Abstract
Background and objectives
Acute apical abscesses are serious endodontic diseases resulting from pulpal infection with opportunistic oral microorganisms. The objective of this study was to identify and compare the oral microbiota in patients (N=18) exhibiting acute apical abscesses, originating from the demographic region in Portland, Oregon. The study hypothesis is that abscesses obtained from this demographic region may contain unique microorganisms not identified in specimens from other regions.
Design
Endodontic abscesses were sampled from patients at the Oregon Health & Science University (OHSU) School of Dentistry. DNA from abscess specimens was subjected to polymerase chain reaction amplification using 16S rRNA gene-specific primers and Cy3-dCTP labeling. Labeled DNA was then applied to microbial microarrays (280 species) generated by the Human Oral Microbial Identification Microarray Laboratory (Forsyth Institute, Cambridge, MA).
Results
The most prevalent microorganisms, found across multiple abscess specimens, include Fusobacterium nucleatum, Parvimonas micra, Megasphaera species clone CS025, Prevotella multisaccharivorax, Atopobium rimae, and Porphyromonas endodontalis. The most abundant microorganisms, found in highest numbers within individual abscesses, include F. nucleatum, P. micra, Streptococcus Cluster III, Solobacterium moorei, Streptococcus constellatus, and Porphyromonas endodontalis. Strong bacterial associations were identified between Prevotella multisaccharivorax, Acidaminococcaceae species clone DM071, Megasphaera species clone CS025, Actinomyces species clone EP053, and Streptococcus cristatus (all with Spearman coefficients >0.9).
Conclusions
Cultivable and uncultivable bacterial species have been identified in endodontic abscesses obtained from the Portland, Oregon demographic region, and taxa identifications correlated well with other published studies, with the exception of Treponema and Streptococcus cristae, which were not commonly identified in endodontic abscesses between the demographic region in Portland, Oregon and other regions.
Keywords: endodontic abscesses, oral microbiota, human oral microbial identification microarrays, anaerobic oral microorganisms, Fusobacterium nucleatum, Streptococcus cristatus
Endodontic disease is initiated by infection with multiple microorganisms and aggravated by an influx of inflammatory cells leading to pulpitis and periapical periodontitis (1–5). The formation of acute apical abscess, an inflammatory response to pulpal infection, is further characterized by pain, sensitivity of the tooth to pressure, and swelling of surrounding tissues (1–5). The induction of an acute inflammatory response leads to bone resorption surrounding the tooth apex. The acute apical abscess is representative of an advanced stage of apical periodontitis and has the potential to expand from the root canal to form cellulitis, by disseminating an inflammatory response generating purulence (1–5).
Siqueira and Rôcas (5) have identified five levels of experimentation analyzing the microorganisms contained within acute apical abscesses: 1) culture methods targeting cultivable bacteria (open-ended or broad-range culture approach), 2) molecular identification tools, including polymerase chain reaction (PCR) and the checkerboard hybridization assay (using genomic probes to detect cultivable bacteria; closed-ended molecular detection approach), 3) broad-range PCR in addition to cloning and sequencing of targeted amplicons (open-ended molecular approach), 4) molecular analysis using PCR or DNA hybridization, including reverse checkerboard hybridization, and the use of human participants in large-scale clinical studies to assess prevalence and association of diverse bacterial strains (close-ended molecular approach), and 5) the use of next-generation sequencing (NGS) for extensive, open-ended analysis of abscess specimens.
On a broad taxonomic level, the most prevalent phyla identified in acute apical abscesses include: 1) the Firmicutes (including representative members within Streptococcus, Parvimonas, Peptostreptococcus and Dialister), 2) Bacteroidetes (including members within Porphyromonas and Prevotella), 3) Fusobacteria (including members within Fusobacterium and Leptotrichia), 4) Actinobacteria (including members within Actinomyces, Atopobium, and Propionibacterium), 5) Spirochaetes (including Treponema), 6) Synergistetes (including Pyramidobacter), and 7) Proteobacteria (including Campylobacter and Eikenella) (1–5).
In acute endodontic infections, the most prevalent microorganisms include members from the phyla Firmicutes, Fusobacteria, and Bacteroidetes, or more specifically, the genera Fusobacterium, Parvimonas, Peptostreptococcus, Dialister, and Atopobium. In chronic endodontic infections, the most prevalent microorganisms include members from the phyla Firmicutes, Bacteroidetes, and Actinobacteria (1–5).
The primary objectives of this study were to identify the most prevalent oral microorganisms contained in endodontic abscesses and to identify important bacterial associations and potential synergistic relationships between species. The study hypothesis is that abscesses obtained from the demographic region in Portland, Oregon may contain unique microorganisms not identified in specimens from other regions. We sought to specifically compare microorganisms originating from the demographic region in Portland, Oregon with those contained in abscesses from Rio de Janeiro, Brazil (6), and found that taxa identifications correlated well, with the exception of Treponema and Streptococcus cristae. The identification of bacterial species in endodontic abscesses, obtained from distinct sites in North and South America, may lead to the assessment of new candidate microorganisms and synergistic relationships associated with the progression of endodontic disease and the development of acute apical abscesses.
Materials and methods
Collection of abscess specimens
Abscess specimens were collected from patients at the OHSU School of Dentistry in 2007–09, following protocols approved by the OHSU Institutional Review Board (IRB #5362). Patients obtaining treatment (adult males and females; age range 16–60 years) had swelling associated with an acute apical abscess. Diagnostic terminology was based on guidelines obtained from the Pulpal & Periapical Diagnostic Terminology, formulated by the American Board of Endodontics (2007) and referenced in Ingle's Endodontics, 6th Edition (4). Specimen collections were obtained as previously described (7) using informed consent, and patient identifiers were anonymously coded. Prior to needle aspiration of the abscess, the oral cavity was rinsed with 0.12% chlorhexidine gluconate (Zila Pharmaceuticals, Phoenix, AZ) for approximately 15 s, and the overlying mucosal surface was disinfected using Povidone–Iodine swabs (Professional Disposables, Inc., Orangeburg, NY) for approximately 30 s. Paper point samples were collected from the site of aspiration and samples placed in growth medium for determination of bacterial contaminants. Clinical aspirates were obtained using a sterile 16-gauge needle and then immediately placed within vials (Port-A-CultTM, Becton Dickinson, Cockeysville, MD) for transport to the laboratory and subsequent freezing (−85°C).
DNA extraction and Human Oral Microbial Identification Microarray (HOMIM) analysis
Abscess specimens were clarified by low-speed centrifugation to remove human cells and debris, and then subjected to DNA extraction (using the PureLink Genomic DNA Mini Kit, Invitrogen, Carlsbad, CA; now Life Technologies). Genomic DNA was electrophoresed in 0.75% agarose gels containing ethidium bromide (0.5 µg/ml) to verify nucleic acid integrity and then processed in the Human Oral Microbial Identification Microarray (HOMIM) Laboratory at the Forsyth Institute (Cambridge, MA). 16S rRNA-based oligonucleotide probes were generated and printed on aldehyde-coated glass slides (8–11). Abscess DNA fragments containing 16S rRNA gene sequences were generated by PCR, using the specimen DNA as template and 16S rRNA universal forward and reverse primers. Positive (16S universal primers) and negative buffer, and primer controls were used in the HOMIM analyses. The HOMD 16S rRNA reference sequence files, including HOMD references and sequences, are available from the Human Oral Microbiome Database.
Data normalization and statistical analysis
Each probe was tested by z test to determine if the average of the signal intensity values was significantly higher than that of local background noise. After filtering, the median of the local background noise was subtracted from the corresponding mean signal intensity and then transformed into log base 2 scale. The background-corrected intensity values in log base 2 scale were subjected to global normalization in gene microarrays (12). In the log base 2 scale, the range of transformed intensity values was clearly quantitative between 0 and 16, and reflective of differences in microbial prevalence. After normalization, the duplicate spots for the same bacterial species were averaged. Prevalence and correlation values were computed when bacteria were detected in at least 50% of the patient specimens. We used R statistical language for all computations (13) and calculated Spearman's rank correlation coefficients to assess associations between species. In cases where two taxa were detected in 100% of the specimens, low correlation values (i.e. close to 0) are possible if their intensity values are not associated. Prevalence values for bacterial species identified in our microarrays were compared with prevalence values determined for corresponding species in the Rio de Janeiro study (6).
Results
Out of 425 unique taxon-specific probes representing 280 species, 81 probes were reactive with the abscess specimen DNA. In positive control reactions, DNA from all abscess specimens generated amplicons using the 16S rRNA universal primers, and negative control reactions without DNA did not generate amplicons. The numbers of bacterial taxa identified within individual abscess specimens ranged from 2 to 30 (mean=15.9; median=17). The single abscess specimen with only two targeted species contained Fusobacterium nucleatum and Parvimonas micra. All other abscesses contained at least six targeted species.
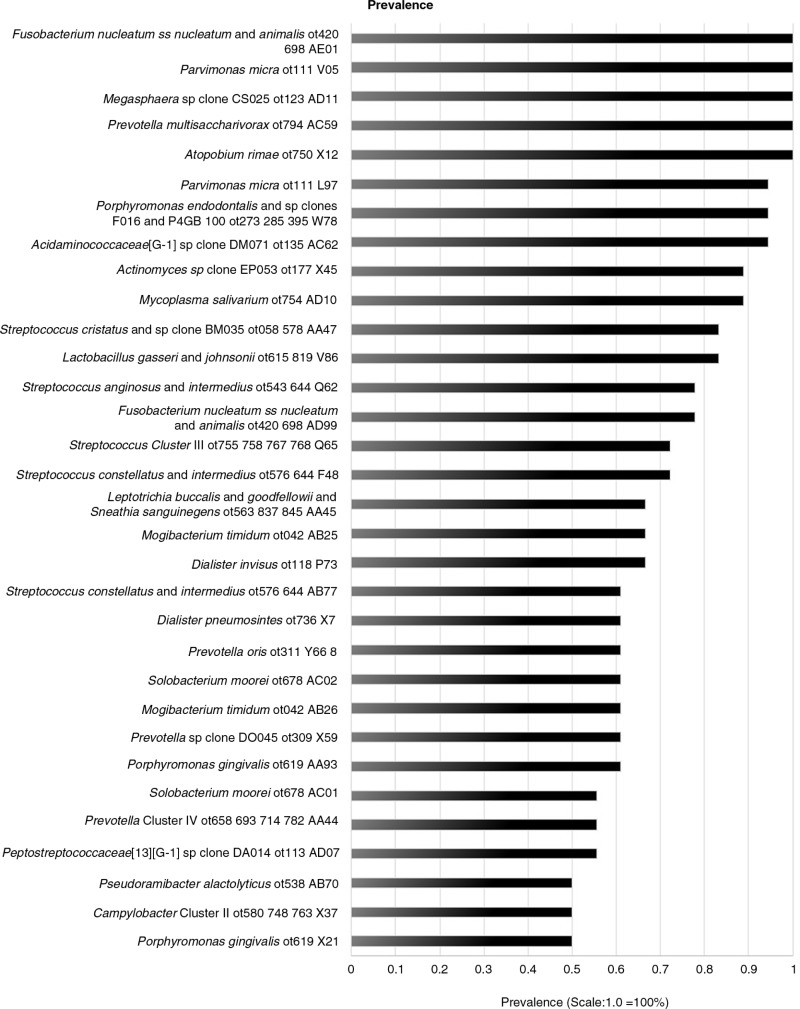
Five taxa, including F. nucleatum, P. micra, Megasphaera species clone CS025, Prevotella multisaccharivorax, and Atopobium rimae, were highly prevalent across abscess specimens and were identified to be present in all specimens (18/18, 100%; Fig. 1). Several additional taxa, including Porphyromonas endodontalis (17/18, 94.4%), Acidaminococcaceae species clone DM071 (17/18, 94.4%), Actinomyces species clone EP053 (16/18, 88.9%), Mycoplasma salivarium (16/18, 88.9%), Streptococcus cristatus (15/18, 83%), and Lactobacillus gasseri (15/18, 83.3%) were identified in >83% of the abscess specimens examined in this study. Taxa with prevalence values >0.39 (out of 81 taxa-specific probes detected) are shown in Fig. 1.
Fig. 1.
Bar graph illustrating prevalence of bacterial species in endodontic abscess specimens (N=18). Prevalence for each bacterial species is defined as the percentage across the entire number of abscess specimens (example: 100% prevalence is equivalent to bacterial species X found in 18 out of 18 abscess specimens). The horizontal axis is set to 0–1 scale; representing 0–100% prevalence. The range of prevalence values displayed in the bar graph is limited to those bacterial species >39%.
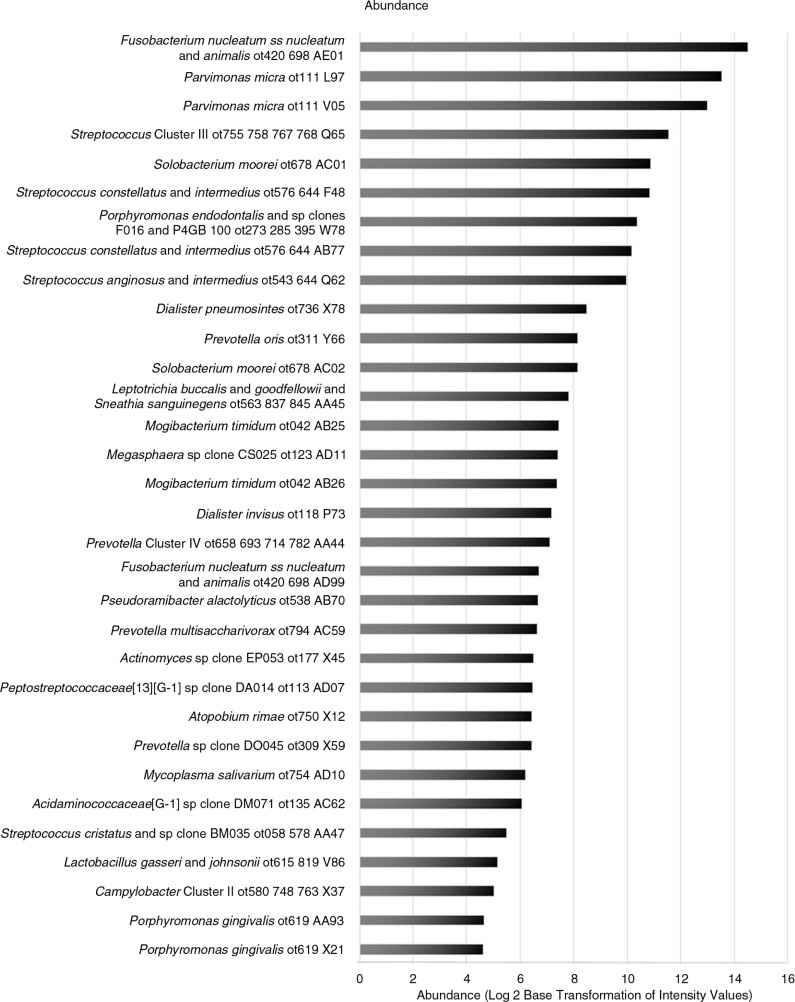
The five taxa with the highest average abundance, measured within individual abscess specimens and then numerically averaged across all abscess specimens, were F. nucleatum, P. micra (two distinct clones), Streptococcus Cluster III, Solobacterium moorei, and Streptococcus constellatus (Fig. 2).
Fig. 2.
Bar graph illustrating abundance of bacterial species in endodontic abscess specimens (N=18). Abundance for each bacterial species is defined as the intensity signal (reflective of the numbers of bacterial species X) within each individual abscess specimen and then averaged for the entire abscess specimen set. Using the log base 2 scale, the range of transformed intensity values in the horizontal axis is 0–16.
F. nucleatum and P. micra were both highly prevalent across abscesses and highly abundant within individual abscess specimens. Interestingly, the other three taxa demonstrating high prevalence across all abscess specimens – Megasphaera species clone CS025, P. multisaccharivorax, and A. rimae – had moderate or lower abundance levels within individual abscesses (Fig. 2). Additional taxa with high prevalence (>83–94% range), including P. endodontalis, Acidaminococcaceae species clone DM071, Actinomyces species clone EP053, M. salivarium, S. cristatus, and L. gasseri, also had moderate or lower abundance levels within individual abscesses (Fig. 2).
The bacteria found most commonly associated with other bacteria, at levels above Spearman rank correlation coefficients of 0.9 were P. multisaccharivorax/ Acidaminococcaceae species clone DM071 (Spearman=0.961), Acidaminococcaceae species clone DM071/Megasphaera species clone CS025 (Spearman=0.926), Megasphaera species clone CS025/Actinomyces species clone EP053 (Spearman=0.924), P. multisaccharivorax/Megasphaera species clone CS025 (Spearman=0.917), Acidaminococcaceae species clone DM071/S. cristatus (Spearman=0.908), and S. cristatus/Actinomyces species clone EP053 (Spearman=0.908). Interestingly, Acidaminococcaceae species clone DM071 is highly associated (Spearman coefficient>0.772) with seven other microorganisms (Table 1) and Megasphaera species clone CS025 is highly associated (Spearman coefficient>0.767) with eight microorganisms (Table 1). P. multisaccharivorax and S. cristatus both have high associations (Spearman coefficients>0.828) with four and three other microorganisms, respectively (Table 1).
Table 1.
Bacterial associations in endodontic abscesses (Spearman's rank correlation coefficient >0.76)
| Bacteria 1 | Bacteria 2 | Spearman |
|---|---|---|
| Acidaminococcaceae[G-1] sp clone DM071_ot135_AC62 | Prevotella multisaccharivorax_ot794_AC59 | 0.961 |
| Acidaminococcaceae[G-1] sp clone DM071_ot135_AC62 | Streptococcus cristatus and sp clone BM035_ot058_578_AA47 | 0.911 |
| Acidaminococcaceae[G-1] sp clone DM071_ot135_AC62 | Lactobacillus gasseri and johnsonii_ot615_819_V86 | 0.893 |
| Acidaminococcaceae[G-1] sp clone DM071_ot135_AC62 | Actinomyces sp clone EP053_ot177_X45 | 0.882 |
| Acidaminococcaceae[G-1] sp clone DM071_ot135_AC62 | Parvimonas micra_ot111_L97 | 0.856 |
| Acidaminococcaceae[G-1] sp clone DM071_ot135_AC62 | Porphyromonas endodontalis and sp clones _F016 and P4GB_100_ot273_285_ 395_ W78 | 0.850 |
| Acidaminococcaceae[G-1] sp clone DM071_ot135_AC62 | Parvimonas micra_ot111_V05 | 0.772 |
| Megasphaera sp clone CS025_ot123_AD11 | Acidaminococcaceae[G-1] sp clone DM071_ot135_AC62 | 0.926 |
| Megasphaera sp clone CS025_ot123_AD11 | Actinomyces sp clone EP053_ot177_X45 | 0.924 |
| Megasphaera sp clone CS025_ot123_AD11 | Prevotella multisaccharivorax_ot794_AC59 | 0.917 |
| Megasphaera sp clone CS025_ot123_AD11 | Streptococcus cristatus and sp clone BM035_ot058_578_AA47 | 0.882 |
| Megasphaera sp clone CS025_ot123_AD11 | Porphyromonas endodontalis and sp clones _F016 and P4GB_100_ot273_285_ 395_ W78 | 0.816 |
| Megasphaera sp clone CS025_ot123_AD11 | Parvimonas micra_ot111_L97 | 0.804 |
| Megasphaera sp clone CS025_ot123_AD11 | Lactobacillus gasseri and johnsonii_ot615_819_V86 | 0.786 |
| Megasphaera sp clone CS025_ot123_AD11 | Parvimonas micra_ot111_V05 | 0.767 |
| Prevotella multisaccharivorax_ot794_AC59 | Lactobacillus gasseri and johnsonii_ot615_819_V86 | 0.886 |
| Prevotella multisaccharivorax_ot794_AC59 | Streptococcus cristatus and sp clone BM035_ot058_578_AA47 | 0.868 |
| Prevotella multisaccharivorax_ot794_AC59 | Actinomyces sp clone EP053_ot177_X45 | 0.865 |
| Prevotella multisaccharivorax_ot794_AC59 | Porphyromonas endodontalis and sp clones _F016 and P4GB_100_ot273_285_395_ W78 | 0.828 |
| Streptococcus cristatus and sp clone BM035_ot058_578_AA47 | Actinomyces sp clone EP053_ot177_X45 | 0.908 |
| Streptococcus cristatus and sp clone BM035_ot058_578_AA47 | Lactobacillus gasseri and johnsonii_ot615_819_V86 | 0.868 |
| Streptococcus cristatus and sp clone BM035_ot058_578_AA47 | Parvimonas micra_ot111_L97 | 0.829 |
| Lactobacillus gasseri and johnsonii_ot615_819_V86 | Actinomyces sp clone EP053_ot177_X45 | 0.793 |
| Mycoplasma salivarium_ot754_AD10 | Fusobacterium nucleatum ss nucleatum and animalis_ot420_698_AE01 | 0.829 |
| Porphyromonas endodontalis and sp clones _F016 and P4GB_100_ot273_285_395_W78 | Lactobacillus gasseri and johnsonii_ot615_819_V86 | 0.775 |
| Streptococcus constellatus and intermedius_ot576_644_F48 | Streptococcus Cluster III_ot755_758_767_768_Q65 | 0.873 |
Discussion
Oral microbiota contained within endodontic abscesses
For the current study, we examined endodontic abscesses for the presence of 280 oral bacterial species (425 taxa-specific probes) using microarray determinations conducted by the Human Oral Microbial Identification Laboratory at the Forsyth Institute (Cambridge, MA). Our findings confirmed and provided new information supporting the earlier research by Siqueira and Rôcas (6), describing the microbiota of endodontic abscesses obtained from the demographic region in Rio de Janeiro, Brazil. In our Portland, Oregon abscess collection, we found similar microbial species as those defined in Siqueira and Rôcas (6) as well as several new strong bacterial associations, including those between F. nucleatum and M. salivarium, and between F. nucleatum and S. cristatus. Studies in Portland, Oregon, and Rio de Janeiro, Brazil, noted that F. nucleatum was the principal bacterial species identified in endodontic abscesses and was used as a comparative baseline for positive bacterial associations. F. nucleatum was present in all abscesses (N=18) in the current study and in 27 out of 44 abscesses (64%) in the study by Siqueira and Rôcas (6). P. micra and Porphyromomas endodontalis were also found in high prevalence in abscess specimens from the demographic regions in Portland, Oregon and Rio de Janeiro, Brazil. Interestingly, Veillonella species, which were detected in many culture-independent studies, but absent in Siqueira and Rôcas (6), were also found in low prevalence (2 out of 18 abscesses) in our study. In addition, Treponema species clones, numbering 25 taxa-specific probes in our study, were detected in very low abundance in many specimens; only three abscess specimens generated moderate signals with the Treponema probes, Treponema maltophilum in two abscesses and Treponema cluster in the third abscess. This is in contrast to the findings of Siqueira and Rôcas (6) and to numerous culture-independent studies that have detected moderate-to-high numbers of the oral treponemes, Treponema denticola and Treponema socranskii, within endodontic abscesses (14). In addition, Enterococcus faecalis was not detected in any of the abscess specimens in our study, consistent with the findings of others that E. faecalis was significantly more associated with asymptomatic, as opposed to symptomatic cases of endodontic disease (15).
In our study, five bacterial taxa were found in 100% of all surveyed abscess specimens (N=18); these species included F. nucleatum and P. micra, both of which were found in high prevalence (64% and 52%, respectively) in the study by Siqueira and Rôcas (6). In addition, Megasphaera species clone CS025, P. multisaccharivorax, and A. rimae were found at 100% prevalence in our study, albeit at moderate-to-lower abundance levels within individual abscesses, but varied from less than 5% (Megasphaera species clone CS025 and P. multisaccharivorax) to 10% prevalence (A. rimae) in the study by Siqueira and Rôcas (6). Strong associations were also found between bacterial taxa whose correlations had not previously been established. Several significant positive associations involving F. nucleatum as the reference strain included F. nucleatum/M. salivarium (ρ=0.829), F. nucleatum/Porphyromonas gingivalis (ρ=0.691), and F. nucleatum/Megasphaera species clone CS025 (ρ=0.684) and were based on Spearman's rank correlation coefficients >0.65.
We have minimized the risk of abscess sample contamination with the use of the chlorhexidine gluconate oral rinse and the disinfection of mucosal surfaces with Povidone–Iodine swabs. Paper point collections, conducted at the site of abscess aspiration, did not produce any microbial growth. Abscess specimens were collected and frozen at −85°C, prior to DNA extraction conducted as a batch in 2010. DNA was then sent to the Forsyth Institute for HOMIM analysis. The 5-year delay between HOMIM analysis and the preparation of this manuscript was due to the time required to obtain funding to recruit and pay biostatisticians qualified to analyze complex microarray data. We acknowledge that another potential negative control, which was not conducted during the experimental phase of this work in 2010, may have been the aspiration of tissue or blood from the mucosa of a healthy subject and the identification of any potential contaminants by microarray analysis. We also acknowledge that differences in the microbiota profiles identified in abscesses obtained from the demographic regions in Portland, Oregon, and Rio de Janeiro, Brazil, may be due to technical differences in the microarray methods, including the design of the 16S rRNA gene probes and the hybridization conditions used to target specific microbial sequences.
Coinfection of F. nucleatum and other microorganisms may work synergistically within endodontic and periodontal disease
Multiple microorganisms initiate endodontic infections, and genera such as Fusobacterium, Treponema, Prevotella, and Streptococcus dominate these lesions, although the range of bacteria present may vary (5, 16–18). Within specific genera, different species have been found to work synergistically, utilizing virulence factors from complementary microorganisms to their advantage (19).
F. nucleatum has been found to enhance the adhesion and invasion of other oral microorganisms (20). Furthermore, coinfection of F. nucleatum with other oral bacteria may inhibit host immune response, allowing for their persistent growth (20). This cooperative growth may explain the presence of M. salivarium, P. gingivalis, or Megasphaera clone with F. nucleatum in endodontic and periodontal disease. In addition, F. nucleatum was found to be associated with S. cristatus, which has been specifically shown to inhibit the pro-inflammatory actions of the former microorganism (21). The synergistic interactions of enhancing virulence and inhibiting host immune response are believed to be contributory determinants of endodontic and periodontal disease.
Microbiota in endodontic abscesses may differ based on geographic considerations
The inter-study variability of microbiota species identified in periodontal and endodontic diseases may be based on the geographic location of the patient pool and specimen acquisition. Several studies have shown that body sites typically colonize different microbes depending on geographic locations. For example, Kemppainem et al. (22) demonstrated that gut flora colonization and diversity in infants at risk for Type I diabetes may be based on country of birth. Furthermore, different single-nucleotide polymorphisms (SNPs) of Mycobacterium tuberculosis were found to occur in different geographic locations (23). Machado de Oliveira et al. (24) and Siqueira and Rôcas (5) also previously noted differences in bacterial community profiles and certain microorganisms from endodontic abscesses in patients from Portland, Oregon, and Rio de Janeiro, Brazil. Although common microbiota profiles were found between these two geographic locations, the prevalence of microbes varied between studies. Interestingly, consistent with a nested PCR analysis conducted using abscess specimens jointly obtained from Portland, Oregon, and Rio d Janeiro, Brazil (25, 26), T. denticola and Tannerella forsythia were not present, or present in extremely low levels in our microarray analyses, and were present at high prevalence in specimens described in the Rio de Janeiro study (6). In addition, it is widely accepted that ethnic and geographic differences exist in periodontal microbiota profiles (27), and we conclude, in part by extrapolation, that geographic location may play a role in the pathogenic microbial causes of endodontic and periodontal infections.
Potential role of streptococci species and S. cristatus in dental disease
S. cristatus has been demonstrated to inhibit host immune response by attenuating interleukin-8 in the inflammatory pathway (21) and has been implicated as opportunistically pathogenic by its role in severe early childhood caries (S-ECC) and by its association with periodontal bacteria (28, 29). F. nucleatum uses a nuclear factor-kappa B (nuclear translocation) inflammatory pathway to induce a pro-inflammatory cascade, which is blocked by S. cristatus (21). S. cristatus is also negatively correlated with P. gingivalis (29), described as a potential causative determinant of adult periodontitis. The pathogenicity of P. gingivalis lies in part by its ability to adhere to oral surfaces with its long fimbriae; S. cristatus appears to inhibit the expression of the fimbrillin (FimA) gene, which is involved in the adhesion and subsequent colonization of P. gingivalis (30). S. cristatus also produces arginine deiminase (ArcA), which represses FimA production and inhibits the colonization of P. gingivalis (31). Through these mechanisms, S. cristatus has been implicated to attenuate the pathogenic activity of P. gingivalis and significantly contributes to decreased inflammatory destruction in periodontal disease, including decreased alveolar bone loss (32).
Furthermore, the ability of S. cristatus to undergo transformation at an increased frequency has been proposed to contribute to the virulence of this microorganism (33).
Implications of mixed microbial infections and concluding remarks
Bacterial interactions within endodontic infections may synergistically or antagonistically affect growth and underscore the complexity of the ecological determinants that support the metabolic efficiency and pathogenic potency of the composite bacterial community. Our study supports the findings of previous research in defining the most prevalent bacteria within endodontic abscesses and identifies strong bacterial associations, some of which may be unique for the demographic region in Portland, Oregon, or patient population. Furthermore, the presence of S. cristatus within endodontic abscesses, which has not been identified in previous endodontic disease studies, may indicate a new role as a member of a pathogenic microbial ecology.
Acknowledgements
NG and EF are noted as equal contributors to this work. This work was originally supported in part by a grant from the American Association of Endodontists Foundation (to CM). NG is a recipient of the 2015 ADA/Dentsply Student Clinician Research Award. EF is a recipient of the Dean's Student Research Fellowship Award from the OHSU School of Dentistry. NG, EF, KK, NK, CC, BP, and RJ are enrolled in the DMD program at the OHSU School of Dentistry. SS is a clinical study coordinator for our program at the OHSU School of Dentistry. RK, CS, TM, and CM receive support from the OHSU School of Dentistry. DC and CM are also supported in part by NIH DE024317. We thank Truman Nielsen for support in conducting earlier portions of the experimentation. We also thank Dr. Bruce J. Paster for providing detailed commentary concerning the HOMIM probe generation, hybridization, and washing procedures.
Conflict of interest and funding
There is no conflict of interest in the present study for any of the authors.
References
- 1.Torabinejad M, Shabahang S. Pulp and periapical pathosis. In: Torabinejad M, Walton RE, editors. Endodontics. Principles and practice. fourth ed. St. Louis: Saunders/Elsevier; 2009. pp. 49–67. [Google Scholar]
- 2.Siqueira JF., Jr Endodontic infections: concepts, paradigms, and perspectives. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2002;94:281–93. doi: 10.1067/moe.2002.126163. [DOI] [PubMed] [Google Scholar]
- 3.Sasaki H, Stashenko P. Interrelationship of the pulp and apical periodontitis. In: Hargreaves KM, Goodis HE, Tay FR, editors. Seltzer and Bender's dental pulp. second ed. Chicago, IL: Quintessence Publishing; 2012. pp. 277–99. [Google Scholar]
- 4.Baumgartner JC, Siqueira JF, Jr, Sedgley CM, Kishen A. Microbiology of endodontic disease. In: Ingle JI, Bakland LK, Baumgartner JC, editors. Ingle's Endodontics. sixth ed. Hamilton, Canada: BC Decker; 2008. pp. 221–308. [Google Scholar]
- 5.Siqueira JF, Jr, Rôcas IN. Microbiology and treatment of acute apical abscesses. Clin Microbiol Rev. 2013;26:255–73. doi: 10.1128/CMR.00082-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siqueira JF, Jr, Rôcas IN. The microbiota of acute apical abscesses. J Dent Res. 2009;88:61–5. doi: 10.1177/0022034508328124. [DOI] [PubMed] [Google Scholar]
- 7.Xia T, Baumgartner JC. Occurrence of Actinomyces in infections of endodontic origin. J Endod. 2003;29:549–52. doi: 10.1097/00004770-200309000-00001. [DOI] [PubMed] [Google Scholar]
- 8.Ahn J, Yang L, Paster BJ, Ganly I, Morris L, Pei Z, et al. Oral microbiome profiles: 16S rRNA pyrosequencing and microarray assay comparison. PLoS One. 2011;6:e22788. doi: 10.1371/journal.pone.0022788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Preza D, Olsen I, Willumsen T, Grinde B, Paster BJ. Diversity and site-specificity of the oral microflora in the elderly. Eur J Clin Microbiol Infect Dis. 2009;28:1033–40. doi: 10.1007/s10096-009-0743-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Preza D, Olsen I, Willumsen T, Boches SK, Cotton SL, Grinde B, et al. Microarray analysis of the microflora of root caries in elderly. Eur J Clin Microbiol Infect Dis. 2009;28:509–17. doi: 10.1007/s10096-008-0662-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colombo AP, Bennet S, Cotton SL, Goodson JM, Kent R, Haffajee AD, et al. Impact of periodontal therapy on the subgingival microbiota of severe periodontitis: comparison between good responders and individuals with refractory periodontitis using the human oral microbe identification microarray. J Periodontol. 2012;83:1279–87. doi: 10.1902/jop.2012.110566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, et al. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.R Development core team. A language and environment for statistical computing, the R Foundation for Statistical Computing. Available from: http://www.R-project.org/ [cited 17 August 2015]
- 14.Baumgartner JC, Khemaleelakul SU, Xia T. Identification of spirochetes (Treponemes) in endodontic infections. J Endod. 2003;29:794–7. doi: 10.1097/00004770-200312000-00002. [DOI] [PubMed] [Google Scholar]
- 15.Rôcas IN, Siqueira JF, Santos KR. Association of Enterococcus faecalis with different forms of periradicular diseases. J Endod. 2004;30:315–20. doi: 10.1097/00004770-200405000-00004. [DOI] [PubMed] [Google Scholar]
- 16.Lin LM, Huang GT. Pathobiology of the periapex. In: Hargreaves K, Berman L, editors. Cohen's pathway to the pulp. tenth ed. St. Louis MO: Mosby Inc; 2011. pp. 529–55. [Google Scholar]
- 17.Siqueira JF, Rôcas IN. Microbiology and treatment of endodontic infections. In: Hargreaves K, Berman L, editors. Cohen's pathway to the pulp. tenth ed. St. Louis MO: Mosby Inc; 2011. pp. 559–604. [Google Scholar]
- 18.Lana MA, Ribeiro-Sobrinho AP, Stehling R, Garcia GD, Silva BK, Hamdan JS, et al. Microorganisms isolated from root canals presenting necrotic pulp and their drug susceptibility in vitro . Oral Microbiol Immunol. 2001;16:100–5. doi: 10.1034/j.1399-302x.2001.016002100.x. [DOI] [PubMed] [Google Scholar]
- 19.Tennert C, Fuhrmann M, Wittmer A, Karygianni L, Altenburger MJ, Pelz K, et al. New bacterial composition in primary persistent/secondary endodontic infections with respect to clinical and radiographic findings. J Endod. 2014;40:670–7. doi: 10.1016/j.joen.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 20.Li Y, Guo H, Wang X, Lu Y, Yang C, Yang P. Coinfection with F. nucleatum can enhance the attachment and invasion of Porphyromonas gingivalis or Aggregatibacter actinomycetemcomitans to human gingival epithelial cells. Arch Oral Biol. 2015;60:1387–93. doi: 10.1016/j.archoralbio.2015.06.017. [DOI] [PubMed] [Google Scholar]
- 21.Zhang G, Rudney JD. Streptococcus cristatus attenuates F. nucleatum-induced cytokine expression by influencing pathways converging on nuclear factor-Kβ. Mol Oral Microbiol. 2011;26:150–63. doi: 10.1111/j.2041-1014.2010.00600.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kemppainem KM, Ardissone AN, Davis-Richardson AG, Fagen JR, Gano KA, Leon-Novelo LG, et al. Early childhood gut microbiomes show strong geographic differences among subjects at high risk for type 1 diabetes. Diabetes Care. 2015;38:329–32. doi: 10.2337/dc14-0850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoshide M, Qian L, Rodrigues C, Warren R, Victor T, Evasco HB, 2nd, et al. Geographical differences associated with single-nucleotide polymorphisms (SNPs) in nine gene targets among resistant clinical isolates of Mycobacterium tuberculosis . J Clin Microbiol. 2014;52:1322–9. doi: 10.1128/JCM.00857-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Machado de Oliveira JC, Siqueira JF, Rôcas IN, Baumgartner JC, Xia T, Peixoto RS, et al. Bacterial community profiles of endodontic abscesses from Brazilian and USA subjects as compared by denaturing gradient gel electrophoresis analysis. Oral Microbiol Immunol. 2007;22:14–18. doi: 10.1111/j.1399-302X.2007.00311.x. [DOI] [PubMed] [Google Scholar]
- 25.Baumgartner JC, Siqueira JF, Xia T, Rôcas IN. Geographical differences in bacteria detected in infections using polymerase chain reaction. J Endod. 2004;30:141–4. doi: 10.1097/00004770-200403000-00004. [DOI] [PubMed] [Google Scholar]
- 26.Rôcas IN, Baumgartner JC, Xia T, Siqueira JF., Jr Prevalence of selected bacterial named species and uncultivated phylotypes in endodontic abscesses from two geographic locations. J Endod. 2006;32:1135–8. doi: 10.1016/j.joen.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 27.Könönen E, Müller HP. Microbiology of aggressive periodontitis. Periodontol 2000. 2014;65:46–78. doi: 10.1111/prd.12016. [DOI] [PubMed] [Google Scholar]
- 28.Yao ES, Lamont RJ, Leu SP, Weinberg A. Interbacterial binding among strains of pathogenic and commensal oral bacterial species. Oral Microbiol Immunol. 1996;11:35–41. doi: 10.1111/j.1399-302x.1996.tb00334.x. [DOI] [PubMed] [Google Scholar]
- 29.Tanner AC, Mathney JM, Kent RL, Chalmers NI, Hughes CV, Loo CY, et al. Cultivable anaerobic microbiota of severe early childhood caries. J Clin Microbiol. 2011;49:1464–74. doi: 10.1128/JCM.02427-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang BY, Wu J, Lamont RJ, Lin X, Xie H. Negative correlation of distributions of Streptococcus cristatus and Porphyromonas gingivalis in subgingival plaque. J Clin Microbiol. 2009;47:3902–6. doi: 10.1128/JCM.00072-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu J, Xie H. Role of arginine deiminase of Streptococcus cristatus in Porphyromonas gingivalis colonization. Antimicrob Agents Chemother. 2010;54:4694–8. doi: 10.1128/AAC.00284-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie H, Hong J, Sharma A, Wang BY. Streptococcus cristatus ArcA interferes with Porphyromonas gingivalis pathogenicity in mice. J Periodontal Res. 2012;47:578–83. doi: 10.1111/j.1600-0765.2012.01469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Correia FF, McKay TL, Farrow MF, Rosan B, DiRienzo JM. Natural transformation of Streptococcus crista . FEMS Microbiol Lett. 1996;143:13–18. doi: 10.1111/j.1574-6968.1996.tb08454.x. [DOI] [PMC free article] [PubMed] [Google Scholar]