratory contaminants were also added. Sequest search engine searches were performed with Thermo Proteome Discoverer software (v.1.3.0.339), using a target and a decoy database to obtain a false-discovery rate (FDR) (strict, 0.01; relaxed, 0.05) and to estimate the number of incorrect peptide spectrum matches exceeding a given threshold (peptide tolerances of 10 ppm and 0.6 Da, respectively, for MS and MS/MS), with parameters for trypsin allowing up to 2 missed cleavages, considering cysteine carbamidomethylation as fixed and methionine oxidation as variable modifications. Validation was based on FDR, using Percolator (a semisupervised learning machine) to discriminate peptide spectrum matches. Only proteins identified with at least 2 high-confidence peptides (FDR of ≤0.01) were considered.
RNA silencing suppression activity assays: GFP imaging, Northern blotting, and RT-qPCR analysis.
Leaves of N. benthamiana plants were coagroinfiltrated with a green fluorescent protein (GFP)-expressing construct together with constructs of the different viral products or adequate controls. The infiltrated patches were visualized under UV light with a Black Ray B 100 AP lamp, and photographs were taken with a Nikon D7000 digital camera.
Total RNA from N. benthamiana tissue was isolated using TRIzol and treated with Turbo DNase (Ambion) to remove contaminating DNA. For Northern blot analysis of mRNAs and small interfering RNAs (siRNAs), approximately 10 and 15 μg of total RNA were resolved on 1.2% agarose (containing 2% formaldehyde) or 15% denaturing polyacrylamide (containing 7 M urea) gels, respectively, and transferred to nylon Hybond-N+ membranes (Amersham) by capillary blotting for the agarose gels or using a transfer apparatus (XCell SureLock; Invitrogen) for the acrylamide gels. Ethidium bromide staining was used to verify equal loading of lanes and monitor the transference processes. After UV cross-linking and prehybridization in UltraHyb buffer (Ambion), blots were hybridized in the same solution with probes specific to the GFP sequence, using a 32P-labeled DNA probe (33) in the case of mRNAs or a 32P-labeled RNA probe (36) in the case of small RNAs. Hybridization signals were detected with a PhosphorImager.
For quantitative real-time reverse transcription-PCR (RT-qPCR) analysis, 1 μg of total RNA was reverse transcribed using the SuperScript III first-strand synthesis system (Invitrogen), and RT-PCRs were performed in a 20-μl volume with gene-specific primers and LightCycler 480 SYBR green I master mix (Roche). A 102-bp GFP fragment was amplified with primers described by Leckie and Stewart (37). Ubiquitin was selected as a reference gene, amplifying a 88-bp fragment using the primers described by Lacomme and coworkers (38) (see Table S1 in the supplemental material). The average cycle threshold (CT) value for triplicate PCRs was normalized to the average CT value for the reference gene, yielding the ΔCT value. An analysis of variance (ANOVA) for at least three independent biological replicates was performed using the Tukey-Kramer test.
Nucleotide sequence accession numbers.
Data corresponding to virus sequences are available in GenBank under accession numbers KU511268, KU511269, KU511270, KU511271, and KU511272.
RESULTS
Sequencing the virome of the sweet potato plant AM-MB2.
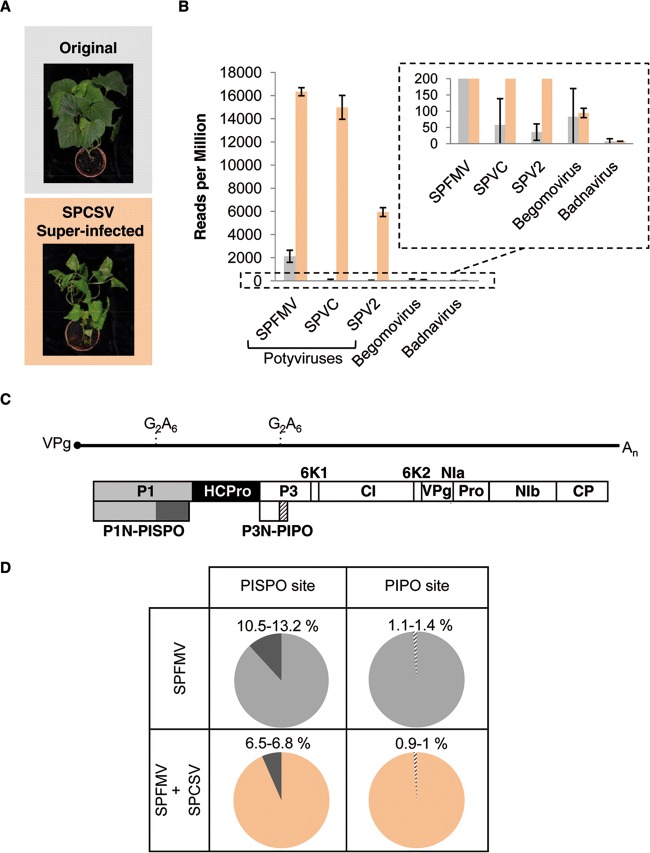
AM-MB2 sweet potato plants, which are naturally infected with several viruses, including SPFMV, were used. With the aim of boosting the load of potyviruses, plants were superinfected with the crinivirus SPCSV, which is known to cause a synergistic effect with potyviruses (39). As shown in Fig. 1A, the superinfection of AM-MB2 plants with SPCSV resulted in a strong enhancement of disease symptoms.
FIG 1.
Synergism between SPFMV and SPCSV. (A) Appearance of AM-MB2 plants propagated vegetatively, with a representative original plant (top) and another superinfected with SPCSV (bottom). Pictures were taken 4 months after whitefly-mediated inoculation of the crinivirus. (B) Normalized number of virus-derived RNA-seq reads in AM-MB2 plants (gray bars) compared to those plants superinfected with SPCSV (salmon bars). Individual viruses are indicated, and for each one the average and standard deviation are plotted. The inset with an extended scale was included to accommodate the large differences between the conditions. (C) Genomic organization of the SPFMV genome assembled from the RNA-seq data. The RNA genome of SPFMV is represented as a solid line flanked by a covalently linked VPg (solid circle) and the poly(A) tail, with the three ORFs corresponding to the polyprotein, PISPO, and PIPO depicted as boxes (details of the conserved G2A6 motifs are shown). Boxes with names represent the different gene products. (D) Percentages of RNA-seq reads with insertion of 1 nucleotide at the slippage sites in SPFMV. No other alterations were observed at these points, and the numbers show the values for two independent samples used for the RNA-seq analysis (see Table S7 in the supplemental material).
Total RNA isolated from samples of the plants coinfected with SPCSV and SPFMV was analyzed by RNA-seq using Illumina technology (SRA references SRR1693374 and SRR1693416) and compared to equivalent samples deriving from AM-MB2 plants not superinfected with the crinivirus (13). Abundant SPFMV-derived reads were found, together with a smaller number of reads from two additional potyviruses, SPV2 and SPVC. In addition, reads corresponding to a begomovirus and a badnavirus were identified, but in insufficient number to obtain complete coverage of the genomes of these viruses. As expected, the number of reads for potyviruses increased notably after superinfection with SPCSV, as shown in Fig. 1B, whereas no major change in the accumulation of reads was observed for the begomovirus and badnavirus.
Total or partial assemblies of the different viruses were deposited in GenBank, and their preliminary annotations are provided in Tables S2 to S6 in the supplemental material.
Boosting of SPV2 and SPVC accumulation in the crinivirus-superinfected samples allowed us to analyze the presence of insertions/deletions in their genomes. As predicted from our previous findings (13), most of the indels in SPV2 and SPVC genomes were found in the G2A6 motifs upstream of trans-frame PISPO and PIPO (see Table S7 in the supplemental material).
The assembly of the genomic sequence from SPFMV (see Table S2 in the supplemental material) spanned 10,814 nt, representing almost all of its genome, except for a short region at the 5′ end of ∼6 nt. The sequence of SPFMV AM-MB2 encoded the canonical potyvirus gene products in the polyprotein, i.e., P1, HCPro, P3, 6K1, CI, 6K2, VPg/NIaPro, NIb, and CP, as well as the predicted P1N-PISPO and P3N-PIPO, in both cases preceded by G2A6 motifs. The genomic organization is summarized in Fig. 1C. Positions in the genome were numbered after alignment with other published SPFMV sequences to facilitate comparison of regions with the reference genome (5) (95.12% identity).
Our data also showed that superinfection with SPCSV resulted in a decrease in the relative proportion of RNA-seq reads with an additional A in the G2A6 motifs at the PISPO site (so-called RNA slippage frequency) in SPFMV, while there was no change in slippage at the PIPO site where a less frequent slippage was observed (Fig. 1D).
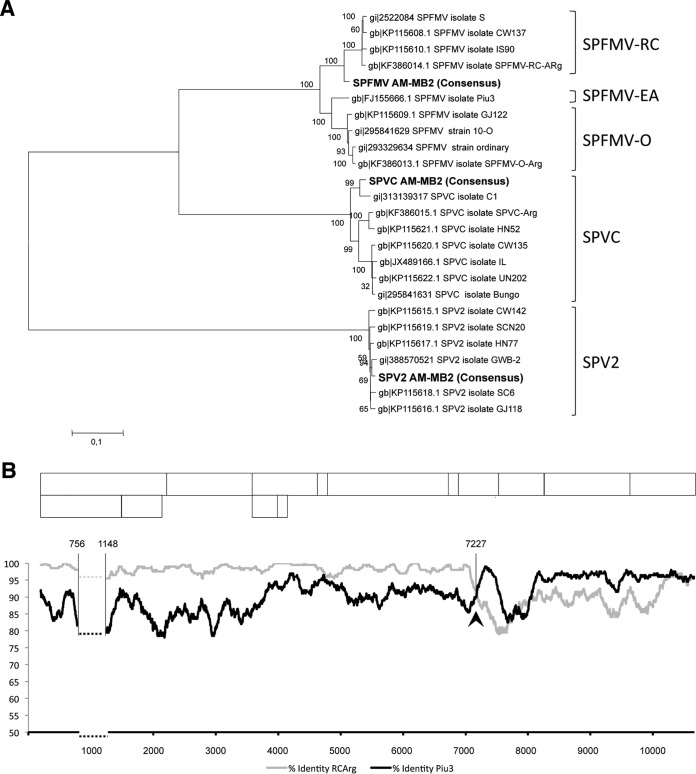
Phylogenetic analyses were performed with all potyviral sequences in AM-MB2 using whole-genome sequences (Fig. 2A) and selected CP sequences of individual viruses. In the case of the SPFMV isolate, a comparison of trees constructed with whole-genome and CP sequences suggested that it can be placed in the group of recombinant isolates, a frequent occurrence among SPFMV isolates (40, 41). Comparison of our sequence with those of two selected isolates (Piu3 and RC-ARg, accession numbers FJ155666.1 and KF386014.1, respectively) is illustrated in Fig. 2B, where a crosspoint upstream from the CP region is noticeable. Application of adequate analysis tools predicted a recombination site located at position 7227 in the NIa region, further confirming the recombinant nature of the SPFMV AM-MB2 isolate.
FIG 2.
Taxonomic characterization of potyviruses from the AM-MB2 sweet potato plant. (A) Phylogenetic analysis of the complete nucleotide sequences corresponding to the three potyviruses found in AM-MB2 (highlighted in bold), compared to full-length sequences of SPFMV, SPVC, and SPV2 available in GenBank, using alignments and the maximum-likelihood algorithm to generate a tree with 1,000 replications for the bootstrapping test. (B) Plot of nucleotide identities between SPFMV isolate AM-MB2 and isolates Piu3 (EA strain) and RC-ARg (RC strain), after comparison with a sliding window of 100 nt. The recombination site detected by the SBP and GARD analysis is indicated with an arrowhead and the position number in the viral genome. The dashed horizontal lines correspond to a highly divergent region (between the indicated positions in the viral genome) with abundant gaps, which was eliminated to facilitate the global comparison. A schematic drawing of the expected gene products of SPFMV is shown above the plot to facilitate comparison of the recombination site.
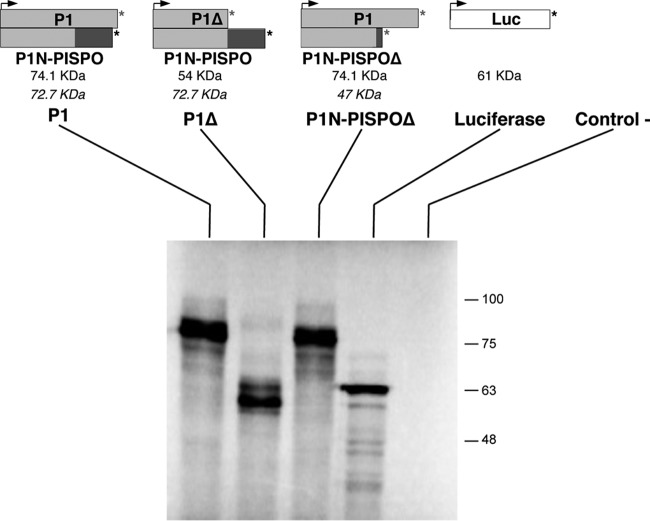
Absence of detectable production of P1N-PISPO by frameshifting in in vitro translation experiments.
To evaluate the putative contribution of translational frameshifting to the production of P1N-PISPO, the SPFMV P1-coding region was cloned after RT-PCR amplification (P1 construct), and appropriate mutations were designed to express truncated proteins in each of the frames. In the P1Δ construct, a stop codon interrupted the main ORF of the polyprotein downstream of the G2A6 motif to generate a 54-kDa variant, whereas in the P1N-PISPOΔ construct, the stop was introduced in the PISPO sequence (−1 frame), leading to a putative 47-kDa product without altering the P1-coding sequence. P1, P1Δ, and P1N-PISPOΔ, together with a luciferase control, were used for in vitro transcription. The analysis showed a band with a mobility corresponding to ∼70 kDa in the P1 sample, compatible with both P1 and P1N-PISPO (expected sizes of 74.1 kDa and 72.7 kDa, respectively) (Fig. 3). However, major protein products derived from translational frameshifting were not detected in the analysis of the two other constructs designed to yield products of different sizes in the P1 and PISPO frames: in P1Δ, the major band was compatible with the expected size of the truncated P1 (54 kDa), while in P1N-PISPOΔ, the pattern was similar to that for the unaltered P1 (compare first and third lanes in Fig. 3, and note the absence of evident products at the expected region for the truncated 47-kDa protein). The results of this experiment support the hypothesis that viral RNA polymerase slippage is the main mechanism that produces the out-of-frame product.
FIG 3.
In vitro expression of P1-related products. Wild-type and variant constructs were designed to express selected proteins. Arrowheads and asterisks indicate the translation initiation sites (AUG) and stop codons, respectively. Truncated forms with stop codons introduced by mutagenesis are indicated by asterisks in gray. The expected sizes (in kDa) of the anticipated translated protein products are indicated below each construct, with italics used to indicate the trans-frame products expected in the case of translational frameshifting. Wheat germ extract was programmed with RNA transcripts, including a luciferase mRNA and water as controls. Each construct is connected to the lanes on the SDS-PAGE analysis of the proteins synthesized after in vitro translation and detected by autoradiography. The electrophoretic mobilities of molecular mass markers are shown at the right.
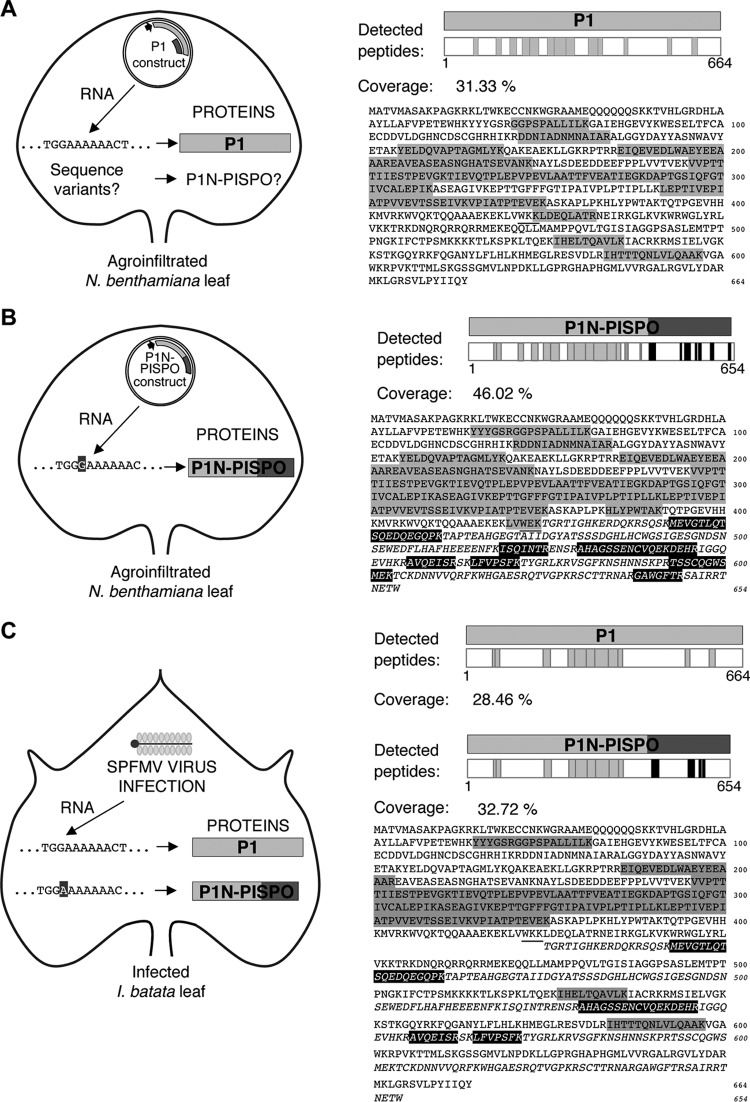
Identification of virus-derived proteins in agroinfiltrated plant tissue by LC-MS/MS analysis.
A construct containing the wild-type P1-coding sequence and a construct designed to produce only P1N-PISPO (by insertion of a nucleotide in the G2A6 motif) were expressed by agroinfiltration in N. benthamiana leaves in the presence of P1b RSS from the unrelated ipomovirus cucumber vein yellowing virus (CVYV) (33). The P1b RSS was included to enhance protein expression levels. Total proteins present in the infiltrated patches were extracted and separated by SDS-PAGE, and the fraction above 50 kDa (to avoid the highly abundant RuBisCO) was excised and analyzed by LC-MS/MS.
The analysis identified more than 180 and 200 proteins in samples expressing the wild-type P1 and the mutant forced to produce P1N-PISPO, respectively. Most of the proteins corresponded to the host plant or to the A. tumefaciens bacteria used as the expression vector. In each sample, the overexpressed viral gene products were readily identified with high scores and coverage: for P1, a score of 97.38, 13 unique peptides, and 31.33% coverage; for P1N-PISPO, a score of 173.38, 25 unique peptides, and 46.02% coverage. Intriguingly, no peptides corresponding to the PISPO frame were found in the sample that expressed the wild-type P1 gene products.
In order to maximize the chance of detecting P1N-PISPO in further analysis of infected samples, a targeted search mode was adopted by using the information about true peptides derived from overexpression. The detected peptides corresponding to the transiently expressed viral gene products; their distributions along the amino acid sequences are shown in Fig. 4A and B, and complete details are provided in Table 1. The application of this targeted approach confirmed the absence of PISPO-derived peptides in plants transiently expressing wild-type P1. In addition, it also confirmed the identity of the expressed P1N-PISPO protein, as an identified peptide overlapped the frameshifted region and showed the amino acid introduced to force the frameshift (sequence, LVWEK [mutation in bold]) (Table 1).
FIG 4.
Identification of viral proteins by LC-MS/MS in plant samples. (A) Detection of peptides corresponding to proteins transiently expressed in N. benthamiana leaves agroinfiltrated with a construct that contains the wild-type P1 sequence of SPFMV under control of the appropriate promoter for overexpression. The plasmid construct is schematically drawn inside a leaf outline, with a detail of the expected RNA sequence corresponding to the G2A6 motif upstream of the PISPO starting point and the detected protein P1. A question mark besides the P1N-PISPO name indicates that no peptides corresponding to the PISPO domain were found in the analysis. The peptides detected are shown (as gray boxes) distributed along the P1 protein sequence in the right panel, and the percentage of coverage is indicated. The sequences of the detected peptides are shaded in gray on the sequence of the P1 gene product, and the frameshift point is underlined. (B) Detection of peptides as in panel A but in the sample agroinfiltrated with the P1N-PISPO construct, which ensures expression of the trans-frame product. The PISPO region is highlighted and is depicted in italic lettering in the sequence, starting at the underlined frameshift point. Peptides found in the analysis are shown as boxes (gray for P1N and black for PISPO) in the graphic and shaded in the sequence below, using a gray background in the case of peptides corresponding to the P1N or a black background with white letters for the PISPO region. (C) Detection of viral peptides in the AM-MB2 I. batatas plant. The virus SPFMV is represented by a schematic virion inside the outline of the sweet potato leaf, and two variants of viral RNAs with the G2A6 or G2A7 motifs are indicated (13). Peptides deriving from the common P1N region, from the P1 C-terminal region, and from PISPO are shown in the protein schemes, using gray for the common P1N part and the rest of P1, while the peptides found in the PISPO frame are represented by black boxes. The sequences of peptides are also highlighted in the sequence, with the two variants shown after the frameshift point (underlined), represented in the upper lines for P1 or in the lower (in italic) for PISPO and using as above a gray or black background in the sequence detail, respectively.
TABLE 1.
SPFMV viral gene products transiently expressed on N. benthamiana plant tissue and detected by LC-MS/MS analysis
| Sample | Viral protein | Scorea | Coverage (%) | No. of unique peptidesb | PSMs (Da)c | Size (aa) | Molecular mass (kDa) | Calculated pI | Peptides |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sequenced | Size (aa) | Position (aa)e |
Missed cleavages | MH+ (Da)f | PSMs | PEPg | ||||||||||
| Initial | Final | |||||||||||||||
| Agroinfiltrated P1 construct | P1 | 97.38 | 31.33 | 13 (10 + 3) | 26 | 664 | 74.1 | 9.17 | GGPSPALLILK | 11 | 71 | 81 | 0 | 1,065.67009 | 2 | 0.02370553 |
| DDNIADNmNAIAR | 13 | 122 | 134 | 0 | 1,448.64372 | 1 | 0.002282131 | |||||||||
| DDNIADNMNAIAR | 13 | 122 | 134 | 0 | 1,432.65166 | 1 | 0.000296073 | |||||||||
| YELDQVAPTAGmLYK | 15 | 155 | 169 | 0 | 1,714.84661 | 2 | 4.98689E−07 | |||||||||
| YELDQVAPTAGMLYK | 15 | 155 | 169 | 0 | 1,698.84929 | 2 | 1.07017E−06 | |||||||||
| EIQEVEDLWAEYEEAAAR | 18 | 186 | 203 | 0 | 2,150.99516 | 2 | 1.30644E−05 | |||||||||
| EAVEASEASNGHATSEVANK | 20 | 204 | 223 | 0 | 2,000.91977 | 1 | 0.009433393 | |||||||||
| VVPTTTIIESTPEVGK | 16 | 246 | 261 | 0 | 1,670.92717 | 2 | 5.91988E−05 | |||||||||
| TIEVQTPLEPVPEVLAATTFVEATIEGK | 28 | 262 | 289 | 0 | 2,982.59946 | 1 | 0.001826707 | |||||||||
| DAPTGSIQFGTIVcALEPIK | 20 | 290 | 309 | 0 | 2,117.09990 | 3 | 4.50031E−07 | |||||||||
| LEPTIVEPIATPVVEVTSSEIVK | 23 | 342 | 364 | 0 | 2,450.36716 | 4 | 0.000277995 | |||||||||
| VPIATPTEVEK | 11 | 365 | 375 | 0 | 1,183.65911 | 2 | 0.002508105 | |||||||||
| KLDEQLATR | 9 | 424 | 432 | 1 | 1,073.59978 | 1 | 0.01176776 | |||||||||
| IHELTQAVLK | 10 | 527 | 536 | 0 | 1,151.67961 | 1 | 0.03467921 | |||||||||
| IHTTTQNLVLQAAK | 14 | 584 | 597 | 0 | 1,537.87314 | 1 | 0.000769235 | |||||||||
| Agroinfiltrated P1N-PISPO construct | P1N-PISPO | 173.38 | 46.02 | 25 (17 + 8) | 58 | 654 | 72.7 | 6.47 | YYYGSR | 6 | 65 | 70 | 0 | 808.36339 | 2 | 0.05871485 |
| GGPSPALLILK | 11 | 71 | 81 | 0 | 1,065.67095 | 2 | 0.01128503 | |||||||||
| RDDNIADNmNAIAR | 14 | 121 | 134 | 1 | 1,604.74381 | 1 | 0.001550777 | |||||||||
| DDNIADNmNAIAR | 13 | 122 | 134 | 0 | 1,448.64348 | 2 | 8.56681E−05 | |||||||||
| YELDQVAPTAGmLYK | 15 | 155 | 169 | 0 | 1,714.83550 | 2 | 2.69142E−05 | |||||||||
| YELDQVAPTAGMLYK | 15 | 155 | 169 | 0 | 1,698.84050 | 2 | 0.00018226 | |||||||||
| EIQEVEDLWAEYEEAAAR | 18 | 186 | 203 | 0 | 2,150.98906 | 5 | 4.48992E−07 | |||||||||
| EAVEASEASNGHATSEVANK | 20 | 204 | 223 | 0 | 2,000.91300 | 3 | 0.002131125 | |||||||||
| VVPTTTIIESTPEVGK | 16 | 246 | 261 | 0 | 1,670.92192 | 4 | 0.0003182 | |||||||||
| TIEVQTPLEPVPEVLAATTFVEATIEGK | 28 | 262 | 289 | 0 | 2,982.59671 | 5 | 0.02217205 | |||||||||
| DAPTGSIQFGTIVcALEPIK | 20 | 290 | 309 | 0 | 2,117.09770 | 4 | 6.45506E−06 | |||||||||
| ASEAGIVKEPTTGFFFGTIPAIVPLPTIPLLK | 32 | 310 | 341 | 1 | 3,324.89353 | 2 | 0.00897333 | |||||||||
| ASEAGIVK | 8 | 310 | 317 | 0 | 774.43566 | 1 | 0.02094499 | |||||||||
| EPTTGFFFGTIPAIVPLPTIPLLK | 24 | 318 | 341 | 0 | 2,569.48111 | 2 | 0.000227663 | |||||||||
| LEPTIVEPIATPVVEVTSSEIVKVPIATPTEVEK | 34 | 342 | 375 | 1 | 3,615.00620 | 2 | 0.008460695 | |||||||||
| LEPTIVEPIATPVVEVTSSEIVK | 23 | 342 | 364 | 0 | 2,450.37339 | 5 | 3.8468E−06 | |||||||||
| VPIATPTEVEK | 11 | 365 | 375 | 0 | 1,183.65825 | 2 | 0.008864285 | |||||||||
| HLYPWTAK | 8 | 384 | 391 | 0 | 1,015.53710 | 1 | 0.1502403 | |||||||||
| LVWEK | 5 | 420 | 424 | 0 | 674.38786 | 1 | 0.14696 | |||||||||
| mEVGTLQTSQEDQEGQPK | 18 | 443 | 460 | 0 | 2,020.91570 | 1 | 9.41641E−07 | |||||||||
| ISQINTR | 7 | 519 | 525 | 0 | 831.46898 | 2 | 0.02658513 | |||||||||
| AHAGSSENcVQEKDEHR | 17 | 530 | 546 | 1 | 1,953.85525 | 2 | 2.32708E−07 | |||||||||
| AVQEISR | 7 | 556 | 562 | 0 | 802.44304 | 1 | 0.02908999 | |||||||||
| LFVPSFK | 7 | 565 | 571 | 0 | 837.48924 | 1 | 0.08113639 | |||||||||
| TSScQGWSmEK | 11 | 593 | 603 | 0 | 1,316.52410 | 1 | 0.005354153 | |||||||||
| GAWGFTR | 7 | 598 | 604 | 0 | 794.39562 | 2 | 0.1082382 | |||||||||
Sum of the SEQUEST scores of the individual peptides.
Number of identified peptide sequences (peptide spectrum matches) for the protein, including those redundantly identified. In parentheses are indicated the number of common peptides (corresponding to the N-terminal part of the protein, upstream of the G2A6 motif) followed by the differential ones (for the alternative C-terminal part of the P1 or PISPO sequence).
Protonated monoisotopic mass of the peptides.
Modifications are indicated by lowercase letters (m, oxidation; c, carbamidomethyl). The alternative C-terminal parts of P1 or PISPO sequences are in bold.
Positions in the predicted genomic-frame polyprotein. Numbers in italic corresponded to the + 2 frame of PISPO continuing P1N.
Protonated monoisotopic mass of the peptides.
Posterior error probability, i.e., probability that the observed PSM is incorrect.
Detection of SPFMV P1N-PISPO in infected sweet potato plants.
Total proteins were extracted from plants infected with SPFMV AM-MB2, and the fraction corresponding to protein products of >50 kDa was subjected to LC-MS/MS analysis. The list of peptides corresponding to viral proteins included several gene products expected to accumulate during infection, such as P1, HCPro, CI, and NIb (all of them with sizes above the exclusion limit selected and translated from the long viral ORF). Detection was also positive for 4 peptides corresponding exclusively to the PISPO region of P1N-PISPO and thus derived from a trans-frame protein that incorporated PISPO (21.3% coverage). Along with these peptides, peptides corresponding to the P1 protein were detected from upstream of the polymerase slippage signal (11 different peptides, 39% coverage of the P1N region), and therefore common to P1 and P1N-PISPO, and from the C-terminal part (2 peptides, 10% coverage). Scores were 116.56 (13 peptides, 28.46% coverage) for P1 and 196.2 (15 peptides, 32.72% coverage) for P1N-PISPO.
Figure 4C summarizes the coverage and location of the peptides corresponding to the two alternative gene products from the P1 region, P1 and P1N-PISPO, which were present in infected sweet potato samples. Details on all identified virus-derived proteins and the individual peptides found in the analysis are provided in Table 2. Considering the exclusion limit (>50 kDa) used for sampling and that no smaller viral proteins, including the abundant CP (35.1 kDa), were detected by this analysis, our results are compatible with the simultaneous expression and accumulation of both P1 and P1N-PISPO (predicted to be 74.1 and 72.7 kDa, respectively).
TABLE 2.
SPFMV viral gene products identified by LC-MS/MS analysis of sweet potato tissue samples infected with SPFMV AM-MB2
| Viral protein | Scorea | Coverage (%) | No. of unique peptidesb | PSMs (Da)c | Size (aa) | Molecular mass (kDa) | Calculated pI | Peptides |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sequenced | Size (aa) | Position (aa)e |
Missed cleavages | MH+ (Da)f | PSMs | PEPg | |||||||||
| Initial | Final | ||||||||||||||
| P1N-PISPO | 196.22 | 32.72 | 15 (11 + 4) | 70 | 654 | 72.7 | 6.47 | YYYGSR | 6 | 65 | 70 | 0 | 808.36315 | 1 | 0.05593261 |
| GGPSPALLILK | 11 | 71 | 81 | 0 | 1,065.66899 | 1 | 0.004198602 | ||||||||
| EIQEVEDLWAEYEEAAAR | 18 | 186 | 203 | 0 | 2,150.99101 | 2 | 3.99548E−09 | ||||||||
| VVPTTTIIESTPEVGK | 16 | 246 | 261 | 0 | 1,670.91252 | 3 | 3.90719E−05 | ||||||||
| TIEVQTPLEPVPEVLAATTFVEATIEGK | 28 | 262 | 289 | 0 | 2,982.58792 | 7 | 3.90915E−06 | ||||||||
| DAPTGSIQFGTIVcALEPIK | 20 | 290 | 309 | 0 | 2,117.08965 | 2 | 5.44594E−08 | ||||||||
| ASEAGIVKEPTTGFFFGTIPAIVPLPTIPLLK | 32 | 310 | 341 | 1 | 3,324.87595 | 4 | 0.000152128 | ||||||||
| ASEAGIVK | 8 | 310 | 317 | 0 | 774.43645 | 1 | 0.03614698 | ||||||||
| EPTTGFFFGTIPAIVPLPTIPLLK | 24 | 318 | 341 | 0 | 2,569.46390 | 6 | 6.45971E−06 | ||||||||
| LEPTIVEPIATPVVEVTSSEIVK | 24 | 342 | 365 | 0 | 2,450.36588 | 1 | 2.7674E−05 | ||||||||
| VPIATPTEVEK | 11 | 365 | 375 | 0 | 1,183.65666 | 3 | 0.01060038 | ||||||||
| mEVGTLQTSQEDQEGQPK | 18 | 443 | 460 | 0 | 2,020.91277 | 15 | 4.60753E−10 | ||||||||
| AHAGSSENcVQEKDEHR | 17 | 530 | 546 | 1 | 1,953.84780 | 9 | 5.28656E−08 | ||||||||
| AVQEISR | 7 | 556 | 562 | 0 | 802.44170 | 3 | 0.2101074 | ||||||||
| LFVPSFK | 7 | 565 | 571 | 0 | 837.48723 | 12 | 0.007931116 | ||||||||
| P1 | 116.56 | 28.46 | 13 (11 + 2) | 35 | 664 | 74.1 | 9.17 | YYYGSR | 6 | 65 | 70 | 0 | 808.36315 | 1 | 0.05593261 |
| GGPSPALLILK | 11 | 71 | 81 | 0 | 1,065.66899 | 1 | 0.004198602 | ||||||||
| EIQEVEDLWAEYEEAAAR | 18 | 186 | 203 | 0 | 2,150.99101 | 2 | 3.99548E−09 | ||||||||
| VVPTTTIIESTPEVGK | 16 | 246 | 261 | 0 | 1,670.91252 | 3 | 3.90719E−05 | ||||||||
| TIEVQTPLEPVPEVLAATTFVEATIEGK | 28 | 262 | 289 | 0 | 2,982.58792 | 7 | 3.90915E−06 | ||||||||
| DAPTGSIQFGTIVcALEPIK | 20 | 290 | 309 | 0 | 2,117.08965 | 2 | 5.44594E−08 | ||||||||
| ASEAGIVKEPTTGFFFGTIPAIVPLPTIPLLK | 32 | 310 | 341 | 1 | 3,324.87595 | 4 | 0.000152128 | ||||||||
| ASEAGIVK | 8 | 310 | 317 | 0 | 774.43645 | 1 | 0.03614698 | ||||||||
| EPTTGFFFGTIPAIVPLPTIPLLK | 24 | 318 | 341 | 0 | 2,569.46390 | 6 | 6.45971E−06 | ||||||||
| LEPTIVEPIATPVVEVTSSEIVK | 24 | 342 | 365 | 0 | 2,450.36588 | 1 | 2.7674E−05 | ||||||||
| VPIATPTEVEK | 11 | 365 | 375 | 0 | 1,183.65666 | 3 | 0.01060038 | ||||||||
| IHELTQAVLK | 10 | 527 | 536 | 0 | 1,151.67827 | 2 | 0.1482055 | ||||||||
| IHTTTQNLVLQAAK | 14 | 584 | 597 | 0 | 1,537.86948 | 2 | 0.2301637 | ||||||||
| CI | 45.28 | 19.75 | 9 | 21 | 643 | 72.2 | 7.25 | STGLPFYLSR | 10 | 1617 | 1626 | 0 | 1,140.60686 | 3 | 5.92849E−05 |
| VLLLEPTRPLAENVHR | 16 | 1630 | 1645 | 0 | 1,857.07461 | 5 | 1.30016E−05 | ||||||||
| EVEFTTQYPVQIK | 13 | 1735 | 1747 | 0 | 1,581.81047 | 3 | 5.24599E−06 | ||||||||
| VGNVEIQTcGSPQKK | 15 | 1810 | 1824 | 1 | 1,644.83823 | 2 | 0.023518 | ||||||||
| GPISYGER | 8 | 1867 | 1874 | 0 | 878.43834 | 3 | 0.004448509 | ||||||||
| GVTGWLSVGEYAK | 13 | 1991 | 2003 | 0 | 1,366.70268 | 1 | 0.000104976 | ||||||||
| YKHEAGFGR | 9 | 2038 | 2046 | 1 | 1,064.52701 | 2 | 3.29513E−05 | ||||||||
| IScINScK | 8 | 2047 | 2054 | 0 | 981.45098 | 2 | 0.02123955 | ||||||||
| VAYTLQTDLYAIPR | 14 | 2055 | 2068 | 0 | 1,623.87663 | 1 | 4.87008E−05 | ||||||||
| TVSSSSFTLNSIATLWR | 17 | 2094 | 2110 | 0 | 1,869.97112 | 3 | 3.39834E−08 | ||||||||
Sum of the SEQUEST scores of the individual peptides. The different viral proteins are listed according to their scores.
Number of identified peptide sequences (peptide spectrum matches) for the protein, including those redundantly identified. In parentheses are indicated the number of common peptides (corresponding to the N-terminal part of the protein, upstream of the G2A6 motif) followed by the differential ones (for the alternative PISPO or C-terminal part of P1 sequences).
Protonated monoisotopic mass of the peptides.
Modifications are indicated by lowercase letters (m, oxidation; c, carbamidomethyl). The alternative for PISPO or the alternative C-terminal part of P1 sequences is in bold.
Positions in the predicted genomic-frame polyprotein. Numbers in italic corresponded to the + 2 frame of PISPO continuing P1N.
Protonated monoisotopic mass of the peptides.
Posterior error probability, i.e., probability that the observed PSM is incorrect.
P1N-PISPO is an RNA silencing suppressor.
Once we determined that P1N-PISPO is produced in plants infected with SPFMV, we investigated putative functions for the novel gene product. Based on our previous experience with proteins encoded at the 5′ ends of potyviral genomes, we tested whether P1N-PISPO exhibits RNA silencing suppression activity. Constructs suitable for the transient expression of selected gene products (Fig. 5A) were coagroinfiltrated with a GFP-expressing construct, and the GFP fluorescence was monitored over time under UV light. Both positive and negative controls were included side-to-side with the tested constructs in the same leaves following the design depicted in Fig. 5B. The experiment was repeated several times, using at least three independent Agrobacterium cultures of each construct.
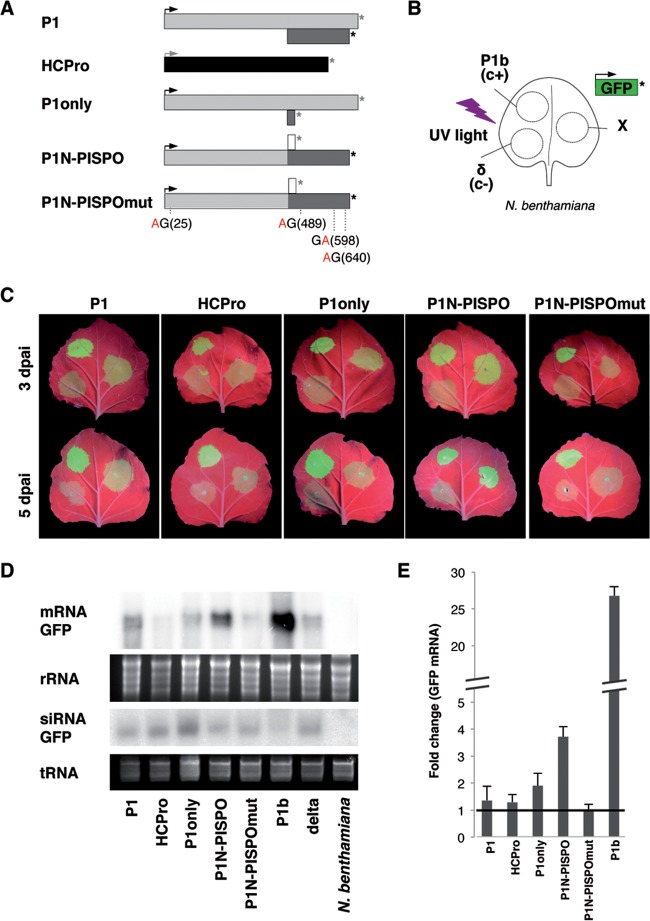
FIG 5.
RNA silencing suppression activity of SPFMV P1N-PISPO. (A) The constructs used are represented with the same conventions as in the other figures for AUG and stop codons. The distribution of the four WG/GW motifs present in the sequence (positions in parentheses) in the mutant in which the four W residues were replaced by A residues is indicated. (B) Patch design used for coagroinfiltration in N. benthamiana leaves of a GFP-expressing construct together with other constructs expressing SPFMV products (indicated by X), a CVYV P1b positive control (c+), or an empty vector (δ, c−). (C) Pictures of representative agroinfiltrated leaves taken at 3 (top row) or 5 (bottom row) days postagroinfiltration (dpai) under UV light. (D) Northern blot analysis of GFP mRNA and siRNA extracted from agroinfiltrated tissue patches at 3 dpai, comparing the different constructs indicated above each lane. The bottom panels show the ethidium bromide staining of the gels as loading controls. (E) Relative accumulation of GFP mRNAs measured by specific RT-qPCR and normalized against the mean value corresponding to the negative control. The average values ± standard deviations from several experiments, each performed with at least three independent Agrobacterium cultures, are plotted. Significant difference in pairwise comparisons and after applying the Tukey-Kramer test were found only for the positive control CVYV P1b (shown with a broken axis to accommodate the large difference) and for the P1N-PISPO samples.
Compared to other individual SPFMV gene products, only P1N-PISPO showed clear RNA silencing suppression activity at 3 and 5 days postagroinfiltration (dpai) in the visual assay (Fig. 5C), which correlated with higher accumulation of GFP mRNA as observed by Northern blotting (Fig. 5D) and RT-qPCR (Fig. 5E). Differences in GFP fluorescence and accumulation of GFP mRNA between the CVYV P1b positive control and P1N-PISPO suggest that the last is a weaker RSS. This was confirmed by the RT-qPCR analysis, which showed around 5-times-higher levels of GFP mRNA in the presence of P1b (Fig. 5E).
As expected from previous results (18, 36) CVYV P1b prevented to certain extent the generation of GFP-derived siRNAs, while a clear similar effect was not observed for P1N-PISPO (Fig. 5D), suggesting that these two viral proteins might use different mechanisms to suppress the RNA silencing, as we propose below.
Constructs expressing P1 (wild-type P1 sequence) and P1 only (P1 with an out-of-frame stop codon precluding any expression of PISPO) failed to exhibit enough noticeable RNA silencing suppression activity in our assays. Interestingly, the construct expressing HCPro also failed to exhibit RSS activity, with quantitative values always below those obtained for P1 (Fig. 5C, D, and E). The same P1 and P1N-PISPO constructs tested for RSS activity were used in the LC-MS/MS experiments described above. Although it is not a quantitative assay, the detection of similar numbers of specific peptides in both cases (Table 1) suggests that P1 and P1N-PISPO accumulated at comparable levels in agroinfiltrated leaves in the presence of a heterologous RSS. Hence, these results support the idea that the absence of noticeable silencing suppression activity by the P1 construct is not due to problems in protein expression, accumulation, or stability.
WG/GW motifs play important roles in interactions with Argonaute (Ago) proteins and small RNA binding necessary for activity of some RSSs (18, 19, 42, 43). All four WG/GW motifs present in P1N-PISPO were mutated to AG/GA. The mutated variant (P1N-PISPOmut) was agroinfiltrated in N. benthamiana leaves, along with the GFP-expressing construct, to evaluate its activity as an RSS. The mutated product failed to counteract the RNA silencing, as shown by GFP fluorescence under UV light, GFP mRNA Northern blots, and RT-qPCR quantification of GFP mRNA (Fig. 5C to E).
DISCUSSION
Among all the known members of the Potyvirus genus (158 species according to the International Committee of Taxonomy of Viruses [ICTV] [2014] release), SPFMV presents several peculiarities. First, its genome is the largest, with an extraordinarily long P1 region (664 to 724 aa), which is surpassed in the Potyviridae family only by the equivalent P1 product of the ipomovirus SPMMV (758 aa), which shares some similarity with SPFMV P1 in the N-terminal region (7). The bioinformatic prediction of the additional ORF PISPO within the P1-coding sequence (4, 44) added a further peculiarity, although it was unknown until now whether PISPO was expressed. Our results shed light on these aspects of SPFMV biology, first by demonstrating that the predicted trans-framed P1N-PISPO product is expressed during SPFMV infection and then by finding that P1N-PISPO contributes to counteract the RNA silencing-based plant defense to viral infection.
These results might be especially relevant in the context of the devastating and widespread sweet potato viral disease (SPVD) (4, 39). In contrast to many other synergisms involving potyviruses (see reference 45 and references therein), sweet potato viral disease is considered atypical because the potyvirus is the partner with a boost in its accumulation. Although we have confirmed this previous observation with our plant material, many important aspects still remain to be explained. For instance, further research is needed to elucidate whether the rather peculiar RNase 3 RSSs from SPCSV (46–48) and P1N-PISPO are involved in the outcome of this complex interaction. In line with that, another interesting but unexpected observation of our work is related to this unusual synergism: we have noticed a specific reduction in RNA slippage frequency at the PISPO site, but not in PIPO, in plants superinfected with SPCSV (Fig. 1). The different roles of P1N-PISPO and P3N-PIPO during viral infection, named suppression of RNA silencing (this report) or viral movement (9–11), respectively, might provide clues not only to explain this observation but also to understand how changes in the relative amounts of these two gene products could affect the outcome of SPVD in the coinfected plants.
The novel potyviral gene product P1N-PISPO (654 aa, 72.7 kDa) combines a P1N part shared with the canonical P1 (N-terminal portion of 422 aa, equivalent to a 46.5-kDa protein fragment) and the PISPO sequence (232 aa, fragment of 26.1 kDa). The product is similar in size to P1 (664 aa, 74.1 kDa) but with notable differences, including, for instance, the predicted isoelectric points of the two proteins: 9.24 for P1 and 6.17 for P1N-PISPO. Interestingly, an isoelectric point of slightly below 7 is a hallmark of P1s displaying RNA silencing suppression activity in other potyvirids (7).
The expression of P1N-PISPO is likely to result from the translation of RNA variants with a G2A7 sequence, generated by polymerase slippage in a G2A6 conserved motif (13). This mechanism of expression is supported by examination of sequences corresponding to different potyviruses present in the virome of the AM-MB2 plant, which showed results consistent with polymerase slippage in equivalent G2A6 motifs for SPV2 and SPVC (see Table S7 in the supplemental material). Moreover, our experiments designed to identify translational frameshifting in WGE failed to show a noticeable production of truncated frameshifted protein products, and the presence of minor products compatible in size could be also generated by T7 polymerase slippage, as shown by others (14, 49, 50). Consistent with our view that viral RNA polymerase slippage is the most likely mechanism of production of out-of-frame products in potyviruses, the transient expression of wild-type P1 sequence out of the viral infection context resulted in the production only of P1-derived peptides when analyzed with the sensitive mass spectrometry technique (Fig. 4A). Altogether, a strong case can be proposed for viral polymerase transcriptional slippage as the mechanism used to produce the trans-frame P1N-PISPO, and likely the same conclusion can be expanded to P3N-PIPO proteins found in all other potyvirids. The combination of our analysis with SPFMV, SPVC, and SPV2 suggests that polymerase slippage might occur in all sweet potato potyviruses where the PISPO sequence was predicted (4).
When facing the challenge of detecting a previously unknown gene product that was predicted bioinformatically, we decided to adopt a straightforward approach using mass spectrometry. Similar techniques have been used previously to identify viral infections in plants, including those produced for potyviruses (51), but to our knowledge, this is the first time that it serves to demonstrate that an out-of-frame gene product is being expressed in infected plant tissues. We believe that this new method is a convenient approach that gives fast and unequivocal proof of protein translation without the need of obtaining specific antibodies, which is a time-consuming procedure that may fail depending on the antigenicity of the target protein. Mass spectrometry allowed unambiguous detection of both P1 and P1N-PISPO together in the same sample, showing that the two gene products coexist in infected plants. Although the methodology is not quantitative, the abundance and number of peptides might provide some indication of the expression levels and the stability of the mature proteins. For example, compared to another large viral product, CI, the high coverage obtained for P1 and P1N-PISPO suggests that these two proteins are quite stable. While we have shown mass spectrometry to be a useful method for identifying novel gene products, further research is required to determine the turnover and subcellular localization of P1N-PISPO during the infection cycle and to evaluate its impact in pathogenicity.
The functions of potyviral P1s have remained elusive for many years, but recent results are revealing the importance of P1 during potyvirus infection (32, 52). In the case of SPFMV and other potyviruses infecting sweet potato, the presence of P1N-PISPO adds a further layer of complexity to efforts to unravel the role(s) of these gene products in the infection context (53). To start addressing this, our experiments show a clear role for P1N-PISPO as an RSS. Interestingly, for all the members of the Potyvirus genus tested so far, the essential RNA silencing suppression activity was found associated with HCPro, which was the first characterized RSS (54, 55). In other members of the Potyviridae family, however, the RNA silencing suppression function is often shifted to P1 products, as has been observed in ipomoviruses (18, 33, 56), tritimoviruses (57), and poaceviruses (58). These RSSs belong to a distinct group of P1 proteins that appears to be evolutionarily separated from typical P1 proteins of members of the genus Potyvirus (7, 59). Our finding that the P1N-PISPO product acts as an RSS would serve to expand this list of known P1-related sources of RNA silencing suppression activity in the family Potyviridae and might help in our understanding of the evolutionary acquisition of this important viral function.
Whereas the P1b protein of the ipomovirus CVYV displays a strong RSS activity that depends on its ability to bind small RNAs (36), the P1 protein of SPMMV suppresses RNA silencing by interfering with RISC activity, specifically through blocking Ago binding via WG/GW hooks (18). Interestingly, a recent study showed that the native SPFMV P1, which shares noticeable sequence similarity with SPMMV P1 (7), does not work as an RSS but gains this functionality when mutations are introduced to create additional WG/GW motifs in positions near the ones present in SPMMV P1 (60). In our experiments, P1 also failed to show a clear RSS activity, confirming the observations of Szabó and coworkers (60).
Our result that an SPFMV P1N-PISPO variant in which all the WG/GW motifs have been mutated loses RNA silencing suppression activity suggests that P1N-PISPO disrupts RNA silencing by a mechanism involving Ago hooks, similar to that of SPMMV P1 (18). The fact that the silencing activities of SPFMV P1N-PISPO and SPMMV P1 are both quite weak further supports the hypothesis that these suppressors share a similar mechanism of action. However, we cannot rule out that the correlation between functionality and presence of WG/GW in SPFMV P1N-PISPO could correspond to Ago-independent disturbances caused, for instance, by conformational alterations derived from mutations of W residues, as might be the case in other RSSs (42).
Giner and coworkers suggested that the expression of a weak RSS in SPMMV could be a viral strategy to cause only mild damage in the host, allowing sweet potato potyvirids to survive in infected perennial plants for an extended period (18). This idea is in agreement with the weak RSS activity of SPFMV P1N-PISPO. We can also speculate that sweet potato potyvirids might not depend exclusively on the weak RNA silencing suppression activities of their P1N-PISPOs found here and that other viral products could help to counteract RNA silencing during infection. In this regard, for instance, the VPg of the potyvirus PVA has been found to counteract RNA silencing (46, 61). Additionally, the levels of expression of HCPro in other potyviruses were reported to be regulated by P1 (62). In the case of SPFMV, although our transient agroinfiltration experiments with individual gene products showed no clear antisilencing activity for P1 or HCPro, preliminary results suggest that P1-HCPro might display some activity. Taking into account that the expression of the P1 construct by agroinfiltration does not produce P1N-PISPO (Fig. 4), contributions of P1 and/or HCPro to silencing suppression during SPFMV infection cannot be ruled out. Indeed, the context of the viral infection is quite different from that in the transient agroinfiltration assays, and therefore further work will be needed to fully understand the modes of action and the relationships of all viral factors that might be participating in counteracting the RNA silencing-based host defenses.
The switch of the antisilencing role from established suppressors such as HC-Pro or P1 (the strength of which could be adjusted through regular mutation/selection processes) to P1N-PISPO with its rather peculiar expression mechanism deserves some attention from an evolutionary point of view. As mentioned above, whereas the main RNA silencing suppression activity lies with HCPro in members of the genus Potyvirus, it is supplied by a P1-type protein in ipomoviruses, tritimoviruses, and poaceviruses, likely highlighting two evolutionary lineages in the family Potyviridae. Thus, we can speculate that the existence of a P1-related RNA silencing suppressor in the potyvirus SPFMV could be the result of a recombination event between a potyvirus and an ipomovirus in sweet potato, which is supported by the notable similarity between the N termini of the P1 proteins from SPFMV and the ipomovirus SPMMV (7).
Importantly, the fact that all members of the potyvirids produce P3N-PIPO suggests that this gene product appeared very early in the evolutionary history of these viruses. Potential strategies developed to deal with risks associated with RNA polymerase slippage, such as genomic modifications in the viral progeny (14, 63) or those derived from mRNA decay (64), could have favored the more recent emergence of P1N-PISPO in a subset of potyviruses. Thus, understanding how potyvirids counteract problems derived from this peculiar gene expression mechanism will certainly deserve further experimental work.
To summarize, our results highlight the enormous genomic flexibility of viruses, which allows them to profit from particular biochemical features of their gene products, such as the slippage capacity of the RNA polymerase, to expand their gene dotation and explore alternative pathways to improve adaptation to a variety of host and environmental conditions.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by Mineco grants AGL2013-42537-R to J.J.L.-M. and BIO2013-49053-R to J.A.G. and by Plant KBBE PCIN-2013-056 to J.A.G. A.M. is a recipient of FPI fellowship BES-2011-045699. D.C.B. is the Royal Society Edward Penley Abraham Research Professor.
We thank Jesús Navas (IHSM-UMA-CSIC La Mayora, Málaga, Spain) for providing us with SPCSV-infected material and the anonymous reviewers for their insightful contributions. We acknowledge the assistance of Eliandre de Oliveira and María Antonia Odena (Proteomics Platform of Barcelona Science Park, UB, a member of the ProteoRed-ISCIII network). We also thank Claire Agius for valuable help.
ADDENDUM IN PROOF
During the preparation of this work, we became aware that another paper on the same subject arrived at equivalent conclusions (Untiveros M, Olspert A, Artola K, Firth AE, Kreuze JF, Valkonen JP, Mol Plant Pathol, http://dx.doi.org/10.1111/mpp.12366, in press).
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.02360-15.
REFERENCES
- 1.Valli A, García JA, López-Moya JJ. 2015. Potyviridae, p 1–10. In Encyclopedia of life sciences. John Wiley & Sons, Ltd., Chichester, United Kingdom. [Google Scholar]
- 2.Revers F, García JA. 2015. Molecular biology of potyviruses. Adv Virus Res 92:101–199. doi: 10.1016/bs.aivir.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 3.Chung BY-W, Miller WA, Atkins JF, Firth AE. 2008. An overlapping essential gene in the Potyviridae. Proc Natl Acad Sci U S A 105:5897–5902. doi: 10.1073/pnas.0800468105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark CA, Davis JA, Abad JA, Cuellar WJ, Fuentes S, Kreuze JF, Gibson RW, Mukasa SB, Tugume AK, Tairo FD, Valkonen JPT. 2012. Sweetpotato viruses: 15 years of progress on understanding and managing complex diseases. Plant Dis 96:168–185. doi: 10.1094/PDIS-07-11-0550. [DOI] [PubMed] [Google Scholar]
- 5.Sakai J, Mori M, Morishita T, Tanaka M, Hanada K, Usugi T, Nishiguchi M. 1997. Complete nucleotide sequence and genome organization of sweet potato feathery mottle virus (S strain) genomic RNA: the large coding region of the P1 gene. Arch Virol 142:1553–1562. doi: 10.1007/s007050050179. [DOI] [PubMed] [Google Scholar]
- 6.Verchot J, Koonin EV, Carrington JC. 1991. The 35-kDa protein from the N-terminus of the potyviral polyprotein functions as a third virus-encoded proteinase. Virology 185:527–535. doi: 10.1016/0042-6822(91)90522-D. [DOI] [PubMed] [Google Scholar]
- 7.Valli A, López-Moya JJ, García JA. 2007. Recombination and gene duplication in the evolutionary diversification of P1 proteins in the family Potyviridae. J Gen Virol 88:1016–1028. doi: 10.1099/vir.0.82402-0. [DOI] [PubMed] [Google Scholar]
- 8.Choi SH, Hagiwara-Komoda Y, Nakahara KS, Atsumi G, Shimada R, Hisa Y, Naito S, Uyeda I. 2013. Quantitative and qualitative involvement of P3N-PIPO in overcoming recessive resistance against Clover yellow vein virus in pea carrying the cyv1 gene. J Virol 87:7326–7337. doi: 10.1128/JVI.00065-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wen RH, Hajimorad MR. 2010. Mutational analysis of the putative pipo of soybean mosaic virus suggests disruption of PIPO protein impedes movement. Virology 400:1–7. doi: 10.1016/j.virol.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 10.Vijayapalani P, Maeshima M, Nagasaki-Takekuchi N, Miller WA. 2012. Interaction of the trans-frame potyvirus protein P3N-PIPO with host protein PCaP1 facilitates potyvirus movement. PLoS Pathog 8:e1002639. doi: 10.1371/journal.ppat.1002639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei T, Zhang C, Hong J, Xiong R, Kasschau KD, Zhou X, Carrington JC, Wang A. 2010. Formation of complexes at plasmodesmata for potyvirus intercellular movement is mediated by the viral protein P3N-PIPO. PLoS Pathog 6:e1000962. doi: 10.1371/journal.ppat.1000962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hisa Y, Suzuki H, Atsumi G, Choi SH, Nakahara KS, Uyeda I. 2014. P3N-PIPO of Clover yellow vein virus exacerbates symptoms in pea infected with White clover mosaic virus and is implicated in viral synergism. Virology 449:200–206. doi: 10.1016/j.virol.2013.11.016. [DOI] [PubMed] [Google Scholar]
- 13.Rodamilans B, Valli A, Mingot A, San León D, Baulcombe D, López-Moya JJ, García JA. 2015. RNA polymerase slippage as a mechanism for the production of frameshift gene products in plant viruses of the Potyviridae family. J Virol 89:6965–6967. doi: 10.1128/JVI.00337-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olspert A, Chung BY-W, Atkins JF, Carr JP, Firth AE. 2015. Transcriptional slippage in the positive-sense RNA virus family Potyviridae. EMBO Rep 16:995–1004. doi: 10.15252/embr.201540509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baulcombe D. 2004. RNA silencing in plants. Nature 431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- 16.Valli A, López-Moya JJ, García JA. 2009. RNA silencing and its suppressors in the plant-virus interplay, p 1–29. In Encyclopedia of life sciences. John Wiley & Sons, Ltd., Chichester, United Kingdom. [Google Scholar]
- 17.Csorba T, Kontra L, Burgyán J. 2015. Viral silencing suppressors: Tools forged to fine-tune host-pathogen coexistence. Virology 479–480:85–103. doi: 10.1016/j.virol.2015.02.028. [DOI] [PubMed] [Google Scholar]
- 18.Giner A, Lakatos L, García-Chapa M, López-Moya JJ, Burgyán J. 2010. Viral protein inhibits RISC activity by argonaute binding through conserved WG/GW motifs. PLoS Pathog 6:e1000996. doi: 10.1371/journal.ppat.1000996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Azevedo J, Garcia D, Pontier D, Ohnesorge S, Yu A, Garcia S, Braun L, Bergdoll M, Hakimi MA, Lagrange T, Voinnet O. 2010. Argonaute quenching and global changes in Dicer homeostasis caused by a pathogen-encoded GW repeat protein. Genes Dev 24:904–915. doi: 10.1101/gad.1908710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wright CF, Morelli MJ, Thébaud G, Knowles NJ, Herzyk P, Paton DJ, Haydon DT, King DP. 2011. Beyond the consensus: dissecting within-host viral population diversity of foot-and-mouth disease virus by using next-generation genome sequencing. J Virol 85:2266–2275. doi: 10.1128/JVI.01396-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Töpfer A, Zagordi O, Prabhakaran S, Roth V, Halperin E, Beerenwinkel N. 2013. Probabilistic inference of viral quasispecies subject to recombination. J Comput Biol 20:113–123. doi: 10.1089/cmb.2012.0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin DP, Lemey P, Lott M, Moulton V, Posada D, Lefeuvre P. 2010. RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics 26:2462–2463. doi: 10.1093/bioinformatics/btq467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin D, Rybicki E. 2000. RDP: detection of recombination amongst aligned sequences. Bioinformatics 16:562–563. doi: 10.1093/bioinformatics/16.6.562. [DOI] [PubMed] [Google Scholar]
- 26.Martin DP, Posada D, Crandall KA, Williamson C. 2005. A modified bootscan algorithm for automated identification of recombinant sequences and recombination breakpoints. AIDS Res Hum Retroviruses 21:98–102. doi: 10.1089/aid.2005.21.98. [DOI] [PubMed] [Google Scholar]
- 27.Smith JM. 1992. Analyzing the mosaic structure of genes. J Mol Evol 34:126–129. [DOI] [PubMed] [Google Scholar]
- 28.Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SDW. 2006. GARD: a genetic algorithm for recombination detection. Bioinformatics 22:3096–3098. doi: 10.1093/bioinformatics/btl474. [DOI] [PubMed] [Google Scholar]
- 29.Delport W, Poon AFY, Frost SDW, Kosakovsky Pond SL. 2010. Datamonkey 2010: a suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics 26:2455–2457. doi: 10.1093/bioinformatics/btq429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanaka Y, Nakamura S, Kawamukai M, Koizumi N, Nakagawa T. 2011. Development of a series of gateway binary vectors possessing a tunicamycin resistance gene as a marker for the transformation of Arabidopsis thaliana. Biosci Biotechnol Biochem 75:804–807. doi: 10.1271/bbb.110063. [DOI] [PubMed] [Google Scholar]
- 31.Nakagawa T, Suzuki T, Murata S, Nakamura S, Hino T, Maeo K, Tabata R, Kawai T, Tanaka K, Niwa Y, Watanabe Y, Nakamura K, Kimura T, Ishiguro S. 2007. Improved Gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci Biotechnol Biochem 71:2095–2100. doi: 10.1271/bbb.70216. [DOI] [PubMed] [Google Scholar]
- 32.Pasin F, Simón-Mateo C, García JA. 2014. The hypervariable amino-terminus of P1 protease modulates potyviral replication and host defense responses. PLoS Pathog 10:e1003985. doi: 10.1371/journal.ppat.1003985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Valli A, Martín-Hernández AM, López-Moya JJ, García JA. 2006. RNA silencing suppression by a second copy of the P1 serine protease of Cucumber vein yellowing ipomovirus, a member of the family Potyviridae that lacks the cysteine protease HCPro. J Virol 80:10055–10063. doi: 10.1128/JVI.00985-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carbonell A, Dujovny G, García JA, Valli A. 2012. The Cucumber vein yellowing virus silencing suppressor P1b can functionally replace HCPro in Plum pox virus infection in a host-specific manner. Mol Plant Microbe Interact 25:151–164. doi: 10.1094/MPMI-08-11-0216. [DOI] [PubMed] [Google Scholar]
- 35.Valli A, Dujovny G, García JA. 2008. Protease activity, self interaction, and small interfering RNA binding of the silencing suppressor P1b from Cucumber vein yellowing ipomovirus. J Virol 82:974–986. doi: 10.1128/JVI.01664-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Valli A, Oliveros JC, Molnar A, Baulcombe D, García JA. 2011. The specific binding to 21-nt double-stranded RNAs is crucial for the anti-silencing activity of Cucumber vein yellowing virus P1b and perturbs endogenous small RNA populations. RNA 17:1148–1158. doi: 10.1261/rna.2510611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leckie BM, Stewart CN Jr. 2011. Agroinfiltration as a technique for rapid assays for evaluating candidate insect resistance transgenes in plants. Plant Cell Rep 30:325–334. doi: 10.1007/s00299-010-0961-2. [DOI] [PubMed] [Google Scholar]
- 38.Lacomme C, Hrubikova K, Hein I. 2003. Enhancement of virus-induced gene silencing through viral-based production of inverted-repeats. Plant J 34:543–553. doi: 10.1046/j.1365-313X.2003.01733.x. [DOI] [PubMed] [Google Scholar]
- 39.Karyeija RF, Kreuze JF, Gibson RW, Valkonen JP. 2000. Synergistic interactions of a potyvirus and a phloem-limited crinivirus in sweet potato plants. Virology 269:26–36. doi: 10.1006/viro.1999.0169. [DOI] [PubMed] [Google Scholar]
- 40.Untiveros M, Fuentes S, Kreuze J. 2008. Molecular variability of Sweet potato feathery mottle virus and other potyviruses infecting sweet potato in Peru. Arch Virol 153:473–483. doi: 10.1007/s00705-007-0019-0. [DOI] [PubMed] [Google Scholar]
- 41.Tugume AK, Cuellar WJ, Mukasa SB, Valkonen JP. 2010. Molecular genetic analysis of virus isolates from wild and cultivated plants demonstrates that East Africa is a hotspot for the evolution and diversification of sweet potato feathery mottle virus. Mol Ecol 19:3139–3156. doi: 10.1111/j.1365-294X.2010.04682.x. [DOI] [PubMed] [Google Scholar]
- 42.Pérez-Cañamás M, Hernández C. 2015. Key importance of small RNA binding for the activity of a glycine-tryptophan (GW) motif-containing viral suppressor of RNA silencing. J Biol Chem 290:3106–3120. doi: 10.1074/jbc.M114.593707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chattopadhyay M, Stupina VA, Gao F, Szarko CR, Kuhlmann MM, Yuan X, Shi K, Simon AE. 2015. Requirement for host RNA silencing components and the virus silencing suppressor when second-site mutations compensate for structural defects in the 3′ UTR. J Virol 89:11603–11618. doi: 10.1128/JVI.01566-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li F, Xu D, Abad J, Li R. 2012. Phylogenetic relationships of closely related potyviruses infecting sweet potato determined by genomic characterization of Sweet potato virus G and Sweet potato virus 2. Virus Genes 45:118–125. doi: 10.1007/s11262-012-0749-2. [DOI] [PubMed] [Google Scholar]
- 45.Syller J. 2012. Facilitative and antagonistic interactions between plant viruses in mixed infections. Mol Plant Pathol 13:204–216. doi: 10.1111/j.1364-3703.2011.00734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cuellar WJ, Kreuze JF, Rajamäki M-L, Cruzado KR, Untiveros M, Valkonen JPT. 2009. Elimination of antiviral defense by viral RNase III. Proc Natl Acad Sci U S A 106:10354–10358. doi: 10.1073/pnas.0806042106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tugume AK, Amayo R, Weinheimer I, Mukasa SB, Rubaihayo PR, Valkonen JP. 2013. Genetic variability and evolutionary implications of RNA silencing suppressor genes in RNA1 of sweet potato chlorotic stunt virus isolates infecting sweetpotato and related wild species. PLoS One 8:e81479. doi: 10.1371/journal.pone.0081479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weinheimer I, Jiu Y, Rajamäki M-L, Matilainen O, Kallijärvi J, Cuellar WJ, Lu R, Saarma M, Holmberg CI, Jäntti J, Valkonen JPT. 2015. Suppression of RNAi by dsRNA-degrading RNaseIII enzymes of viruses in animals and plants. PLoS Pathog 11:e1004711. doi: 10.1371/journal.ppat.1004711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Molodtsov V, Anikin M, McAllister WT. 2014. The presence of an RNA:DNA hybrid that is prone to slippage promotes termination by T7 RNA polymerase. J Mol Biol 426:3095–3107. doi: 10.1016/j.jmb.2014.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wons E, Furmanek-Blaszk B, Sektas M. 2015. RNA editing by T7 RNA polymerase bypasses InDel mutations causing unexpected phenotypic changes. Nucleic Acids Res 43:3950–3963. doi: 10.1093/nar/gkv269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Luo H, Wylie SJ, Jones MGK. 2010. Identification of plant viruses using one-dimensional gel electrophoresis and peptide mass fingerprints. J Virol Methods 165:297–301. doi: 10.1016/j.jviromet.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 52.Martinez F, Daros JA. 2014. Tobacco etch virus protein P1 traffics to the nucleolus and associates with the host 60S ribosomal subunits during infection. J Virol 88:10725–10737. doi: 10.1128/JVI.00928-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ivanov KI, Eskelin K, Lohmus A, Makinen K. 2014. Molecular and cellular mechanisms underlying potyvirus infection. J Gen Virol 95:1415–1429. doi: 10.1099/vir.0.064220-0. [DOI] [PubMed] [Google Scholar]
- 54.Anandalakshmi R, Pruss GJ, Ge X, Marathe R, Mallory AC, Smith TH, Vance VB. 1998. A viral suppressor of gene silencing in plants. Proc Natl Acad Sci U S A 95:13079–13084. doi: 10.1073/pnas.95.22.13079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kasschau KD, Carrington JC. 1998. A counterdefensive strategy of plant viruses: suppression of posttranscriptional gene silencing. Cell 95:461–470. doi: 10.1016/S0092-8674(00)81614-1. [DOI] [PubMed] [Google Scholar]
- 56.Mbanzibwa DR, Tian Y, Mukasa SB, Valkonen JP. 2009. Cassava brown streak virus (Potyviridae) encodes a putative Maf/HAM1 pyrophosphatase implicated in reduction of mutations and a P1 proteinase that suppresses RNA silencing but contains no HC-Pro. J Virol 83:6934–6940. doi: 10.1128/JVI.00537-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Young BA, Stenger DC, Qu F, Morris TJ, Tatineni S, French R. 2012. Tritimovirus P1 functions as a suppressor of RNA silencing and an enhancer of disease symptoms. Virus Res 163:672–677. doi: 10.1016/j.virusres.2011.12.019. [DOI] [PubMed] [Google Scholar]
- 58.Tatineni S, Qu F, Li R, Morris TJ, French R. 2012. Triticum mosaic poacevirus enlists P1 rather than HC-Pro to suppress RNA silencing-mediated host defense. Virology 433:104–115. doi: 10.1016/j.virol.2012.07.016. [DOI] [PubMed] [Google Scholar]
- 59.Rodamilans B, Valli A, García JA. 2013. Mechanistic divergence between P1 proteases of the family Potyviridae. J Gen Virol 94:1407–1414. doi: 10.1099/vir.0.050781-0. [DOI] [PubMed] [Google Scholar]
- 60.Szabó EZ, Manczinger M, Göblös A, Kemény L, Lakatos L. 2012. Switching on RNA silencing suppressor activity by restoring argonaute binding to a viral protein. J Virol 86:8324–8327. doi: 10.1128/JVI.00627-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rajamäki ML, Streng J, Valkonen JP. 2014. Silencing suppressor protein VPg of a potyvirus interacts with the plant silencing-related protein SGS3. Mol Plant Microbe Interact 27:1199–1210. doi: 10.1094/MPMI-04-14-0109-R. [DOI] [PubMed] [Google Scholar]
- 62.Tena Fernández F, González I, Doblas P, Rodríguez C, Sahana N, Kaur H, Tenllado F, Praveen S, Canto T. 2013. The influence of cis-acting P1 protein and translational elements on the expression of potato virus Y helper-component proteinase (HCPro) in heterologous systems and its suppression of silencing activity. Mol Plant Pathol 14:530–541. doi: 10.1111/mpp.12025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shabman RS, Jabado OJ, Mire CE, Stockwell TB, Edwards M, Mahajan M, Geisbert TW, Basler CF. 2014. Deep sequencing identifies noncanonical editing of Ebola and Marburg virus RNAs in infected cells. mBio 5:e02011. doi: 10.1128/mBio.02011-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Garcia D, Garcia S, Voinnet O. 2014. Nonsense-mediated decay serves as a general viral restriction mechanism in plants. Cell Host Microbe 16:391–402. doi: 10.1016/j.chom.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.