ABSTRACT
Human T-cell leukemia virus type 1 (HTLV-1) and type 2 (HTLV-2) are highly related retroviruses that transform T cells in vitro but have distinct pathological outcomes in vivo. HTLV-1 encodes a protein from the antisense strand of its proviral genome, the HTLV-1 basic leucine zipper factor (HBZ), which inhibits Tax-1-mediated viral transcription and promotes cell proliferation, a high proviral load, and persistence in vivo. In adult T-cell leukemia/lymphoma (ATL) cell lines and patient T cells, hbz is often the only viral gene expressed. The antisense strand of the HTLV-2 proviral genome also encodes a protein termed APH-2. Like HBZ, APH-2 is able to inhibit Tax-2-mediated viral transcription and is detectable in most primary lymphocytes from HTLV-2-infected patients. However, unlike HBZ, the loss of APH-2 in vivo results in increased viral replication and proviral loads, suggesting that HBZ and APH-2 modulate the virus and cellular pathways differently. Herein, we examined the effect of APH-2 on several known HBZ-modulated pathways: NF-κB (p65) transactivation, transforming growth factor β (TGF-β) signaling, and interferon regulatory factor 1 (IRF-1) transactivation. Like HBZ, APH-2 has the ability to inhibit p65 transactivation. Conversely, HBZ and APH-2 have divergent effects on TGF-β signaling and IRF-1 transactivation. Quantitative PCR and protein half-life experiments revealed a substantial disparity between HBZ and APH-2 transcript levels and protein stability, respectively. Taken together, our data further elucidate the functional differences between HBZ and APH-2 and how these differences can have profound effects on the survival of infected cells and, ultimately, pathogenesis.
IMPORTANCE Human T-cell leukemia virus type 1 (HTLV-1) and type 2 (HTLV-2) are highly related retroviruses that have distinct pathological outcomes in infected hosts. Functional comparisons of HTLV-1 and HTLV-2 proteins provide a better understanding about how HTLV-1 infection is associated with disease and HTLV-2 infection is not. The HTLV genome antisense-strand genes hbz and aph-2 are often the only viral genes expressed in HTLV-infected T cells. Previously, our group found that HTLV-1 HBZ and HTLV-2 APH-2 had distinct effects in vivo and hypothesized that the differences in the interactions of HBZ and APH-2 with important cell signaling pathways dictate whether cells undergo proliferation, apoptosis, or senescence. Ultimately, these functional differences may affect how HTLV-1 causes disease but HTLV-2 generally does not. In the current study, we compared the effects of HBZ and APH-2 on several HTLV-relevant cellular pathways, including the TGF-β signaling, NF-κB activation, and IRF-1 transactivation pathways.
INTRODUCTION
Human T-cell leukemia virus type 1 (HTLV-1) is a complex oncogenic deltaretrovirus that infects an estimated 15 million to 25 million people worldwide, with areas of endemic infection being found in southwestern Japan, Africa, South America, and the Caribbean Basin (1). Approximately 2 to 5% of HTLV-1-infected individuals develop disease after a long clinical latency period upwards of 4 decades. HTLV-1 is the causative infectious agent of a highly aggressive CD4+ T-cell malignancy, adult T-cell leukemia/lymphoma (ATL) (2, 3), and a neurodegenerative disease, HTLV-1-associated myelopathy/tropical spastic paraparesis (HAM/TSP) (4, 5). ATL is refractory to current chemotherapies, and even aggressive treatments provide only a meager increase in survival of 8 to 10 months (6–8).
Human T-cell leukemia virus type 2 (HTLV-2) is a related retrovirus, sharing a similar genomic structure with HTLV-1. The genomes of both viruses encode the retroviral structural and enzymatic genes (gag, pol, and env), regulatory genes (tax and rex), and accessory genes important for viral infection and persistence (6). HTLV-1 and HTLV-2 are transmitted most efficiently through cell-to-cell contact (9, 10), and both viruses are capable of transforming T cells in vitro (11–15). Despite strong genomic similarities, HTLV-2 has not been closely associated with disease and has been linked to only a few cases of neurological disorders (16–18).
The proviral genomes of HTLV-1 and HTLV-2 encode gene products from their antisense strands. The HTLV-1 basic leucine zipper factor (HBZ) localizes to the nucleus and represses Tax-1 transactivation by binding the cellular cofactors CREB and p300, preventing them from interacting with Tax-1 (19–21). HBZ contains an N-terminal transactivation domain (which is responsible for its effects on p300/CBP), a central modulatory domain, and a C-terminal bZIP domain (which is responsible for its effects on the JunD, JunB, c-Jun, and ATF/CREB proteins) (19–24). Unlike Tax-1, hbz is expressed in all ATL cell lines and in HTLV-1-infected individuals (25, 26). Studies using infectious molecular clones deficient in HBZ protein expression revealed that HBZ silencing had no effect on HTLV-1 in vitro immortalization (27). However, using the rabbit model of infection, HBZ was required for efficient HTLV-1 infection and persistence (27). These studies and others have provided evidence that HBZ is a secondary oncogene that plays a key role in cell proliferation (25, 26, 28, 29) and cell survival (29, 30). The antisense-strand protein of HTLV-2 (APH-2) has been detected in most HTLV-2-infected samples (31, 32). Like HBZ, APH-2 is a nuclear protein that represses Tax-2 transactivation through its interaction with CREB (32, 33). APH-2 lacks an activation domain and a canonical bZIP domain; however, it has a noncanonical bZIP region (which is responsible for its interactions and effects on c-Jun and JunB) and a C-terminal CREB-binding motif (which is responsible for its interactions with CREB) (32–34). Studies with infectious molecular clones deficient in APH-2 protein expression revealed that, like the effect of HBZ silencing on HTLV-1, APH-2 silencing had no effect on HTLV-2 in vitro immortalization (33). In contrast, using a rabbit model of infection, APH-2 was found to be dispensable for HTLV-2 infection and persistence. Interestingly, the APH-2-knockout virus was able to replicate significantly better than wild-type HTLV-2 in rabbits, which suggested that APH-2 dampens HTLV-2 replication in vivo (33).
Comparative studies of the HTLV-1 and HTLV-2 gene products have allowed a better understanding of the mechanisms of disease development associated with HTLV-1 infection. Indeed, studies comparing HTLV-1 Tax-1 and HTLV-2 Tax-2, as well as related accessory gene products, have identified distinct protein domains or activities associated with virus replication and HTLV-induced T-cell transformation (11, 12, 35–41). On the basis of the genomic and general protein similarities between HBZ and APH-2 and given the known differences in their effects on cells, we hypothesized that comparative studies of HBZ and APH-2 would provide a better understanding of the role of HBZ in viral persistence and, potentially, ATL development. A limited number of studies have compared the effects of APH-2 on known HBZ-modulated pathways (the findings are summarized in Table 1). Herein, we expanded those studies to evaluate APH-2 and HBZ modulation of transforming growth factor β (TGF-β) signaling, p65-mediated transactivation, and interferon regulatory factor 1 (IRF-1)-mediated transactivation, to compare how each protein modulates the immune response, affects cell growth, and potentiates the survival of infected cells. Finally, our detailed comparisons of HBZ and APH-2 protein stability and RNA transcript abundance revealed differences in how HBZ and APH-2 act on the cell signaling pathways leading to disease (HTLV-1) or not (HTLV-2) during natural infection in vivo.
TABLE 1.
Overview of HBZ and APH-2 regulatory functions
| Function | Finding for: |
|
|---|---|---|
| HBZ | APH-2 | |
| Expression in HTLV-infected cells | Yes (100%) | Yes (most but not all cases) |
| Required for in vitro immortalization | No | No |
| Required for efficient in vivo infection and persistence | Yes | No |
| HTLV 5′ LTRa transcription | Inhibits | Inhibits |
| Promotion of T-cell proliferation | Yes | No |
| AP-1 transcription | Inhibits | Activates |
| JunD transcription | Activates | Activates |
| TGF-β signaling | Activates | ? |
| Modulation of classical NF-κB pathway | Inhibits | ? |
| Modulation of IRF-1 pathway | Inhibits | ? |
LTR, long terminal repeat.
MATERIALS AND METHODS
Cell lines and culture.
HEK293T and HepG2 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS; Gemini Bio-Products, West Sacramento, CA), 2 mM glutamine, penicillin (100 U/ml), and streptomycin (100 μg/ml). Peripheral blood lymphocyte (PBL) lines (early-passage HTLV-1- and HTLV-2-immortalized peripheral blood mononuclear cells [PBMCs]) were maintained in RPMI 1640 medium supplemented with 20% FBS, 20 U/ml recombinant human interleukin-2 (rhIL-2; Roche Applied Biosciences, Indianapolis, IN), 2 mM glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin. Jurkat cells were maintained in RPMI 1640 medium supplemented with 10% FBS, 2 mM glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin. All cells were grown at 37°C in a humidified atmosphere of 5% CO2 and air. Human PBMCs were isolated using Ficoll-Paque Plus (GE Healthcare Life Sciences, Pittsburgh, PA), and naive T cells were enriched using a Dynabeads Untouched human T-cell kit (Life Technologies, Grand Island, NY) according to the manufacturers' instructions.
Plasmids and cloning.
Plasmid DNA was purified on maxiprep columns according to the manufacturer's protocol (Qiagen, Germantown, MD). The FLAG-tagged HBZ and APH-2 expression vectors had HBZ or APH-2 cDNA inserted downstream of the cytomegalovirus (CMV) promoter and the FLAG-6×His epitope tag for mammalian cell expression of FLAG-tagged HBZ and APH-2 proteins. The S-tagged HBZ and APH-2 expression vectors contained HBZ or APH-2 cDNA inserted into a pTriEx-4 Neo vector (Novagen, Reno, NV) for mammalian cell expression of S-tagged HBZ and APH-2 proteins. The pME-HBZ and pME-APH-2 cDNA expression plasmids were generated and described previously (27, 33). The p65 expression plasmid and κB luciferase (luc) reporter plasmid were described previously (42). The TGF-β-responsive luciferase reporter plasmid 9×CAGA-luc was kindly provided by Masao Matsuoka (Kyoto University, Kyoto, Japan) and described previously (43). The IRF-1 expression plasmid and IRF-1 luciferase reporter plasmid (IRF-1–luc) were graciously provided by John Yim (Beckman Research Institute, Duarte, CA) and described previously (44). The transfection efficiency control plasmid TK-renilla was described previously (41). HBZ and APH-2 cDNAs were cloned into the pCDH-CMV-MCS-EF1-copGFP lentiviral expression vector (System Biosciences [SBI], Mountain View, CA) to create pCDH-HBZ and pCDH-APH-2, respectively, for the transduction of Jurkat cells. HBZ and APH-2 cDNAs were cloned into the pCDH-MSCV-MCS-EF1-copGFP lentiviral expression vector (SBI) to create pMSCV-HBZ and pMSCV-APH-2, respectively, for the transduction of primary T cells.
Transient transfections.
HEK293T cells were transfected with titrating amounts of FLAG-HBZ, FLAG–APH-2, or the control expression vector (approximately 2,000 ng total DNA per well) in a 6-well dish using the Lipofectamine 2000 transfection reagent (Life Technologies) according to the manufacturer's instructions. When indicated, 200 ng of the p65 and IRF-1 expression vectors was used. Immunoblot analysis was performed 48 h after transfection to compare the levels of transfected proteins.
Reporter gene assays.
HEK293T, Jurkat, and HepG2 cells were transfected using the Lipofectamine 2000 transfection reagent according to the manufacturer's instructions. Each transfection experiment was performed at least in triplicate; data are presented as the means with standard deviations. In general, HEK293T cells were transfected in a 6-well dish with 20 ng TK-renilla (the transfection control), 200 ng a luciferase construct (IRF-1–luc or κB-luc), limiting amounts of p65 (50 ng) or IRF-1 (25 ng), and titrating amounts of FLAG-HBZ, FLAG–APH-2, or the control expression vector (approximately 2,000 ng total DNA per well). Jurkat cells were transfected in a 6-well dish with 200 ng TK-renilla (the transfection control), 500 ng the luciferase construct (κB-luc), 500 ng p65, and titrating amounts of FLAG-HBZ, FLAG–APH-2, or the control expression vector (approximately 4,000 ng total DNA per well). HepG2 cells were transfected in a 6-well dish with 200 ng TK-renilla (the transfection control), 500 ng the luciferase construct (9×CAGA-luc), and titrating amounts of FLAG-HBZ, FLAG–APH-2, or the control expression vector (approximately 2,000 ng total DNA per well). At 24 h posttransfection, exogenous TGF-β (10 ng/ml; R&D Systems, Minneapolis, MN) was added to the HepG2 cell culture medium. HEK293T cells were harvested at 24 h posttransfection, and Jurkat and HepG2 cells were harvested at 48 h posttransfection. The cells were placed in passive lysis buffer (Promega, Madison, WI). The relative amounts of firefly and renilla luciferase were measured by a FilterMax F5 multimode microplate reader using a dual-luciferase reporter assay system (Promega) according to the manufacturer's instructions. Assays were performed under each condition in duplicate. Extracts were also subjected to immunoblotting to verify the presence of equivalent protein levels.
Immunoblotting.
Cell lysates were harvested and placed in passive lysis buffer (Promega) containing protease inhibitor cocktail (Roche) and quantitated using an ND-1000 NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA). Equivalent amounts of protein were separated in Mini-Protean TGX precast 4 to 20% gels (Bio-Rad Laboratories, Hercules, CA) and transferred to nitrocellulose membranes. Membranes were blocked in phosphate-buffered saline (PBS) containing 5% milk and 0.1% Tween 20 and incubated with primary antibody. The following antibodies were used: anti-HBZ (1:1,000) (27), anti-APH-2 (1:1,000) (33), anti-FLAG clone M2 (1:1,000; Agilent, Santa Clara, CA), anti-p65 (1:250; Santa Cruz Biotechnology, Dallas, TX), anti-IRF-1 (1:250; Santa Cruz Biotechnology), anti-ubiquitin (1:250; Santa Cruz Biotechnology), and anti-β-actin (1:5,000; Sigma, St. Louis, MO). The secondary antibodies used were horseradish peroxidase-labeled goat anti-rabbit and goat anti-mouse immunoglobulin antibodies (1:5,000; Santa Cruz Biotechnology). The blots were developed using the ImmunoCruz luminol reagent (Santa Cruz Biotechnology). Images were taken using an Amersham Imager 600 imaging system (GE Healthcare Life Sciences, Piscataway, NJ), and densitometric data were calculated using the ImageQuant TL program (GE Healthcare Life Sciences).
Quantitative RT-PCR (qRT-PCR).
Total RNA was isolated from 106 cells per condition using an RNeasy minikit (Qiagen) according to the manufacturer's instructions. Isolated RNA was quantitated and DNase treated using recombinant DNase I (Roche). Reverse transcription (RT) was performed using a SuperScript first-strand synthesis system for RT-PCR (Life Technologies) according to the manufacturer's instructions. The instrumentation and general principles of the CFX96 Touch real-time PCR detection system (Bio-Rad) are described in detail in the operator's manual. PCR amplification was carried out in 96-well plates with optical caps. The final reaction volume was 20 μl and consisted of 10 μl iQ SYBR green Supermix (Bio-Rad), 300 nM each specific primer, and 2 μl of cDNA template. For each run, standard cDNA, sample cDNA, and a no-template control were all assayed in triplicate. The reaction conditions were 95°C for 5 min, followed by 40 cycles of 94°C for 30 s, 56°C for 30 s, and 72°C for 45 s. Primer pairs for the specific detection of viral mRNA species (gag/pol, tax/rex, hbz, aph-2), platelet-derived growth factor beta polypeptide (pdgfb), and human glyceraldehyde-3-phosphate dehydrogenase (hgapdh) were described previously (45–47). Data from triplicate experiments are presented in histogram form as means with standard deviations. The total copy number for each viral gene was determined using a plasmid DNA standard curve and normalized to 106 copies of hGAPDH mRNA.
Cycloheximide pulse-chase experiments.
HEK293T cells were transiently transfected with either the untagged (pME) or the S-tagged (pTriEx) HBZ or APH-2 vector using the Lipofectamine 2000 reagent according to the manufacturer's instructions. Forty-eight hours later, the cells were treated with 100 μg/ml cycloheximide (a translation elongation inhibitor; Sigma) and then harvested at different time points. Cell lysates were harvested in NP-40 lysis buffer containing protease inhibitor cocktail (Roche) and quantitated using a Pierce bicinchoninic acid protein assay kit (Thermo Fisher Scientific). Extracted proteins were separated by SDS-PAGE and blotted with the appropriate antibodies as described in “Immunoblotting” above. Densitometric data were calculated using the ImageQuant TL program (GE Healthcare Life Sciences).
Infection and packaging of lentivirus vectors.
HEK293T cells were transfected with the appropriate pCDH-lentiviral vector expressing HBZ or APH-2, plus DNA vectors encoding HIV Gag/Pol and vesicular stomatitis virus glycoprotein G in 10-cm dishes with the Lipofectamine 2000 reagent according to the manufacturer's instructions. Media containing the lentiviral particles were collected 72 h later and filtered through 0.45-μm-pore-size filters (Thermo Fisher Scientific). Lentiviral particles were then concentrated using ultracentrifugation in a Sorvall SW-41 swinging bucket rotor according to standard procedures. Jurkat cells were infected with the pCDH-CMV-MCS-EF1-copGFP lentivirus and naive human T cells were infected with the pCDH-MSCV-MCS-EF1-copGFP lentivirus by spinoculation at 2,000 × g for 2 h at room temperature. After 3 days, Jurkat cells were sorted for green fluorescent protein (GFP) expression. After 5 days, T cells were stained for Foxp3 expression according to the manufacturer's protocol (Foxp3/transcription factor staining buffer set; eBioscience, San Diego, CA) and then sorted for GFP and Foxp3.
S-tag-affinity pulldown assays.
Cell lysates were prepared with 1× passive lysis buffer (Promega) in the presence of protease inhibitor (Roche). S-tag purification was performed by rocking cell lysates with S beads (Novagen) overnight at 4°C. The S beads were washed twice with low-salt radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 100 mM sodium pyrophosphate, 10 mM EDTA, 50 mM Tris-HCl [pH 8], 0.1% SDS, 12 mM deoxycholic acid, 10% glycerol, 1% Nonidet P-40) and twice with high-salt RIPA buffer (1 M NaCl, 100 mM sodium pyrophosphate, 10 mM EDTA, 50 mM Tris-HCl [pH 8], 0.1% SDS, 12 mM deoxycholic acid, 10% glycerol, 1% Nonidet P-40). An equal volume of 2× SDS-sample buffer was added, and proteins were extracted by heating at 95°C for 10 min.
Immunoprecipitation.
Cells were washed with 1× PBS and then incubated with gentle rocking at 4°C for 30 min in NP-40 lysis buffer. Cells were centrifuged at maximum speed for 10 min at 4°C. To preclear the samples, protein A/G Plus agarose beads (catalog number sc-2003; Santa Cruz Biotechnology) were added and the samples were rocked for an hour at 4°C. The beads were spun down, and the supernatant was divided for testing under the different conditions. Antibody (1 to 2 μg or no antibody for the direct load) was added to each sample, and the samples were rocked overnight at 4°C. The antibodies used were as follows: control rabbit IgG (Santa Cruz Biotechnology), rabbit anti-p65 (Santa Cruz Biotechnology), and rabbit anti-APH-2 antiserum (33). A/G beads were added, and the mixture was rocked at 4°C for 2 h. The beads were spun down and washed three times in NP-40 lysis buffer. An equal volume of 2× SDS-sample buffer was added, and proteins were extracted by heating at 95°C for 10 min. For detection by FLAG antibody, cells were washed with 1× PBS and then incubated with gentle rocking at 4°C for 30 min in NP-40 lysis buffer. Cells were centrifuged at maximum speed for 10 min at 4°C. FLAG beads were added, and the samples were rocked overnight at 4°C. The beads were spun down and washed three times in PBS. An equal volume of 2× SDS-sample buffer was added, and proteins were extracted by heating at 95°C for 10 min.
DNA affinity precipitation assay.
HEK293T cells in 10-cm dishes (two dishes per condition) were transfected with the FLAG–APH-2, FLAG-HBZ, or FLAG control expression vector by use of the Lipofectamine 2000 reagent according to the manufacturer's instructions. After 24 h, 10 μM MG132 was added to the cells. After 24 h, the cells were lysed in 1 ml of biotin binding buffer (25 mM Tris-HCl, pH 8.0, 100 mM NaCl, 0.5% Triton X-100, 1 mM EDTA, complete protease inhibitor cocktail), and cell debris was removed by centrifugation for 15 min. Lysates were incubated with 10 μg/ml poly(dI-dC) (Sigma) and biotinylated oligonucleotide for 1 h at 4°C. Streptavidin-Sepharose beads (Thermo Fisher Scientific) were added to the mixture, and the mixture was further incubated for 1 h. The beads were washed four times with biotin binding buffer. An equal volume of 2× SDS-sample buffer was added, and proteins were extracted by heating at 95°C for 10 min. 5′-biotinylated oligonucleotides (specific for the 6× IRF binding element [IRF-E] site [5′-GAAACCGAAACT-3′] or a nonspecific sequence [5′-CGCTTGATGACTCAGCCGGAA-3′]) were annealed with their complementary oligonucleotide.
RESULTS
The APH-2 protein is less stable than the HBZ protein.
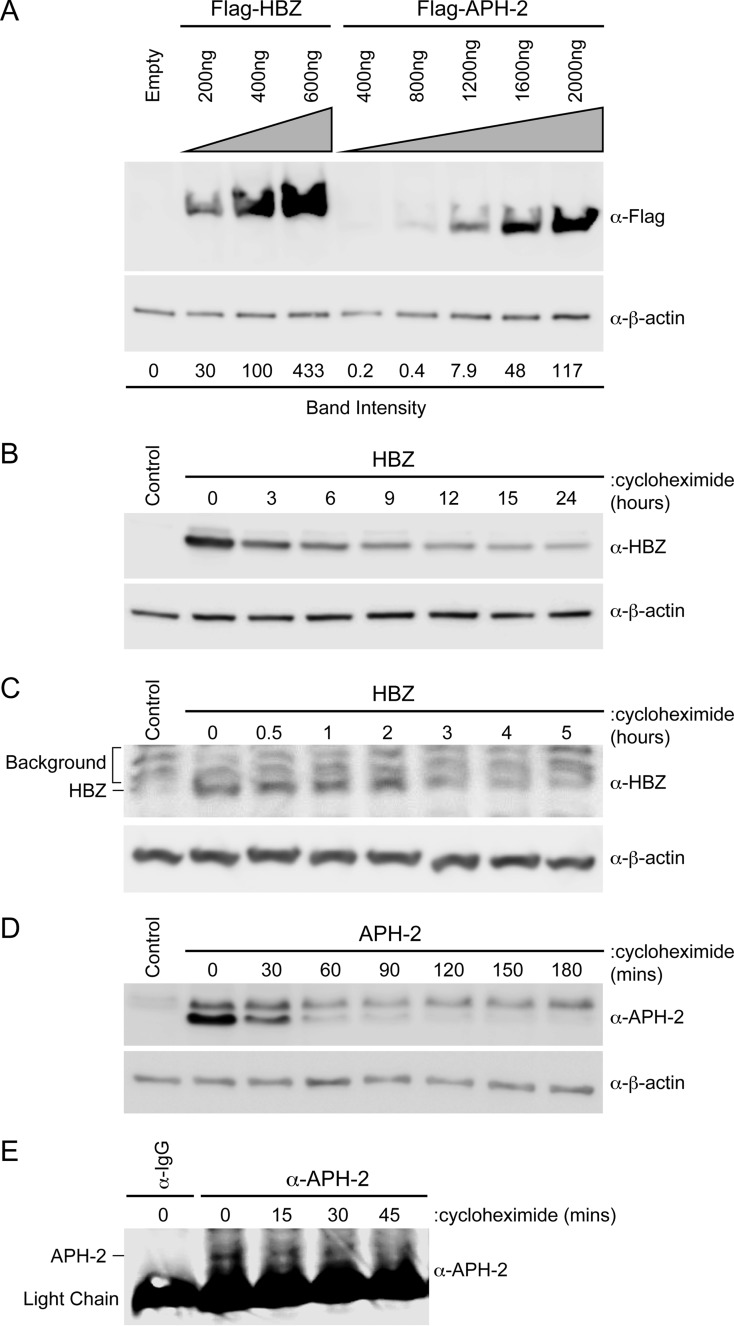
In order to conduct functional comparisons of HBZ and APH-2, we first measured the levels of expression of the two proteins by immunoblot analyses (Fig. 1A). Both HBZ and APH-2 were cloned into a CMV-driven expression vector containing an N-terminal FLAG epitope, creating FLAG-HBZ and FLAG–APH-2, respectively. HEK293T cells were transfected with the FLAG-HBZ (200, 400, or 600 ng), FLAG–APH-2 (400, 800, 1,200, 1,600, or 2,000 ng), or control expression vector as indicated, and immunoblot analysis was performed. The amount of FLAG protein relative to the amount of β-actin under each condition was quantified using densitometry. The value for the condition with 400 ng of transfected FLAG-HBZ was set at 100. The transfection of equivalent amounts of FLAG-HBZ or FLAG–APH-2 DNA did not equate to the production of equivalent amounts of protein. Approximately 5-fold more FLAG–APH-2 DNA was needed to reach a comparable steady-state level of protein (Fig. 1A). Given the disparity between the transient expression levels of HBZ and APH-2, cycloheximide chase experiments were performed to determine the half-lives of both the HBZ and APH-2 proteins. In a previous study by Yoshida et al., the HBZ half-life was approximated to be less than 12 h (48); however, no study to date has examined the half-life of APH-2. HEK293T cells were transfected with untagged APH-2 and HBZ expression constructs and then treated with the translation elongation inhibitor cycloheximide. Cell lysates were collected at defined time points, and the expression levels of both HBZ and APH-2 were examined by immunoblot analysis. The calculated half-life of HBZ was approximately 6.4 h (Fig. 1B), whereas the calculated half-life of APH-2 was approximately 33 min (Fig. 1D). Similar results were obtained using epitope-tagged HBZ and APH-2 constructs in HEK293T cells (data not shown). The half-lives of HBZ and APH-2 were also measured in cells of the more physiologically relevant transformed T-cell line Jurkat. The calculated half-life of HBZ was approximately 2 to 3 h (Fig. 1C), whereas the calculated half-life of APH-2 was approximately 20 min (Fig. 1E). Using the transcriptional inhibitor actinomycin D, we also determined the stability of hbz and aph-2 mRNA transcripts to be >12 h (data not shown). These results indicated that the difference in protein levels is largely due to differences in HBZ and APH-2 protein stability. Consequently, we used titrating amounts of both FLAG-HBZ and FLAG–APH-2 expression vectors in our subsequent studies. These vectors allowed us to examine the effects of HBZ and APH-2 when similar levels of either transcript or protein were present.
FIG 1.
The APH-2 protein was less stable than the HBZ protein. (A) HEK293T cells were transfected with titrating amounts of FLAG-HBZ, FLAG–APH-2, or the control expression vector (Empty), as indicated. Immunoblot analysis was performed 48 h after transfection to compare the levels of transfected HBZ and APH-2. β-Actin expression was used as a loading control. The amount of FLAG-tagged HBZ or APH-2 relative to the amount of β-actin was measured under each condition. (B and D) HEK293T cells were transfected with 6 μg HBZ (B) or APH-2 (D) expression plasmid in 10-cm dishes. Cells were trypsinized and at 48 h after transfection were replated into 6-well dishes until they reached approximately 80 to 90% confluence. The cells were then treated with 100 μg/ml cycloheximide for the indicated times. Cells were collected by centrifugation, washed in PBS, and lysed using NP-40 lysis buffer. Immunoblot analysis was performed to detect HBZ and APH-2 expression levels. β-Actin was used as a loading control. (C and E) Jurkat cells stably transduced with lentiviral vectors expressing HBZ (C) or APH-2 (E) were treated with 100 μg/ml cycloheximide for the indicated times. Cells were collected by centrifugation, washed in PBS, and lysed using NP-40 lysis buffer. Immunoblot analysis was performed to detect HBZ expression levels. β-Actin was used as a loading control. APH-2 was immunoprecipitated with a control normal rabbit IgG or APH-2 rabbit antiserum. The amount of immunoprecipitated APH-2 was then measured by immunoblot analysis.
APH-2, unlike HBZ, did not enhance TGF-β signaling.
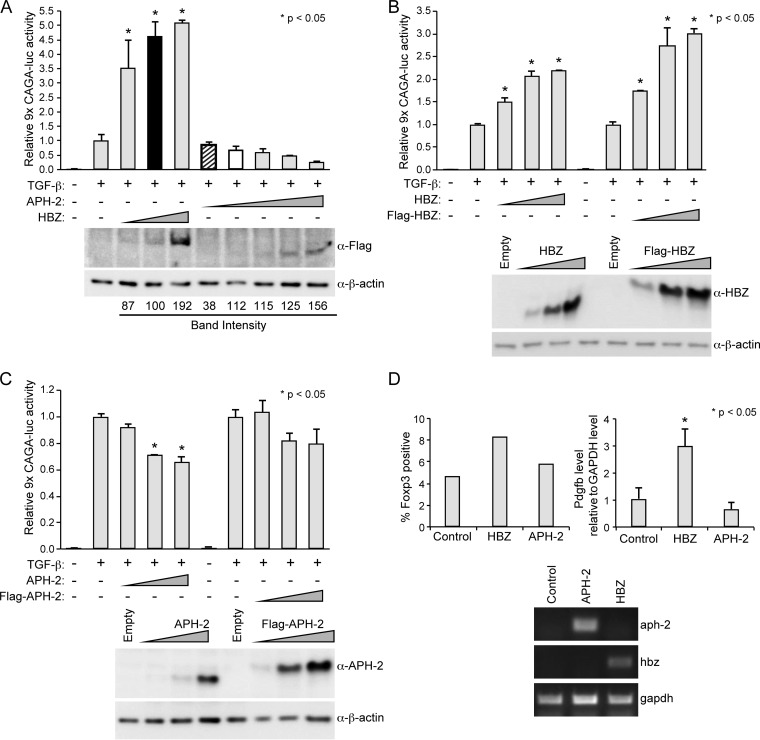
HBZ augments TGF-β signaling by physically enhancing the interaction between the cellular transcription factors Smad3 and p300, leading to the activation of TGF-β target genes (43). Studies have reported that the exogenous expression of HBZ in T cells led to an increase in various TGF-β-responsive genes, including foxp3 (43, 49). Foxp3 is the master regulator of regulatory T cells (Tregs). The ability of HTLV-1 to convert infected T cells into Tregs is believed to be critical for viral persistence. Given the significant effect of HBZ on TGF-β signaling, we examined the consequences of APH-2 expression on TGF-β signaling using a reporter gene assay. HepG2 cells were transfected with a TGF-β-responsive luciferase reporter plasmid (9×CAGA-luc), the TK-renilla control, and titrating amounts of FLAG-HBZ, FLAG–APH-2, or the control expression vector. Twenty-four hours later, the transfected cells were treated with or without 10 ng/ml exogenous TGF-β for an additional 24 h. In the absence of TGF-β, neither HBZ nor APH-2 had any effect on 9×CAGA-luc reporter activation (data not shown). In the presence of TGF-β, the 9×CAGA-luc reporter was activated approximately 70-fold compared to the level of activation for the negative control (no TGF-β); this value was set at 1 (Fig. 2A). In the presence of increasing amounts of HBZ protein, we detected a dose-dependent increase in TGF-β signaling, whereas in the presence of increasing amounts of APH-2 protein, we observed a dose-dependent decrease in TGF-β signaling. When we compared conditions with equivalent levels of transfected HBZ DNA (Fig. 2A, black bar) and APH-2 DNA (Fig. 2A, bar with diagonal stripes), APH-2 had no effect on TGF-β signaling (P = 0.9443), whereas HBZ significantly enhanced TGF-β signaling (P < 0.0001). Likewise, when we expressed similar levels of the HBZ and APH-2 proteins (Fig. 2A, white bar), APH-2 did decrease TGF-β signaling, but this effect was not statistically significant (P = 0.2976). Similar results were obtained using untagged HBZ (Fig. 2B) and APH-2 (Fig. 2C) expression vectors, which indicated that the FLAG tag did not interfere with the effects of HBZ and APH-2 on TGF-β signaling. Numerous attempts to successfully measure TGF-β signaling in Jurkat cells using our reporter gene construct (9×CAGA-luc) were unsuccessful for unknown reasons. However, we did examine the effects of HBZ and APH-2 on the levels of two TGF-β downstream targets, Foxp3 and Pdgfb, in human naive T cells. Naive T cells were transduced with HBZ, APH-2, or the control lentiviral vector. Infected cells were then stained for Foxp3 and sorted for GFP vector and Foxp3 expression (Fig. 2D, top left). As expected, the amount of Foxp3-expressing cells roughly doubled in the presence of HBZ, whereas the APH-2 protein had little to no effect on Foxp3 levels. Quantitative RT-PCR was also performed to examine the level of the pdgfb transcript in each cell population (Fig. 2D, top right). HBZ-expressing cells had an increased level of the pdgfb transcript, as expected, while APH-2-expressing cells had no change in the pdgfb level relative to that for the control. Due to the limited amount of sample and low protein expression levels, we were unable to detect APH-2 and HBZ protein expression in naive T cells. However, hbz and aph-2 mRNA expression was confirmed (Fig. 2D, bottom). Taken together, our results indicated that, unlike HBZ, APH-2 is not able to enhance TGF-β signaling and subsequently had no effect on Foxp3 or Pdgfb levels.
FIG 2.
APH-2, unlike HBZ, did not enhance TGF-β signaling. (A to C) HepG2 cells were transfected with 200 ng TK-renilla control, 500 ng 9×CAGA-luc reporter (a TGF-β-responsive reporter), and titrating amounts of FLAG-HBZ, FLAG–APH-2, HBZ, APH-2, or the control expression vector, as indicated. (Top) At 24 h posttransfection, the cells were treated with 10 ng/ml exogenous TGF-β for 24 h. Cell lysates were then collected and luciferase levels were measured; for each condition, relative luciferase activity is shown as the mean fold change from that obtained with TGF-β treatment, which was set at 1. Diagonal striped bar, equivalent FLAG-tagged APH-2 and HBZ (black bar) transfected DNA levels; white bar, similar levels of protein expression. (Bottom) Immunoblot analysis was performed to detect the expression levels of HBZ and APH-2 under each condition relative to the expression level of the loading control, β-actin. A generalized linear model was used to study the differences between the results obtained with the various treatments and those obtained with TGF-β alone. Dunnett's method was used to control type I error. *, a statistically significant P value of <0.05 compared to the results obtained with TGF-β treatment alone. (D) Human T cells were transduced with lentiviral vectors expressing HBZ, APH-2, or the empty vector, as described in Materials and Methods. (Top left) The percentages of transduced cells expressing Foxp3 (as measured by GFP) are depicted in histogram form. (Top right) Quantitative RT-PCR for pdgfb and gapdh was performed with mRNA isolated from lentivirus-transduced human T cells. The total Pdgfb level was determined using the ΔΔCT threshold cycle (CT) method (66) and normalized to the GAPDH level. Data from triplicate experiments are presented in histogram form and represent means and standard deviations. (Bottom) The levels of the hbz, aph-2, and gapdh transcripts in transduced human T cells were measured by RT-PCR analysis.
HBZ and APH-2 repressed p65 transactivation.
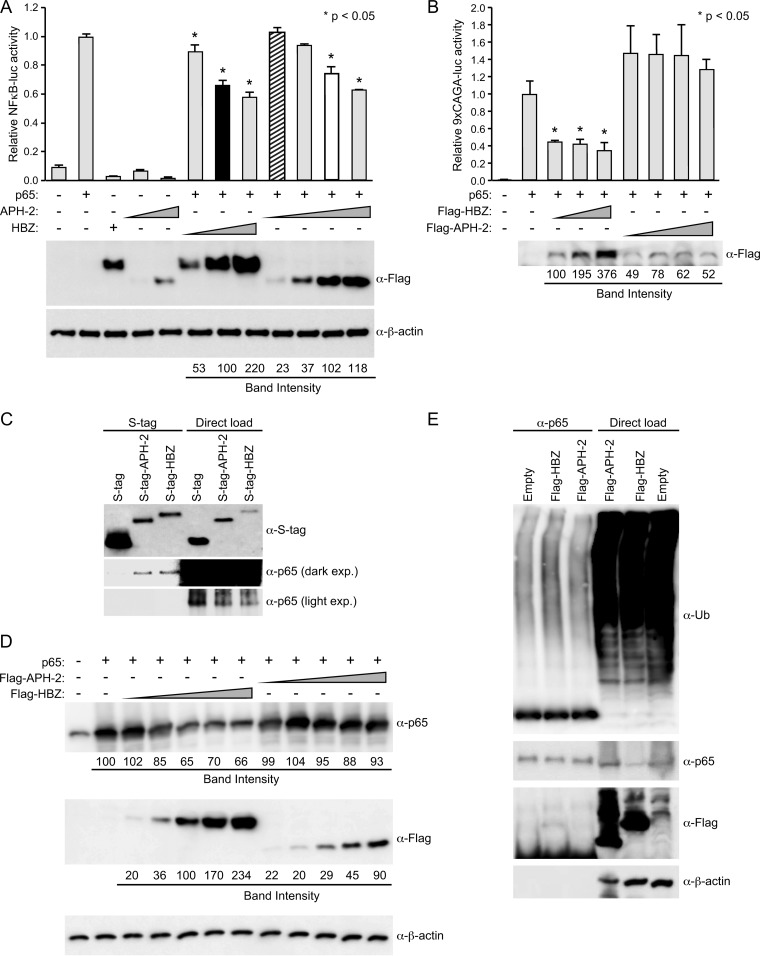
HTLV primarily replicates through mitosis of infected cells as opposed to infection of new cells. HTLV also induces the classical NF-κB pathway through expression of the regulatory proteins Tax-1 and Tax-2 (50). Although this pathway induces cellular growth, hyperactivation of the NF-κB pathway by Tax can lead to a phenomenon known as Tax-induced senescence (TIS) (51–53). In the case of HTLV-1, TIS can be alleviated by HBZ through the downregulation of NF-κB, more specifically, through binding and degradation of the classical NF-κB component p65 (47, 53). The balance between HBZ and Tax-1 enables continuous cell growth and evasion of TIS. However, the effect of APH-2 on NF-κB transactivation is currently unknown. To address this, HEK293T cells were transfected with an NF-κB-responsive luciferase reporter plasmid (κB-luc), the TK-renilla control, a p65 expression vector, and titrating amounts of FLAG-HBZ, FLAG–APH-2, or the control expression vector. Luciferase activity was measured after 24 h. In the presence of exogenous p65, the κB-luc reporter was activated approximately 10-fold more than the negative control; this value was set at 1 (Fig. 3A). In the presence of increasing amounts of HBZ protein, we detected a dose-dependent decrease in p65 transactivation. Similarly, in the presence of increasing amounts of APH-2 protein, we also observed a dose-dependent decrease in p65 transactivation. While equivalent levels of transfected APH-2 DNA (Fig. 3A, bar with diagonal stripes) had no significant effect on p65 transactivation (P = 0.8808), similar levels of APH-2 protein (Fig. 3A, white bar) significantly decreased the level of p65 transactivation (P < 0.0001). This experiment was also performed in Jurkat cells (Fig. 3B). In Jurkat cells, the presence of increasing amounts of HBZ protein again caused a dose-dependent decrease in the level of p65 transactivation. However, the presence of increasing amounts of APH-2 protein did not result in a significant increase or decrease in the level of p65 transactivation. Taken together, these results indicated that both HBZ and APH-2 repress p65 transactivation in HEK293T cells. However, in the T-cell line Jurkat, which is more similar to the HTLV target cell in vivo, only HBZ repressed p65 transactivation. Previous reports demonstrated that HBZ represses p65-mediated transcription through a direct physical association between HBZ and p65 via the activation and bZIP domains of HBZ, which inhibits the binding of p65 to DNA sequences (47). The transactivation domain and bZIP domain of HBZ were also shown to be involved in p65 ubiquitination by HBZ. Using an S-tag-affinity pulldown assay, we found that, like HBZ, APH-2 could interact with p65, despite the absence of transactivation and bZIP domains (Fig. 3C). However, unlike HBZ, APH-2 did not drastically decrease p65 levels (Fig. 3D) or increase p65 ubiquitination (Fig. 3E). Our results suggested that the ability of APH-2 to repress p65 depends on the cellular environment/protein bioavailability; repression likely occurs via a separate mechanism but a mechanism possibly related to HBZ repression of p65.
FIG 3.
HBZ and APH-2 repressed p65 transactivation. (A) HEK293T cells were transfected with 20 ng TK-renilla control, 200 ng κB-luciferase reporter, 50 ng p65 expression plasmid, and titrating amounts of FLAG-HBZ, FLAG–APH-2, or the control expression vector, as indicated. (Top) At 24 h posttransfection, cell lysates were collected and luciferase levels were measured; for each condition, relative luciferase activity is shown as the mean fold change from the luciferase activity obtained with p65 expression, which was set at 1. Diagonal striped bar, equivalent FLAG-tagged APH-2 and HBZ (black bar) transfected DNA levels; white bar, similar levels of protein expression. A generalized linear model was used to study the differences between treatments versus p65 expression. Dunnett's method was used to control type I error. *, a statistically significant P value of <0.05 compared to the result for p65 expression. (Bottom) Immunoblot analysis was performed to detect the expression levels (FLAG) of HBZ and APH-2 under each condition relative to that obtained with the loading control, β-actin. (B) Jurkat cells were transfected with 200 ng TK-renilla, 500 ng κB-luciferase reporter, 500 ng p65 expression plasmid, and titrating amounts of FLAG-HBZ, FLAG–APH-2, or the control expression vector, as indicated. At 48 h posttransfection, cells were collected by centrifugation and washed in PBS. A portion of the cells (1/10) was used to measure luciferase levels; relative luciferase activity for each condition is shown as the mean fold change from the luciferase activity obtained with p65 expression, which was set at 1. The remainder of the cells was lysed using NP-40 lysis buffer and subjected to FLAG immunoprecipitation as described in Materials and Methods. The amounts of the immunoprecipitated proteins were then measured by immunoblot analysis (with FLAG antibody). (C) HEK293T cells were cotransfected with p65 and the empty, S-tagged APH-2, or S-tagged HBZ vector. Tagged proteins were purified by S-tag-affinity purification 48 h after transfection. Pulldowns were examined by immunoblot analysis using anti-S-tag and anti-p65 antibodies, as indicated. Five percent of the direct load was used for immunoblot analysis. exp., exposure. (D) HEK293T cells were cotransfected with p65 and titrating amounts of FLAG-HBZ, FLAG–APH-2, or the control expression vector, as indicated. Immunoblot analysis was performed 48 h after transfection to compare the levels of transfected FLAG-tagged HBZ, FLAG-tagged APH-2, and p65. β-Actin expression was used as a loading control. The amount of FLAG-tagged HBZ or APH-2, as well as p65, under each condition relative to the amount of β-actin was measured. (E) HEK293T cells were cotransfected with p65 and FLAG-HBZ, FLAG–APH-2, or the control expression vector, as indicated. At 24 h after transfection, the cells were treated with 10 μM MG132. Coimmunoprecipitation was performed 24 h after MG132 treatment, as described in Materials and Methods. Immunoprecipitated proteins were examined by immunoblot analysis using anti-ubiquitin (α-Ub), anti-p65, and anti-FLAG antibodies, as indicated. Five percent of the direct load was used for immunoblot analysis.
HBZ, but not APH-2, repressed IRF-1 transactivation.
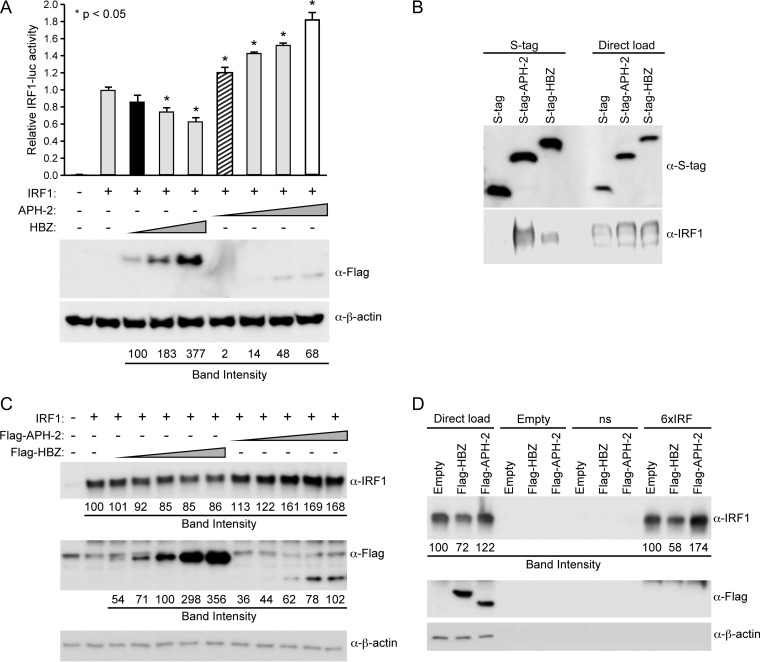
IRF-1 is a component of the innate immune response and is a tumor suppressor protein (44, 54) that is required for the induction of apoptosis in response to DNA damage (55). Interestingly, the loss of IRF-1 expression has been observed in several cases of leukemia (56, 57). In 2011, Mukai and Ohshima found that HBZ interacted with IRF-1, inhibited its DNA binding ability, and induced its degradation and thereby significantly reduced the number of cells undergoing apoptosis (30). To examine the effects of APH-2 on IRF-1, HEK293T cells were transfected with an IRF-1-responsive luciferase reporter vector (IRF-1–luc), the TK-renilla control, an IRF-1 expression vector, and titrating amounts of FLAG-HBZ, FLAG–APH-2, or the control expression vector. Luciferase activity was measured after 24 h. In the presence of exogenous IRF-1, the IRF-1–luc reporter was activated approximately 200-fold compared to the level of activation of the negative control; this value was set at 1 (Fig. 4). In the presence of increasing amounts of HBZ protein, we detected a dose-dependent decrease in the level of IRF-1 transactivation, whereas in the presence of increasing amounts of APH-2 protein, we detected a dose-dependent increase in the level of IRF-1 transactivation. Both equivalent levels of transfected APH-2 DNA (Fig. 4A, bar with diagonal stripes; P = 0.0105) and similar levels of APH-2 protein (Fig. 4A, white bar; P < 0.0001) significantly increased the level of IRF-1 transactivation. We observed that the IRF-1 expression plasmid failed to activate the IRF-1 luciferase reporter in Jurkat cells (data not shown). Similar to the findings described in previous reports, we found that HBZ interacted with IRF-1 using an S-tag-affinity pulldown assay (Fig. 4B). We also found that APH-2 associated with IRF-1 (Fig. 4B). When we compared the effects of HBZ and APH-2 on IRF-1 levels, we confirmed the findings presented in a previous report by Mukai and Ohshima that HBZ decreases IRF-1 steady-state levels and found that APH-2 increased IRF-1 levels in HEK293T cells (Fig. 4C) (30). Using biotinylated nucleotide probes, we examined the binding of IRF-1 in the presence of either HBZ or APH-2. HBZ decreased while APH-2 increased the level of IRF-1 binding to the biotinylated consensus IRF-1 probe (Fig. 4D). Our results indicated that, unlike HBZ (which interacted with and degraded IRF-1) (30), APH-2 interacts with IRF-1 and enhances its DNA binding and steady-state expression levels.
FIG 4.
HBZ, but not APH-2, repressed IRF-1 transactivation. (A) HEK293T cells were transfected with 20 ng TK-renilla control, 200 ng IRF-1 luciferase reporter, 25 ng IRF-1 expression plasmid, and titrating amounts of FLAG-HBZ, FLAG–APH-2, or the control expression vector, as indicated. (Top) At 24 h posttransfection, cell lysates were collected and luciferase levels were measured; for each condition, relative luciferase activity is shown as the mean fold change from the value obtained with IRF-1 expression, which was set at 1. Diagonal striped bar, equivalent FLAG-tagged APH-2 and HBZ (black bar) transfected DNA levels; white bar, similar levels of protein expression. A generalized linear model was used to study the differences between treatments and IRF-1 expression. Dunnett's method was used to control type I error. *, a statistically significant P value of <0.05 compared to the results obtained with IRF-1 expression. (Bottom) Immunoblot analysis was performed to detect the expression levels (FLAG) of HBZ and APH-2 under each condition relative to the expression level of the loading control, β-actin. (B) HEK293T cells were cotransfected with IRF-1 and the empty, S-tagged APH-2, or S-tagged HBZ vector. Cells were treated with 10 μM MG132 for 24 h, and tagged proteins were purified by S-tag-affinity purification at 48 h after transfection. Pulldowns were examined by immunoblot analysis using anti-S-tag and anti-IRF-1 antibodies, as indicated. Five percent of the direct load was used for immunoblot analysis. (C) HEK293T cells were cotransfected with IRF-1 and titrating amounts of FLAG-HBZ, FLAG–APH-2, or the control expression vector, as indicated. Immunoblot analysis was performed 48 h after transfection to compare the levels of transfected FLAG-tagged HBZ, FLAG-tagged APH-2, and IRF-1. β-Actin expression was used as a loading control. The amount of FLAG-tagged HBZ or APH-2, as well as IRF-1, under each condition was measured relative to the amount of β-actin. (D) HEK293T cells were cotransfected with IRF-1 and the FLAG-HBZ, FLAG–APH-2, or control expression vector. After 24 h, cells were treated with 10 μM MG132 for 24 h. Whole-cell lysates were used for DNA affinity precipitation assays. Oligonucleotides that were biotinylated at the 5′ end and that contained a nonspecific sequence (ns) or an IRF-E sequence (6×IRF), as well as nonbiotinylated oligonucleotides (Empty), were used. The complexes pulled down were eluted and analyzed by immunoblotting using IRF-1, FLAG, and β-actin antibodies. The amount of IRF-1 in the direct load was measured relative to the amount of β-actin. The total amount of IRF-1 bound to DNA in the pulldown was also measured.
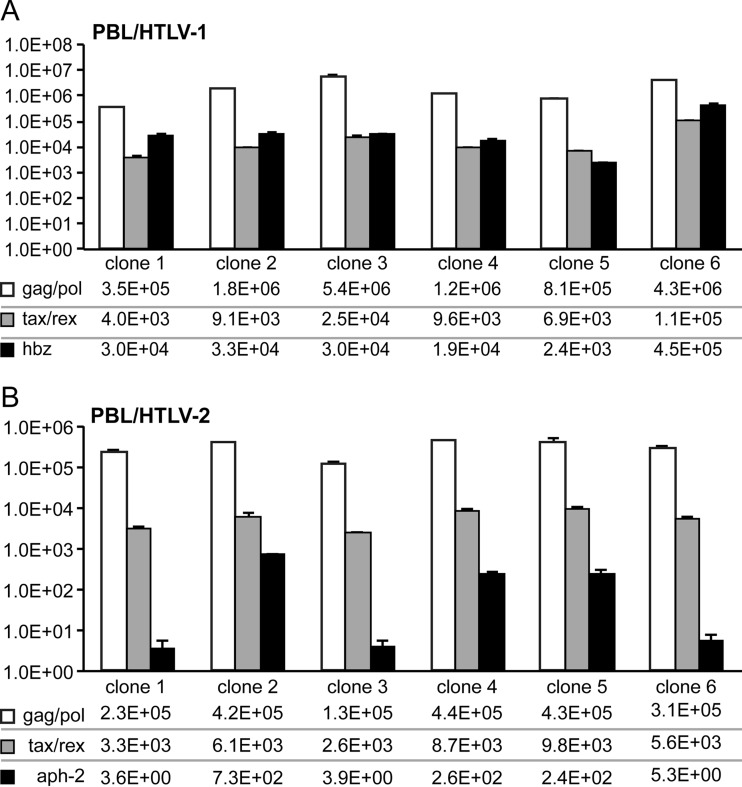
aph-2 transcript levels were consistently lower than hbz transcript levels in HTLV-infected PBL lines.
Given the difference in protein stability between APH-2 and HBZ (Fig. 1), we used quantitative RT-PCR (qRT-PCR) to measure the relative aph-2 and hbz RNA transcript levels in HTLV-infected PBL lines (Fig. 5). Total RNAs were collected and isolated from six separate clones of the immortalized primary human T-cell lines PBL/HTLV-1 and PBL/HTLV-2. RNA was reverse transcribed, and quantitative PCR analysis was performed to determine the numbers of copies of the gag/pol, tax/rex, hbz, and aph-2 transcripts relative to 106 copies of gapdh. In the immortalized PBL/HTLV-2 cell lines, aph-2 mRNA levels were consistently lower than hbz mRNA levels in the PBL/HTLV-1 cell lines. This finding indicated that HTLV-2 does not compensate for APH-2 protein instability with an increase in the aph-2 transcript level and subsequent protein expression.
FIG 5.
aph-2 transcript levels were consistently lower than hbz transcript levels in HTLV-infected PBL lines. Quantitative RT-PCR was performed on mRNA isolated from an HTLV-1-infected PBL line (PBL/HTLV-1) (A) and an HTLV-2-infected PBL line (PBL/HTLV-2) (B), as described in Materials and Methods. The primer pairs used for the specific detection of viral mRNA species were described previously (45, 46). Data from triplicate experiments are presented in histogram form with means and standard deviations. The total numbers of copies given beneath the histograms were determined using plasmid DNA standards and normalized to 106 copies of gapdh mRNA.
DISCUSSION
HTLV-1 and HTLV-2 share similar genomic structures and the ability to transform T cells in vitro. However, HTLV-1 infection can lead to ATL development, while HTLV-2 infection is nonpathogenic. Comparative studies of the HTLV-1 and HTLV-2 gene products allow a better understanding of the mechanisms of disease development in HTLV-1 infection. For example, studies comparing HTLV-1 Tax-1 and HTLV-2 Tax-2 have highlighted the importance of activation of the alternative NF-κB pathway or interactive proteins of the PDZ domain in HTLV-1-induced T-cell transformation (58). We hypothesized that comparative studies of the HTLV-1 protein HBZ and its HTLV-2 counterpart, APH-2, would clarify the role of these viral genes in cell proliferation and disease development.
In our pursuit of functional comparisons of HBZ and APH-2, we utilized FLAG-tagged vectors, which allowed us to examine the proteins at equivalent levels. Upon transfection of FLAG-tagged APH-2 and HBZ expression vectors into HEK293T cells, we quickly noted that more APH-2 DNA was needed to generate steady-state-equivalent levels of APH-2 than for HBZ DNA to generate steady-state levels of HBZ under similar conditions (Fig. 1A). By a cycloheximide pulse-chase protein stability assay, we found that the HBZ protein half-life was 6.4 h in HEK293T cells and 2 to 3 h in Jurkat cells, while the APH-2 protein half-life was a mere 33 min in HEK293T cells and 20 min in Jurkat cells (Fig. 1B to E). We also examined aph-2 and hbz mRNA levels in HTLV-1- and HTLV-2-infected PBL lines (Fig. 5). The aph-2 levels were decreased at least 2 log fold compared to the hbz levels, which indicated that HTLV-2 did not compensate for APH-2 protein instability via an increase in the aph-2 mRNA level and subsequent protein levels. When HTLV-2-infected cells were compared to HTLV-1-infected cells, not only was less aph-2 mRNA present in HTLV-2-infected cells, but also the levels of APH-2 protein remained lower than the levels of HBZ protein in HTLV-1-infected cells due to protein instability. Taken together, our results suggested that studies comparing HBZ and APH-2 should be performed and the results should be interpreted carefully, given that the relative abundance of APH-2 is much lower than that of HBZ in natural infection.
Activation of TGF-β signaling resulted in the induction of Foxp3, which is the master switch of Tregs. HBZ was reported to enhance TGF-β signaling and subsequent Foxp3 expression (43). Using a TGF-β-responsive luciferase construct (9×CAGA-luc), we confirmed that HBZ enhanced TGF-β signaling, while APH-2 slightly repressed TGF-β signaling (Fig. 2A). It is noteworthy that the level of APH-2 repression of TGF-β signaling was not statistically significant. This finding indicated that while overexpression of APH-2 represses TGF-β signaling, during natural HTLV-2 infection the lower levels of the APH-2 transcript and protein (compared to those of the HBZ transcript and protein) would be expected to repress TGF-β signaling minimally. Similar effects on TGF-β signaling were obtained using untagged APH-2 and HBZ expression vectors, indicating that the FLAG tag did not have detrimental effects on APH-2 or HBZ (Fig. 2B and C).
In a recent publication, HBZ-induced Foxp3 expression was demonstrated to be labile (59). However, other studies showed that Foxp3 expression in Tregs was stabilized by the cellular protein SOCS1 (60). The SOCS1 protein is upregulated by HTLV-1 infection, specifically, by Tax-1 (61, 62). Previous observations suggested that ATL cells, which act phenotypically like Tregs, have high levels of expression of Foxp3 and maintain Tax expression (63). Our studies indicated that the expression of APH-2 was unable to significantly induce the expression of either TGF-β target gene, foxp3 or pdgfb, in naive human T cells, while HBZ enhanced TGF-β signaling and subsequent foxp3 and pdgfb expression (Fig. 2D). Taken together, our data supported and were consistent with the conclusion that HBZ facilitates a Treg phenotype for HTLV-1 infection, whereas APH-2 does not.
Regulation of the classical NF-κB pathway is essential during HTLV-1 cellular transformation and is highly regulated by both the Tax and HBZ proteins. Tax activation of the classical NF-κB pathway induces the growth of HTLV-1-infected cells; however, too much Tax activation can lead to senescence. HBZ modulates NF-κB activation through binding to p65 and diminishes its DNA binding capacity. HBZ also induces p65 degradation. Suppression of the classical NF-κB pathway by HBZ allows the alternative NF-κB pathway to predominate in HTLV-1-infected cells, a phenomenon hypothesized to be critical for oncogenesis (47). Like HBZ, we found that APH-2 had the ability to repress p65 transactivation (Fig. 3A) and interact with p65 (Fig. 3C); however, APH-2 did not readily decrease the level of p65 expression (Fig. 3D) or induce the ubiquitination of p65 (Fig. 3E) like HBZ did. This would suggest that APH-2 might decrease the level of p65 transactivation though a mechanism that is different from but possibly related to that used by HBZ.
While both APH-2 and HBZ decreased the level of p65 transactivation in HEK293T cells, the effects of APH-2 on p65 transactivation were less clear in T cells (Fig. 3B). Due to the inherent difficulties in transient transfection of lymphoid cells coupled with the short half-life of the APH-2 protein in Jurkat cells (Fig. 1E), we were unable to adequately express the APH-2 protein at levels similar to our lowest detectable level of HBZ protein, with the level of APH-2 expressed being 49 to 78% of the level of HBZ expressed. Nonetheless, APH-2 failed to suppress NF-κB activation by p65. This finding suggested that T cells may lack an essential cellular component necessary for APH-2 to inhibit NF-κB activation. It is also important to note that Tax-2 fails to efficiently activate the alternative NF-κB pathway due to its domain structure relative to that of Tax-1 (12, 64, 65) but also possibly due to the inability of APH-2 to suppress the classical NF-κB pathway and drive activation of the alternative pathway. The differences in the pathological outcomes seen between HTLV-1 and HTLV-2 may be influenced by the differences in viral regulation of the NF-κB pathway.
IRF-1, a known tumor suppressor that is downregulated in several cases of leukemia (44, 54, 56, 57), was recently shown to be negatively regulated by HBZ (30). In this study, we found that both APH-2 and HBZ interacted with IRF-1 (Fig. 4B). However, APH-2, unlike HBZ, enhanced the transcriptional activity of IRF-1 (Fig. 4A), increased IRF-1 expression (Fig. 4C), and improved IRF-1 binding to DNA (Fig. 4D). This observation indicated that HTLV-2-infected cells are more prone to IRF-1-mediated apoptosis and therefore have less of an ability to become transformed. Interestingly, previous studies found the APH-2-knockout virus replicated better than wild-type virus in rabbits, which further supports a role for APH-2 in vivo. However, given the short half-life and low level of bioavailability of APH-2 in the HTLV-2-infected cell, the effects of APH-2 on IRF-1 are likely minimal.
Our comparative studies have expanded our understanding of the role of genome antisense strand proteins during HTLV infection. APH-2, unlike HBZ, did not enhance TGF-β signaling and therefore might direct infected CD4+ cells away from Treg differentiation and the Treg phenotype. Like HBZ, APH-2 was able to inhibit classical NF-κB activation. However, protein stability and bioavailability, as well as the cellular environment, might hinder the ability of APH-2 to significantly affect NF-κB signaling during infection. APH-2 and HBZ also have contrasting effects on IRF-1 transcriptional activity. APH-2 enhanced IRF-1 transcriptional activation, which created a cellular environment favorable for apoptosis. Our comparative approach revealed distinct differences in how HBZ and APH-2 interact with important cell signaling pathways, and these differences are consistent with the different pathological outcomes from infection with these two highly related retroviruses.
ACKNOWLEDGMENTS
We thank Kate Hayes-Ozello for editorial comments on the manuscript and Tim Vogt for figure preparation.
REFERENCES
- 1.Proietti FA, Carneiro-Proietti AB, Catalan-Soares BC, Murphy EL. 2005. Global epidemiology of HTLV-I infection and associated diseases. Oncogene 24:6058–6068. doi: 10.1038/sj.onc.1208968. [DOI] [PubMed] [Google Scholar]
- 2.Hinuma Y, Nagata K, Hanaoka M, Nakai M, Matsumoto T, Kinoshita K-I, Shirakawa S, Miyoshi I. 1981. Adult T-cell leukemia: antigen in an ATL cell line and detection of antibodies to the antigen in human sera. Proc Natl Acad Sci U S A 78:6476–6480. doi: 10.1073/pnas.78.10.6476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoshida M, Miyoshi I, Hinuma Y. 1982. Isolation and characterization of retrovirus from cell lines of human adult T-cell leukemia and its implication in the disease. Proc Natl Acad Sci U S A 79:2031–2035. doi: 10.1073/pnas.79.6.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gessain A, Barin F, Vernant JC, Gout O, Maurs L, Calender A, de The G. 1985. Antibodies to human T-lymphotropic virus type-I in patients with tropical spastic paraparesis. Lancet ii:407–410. [DOI] [PubMed] [Google Scholar]
- 5.Osame M, Usuku K, Izumo S, Ijichi N, Amitani H, Igata A, Matsumoto M, Tara M. 1986. HTLV-I associated myelopathy, a new clinical entity. Lancet i:1031–1032. [DOI] [PubMed] [Google Scholar]
- 6.Lairmore M, Franchini G. 2007. Human T-cell leukemia virus types 1 and 2, p 2071–2106. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 7.Marcais A, Suarez F, Sibon D, Frenzel L, Hermine O, Bazarbachi A. 2013. Therapeutic options for adult T-cell leukemia/lymphoma. Curr Oncol Rep 15:457–464. doi: 10.1007/s11912-013-0332-6. [DOI] [PubMed] [Google Scholar]
- 8.Shimoyama M. 1991. Diagnostic criteria and classification of clinical subtypes of adult T-cell leukemia-lymphoma. A report from the Lymphoma Study Group (1984-1987). Br J Haematol 79:437. [DOI] [PubMed] [Google Scholar]
- 9.Miyoshi I, Yoshimoto S, Kubonishi I, Taguchi H, Shiraishi Y, Ohtsuki Y, Akagi T. 1981. Transformation of normal human cord lymphocytes by co-cultivation with a lethally irradiated human T-cell line carrying type C particles. Gann 72:997–998. [PubMed] [Google Scholar]
- 10.Yamamoto N, Okada M, Koyanagi Y, Kannagi Y, Kannagi M, Hinuma Y. 1982. Transformation of human leukocytes by cocultivation with an adult T cell leukemia virus producer cell line. Science 217:737–739. doi: 10.1126/science.6980467. [DOI] [PubMed] [Google Scholar]
- 11.Robek MD, Ratner L. 1999. Immortalization of CD4+ and CD8+ T lymphocytes by human T-cell leukemia virus type 1 Tax mutants expressed in a functional molecular clone. J Virol 73:4856–4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ross TM, Narayan M, Fang ZY, Minella AC, Green PL. 2000. Human T-cell leukemia virus type 2 Tax mutants that selectively abrogate NFκB or CREB/ATF activation fail to transform primary human T cells. J Virol 74:2655–2662. doi: 10.1128/JVI.74.6.2655-2662.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xie L, Green PL. 2005. Envelope is a major viral determinant of the distinct in vitro cellular transformation tropism of human T-cell leukemia virus type 1 (HTLV-1) and HTLV-2. J Virol 79:14536–14545. doi: 10.1128/JVI.79.23.14536-14545.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamamoto N, Matsumoto T, Koyanagi Y, Tanaka Y, Hinuma Y. 1982. Unique cell lines harbouring both Epstein-Barr virus and adult T-cell leukaemia virus, established from leukaemia patients. Nature 299:367–369. doi: 10.1038/299367a0. [DOI] [PubMed] [Google Scholar]
- 15.Ye J, Xie L, Green PL. 2003. Tax and overlapping Rex sequences do not confer the distinct transformation tropisms of human T-cell leukemia virus types 1 and 2. J Virol 77:7728–7735. doi: 10.1128/JVI.77.14.7728-7735.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Araujo A, Hall WW. 2004. Human T-lymphotropic virus type II and neurological disease. Ann Neurol 56:10–19. doi: 10.1002/ana.20126. [DOI] [PubMed] [Google Scholar]
- 17.Biglione MM, Pizarro M, Salomon HE, Berria MI. 2003. A possible case of myelopathy/tropical spastic paraparesis in an Argentinian woman with human T lymphocyte virus type II. Clin Infect Dis 37:456–458. doi: 10.1086/376620. [DOI] [PubMed] [Google Scholar]
- 18.Murphy EL, Fridey J, Smith JW, Engstrom J, Sacher RA, Miller K, Gibble J, Stevens J, Thomson R, Hansma D, Kaplan J, Khabbaz R, Nemo G. 1997. HTLV-associated myelopathy in a cohort of HTLV-I and HTLV-II-infected blood donors. The REDS Investigators. Neurology 48:315–320. [DOI] [PubMed] [Google Scholar]
- 19.Clerc I, Polakowski N, Andre-Arpin C, Cook P, Barbeau B, Mesnard JM, Lemasson I. 2008. An interaction between the human T cell leukemia virus type 1 basic leucine zipper factor (HBZ) and the KIX domain of p300/CBP contributes to the down-regulation of tax-dependent viral transcription by HBZ. J Biol Chem 283:23903–23913. doi: 10.1074/jbc.M803116200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaudray G, Gachon F, Basbous J, Biard-Piechaczyk M, Devaux C, Mesnard J. 2002. The complementary strand of the human T-cell leukemia virus type 1 RNA genome encodes a bZIP transcription factor that down-regulates viral transcription. J Virol 76:12813–12822. doi: 10.1128/JVI.76.24.12813-12822.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lemasson I, Lewis MR, Polakowski N, Hivin P, Cavanagh MH, Thebault S, Barbeau B, Nyborg JK, Mesnard JM. 2007. Human T-cell leukemia virus type 1 (HTLV-1) bZIP protein interacts with the cellular transcription factor CREB to inhibit HTLV-1 transcription. J Virol 81:1543–1553. doi: 10.1128/JVI.00480-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Basbous J, Arpin C, Gaudray G, Piechaczyk M, Devaux C, Mesnard J. 2003. HBZ factor of HTLV-1 dimerizes with transcription factors JunB and c-Jun and modulates their transcriptional activity. J Biol Chem 278:43620–43627. doi: 10.1074/jbc.M307275200. [DOI] [PubMed] [Google Scholar]
- 23.Matsumoto J, Ohshima T, Isono O, Shimotohno K. 2005. HTLV-1 HBZ suppresses AP-1 activity by impairing both the DNA-binding ability and the stability of c-Jun protein. Oncogene 24:1001–1010. doi: 10.1038/sj.onc.1208297. [DOI] [PubMed] [Google Scholar]
- 24.Thebault S, Basbous J, Hivin P, Devaux C, Mesnard JM. 2004. HBZ interacts with JunD and stimulates its transcriptional activity. FEBS Lett 562:165–170. doi: 10.1016/S0014-5793(04)00225-X. [DOI] [PubMed] [Google Scholar]
- 25.Arnold J, Zimmerman B, Li M, Lairmore MD, Green PL. 2008. Human T-cell leukemia virus type-1 antisense-encoded gene, Hbz, promotes T lymphocyte proliferation. Blood 112:3788–3797. doi: 10.1182/blood-2008-04-154286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Satou Y, Yasunaga J, Yoshida M, Matsuoka M. 2006. HTLV-I basic leucine zipper factor gene mRNA supports proliferation of adult T cell leukemia cells. Proc Natl Acad Sci U S A 103:720–725. doi: 10.1073/pnas.0507631103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arnold J, Yamamoto B, Li M, Phipps AJ, Younis I, Lairmore MD, Green PL. 2006. Enhancement of infectivity and persistence in vivo by HBZ, a natural antisense coded protein of HTLV-1. Blood 107:3976–3982. doi: 10.1182/blood-2005-11-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gazon H, Lemasson I, Polakowski N, Cesaire R, Matsuoka M, Barbeau B, Mesnard JM, Peloponese JM Jr. 2012. Human T-cell leukemia virus type 1 (HTLV-1) bZIP factor requires cellular transcription factor JunD to upregulate HTLV-1 antisense transcription from the 3′ long terminal repeat. J Virol 86:9070–9078. doi: 10.1128/JVI.00661-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hagiya K, Yasunaga J, Satou Y, Ohshima K, Matsuoka M. 2011. ATF3, an HTLV-1 bZip factor binding protein, promotes proliferation of adult T-cell leukemia cells. Retrovirology 8:19. doi: 10.1186/1742-4690-8-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mukai R, Ohshima T. 2011. Dual effects of HTLV-1 bZIP factor in suppression of interferon regulatory factor 1. Biochem Biophys Res Commun 409:328–332. doi: 10.1016/j.bbrc.2011.05.014. [DOI] [PubMed] [Google Scholar]
- 31.Douceron E, Kaidarova Z, Miyazato P, Matsuoka M, Murphy EL, Mahieux R. 2012. HTLV-2 APH-2 expression is correlated with proviral load but APH-2 does not promote lymphocytosis. J Infect Dis 205:82–86. doi: 10.1093/infdis/jir708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Halin M, Douceron E, Clerc I, Journo C, Ko NL, Landry S, Murphy EL, Gessain A, Lemasson I, Mesnard JM, Barbeau B, Mahieux R. 2009. Human T-cell leukemia virus type 2 produces a spliced antisense transcript encoding a protein that lacks a classic bZIP domain but still inhibits Tax2-mediated transcription. Blood 114:2427–2438. doi: 10.1182/blood-2008-09-179879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yin H, Kannian P, Dissinger N, Haines R, Niewiesk S, Green PL. 2012. HTLV-2 APH-2 is dispensable for in vitro immortalization, but functions to repress early viral replication in vivo. J Virol 86:8412–8421. doi: 10.1128/JVI.00717-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marban C, McCabe A, Bukong TN, Hall WW, Sheehy N. 2012. Interplay between the HTLV-2 Tax and APH-2 proteins in the regulation of the AP-1 pathway. Retrovirology 9:98. doi: 10.1186/1742-4690-9-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doueiri R, Anupam R, Kvaratskhelia M, Green KB, Lairmore MD, Green PL. 2012. Comparative host protein interactions with HTLV-1 p30 and HTLV-2 p28: insights into difference in pathobiology of human retroviruses. Retrovirology 9:64. doi: 10.1186/1742-4690-9-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feuer G, Green PL. 2005. Comparative biology of human T-cell lymphotropic virus type 1 (HTLV-1) and HTLV-2. Oncogene 24:5996–6004. doi: 10.1038/sj.onc.1208971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nicot C, Dundr JM, Johnson JR, Fullen JR, Alonzo N, Fukumoto R, Princler GL, Derse D, Misteli T, Franchini G. 2004. HTLV-1-encoded p30II is a post-transcriptional negative regulator of viral replication. Nat Med 10:197–201. doi: 10.1038/nm984. [DOI] [PubMed] [Google Scholar]
- 38.Ross TM, Pettiford SM, Green PL. 1996. The tax gene of human T-cell leukemia virus type 2 is essential for transformation of human T lymphocytes. J Virol 70:5194–5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Valeri VW, Hryniewicz A, Andresen V, Jones K, Fenizia C, Bialuk I, Chung HK, Fukumoto R, Washington Parks R, Ferrari MG, Nicot C, Cecchinato V, Ruscetti F, Franchini G. 2010. Requirement of the human T-cell leukemia virus p12 and p30 genes for infectivity of human dendritic cells and macaques but not rabbits. Blood 116:3809–3817. doi: 10.1182/blood-2010-05-284141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamamoto B, Li M, Kesic M, Younis I, Lairmore MD, Green PL. 2008. Human T-cell leukemia virus type 2 post-transcriptional control protein p28 is required for viral infectivity and persistence in vivo. Retrovirology 5:38. doi: 10.1186/1742-4690-5-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Younis I, Khair L, Dundr M, Lairmore MD, Franchini G, Green PL. 2004. Repression of human T-cell leukemia virus type 1 and 2 replication by a viral mRNA-encoded posttranscriptional regulator. J Virol 78:11077–11083. doi: 10.1128/JVI.78.20.11077-11083.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McKinsey TA, Brockman JA, Scherer DC, Al-Murrani SW, Green PL, Ballard DW. 1996. Inactivation of IkappaBbeta by the Tax protein of human T-cell leukemia virus type 1: a potential mechanism for constitutive induction of NF-kappaB. Mol Cell Biol 16:2083–2090. doi: 10.1128/MCB.16.5.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao T, Satou Y, Sugata K, Miyazato P, Green PL, Imamura T, Matsuoka M. 2011. HTLV-1 bZIP factor enhances TGF-beta signaling through p300 coactivator. Blood 118:1865–1876. doi: 10.1182/blood-2010-12-326199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao J, Senthil M, Ren B, Yan J, Xing Q, Yu J, Zhang L, Yim JH. 2010. IRF-1 transcriptionally upregulates PUMA, which mediates the mitochondrial apoptotic pathway in IRF-1-induced apoptosis in cancer cells. Cell Death Differ 17:699–709. doi: 10.1038/cdd.2009.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kannian P, Yin H, Doueiri R, Lairmore MD, Fernandez S, Green PL. 2012. Distinct transformation tropism exhibited by human T lymphotropic virus type 1 (HTLV-1) and HTLV-2 is the result of postinfection T cell clonal expansion. J Virol 86:3757–3766. doi: 10.1128/JVI.06900-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li M, Green PL. 2007. Detection and quantitation of HTLV-1 and HTLV-2 mRNA species by real-time RT-PCR. J Virol Methods 142:159–168. doi: 10.1016/j.jviromet.2007.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao T, Yasunaga J, Satou Y, Nakao M, Takahashi M, Fujii M, Matsuoka M. 2009. Human T-cell leukemia virus type 1 bZIP factor selectively suppresses the classical pathway of NF-kappaB. Blood 113:2755–2764. doi: 10.1182/blood-2008-06-161729. [DOI] [PubMed] [Google Scholar]
- 48.Yoshida M, Satou Y, Yasunaga J, Fujisawa J, Matsuoka M. 2008. Transcriptional control of spliced and unspliced human T-cell leukemia virus type 1 bZIP factor (HBZ) gene. J Virol 82:9359–9368. doi: 10.1128/JVI.00242-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Satou Y, Yasunaga J, Zhao T, Yoshida M, Miyazato P, Takai K, Shimizu K, Ohshima K, Green PL, Ohkura N, Yamaguchi T, Ono M, Sakaguchi S, Matsuoka M. 2011. HTLV-1 bZIP factor induces T-cell lymphoma and systemic inflammation in vivo. PLoS Pathog 7:e1001274. doi: 10.1371/journal.ppat.1001274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Romanelli MG, Diani E, Bergamo E, Casoli C, Ciminale V, Bex F, Bertazzoni U. 2013. Highlights on distinctive structural and functional properties of HTLV Tax proteins. Front Microbiol 4:271. doi: 10.3389/fmicb.2013.00271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu M, Yang L, Zhang L, Liu B, Merling R, Xia Z, Giam CZ. 2008. Human T-cell leukemia virus type 1 infection leads to arrest in the G1 phase of the cell cycle. J Virol 82:8442–8455. doi: 10.1128/JVI.00091-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang L, Kotomura N, Ho YK, Zhi H, Bixler S, Schell MJ, Giam CZ. 2011. Complex cell cycle abnormalities caused by human T-lymphotropic virus type 1 Tax. J Virol 85:3001–3009. doi: 10.1128/JVI.00086-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhi H, Yang L, Kuo YL, Ho YK, Shih HM, Giam CZ. 2011. NF-kappaB hyper-activation by HTLV-1 Tax induces cellular senescence, but can be alleviated by the viral anti-sense protein HBZ. PLoS Pathog 7:e1002025. doi: 10.1371/journal.ppat.1002025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tanaka N, Ishihara M, Kitagawa M, Harada H, Kimura T, Matsuyama T, Lamphier MS, Aizawa S, Mak TW, Taniguchi T. 1994. Cellular commitment to oncogene-induced transformation or apoptosis is dependent on the transcription factor IRF-1. Cell 77:829–839. doi: 10.1016/0092-8674(94)90132-5. [DOI] [PubMed] [Google Scholar]
- 55.Tamura T, Ishihara M, Lamphier MS, Tanaka N, Oishi I, Aizawa S, Matsuyama T, Mak TW, Taki S, Taniguchi T. 1997. DNA damage-induced apoptosis and Ice gene induction in mitogenically activated T lymphocytes require IRF-1. Leukemia 11(Suppl 3):S439–S440. [PubMed] [Google Scholar]
- 56.Boultwood J, Fidler C, Lewis S, MacCarthy A, Sheridan H, Kelly S, Oscier D, Buckle VJ, Wainscoat JS. 1993. Allelic loss of IRF1 in myelodysplasia and acute myeloid leukemia: retention of IRF1 on the 5q- chromosome in some patients with the 5q- syndrome. Blood 82:2611–2616. [PubMed] [Google Scholar]
- 57.Willman CL, Sever CE, Pallavicini MG, Harada H, Tanaka N, Slovak ML, Yamamoto H, Harada K, Meeker TC, List AF, Taniguchi T. 1993. Deletion of Irf-1, mapping to chromosome 5q311 in human leukemia and preleukemic myelodysplasia. Science 259:968–971. doi: 10.1126/science.8438156. [DOI] [PubMed] [Google Scholar]
- 58.Higuchi M, Tsubata C, Kondo R, Yoshida S, Takahashi M, Oie M, Tanaka Y, Mahieux R, Matsuoka M, Fujii M. 2007. Cooperation of NF-kappaB2/p100 activation and the PDZ domain binding motif signal in human T-cell leukemia virus type 1 (HTLV-1) Tax1 but not HTLV-2 Tax2 is crucial for interleukin-2-independent growth transformation of a T-cell line. J Virol 81:11900–11907. doi: 10.1128/JVI.00532-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yamamoto-Taguchi N, Satou Y, Miyazato P, Ohshima K, Nakagawa M, Katagiri K, Kinashi T, Matsuoka M. 2013. HTLV-1 bZIP factor induces inflammation through labile Foxp3 expression. PLoS Pathog 9:e1003630. doi: 10.1371/journal.ppat.1003630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Takahashi R, Nishimoto S, Muto G, Sekiya T, Tamiya T, Kimura A, Morita R, Asakawa M, Chinen T, Yoshimura A. 2011. SOCS1 is essential for regulatory T cell functions by preventing loss of Foxp3 expression as well as IFN-gamma and IL-17A production. J Exp Med 208:2055–2067. doi: 10.1084/jem.20110428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Charoenthongtrakul S, Zhou QJ, Shembade N, Harhaj NS, Harhaj EW. 2011. Human T cell leukemia virus type 1 Tax inhibits innate antiviral signaling via NF-kappa B-dependent induction of SOCS1. J Virol 85:6955–6962. doi: 10.1128/JVI.00007-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oliere S, Hernandez E, Lezin A, Arguello M, Douville R, Nguyen TLA, Olindo S, Panelatti G, Kazanji M, Wilkinson P, Sekaly RP, Cesaire R, Hiscott J. 2010. HTLV-1 evades type I interferon antiviral signaling by inducing the suppressor of cytokine signaling 1 (SOCS1). PLoS Pathog 6:e1001177. doi: 10.1371/journal.ppat.1001177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen S, Ishii N, Ine S, Ikeda S, Fujimura T, Ndhlovu LC, Soroosh P, Tada K, Harigae H, Kameoka J, Kasai N, Sasaki T, Sugamura K. 2006. Regulatory T cell-like activity of Foxp3+ adult T cell leukemia cells. Int Immunol 18:269–277. [DOI] [PubMed] [Google Scholar]
- 64.Ross TM, Minella AC, Fang ZY, Pettiford SM, Green PL. 1997. Mutational analysis of human T-cell leukemia virus type 2 Tax. J Virol 71:8912–8917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xie L, Yamamoto B, Haoudi A, Semmes OJ, Green PL. 2006. PDZ binding motif of HTLV-1 Tax promotes virus-mediated T-cell proliferation in vitro and persistence in vivo. Blood 107:1980–1988. doi: 10.1182/blood-2005-03-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−delta delta C(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]