ABSTRACT
Porcine reproductive and respiratory syndrome virus (PRRSV) nonstructural protein 1β (nsp1β) is a multifunctional viral protein, which is involved in suppressing the host innate immune response and activating a unique −2/−1 programmed ribosomal frameshifting (PRF) signal for the expression of frameshifting products. In this study, site-directed mutagenesis analysis showed that the R128A or R129A mutation introduced into a highly conserved motif (123GKYLQRRLQ131) reduced the ability of nsp1β to suppress interferon beta (IFN-β) activation and also impaired nsp1β's function as a PRF transactivator. Three recombinant viruses, vR128A, vR129A, and vRR129AA, carrying single or double mutations in the GKYLQRRLQ motif were characterized. In comparison to the wild-type (WT) virus, vR128A and vR129A showed slightly reduced growth abilities, while the vRR129AA mutant had a significantly reduced growth ability in infected cells. Consistent with the attenuated growth phenotype in vitro, pigs infected with nsp1β mutants had lower levels of viremia than did WT virus-infected pigs. Compared to the WT virus in infected cells, all three mutated viruses stimulated high levels of IFN-α expression and exhibited a reduced ability to suppress the mRNA expression of selected interferon-stimulated genes (ISGs). In pigs infected with nsp1β mutants, IFN-α production was increased in the lungs at early time points postinfection, which was correlated with increased innate NK cell function. Furthermore, the augmented innate response was consistent with the increased production of IFN-γ in pigs infected with mutated viruses. These data demonstrate that residues R128 and R129 are critical for nsp1β function and that modifying these key residues in the GKYLQRRLQ motif attenuates virus growth ability and improves the innate and adaptive immune responses in infected animals.
IMPORTANCE PRRSV infection induces poor antiviral innate IFN and cytokine responses, which results in weak adaptive immunity. One of the strategies in next-generation vaccine construction is to manipulate viral proteins/genetic elements involved in antagonizing the host immune response. PRRSV nsp1β was identified to be a strong innate immune antagonist. In this study, two basic amino acids, R128 and R129, in a highly conserved GKYLQRRLQ motif were determined to be critical for nsp1β function. Mutations introduced into these two residues attenuated virus growth and improved the innate and adaptive immune responses of infected animals. Technologies developed in this study could be broadly applied to current commercial PRRSV modified live-virus (MLV) vaccines and other candidate vaccines.
INTRODUCTION
Porcine reproductive and respiratory syndrome (PRRS), a disease described in the United States in 1987 (1) and in Europe in 1990 (2), has caused tremendous economic losses to the swine industry since its appearance. Hallmark symptoms of PRRS are mild to severe respiratory disease in infected newborn and growing pigs and reproductive failure in pregnant sows. The etiologic agent, PRRS virus (PRRSV), was first discovered in The Netherlands in 1991 (2). In the United States, PRRSV was first isolated and characterized in 1992 (3, 4). Generally, infection of pigs by most PRRSV strains dampens the host innate immune response (5, 6). This initial suppression of the host innate immune response leads to the delayed induction of protective cellular and humoral immunity (7, 8), which provides a window of time that allows PRRSV to replicate, be shed, and be transmitted to other contact-naive animals. Therefore, strategies for vaccine development are directed at constructing a PRRS vaccine capable of inducing high levels of innate and adaptive immune responses.
PRRSV is an enveloped, positive-stranded RNA virus that belongs to the order Nidovirales, family Arteriviridae, which includes equine arteritis virus (EAV), mouse lactate dehydrogenase-elevating virus (LDV), simian hemorrhagic fever virus (SHFV), and several recently discovered monkey arteriviruses that are only distantly related to SHFV (9). The PRRSV genome is ∼15 kb in length and contains at least 11 open reading frames (ORFs). The 3′ end of the genome encodes four membrane-associated glycoproteins (GP2a, GP3, GP4, and GP5), three unglycosylated membrane proteins (E, ORF5a, and M), and a nucleocapsid protein (N) (10–19). The replicase-associated ORF1a and ORF1b genes are situated at the 5′ end and represent nearly 75% of the viral genome. ORF1a and ORF1b encode two large polyproteins, pp1a and pp1ab, with the expression of the latter depending on a −1 ribosomal frameshifting signal in the ORF1a/ORF1b overlap region. Following their synthesis from the genomic mRNA template, the pp1a and pp1ab replicase polyproteins are processed into at least 14 nonstructural proteins by a complex proteolytic cascade that is directed by four proteinase domains encoded in ORF1a, which include two papain-like proteinases (PLP1α and PLP1β) located in nsp1α and nsp1β, a papain-like proteinase (PLP2) domain located at the N terminus of nsp2, and a serine proteinase located in nsp4. PLP1α autocleaves between nsp1α and nsp1β, PLP1β autocleaves between nsp1β/and nsp2, and PLP2 cleaves between nsp2 and nsp3, which mediates the rapid release of nsp1α, nsp1β, and nsp2 from the polyprotein (20). Recently, two novel PRRSV proteins, nsp2TF and nsp2N, were identified (21). nsp2TF and nsp2N are expressed by a novel −2/−1 programmed ribosomal frameshifting (PRF) mechanism, which accesses the alternative ORF (TF) through a frameshifting site that overlaps the nsp2-encoding region. Both nsp2TF and nsp2N share the N-terminal two-thirds sequence of nsp2, which contains the PLP2 domain.
Previous studies from our laboratory and others identified PRRSV nsp1β to be a strong innate immune antagonist (22–24). PRRSV nsp1β has strong inhibitory effects on type I interferon (IFN) production and signaling pathways that lead to the expression of interferon-stimulated genes (ISGs). Interestingly, this protein was recently identified to also function as a transactivator for the expression of the −2/−1 PRF products nsp2TF and nsp2N (25). Embedded in nsp1β's papain-like autoproteinase domain (PLP1β), a highly conserved GKYLQRRLQ motif was identified to be critical for −2/−1 PRF transactivation and the innate immune suppression function of the virus (25, 26). Based on crystal structure analysis, three basic residues (K124, R128, and R129) in the GKYLQRRLQ motif are exposed on the surface of the protein (25). In this study, we further investigated the function of the basic residues K124, R128, and R129 involved in the modulation of host immune responses. Recombinant viruses carrying mutations in these basic residues were created and characterized in cell culture systems as well as in a nursery piglet model.
MATERIALS AND METHODS
Cells and viruses.
HEK-293T cells and MARC-145 cells were maintained in minimum essential medium (MEM) (Gibco) supplemented with 10% fetal bovine serum and antibiotic (streptomycin [100 μg/ml]) at 37°C with 5% CO2. BHK-21 cells were cultured in minimum essential medium supplemented with 5% fetal bovine serum and antibiotic (streptomycin [100 μg/ml]). As described previously, porcine alveolar macrophages (PAMs) were obtained from lung lavage samples of 6-week-old PRRSV-naive piglets (27). The Sendai virus (SeV) Cantell strain, grown in embryonated chicken eggs, was used for stimulation of the type I IFN response in the cell culture system. Type 2 PRRSV isolate SD95-21 (GenBank accession no. KC469618) and its nsp1β mutants were used for subsequent experiments.
Antibodies.
To detect the expression of nsp1β and its mutants, monoclonal antibody (MAb) 123-128 (25) or anti-FLAG M2 MAb (Sigma-Aldrich, St. Louis, MO) was used. MAb 140-68 (25), which specifically recognizes the common N-terminal PLP2 domain of nsp2, nsp2TF, and nsp2N, was used to detect the expression of nsp2-related proteins. A rabbit polyclonal antibody against nsp2TF (PAb-TF) (25) was utilized to immunoprecipitate and detect nsp2TF. In addition, an anti-β-tubulin MAb (abm Inc., BC, Canada) was used to detect the expression of the housekeeping gene β-tubulin. An antibody mixture of MAb M2 against FLAG and anti-β-tubulin was used for the simultaneous detection of the expression of FLAG-tagged nsp1β and β-tubulin in Western blots (WBs), while an antibody mixture of MAb 123-128 and anti-β-tubulin was used for the simultaneous detection of the expression of nsp1β and β-tubulin in Western blots.
Plasmids.
By using the nsp1β-expressing plasmid (p3xFLAG-NA-nsp1β) that we generated previously (26), specific mutations, K124A, R128A, R129A, or RR129AA (double mutation of R128A and R129A), in the GKYLQRRLQ motif region (amino acids 123 to 131) of nsp1β were introduced by site-directed mutagenesis using a QuikChange site-directed mutagenesis kit (Agilent Technologies Inc., Santa Clara, CA) according to the manufacturer's instructions. A vaccinia virus-T7 polymerase system (pL-NA-nsp1β-2) expressing nsp1β-nsp2 of SD95-21 virus was described previously (26). Specific mutations (K124A, R128A, R129A, or RR129AA) were introduced into the nsp1β region of pL-NA-nsp1β-2 by using a QuikChange site-directed mutagenesis kit. To generate full-length PRRSV cDNA clones containing these specific mutations (R128A, R129A, or RR129AA), a shuttle plasmid carrying the region between two unique restriction sites (SphI and ScaI) of the full-length cDNA clone of PRRSV (pCMV-SD95-21) was constructed by using a Zero Blunt PCR cloning kit (Invitrogen, Carlsbad, CA). A QuikChange site-directed mutagenesis kit (Agilent Technologies Inc., Santa Clara, CA) was employed to introduce the specific mutations into the shuttle plasmid. The region between SphI and ScaI of pCMV-SD95-21 was replaced by the corresponding regions of the shuttle plasmids containing the specific mutations. The mutated full-length cDNA clones were designated pCMV-SD95-21-R128A, pCMV-SD95-21-R129A, and pCMV-SD95-21-RR129AA. DNA sequencing was further performed to verify the introduced mutations. For in vitro luciferase reporter assays, two reporter plasmids, p125-Luc and pISRE-Luc, were used as described previously (26).
Luciferase reporter assay.
HEK-293T cells were seeded at 0.5 × 105 cells/ml into 24-well plates 1 day before transfection. DNA transfection was conducted by using FuGENE HD transfection reagent (Promega, Madison, WI). Briefly, cells were cotransfected with 0.5 μg plasmid DNA expressing wild-type (WT) nsp1β (or its mutants) and 0.5 μg luciferase reporter plasmid DNA of p125-Luc or pISRE-Luc. At 24 h posttransfection, cells were mock treated, stimulated with SeV inoculated at 100 hemagglutination (HA) units/ml/well for 16 h, or treated with IFN-β at 2,000 IU/ml/well for 16 h. Cells were lysed and used for reporter gene assays using the dual-luciferase reporter system (Promega, Madison, WI) according to the manufacturer's instructions. Firefly luciferase activities were measured with FLUOstar Omega reader (BMG Labtech, Cary, NC).
Vaccinia virus-T7 polymerase expression system.
nsp1β-nsp2 and its mutants were expressed by using a vaccinia virus-T7 polymerase system (28) as described previously (26). Briefly, HEK-293T cells (1 × 106 cells/well) were seeded into 6-well plates 1 day before infection. Cells in each well were infected with a vaccinia virus expressing T7 polymerase at a multiplicity of infection (MOI) of 10. At 1 h postinfection (hpi), cells were transfected with 2 μg DNA of pL-NA-nsp1β-2 or its mutants by using FuGENE HD transfection reagent (Promega, Madison, WI). At 18 h posttransfection, the cell lysate from each well of the 6-well plate was harvested and subjected to Western blot analysis using antibodies against nsp1β (MAb 123-128) and nsp2 (MAb 140-68). In addition, the cell lysate was used for immunoprecipitation (IP) assays to evaluate the expression of the −2 PRF product with an antibody that specifically recognizes nsp2TF (PAb-TF).
Western blot analysis.
Western blot analysis was performed to evaluate protein expression according to methods described previously (20, 26). Briefly, cell lysates were prepared by harvesting virus-infected or plasmid DNA-transfected cells with radioimmunoprecipitation assay (RIPA) buffer. The cell lysate was mixed with an equal volume of Laemmli sample buffer and heated at 95°C for 6 min. After being separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), proteins were transferred onto a nitrocellulose membrane. The membrane was blocked with 5% skim milk in PBST (phosphate-buffered saline [PBS] with 0.05% Tween 20) at 4°C overnight and then incubated with primary antibody at the appropriate dilution at room temperature for 1 h. After washing three times with PBST, IRDye 800CW goat anti-mouse IgG(H+L) and/or IRDye 680RD goat anti-rabbit IgG(H+L) (Li-Cor Biosciences, Lincoln, NE) secondary antibody was added, and the membrane was incubated for an additional 1 h at room temperature. The target proteins were visualized and quantified by using a digital image system (Odyssey infrared imaging system; Li-Cor Biosciences, Lincoln, NE). For quantification of the target proteins, the expression levels were normalized to the expression level of β-tubulin, which is a housekeeping gene used as a loading control.
Recovery of recombinant viruses from infectious cDNA clones.
The procedure for generating recombinant viruses was described previously (26). BHK-21 cells at 70 to 80% confluence were transfected with 2 μg of the type 2 PRRSV full-length cDNA clone of pCMV-SD95-21 or full-length cDNA clones containing nsp1β mutations. Transfection was performed by using FuGENE HD reagent (Promega, Madison, WI). At 48 h posttransfection, the cell culture supernatant was harvested and passaged on MARC-145 cells. After 48 to 60 h of incubation, indirect immunofluorescence assays were performed to confirm the viability of recombinant viruses by using MAb SDOW17 (PRRSV N protein-specific monoclonal antibody) (29). The recombinant viruses were serially passaged on MARC-145 cells, and passage 3 and 4 viruses were used for further analysis.
Sequencing of nsp1β mutation regions.
To determine the stability of each mutation, cell culture supernatants from recombinant virus-infected cells or serum samples collected from experimentally infected animals (at 14, 21, and 35 days postinfection [dpi]) were used for viral RNA extraction using the QIAamp viral RNA kit (Qiagen). The nsp1β coding region containing the corresponding mutations was amplified by reverse transcription-PCR (RT-PCR), and PCR products were subjected to DNA sequencing at Genewiz Inc. (South Plainfield, NJ).
Virus growth kinetics and plaque assay.
WT and mutant viruses at passage 3 were used to characterize virus growth properties in vitro. Confluent MARC-145 cells were inoculated with the WT virus or nsp1β mutants at an MOI of 0.01. The cell culture supernatant was harvested at 12, 24, 36, 48, 60, and 72 h postinfection. The virus titer was measured by a microtitration assay using MARC-145 cells in 96-well plates and calculated as 50% tissue culture infective doses (TCID50) per milliliter according to the method of Reed and Muench (30). To determine the plaque morphology of the WT virus and nsp1β mutants, a plaque assay was conducted by using MARC-145 cells as described previously (31).
Pig groups and sample collection and preparation.
A total of 45 specific-pathogen-free (SPF) pigs were obtained from the swine farm of The Ohio State University. Pigs were randomly divided into 5 groups (n = 9) (Table 1). Pigs were mock infected (group 1) or infected with 4 × 106 TCID50 of WT PRRSV (group 2), the vR128A mutant (group 3), the vR129A mutant (group 4), or the vRR129AA mutant (group 5). Pigs were inoculated by both the intranasal (i.n.) and intramuscular (i.m.) routes with 1 ml (1 × 106 TCID50) of the virus injected into each nostril and into each side of the neck. Pigs were observed daily, and blood samples were collected at 0, 1, 2, 5, 7, 14, 21, 28, and 35 dpi. Three pigs from each group were sequentially euthanized at 7, 21, and 35 dpi (Table 1). During necropsy, lungs were evaluated for gross lesions by using a method described previously (32), and bronchoalveolar lavage fluid (BALF) and lung tissue samples were collected as described previously (33). The pig experiment was performed according to protocols approved by the Institutional Animal Care and Use Committee (IACUC) of The Ohio State University.
TABLE 1.
Experimental design for testing of nsp1β mutants in nursery pigs
| Procedure | dpi | Pigs in treatment group |
||||
|---|---|---|---|---|---|---|
| Negative control (group 1) | WTe (group 2) | R128A (group 3) | R129A (group 4) | RR129AA (group 5) | ||
| Blood collectiona | 0 | 1–9c | 10–18 | 19–27 | 28–36 | 37–45 |
| 1 | 1–9 | 10–18 | 19–27 | 28–36 | 37–45 | |
| 2 | 1–9 | 10–18 | 19–27 | 28–36 | 37–45 | |
| 5 | 1–9 | 10–18 | 19–27 | 28–36 | 37–45 | |
| 14 | 4–9 | 14–18 | 22–27 | 31–36 | 40–45 | |
| 28 | 7–9 | 17, 18 | 25–27 | 34–36 | 43–45 | |
| Animal terminationb | 7 | 1–3d | 10, 11, 13 | 19–21 | 28–30 | 37–39 |
| 21 | 4–6 | 14–16 | 22–24 | 31–33 | 40–42 | |
| 35 | 7–9 | 17, 18 | 25–27 | 34–36 | 43–45 | |
At 0, 1, 2, 5, 14, and 28 dpi, plasma and PBMCs were collected.
At 7, 21, and 35 dpi, 3 pigs from each group were terminated, and their plasma, PBMCs, bronchoalveolar lavage fluid, and lung tissue samples were collected.
Pigs for blood collection at the indicated time point.
Pigs terminated at the indicated time point.
Pig 12 died before 7 dpi.
Real-time RT-PCR quantification of viral load in infected animals.
For the determination of viral RNA load, serum, BALF, and lung lysate samples were examined by using real-time quantitative RT-PCR (qRT-PCR). Briefly, viral genomic RNA was extracted by using a MagMAX-96 viral RNA isolation kit (Life Technologies) according to the manufacturer's instructions. The viral RNA level was determined by quantitative RT-PCR using an iTaq Universal SYBR green one-step kit (Bio-Rad, Hercules, CA), and the RNA copy numbers were calculated based on an RNA standard curve. A pair of primers, PRRS-qF1 (CCATTTCCTTGACACAGTCG) and PRRS21-qR2 (GACCGCGTAGATGCTACTTAGG), located at the viral genomic region (nucleotides [nt] 14043 to 14130), was designed for real-time RT-PCR. The RNA standard was prepared by in vitro transcription. Briefly, the viral genomic region (nt 13918 to 14246) was amplified by RT-PCR using primers T7-GP5F (TCTAGATAATACGACTCACTATAGGGAACTTGACGCTATGTGAGCTG [underlining indicates the T7 promoter]) and GP5R (TAGAGTCTGCCCTTAGTGTCCA). The PCR product was purified and subjected to in vitro transcription by using a MEGAscript T7 transcription kit (Invitrogen, Carlsbad, CA). The purified RNA product was used as the quantification standard.
Quantitative analysis of mRNA.
Porcine alveolar macrophages were infected with the WT virus or nsp1β mutants at an MOI of 1. At 12 h postinfection, PAMs were harvested with TRIzol LS (Ambion, Foster City, CA) and subjected to total RNA extraction according to the manufacturer's instructions. After removal of contaminating genomic DNA with a Turbo DNA-free kit (Invitrogen, Carlsbad, CA), 1 μg total RNA was used to synthesize first-strand cDNA by using a SuperScript Vilo cDNA synthesis kit (Invitrogen, Carlsbad, CA). Subsequently, real-time PCR was performed to quantify the expression of mRNAs of ISG15, IFIT1, IFITM1, and β-tubulin by using predesigned primer/probe sets (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions. The amounts of ISG15, IFIT1, and IFITM1 mRNAs were normalized to the amount of endogenous β-tubulin mRNA.
Analysis of the swine cytokine response.
Porcine alveolar macrophages were infected with the WT virus or nsp1β mutants at an MOI of 1. At 12 h postinfection, the cell culture supernatant was harvested for analysis of IFN-α expression using a ProcartaPlex Porcine IFN alpha Simplex kit (eBioscience, San Diego, CA). In addition, Serum, BALF, and lung lysate samples were used for measuring the levels of the secreted cytokines IFN-α, IFN-γ, interleukin-6 (IL-6), and IL-10 by an enzyme-linked immunosorbent assay (ELISA), as described previously (34).
Pig NK cell cytotoxic assay.
To determine pig natural killer (NK) cell-mediated cytotoxicity, an immunofluorescence-based assay was performed by using a modified method described previously (34–36). The assay was conducted by using a 7-aminoactinomycin D (7-AAD)/carboxyfluorescein succinimidyl ester (CFSE) cell-mediated cytotoxicity assay kit (Cayman Chemical, Ann Arbor, MI). Briefly, peripheral blood mononuclear cells (PBMCs) isolated from pigs were used as the source of NK cells (effectors) against K562 (human myeloblastoid cell line) target cells. The target cells were labeled with CFSE according to the manufacturer's recommendations. Effector and target cells were incubated at different effector-to-target cell (E:T) ratios at 37°C overnight. The frequency of CFSE-labeled apoptotic K562 cells was measured by staining the cocultured target cells with 7-AAD nuclear dye. Specific NK cell cytotoxicity was measured by flow cytometry by acquiring 10,000 CFSE-labeled events and further gated for the frequency of CFSE and 7-AAD (green and red) double-stained cells, which indicates the frequency of lysed target cells. Appropriate controls included K562 cells labeled or unlabeled with CFSE and apoptosis-induced K562 cells (treated with UV at 254 nm for 30 min and then incubated for 6 to 8 h at 37°C). The percentage of NK cell-specific lysis was calculated by using the following formula: number of double-positive K562 cells/number of CFSE-positive K562 cells multiplied by 100.
Flow cytometry analysis.
Immunophenotyping of PBMCs was performed as previously described (33, 37). Briefly, PBMCs were first surface labeled with pig lymphocyte-specific fluorochrome-conjugated MAbs (CD3ε-peridinin chlorophyll protein [PerCP], CD4α-allophycocyanin [APC], and CD8α-fluorescein isothiocyanate [FITC]). For intracellular IFN-γ staining, GolgiPlug (BD Biosciences, San Jose, CA, USA) and brefeldin A (catalog number B7651; Sigma-Aldrich, St. Louis, MO) were added during the last 12 h of incubation of PBMCs treated with or without the respective virus as a stimulant at an MOI of 1. The surface-immunostained cells were fixed with 1% paraformaldehyde and permeabilized with a cell permeabilization buffer (85.9% deionized water, 11% PBS without Ca2+ or Mg2+, 3% formaldehyde solution, and 0.1% saponin) overnight at 4°C. Cells were washed and stained with fluorochrome-conjugated anti-pig IFN-γ or the isotype control MAb (BD Biosciences, East Rutherford, NJ) in 0.1% saponin-containing fluorescence-activated cell sorter (FACS) buffer. Finally, 25,000 immunostained cells were acquired using a FACSAria II flow cytometer (BD Biosciences, East Rutherford, NJ) and the data were analyzed using FlowJo software (Tree Star, Ashland, OR, USA). All cell population frequencies are presented as percentages of specific lymphocytes in PBMCs.
Statistical analysis.
All the data were expressed as the means of results from 3 to 9 pigs ± standard errors of the means (SEM). Statistical analyses were performed by using one-way analysis of variance (ANOVA) followed by Tukey's post hoc test using GraphPad InStat Prism software (version 5.0) to establish variations between the indicated pig groups. Statistical significance was assessed at P values of <0.05, <0.01, and <0.001.
RESULTS
Identification of critical residues in the GKYLQRRLQ motif for PRRSV nsp1β function.
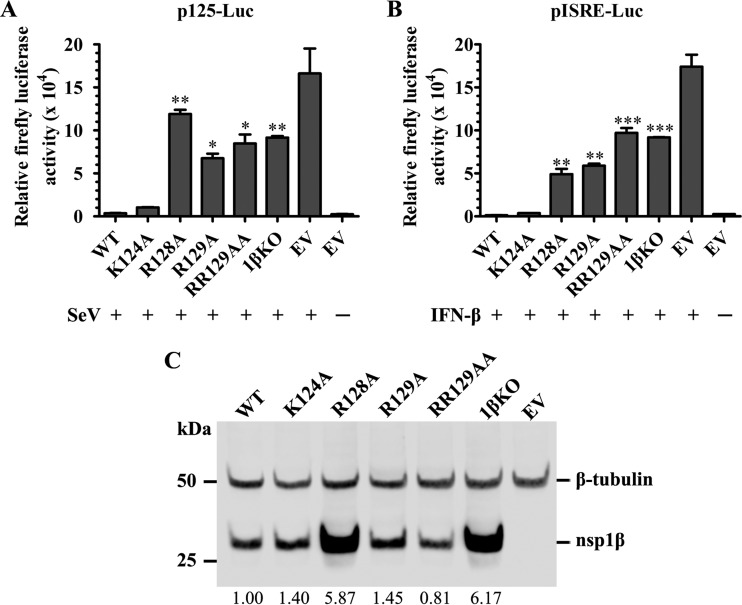
In our previous study (26), we identified a highly conserved GKYLQRRLQ motif in PRRSV nsp1β that is critical for the innate immune suppression function of this protein. Protein structural analysis showed that three basic residues (K124, R128, and R129) in the GKYLQRRLQ motif (boldface type indicates the positions of the three basic residues) are exposed on the surface of nsp1β (25). In this study, we further investigated the function of these three basic residues. A panel of nsp1β mutants was generated. Each of the nsp1β genes carries a single alanine substitution at amino acid K124 (K124A), R128 (R128A), and R129 (R129A) or a double alanine substitution at R128/R129 (RR129AA). A previously created mutant, nsp1βKO (1βKO) (R124/R128-to-A124/A128 double substitution) (26), was also included in the analysis. These nsp1β mutants were cloned into the plasmid vector p3xFLAG-Myc-CMV-23, in which gene expression is under the control of a cytomegalovirus (CMV) promoter, and expressed as a 3xFLAG-tagged protein. Initially, this panel of nsp1β mutants was analyzed by an IFN-β promoter-driven luciferase reporter assay. HEK-293T cells were cotransfected with a plasmid expressing wild-type (WT) or mutated nsp1β and a reporter plasmid (p125-Luc) that expresses the firefly luciferase reporter gene under the control of the IFN-β promoter. The empty vector p3xFLAG-Myc-CMV-23 was included in the analysis as a control. At 24 h posttransfection, cells were mock infected or infected with Sendai virus (SeV). Cells were harvested to test the luciferase activities at 16 h postinfection (hpi). As shown in Fig. 1A, SeV infection induced a high level of luciferase reporter expression in cells transfected with the empty vector, but the luciferase expression level was ∼46- to 16-fold lower in cells expressing WT nsp1β and the K124A mutant. In contrast, ∼33-fold-, 19-fold-, 24-fold-, and 26-fold-higher levels of reporter expression were detected in cells expressing the R128A, R129A, RR129AA, and 1βKO mutants, respectively, than in cells expressing WT nsp1β. We further determined whether these mutations had an effect on the ability of nsp1β to suppress the IFN-dependent signaling pathway for interferon-stimulated gene (ISG) expression. The panel of nsp1β mutants was analyzed by using an interferon-stimulated response element (ISRE) promoter-driven luciferase reporter assay. Results similar to those shown in Fig. 1A were generated. In comparison to those cells expressing WT nsp1β, there were about 38-fold-, 46-fold-, 75-fold-, and 71-fold-higher levels of reporter expression detected in cells expressing the R128A, R129A, RR129AA, and 1βKO mutants, respectively (Fig. 1B). These results suggest that R128 and R129 are critical to the IFN antagonist function of nsp1β. In contrast, K124 appeared not to significantly affect the IFN antagonist function of nsp1β.
FIG 1.
Mutations in the GKYLQRRLQ motif impair nsp1β's inhibitory effect on type I interferon production and signaling. (A and B) HEK-293T cells in a 24-well plate were cotransfected with a plasmid expressing WT nsp1β or the nsp1β mutant, the p125-Luc reporter plasmid expressing firefly luciferase under the control of the IFN-β promoter (A) or pISRE-Luc expressing firefly luciferase derived from the interferon-stimulated response element (ISRE) (B). An empty vector (EV) was used as a control. At 24 h posttransfection, cells were stimulated with SeV at 100 HA units/ml or stimulated with IFN-β at 2,000 IU/ml for 16 h. Cell lysates were harvested for measurement of luciferase activity. (C) The expression level of nsp1β was evaluated by Western blot analysis using nsp1β-specific MAb 123-128. β-Tubulin was detected as a loading control. The membrane was incubated with a mixture of primary anti-FLAG M2 MAb (Sigma-Aldrich, St. Louis, MO) and MAb against β-tubulin. The secondary antibody IRDye 800CW goat anti-mouse IgG(H+L) (Li-Cor Biosciences, Lincoln, NE) was used for visualizing the target proteins with a digital image system (Odyssey infrared imaging system; Li-Cor Biosciences, Lincoln, NE). The expression of nsp1β was quantified and normalized to the expression level of β-tubulin, and the relative expression levels are shown under each band. Statistical significance between the wild-type virus-infected group and the mutant virus-infected group was determined by one-way ANOVA and Tukey's test and is indicated with asterisks (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
The expression level of nsp1β was evaluated by Western blot analysis using MAb M2 against the FLAG tag. The results confirmed the expression of nsp1β in WT- and mutant-transfected cells used in luciferase assays (Fig. 1C). In our previous study, we showed that a double mutation of K124/R128 to A124/A128 caused increased levels of nsp1β expression in comparison to those of WT nsp1β and other mutants, which suggested that nsp1β may suppress its “self-expression” (26). In this study, individual substitutions introduced at K124, R128, and R129 showed that only the R128A substitution affects the ability of nsp1β to suppress self-expression in vitro. The detailed mechanism for the ability of nsp1β to suppress self-expression and whether such a property relates to the innate immune suppression function of the virus need to be further studied (see Discussion, below).
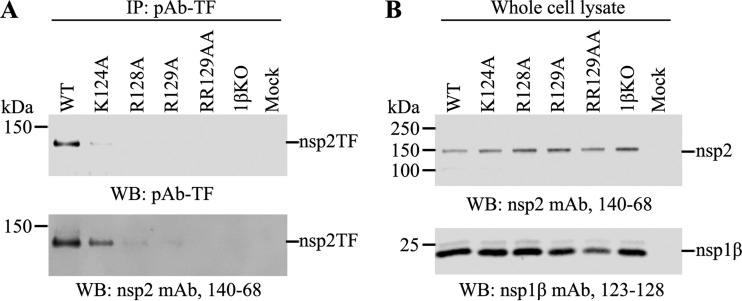
We further determined whether mutations introduced into the GKYLQRRLQ motif also affect the transactivator function of nsp1β. A vaccinia virus-T7 polymerase system expressing the nsp1β-nsp2 (nsp1β-2) region was used to analyze the expression of nsp2 and PRF products. The expression of the −2 PRF product (nsp2TF) was determined by immunoprecipitation (IP) and Western blot (WB) assays. Equal amounts of lysates of transfected cells expressing WT nsp1β-2 or its mutants (K124A, R128A, R129A, 1βKO, and RR129AA) were subjected to immunoprecipitation using a polyclonal antibody (PAb-TF) that specifically recognizes the C-terminal peptide of nsp2TF. Subsequently, Western blot analysis was performed by using PAb-TF and MAb 140-68, which recognizes the N-terminal PLP2 domain of the protein. As shown in Fig. 2A, the nsp2TF product was detected only in cells expressing WT nsp1β-2 or the K124A mutant but not in cells expressing the R128A, R129A, RR129AA, and 1βKO mutants. In contrast, the expression of full-length nsp2 and nsp1β was detected in the WT and all nsp1β-2 mutants (Fig. 2B). The results indicate that residues R128 and R129 are critical for the transactivator function of nsp1β in activating the −2 PRF signal. K124 did not show a significant effect on the nsp1β function in PRF transactivation.
FIG 2.
Mutations at R128 and R129 in the GKYLQRRLQ motif impair the expression of nsp2TF in the vaccinia virus-T7 expression system. HEK-293T cells were infected with vaccinia virus expressing T7 polymerase at an MOI of 10 and then transfected with pLnsp1β-2 constructs at 1 h postinfection. The cell lysates were harvested at 18 h posttransfection. (A) nsp2TF was immunoprecipitated by a polyclonal antibody (PAb-TF) that specifically recognizes the C-terminal region of nsp2TF using equal amounts of the cell lysate. Immunoprecipitated proteins were detected by WB using PAb-TF (top) and MAb 140-68 recognizing the common N-terminal region of nsp2-related proteins (bottom). (B) Western blot detection of the expression of nsp2 (top) and nsp1β (bottom) using specific MAbs.
In vitro characterization of recombinant viruses containing mutations in the GKYLQRRLQ motif.
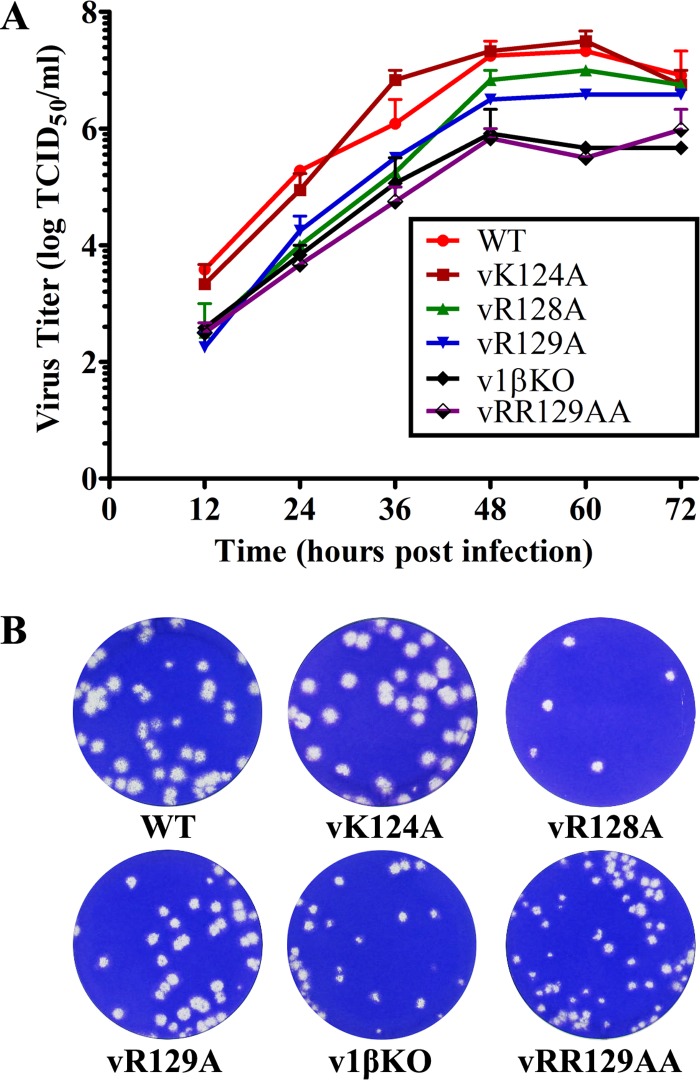
To further investigate whether the specific substitutions introduced into the nsp1β GKYLQRRLQ motif of the virus could improve innate immune responses in PRRSV-infected cells, we created a panel of recombinant viruses using a reverse-genetics approach. Three viable recombinant viruses were generated, vSD95-21-R128A (vR128A), vSD95-21-R129A (vR129A), and vSD95-21-R128A/R129A (vRR129AA), carrying single or double mutations at residues R128 and R129 of the GKYLQRRLQ motif. For comparison, recombinant viruses with a mutation at K124 (vSD95-21-K124A [vK124A]) and a double mutation at K124/R128 (vSD95-21-K124A/R128A [v1βKO]) and the WT virus vSD95-21 were also recovered by the reverse-genetics approach. The stability of these mutations introduced into the virus was determined by serially passaging each virus 5 times in MARC-145 cells, and sequence analysis of passage 3 and passage 5 viruses showed that all of the introduced mutations were stably maintained in the mutant viruses. The growth properties of these mutants (passage 3) were compared with those of the WT parental virus. In comparison to the WT virus (peak virus titer of 107.33 TCID50/ml), vK124A showed a similar growth ability, while vR128A and vR129A showed certain levels of reduced growth ability (peak virus titers of 107.0 TCID50/ml and 106.58 TCID50/ml, respectively). In contrast, the vRR129AA and v1βKO mutants had significantly reduced growth abilities, with ∼1- to 1.5-log decreases in virus titers throughout the time course of the study (Fig. 3A). Plaque assay results consistently showed that v1βKO and vRR129AA developed smaller plaques than did the WT virus (Fig. 3B).
FIG 3.
In vitro characterization of recombinant viruses containing nsp1β mutations. (A) Multiple-step virus growth curve. Each data point shown represents the mean value from duplicates, and error bars show SEM. (B) Plaque morphology of WT and recombinant viruses containing mutations in the GKYLQRRLQ motif of nsp1β.
Expression of innate immune genes in nsp1β mutant virus-infected cells.
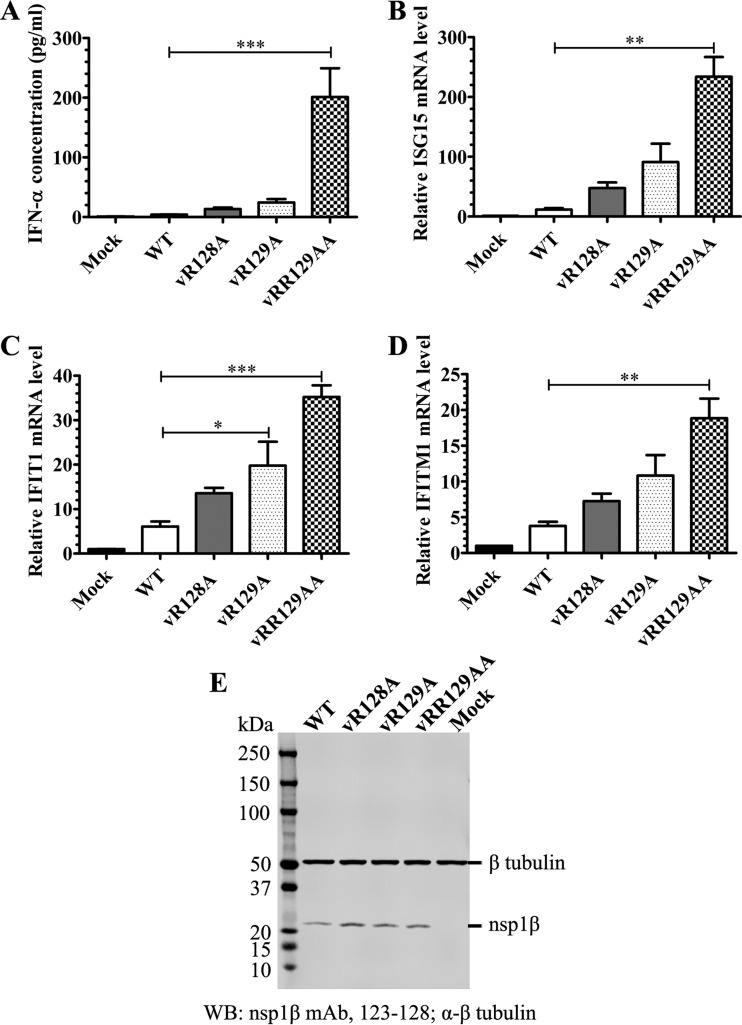
As we determined that the R128A and/or R129A mutation introduced into the GKYLQRRLQ motif reduced the ability of nsp1β to suppress the innate immune response (Fig. 1), we further analyzed whether these mutations could alter the inhibitory effect of PRRSV on type I IFN production and signaling. Since K124A did not show much of an effect on the function of nsp1β in PRF transactivation and innate immune suppression, recombinant viruses containing the K124A substitution (vK124A and v1βKO) were not further analyzed in the following experiments (for more about the characteristics of v1βKO, see Discussion, below). Initially, IFN-α expression was evaluated in nsp1β mutants or WT virus-infected porcine alveolar macrophages (PAMs) by using a ProcartaPlex Porcine IFN alpha Simplex kit (eBioscience, San Diego, CA). PAMs were initially infected with equal amounts (MOI = 1) of the WT or an nsp1β mutant. At 12 h postinfection, the IFN-α concentration in the cell culture supernatant of virus-infected PAMs was evaluated. All of the nsp1β mutants showed an improved ability to induce the production of IFN-α, which is indicated by the 3.3-fold-higher (vR128A), 6.1-fold-higher (vR129A), and 50-fold-higher (vRR129AA) concentrations of IFN-α in the supernatants of cells infected with mutant viruses than in the supernatant of WT virus-infected cells (Fig. 4A). Of note, vRR129AA showed the strongest stimulation of IFN-α production. In addition, the expression of nsp1β was detected by Western blotting, indicating successful viral replication of WT virus and nsp1β mutants in PAMs (Fig. 4E). Subsequently, we analyzed whether these mutations could alter the inhibitory effect of PRRSV on the production of ISGs. At 12 h postinfection, the mRNA expression levels of three selected ISGs, ISG15, IFIT1, and IFITM1, were assessed by quantitative real-time RT-PCR (qRT-PCR) using predesigned primer/probe sets (Applied Biosystems, Foster City, CA). Consistent with their improved ability for IFN-α induction, all of the nsp1β mutants stimulated higher mRNA expression levels of ISGs. As indicated in Fig. 4B, ∼4.1-fold-higher (vR128A), 7.8-fold-higher (vR129A), and 20-fold-higher (vRR129AA) mRNA expression levels of ISG15 were detected in mutant virus-infected cells than that in WT virus-infected cells, although the increases in vR128A- and vR129A-infected cells are not statistically significant. Similarly, the mRNA expression levels of IFIT1 and IFITM1 were increased in mutant virus-infected cells in comparison to those in WT virus-infected cells (Fig. 4C and D).
FIG 4.
Mutations in the GKYLQRRLQ motif attenuate the ability of PRRSV to suppress the expression of IFN-α and ISGs. Porcine alveolar macrophages seeded into a 24-well plate were infected with the WT virus or nsp1β mutants at an MOI of 1.0, and cell culture supernatants were harvested at 12 h postinfection. (A) IFN-α production was quantified by using a ProcartaPlex Porcine IFN alpha Simplex kit (eBioscience, San Diego, CA). Each data point shown represents the mean value from three independent experiments with duplicates, and error bars show SEM. (B) The mRNA expression level of ISG15 was evaluated by quantitative real-time RT-PCR and normalized to the level of endogenous β-tubulin mRNA. (C) The mRNA expression level of IFIT1 was evaluated by quantitative real-time RT-PCR and normalized to the level of endogenous β-tubulin mRNA. (D) The mRNA expression level of IFITM1 was evaluated by quantitative real-time RT-PCR and normalized to the level of endogenous β-tubulin mRNA. Values in panels B to D are expressed as means ± SEM from three independent experiments. (E) The expression level of nsp1β at 12 h postinfection was determined by Western blot analysis with nsp1β-specific MAb 123-128. β-Tubulin was detected as a loading control. The membrane was incubated with a mixture of primary MAb 123-128 and MAb against β-tubulin. The secondary antibody IRDye 800CW goat anti-mouse IgG(H+L) (Li-Cor Biosciences, Lincoln, NE) was used to visualize the target proteins with a digital image system (Odyssey infrared imaging system; Li-Cor Biosciences, Lincoln, NE). Statistical significance between the wild-type virus-infected group and mutant virus-infected groups was determined by one-way ANOVA and Tukey's test and is indicated with asterisks (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
In vivo characterization of nsp1β mutants.
Subsequently, we obtained five groups of 4-week-old pigs to determine whether the R128- and R129-related mutants could improve specific immune responses in PRRSV-infected pigs. As shown in Table 1, each group of pigs (n = 9) was infected with the WT virus or an nsp1β mutant or mock infected with cell culture medium as a negative control. Serum, BALF, and lung lysate samples were collected and stored at −80°C for further analysis. We did not observe any noticeable clinical PRRS symptoms and fever in any of the WT virus- or nsp1β mutant-infected pigs.
Viral load in serum and tissue samples.
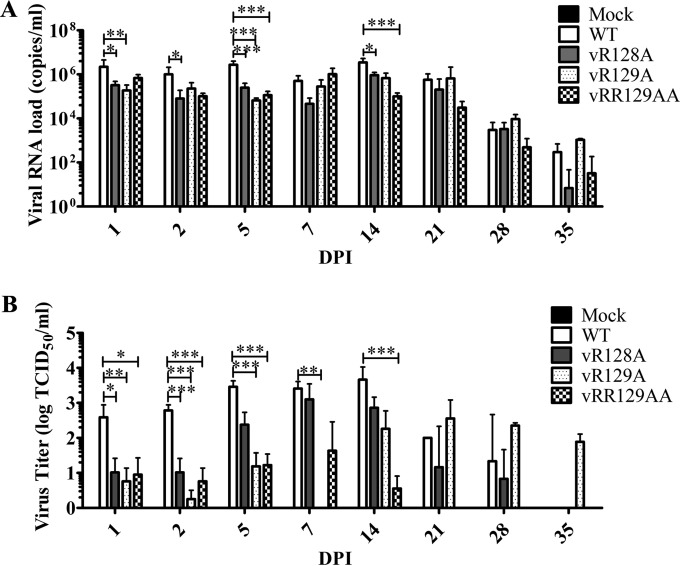
Initially, we measured viral RNA loads in serum samples using real-time qRT-PCR. In comparison with the group of pigs infected with the WT virus, pigs infected with the vR128A and vRR129AA mutants showed consistently lower viral RNA loads throughout the entire time course of the experiment, while pigs infected with the vR129A mutant exhibited lower viral RNA loads from 1 dpi to14 dpi (Fig. 5A). Statistically significantly lower levels of viral RNA were obtained at 1, 2, 5, and 14 dpi in pigs infected with the vR128A mutant than in pigs infected with the WT virus (Fig. 5A). At most of the time points during the later stage of infection (14 to 35 dpi), the mean viral RNA load in pigs infected with the vRR129AA mutant was the lowest among all the infected pigs (Fig. 5A). Surprisingly, vR129A mutant-infected pigs showed mean viral RNA loads that were similar to those of the group of pigs infected with the WT virus at 21 dpi and exhibited higher (but not statistically significant) viral RNA loads than those of pigs infected with the WT virus at 28 dpi and 35 dpi.
FIG 5.
Comparison of viral loads in serum samples from pigs inoculated with the WT virus and nsp1β mutants. Pigs were uninfected (mock) or infected with WT PRRSV (WT) or the three indicated mutants (vR128A, vR129A, vRR129AA). Serum samples were collected on the indicated days postinfection. (A) Viral loads in serum samples quantified by quantitative RT-PCR and calculated as viral RNA copies per milliliter. (B) Infectious virus titer in serum samples determined by a microtitration assay and calculated as log TCID50 per milliliter. Statistical significance between the wild-type virus-infected group and mutant virus-infected groups was determined by one-way ANOVA and Tukey's test and are indicated with asterisks (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Since qRT-PCR does not distinguish between viable and nonviable forms of the virus, we further quantified infectious virus particles in serum samples. The infectious virus titer was measured by a microtitration assay using MARC-145 cells. At 1, 2, and 5 dpi, infectious virus titers in groups of pigs infected with nsp1β mutants were ∼1 to 3 logs lower (statistically significant) than the virus titers in pigs infected with the WT virus (Fig. 5B), which is consistent with viral loads quantified by qRT-PCR (Fig. 5A). The infectious virus titer in many pigs infected with nsp1β mutants was lower than the detection limit (101.67 TCID50/ml) of the microtitration assay throughout the time course of infection. A rebounded virus titer was observed for vR129A mutant-infected pigs at 21, 28, and 35 dpi, which is consistent with the viral RNA load result generated by qRT-PCR. These results indicate that the growth abilities of vR128A and vRR129AA were attenuated in vivo and that the growth ability of vR129A was attenuated at an early stage of infection but reverted to the WT phenotype at a certain level during the later stage of infection (Table 2).
TABLE 2.
Sequence analysis of the nsp1β coding region in viruses recovered from infected pigs
| Group | Pig | Designed mutation | Position of 2nd-site mutation, nucleotide substitution, amino acid substitutiona |
|---|---|---|---|
| 14 dpi | |||
| vR129A | 31 | Reversion (GCG to AGG) | |
| 32 | Stable | 122, UCU to CCU, Ser to Pro | |
| 33 | Stable | 122, UCU to CCU, Ser to Pro | |
| 34 | Stable | 122, UCU to CCU, Ser to Pro | |
| 35 | Stable | 122, UCU to CCU, Ser to Pro | |
| 36 | Stable | 122, UCU to CCU, Ser to Pro | |
| 21 dpi | |||
| WT | 14 | 9, GAC to GGC, Asp to Gly | |
| 15 | 9, GAC to GGC, Asp to Gly | ||
| 16 | 9, GAC to GGC, Asp to Gly | ||
| vR128A | 22 | Stable | 9, GAC to GGC, Asp to Gly |
| 122, UCU to CCU, Ser to Pro | |||
| 23 | Stable | 9, GAC to GGC, Asp to Gly | |
| 122, UCU to CCU, Ser to Pro | |||
| 24 | Stable | 9, GAC to GGC, Asp to Gly | |
| 109, CAU to C(A/G)U, His to His/Arg | |||
| 122, UCU to CCU, Ser to Pro | |||
| 141, CUA to C(U/C)A, Leu to Leu/Pro | |||
| vR129A | 31 | Reversion (GCG to AGG) | 9, GAC to GGC, Asp to Gly |
| 32 | Stable | 9, GAC to GGC, Asp to Gly | |
| 87, GAA to GA(A/G) | |||
| 122, UCU to CCU, Ser to Pro | |||
| 33 | Stable | 9, GAC to GGC, Asp to Gly | |
| 122, UCU to CCU, Ser to Pro | |||
| 169, UCU to (C/U)CU, Ser to Ser/Pro | |||
| vRR129AA | 40 | Stable | |
| 41 | Stable | ||
| 42 | Stable | ||
| 35 dpi | |||
| WT | 17 | 9, GAC to GGC, Asp to Gly | |
| 126, CUA to CU(A/G) | |||
| 18 | 9, GAC to GGC, Asp to Gly | ||
| vR128A | 27 | Stable | 9, GAC to GGC, Asp to Gly |
| 109, CAU to CGU, His to Arg | |||
| 122, UCU to CCU, Ser to Pro | |||
| vR129A | 34 | Stable | 9, GAC to GGC, Asp to Gly |
| 122, UCU to CCU, Ser to Pro | |||
| 132, UUU to (G/U)UU, Phe to Val | |||
| 34 | 141, CUA to CU(A/G) | ||
| 169, UCU to (C/U)CU, Ser to Ser/Pro | |||
| 35 | Stable | 9, GAC to GGC, Asp to Gly | |
| 122, UCU to CCU, Ser to Pro | |||
| 36 | Stable | 9, GAC to GGC, Asp to Gly | |
| 122, UCU to CCU, Ser to Pro | |||
| 169, UCU to CCU, Ser to Pro | |||
| vRR129AA | 44 | Stable | 9, GAC to GGC, Asp to Gly |
| 45 | Stable | 9, GAC to GGC, Asp to Gly |
Numbers refer to codon positions in the nsp1β coding region.
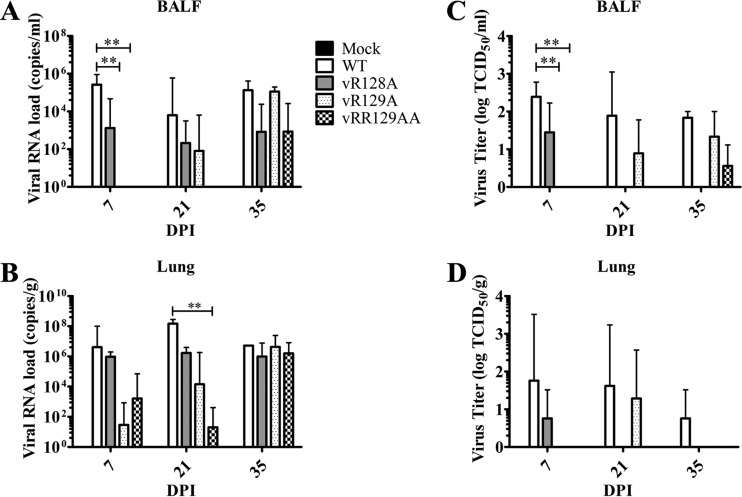
Since PAMs serve as the primary target cells for PRRSV, we further evaluated the viral load in lung lysates and BALF collected at 7, 21, and 35 dpi. The viral RNA load was quantified by qRT-PCR, and the infectious virus titer was determined by a microtitration assay. Results from both qRT-PCR and microtitration assays showed that the viral loads in lung lysates and BALF from all groups of pigs infected with nsp1β mutants were consistently lower than those in pigs infected with the WT virus at 7 and 21 dpi (Fig. 6), although some of the differences were not statistically significant. At 35 dpi, lower mean viral loads and infectious virus titers were detected in lung samples from pigs infected with the vR128A and vRR129AA mutants than in pigs infected with the WT virus (Fig. 6). The mean viral loads in lung samples and BALF from vR129A mutant-infected pigs were similar to those in WT virus-infected pigs at 35 dpi. These results suggest that nsp1β mutants have an attenuated replication ability in the lungs of infected pigs, but vR129A showed a reversion to the WT phenotype at certain levels during the later stage of infection.
FIG 6.
Comparison of viral loads in BALF and lung samples from pigs inoculated with the WT virus and nsp1β mutants. Pigs were uninfected (mock) or infected with WT PRRSV (WT) or the three indicated mutants (vR128A, vR129A, and vRR129AA). Lungs harvested on the day of necropsy (7, 21, and 35 dpi) were used for collecting BALF and lung lysates. (A and B) Viral loads in BALF (A) and lung lysate (B) samples were quantified by quantitative RT-PCR and calculated as viral RNA copies per milliliter of BALF or viral RNA copies per gram of lung. (C and D) Infectious virus titers in BALF (C) and lung lysate (D) samples were determined by a microtitration assay and calculated as log TCID50 per milliliter or log TCID50 per gram. A key explaining the treatment groups in panels B to D is given in panel A. Statistical significance between the wild-type virus-infected group and mutant virus-infected groups was determined by one-way ANOVA and Tukey's test and is indicated with asterisks (**, P < 0.01).
Genetic stability of nsp1β mutants in pigs.
Genetic stability is one of the important criteria for selecting vaccine candidates. Initially, serum samples from 3 pigs per group terminated at 21 dpi were used to determine the stability of the introduced mutations. The nsp1β coding region was amplified by RT-PCR, and the PCR product was subjected to DNA sequencing analysis. As shown in Table 2, no second-site mutation and also no reversion were found in the nsp1β coding region of the virus isolated from serum samples of pigs infected by the vRR129AA mutant virus. In the group of pigs infected with the vR128A mutant, the designed mutation was maintained in the viruses recovered from all three tested pigs, but several second-site mutations were observed, including the replacement of Asp9 by Gly and the replacement of Ser122 by Pro in all three pigs and the replacement of His109 and Leu141 by Arg and Pro, respectively, in one pig. In the group of pigs infected with the vR129A mutant, the designed mutation in one of the three tested pigs reversed from Ala back to Arg (in the WT virus), and the R129A mutation was maintained in the other two pigs. Similar to the vR128A group, the second-site mutation of Ser122 to Pro was detected in the two pigs that maintained the designed mutation; an additional mutation of Ser169 to Pro occurred in one of the two pigs, and the replacement of Asp9 by Gly was observed in all three pigs. Since a reversion occurred in one of the vR129A mutant-infected pigs at 21 dpi, we further analyzed serum samples from all 6 pigs infected with vR129A at 14 dpi. Remarkably, the pig with an Ala129-to-Arg reversion at 21 dpi had already obtained the reversion at 14 dpi. However, the designed R129A mutation was maintained in the other five pigs. It is worth noting that the second-site mutation of Ser122 to Pro occurred in all of the pigs that maintained the designed mutations (R128A and R129A) at 14, 21, and 35 dpi, suggesting that this substitution may compromise the effect of the designed mutations on virus growth ability in vivo. Interestingly, the mutation of Asp9 to Gly was detected not only in pigs infected with all mutants but also in pigs infected with the WT virus. This mutation may relate to the in vivo fitness of PRRSV, which was most likely not caused by our designed mutations. In addition, using serum samples at 35 dpi that were determined to be PRRSV RNA positive by real-time RT-PCR, no reversion was observed by sequencing analysis. We searched PRRSV full-length genome sequences available in GenBank (as of 27 July 2015), and all of the second-site substitutions that were observed here can be found in field strains; for example, the PRRSV P129 strain contains Pro122. Taken together, among the three nsp1β mutants, vRR129AA maintained the best genetic stability in vivo, with no reversion and fewer second-site mutations being detected in the nsp1β coding region.
Innate immune response in PRRSV nsp1β mutant-infected pigs.
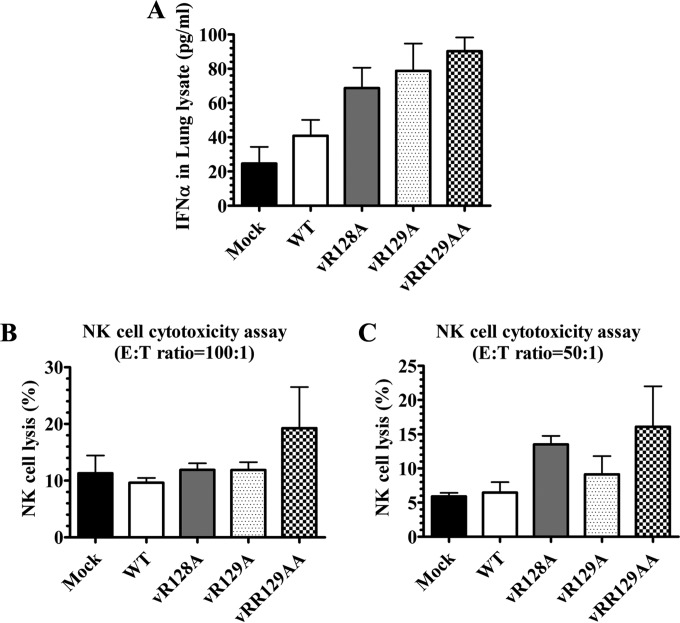
To determine whether the mutations introduced into the nsp1β region could improve the innate immune response, we initially measured IFN-α expression in infected and control pigs during the early stage of infection. Compared to WT virus-infected pigs, 1.5-fold-higher (but not significant) levels of IFN-α were observed in serum samples of vRR129AA-infected pigs at 1 dpi (data not shown). Since the virus replicates primarily in alveolar macrophages, we also inoculated the pigs by the intranasal (i.n.) route, and as the immune response in the lung is important, we measured IFN-α levels in both BALF (representing airways) and lung lysates (representing the lung parenchyma, a local site of PRRSV infection). The IFN-α levels in BALF at 7 dpi were comparable among all pig groups, while in pigs inoculated with the vRR129AA and vR129A mutants, there was an increased level (but not statistically significant) of IFN-α production compared to that in WT virus-infected pigs at 21 and 35 dpi (data not shown). At 7 dpi, higher levels of IFN-α were observed in the lung lysates of pigs inoculated with nsp1β mutant viruses than in those of WT virus-infected pigs (Fig. 7A).
FIG 7.
Comparison of IFN-α production levels and NK cell cytotoxicity in pigs inoculated with the WT virus and nsp1β mutants. Pigs were uninfected (mock) or infected with WT PRRSV (WT) or the three indicated mutants (vR128A, vR129A, and vRR129AA). (A) Lung samples collected at 7 dpi were used to prepare lung lysates, and IFN-α levels were analyzed by an ELISA. (B and C) PBMCs (NK effectors) were harvested on the day of necropsy (7 dpi), and cells were cocultured with target cells (K562) at an E:T ratio of 100:1 (B) or 50:1 (C). After overnight incubation, the NK cell-specific cytotoxic activity was determined by flow cytometry. Each data point represents the mean value ± SEM of data from 3 pigs. Statistical significance between the wild-type virus-infected group and mutant virus-infected groups was determined by one-way ANOVA and Tukey's test.
IFN-α is critical for natural killer (NK) cell-mediated cytotoxic function. To determine whether the nsp1β mutations impaired the IFN-α antagonist function of the virus, PBMCs from mutant and WT virus-infected pigs were used as a source of NK cells to evaluate NK cell function in an NK cell cytotoxicity assay. At both E:T ratios used (100:1 and 50:1), the vRR129AA mutant-infected pigs had increased NK cell cytotoxic function at 7 dpi (Fig. 7B and C). This result is consistent with the increased level of IFN-α production in the serum and lung lysates of vRR129AA-infected pigs.
Adaptive immune response in PRRSV nsp1β mutant-infected pigs.
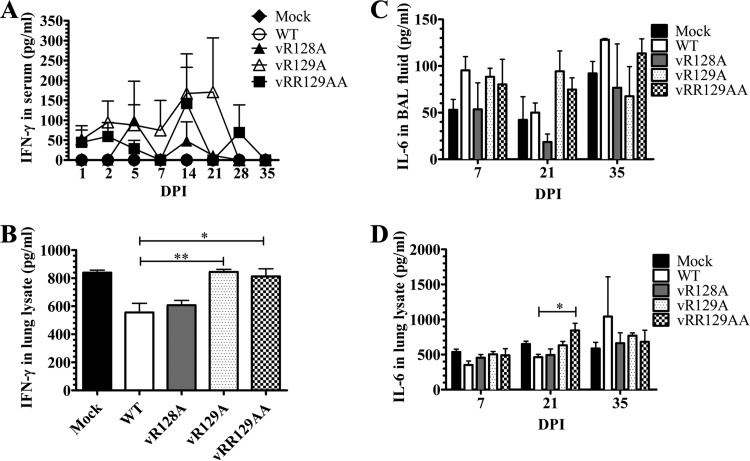
A strong innate immune response following virus infection augments cell-mediated adaptive immunity. Therefore, we analyzed the production of an important Th1 cytokine, IFN-γ, in WT virus- and nsp1β mutant-infected pigs. The production of IFN-γ in the serum of WT virus-infected pigs was undetectable throughout the time course of the study (0 to 35 dpi), while in the serum of nsp1β mutant-infected pigs, spurts of IFN-γ secretion (100 to 150 pg/ml) were detected on multiple days postinfection, with IFN-γ being detected in vR129A mutant-infected pigs at 7 to 21 dpi and in vRR129AA mutant-infected pigs at 14 and 28 dpi (Fig. 8A). In vR128A mutant-infected pigs, increased IFN-γ levels were detected in serum at 5 and 14 dpi (Fig. 8A). Such an early response of IFN-γ in nsp1β mutant-infected pigs might be due to the rescue of adaptive immunity mediated by the induction of IFN-α secretion and NK cell function by these mutants. A similar increase in IFN-γ secretion (but not statistically significant) was detected in the BALF of vR128A-, vR129A-, and vRR129AA-infected pigs, observed only at 7 dpi (data not shown). However, the increased levels of IFN-γ production in the lung lysates of vR129A- and vRR129AA-infected pigs at 7 dpi were significantly higher than those in WT virus-infected pigs (Fig. 8B).
FIG 8.
Comparison of IFN-γ production levels in pigs inoculated with the WT virus and nsp1β mutants. Pigs were uninfected (mock) or infected with WT PRRSV (WT) or the three indicated mutants (vR128A, vR129A, and vRR129AA). Blood samples were collected on the indicated days postinfection, and BALF and lung lysates were prepared by using lungs harvested on the day of necropsy (7, 21, and 35 dpi). IFN-γ levels in serum (A) and lung lysates (B) and IL-6 levels in BALF (C) and lung lysates (D) were analyzed by an ELISA. Each data point represents the mean value ± SEM of data from 3 pigs. Statistical significance between the wild-type virus-infected group and mutant virus-infected groups was determined by one-way ANOVA and Tukey's test and is indicated with asterisks (*, P < 0.05; **, P < 0.01).
Production of the proinflammatory cytokine IL-6 suggests an inflammatory reaction in the lungs of pigs (34). The levels of IL-6 were higher (but not significant) at 21 dpi in the BALF of vR129A- and vRR129AA-infected pigs than in the BALF of WT virus-infected pigs (Fig. 8C). At 21 dpi, significantly higher levels of IL-6 production were detected in the lung lysates of vRR129AA-infected pigs than in WT virus-infected pigs (Fig. 8D). These data suggest that the induction of IL-6 production by the vRR129AA mutant in pigs appears to be responsible for augmenting IFN-γ production, an indicator of adaptive immunity.
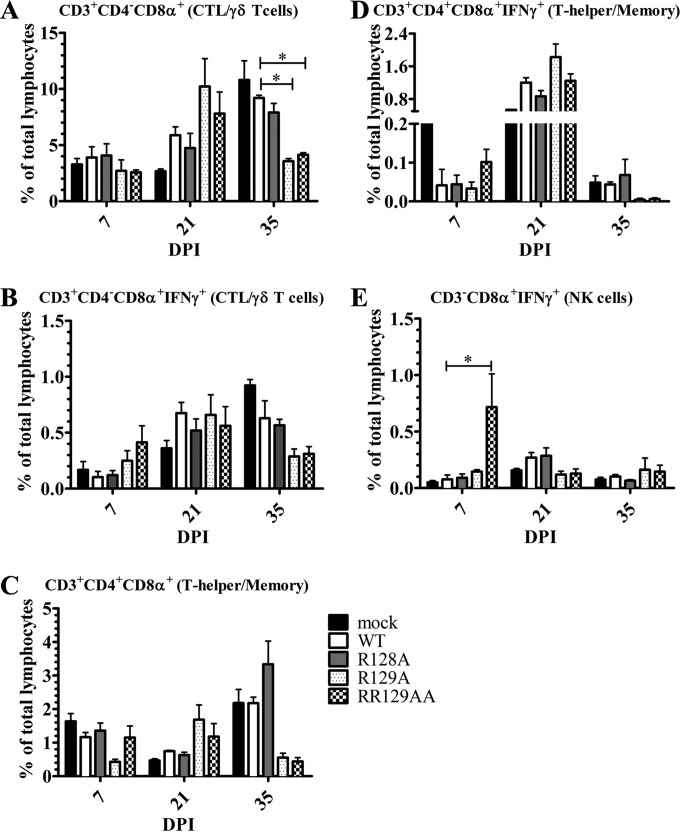
We further evaluated the frequencies of different T cell subpopulations in PBMCs, expressed as the percentage of CD3+ or CD3− cells. Pig T cells expressing the CD3+ CD4− CD8α+ combination of phenotypic markers are either cytotoxic T cells (CTLs) or γδ T cells, and CD3+ CD4− CD8αβ+ cells are exclusively CTLs (38, 39). The porcine immune system has a unique frequency of CD3+ CD4+ CD8α+ T cells, which have memory, cytotoxic, and T helper cell properties (40, 41). To determine the antigen-specific activation of the T cell response, IFN-γ-secreting lymphocyte subsets were elucidated by restimulating PBMCs with the same virus in vitro. For each experiment with phenotypic marker staining, the respective isotype controls were included to eliminate the background. The specific population of cells was identified based on a combination of phenotypic cell surface markers, which include CD3+ CD4− CD8α+ (CTLs or γδ T cells), CD3+ CD4+ CD8α+ (T helper/memory), and CD3− CD4− CD8α+ (NK) cells (38, 40–43). Subsequently, the cells were fixed and stained for intracellular IFN-γ and gated for their respective activated (IFN-γ+) phenotype. Compared to those in WT virus-infected pigs, the frequencies of total CTLs/γδ T cells in vR129A- and vRR129AA-infected pigs were increased (but not statistically significantly) at 21 dpi and then reduced significantly at 35 dpi (Fig. 9A), while the frequencies of activated (IFN-γ+) CTLs/γδ T cells in the same mutant-infected pigs were increased at 7 dpi and then decreased at 35 dpi (but not statistically significantly) (Fig. 9B). In comparisons of WT virus-infected pigs with vR129A- and vRR129AA-infected pigs, a trend of frequency changes similar to that of CTLs/γδ T cells was observed in total and activated T helper/memory cells from pigs at all three days postinfection (Fig. 9C and D). The frequency of NK cells was significantly increased only in vRR129AA-infected pigs at 7 dpi (Fig. 9E). These data suggest that NK cells and the two important T cell subsets were activated particularly in PRRSV vRR129AA mutant-infected pigs, suggesting virus-specific activation of innate and adaptive immunity.
FIG 9.
T helper and memory T cell responses in pigs infected with nsp1β mutants. PBMCs collected at 7, 21, and 35 dpi were unstimulated or restimulated with the respective WT or mutant viruses that were used to infect pigs. Cells were immunostained using the pig-specific markers CD3, CD4, and CD8α, followed by intracellular IFN-γ detection. Each lymphocyte subset was grouped based on the combination of cellular markers: CD3+ CD4− CD8α+ (CTLs/γδ T cells) (A), CD3+ CD4− CD8α+ IFN-γ+ (activated CTLs/γδ T cells) (B), CD3+ CD4+ CD8α+ (T helper/memory cells) (C), CD3+ CD4+ CD8α+ IFN-γ+ (activated T helper/memory cells) (D), and CD3− CD8α+ IFN-γ+ (activated NK cells) (E). A key explaining the treatment groups in panels A, B, D, and E is given in panel C. Each bar is the mean value ± SEM of data from 3 pigs. Statistical significance between wild-type and mutant virus-infected pig groups was determined by one-way ANOVA followed by Tukey's t test and is indicated with an asterisk (*, P < 0.05).
DISCUSSION
Many studies have demonstrated that a potent innate immune response induced by microbial infection/vaccination will lead to the generation of sufficient adaptive immunity, which subsequently clears pathogen infection from the host completely (44, 45). However, PRRSV infection generally induces poor antiviral innate IFN and cytokine responses, which result in weak adaptive immunity (46–50). One of the key steps in new PRRS vaccine construction is to develop strategies to target these initial immune response events to enhance virus-specific immunity. Previous studies of other viral pathogens showed that recombinant viruses generated with targeted mutations (deletions) in genes encoding immune antagonists are excellent candidates for modified live-virus (MLV) vaccines (51–54). The (selected) recombinant viruses normally grow well in tissue culture, while in infected animals, they are attenuated but still replicate to sufficient amounts for stimulating robust immune responses. Several PRRSV proteins have been identified as antagonists of type I IFN induction (and signaling), and nsp1β was determined to have the strongest inhibitory effect among these proteins (22–24, 26). Therefore, in this study, our vaccine development strategy is to generate recombinant viruses with targeted mutations in the nsp1β regions.
Our previous study identified a highly conserved GKYLQRRLQ motif in nsp1β that is critical for its inhibitory effect on type I IFN production and signaling. Based on the crystal structure of nsp1β, three basic residues (K124, R128, and R129) in the GKYLQRRLQ motif (boldface type indicates the positions of the three basic residues) are exposed on the surface of the protein. In our previous study, the K124A/R128A double mutation impaired the IFN antagonist function of nsp1β (26). In this study, we further tested each of these three individual residues. In comparison to WT nsp1β, nsp1β mutants carrying alanine substitutions at R128 and R129 showed a significantly reduced antagonism effect on reporter gene expression under the control of the IFN-β promoter (p125-Luc); however, the K124A mutant still had an inhibitory effect on reporter gene expression similar to that of WT nsp1β. Similar results were observed in a luciferase reporter assay (pISRE-Luc) utilized to examine the inhibitory effect on type I IFN signaling. Both WT nsp1β and the K124A mutant severely suppressed luciferase reporter expression, in contrast to the significantly higher levels of luciferase expression in cells transfected with nsp1β mutants that contain mutations at R128 and/or R129 (Fig. 1B). These results indicate that the R128 and R129 residues, but not K124, are critical for nsp1β's function in antagonizing type I IFN production and signaling.
As we discussed previously (26), suppression of host cellular gene expression, including nsp1β self-expression, could be a mechanism of its immune antagonist function. An obviously higher level of nsp1β expression was detected for the R128A and K124A/R128A (1βKO) mutants than for the WT by Western blot analysis (Fig. 1C). Interestingly, the combination of the R129A and R128A substitutions (RR129AA) appeared to restore the ability of nsp1β to suppress its self-expression. The single alanine substitution at residue R129 did not impair the ability of nsp1β to suppress its self-expression, although it attenuated the ability of nsp1β to suppress type I IFN expression. These data make us speculate that different mechanisms may be utilized by the R128 and R129 residues to evade host innate immune defense, which needs to be further elucidated.
As discussed above, besides its function as an innate immune antagonist, nsp1β was recently identified as a transactivator for the expression of the −2/−1 PRF products nsp2TF and nsp2N. Both nsp2TF and nsp2N share the N-terminal two-thirds sequence of nsp2, which contains the PLP2 domain. PLP2 was also identified as an innate immune antagonist that is capable of removing ubiquitin (Ub) and Ub-like modifiers like ISG15 from host cell substrates (55–58). In testing the effect of nsp1β during viral infection (see below), it remains to be established to what extent nsp1β directly modulates the innate immune response or whether it does so by stimulating the expression of nsp2TF and nsp2N.
In this study, we generated recombinant viruses of nsp1β mutants using a reverse-genetics approach, including vK124A, vR128A, vR129A, and vRR129AA. For comparison, our previously constructed mutant, v1βKO (containing the K124A/R128A double mutation), was included for characterization of virus growth in cell culture. In comparison to the WT virus, all nsp1β mutants except vK124A had attenuated growth ability in cell culture, but their peak viral titers all reached >5 log TCID50/ml, which is acceptable for subsequent application in animals. Multiple-step virus growth curves showed that vR128A and vR129A had slightly slower growth kinetics, while v1βKO and vRR129AA showed ∼1- to 1.5-log-lower virus titers than that of the parental virus at all tested time points (Fig. 3A). It is worth noting that v1βKO, containing the K124A/R128A double mutation, had a lower virus titer than that of the mutant virus with the single R128A mutation, but the single K124A mutation did not have much of an effect on virus growth ability in cell culture. In addition, none of the mutations affected the release of nsp1β from the nsp1β-2 polyprotein (Fig. 2B), suggesting that the reduced growth rate (viral titer) of the nsp1β mutants may not be directly caused by a basic defect in replicase polyprotein proteolysis. We speculate that R128A, R129A, or the combined K124A/R128A or R128A/R129A mutation may change the nsp1β protein or RNA structure, which in turn affects virus replication ability. The in-depth mechanism of these mutations that affect virus growth ability requires more studies in the future. When tested with the vaccinia virus-T7 expression system, the expression of nsp2TF was impaired by alanine substitutions at residues 128 and 129 using nsp1β-2 expression constructs (Fig. 2A). Under virus infection conditions, these two residues are also essential for the PRF transactivator function of nsp1β (data not shown). Taken together, our data indicate that the three basic amino acids exposed on the nsp1β surface appear to have different functions, and the detailed mechanism needs to be further elucidated.
Subsequently, these mutants were characterized in nursery pigs. Since the K124A mutant did not have much of an effect on the innate suppression function of nsp1β, and the recombinant virus containing the K124A mutation did not have much of an effect on virus replication, this mutant was excluded in the present animal study. In a previous study, we evaluated 1βKO and WT viruses in pigs. 1βKO showed an overattenuated phenotype, with virus growing to an extremely low titer (∼5 × 104 RNA copies/ml in serum), which is 2 to 3 logs lower than that of the wild-type virus. As a consequence, pigs did not seroconvert until 28 dpi. The initial IFN-α response was very limited (8.4 pg/ml serum at 3 dpi); in contrast, we detected an IFN-α response (100.9 pg/ml serum at 3 dpi) in WT virus-infected pigs. As discussed above, a single mutation at the K124 residue did not seem to have much of an effect on the in vitro growth ability of the virus, but combined mutations at R128 significantly impaired the virus growth ability in vitro and in vivo. The in-depth mechanism of the involvement of K124 in viral replication and its function in relation to the combined effect of the R128 mutation need to be further studied. Since our previous data showed an overattenuated phenotype of 1βKO, in the present study, we focused on characterizing the other three nsp1β mutants (vR128A, vR129A, and vRR129AA) in nursery pigs. Active virus replication was observed in all of the virus-infected pigs. The viremia data indicate that, consistent with in vitro results, nsp1β mutants were also attenuated in pigs. At all time points, vR128A- and vRR129AA-infected pigs had consistently lower mean viral RNA loads and infectious virus titers than did WT virus-infected pigs. The pigs in the vR129A group also had lower levels of viremia than that of the WT virus group at early time points; however, they showed even higher levels of viremia than that of the WT group at 35 dpi. These results suggest that vR129A could be reversed back to the WT virus. Subsequently, sequence analysis of the nsp1β coding region was performed to confirm the stability of the mutations introduced into the virus. In viruses recovered at 21 dpi, the designed alanine substitutions were stably maintained, except for a reversion identified in one of the pigs (pig 31) infected with vR129A. We further sequenced the nsp1β coding region in the viruses recovered from vR129A-infected pigs at 14 dpi, and the results showed that reversion occurred only in pig 31 and not the other five tested pigs. Unexpectedly, no reversion in the nsp1β coding region was identified in the vR129A group of pigs at 35 dpi, although the vR129A group of pigs showed even higher virus titers than those of the WT group of pigs. We speculate that spontaneous mutations in other regions of the virus genome may compensate for the viral replication ability in vivo. Remarkably, for the viruses that stably maintained the designed mutations, vR128A and vR129A, in infected pigs, a specific second-site mutation of Ser122 to Pro122 was consistently identified. We analyzed the sequence of nsp1β from in vitro expression plasmids and original recombinant viruses that grew in MARC-145 cells (before inoculation into pigs), and the results showed that Ser122 was stably maintained in vR128A, vR129A, and vRR129AA. The data suggest that Pro122 may contribute to virus fitness in vivo and that the Ser122-to-Pro122 substitution may compromise the side effect of our designed mutations on virus replication ability in animals. Whether the Ser122-to-Pro122 substitution has an effect on the function of nsp1β needs to be further studied.
Given the impaired ability of the vR128A, vR129A, and vRR129AA mutants to antagonize the innate immune response in vitro, we further assessed the ability of these viruses to induce a host immune response in vivo. After immunization, no clinical symptoms and adverse side effects were observed in the WT and mutant virus-infected pigs. In addition, when three pigs per group were terminated at 7, 21, and 35 dpi, no obvious lung lesions were observed. This result is expected, since the WT virus SD95-21 (backbone of nsp1β mutants) has 99.5% nucleotide identity to VR2332, the parental virus of the PRRS MLV vaccine (Ingelvac PRRS MLV; Boehringer Ingelheim Vetmedica Inc.), and 99.6% identity to Ingelvac PRRS MLV. In addition, SD95-21 virus was adapted to grow in MARC-145 cells. In previous studies, pigs infected with PRRS MLV of the VR2332 strain showed very mild or undetectable clinical symptoms, gross lesions, or histopathological lesions (59). The goal of our vaccine development strategy is to improve the ability of current MLV vaccines to stimulate higher-level innate and cell-mediated immune responses.
Since type I IFNs are the principal cytokines for innate immunity against viral infections, IFN-α was selected as a representative to assess the ability of PRRSV to induce the host innate immune response. Consistent with our data generated in an in vitro expression system, nsp1β mutants induced higher levels of IFN-α than did the WT virus at early time points postinfection. In comparison to WT virus-infected pigs, high cytotoxic activity of NK cells was also observed in pigs infected with nsp1β mutants. These results suggest that PRRSV nsp1β plays a crucial role in suppressing host innate immunity, and modifying this protein could effectively improve the ability of PRRSV to stimulate the host innate immune response. Furthermore, these mutants induced earlier and higher-level IFN-γ expression than did the WT virus. As an important factor in adaptive immunity against viral infection, IFN-γ increases antigen presentation and promotes Th1 cell differentiation (60). The increased expression of IFN-γ in mutant virus-infected pigs indirectly indicates the activation of the Th1 cell-mediated immune response. This not only was observed in serum of mutant-infected pigs but also was indicated by the increased activation of CTLs/γδ T cells, T helper/memory cells, and NK cells. Previously, an increased frequency of activated T helper/memory cells in pigs was shown to be beneficial in the clearance of Aujeszky's disease virus, African swine fever virus, classical swine fever virus, and PRRSV infections (33, 41, 61–64). Taken together, besides the induction of higher-level innate immune responses, these mutants may also have a stronger ability to augment Th1 cell-mediated adaptive immunity than the WT virus. As a candidate vaccine, one would expect its ability to stimulate a significant humoral response in animals. In fact, we performed both a virus-neutralizing antibody assay and an ELISA using serum samples, but the results showed no significant difference among pig groups infected with the WT virus and the three nsp1β mutants (data not shown). It is a well-established phenomenon that a strong Th1 response suppresses the Th2 response (humoral response) and vice versa, which was demonstrated previously in mice (65, 66) and pigs (33, 64, 67). Therefore, our data suggest that the lack of an improved virus-neutralizing antibody response in nsp1β mutant-infected pigs could be partially caused by the strong Th1 response. Future virus challenge studies of nsp1β mutant-vaccinated pigs may reveal benefits of increased Th1 responses in virus clearance.
Based on our data, the vRR129AA mutant could be a potential vaccine candidate, and this attenuation strategy could be easily applied to improve current vaccines. This conclusion is based on multiple observations. First, vRR129AA can grow to sufficient virus titers in cell culture (>5 log TCID50/ml), which facilitates large-scale vaccine production. Second, in comparison to the WT virus, vRR129AA induced an early and strong innate immune response, which was supported by elevated levels of IFN-α expression and NK cell cytotoxic activity. The improved innate immune response appears to augment cell-mediated adaptive immunity, indicated by the early induction of IFN-γ expression by both NK cells and T cells, followed by the depletion of activated T cell subsets, as presented in phenotypic analyses of PBMCs. Another important aspect is that this mutant appeared to be quickly cleared from virus-infected pigs and showed better genetic stability than nsp1β mutants containing a single alanine substitution (vR128A and vR129A). Nevertheless, the protection efficacy of this potential vaccine candidate needs to be further assessed in animal challenge studies. Finally, residues R128 and R129 are highly conserved in all available PRRSV strains, as described previously (25, 26); therefore, the technology described in this study can be easily applied to current commercial vaccines and other candidate vaccines.
ACKNOWLEDGMENTS
We thank Eric J. Snijder (Leiden University Medical Center, Leiden, The Netherlands) for helpful discussion and Russell Ransburgh and Elizabeth Poulsen (Kansas State University, Manhattan, KS, USA) for technical assistance.
Funding Statement
This project was supported by Agriculture and Food Research Initiative competitive grant no. 2012-67015-21823 from the USDA National Institute of Food and Agriculture and by the Kansas State University research startup fund. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Keffaber KK. 1989. Reproductive failure of unknown etiology. Am Assoc Swine Pract Newsl 1:1–9. [Google Scholar]
- 2.Wensvoort G, Terpstra C, Pol JM, ter Laak EA, Bloemraad M, de Kluyver EP, Kragten C, van Buiten L, den Besten A, Wagenaar F, Broekhuijsen , Moonen PLJM, Zetstra T, de Boer EA, Tibben HJ, de Jong MF, van't Veld P, Greenland GJR, van Gennep JA, Voets MT, Verheijden JHM, Braamskamp J. 1991. Mystery swine disease in The Netherlands: the isolation of Lelystad virus. Vet Q 13:121–130. doi: 10.1080/01652176.1991.9694296. [DOI] [PubMed] [Google Scholar]
- 3.Benfield DA, Nelson E, Collins JE, Harris L, Goyal SM, Robison D, Christianson WT, Morrison RB, Gorcyca D, Chladek D. 1992. Characterization of swine infertility and respiratory syndrome (SIRS) virus (isolate ATCC VR-2332). J Vet Diagn Invest 4:127–133. doi: 10.1177/104063879200400202. [DOI] [PubMed] [Google Scholar]
- 4.Collins JE, Benfield DA, Christianson WT, Harris L, Hennings JC, Shaw DP, Goyal SM, McCullough S, Morrison RB, Joo HS, Gorcyca D, Chladek D. 1992. Isolation of swine infertility and respiratory syndrome virus (isolate ATCC VR-2332) in North America and experimental reproduction of the disease in gnotobiotic pigs. J Vet Diagn Invest 4:117–126. doi: 10.1177/104063879200400201. [DOI] [PubMed] [Google Scholar]
- 5.Albina E, Madec F, Cariolet R, Torrison J. 1994. Immune response and persistence of the porcine reproductive and respiratory syndrome virus in infected pigs and farm units. Vet Rec 134:567–573. doi: 10.1136/vr.134.22.567. [DOI] [PubMed] [Google Scholar]
- 6.Dwivedi V, Manickam C, Binjawadagi B, Linhares D, Murtaugh MP, Renukaradhya GJ. 2012. Evaluation of immune responses to porcine reproductive and respiratory syndrome virus in pigs during early stage of infection under farm conditions. Virol J 9:45. doi: 10.1186/1743-422X-9-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murtaugh MP, Xiao Z, Zuckermann F. 2002. Immunological responses of swine to porcine reproductive and respiratory syndrome virus infection. Viral Immunol 15:533–547. doi: 10.1089/088282402320914485. [DOI] [PubMed] [Google Scholar]
- 8.Renukaradhya GJ, Dwivedi V, Manickam C, Binjawadagi B, Benfield D. 2012. Mucosal vaccines to prevent porcine reproductive and respiratory syndrome: a new perspective. Anim Health Res Rev 13:21–37. doi: 10.1017/S1466252312000023. [DOI] [PubMed] [Google Scholar]
- 9.Snijder EJ, Kikkert M, Fang Y. 2013. Arterivirus molecular biology and pathogenesis. J Gen Virol 94:2141–2163. doi: 10.1099/vir.0.056341-0. [DOI] [PubMed] [Google Scholar]
- 10.Meulenberg JJ, Petersen-den Besten A, De Kluyver EP, Moormann RJ, Schaaper WM, Wensvoort G. 1995. Characterization of proteins encoded by ORFs 2 to 7 of Lelystad virus. Virology 206:155–163. doi: 10.1016/S0042-6822(95)80030-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meulenberg JJ, Petersen-den Besten A, de Kluyver EP, Moormann RJ, Schaaper WM, Wensvoort G. 1995. Characterization of structural proteins of Lelystad virus. Adv Exp Med Biol 380:271–276. doi: 10.1007/978-1-4615-1899-0_43. [DOI] [PubMed] [Google Scholar]
- 12.Mounir S, Mardassi H, Dea S. 1995. Identification and characterization of the porcine reproductive and respiratory virus ORFs 7, 5 and 4 products. Adv Exp Med Biol 380:317–320. doi: 10.1007/978-1-4615-1899-0_51. [DOI] [PubMed] [Google Scholar]
- 13.Bautista EM, Meulenberg JJ, Choi CS, Molitor TW. 1996. Structural polypeptides of the American (VR-2332) strain of porcine reproductive and respiratory syndrome virus. Arch Virol 141:1357–1365. doi: 10.1007/BF01718837. [DOI] [PubMed] [Google Scholar]
- 14.Mardassi H, Massie B, Dea S. 1996. Intracellular synthesis, processing, and transport of proteins encoded by ORFs 5 to 7 of porcine reproductive and respiratory syndrome virus. Virology 221:98–112. doi: 10.1006/viro.1996.0356. [DOI] [PubMed] [Google Scholar]
- 15.Meng XJ, Paul PS, Morozov I, Halbur PG. 1996. A nested set of six or seven subgenomic mRNAs is formed in cells infected with different isolates of porcine reproductive and respiratory syndrome virus. J Gen Virol 77(Part 6):1265–1270. [DOI] [PubMed] [Google Scholar]
- 16.Meulenberg JJ, Petersen-den Besten A. 1996. Identification and characterization of a sixth structural protein of Lelystad virus: the glycoprotein GP2 encoded by ORF2 is incorporated in virus particles. Virology 225:44–51. doi: 10.1006/viro.1996.0573. [DOI] [PubMed] [Google Scholar]
- 17.Snijder EJ, van Tol H, Pedersen KW, Raamsman MJ, de Vries AA. 1999. Identification of a novel structural protein of arteriviruses. J Virol 73:6335–6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu WH, Fang Y, Farwell R, Steffen-Bien M, Rowland RR, Christopher-Hennings J, Nelson EA. 2001. A 10-kDa structural protein of porcine reproductive and respiratory syndrome virus encoded by ORF2b. Virology 287:183–191. doi: 10.1006/viro.2001.1034. [DOI] [PubMed] [Google Scholar]
- 19.Johnson CR, Griggs TF, Gnanandarajah J, Murtaugh MP. 2011. Novel structural protein in porcine reproductive and respiratory syndrome virus encoded by an alternative ORF5 present in all arteriviruses. J Gen Virol 92:1107–1116. doi: 10.1099/vir.0.030213-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y, Tas A, Sun Z, Snijder EJ, Fang Y. 2015. Proteolytic processing of the porcine reproductive and respiratory syndrome virus replicase. Virus Res 202:48–59. doi: 10.1016/j.virusres.2014.12.027. [DOI] [PubMed] [Google Scholar]
- 21.Fang Y, Treffers EE, Li Y, Tas A, Sun Z, van der Meer Y, de Ru AH, van Veelen PA, Atkins JF, Snijder EJ, Firth AE. 2012. Efficient −2 frameshifting by mammalian ribosomes to synthesize an additional arterivirus protein. Proc Natl Acad Sci U S A 109:E2920–E2928. doi: 10.1073/pnas.1211145109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Z, Lawson S, Sun Z, Zhou X, Guan X, Christopher-Hennings J, Nelson EA, Fang Y. 2010. Identification of two auto-cleavage products of nonstructural protein 1 (nsp1) in porcine reproductive and respiratory syndrome virus infected cells: nsp1 function as interferon antagonist. Virology 398:87–97. doi: 10.1016/j.virol.2009.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beura LK, Sarkar SN, Kwon B, Subramaniam S, Jones C, Pattnaik AK, Osorio FA. 2010. Porcine reproductive and respiratory syndrome virus nonstructural protein 1beta modulates host innate immune response by antagonizing IRF3 activation. J Virol 84:1574–1584. doi: 10.1128/JVI.01326-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim O, Sun Y, Lai FW, Song C, Yoo D. 2010. Modulation of type I interferon induction by porcine reproductive and respiratory syndrome virus and degradation of CREB-binding protein by non-structural protein 1 in MARC-145 and HeLa cells. Virology 402:315–326. doi: 10.1016/j.virol.2010.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, Treffers EE, Napthine S, Tas A, Zhu L, Sun Z, Bell S, Mark BL, van Veelen PA, van Hemert MJ, Firth AE, Brierley I, Snijder EJ, Fang Y. 2014. Transactivation of programmed ribosomal frameshifting by a viral protein. Proc Natl Acad Sci U S A 111:E2172–E2181. doi: 10.1073/pnas.1321930111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y, Zhu L, Lawson SR, Fang Y. 2013. Targeted mutations in a highly conserved motif of the nsp1beta protein impair the interferon antagonizing activity of porcine reproductive and respiratory syndrome virus. J Gen Virol 94:1972–1983. doi: 10.1099/vir.0.051748-0. [DOI] [PubMed] [Google Scholar]
- 27.Zeman D, Neiger R, Yaeger M, Nelson E, Benfield D, Leslie-Steen P, Thomson J, Miskimins D, Daly R, Minehart M. 1993. Laboratory investigation of PRRS virus infection in three swine herds. J Vet Diagn Invest 5:522–528. doi: 10.1177/104063879300500404. [DOI] [PubMed] [Google Scholar]
- 28.Fuerst TR, Niles EG, Studier FW, Moss B. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci U S A 83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nelson EA, Christopher-Hennings J, Drew T, Wensvoort G, Collins JE, Benfield DA. 1993. Differentiation of U.S. and European isolates of porcine reproductive and respiratory syndrome virus by monoclonal antibodies. J Clin Microbiol 31:3184–3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am J Epidemiol 27:493–497. [Google Scholar]
- 31.Fang Y, Rowland RR, Roof M, Lunney JK, Christopher-Hennings J, Nelson EA. 2006. A full-length cDNA infectious clone of North American type 1 porcine reproductive and respiratory syndrome virus: expression of green fluorescent protein in the Nsp2 region. J Virol 80:11447–11455. doi: 10.1128/JVI.01032-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Halbur PG, Paul PS, Frey ML, Landgraf J, Eernisse K, Meng XJ, Lum MA, Andrews JJ, Rathje JA. 1995. Comparison of the pathogenicity of two US porcine reproductive and respiratory syndrome virus isolates with that of the Lelystad virus. Vet Pathol 32:648–660. doi: 10.1177/030098589503200606. [DOI] [PubMed] [Google Scholar]
- 33.Binjawadagi B, Dwivedi V, Manickam C, Ouyang K, Torrelles JB, Renukaradhya GJ. 2014. An innovative approach to induce cross-protective immunity against porcine reproductive and respiratory syndrome virus in the lungs of pigs through adjuvanted nanotechnology-based vaccination. Int J Nanomedicine 9:1519–1535. doi: 10.2147/IJN.S59924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Renukaradhya GJ, Alekseev K, Jung K, Fang Y, Saif LJ. 2010. Porcine reproductive and respiratory syndrome virus-induced immunosuppression exacerbates the inflammatory response to porcine respiratory coronavirus in pigs. Viral Immunol 23:457–466. doi: 10.1089/vim.2010.0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garcia-Iglesias T, Del Toro-Arreola A, Albarran-Somoza B, Del Toro-Arreola S, Sanchez-Hernandez PE, Ramirez-Duenas MG, Balderas-Pena LM, Bravo-Cuellar A, Ortiz-Lazareno PC, Daneri-Navarro A. 2009. Low NKp30, NKp46 and NKG2D expression and reduced cytotoxic activity on NK cells in cervical cancer and precursor lesions. BMC Cancer 9:186. doi: 10.1186/1471-2407-9-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lecoeur H, Fevrier M, Garcia S, Riviere Y, Gougeon ML. 2001. A novel flow cytometric assay for quantitation and multiparametric characterization of cell-mediated cytotoxicity. J Immunol Methods 253:177–187. doi: 10.1016/S0022-1759(01)00359-3. [DOI] [PubMed] [Google Scholar]
- 37.Dwivedi V, Manickam C, Patterson R, Dodson K, Murtaugh M, Torrelles JB, Schlesinger LS, Renukaradhya GJ. 2011. Cross-protective immunity to porcine reproductive and respiratory syndrome virus by intranasal delivery of a live virus vaccine with a potent adjuvant. Vaccine 29:4058–4066. doi: 10.1016/j.vaccine.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Talker SC, Kaser T, Reutner K, Sedlak C, Mair KH, Koinig H, Graage R, Viehmann M, Klingler E, Ladinig A, Ritzmann M, Saalmuller A, Gerner W. 2013. Phenotypic maturation of porcine NK- and T-cell subsets. Dev Comp Immunol 40:51–68. doi: 10.1016/j.dci.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 39.Sinkora M, Butler JE. 2009. The ontogeny of the porcine immune system. Dev Comp Immunol 33:273–283. doi: 10.1016/j.dci.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zuckermann FA, Husmann RJ. 1996. Functional and phenotypic analysis of porcine peripheral blood CD4/CD8 double-positive T cells. Immunology 87:500–512. [PMC free article] [PubMed] [Google Scholar]
- 41.Denyer MS, Wileman TE, Stirling CM, Zuber B, Takamatsu HH. 2006. Perforin expression can define CD8 positive lymphocyte subsets in pigs allowing phenotypic and functional analysis of natural killer, cytotoxic T, natural killer T and MHC un-restricted cytotoxic T-cells. Vet Immunol Immunopathol 110:279–292. doi: 10.1016/j.vetimm.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 42.Costers S, Lefebvre DJ, Goddeeris B, Delputte PL, Nauwynck HJ. 2009. Functional impairment of PRRSV-specific peripheral CD3+CD8high cells. Vet Res 40:46. doi: 10.1051/vetres/2009029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dwivedi V, Manickam C, Patterson R, Dodson K, Weeman M, Renukaradhya GJ. 2011. Intranasal delivery of whole cell lysate of Mycobacterium tuberculosis induces protective immune responses to a modified live porcine reproductive and respiratory syndrome virus vaccine in pigs. Vaccine 29:4067–4076. doi: 10.1016/j.vaccine.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kamijuku H, Nagata Y, Jiang X, Ichinohe T, Tashiro T, Mori K, Taniguchi M, Hase K, Ohno H, Shimaoka T, Yonehara S, Odagiri T, Tashiro M, Sata T, Hasegawa H, Seino KI. 2008. Mechanism of NKT cell activation by intranasal coadministration of alpha-galactosylceramide, which can induce cross-protection against influenza viruses. Mucosal Immunol 1:208–218. doi: 10.1038/mi.2008.2. [DOI] [PubMed] [Google Scholar]
- 45.Guillonneau C, Mintern JD, Hubert FX, Hurt AC, Besra GS, Porcelli S, Barr IG, Doherty PC, Godfrey DI, Turner SJ. 2009. Combined NKT cell activation and influenza virus vaccination boosts memory CTL generation and protective immunity. Proc Natl Acad Sci U S A 106:3330–3335. doi: 10.1073/pnas.0813309106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Albina E, Piriou L, Hutet E, Cariolet R, L'Hospitalier R. 1998. Immune responses in pigs infected with porcine reproductive and respiratory syndrome virus (PRRSV). Vet Immunol Immunopathol 61:49–66. doi: 10.1016/S0165-2427(97)00134-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Reeth K, Labarque G, Nauwynck H, Pensaert M. 1999. Differential production of proinflammatory cytokines in the pig lung during different respiratory virus infections: correlations with pathogenicity. Res Vet Sci 67:47–52. doi: 10.1053/rvsc.1998.0277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Buddaert W, Van Reeth K, Pensaert M. 1998. In vivo and in vitro interferon (IFN) studies with the porcine reproductive and respiratory syndrome virus (PRRSV). Adv Exp Med Biol 440:461–467. doi: 10.1007/978-1-4615-5331-1_59. [DOI] [PubMed] [Google Scholar]
- 49.Miller LC, Laegreid WW, Bono JL, Chitko-McKown CG, Fox JM. 2004. Interferon type I response in porcine reproductive and respiratory syndrome virus-infected MARC-145 cells. Arch Virol 149:2453–2463. doi: 10.1007/s00705-004-0377-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luo R, Xiao S, Jiang Y, Jin H, Wang D, Liu M, Chen H, Fang L. 2008. Porcine reproductive and respiratory syndrome virus (PRRSV) suppresses interferon-beta production by interfering with the RIG-I signaling pathway. Mol Immunol 45:2839–2846. doi: 10.1016/j.molimm.2008.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Donelan NR, Basler CF, Garcia-Sastre A. 2003. A recombinant influenza A virus expressing an RNA-binding-defective NS1 protein induces high levels of beta interferon and is attenuated in mice. J Virol 77:13257–13266. doi: 10.1128/JVI.77.24.13257-13266.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Talon J, Salvatore M, O'Neill RE, Nakaya Y, Zheng H, Muster T, Garcia-Sastre A, Palese P. 2000. Influenza A and B viruses expressing altered NS1 proteins: a vaccine approach. Proc Natl Acad Sci U S A 97:4309–4314. doi: 10.1073/pnas.070525997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Valarcher JF, Furze J, Wyld S, Cook R, Conzelmann KK, Taylor G. 2003. Role of alpha/beta interferons in the attenuation and immunogenicity of recombinant bovine respiratory syncytial viruses lacking NS proteins. J Virol 77:8426–8439. doi: 10.1128/JVI.77.15.8426-8439.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zust R, Cervantes-Barragan L, Kuri T, Blakqori G, Weber F, Ludewig B, Thiel V. 2007. Coronavirus non-structural protein 1 is a major pathogenicity factor: implications for the rational design of coronavirus vaccines. PLoS Pathog 3:e109. doi: 10.1371/journal.ppat.0030109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Frias-Staheli N, Giannakopoulos NV, Kikkert M, Taylor SL, Bridgen A, Paragas J, Richt JA, Rowland RR, Schmaljohn CS, Lenschow DJ, Snijder EJ, Garcia-Sastre A, Virgin HW IV. 2007. Ovarian tumor domain-containing viral proteases evade ubiquitin- and ISG15-dependent innate immune responses. Cell Host Microbe 2:404–416. doi: 10.1016/j.chom.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun Z, Chen Z, Lawson SR, Fang Y. 2010. The cysteine protease domain of porcine reproductive and respiratory syndrome virus nonstructural protein 2 possesses deubiquitinating and interferon antagonism functions. J Virol 84:7832–7846. doi: 10.1128/JVI.00217-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun Z, Li Y, Ransburgh R, Snijder EJ, Fang Y. 2012. Nonstructural protein 2 of porcine reproductive and respiratory syndrome virus inhibits the antiviral function of interferon-stimulated gene 15. J Virol 86:3839–3850. doi: 10.1128/JVI.06466-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Kasteren PB, Bailey-Elkin BA, James TW, Ninaber DK, Beugeling C, Khajehpour M, Snijder EJ, Mark BL, Kikkert M. 2013. Deubiquitinase function of arterivirus papain-like protease 2 suppresses the innate immune response in infected host cells. Proc Natl Acad Sci U S A 110:E838–E847. doi: 10.1073/pnas.1218464110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Manickam C, Dwivedi V, Patterson R, Papenfuss T, Renukaradhya GJ. 2013. Porcine reproductive and respiratory syndrome virus induces pronounced immune modulatory responses at mucosal tissues in the parental vaccine strain VR2332 infected pigs. Vet Microbiol 162:68–77. doi: 10.1016/j.vetmic.2012.08.021. [DOI] [PubMed] [Google Scholar]
- 60.Schroder K, Hertzog PJ, Ravasi T, Hume DA. 2004. Interferon-gamma: an overview of signals, mechanisms and functions. J Leukoc Biol 75:163–189. [DOI] [PubMed] [Google Scholar]
- 61.De Bruin TG, Van Rooij EM, De Visser YE, Bianchi AT. 2000. Cytolytic function for pseudorabies virus-stimulated porcine CD4+ CD8dull+ lymphocytes. Viral Immunol 13:511–520. doi: 10.1089/vim.2000.13.511. [DOI] [PubMed] [Google Scholar]
- 62.Franzoni G, Kurkure NV, Essler SE, Pedrera M, Everett HE, Bodman-Smith KB, Crooke HR, Graham SP. 2013. Proteome-wide screening reveals immunodominance in the CD8 T cell response against classical swine fever virus with antigen-specificity dependent on MHC class I haplotype expression. PLoS One 8:e84246. doi: 10.1371/journal.pone.0084246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Takamatsu HH, Denyer MS, Lacasta A, Stirling CM, Argilaguet JM, Netherton CL, Oura CA, Martins C, Rodriguez F. 2013. Cellular immunity in ASFV responses. Virus Res 173:110–121. doi: 10.1016/j.virusres.2012.11.009. [DOI] [PubMed] [Google Scholar]
- 64.Binjawadagi B, Dwivedi V, Manickam C, Ouyang K, Wu Y, Lee LJ, Torrelles JB, Renukaradhya GJ. 2014. Adjuvanted poly(lactic-co-glycolic) acid nanoparticle-entrapped inactivated porcine reproductive and respiratory syndrome virus vaccine elicits cross-protective immune response in pigs. Int J Nanomedicine 9:679–694. doi: 10.2147/IJN.S56127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang S, Fan Y, Brunham RC, Yang X. 1999. IFN-gamma knockout mice show Th2-associated delayed-type hypersensitivity and the inflammatory cells fail to localize and control chlamydial infection. Eur J Immunol 29:3782–3792. doi:. [DOI] [PubMed] [Google Scholar]
- 66.Song Z, Wu H, Ciofu O, Kong KF, Hoiby N, Rygaard J, Kharazmi A, Mathee K. 2003. Pseudomonas aeruginosa alginate is refractory to Th1 immune response and impedes host immune clearance in a mouse model of acute lung infection. J Med Microbiol 52:731–740. doi: 10.1099/jmm.0.05122-0. [DOI] [PubMed] [Google Scholar]
- 67.Diaz MA, Villalobos N, de Aluja A, Rosas G, Gomez-Conde E, Hernandez P, Larralde C, Sciutto E, Fragoso G. 2003. Th1 and Th2 indices of the immune response in pigs vaccinated against Taenia solium cysticercosis suggest various host immune strategies against the parasite. Vet Immunol Immunopathol 93:81–90. doi: 10.1016/S0165-2427(03)00071-0. [DOI] [PubMed] [Google Scholar]