ABSTRACT
Viruses that generate double-stranded RNA (dsRNA) during replication must overcome host defense systems designed to detect this infection intermediate. All positive-sense RNA viruses studied to date modify host membranes to help facilitate the sequestration of dsRNA from host defenses and concentrate replication factors to enhance RNA production. Flock House virus (FHV) is an attractive model for the study of these processes since it is well characterized and infects Drosophila cells, which are known to have a highly effective RNA silencing system. During infection, FHV modifies the outer membrane of host mitochondria to form numerous membrane invaginations, called spherules, that are ∼50 nm in diameter and known to be the site of viral RNA replication. While previous studies have outlined basic structural features of these invaginations, very little is known about the mechanism underlying their formation. Here we describe the optimization of an experimental system for the analysis of FHV host membrane modifications using crude mitochondrial preparations from infected Drosophila cells. These preparations can be programmed to synthesize both single- and double-stranded FHV RNA. The system was used to demonstrate that dsRNA is protected from nuclease digestion by virus-induced membrane invaginations and that spherules play an important role in stimulating RNA replication. Finally, we show that spherules generated during FHV infection appear to be dynamic as evidenced by their ability to form or disperse based on the presence or absence of RNA synthesis.
IMPORTANCE It is well established that positive-sense RNA viruses induce significant membrane rearrangements in infected cells. However, the molecular mechanisms underlying these rearrangements, particularly membrane invagination and spherule formation, remain essentially unknown. How the formation of spherules enhances viral RNA synthesis is also not understood, although it is assumed to be partly a result of evading host defense pathways. To help interrogate some of these issues, we optimized a cell-free replication system consisting of mitochondria isolated from Flock House virus-infected Drosophila cells for use in biochemical and structural studies. Our data suggest that spherules generated during Flock House virus replication are dynamic, protect double-stranded RNA, and enhance RNA replication in general. Cryo-electron microscopy suggests that the samples are amenable to detailed structural analyses of spherules engaged in RNA synthesis. This system thus provides a foundation for understanding the molecular mechanisms underlying spherule formation, maintenance, and function during positive-sense viral RNA replication.
INTRODUCTION
For viruses that replicate via double-stranded RNA (dsRNA) intermediates, the multiple cytosolic defense pathways directed against this intermediate represent a significant hurdle that must be overcome for effective propagation. This is achieved through a variety of mechanisms. For example, viruses with dsRNA genomes assemble particles around positive-sense RNA [(+) RNA] and generate dsRNA only within enclosed capsids (1–3). In contrast, (+) RNA viruses replicate their genomes in association with highly modified host membranes (reviewed in references 4 and 5), generating convoluted single- and double-membrane vesicles (e.g., coronaviruses and hepaciviruses) or relatively simple 50-to-70-nm-diameter membrane invaginations called spherules (first discovered in alphaviruses [6, 7]). In both cases, these membrane modifications are sufficient to protect viral RNA (vRNA) from nuclease degradation and, by inference, from host defense systems (8–10).
Flock House virus (FHV; Nodaviridae) is a nonenveloped, bipartite, (+) RNA virus that replicates its genome in association with the outer mitochondrial membranes (OMM) of infected cells (11). Protein A, the viral RNA-dependent RNA polymerase (pol), is directed to the OMM through an N-terminal targeting sequence. The process of progeny RNA synthesis leads to formation of spherules by a currently unknown mechanism(s) (11, 12). Electron tomography (ET) of fixed sections from FHV-infected Drosophila cells revealed basic architectural features of these spherules (13). They are ∼50 nm in diameter and connected to the cytoplasm through an ∼10-nm-wide “neck.” It was also confirmed that they contain protein A and represent the sites of viral RNA synthesis. The FHV-induced spherules are structurally similar to those formed by the plant virus brome mosaic virus (BMV), which generates invaginations ∼60 nm in diameter on perinuclear endoplasmic reticulum (ER) membranes (14, 15). The similarities were assumed to indicate that FHV invaginations are formed in a manner comparable to those seen in BMV (13).
In BMV, spherules can be induced in the absence of any other viral factors through the expression of protein 1a, a multifunctional, nonstructural protein that has 5′ RNA capping and RNA NTPase/helicase domains (8, 16). Since 1a is sufficient for development of ER invaginations, it is thought to line their internal surface, where it helps to maintain membrane curvature, and is responsible for the recruitment of the BMV replicase (2a polymerase [2apol]) and vRNA for replication (4, 8, 14). Although 2apol is recruited before spherule formation, it is thought that vRNA is recruited into preformed invaginations where it becomes resistant to RNase digestion (8, 17, 18). While 1a is the only viral factor required for spherule formation, it has recently been demonstrated that host reticulon and ESCRT (endosomal sorting complexes required for transport) proteins play an important role in the formation and maintenance of the energetically unfavorable membrane curvatures required for functional invaginations (19, 20).
There are significant differences between BMV and FHV in terms of their fundamental mechanisms of RNA synthesis. While BMV makes use of a viral replicase (2apol) in addition to an accessory protein (1a) that is thought to be present in spherules at a 20-fold excess over 2apol (8), FHV expresses only a single replication protein (protein A), which, although sufficient for replication and spherule formation, is unable to form invaginations without active RNA synthesis (12, 21). This suggests that at least some details of the mechanisms of FHV spherule formation are different from those used by BMV (8, 22, 23). Moreover, it suggests the potential for a direct role of vRNA in FHV spherule formation that is absent in BMV. The significance of these observations has yet to be explored.
As a first step toward exploring whether there are any unique features of FHV spherule formation, we have optimized a cell-free replication system consisting of crude mitochondria from FHV-infected Drosophila cells. This system contains all of the necessary components for vRNA replication. Our data suggest that, in FHV, spherules can disappear and reform relatively quickly in response to cessation or reinitiation of RNA replication in vitro. In addition, we show that disruption of membranes inhibits efficient RNA synthesis and that, following spherule formation, dsRNA, but not ssRNA, is protected from nuclease degradation. These results confirm the role that spherules play in the concentration of factors for RNA replication and their function in protection of dsRNA from host defense systems.
MATERIALS AND METHODS
Preparation of isolated mitochondria containing FHV protein A.
Drosophila S2 cells (5 × 106 cells/ml) were infected with gradient-purified FHV (multiplicity of infection [MOI] of 5) and incubated at 28°C for 14 h. Cells were harvested at 1,500 × g for 4 min at 4°C and washed once in lysis buffer (15 mM HEPES [pH 7.4], 5 mM KCl, 1 mM CaCl2, 1 mM MnCl2, 0.5 mM dithiothreitol [DTT], 0.28 M sucrose, protease inhibitor cocktail set V [Millipore]). Cells were then resuspended in 1 ml lysis buffer per 2.5 × 108 cells and homogenized using 20 passes through a 25-gauge needle. Unlysed cells and debris were pelleted at 1,500 × g for 4 min at 4°C and lysed again in half the volume of lysis buffer. Debris was pelleted again, and the supernatants were pooled to generate a cleared lysate. This was centrifuged at 10,000 × g for 10 min at 4°C to obtain the crude mitochondrial pellet, which was gently resuspended in 1 ml mitochondrial resuspension buffer (50 mM Tris [pH 8.0], 100 mM NaCl, 1 mM DTT, 1 mM MnCl2, 20 mM MgCl2, 15% glycerol [vol/vol]) per 5 × 108 input cells. Crude mitochondria were washed 3 times by pelleting and resuspension in mitochondrial resuspension buffer before being stored in aliquots at −20°C.
FHV polymerase assays.
The standard assay (50 μl) contained 8 μl mitochondrial extract diluted into replication buffer to a final concentration of 50 mM Tris (pH 8.2), 18 mM MgCl2, 30 mM KCl, 16 mM NaCl, 2.4% (vol/vol) glycerol, 10 μg/ml actinomycin D, 30 U RNaseOUT (Life Technologies), 1 mM ATP/GTP/UTP, 0.8 mM CTP, 0.2 mM [α-33P] CTP (specific activity, 3,000 Ci/mmol [PerkinElmer]), and 2.5 ng/μl purified FHV vRNA. Due to the presence of both magnesium and manganese in the mitochondrial resuspension buffer, all reaction mixtures contained an additional 0.16 mM Mn2+ and 3.2 mM Mg2+. Reaction mixtures were incubated at 30°C for 90 min before purification of RNA using TRIzol LS (Life Technologies). Total RNA was resuspended in a 10-μl solution (7 μl RNase-free water plus 3 μl gel loading buffer II [Ambion]), heated to 55°C for 10 min, cooled on ice, and resolved on 1.5% native agarose gels. Gels were covered with one layer of Saran plastic wrap and dried on Whatman 3MM chromatography paper (GE Healthcare), and RNA products were detected using film or imaging using storage phosphor screens (GE Healthcare) and a Storm 820 phosphorimager (Amersham Biosciences).
Assignment of various RNA species.
Total RNA generated in polymerase assays was treated with micrococcal nuclease (New England BioLabs), RNase H (New England BioLabs), or RNase A (Roche Diagnostics) according to the manufacturers' instructions. In addition, total RNA products were resolved alongside ssRNA1 and ssRNA2 generated by in vitro transcription as previously described (24) in the presence of 0.1 mM [α-33P]CTP.
Effect of detergent on RNA replication.
RNA polymerase reaction mixtures were prepared as normal in the absence of detergent or with the addition of either 1% (vol/vol) NP-40 or 1% (vol/vol) Triton X-100 and incubated for 90 min before being purified and analyzed.
Accessibility of viral RNA to nucleases during replication.
RNA polymerase assay mixtures were incubated for 90 min before the addition of micrococcal nuclease with or without detergents (1% [vol/vol] NP-40 or 1% [vol/vol] Triton X-100) and incubated at 37°C for 30 min prior to RNA purification and analysis.
Cryo-electron microscopy (cryo-EM) of isolated mitochondria.
Undiluted isolated mitochondria (3 μl) in mitochondrial resuspension buffer without glycerol were placed on 2.0/0.5 C-flat grids (Protochips) that had been plasma cleaned for 5 s at 20 mA using a Gatan Solarus cleaning system and vitrified in liquid ethane with a manual cryo-plunger using a blot time of ∼2 to ∼3 s. Images were acquired using an FEI Tecnai F20 Twin transmission electron microscope operating at 200 kV and equipped with a Tietz complementary metal-oxide semiconductor (CMOS) digital camera (Tietz F416) utilizing the Leginon software system (25) for automated data collection. Images were recorded at ×62,000 magnification (2.73 Å/pixel at the specimen level) with a nominal underfocus range of 2 to 4 μm and a dose rate of between 20 and 30 e-/Å2.
The same process was followed for imaging of mitochondria following the reinitiation of RNA replication. For these experiments, one part crude mitochondria was mixed with one part of a 2× replication reaction mixture (as described above, without radiolabeled CTP) and incubated at 30°C for 30 to 90 min before being placed on the grid and vitrified.
Fixation, staining, sectioning, and transmission electron microscopy of mitochondria in FHV-infected Drosophila cells.
Drosophila S2 cells were infected with gradient-purified FHV (MOI = 10). Twenty hours after infection, cells were chemically fixed on 35-by-10-mm glass-bottom petri dishes (Ted Pella) with 2% glutaraldehyde (in 100 mM sodium cacodylate buffer; pH 7.4) for 1 h on ice. Cells were stained in the dish with 1% osmium tetroxide for 30 min on ice and washed with water followed by 2% uranyl acetate for 1 h on ice. Cells were then dehydrated in an ethanol/water gradient and embedded in Durcupan resin in dish. Sections (100 nm) were cut (Leica UC7 Ultramicrotome) and placed on Formvar-coated copper slot grids (Electron Microscopy Sciences). Sections were poststained in 2% uranyl acetate (1 h) and Sato's lead (2 min) before imaging was performed using a Philips CM100 transmission electron microscope at 100 kV. Images were captured using a Gatan Orius charge-coupled-device (CCD) camera.
RESULTS
Optimization of FHV RNA replication system.
We optimized a FHV RNA replication system described in two previous reports: the first outlined a cell-free replication system consisting of a lysate generated from FHV-infected Drosophila cells (26), and the second described the biochemical characterization of a protein A-maltose binding protein (MBP) fusion construct expressed in Escherichia coli (27). Our assay relied on isolation of crude mitochondria from FHV-infected Drosophila S2 cells by differential centrifugation. These mitochondria (in mitochondrial resuspension buffer consisting of 50 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM DTT, 1 mM MnCl2, 20 mM MgCl2, and 15% glycerol) were then placed in buffer containing the components necessary for RNA replication (see Materials and Methods for details) and incubated at 30°C for 90 min. Total RNA was extracted and analyzed by gel electrophoresis.
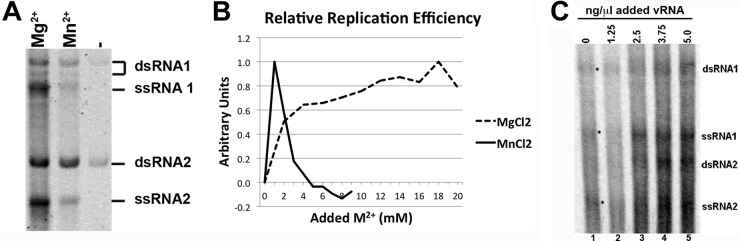
Following gel electrophoresis, radiolabeled progeny bands were identified as double-stranded and single-stranded species of RNA 1 and RNA 2 based on previously published observations (26) and our own RNase digestion reactions (Fig. 1). Interestingly, dsRNA1 consistently appeared as a doublet throughout this study, though the reason for the presence of two species remains unknown. In the absence of exogenously added FHV RNA, radiolabeled viral progeny RNA was faintly detectable (Fig. 2C, lane 1). This was consistent with previous results by Wu and Kaesberg (26), who had shown that vRNA copurifies with the replication complex in a crude cell lysate. In addition, they had demonstrated that purified FHV RNA represents a suitable exogenous template for generating single- and double-stranded progeny RNA1 and -2. Because our system generated progeny RNA that was only faintly detectable, we routinely added purified FHV RNA (2.5 ng/μl) to the mitochondrial extract prior to incubation at 30°C for 90 min. This increased the amount of radiolabeled RNA produced and thereby facilitated accurate quantification.
FIG 1.
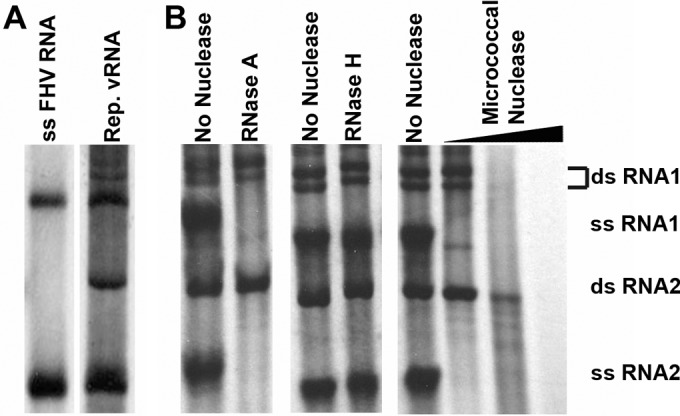
Assignment of radiolabeled FHV RNA replication products. Total radiolabeled RNA products were resolved alongside radiolabeled in vitro-transcribed single-stranded FHV RNA1 and RNA 2 (A) or nuclease-digested FHV replication (Rep.) products (B). RNase A digests ssRNA, RNase H digests DNA:RNA hybrids, and micrococcal nuclease digests single-stranded and double-stranded RNA and DNA. The doublet for dsRNA1 was assigned based on resistance to RNase A digestion and increased size in comparison to ssRNA1 and was consistently observed throughout all experiments.
FIG 2.
Determination of optimal divalent cation concentration for FHV RNA replication. (A) Total RNA (1 μg) from reaction mixtures supplemented with 18 mM Mg2+ (“Mg2+”) or with 1 mM Mn2+ (“Mn2+”) or without added divalent cations (“−”) was resolved on a 1.5% agarose gel. Radioactive vial progeny RNA was visualized by phosphorimaging. (B) The efficiency of RNA replication in the presence of various concentrations of added Mg2+ or Mn2+ was measured using densitometry analysis of purified radiolabeled RNA resolved on 1.5% agarose gels. Arbitrary unit values were calculated by assigning the reaction with no added divalent cation a value of 0 and the reaction with the strongest signal a value of 1. Panels A and B show data from single experiments that are representative of four and five experimental repeats, respectively. Although the peak at 18 mM Mg2+ was less pronounced in some replicates, this concentration of added magnesium consistently generated the most RNA product. All reaction mixtures contained an additional 0.16 mM Mn2+ and 3.2 mM Mg2+ as a consequence of the use of the mitochondrial resuspension buffer containing 1 mM Mn2+ and 20 mM Mg2+ (see Materials and Methods for details). (C) Radiolabeled RNA generated in the presence of various amounts of exogenously added purified FHV RNA template. Asterisks in lane 1 indicate faintly visible progeny RNA in the absence of added template.
Using these crude mitochondrial preparations, a wide range of parameters was tested in order to identify optimal reaction conditions. These included the evaluation of compositional differences in the two previously reported systems (summarized in Table 1). The analysis revealed that, of the parameters whose values were in disagreement, only the type and concentration of divalent cations had a significant impact on RNA replication efficiency. When the effect of Mg2+ and Mn2+ on RNA replication was assessed, resolution of the two reaction mixtures on 1.5% agarose gels generated identical RNA products (Fig. 2A). Due to the presence of both divalent cations in the mitochondrial resuspension buffer, all reaction mixtures contained an additional 0.16 mM Mn2+ and 3.2 mM Mg2+ which were likely responsible for the small amount of residual activity in the control reaction (Fig. 2A).
TABLE 1.
Comparison of the reaction conditions optimized in this study with those already publisheda
| Study | Buffer | Monovalent salt | Divalent salt | Act. D | Detection | [NTP] | Template |
|---|---|---|---|---|---|---|---|
| Wu and Kaesberg (26) | Tris acetate (pH 8.2) | 15 mM potassium acetate | 15 mM MgAc2 | + | [α-32P] CTP | 1 mM | Copurified/purified vRNAb |
| Wu et al. (27) | HEPES (pH 8.0) | 100 mM NaCl | 1 mM MnCl2 | − | DIG-11-UTP | 250 μM | ∼200-nt (+) sense genome fragments |
| This study | Tris acetate (pH 8.2) | 30 mM KCl | 18 mM MgCl2 | + | [α-33P]CTP | 1 mM | Copurified/purified vRNAb |
Act. D, actinomycin D; nt, nucleotide; NTP, nucleoside triphosphate.
Template RNA was either viral RNA copurified with mitochondrial fractions or viral RNA purified from virus particles or a combination of the two.
To determine which concentration produced the largest amount of progeny RNA, total RNAs from reaction mixtures containing various amounts of added divalent cations (0 to 20 mM for Mg2+ and 0 to 9 mM for Mn2+) were resolved and visualized using a phosphorimager. Densitometry measurements showed that optimal RNA replication occurred with the addition of 18 mM Mg2+ and 1 mM Mn2+ (Fig. 2B). The Mn2+ value is in agreement with previous biochemical characterizations (27, 28), while the Mg2+ concentration was similar to the amount previously used in cell extracts (26). Specific radioactivity (counts per minute per nanogram of RNA) of total RNA generated in the presence of 18 mM Mg2+ or 1 mM Mn2+ (Fig. 2A) showed that reaction mixtures containing Mg2+ generated approximately 2.2-fold (±0.3-fold over four experiments) more radiolabeled RNA than those containing Mn2+ (data not shown). When the activity that was the result of the small amount of divalent cations present in the mitochondrial resuspension buffer was taken into account, 4.5-fold (± 0.9-fold) more radiolabeled RNA was generated in the presence of Mg2+ than in the presence of Mn2+.
Intact membranes are required for efficient RNA synthesis.
Since vRNA replication is required for the formation of spherules in OMMs (12) and since glycerophospholipids have been shown to stimulate complete FHV vRNA replication in vitro (29), the effect of membrane destabilization on RNA replication was also assessed. FHV RNA replication reaction mixtures were prepared in the presence or absence of 1% NP-40 or 1% Triton X-100 and analyzed as before (Fig. 3). Unlike the control reactions, which generated significant amounts of all FHV replication products, reaction mixtures containing either detergent produced only small amounts of dsRNA1 and dsRNA2 (∼4-fold and 9-fold less than the control, respectively) whereas no ssRNA was detected.
FIG 3.
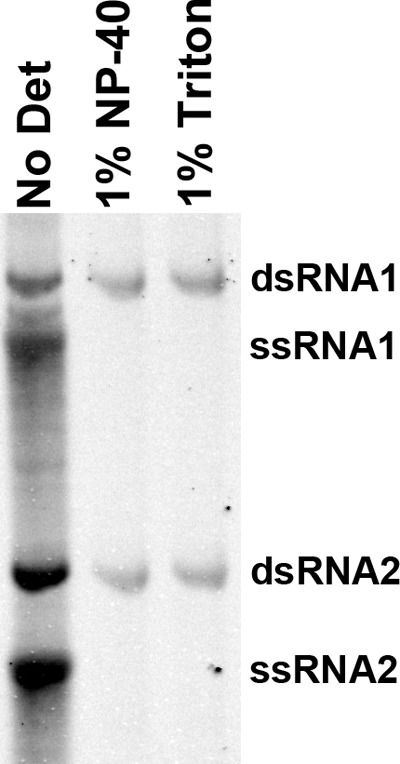
Effect of membrane disruption on FHV RNA replication. RNA replication reactions were carried out in the presence and absence of detergent. Purified total RNA was resolved on a 1.5% agarose gel before radiolabeled RNA was imaged using a phosphorimager. No Det, no detergent; 1% Triton, 1% Triton X-100. This figure is representative of results of three experimental repeats.
Intact membranes protect dsRNA, but not ssRNA, from nuclease degradation.
One of the stated functions of virus-induced spherules is the sequestration of dsRNA away from cytosolic host defenses. In order to experimentally assess this function in FHV, replication reaction mixtures were incubated for 90 min to generate significant amounts of radiolabeled RNA and then treated with micrococcal nuclease (MN) in the presence and absence of detergent (Fig. 4). In the absence of detergent, only ssRNA was degraded, with significant amounts of both dsRNA1 and dsRNA2 remaining intact (lane 2). This demonstrates that intact membranes protect dsRNA, but not ssRNA, in the context of active RNA replication. The addition of detergent alone, in the absence of MN, consistently resulted in a decrease in intensity of both ssRNA1 and dsRNA1. While we have been unable to determine the cause for this loss of signal, it appeared consistently over six experimental repeats and both RNA1 species were still detectable with extended exposures (lanes 9 and 11).
FIG 4.
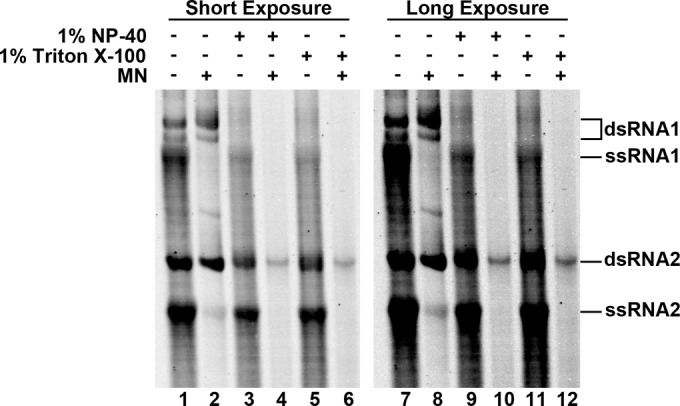
Protection of dsRNA from nuclease digestion. FHV RNA replication reactions were allowed to generate significant amounts of radiolabeled progeny before the reaction mixtures were treated with micrococcal nuclease (MN) in the presence or absence of detergent (1% NP-40 or 1% Triton X-100) as indicated. Radiolabeled RNA was imaged using a phosphorimager after total purified RNA was resolved on a 1.5% agarose gel. A longer exposure is included to show RNA1 in the presence of detergent and absence of MN.
In the presence of MN and either 1% NP-40 or 1% Triton X-100, all radiolabeled RNA species were digested (Fig. 4, lanes 4 and 6). In most cases, the small amounts of dsRNA2 that remained represented only a small proportion of the total originally present (compare adjacent control lanes). Earlier experiments had also shown that dsRNA2 was the last species to be completely digested when purified vRNA products were incubated with MN (Fig. 1), suggesting that it is inherently more resistant to MN degradation than the three other species.
Spherules form in mitochondrial membranes following reinitiation of RNA replication.
Although there has been significant advancement in our understanding of the structure of virus-induced membrane modifications, the precise organization of these structures and how they enable viral replication proteins to support genome synthesis are still unclear. Neither high-resolution nor moderate-resolution structural information is available for RNA polymerases embedded in membranes, and the molecular details of how they replicate RNA in this context remain obscure. As a result, there are many outstanding questions relating to viral RNA replication in the context of host membranes. How is the replicase positioned in the membrane? How is RNA transported in and out of spherules? Why does spherule formation in FHV require active RNA synthesis (12)? As a first step toward interrogating these questions, we visualized the spherules generated in isolated mitochondrial membranes using cryo-electron microscopy (cryo-EM). This technique has a number of important advantages over the traditional techniques used in previous studies. Cryo-EM allows preservation and analysis of specimens in a frozen-hydrated state, eliminating the need for fixation and staining, which are known to cause artifacts, particularly in the case of structural preservation of cellular membranes (30). In addition, isolated mitochondria are small enough to be imaged whole, without the need for sectioning (31).
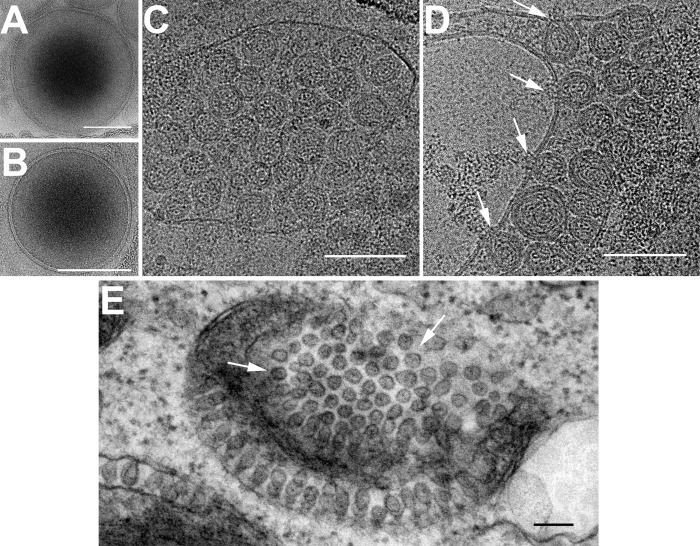
In the absence of RNA replication, the results of cryo-EM of mitochondrial isolates from infected and uninfected cells were indistinguishable (Fig. 5A and B). Both samples contained spherical, double-membraned structures as expected for mitochondria (31), but no spherules were observed in samples taken from infected cells. Since most of the replication templates, as well as the substrate, were presumably lost during mitochondrial purification, the lack of membrane invaginations was consistent with previous observations showing that spherule formation requires active RNA replication (12). In order to induce spherule formation, mitochondria from infected cells were preincubated in an RNA replication mixture for up to 90 min prior to imaging. The mitochondria were fragile following replication reinitiation, but numerous spherules were detectable in mitochondrial fragments after the incubation (Fig. 5C and D). They typically appeared as closed circles highly reminiscent of spherules that were perpendicular to the image plane in mitochondrial slices obtained after fixation, sectioning, and staining of infected Drosophila cells (white arrows in Fig. 5E). All of the spherules consisted of spherical double membranes that encapsulated swirls of density inside the lumen. Several views of the spherules confirmed that they are invaginations of the membrane that leave an opening (neck) approximately 10 nm wide, and the internal density appeared in some cases to be spilling out of the neck (white arrows in Fig. 5D). Although the spherules were not as uniform in size as those described previously (13), they had a similar overall structure (body and neck) and similar dimensions (∼35 to 65 nm in diameter).
FIG 5.
Electron microscopy of crude mitochondria isolated from mock- and FHV-infected Drosophila cells. (A and B) Cryo-electron microscopy of mitochondria isolated from uninfected (A) and FHV-infected (B) Drosophila cells showing the characteristic double mitochondrial membrane. (C and D) Mitochondria isolated from FHV-infected cells develop spherules after preincubation in RNA replication mix. White arrows in panel D indicate areas where density is seen coming out of the neck region of spherules. (E) Transmission electron microscopy (TEM) of fixed and stained sections from FHV-infected Drosophila cells showing mitochondrial membranes containing spherules perpendicular to the image plane (white arrows). Scale bars = 100 nm.
DISCUSSION
All viruses that generate dsRNA must overcome cytosolic defense systems that are highly effective at recognizing this essential replication intermediate. FHV provides an interesting opportunity to study these defense systems since it is very well characterized in terms of its replication biology and naturally infects Drosophila cells, which have a sophisticated RNA silencing system. In order to produce active infection, FHV requires the activity of protein B2, which is known to block cleavage of dsRNA by Dicer and prevent the incorporation of small interfering RNAs (siRNAs) into the RNA-induced silencing complex (32, 33). In addition, like that of all (+) RNA viruses studied to date, FHV RNA replication takes place in association with host membranes, which is thought to aid replication by concentrating essential factors where they are needed and to sequester dsRNA away from host defense systems. Here we describe the development of an in vitro experimental system for the study of FHV-induced membrane invaginations that uses crude preparations of mitochondria from infected Drosophila cells and its application in the study of the role of spherules in FHV RNA replication.
Of interest during the optimization of this system was a discrepancy in the literature regarding the divalent cation required for optimal protein A activity. Two previous studies had reported reaction conditions for FHV replication: the first included 15 mM Mg2+ in reactions using cell extracts (26, 34) in contrast to the 1 mM Mn2+ suggested by a more recent biochemical study performed using a purified protein A-MBP fusion protein (27). Our analysis suggests that, whereas protein A is able to generate the same RNA species in the presence of either metal ion, inclusion of magnesium in reaction mixtures results in the production of significantly more progeny RNA (Fig. 2). There are two main differences between this study and that by Wu and colleagues (27) that could account for the apparent differences in replication efficiency. First, the sources and contexts of protein A are different, consisting of wild-type protein embedded in OMM in this study in contrast to a purified, heterologously expressed protein A-MBP fusion protein. Subtle differences resulting from the addition of an MBP tag and the removal of associated membranes during purification or bacterial expression could explain the change in divalent cation preference—especially considering that it is known that lipid composition has a significant effect on RNA replication rates in FHV (29). Second, the choice of reaction template (full-length vRNA versus ∼200-nucleotide [nt] FHV RNA fragments in the study by Wu et al.) could highlight differential preferences based on the stage of replication being analyzed. This is the case in BMV, where manganese can substitute for magnesium during replication initiation but not during subsequent steps (35). Similarly, both magnesium and manganese have been implicated in various stages of RNA replication in hepatitis C virus and it has been suggested that Mn2+ is preferred for initiation (36, 37). While it is unclear which of these factors are involved (and to what extent), it is most likely that the differences in replication rates are due to changes in cation preferences at different stages of replication, as BMV and hepatitis C virus exhibit similar patterns of inhibition in the presence of increasing amounts of Mn2+ (Fig. 2A) (35, 37). This suggests that Mg2+, and not Mn2+, is the preferred catalytic metal ion required for maximum protein A activity, although Mn2+ may be able to substitute during replication initiation and/or dsRNA synthesis.
The development of this in vitro experimental system provided an opportunity to gain a greater understanding of both the formation and role of spherules in FHV RNA replication. Previous studies had established that complete RNA replication of FHV RNA is stimulated by the addition of glycerophospholipids (29); however, the requirement of intact membrane surfaces for protein A activity had yet to be demonstrated. Upon disruption of membranes, there was a significant loss of dsRNA production whereas ssRNA synthesis was completely inhibited. Earlier studies using cell extracts showed that template RNA can remain associated with protein A during the purification process and can act as a template for RNA synthesis once replication is reinitiated (26, 34). In addition to nuclease, 0.3% dodecyl maltoside was required to completely remove that endogenous template, suggesting a weak RNA-membrane interaction, potentially mediated by protein A. It is likely that these RNAs represent replication complexes stalled during membrane isolation as a result of the removal of cofactors and substrates, and it follows that the dsRNA products detected in the presence of detergent probably represent the reinitiation and completion of these reactions. The complete lack of ssRNA products indicates that while protein A remains enzymatically active, it is unable to initiate subsequent rounds of synthesis using these products. The presence of detergent could also have an effect on protein A self-interaction, which is important for protein A activity (38). In the event that self-interaction is specifically required for ssRNA production, the detergent in these reaction mixtures could therefore be inhibitory. Considering that protein A is capable of dsRNA synthesis under these conditions, a more likely explanation for the absence of ssRNA in these reaction mixtures is that intact membranes are required for ensuring a close association of dsRNA and protein A or for directly triggering the shift from dsRNA to ssRNA synthesis. This is in agreement with the hypothesis that one of the functions of the membrane modifications induced during (+) ssRNA replication is to concentrate factors and thereby enhance replication.
In addition to the concentration of factors to enhance replication, the association of RNA polymerization with host membranes also plays a role in sequestering dsRNA intermediates away from host defense systems. Our data show that ssRNA is readily accessible to nucleases regardless of the presence of detergent whereas the disruption of membranes is required for dsRNA digestion. This is not surprising, since dsRNA is directly targeted by host RNA interference (RNAi) defense systems whereas ssRNA is less likely to elicit intracellular defense responses. Additional protection of dsRNA is provided by B2, which is known to suppress host RNAi pathways by blocking cleavage of dsRNA by Dicer and preventing the incorporation of siRNAs into the RNA-induced silencing complex (32, 39). Earlier studies have shown that B2 directly interacts with protein A and that, in the absence of B2, the first 400 bp of dsRNA produced during (+) RNA1 synthesis are especially susceptible to degradation by Dicer (39). The functional importance of B2 is underscored by the fact that it is absolutely required for successful infection in vivo (33). Taken together, these data suggest that while the majority of dsRNA is protected within spherules, short stretches may be briefly accessible to RNAi machinery during replication. Host detection of these dsRNAs is likely prevented by the B2 retained at the site of replication through its interaction with protein A. B2 is, however, expressed at much higher levels than protein A (40), leading to uncertainty regarding the role of B2 if relatively small amounts are required to protect dsRNA. There are two possible reasons for this inconsistency. First, the additional B2 could represent an extra level of defense against a system that, if activated, has the potential to rapidly and completely shut down FHV replication. Second, it is our experience that RNase III is capable of digesting both ssRNA1 and ssRNA2, suggesting that there may be elements of secondary structure in both ssRNA1 and ssRNA2 that can serve as the templates for Dicer. As a consequence, this could be an additional important area of activity for B2: protection of regions of secondary structure in progeny ssRNA from entry in the RNAi pathway.
The development of this in vitro experimental system also presented an opportunity to visualize spherules by cryo-EM in the absence of stains and fixatives, which was of particular interest since all the EM studies completed to date have used fixed and sectioned samples (11–13). While it is known that active RNA replication is required for spherule formation (12), very little is known about how these structures form or if they persist when RNA replication is inhibited. Our observations that mitochondria isolated from infected cells no longer show any signs of invaginations and that spherules reform following the reinitiation of RNA replication (Fig. 5C and D) indicate that these structures are not stable once formed and are likely to be more dynamic than previously thought, at least in FHV. Whether these structures are more stable in other viruses is yet to be seen. It is possible that similar host factors are involved in the development and maintenance of membrane curvature in BMV (19, 20) and FHV, although the lack of an accessory replication protein analogous to BMV 1a could imply that FHV is reliant on additional host factors.
Our ability to preserve mitochondrial spherules in the frozen-hydrated state sets the stage for analysis of their structure by cryo-electron tomography (cryoET). CryoET provides an opportunity to investigate the structure of macromolecular complexes and organelles in an essentially undisturbed context, as it eliminates the need for fixation and staining. CryoET, especially combined with subtomogram averaging, can achieve resolutions below 5 nm (41–43) and has previously been used to determine the structure of the mitochondrial F1F0 ATP synthase dimer in situ at an estimated resolution of 3.7 nm (44). Moreover, the higher resolution of cryoET structures should permit more detailed insights into the supramolecular organization of FHV protein A in the membrane, provide clues regarding its orientation and oligomeric state, and identify the structure of viral dsRNA inside the spherules.
Since it is well known that dsRNA is present within spherules (45–47) and since our data demonstrate that spherules protect dsRNA from nuclease degradation (Fig. 3), it is probable that the swirls of density observed within the spherules represent dsRNA. The fact that this density was observed in all the spherules that were imaged suggests that dsRNA could play a role in the maintenance of their structure and/or their formation. These data also confirm the role of active RNA replication in the formation of spherules during FHV infection and suggest that active replication is an ongoing mechanism for their maintenance—since they disappear once replication is halted. To our knowledge, this is the first time that these characteristics have been observed in virus replication-induced membrane modifications.
The dynamics of the invaginations generated by FHV and the direct role of RNA replication in their formation are in stark contrast to those of the structures generated by BMV, which form the basis for the current spherule formation model for FHV. BMV requires a single protein for the formation of spherules, protein 1a, which is thought to line the internal surface of ER invaginations (8, 16). The lack of additional factors (including the viral polymerase and active viral replication) suggests that, in BMV at least, there is no external signal required for spherule formation and therefore that these structures are likely to be more stable. In FHV, the lack of an accessory protein whose function is to form and maintain spherules has led to the hypothesis that protein A lines the surface in much the same way that 1a does in BMV (13). However, as we have shown here, these structures are dynamic in FHV and it appears that dsRNA could play an important role in structural maintenance and spherule formation. This does not preclude the possibility that the two viruses manipulate the same host mechanisms during spherule formation. There may, however, be some differences in the ways in which FHV uses these systems to produce spherules with similar though less-rigid structures. Taken together, these observations suggest that, while there are substantial similarities in the overall structures of FHV and BMV spherules, there could be significant differences in the fundamental properties of these structures and in the ways that they are maintained.
ACKNOWLEDGMENTS
Cryo-EM was conducted at the National Resource for Automated Molecular Microscopy, which is supported by the National Institutes of Health through the National Center for Research Resource P41 program (RR17573).
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Roy P. 2008. Bluetongue virus: dissection of the polymerase complex. J Gen Virol 89:1789–1804. doi: 10.1099/vir.0.2008/002089-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Desselberger U. 2014. Rotaviruses. Virus Res 190:75–96. doi: 10.1016/j.virusres.2014.06.016. [DOI] [PubMed] [Google Scholar]
- 3.Gridley CL, Patton JT. 2014. Regulation of rotavirus polymerase activity by inner capsid proteins. Curr Opin Virol 9:31–38. doi: 10.1016/j.coviro.2014.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.den Boon JA, Diaz A, Ahlquist P. 2010. Cytoplasmic viral replication complexes. Cell Host Microbe 8:77–85. doi: 10.1016/j.chom.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harak C, Lohmann V. 2015. Ultrastructure of the replication sites of positive-strand RNA viruses. Virology 479–480:418–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grimley PM, Berezesky IK, Friedman RM. 1968. Cytoplasmic structures associated with an arbovirus infection: loci of viral ribonucleic acid synthesis. J Virol 2:1326–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Froshauer S, Kartenbeck J, Helenius A. 1988. Alphavirus RNA replicase is located on the cytoplasmic surface of endosomes and lysosomes. J Cell Biol 107:2075–2086. doi: 10.1083/jcb.107.6.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwartz M, Chen J, Janda M, Sullivan M, den Boon J, Ahlquist P. 2002. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol Cell 9:505–514. doi: 10.1016/S1097-2765(02)00474-4. [DOI] [PubMed] [Google Scholar]
- 9.van Hemert MJ, van den Worm SH, Knoops K, Mommaas AM, Gorbalenya AE, Snijder EJ. 2008. SARS-coronavirus replication/transcription complexes are membrane-protected and need a host factor for activity in vitro. PLoS Pathog 4:e1000054. doi: 10.1371/journal.ppat.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kovalev N, Pogany J, Nagy PD. 2014. Template role of double-stranded RNA in tombusvirus replication. J Virol 88:5638–5651. doi: 10.1128/JVI.03842-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller DJ, Schwartz MD, Ahlquist P. 2001. Flock house virus RNA replicates on outer mitochondrial membranes in Drosophila cells. J Virol 75:11664–11676. doi: 10.1128/JVI.75.23.11664-11676.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kopek BG, Settles EW, Friesen PD, Ahlquist P. 2010. Nodavirus-induced membrane rearrangement in replication complex assembly requires replicase protein A, RNA templates, and polymerase activity. J Virol 84:12492–12503. doi: 10.1128/JVI.01495-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kopek BG, Perkins G, Miller DJ, Ellisman MH, Ahlquist P. 2007. Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PLoS Biol 5:e220. doi: 10.1371/journal.pbio.0050220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janda M, Ahlquist P. 1993. RNA-dependent replication, transcription, and persistence of brome mosaic virus RNA replicons in S. cerevisiae. Cell 72:961–979. doi: 10.1016/0092-8674(93)90584-D. [DOI] [PubMed] [Google Scholar]
- 15.Restrepo-Hartwig MA, Ahlquist P. 1996. Brome mosaic virus helicase- and polymerase-like proteins colocalize on the endoplasmic reticulum at sites of viral RNA synthesis. J Virol 70:8908–8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.den Boon JA, Chen J, Ahlquist P. 2001. Identification of sequences in brome mosaic virus replicase protein 1a that mediate association with endoplasmic reticulum membranes. J Virol 75:12370–12381. doi: 10.1128/JVI.75.24.12370-12381.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X, Lee WM, Watanabe T, Schwartz M, Janda M, Ahlquist P. 2005. Brome mosaic virus 1a nucleoside triphosphatase/helicase domain plays crucial roles in recruiting RNA replication templates. J Virol 79:13747–13758. doi: 10.1128/JVI.79.21.13747-13758.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu L, Westler WM, den Boon JA, Wang X, Diaz A, Steinberg HA, Ahlquist P. 2009. An amphipathic alpha-helix controls multiple roles of brome mosaic virus protein 1a in RNA replication complex assembly and function. PLoS Pathog 5:e1000351. doi: 10.1371/journal.ppat.1000351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diaz A, Wang X, Ahlquist P. 2010. Membrane-shaping host reticulon proteins play crucial roles in viral RNA replication compartment formation and function. Proc Natl Acad Sci U S A 107:16291–16296. doi: 10.1073/pnas.1011105107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diaz A, Zhang J, Ollwerther A, Wang X, Ahlquist P. 2015. Host ESCRT proteins are required for bromovirus RNA replication compartment assembly and function. PLoS Pathog 11:e1004742. doi: 10.1371/journal.ppat.1004742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ball LA. 1995. Requirements for the self-directed replication of flock house virus RNA 1. J Virol 69:720–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kao CC, Ahlquist P. 1992. Identification of the domains required for direct interaction of the helicase-like and polymerase-like RNA replication proteins of brome mosaic virus. J Virol 66:7293–7302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Reilly EK, Tang N, Ahlquist P, Kao CC. 1995. Biochemical and genetic analyses of the interaction between the helicase-like and polymerase-like proteins of the brome mosaic virus. Virology 214:59–71. doi: 10.1006/viro.1995.9954. [DOI] [PubMed] [Google Scholar]
- 24.Schneemann A, Marshall D. 1998. Specific encapsidation of nodavirus RNAs is mediated through the C terminus of capsid precursor protein alpha. J Virol 72:8738–8746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. 2005. Automated molecular microscopy: the new Leginon system. J Struct Biol 151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 26.Wu S-X, Kaesberg P. 1991. Synthesis of template-sense, single-stranded flockhouse virus RNA in a cell-free replication system. J Virol 183:392–396. doi: 10.1016/0042-6822(91)90153-3. [DOI] [PubMed] [Google Scholar]
- 27.Wu W, Wang Z, Xia H, Liu Y, Qiu Y, Liu Y, Hu Y, Zhou X. 2014. Flock House virus RNA polymerase initiates RNA synthesis de novo and possesses a terminal nucleotidyl transferase activity. PLoS One 9:e86876. doi: 10.1371/journal.pone.0086876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z, Qiu Y, Liu Y, Qi N, Si J, Xia X, Wu D, Hu Y, Zhou X. 2013. Characterization of a nodavirus replicase revealed a de novo initiation mechanism of RNA synthesis and terminal nucleotidyltransferase activity. J Biol Chem 288:30785–30801. doi: 10.1074/jbc.M113.492728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu S-X, Ahlquist P, Kaesberg P. 1992. Active complete in vitro replication of nodavirus RNA requires glycerophospholipid. Proc Natl Acad Sci U S A 89:11136–11140. doi: 10.1073/pnas.89.23.11136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murk JL, Posthuma G, Koster AJ, Geuze HJ, Verkleij AJ, Kleijmeer MJ, Humbel BM. 2003. Influence of aldehyde fixation on the morphology of endosomes and lysosomes: quantitative analysis and electron microscopy. J Microsc 212:81–90. doi: 10.1046/j.1365-2818.2003.01238.x. [DOI] [PubMed] [Google Scholar]
- 31.Nicastro D, Frangakis AS, Typke D, Baumeister W. 2000. Cryo-electron tomography of neurospora mitochondria. J Struct Biol 129:48–56. doi: 10.1006/jsbi.1999.4204. [DOI] [PubMed] [Google Scholar]
- 32.Chao JA, Lee JH, Chapados BR, Debler EW, Schneemann A, Williamson JR. 2005. Dual modes of RNA-silencing suppression by Flock House virus protein B2. Nat Struct Mol Biol 12:952–957. doi: 10.1038/nsmb1005. [DOI] [PubMed] [Google Scholar]
- 33.Galiana-Arnoux D, Dostert C, Schneemann A, Hoffmann JA, Imler JL. 2006. Essential function in vivo for Dicer-2 in host defense against RNA viruses in drosophila. Nat Immunol 7:590–597. doi: 10.1038/ni1335. [DOI] [PubMed] [Google Scholar]
- 34.Saunders K, Kaesberg P. 1985. Template-dependent RNA polymerase from black beetle virus-infected Drosophila melanogaster cells. Virology 147:337–381. [DOI] [PubMed] [Google Scholar]
- 35.Sun J-H, Adkins S, Faurote G, Kao CC. 1996. Initiation of (-)-strand RNA synthesis catalyzed by the BMV RNA-dependent RNA polymerase: synthesis of oligonucleotides. Virology 226:1–12. doi: 10.1006/viro.1996.0622. [DOI] [PubMed] [Google Scholar]
- 36.Luo G, Hamatake RK, Mathis DM, Racela J, Rigat KL, Lemm J, Colonno RJ. 2000. De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J Virol 74:851–863. doi: 10.1128/JVI.74.2.851-863.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uchil PD, Satchidanandam V. 2003. Characterization of RNA synthesis, replication mechanism, and in vitro RNA-dependent RNA polymerase activity of Japanese encephalitis virus. Virology 307:358–371. doi: 10.1016/S0042-6822(02)00130-7. [DOI] [PubMed] [Google Scholar]
- 38.Dye BT, Miller DJ, Ahlquist P. 2005. In vivo self-interaction of nodavirus RNA replicase protein a revealed by fluorescence resonance energy transfer. J Virol 79:8909–8919. doi: 10.1128/JVI.79.14.8909-8919.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aliyari R, Wu Q, Li HW, Wang XH, Li F, Green LD, Han CS, Li WX, Ding SW. 2008. Mechanism of induction and suppression of antiviral immunity directed by virus-derived small RNAs in Drosophila. Cell Host Microbe 4:387–397. doi: 10.1016/j.chom.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Friesen PD, Rueckert RR. 1981. Synthesis of black beetle virus proteins in cultured Drosophila cells: differential expression of RNAs 1 and 2. J Virol 37:876–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barber CF, Heuser T, Carbajal-Gonzalez BI, Botchkarev VV Jr, Nicastro D. 2012. Three-dimensional structure of the radial spokes reveals heterogeneity and interactions with dyneins in Chlamydomonas flagella. Mol Biol Cell 23:111–120. doi: 10.1091/mbc.E11-08-0692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kudryashev M, Cyrklaff M, Wallich R, Baumeister W, Frischknecht F. 2010. Distinct in situ structures of the Borrelia flagellar motor. J Struct Biol 169:54–61. doi: 10.1016/j.jsb.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 43.Liu J, Lin T, Botkin DJ, McCrum E, Winkler H, Norris SJ. 2009. Intact flagellar motor of Borrelia burgdorferi revealed by cryo-electron tomography: evidence for stator ring curvature and rotor/C-ring assembly flexion. J Bacteriol 191:5026–5036. doi: 10.1128/JB.00340-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davies KM, Anselmi C, Wittig I, Faraldo-Gomez JD, Kuhlbrandt W. 2012. Structure of the yeast F1Fo-ATP synthase dimer and its role in shaping the mitochondrial cristae. Proc Natl Acad Sci U S A 109:13602–13607. doi: 10.1073/pnas.1204593109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hatta T, Francki RIB. 1981. Cytopathic structures associated with tonoplasts of plant cells infected with cucumber mosaic and tomato aspermy viruses. J Gen Virol 53:343–346. doi: 10.1099/0022-1317-53-2-343. [DOI] [Google Scholar]
- 46.Di Franco A, Russo M, Martelli GP. 1984. Ultrastructure and origin of cytoplasmic multivesicular bodies induced by carnation Italian ringspot virus. J Gen Virol 65:1233–1237. doi: 10.1099/0022-1317-65-7-1233. [DOI] [Google Scholar]
- 47.Lee J-Y, Marshal JA, Bowden DS. 1994. Characterization of rubella virus replication complexes using antibodies to double-stranded RNA. Virology 200:307–312. doi: 10.1006/viro.1994.1192. [DOI] [PubMed] [Google Scholar]