ABSTRACT
Endogenous retroviruses (ERVs), the majority of which exist as degraded remnants of ancient viruses, comprise approximately 8% of the human genome. The youngest human ERVs (HERVs) belong to the HERV-K(HML-2) subgroup and were endogenized within the past 1 million years. The viral envelope protein (ENV) facilitates the earliest events of endogenization (cellular attachment and entry), and here, we characterize the requirements for HERV-K ENV to mediate infectious cell entry. Cell-cell fusion assays indicate that a minimum of two events are required for fusion, proteolytic processing by furin-like proteases and exposure to acidic pH. We generated an infectious autonomously replicating recombinant vesicular stomatitis virus (VSV) in which the glycoprotein was replaced by HERV-K ENV. HERV-K ENV imparts an endocytic entry pathway that requires dynamin-mediated membrane scission and endosomal acidification but is distinct from clathrin-dependent or macropinocytic uptake pathways. The lack of impediments to the replication of the VSV core in eukaryotic cells allowed us to broadly survey the HERV-K ENV-dictated tropism. Unlike extant betaretroviral envelopes, which impart a narrow species tropism, we found that HERV-K ENV mediates broad tropism encompassing cells from multiple mammalian and nonmammalian species. We conclude that HERV-K ENV dictates an evolutionarily conserved entry pathway and that the restriction of HERV-K to primate genomes reflects downstream stages of the viral replication cycle.
IMPORTANCE Approximately 8% of the human genome is of retroviral origin. While many of those viral genomes have become inactivated, some copies of the most recently endogenized human retrovirus, HERV-K, can encode individual functional proteins. Here, we characterize the envelope protein (ENV) of the virus to define how it mediates infection of cells. We demonstrate that HERV-K ENV undergoes a proteolytic processing step and triggers membrane fusion in response to acidic pH—a strategy common to many viral fusogens. Our data suggest that the infectious entry pathway mediated by this ENV requires endosomal acidification and the GTPase dynamin but does not require clathrin-dependent uptake. In marked contrast to other betaretroviruses, HERV-K ENV imparts broad species tropism in cultured cells. This work provides new insights into the entry pathway of an extinct human virus and provides a powerful tool to further probe the endocytic route by which HERV-K infects cells.
INTRODUCTION
Endogenous retroviruses (ERVs) comprise approximately 8% of the human genome (1). Such ERVs provide a physical record of ancient infections by once exogenous retroviruses; however, the degraded states of most sequences largely obscure their biological properties. Consequently, relatively little is known about the earliest events of endogenization, including how the viruses initially entered the germ line to become vertically transmitted elements. The process of endogenization begins with cellular attachment and viral entry, which is mediated by the envelope protein (ENV). ERV sequences accumulate mutations over time, and consequently, ENVs from the most recently endogenized ERVs are likely to most closely recapitulate the properties of their ancient progenitor viruses.
The most recently endogenized human endogenous retroviruses (HERVs) belong to the HERV-K(HML-2) group. Multiple independent endogenization events have given rise to the approximately 90 proviral copies and 1,000 solo long terminal repeats (LTRs) that are present in reference human genomes (2). While the presence of HERV-K(HML-2) sequences in Old World primates suggests that the group is approximately 30 to 35 million years old (3, 4), the youngest human-specific copies are thought to have been inserted as recently as 100,000 to 250,000 years ago (5–7). As a result, largely intact HERV-K proviruses are present in the human genome, and some loci are capable of producing functional proteins (8–15). This includes the env gene of HERV-K 108, which mediates infection of a pseudotyped lentivirus (15). While no single provirus can produce infectious virions, infectious HERV-K molecular clones have been generated from reconstructed ancestral consensus sequences (16, 17).
The gag, pol, and env genes of HERV-K(HML-2) are similar to those of viruses belonging to the genus Betaretrovirus of the family Retroviridae. Other betaretroviruses with this arrangement include mouse mammary tumor virus (MMTV), Jaagsiekte sheep retrovirus (JSRV), and enzootic nasal tumor virus (ENTV). Such beta-type envelopes are processed by furin or furin-like proteases into surface (SU) and transmembrane (TM) subunits, which are noncovalently associated (18). SU contains the receptor-binding site, and TM contains the fusion machinery, along with a transmembrane domain and cytoplasmic tail of variable length. All betaretroviral ENVs tested (those of MMTV, JSRV, and ENTV) require endosomal uptake and acidification to mediate membrane fusion and infection (19–21).
Beta-type envelopes strongly influence viral tropism. MMTV uses the transferrin receptor of certain species of mice and rats (19, 22–24). JSRV uses Hyal2 from a variety of mammals, but not rodents (25), whereas ENTV uses only Hyal2 of ovine cells (25, 26). Recent work demonstrated that HERV-K ENV imparts broad tropism among mammalian cells (27). Endogenous beta-type envelopes are present in mammals, whereas gamma types are found throughout metazoans (18, 28). However, betaretroviral gag-pol regions and gamma-type env genes are present in nonmammalian species (29–31), suggesting that there may be an ENV-dictated species-specific tropism restriction for beta-type envelopes.
ENV dictates the initial events that lead to endogenization, from cell binding to membrane fusion. Here, we define the requirements for entry and envelope tropism of the ancient progenitor of HERV-K, using a reconstructed ancestral HERV-K ENV. We show that HERV-K ENV requires proteolytic processing and acidic pH to mediate membrane fusion. Using a replication-competent recombinant vesicular stomatitis virus (VSV) in which the attachment and fusion glycoprotein G was replaced by HERV-K ENV, we demonstrate that HERV-K enters cells via an endocytic pathway requiring dynamin-mediated membrane scission and endosomal acidification. We further demonstrate that the HERV-K entry pathway is distinct from clathrin-mediated endocytosis or macropinocytosis. We extend the findings of broad species tropism by demonstrating that HERV-K ENV mediates entry into a broad range of nonmammalian cells. We suggest that restricted species tropism is not a general property of beta-type ENVs, and therefore, the lack of these endogenous env genes outside of mammals and of HERV-K(HML-2) outside of Old World primates requires alternative explanations.
MATERIALS AND METHODS
Cell lines, plasmids, and viruses.
Human embryonic kidney 293T cells (ATCC CRL-3216; American Type Culture Collection, Manassas, VA), human hepatocellular carcinoma Huh7.5 cells (32), African green monkey kidney Vero cells (ATCC CCL-81), African green monkey kidney Bs-c-1 cells (ATCC CCL-26), Syrian golden hamster kidney BSRT7 cells (a kind gift from K. Conzelmann [33]), Crandell-Rees feline kidney CRFK cells (ATCC CCL-94), Mexican free-tailed bat lung Tb1.lu cells (ATCC CCL-88), and chicken embryo DF-1 cells (ATCC CRL-12203) were maintained at 37°C and 5% CO2 in Dulbecco's modified Eagle medium (DMEM) (Corning Life Sciences, Tewksbury, MA) supplemented with 10% fetal bovine serum (FBS) (Tissue Culture Biologicals, Tulare, CA). Russel's viper heart VH2 cells stably expressing human NPC1 (34) were maintained at 34°C and 5% CO2 in DMEM supplemented with 10% FBS.
A codon-optimized version of the env gene of the Phoenix reconstructed ancestral HERV-K(HML-2) (17) was ordered from GenScript (Piscataway, NJ). To generate HERVK+ (see below), HERVK ENV was truncated after amino acid T658, and the entire VSV-G cytoplasmic tail (amino acid sequence, RVGIYLCIKLKHTKKRQIYTDIEMNRLGK) was added using overlap extension PCR. Both HERVK and HERVK+ were cloned into the pGEM3 expression vector using KpnI and XbaI sites and into the pCAGGS expression vector using KpnI and NheI sites. Furin cleavage site (CS) mutants were generated in pGEM-HERVK and pGEM-HERVK+ to alter the cleavage site sequence from RSKR to SSKS, using site-directed mutagenesis with the following primer: GGGCGTCCTGAACAGCTCTAAAAGCTTCATCTTTACCC, where the underlined nucleotides were changed. pVSV-HERVK and pVSV-HERVK+ were generated by cloning the respective env sequences into pVSV-ΔG-eGFP (35) using MluI and NotI sites. To generate pVSV-PIV5, a DNA fragment containing parainfluenzavirus PIV5 F and PIV5 HN (both strain W3A) separated by the VSV intergenic sequence (TTTATGAAAAAAACTAACAGCAATC) was generated by overlap extension PCR and cloned into pVSV-ΔG-eGFP as described above. PIV5 F and HN were cloned from pCAGGS-PIV5F and pCAGGS-PIV5HN (a kind gift from Robert A. Lamb).
VSV-HERVK, VSV-HERVK+, and VSV-PIV5 were generated through transfection of the genomic plasmid, along with helper plasmids encoding VSV N, P, L, and G, as previously described (36). VSV-eGFP (enhanced green fluorescent protein) and VSV-EBOV (Ebola virus) were previously generated (35, 37). All the viruses were propagated on BSRT7 cells in DMEM supplemented with 2% FBS and penicillin-streptomycin/kanamycin.
Cell-cell fusion.
BSRT7 cells were seeded in 12-well plates and infected with VVT7.3 (38), a vaccinia virus encoding the T7 RNA polymerase, at a multiplicity of infection (MOI) of 3. The cells were subsequently transfected with 2 μg per well of pGEM3-HERVK, pGEM3-HERVK+, pGEM3-HERVK-CS−, pGEM3-HERVK+-CS−, or pGEM3 empty vector using Lipofectamine 2000 transfection reagent (Life Technologies, Grand Island, NY). Five hours posttransfection, the medium was changed to DMEM supplemented with 2% FBS, 50 mM HEPES, and 2.5 μg ml−1 AraC (to prevent expression of vaccinia virus glycoproteins). Eighteen hours posttransfection, the cells were treated with phosphate-buffered saline (PBS) at the indicated pH for 20 min at 37°C, at which point the cells were washed and standard growth medium was added. The cells were incubated for 4 h at 37°C and subsequently imaged using a 10× objective on a Nikon eclipse TE300 microscope (Nikon Instruments, Melville, NY) equipped with a Spot RT camera (Diagnostic Instruments, Sterling Heights, MI). For the cleavage site mutants, NucBlue live-cell stain (Life Technologies) was added prior to imaging to ease visualization of syncytia.
SDS-PAGE and Western blots.
Cell lysates from BSRT7 cells transfected as explained above were run on a 10% (wt/vol) polyacrylamide gel and transferred to a nitrocellulose membrane. The membrane was blocked in 5% (wt/vol) nonfat dry milk in PBS plus 0.1% Tween 20 and probed with an anti-HERV-K ENV antibody (HERM-1811-5; Austral Biologicals, San Ramon, CA) at a 1:1,000 dilution, followed by goat anti-mouse horseradish peroxidase (HRP) (Sigma-Aldrich, St. Louis, MO) at a 1:5,000 dilution. Antibodies were diluted in 5% nonfat dry milk in PBS plus 0.1% Tween 20. The membranes were incubated with ECL reagent (Thermo Scientific, Waltham, MA), and signal was detected by film (Denville Scientific, South Plainfield, NJ).
The purified virus was run on a 10% (wt/vol) acrylamide 0.13% (wt/vol) bis-acrylamide gel and transferred to a nitrocellulose membrane or incubated with Sypro-Ruby (Thermo Fisher, Waltham, MA). The membranes were blocked with Li-Cor blocking buffer (Li-Cor, Lincoln, NE) and incubated with anti-HERV-K ENV antibody (1:1,000 dilution), anti-VSV-G cytoplasmic tail antibody (V5507: Sigma-Aldrich) (1:20,000 dilution), or anti-VSV-M antibody 23H12 (a kind gift from D. S. Lyles [39]) in Li-Cor buffer, followed by incubation with anti-mouse antibody conjugated to IRDye 680RD (Li-Cor) (1:20,000 dilution). Fluorescent signals were detected using a Li-Cor Odyssey CLx imager. Gels were incubated with Sypro-Ruby reagent overnight and treated according to the manufacturer's instructions, and fluorescent signal was detected using a Typhoon 9400 variable-mode imager (General Electric Healthcare, Piscataway, NJ).
Virus growth, purification, and titration.
Viruses were grown in BSRT7 cells. The viral supernatant was used directly for infectivity experiments or further purified on a 15 to 45% sucrose gradient, and the titer was determined by plaque assay on BSRT7 cells as previously described (40). Green fluorescent protein (GFP)-expressing fluorescent plaques were imaged using a Typhoon 9400 imager.
To determine the relative infectivities of VSV-eGFP, VSV-HERVK, and VSV-HERVK+, purified virus was diluted to the same protein concentration, and the number of PFU per microgram was determined by plaque assay on BSRT7 cells.
Lentiviral pseudotypes were produced by transfecting 293T cells in T75 tissue culture flasks with 12 μg pNL-EGFP/CMV-WPREDU3 (41); 8 μg pCD/NL-BH*DDD (42); and 8 μg of pCAGGS-VSVG, pCAGGS-HERVK, or pCAGGS-HERVK+ using polyethylenimine (PEI) transfection reagent (Sigma-Aldrich). The viral supernatant was collected 2 days posttransfection.
Electron microscopy.
Purified VSV-eGFP, VSV-HERVK, or VSV-HERVK+ was deposited onto carbon-coated copper grids and stained with 1% (wt/vol) phosphotungstic acid (PTA) in H2O (pH 7.5). Virus particles were viewed using a Technai G2 Spirit BioTwin transmission electron microscope (FEI, Hillsboro, OR).
Endosomal uptake inhibition assay.
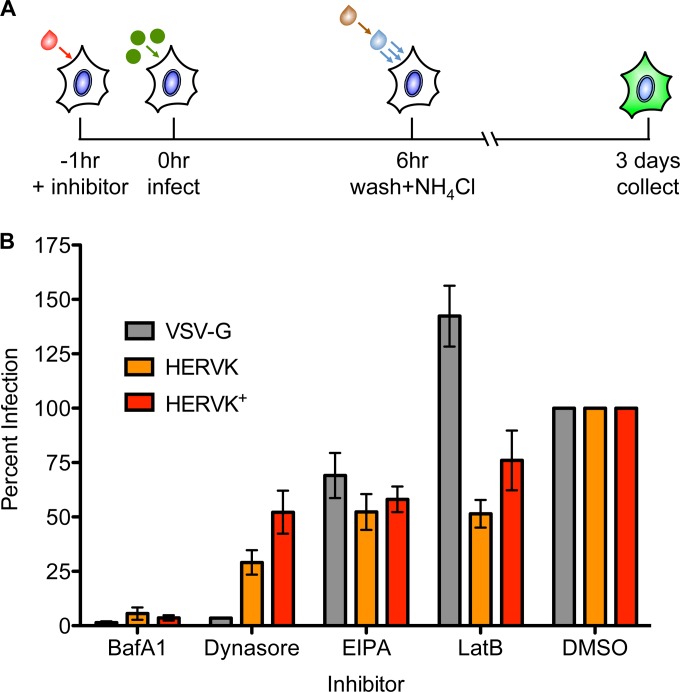
Bs-c-1 or CRFK cells were pretreated for 1 h with the following compounds at the indicated concentrations: 0.1 μM bafilomycin A1 (BafA1; Sigma-Aldrich B1793), 120 μM dynasore (Sigma-Aldrich D7693), 25 μM 5-(N-ethyl-N-isopropyl)amiloride (EIPA; Sigma-Aldrich A3085), and 6 μM latrunculin B (LatB; Sigma-Aldrich L5288). All the compounds were reconstituted in dimethyl sulfoxide (DMSO). Pretreated cells were inoculated with the indicated virus at an MOI of 0.5 for 1 h at 37°C. The cells were washed, and the medium with inhibitor was replaced. Six to 8 h postinfection, cells were either prepared for imaging or collected for flow cytometric analysis. For fluorescence microscopic imaging, cells were washed 3 times with PBS, fixed with 2% paraformaldehyde (PFA) in PBS, and subsequently stained with 4′,6-diamidino-2-phenylindole (DAPI) for 30 min at room temperature. Samples were imaged using an ImageXpress Micro automated microscope (Applied Precision, Issaquah, WA). The images were processed using ImageJ software (U.S. National Institutes of Health, Bethesda, MD). For cytometric analysis, cells were collected and fixed in 2% PFA. Fluorescence was measured using a modified FACSCalibur instrument (Cytek Development, Freemont, CA), and the percentage of GFP-positive cells was quantified using FlowJo software (Tree Star Industries, Ashland, OR). The data are represented as percent GFP-positive cells normalized to DMSO-treated control cells.
For inhibitor studies using lentiviral pseudotypes, cells were pretreated as described above and incubated with GFP-expressing lentivirus carrying the indicated envelope protein at an approximate MOI of 0.05 for 6 h at 37°C in the presence of inhibitor. The cells were washed with DMEM, and the medium was replaced with DMEM with 10 mM ammonium chloride (NH4Cl). Cells were collected 3 days postinfection and analyzed by flow cytometry as described above.
Infectivity and tropism.
The indicated cell lines were incubated with VSV-HERVK+ or VSV-eGFP at an MOI of 1, based on titers in Vero cells, for 1 h at 37°C. The virus was removed, the cells were washed, and the growth medium was replaced. The cells were incubated at 37°C for 6 h, except for VH2-NPC1 cells, which were incubated at 34°C for 8 h. The cells were washed once with PBS, fixed with 2% PFA, and subsequently stained with DAPI for 30 min at room temperature. Samples were imaged using an ImageXpress Micro automated microscope. The images were processed using ImageJ software.
RESULTS
Membrane fusion catalyzed by HERV-K ENV requires proteolytic processing and acidic pH.
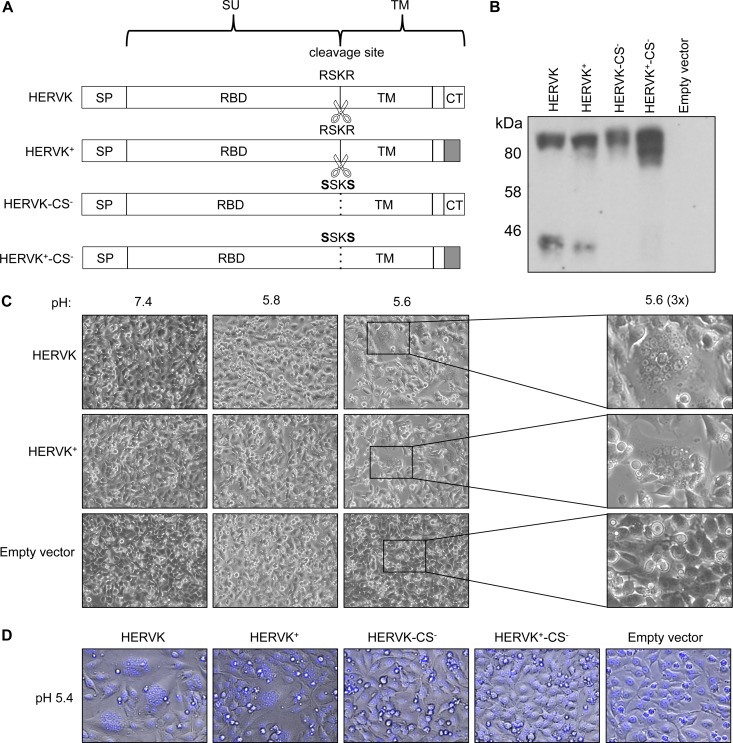
To study HERV-K ENV, we assembled a codon-optimized version of the Phoenix env consensus sequence (17) (referred to here as HERVK). We also generated a variant in which the cytoplasmic tail was truncated and replaced by that of VSV-G (referred to here as HERVK+) to facilitate its incorporation into VSV particles (43–45). As controls, we designed two further variants that should block HERV-K-mediated fusion by engineering the predicted furin cleavage site, RSKR, to SSKS, yielding HERVK-CS− and HERVK+-CS−, respectively (Fig. 1A) (15, 46). To determine the effects of the modifications on HERV-K ENV expression, we transfected the respective plasmids into BSRT7 cells previously infected with a recombinant vaccinia virus expressing T7 RNA polymerase as a source of transcriptase. Each of the variants was expressed, as shown by their reactivity with an antibody against the HERV-K TM domain in a Western blot (Fig. 1B). As expected, HERVK and HERVK+ were processed, yielding uncleaved (∼95-kDa) and cleaved (∼40-kDa) forms, and mutation of the furin cleavage site ablated the cleaved form for HERVK-CS− and HERVK+-CS−.
FIG 1.
HERV-K ENV requires proteolytic processing and acidic pH to mediate membrane fusion. (A) Schematic of HERV-K envelopes tested in this study. A codon-optimized Phoenix env sequence was generated (HERVK) and subsequently altered to switch the cytoplasmic tail with that of VSV-G (HERVK+) and to further remove the furin cleavage site (HERVK-CS− and HERVK+-CS−). The sequences of the WT (RSKR) and mutated (SSKS) cleavage sites are indicated. White, HERV-K sequence; gray, VSV-derived sequence; SU, surface subunit; TM, transmembrane subunit; SP, signal peptide; RBD, receptor-binding domain; CT, cytoplasmic tail. (B) Western blot of cell lysates expressing HERV-K ENVs. BSRT7 cells expressing HERV-K ENV variants from panel A were examined by Western blotting using an anti-HERVK ENV antibody. The positions of molecular mass markers are shown. Upper band, unprocessed ENV; lower band, TM subunit. (C) Cell-cell fusion assay. BSRT7 cells were transfected as in panel B and then treated with buffer at the indicated pH. (Right) Magnification (×3) of the boxed regions from pH 5.6-treated cells to show syncytia. Syncytia are visible in cells transfected with both envelopes at pH 5.6, but not at pH 5.8 or 7.4. (D) Cell fusion assay with envelopes lacking furin cleavage. BSRT7 cells were transfected with the indicated envelopes, and the cell fusion assay was performed as described for panel C with buffer at pH 5.4. NucBlue nuclear stain (blue) was added to ease visualization of syncytia. Syncytia are visible for HERVK and HERVK+, but not for HERVK-CS− or HERVK+-CS−.
To determine whether the expressed ENVs were fusogenic, transfected cells were exposed to the indicated pH, and cell-cell fusion was visualized by microscopy (Fig. 1C). Cell-cell fusion, as evidenced by syncytium formation, required an acidic pH of 5.6 for both HERVK and HERVK+. The extents of fusion were similar for HERVK and HERVK+ and were abrogated by mutation of the furin cleavage site (Fig. 1D). Collectively, these data demonstrate that proteolytic processing and acidic pH are required for HERV-K ENV-catalyzed membrane fusion, whereas its autologous cytoplasmic tail is not critical for fusion.
HERV-K ENV is incorporated into infectious VSV particles.
Retroviral envelopes are the major determinant of viral entry and receptor usage (47). To study HERV-K ENV in the context of an infectious virus, therefore, we generated a recombinant VSV in which the glycoprotein G was replaced by HERV-K ENV (VSV-HERVK) (Fig. 2A). The resulting virus grew to titers of 3 × 105 PFU ml−1 and was markedly attenuated in its growth in cells compared to wild-type (WT) VSV, as also evidenced by its small-plaque phenotype (Fig. 2A). Replacement of the cytoplasmic tail of HERVK with that of VSV-G to produce VSV-HERVK+ increased the viral yield to 3 × 107 PFU ml−1 and increased the plaque size (Fig. 2A). The relative infectivity of the viruses was determined by measuring the infectious titer per microgram of protein of purified virions. WT VSV (1.1 × 109 PFU μg−1) was 37-fold more infectious per microgram than VSV-HERVK (2.9 ×107 PFU μg−1) and 3-fold more infectious than VSV-HERVK+ (3.3 × 108 PFU μg−1). This suggests that the HERV-K variants produce more defective particles, likely reflecting differences in glycoprotein incorporation.
FIG 2.
Generation and characterization of VSV-HERVK and VSV-HERVK+. (A) Genomic organization of VSV-eGFP, VSV-HERVK, and VSV-HERVK+. The negative-sense RNA genomes are shown in the 3′-to-5′ orientation. The five viral genes are shown for VSV-eGFP: N, nucleocapsid; P, phosphoprotein; M, matrix; G, glycoprotein; L, large polymerase. The noncoding leader (le) and trailer (tr) genomic regions are also indicated. Each virus contains the eGFP gene, which serves as a marker for infection. To generate VSV-HERVK and VSV-HERVK+, the glycoprotein gene of VSV-eGFP was replaced with the gene encoding the HERVK or HERVK+ENV, respectively. Viruses generated from these genomic plasmids were replication competent. Representative plaque assays in BSRT7 cells are shown for each virus, as well as the endpoint titers. (B) SDS-PAGE analysis of purified virions. (Left) Sypro Ruby-stained gel showing all the viral proteins. Molecular mass markers are indicated on the left, and the positions of viral proteins are indicated on the right. For both VSV-HERVK and VSV-HERVK+, the SU subunit is indicated (approximately 50 kDa). TM runs in the same position as VSV N (approximately 42 kDa). (Right) Western blots (WB) probed with antibodies against HERV-K TM, VSV-M, and VSV-G cytoplasmic tail. The lower band in the bottom right gel corresponds to the TM of HERVK+, which reacts with the VSV-G cytoplasmic-tail antibody. (C) Electron micrographs of purified particles stained with 1% PTA. Glycoprotein spikes are visible coating the surfaces of the virions. (Left) Magnified portions of the virions corresponding to the boxed areas on the right.
Using SDS-PAGE and Western blotting of purified virions, we show that HERVK and HERVK+ ENVs are present in purified VSV virions (Fig. 2B). As expected, HERVK incorporation appears less efficient than that of HERVK+, as evidenced by their relative abundances compared to internal components of viral particles (Fig. 2B). Electron micrographs of purified virions showed that the particles have a morphology consistent with that of WT VSV and show visible ENV spikes on the surfaces of both viruses (Fig. 2C). Since HERVK+ displays cell-cell fusion properties similar to those of HERVK and the recombinant VSV-HERVK+ yields a 100-fold increase in infectious virus, we elected to use the virus to further characterize the entry pathway mediated by HERV-K.
HERV-K ENV-mediated cell entry requires endosomal acidification and dynamin.
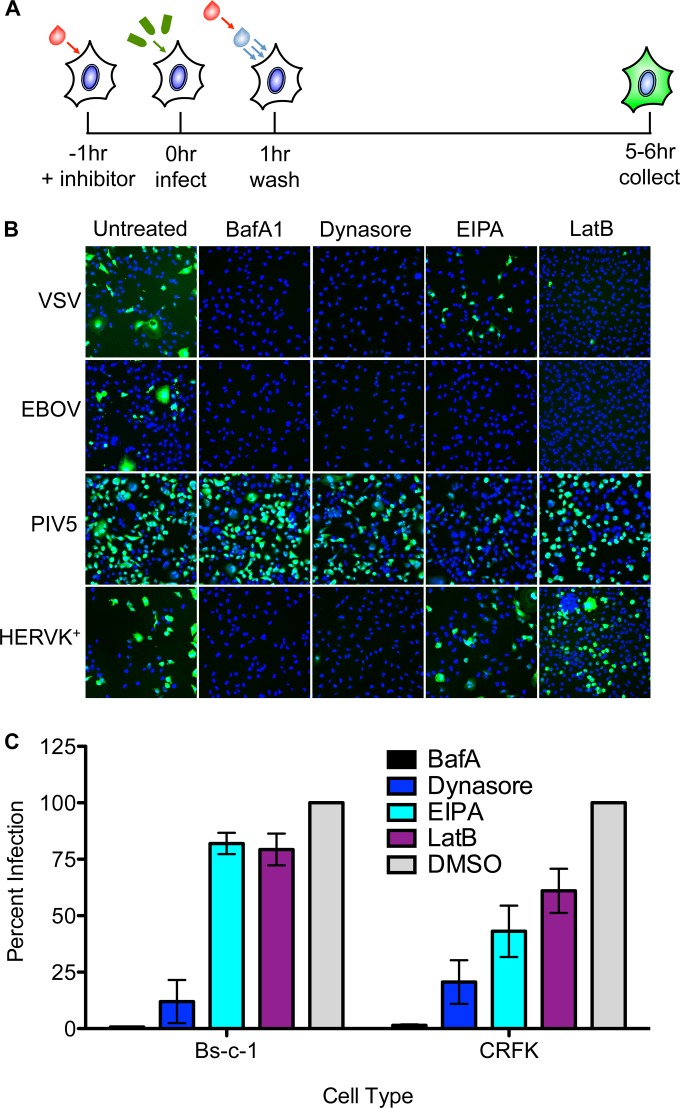
We next investigated whether HERV-K requires endocytic uptake to mediate productive infection. For this purpose, we chose BafA1, an inhibitor of the vacuolar ATPase that prevents endosomal acidification; dynasore, an inhibitor of the GTPase dynamin required for membrane scission; EIPA, an inhibitor of macropinocytosis; and LatB, an inhibitor of actin polymerization. We compared the effects of those inhibitors on HERV-K ENV-mediated infection with their effects on a selected panel of recombinant VSV with known entry pathways. Specifically, we chose WT VSV, which enters via clathrin-mediated endocytosis and requires actin to complete the process of internalization and is thus sensitive to BafA1, dynasore, and LatB (40, 48). VSV-EBOV, which bears the Ebola virus glycoprotein, enters via a dynamin-dependent macropinocytic pathway, rendering its infection sensitive to BafA1, EIPA, dynasore, and LatB (49). In contrast, VSV-PIV5, which bears the F and HN glycoproteins of parainfluenzavirus 5 and fuses at the plasma membrane (50), should be insensitive to any of the inhibitors. We previously demonstrated that VSV virions bearing nonfunctional glycoproteins with single point mutations in the fusion loop are completely noninfectious, indicating that the background level of infectivity in these experiments is negligible (51). We measured infection of Bs-c-1 cells by the expression of eGFP expressed from the respective viral genomes and visualized by fluorescence microscopy (Fig. 3A). As expected, cells infected with VSV, VSV-EBOV, and VSV-PIV5 exhibited the anticipated sensitivity to the various inhibitors (Fig. 3B). Infection of cells by VSV-HERVK+ was sensitive to BafA1 and dynasore, but not to EIPA or LatB, indicating that HERV-K ENV-mediated entry requires endosomal acidification and dynamin but does not require macropinocytosis or actin polymerization. The LatB insensitivity of VSV-HERVK+ contrasts with the LatB sensitivity of VSV and other VSV pseudotypes that depend upon clathrin-mediated endocytosis for their uptake, where the size of the particle dictates a need for the actin machinery to complete the process of internalization (48).
FIG 3.
HERV-K ENV-mediated entry requires endosomal acidification and dynamin. (A) Schematic of the experimental procedure. Cells were pretreated with the indicated endocytic inhibitors and infected with VSV bearing the indicated glycoproteins at an MOI of 1 in the presence of inhibitor. The cells were washed 1 h postinfection and incubated in the presence of medium with inhibitors prior to fixation at 6 to 8 h postinfection. (B and C) eGFP expression was monitored by fluorescence microscopy (B) or flow cytometry (C). (B) Fluorescence microscopy of GFP-expressing infected cells (green). Cell nuclei were stained with DAPI (blue). (C) Inhibition of VSV-HERVK+ infection in Bs-c-1 and CRFK cell lines, measured by flow cytometry. The percent GFP-positive infected cells were normalized to vehicle control (DMSO)-treated cells. Approximately 50% of the cells were infected in the DMSO-treated control group for both cell lines. The errors bars represent standard errors of the mean.
To determine if dependence upon endosomal acidification and dynamin is specific to Bs-c-1 cells or whether it is a general requirement for HERV-K entry, we tested the sensitivity of VSV-HERVK+ to these inhibitors in CRFK cells, a feline kidney cell line that is known to support infection by viruses bearing HERV-K ENV (16, 46). Bs-c-1 and CRFK cells were treated and infected with VSV-HERVK+ as described above, and eGFP expression was measured by flow cytometry (Fig. 3C). The results in the Bs-c-1 cells mirror those of the previous experiment, with a decrease in infectivity in cells treated with BafA1 and dynasore but not in cells treated with EIPA or LatB. In CRFK cells, the virus remained sensitive to BafA1 and dynasore and exhibited partial sensitivity to EIPA and LatB. This may reflect a difference in uptake between the two cell types but confirms the requirement for dynamin and the vacuolar ATPase in uptake.
While there are many advantages to using recombinant VSV to study virus entry, retroviruses are spherical and incorporate smaller amounts of their respective envelope proteins. Previous work has shown that a functional HERV-K ENV can mediate infection of pseudotyped lentiviruses (46). We therefore elected to use pseudotypes of the retrovirus HIV-1 to investigate whether HERV-K ENV dictates the same entry requirements for lentiviruses as for VSV. For this purpose, we also selected CRFK cells, as they lack any postentry block to HIV-1 infection (52). The cells were treated with the endocytic inhibitors and infected with HIV-1 pseudotypes bearing VSV-G, HERVK, or HERVK+. Cells were collected 3 days postinfection, and eGFP expression was analyzed by flow cytometry (Fig. 4A). The trends in sensitivity to the inhibitors were consistent with the recombinant VSV experiments, with the exception that VSV-G was no longer sensitive to LatB (Fig. 4B). Since lentiviruses are spherical and their diameter (120 nm) is less than that of a clathrin-coated pit (53), in contrast to VSV, uptake of these particles should not require actin to complete the process of internalization (48) and therefore will be insensitive to LatB treatment. Both HERVK and HERVK+ were sensitive to BafA1 and dynasore. The HERVK+-pseudotyped lentiviruses did not show the same magnitude of sensitivity to dynasore treatment as VSV-HERVK+; however, this may be due to the differences in timing of the experiments necessitated by the inherent temporal differences between entry and gene expression in the two viral systems. Together, these data indicate that the HERV-K envelope mediates entry via a pathway that requires endosomal acidification and dynamin.
FIG 4.
Retroviral particles bearing HERV-K ENV require endosomal acidification and dynamin. (A) Schematic of the experimental procedure. CRFK cells were treated with inhibitors and infected with pseudotyped lentiviruses bearing the indicated glycoproteins. The cells were washed 6 h postinfection, and medium with ammonium chloride was added to prevent entry of any surface-bound particles. Cells were collected 3 days postinfection, and GFP expression was analyzed by flow cytometry. (B) Effects of endocytic inhibitors on infection by pseudotyped lentiviruses. The percentage of GFP-positive infected cells was normalized to vehicle control (DMSO)-treated cells. Approximately 5% of the cells were infected in the DMSO-treated cells for each virus. The error bars represent standard errors of the mean.
HERV-K ENV has broad species tropism.
Beta-type ENVs are known to have restricted species tropism based on receptor usage; however, the range of cell types tested has been limited to those in which specific retroviruses can replicate (19, 22–26). Since the VSV core can replicate in all eukaryotic cells in culture, VSV-HERVK provides a unique reagent to interrogate more broadly the ability of HERV-K ENV to mediate entry into those cells.
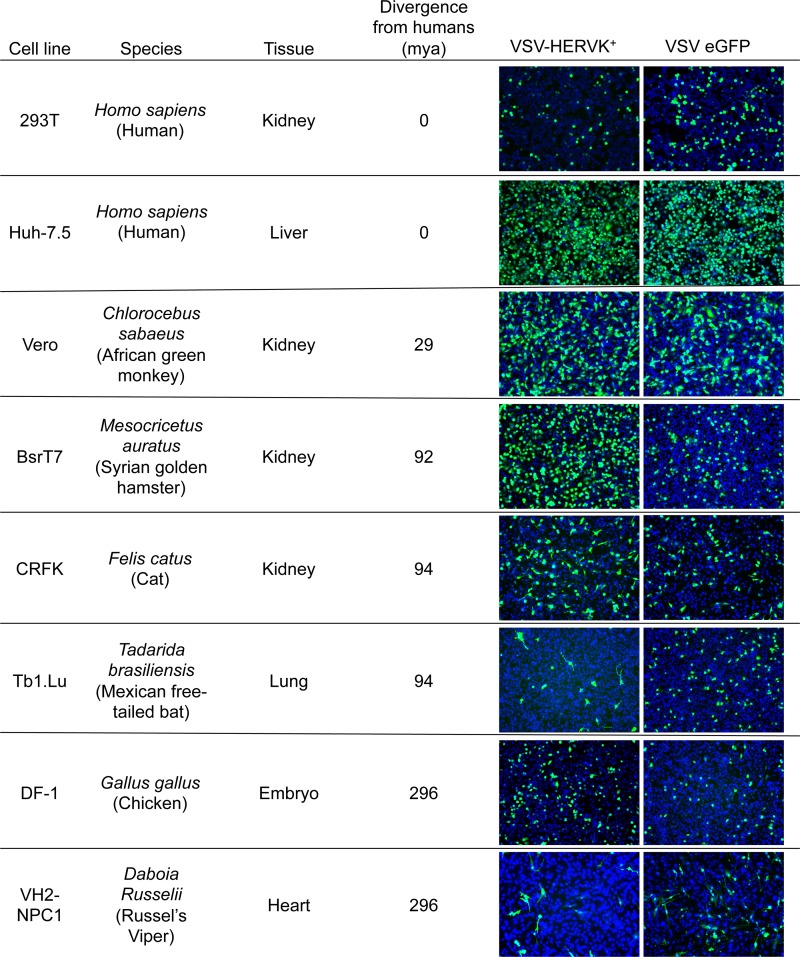
Cell lines from a variety of species and tissues, representing the major clades of amniotes (from reptile to human), were infected with VSV-HERVK+ or VSV-eGFP. They included the human kidney and liver cell lines 293T and Huh-7.5, an African green monkey kidney cell line (Vero), a Syrian golden hamster kidney cell line (BSRT7), a cat kidney cell line (CRFK), a Mexican free-tailed bat lung cell line (Tb1.Lu), a chicken embryo cell line (DF-1), and a Russel's viper heart cell line (VH2-NPC1). The cells were infected with VSV-HERVK or VSV-eGFP at an MOI of 1 (based on titers from Vero cells) and imaged by fluorescence microscopy. HERV-K ENV mediated infection of all the mammalian cell lines tested (27) (Fig. 5). VSV-HERVK+ was also capable of infecting the nonmammalian DF-1 (chicken) and VH2 (viper) cell lines (Fig. 5). The cell lines tested also spanned several tissue types, indicating that HERV-K ENV also has broad tissue tropism. These results indicate that HERV-K ENV imparts broad species tropism to the virus and suggest either that the entry pathway dictated by HERV-K ENV is evolutionarily conserved throughout amniotes or that it can utilize multiple, redundant pathways.
FIG 5.
HERV-K ENV imparts broad tropism. Cell lines from a variety of species spanning multiple classes of animals and tissue types were infected with VSV-HERVK+ and VSV-eGFP at an MOI of 1 based on the titer in Vero cells. Infection was detected by expression of eGFP (green), and cells were stained with DAPI (blue). Cell line species are indicated with Latin and common (in parentheses) names, along with the source tissue for the cells and the time of divergence from the last common ancestor between humans and the indicated species in millions of years (mya).
DISCUSSION
The major findings of this study are (i) that membrane fusion mediated by HERV-K ENV requires its proteolytic processing and is triggered by acidic pH; (ii) that virions bearing HERV-K ENV are endocytosed through a dynamin-dependent, actin-independent pathway; and (iii) that HERV-K ENV mediates entry into a broad range of cell types in culture. These findings imply that the presence of HERV-K in a limited number of species is unrelated to the tropism of the envelope protein itself and that the productive uptake pathway is not via clathrin-dependent endocytosis.
We determined that the HERV-K envelope requires two distinct cellular processes to initiate membrane fusion (Fig. 1). The first is proteolytic processing, likely by furin-like proteases in the producer cell, to enter a fusion-competent state. In a second step, fusion is triggered in the target cell upon exposure to endosomal pH. These requirements match those for extant beta-type envelopes, suggesting the properties are both ancient and conserved within this envelope class (19–21). Replacing the cytoplasmic tail of HERV-K ENV with that of VSV-G had no apparent impact on the ability of the envelope to be cleaved by cellular proteases or to mediate fusion at acidic pH. This need for acidic pH to trigger HERV-K-mediated membrane fusion implies that surface expression of HERV-K ENV alone is insufficient for cell-cell fusion, suggesting that if there is a physiologic role for HERV-K ENV involving membrane fusion, an additional trigger is required.
Chemical inhibitor studies using infectious VSV and lentiviral pseudotypes demonstrated that HERV-K entry is dependent upon endosomal uptake, dynamin-mediated membrane scission, and endosomal acidification (Fig. 3 and 4). The route of entry is independent of macropinocytosis and actin polymerization. Since the morphology of the VSV particle dictates the requirement for actin to complete the process of internalization when entering via a clathrin-mediated process (40, 48), the lack of a requirement for actin polymerization indicates that HERV-K ENV does not enter via the same clathrin-mediated entry pathway as VSV. Together, these data suggest that HERV-K ENV dictates an entry pathway that is dynamin dependent but clathrin and macropinocytosis independent, which is distinct from what we have observed with other recombinant VSVs (49, 54). Available evidence supports the notion that MMTV also enters cells via clathrin-independent endocytic uptake; however, the requirement for dynamin or macropinocytosis is unknown (19).
For HERV-K ENV-mediated infection, we observed a dependence on dynamin and endosomal acidification in all cell types tested but found variation in sensitivity to inhibitors of macropinocytosis and actin polymerization. Although we do not know the basis for this difference, it is possible that HERV-K may exhibit different entry requirements in different cell types or that it has a flexible entry route, potentially utilizing multiple cellular uptake pathways. Lentiviral pseudotypes bearing both the full-length and cytoplasmic-tail-switched ENVs show the same entry requirements as VSV-HERVK+, indicating that VSV-HERVK+ faithfully recapitulates the entry pathway of HERV-K in the context of a retrovirus. Moreover, this shows that switching the cytoplasmic tail of HERV-K ENV does not affect the basic properties of the envelope, suggesting that the sequence of the cytoplasmic tail is not essential for its entry-mediating functionality.
Understanding ERV envelope tropism at both the cellular and species levels underlies efforts to determine the origins of the extant progenitor virus and the series of events that culminated in its presence in the genome. Unlike the relatively narrow species tropism of extant beta-type envelopes, HERV-K ENV imparted exceptionally broad species and tissue tropism. These data extend a recent report that also found broad tropism of HERV-K ENV in mammalian cells (27) and provide an example of a beta-type envelope that is capable of mediating infection of nonmammalian cells. The broad tropism in mammals is consistent with the known distribution of exogenous betaretroviruses and endogenous betaretrovirus-like sequences in a wide range of mammals. However, while there are ERVs in birds and reptiles that have betaretrovirus-like gag-pol regions, the env genes of these ERVs are more similar to gamma-type env genes, and no true beta-type env has been identified outside of mammals (18, 28–31). This suggests that restricted tropism is not a general property of beta-type envelopes and that the lack of endogenous beta-type envelopes outside of mammals is not explained solely by ENV tropism.
The broad tropism and entry requirements of HERV-K ENV are reminiscent of foamy viruses. Foamy viruses, belonging to the retrovirus genus Spumavirus, also have exceptionally broad ENV-mediated tissue and species tropism. The prototype foamy virus (PFV) ENV is capable of mediating infection of cells from mammals, birds, and reptiles, including cells from many different tissue types (55–57). Most foamy virus ENVs also require acidic pH for fusion, reaching maximal cell-cell fusion at approximately pH 5.5 (58). While the specific route of uptake is unknown, all foamy viruses tested require endosomal acidification to mediate viral entry (58). Further investigation will be required to determine whether HERV-K and foamy viruses enter cells via similar mechanisms.
The evidence presented here suggests that HERV-K(HML-2) ENV utilizes host factors that are ubiquitously expressed in cultured cells and highly conserved across 300 million years of evolution. While multiple independent HERV-K infection events have resulted in endogenized viruses, we found no evidence for an ENV-mediated requirement for gamete or gamete progenitor cells. Rather, the broad cellular tropism of the virus likely accounts for its propensity to become endogenized. The absence of HERV-K(HML-2) sequences outside of Old World primate genomes cannot, therefore, be explained by ENV-dictated tropism. The restriction factors TRIM5α and tetherin appear to be ineffective against HERV-K, although it is sensitive to APOBEC 3F (16, 59). Perhaps the restricted presence of HERV-K(HML-2) is achieved by other restriction factors, or it may reflect the stochastic nature of endogenization.
HERV-K(HML-2) exists in the human genome as many copies, which were likely derived from multiple insertion events (2). Here, we test a single HERV-K clone, Phoenix (17). This clone was generated as a putative ancestor to the human-specific HERV-K elements, suggesting that the properties that we describe here are likely conserved among HERV-K elements. Consistent with this work, a study using a different reconstructed ancestral HERV-K ENV also demonstrated broad tropism (27). It is possible, however, that the tropism and/or entry pathway may not be shared by all HERV-K ENVs, and additional work will be required to resolve this.
In addition to the knowledge gained regarding the entry pathway mediated by HERV-K ENV and the host range and tissue tropism, the present study has provided new tools and reagents that should aid in further dissection of the entry pathway of HERV-K ENV, which may shed light on any potential physiological role of the envelope. The generation of an autonomously replicating virus that depends upon HERV-K ENV for infection (VSV-HERVK+) and grows to high titer in cell culture will likely prove invaluable in genetic screens to define the viral and host determinants of HERV-K-mediated infection. This VSV-HERVK virus may also serve as a useful reagent for structural studies of the HERV-K glycoprotein.
ACKNOWLEDGMENTS
We thank S. Piccinotti for helpful discussions and K. R. McCarthy for critical comments on the manuscript.
This work was funded by NIH grants AI081842 and AI109740 to S.P.J.W. S.P.J.W. is a Burroughs Wellcome investigator in the pathogenesis of infectious disease. L.R. is the recipient of an Albert J. Ryan graduate student fellowship.
REFERENCES
- 1.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann Y, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Raymond C, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowki J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ, Szustakowki J, International Human Genome Sequencing Consortium. 2001. Initial sequencing and analysis of the human genome. Nature 409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Subramanian RP, Wildschutte JH, Russo C, Coffin JM. 2011. Identification, characterization, and comparative genomic distribution of the HERV-K (HML-2) group of human endogenous retroviruses. Retrovirology 8:90. doi: 10.1186/1742-4690-8-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steinhuber S, Brack M, Hunsmann G, Schwelberger H, Dierich MP, Vogetseder W. 1995. Distribution of human endogenous retrovirus HERV-K genomes in humans and different primates. Hum Genet 96:188–192. doi: 10.1007/BF00207377. [DOI] [PubMed] [Google Scholar]
- 4.Reus K, Mayer J, Sauter M, Zischler H, Muller-Lantzsch N, Meese E. 2001. HERV-K(OLD): ancestor sequences of the human endogenous retrovirus family HERV-K(HML-2). J Virol 75:8917–8926. doi: 10.1128/JVI.75.19.8917-8926.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jha AR, Nixon DF, Rosenberg MG, Martin JN, Deeks SG, Hudson RR, Garrison KE, Pillai SK. 2011. Human endogenous retrovirus K106 (HERV-K106) was infectious after the emergence of anatomically modern humans. PLoS One 6:e20234. doi: 10.1371/journal.pone.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marchi E, Kanapin A, Magiorkinis G, Belshaw R. 2014. Unfixed endogenous retroviral insertions in the human population. J Virol 88:9529–9537. doi: 10.1128/JVI.00919-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turner G, Barbulescu M, Su M, Jensen-Seaman MI, Kidd KK, Lenz J. 2001. Insertional polymorphisms of full-length endogenous retroviruses in humans. Curr Biol 11:1531–1535. doi: 10.1016/S0960-9822(01)00455-9. [DOI] [PubMed] [Google Scholar]
- 8.Boller K, Schonfeld K, Lischer S, Fischer N, Hoffmann A, Kurth R, Tonjes RR. 2008. Human endogenous retrovirus HERV-K113 is capable of producing intact viral particles. J Gen Virol 89:567–572. doi: 10.1099/vir.0.83534-0. [DOI] [PubMed] [Google Scholar]
- 9.Lower R, Lower J, Frank H, Harzmann R, Kurth R. 1984. Human teratocarcinomas cultured in vitro produce unique retrovirus-like viruses. J Gen Virol 65:887–898. doi: 10.1099/0022-1317-65-5-887. [DOI] [PubMed] [Google Scholar]
- 10.Simpson GR, Patience C, Lower R, Tonjes RR, Moore HD, Weiss RA, Boyd MT. 1996. Endogenous D-type (HERV-K) related sequences are packaged into retroviral particles in the placenta and possess open reading frames for reverse transcriptase. Virology 222:451–456. doi: 10.1006/viro.1996.0443. [DOI] [PubMed] [Google Scholar]
- 11.Contreras-Galindo R, Kaplan MH, Leissner P, Verjat T, Ferlenghi I, Bagnoli F, Giusti F, Dosik MH, Hayes DF, Gitlin SD, Markovitz DM. 2008. Human endogenous retrovirus K (HML-2) elements in the plasma of people with lymphoma and breast cancer. J Virol 82:9329–9336. doi: 10.1128/JVI.00646-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Contreras-Galindo R, Kaplan MH, Contreras-Galindo AC, Gonzalez-Hernandez MJ, Ferlenghi I, Giusti F, Lorenzo E, Gitlin SD, Dosik MH, Yamamura Y, Markovitz DM. 2012. Characterization of human endogenous retroviral elements in the blood of HIV-1-infected individuals. J Virol 86:262–276. doi: 10.1128/JVI.00602-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Contreras-Galindo R, Kaplan MH, Dube D, Gonzalez-Hernandez MJ, Chan S, Meng F, Dai M, Omenn GS, Gitlin SD, Markovitz DM. 2015. Human endogenous retrovirus type K (HERV-K) particles package and transmit HERV-K-related sequences. J Virol 89:7187–7201. doi: 10.1128/JVI.00544-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhardwaj N, Montesion M, Roy F, Coffin JM. 2015. Differential expression of HERV-K (HML-2) proviruses in cells and virions of the teratocarcinoma cell line Tera-1. Viruses 7:939–968. doi: 10.3390/v7030939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dewannieux M, Blaise S, Heidmann T. 2005. Identification of a functional envelope protein from the HERV-K family of human endogenous retroviruses. J Virol 79:15573–15577. doi: 10.1128/JVI.79.24.15573-15577.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee YN, Bieniasz PD. 2007. Reconstitution of an infectious human endogenous retrovirus. PLoS Pathog 3:e10. doi: 10.1371/journal.ppat.0030010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dewannieux M, Harper F, Richaud A, Letzelter C, Ribet D, Pierron G, Heidmann T. 2006. Identification of an infectious progenitor for the multiple-copy HERV-K human endogenous retroelements. Genome Res 16:1548–1556. doi: 10.1101/gr.5565706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henzy JE, Coffin JM. 2013. Betaretroviral envelope subunits are noncovalently associated and restricted to the mammalian class. J Virol 87:1937–1946. doi: 10.1128/JVI.01442-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang E, Obeng-Adjei N, Ying Q, Meertens L, Dragic T, Davey RA, Ross SR. 2008. Mouse mammary tumor virus uses mouse but not human transferrin receptor 1 to reach a low pH compartment and infect cells. Virology 381:230–240. doi: 10.1016/j.virol.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Côté M, Kucharski TJ, Liu S-LL. 2008. Enzootic nasal tumor virus envelope requires a very acidic pH for fusion activation and infection. J Virol 82:9023–9034. doi: 10.1128/JVI.00648-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bertrand P, Côté M, Zheng Y-MM, Albritton LM, Liu S-LL. 2008. Jaagsiekte sheep retrovirus utilizes a pH-dependent endocytosis pathway for entry. J Virol 82:2555–2559. doi: 10.1128/JVI.01853-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang E, Albritton L, Ross SR. 2006. Identification of the segments of the mouse transferrin receptor 1 required for mouse mammary tumor virus infection. J Biol Chem 281:10243–10249. doi: 10.1074/jbc.M511572200. [DOI] [PubMed] [Google Scholar]
- 23.Ross SR, Schofield JJ, Farr CJ, Bucan M. 2002. Mouse transferrin receptor 1 is the cell entry receptor for mouse mammary tumor virus. Proc Natl Acad Sci U S A 99:12386–12390. doi: 10.1073/pnas.192360099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Demogines A, Abraham J, Choe H, Farzan M, Sawyer SL. 2013. Dual host-virus arms races shape an essential housekeeping protein. PLoS Biol 11:e1001571. doi: 10.1371/journal.pbio.1001571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dirks C, Duh F-MM, Rai SK, Lerman MI, Miller AD. 2002. Mechanism of cell entry and transformation by enzootic nasal tumor virus. J Virol 76:2141–2149. doi: 10.1128/jvi.76.5.2141-2149.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Hoeven NS, Miller AD. 2005. Improved enzootic nasal tumor virus pseudotype packaging cell lines reveal virus entry requirements in addition to the primary receptor Hyal2. J Virol 79:87–94. doi: 10.1128/JVI.79.1.87-94.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kramer P, Lausch V, Volkwein A, Hanke K, Hohn O, Bannert N. 2016. The human endogenous retrovirus K(HML-2) has a broad envelope-mediated cellular tropism and is prone to inhibition at a post-entry, pre-integration step. Virology 487:121–128. doi: 10.1016/j.virol.2015.10.014. [DOI] [PubMed] [Google Scholar]
- 28.Henzy JE, Johnson WE. 2013. Pushing the endogenous envelope. Philos Trans R Soc Lond B Biol Sci 368:20120506. doi: 10.1098/rstb.2012.0506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huder JB, Böni J, Hatt J-M, Soldati G, Lutz H, Schüpbach J. 2002. Identification and characterization of two closely related unclassifiable endogenous retroviruses in pythons (Python molurus and Python curtus). J Virol 76:7607–7615. doi: 10.1128/JVI.76.15.7607-7615.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henzy JE, Gifford RJ, Johnson WE, Coffin JM. 2014. A novel recombinant retrovirus in the genomes of modern birds combines features of avian and mammalian retroviruses. J Virol 88:2398–2405. doi: 10.1128/JVI.02863-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bolisetty M, Blomberg J, Benachenhou F, Sperber G. 2012. Unexpected diversity and expression of avian endogenous retroviruses. mBio 3:e00344-12. doi: 10.1128/mBio.00344-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blight KJ, McKeating JA, Rice CM. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J Virol 76:13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buchholz UJ, Finke S, Conzelmann KK. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J Virol 73:251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller EH, Obernosterer G, Raaben M, Herbert AS, Deffieu MS, Krishnan A, Ndungo E, Sandesara RG, Carette JE, Kuehne AI, Ruthel G, Pfeffer SR, Dye JM, Whelan SP, Brummelkamp TR, Chandran K. 2012. Ebola virus entry requires the host-programmed recognition of an intracellular receptor. EMBO J 31:1947–1960. doi: 10.1038/emboj.2012.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wong AC, Sandesara RG, Mulherkar N, Whelan SP, Chandran K. 2010. A forward genetic strategy reveals destabilizing mutations in the Ebolavirus glycoprotein that alter its protease dependence during cell entry. J Virol 84:163–175. doi: 10.1128/JVI.01832-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Whelan SP, Ball LA, Barr JN, Wertz GT. 1995. Efficient recovery of infectious vesicular stomatitis virus entirely from cDNA clones. Proc Natl Acad Sci U S A 92:8388–8392. doi: 10.1073/pnas.92.18.8388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whelan SP, Barr JN, Wertz GW. 2000. Identification of a minimal size requirement for termination of vesicular stomatitis virus mRNA: implications for the mechanism of transcription. J Virol 74:8268–8276. doi: 10.1128/JVI.74.18.8268-8276.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fuerst TR, Niles EG, Studier FW, Moss B. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci U S A 83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lyles DS, Puddington L, McCreedy BJ Jr. 1988. Vesicular stomatitis virus M protein in the nuclei of infected cells. J Virol 62:4387–4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cureton DK, Massol RH, Saffarian S, Kirchhausen TL, Whelan SP. 2009. Vesicular stomatitis virus enters cells through vesicles incompletely coated with clathrin that depend upon actin for internalization. PLoS Pathog 5:e1000394. doi: 10.1371/journal.ppat.1000394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ricks DM, Kutner R, Zhang XY, Welsh DA, Reiser J. 2008. Optimized lentiviral transduction of mouse bone marrow-derived mesenchymal stem cells. Stem Cells Dev 17:441–450. doi: 10.1089/scd.2007.0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang XY, La Russa VF, Reiser J. 2004. Transduction of bone-marrow-derived mesenchymal stem cells by using lentivirus vectors pseudotyped with modified RD114 envelope glycoproteins. J Virol 78:1219–1229. doi: 10.1128/JVI.78.3.1219-1229.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson JE, Rodgers W, Rose JK. 1998. A plasma membrane localization signal in the HIV-1 envelope cytoplasmic domain prevents localization at sites of vesicular stomatitis virus budding and incorporation into VSV virions. Virology 251:244–252. doi: 10.1006/viro.1998.9429. [DOI] [PubMed] [Google Scholar]
- 44.Johnson JE, Schnell MJ, Buonocore L, Rose JK. 1997. Specific targeting to CD4+ cells of recombinant vesicular stomatitis viruses encoding human immunodeficiency virus envelope proteins. J Virol 71:5060–5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Owens RJ, Rose JK. 1993. Cytoplasmic domain requirement for incorporation of a foreign envelope protein into vesicular stomatitis virus. J Virol 67:360–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hanke K, Kramer P, Seeher S, Beimforde N, Kurth R, Bannert N. 2009. Reconstitution of the ancestral glycoprotein of human endogenous retrovirus K and modulation of its functional activity by truncation of the cytoplasmic domain. J Virol 83:12790–12800. doi: 10.1128/JVI.01368-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lindemann D, Steffen I, Pohlmann S. 2013. Cellular entry of retroviruses. Adv Exp Med Biol 790:128–149. doi: 10.1007/978-1-4614-7651-1_7. [DOI] [PubMed] [Google Scholar]
- 48.Cureton DK, Massol RH, Whelan SP, Kirchhausen T. 2010. The length of vesicular stomatitis virus particles dictates a need for actin assembly during clathrin-dependent endocytosis. PLoS Pathog 6:e1001127. doi: 10.1371/journal.ppat.1001127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mulherkar N, Raaben M, de la Torre JC, Whelan SP, Chandran K. 2011. The Ebola virus glycoprotein mediates entry via a non-classical dynamin-dependent macropinocytic pathway. Virology 419:72–83. doi: 10.1016/j.virol.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith EC, Popa A, Chang A, Masante C, Dutch RE. 2009. Viral entry mechanisms: the increasing diversity of paramyxovirus entry. FEBS J 276:7217–7227. doi: 10.1111/j.1742-4658.2009.07401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stanifer ML, Cureton DK, Whelan SP. 2011. A recombinant vesicular stomatitis virus bearing a lethal mutation in the glycoprotein gene uncovers a second site suppressor that restores fusion. J Virol 85:8105–8115. doi: 10.1128/JVI.00735-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McEwan WA, Schaller T, Ylinen LM, Hosie MJ, Towers GJ, Willett BJ. 2009. Truncation of TRIM5 in the Feliformia explains the absence of retroviral restriction in cells of the domestic cat. J Virol 83:8270–8275. doi: 10.1128/JVI.00670-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gonda MA, Wong-Staal F, Gallo RC, Clements JE, Narayan O, Gilden RV. 1985. Sequence homology and morphologic similarity of HTLV-III and visna virus, a pathogenic lentivirus. Science 227:173–177. doi: 10.1126/science.2981428. [DOI] [PubMed] [Google Scholar]
- 54.Piccinotti S, Kirchhausen T, Whelan SP. 2013. Uptake of rabies virus into epithelial cells by clathrin-mediated endocytosis depends upon actin. J Virol 87:11637–11647. doi: 10.1128/JVI.01648-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hill CL, Bieniasz PD, McClure MO. 1999. Properties of human foamy virus relevant to its development as a vector for gene therapy. J Gen Virol 80:2003–2009. doi: 10.1099/0022-1317-80-8-2003. [DOI] [PubMed] [Google Scholar]
- 56.Mergia A, Leung NJ, Blackwell J. 1996. Cell tropism of the simian foamy virus type 1 (SFV-1). J Med Primatol 25:2–7. doi: 10.1111/j.1600-0684.1996.tb00185.x. [DOI] [PubMed] [Google Scholar]
- 57.Russell DW, Miller AD. 1996. Foamy virus vectors. J Virol 70:217–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Picard-Maureau M, Jarmy G, Berg A, Rethwilm A, Lindemann D. 2003. Foamy virus envelope glycoprotein-mediated entry involves a pH-dependent fusion process. J Virol 77:4722–4730. doi: 10.1128/JVI.77.8.4722-4730.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lemaitre C, Harper F, Pierron G, Heidmann T, Dewannieux M. 2014. The HERV-K human endogenous retrovirus envelope protein antagonizes Tetherin antiviral activity. J Virol 88:13626–13637. doi: 10.1128/JVI.02234-14. [DOI] [PMC free article] [PubMed] [Google Scholar]