Summary
Interleukin-17 (IL-17) and IL-17 receptor (IL-17R) signaling are essential for regulating mucosal host defense against many invading pathogens. Commensal bacteria, especially segmented filamentous bacteria (SFB), are a crucial factor that drives T helper 17 (Th17) cell development in the gastrointestinal tract. In this study, we demonstrate that Th17 cells controlled SFB burden. Disruption of IL-17R signaling in the enteric epithelium resulted in SFB dysbiosis due to reduced expression of α-defensins, Pigr and Nox1. When subjected to experimental autoimmune encephalomyelitis, IL-17R signaling deficient mice demonstrated earlier disease onset and worsened severity that was associated with increased intestinal Csf2 expression and elevated systemic GM-CSF cytokine concentrations. Conditional deletion of IL-17R in the enteric epithelium demonstrated that there was a reciprocal relationship between the gut microbiota and enteric IL-17R signaling that controlled dysbiosis, constrained Th17 development, and regulated the susceptibility to autoimmune inflammation.
Introduction
Interleukin-17 (IL-17)-producing cells are present in diverse species from tunicates to mammals, and in non-vertebrates IL-17 is induced by infection in hemocytes (Roberts et al., 2008). In mammals, IL-17-producing Th17 (CD4+ T cells producing IL-17 and IL-22) cells develop in the intestine in response to commensal microbiota (Ivanov et al., 2009; Ivanov et al., 2008). A critical component of the commensal microbiota that drive Th17 cells in mice are segmented filamentous bacteria (SFB) (Gaboriau-Routhiau et al., 2009; Ivanov et al., 2009). Although Th17 cells have been implicated in autoimmunity, these cells also provide the host with an evolutionary advantage by conferring serotype-independent immunity against extracellular pathogens (Chen et al., 2011; Malley et al., 2006; Wüthrich et al., 2011). In support of this, it has been recently reported that SFB induce antigen-specific Th17 responses in the intestine (Yang et al., 2014). Given the link between commensal microbiota and Th17 development, there is intense interest in understanding role of the gut microbiome in health and disease. The role of innate immune signaling system in recognizing commensal bacteria as well as pathogens is well characterized (Bosch et al., 2009; Franzenburg et al., 2012). However, a similar role of the adaptive immune system in regulating commensal colonization is poorly understood.
Th17 immune responses have been implicated in many human autoimmune diseases including arthritis, multiple sclerosis and inflammatory bowel diseases (IBD) (Dardalhon et al., 2008). However, a recent clinical trial providing a neutralizing IL-17A monoclonal antibody in IBD patients was not protective and was associated with increased adverse events including Candida albicans infection (Hueber et al., 2012), suggesting that enteric IL-17 responses may be beneficial in the gut. In contrast, IL-17 has been shown to play a more pathogenic role in certain autoimmune disorders. Patients with multiple sclerosis (MS) have high IL-17 expression in demyelinating lesions (Lock et al., 2002). Indeed, mice lacking IL-17A and IL-17F signaling (Il17rc−/− or Th17 differentiation (Batf−/−, Il23p19−/−) have milder experimental autoimmune encephalomyelitis (EAE), a mouse model of MS (Hu et al., 2010; McGeachy et al., 2009; Schraml et al., 2009). Thus, we sought to determine the role of enteric IL-17R signaling in regulating commensal microbiota, enteric inflammation, as well as autoimmune inflammation.
Despite recent advances in understanding the role of IL-17 in host immunity, its role in regulating enteric immune responses, as well as its impact on the commensal microbiome has not been well studied. Using both global and intestinal epithelial cell specific IL-17R deficient mice, we found that IL-17RA and IL-17RC were required for regulating overgrowth of commensal bacteria including SFB. IL-17RA was required for the expression of Nox1 (an apical NADPH oxidase), α-defensins, and polymeric Immunoglobulin receptor (pIgR), which is required for the transcytosis of secretory immunoglobulin A (sIgA). Abrogation of IL-17R-dependent regulation of the commensal microbiota led to increased colonization with SFB, higher degrees of systemic inflammation and more severe autoimmune inflammation. Our study provides an explanation of the differential roles of IL-17 in different disease models.
Results
Regulation of Th17 lineage commitment by IL-17 receptor family members
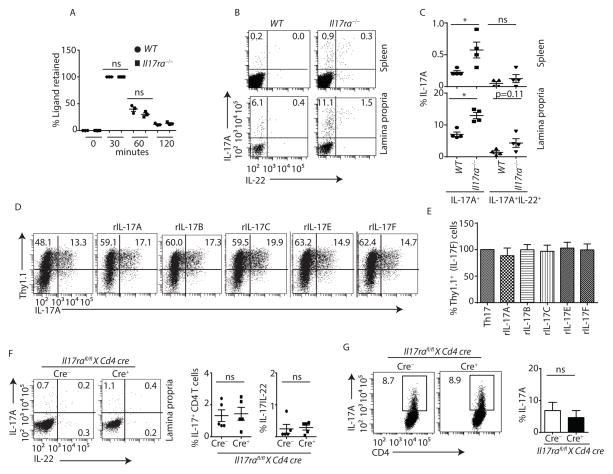
It is has been previously demonstrated that mice deficient in IL-17R signaling have enhanced IL-17 responses. This has been attributed to ligand-dependent regulation of IL-17 producing cells and IL-17R dependent clearance of ligand (Nagata et al., 2008; Ye et al., 2001b). However, we found the in vivo half-life of administered recombinant IL-17A was nearly identical in the serum of both wild-type (WT) and Il17ra−/− mice (Figure 1A), suggesting that enhanced IL-17 responses in Il17ra−/− mice were not entirely due to receptor-mediated clearance of ligand. In support of this concept, we observed a marked expansion of CD4+ T cells that expressed IL-17 in the lamina propria and spleens of Il17ra−/− mice as compared to age and gender (male) matched co-housed WT mice (Figure 1B and 1C).
Figure 1. Exacerbated Th17 responses in Il17ra−/− mice are not due to a T cell intrinsic defect.
(A) WT (n=3) and Il17ra−/− (n=3) mice were treated with recombinant IL-17A, and serum IL-17A concentration was determined by ELISA.
(B) Lymphocytes were isolated from lamina propria and spleen of co-housed gender matched WT (n=4) and Il17ra−/− (n=4) mice, and stained with anti-CD3, anti-CD4, anti-IL-17A and anti-IL-22 antibodies. CD4 T cells (gated on CD3+CD4+) producing IL-17A and IL-22 were analyzed by flow cytometer.
(C) Percentages of IL-17A or both IL-17A and IL-22-producing CD4 T cells in the spleen and lamina propria of multiple WT (n=4) and Il17ra−/− (n=4) mice were shown.
(D and E) Naïve CD4 T cells from Thy1.1-Il17F reporter mice (n=5) were polarized to Th17 cells in presence of indicated IL-17 ligands. After day 4, cells were stained with anti-CD3, anti-CD4, anti-Thy1.1 and anti-IL-17A antibodies. CD4 T cells (gated on CD3+CD4+) producing IL-17A and IL-17F (Thy1.1+) were analyzed by flow cytometer. Data shown are frequency (D) and percentage (E) changes in IL-17F (Thy1.1+) producing cells in response to various Th17 ligands stimulation as compared to control Th17 cells (n=5).
(F) Lymphocytes were isolated from lamina propria of Il17rafl/fl x Cd4 cre+ (n=5) and littermate cre− (n=5) mice (6 week age), and stained with anti-CD3, anti-CD4, anti-IL-17A and anti-IL-22 antibodies. CD4 T cells (gated on CD3+CD4+) producing IL-17A or IL-17 and IL-22 were analyzed by flow cytometer (left panel). Percentages of IL-17 or both IL-17 and IL-22-producing CD4+ T cells from multiple mice were shown in right panel.
(G) Naïve CD4 T cells from Il17rafl/fl x Cd4 cre+ (n=4) and littermate cre− (n=3) mice were polarized to Th17 cells. After 4 days culture, cells were stained with anti-CD3, anti-CD4 and anti-IL-17A antibodies. CD4 T cells (gated on CD3+CD4+) producing IL-17A were analyzed by flow cytometer (left panel). Percentages of IL-17 producing CD4+ T cells from multiple mice were shown in right panel.
Figure 1C, 1D, 1E and 1F were generated from 2 independent experiments. Each symbol indicates experiments from a separate animal. Data presented as mean ± SEM on relevant graphs. *P ≤ 0.05 (Mann-Whitney test, Two-tailed). See also Figure S1 and S2.
As it has been reported that IL-17A or IL-17F may negatively constrain Th17 responses in T cells (Nagata et al., 2008; Smith et al., 2008), we determined whether IL-17 receptor expression affected naïve T cell differentiation toward the Th17 lineage. We observed no expression of IL-17RC on CD4+ T cells, and we observed similar frequencies of Th17 cells between WT and Il17ra−/− mice when naïve T-cells were polarized to Th17 cells ex vivo (Figure S1A and S1B). These observations suggested that IL-17RA expression did not intrinsically control Th17 differentiation. Moreover, the addition of the IL-17RA ligands, IL-17A, IL-17B, IL-17C, IL-17E (also known as IL-25), or IL-17F to WT naïve CD4+ T cells had no effect on differentiation of naïve T cells towards Th17 cells (Figure 1D and 1E) despite the fact that ligand concentration was stable and persistent on day 4 of the Th17 differentiation process (Figure S1C). Furthermore, to eliminate the possibility of a T cell intrinsic defect in the observed expansion of Th17 cells in Il17ra−/− mice, we generated CD4 specific IL-17RA null mice by crossing Il17rafl/fl mice with Cd4 cre mice (Figure S2A and S2B). CD4+ T cells from Il17rafl/fl x Cd4 cre+ mice did not show expanded Th17 responses in the lamina propria, in contrast to what was observed in Il17ra−/− mice (Figure 1F). Furthermore, we observed similar frequencies of Th17 cells between Il17rafl/fl x Cd4 cre+ and littermate cre− (Il17rafl/f) mice when naïve T cells were polarized to Th17 cells ex vivo (Figure 1G). These data show that the expansion of Th17 cells in Il17ra−/− mice was not T cell intrinsic.
Th17 cell expansion in Il17ra−/− mice is due to commensal dysbiosis
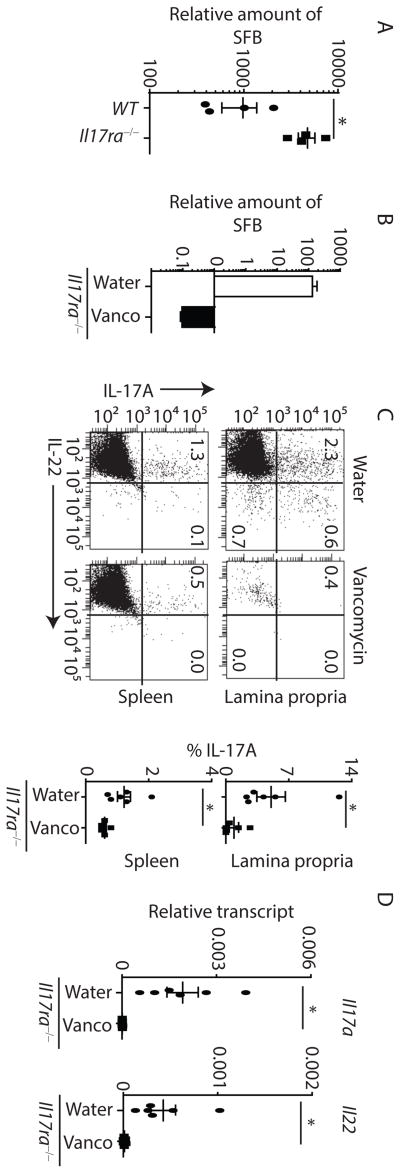
Given the role of commensal microbiota, specifically SFB, in regulating the development of Th17 cells (Gaboriau-Routhiau et al., 2009; Ivanov et al., 2009), we assessed SFB composition in IL-17RA deficient mice. As compared to co-housed gender (male) matched WT mice, fecal contents of Il17ra−/− mice showed substantial SFB overgrowth (Figure 2A). To further confirm this, we generated SFB-free Il17rafl/fl x EIIa cre+ mice. This cre transgene is under the control of the adenovirus EIIa promoter, which deletes in germ cells, thus results in global deletion of Il17ra. Efficient deletion was confirmed by lack of Il17ra expression in the terminal ileum of Il17rafl/fl x EIIa cre+ mice (Figure S3A). These mice were transplanted with high SFB containing feces from Rorc−/− mice and monitored for SFB colonization and Th17 responses. As expected, re-establishment of SFB into Il17rafl/fl x EIIa cre+ mice resulted in substantially higher SFB colonization as well as increased expression of Il17a and Il22 in the terminal ileum as compared to co-housed littermate control cre− mice (Figure S3B and S3C). To determine if the microbiota regulates the expansion of Th17 cells, we used oral vancomycin to deplete gram-positive bacteria, including SFB, in Il17ra−/− mice. Loss of SFB in Il17ra−/− mice resulted in a marked contraction of Th17 cells (Figure 2B to 2D) demonstrating that the microbiota regulates the expansion of Th17 cells in these mice. We next determined the frequency of SFB specific CD4+ T cells in WT and Il17ra−/− mice. To this end, we co-stained SFB-tetramer with anti-CD3 and anti-CD4 antibodies. Indeed, the absolute number of SFB-specific CD4+ T cells was significantly increased in Il17ra−/− mice (Figure S3D and S3E). These data suggested that microbiota possibly SFB regulates the expansion of Th17 cells in Il17ra−/− mice.
Figure 2. Expanded Th17 responses in ll17ra−/− mice is microbiota (SFB)-dependent.
(A) Fecal contents of co-housed age and gender matched WT (n=4) and Il17ra−/− (n=4) mice were analyzed for SFB colonization by qPCR.
(B) SFB-colonized Il17ra−/− mice were treated with vancomycin (Vanco) or control water for 30 days. After 30 days, fecal contents were analyzed for SFB colonization by qPCR.
(C) Lymphocytes were isolated from lamina propria and spleen of vancomycin (Vanco) (n=6) or control water (n=6) treated Il17ra−/− mice. CD4 T cells (gated on CD3+CD4+) producing IL-17A and IL-22 were analyzed by flow cytometer (left panel). Percentages of IL-17 producing CD4 T cells from multiple mice shown in right panel.
(D) Il17ra−/− (n=6) mice were treated with vancomycin or control water for 30 days. After 30 days, terminal ileums were analyzed for the expression of Il17a and Il22 by qPCR.
Figure 2C and 2D were generated and pooled from 2 independent experiments. Figure 2B is representative of two independent experiments. Each symbol indicates an experiment from a separate animal. Data presented as mean ± SEM on relevant graphs. *P ≤ 0.05 (Mann-Whitney test, Two-tailed). See also Figure S3.
CD4+ T cell-derived IL-17 regulates SFB-dysbiosis
High SFB colonization in Il17ra−/− mice led us to investigate the role of T cell derived IL-17A and IL-17F in regulating SFB overgrowth. We monitored the degree of SFB colonization in T cell sufficient C57BL/6 mice. SFB-free C57BL/6 Jackson (Jax) mice were transplanted with high SFB containing Rorc−/− mouse feces and monitored for SFB colonization and Th17 responses at various time points (Figure S4A to S4C). C57BL/6 (Jax) mice were effectively colonized by SFB, but by day 30, the SFB burden was contracted. The degree of SFB colonization at this contraction phase was consistent with that observed in adult Taconic C57BL/6 mice (Figure S4A), and also consistent with prior literature using Taconic C57BL/6 mice (Ivanov et al., 2009). This SFB expansion and contraction were also associated with expansion and contraction of IL-17-producing cells in the lamina propria (Figure S4B and S4C). As IL-17 is produced by both group 3 innate lymphoid cells (ILC3s) and T cells (CD4+ T cells, γδ T cells), we next determined the role of T cells in regulation of SFB colonization. Indeed, CD4+ T cells were the major producer of IL-17 in the lamina propria of Il17ra−/− mice when analyzed by flow cytometry (Figure S4D). To determine the role of CD4+ T cells in SFB colonization, we gavaged age and gender (male) matched SFB-free Jax C57BL/6 and Jax Tcrb−/− mice with SFB containing fecal contents from Rorc−/− mice. These mice were co-housed for 2 weeks and then separated for an additional 4 weeks. Tcrb−/− mice showed substantially higher degrees of SFB colonization in the terminal ileum as compared to WT mice (Figure S4E). Additionally, mice with a conditional deletion of Stat3 in CD4+ T cells (Stat3fl/fl x Cd4 cre+ mice) that lack Th17 or Th22 cells, also had higher SFB colonization compared to co-housed littermate cre− control mice (Figure S4F and S4G). To further investigate the role for IL-17R in the control of SFB colonization, we injected anti-IL17RA or isotype antibody into SFB-colonized co-housed WT mice and analyzed SFB colonization in the feces before and after treatment. WT mice that received the isotype control antibody showed a contraction of the SFB burden over the seven day time-course, whereas mice that received anti-IL-17RA treatment showed an increase in SFB colonization (Figure S4H). These findings further implicate T cell derived IL-17A and IL-17F as a key mediator of immunity against SFB.
Intestinal IL-17R signaling regulates SFB
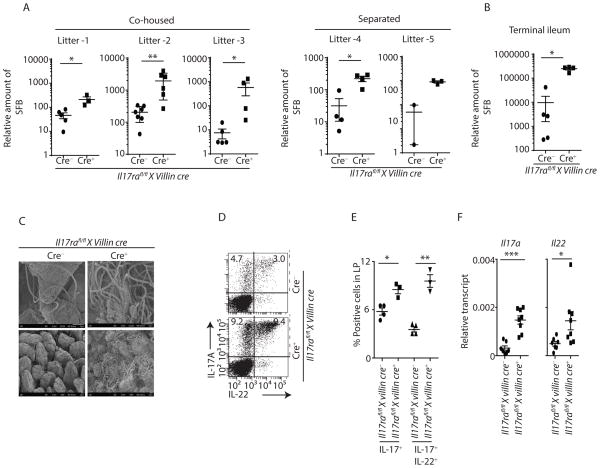
Next, we sought to determine if epithelial specific expression of IL-17RA was required for control of SFB. Thus, we generated mice with a deletion of IL-17RA by crossing Il17rafl/fl mice to villin-cre mice (Figure S5A and S5B). These mice were initially SFB-free. SFB inoculation (by oral gavage) into these mice resulted in substantial SFB growth in Il17rafl/fl x villin cre+ mice compared to co-housed cre− littermate control mice as well as cre+ mice heterozygous for the Il17ra floxed allele (Figure S5C and data not shown). Furthermore, offspring of SFB+ Il17rafl/fl x villin cre+ male and Il17rafl/fl x villin cre− female breeders recapitulated a similar SFB phenotype, where cre+ mice from each individual litter had substantially increased SFB burden as compared to littermate co-housed cre− mice (Figure 3A left panel). Thus, cre status of the offspring whether they are littermate/co-housed or littermate co-housed/separated is the dominant factor for SFB colonization in our floxed mice. This suggested a critical role of IL-17R signaling in regulating SFB colonization. To determine the SFB persistence per genotype, we co-housed littermate cre+ and cre− mice for 6 weeks and then separated them for an additional 4 weeks. As expected, Il17rafl/fl x villin cre+ mice maintained a high SFB burden as compared to littermate cre− mice after the separation period (Figure 3A right panel). SFB overgrowth in Il17rafl/fl x villin cre+ mice compared to co-housed littermate cre− control mice was also confirmed by qPCR and scanning electron microscopy (SEM) analyses of the terminal ileum (Figure 3B and 3C). Consistent with high SFB colonization, Il17rafl/fl x villin cre+ mice also showed elevated Th17 responses in the lamina propria as compared to co-housed cre− littermate controls (Figure 3D and 3E). In addition, the terminal ileum of Il17rafl/fl x villin cre+ mice had substantially higher Il17a and Il22 expression as compared to co-housed cre− littermate control mice (Figure 3F). IL-17RA is the receptor for IL-17A, IL-17F, IL17C and IL-17E. Thus, in order to confirm the role of IL-17A and IL-17F in SFB colonization, we generated IL-17A and IL-17F specific Il17rcfl/fl x villin cre+ knockout mice (Figure S5D). Indeed, consistent with Il17rafl/fl x villin cre+ data (Figure 3A–C), Il17rcfl/fl x villin cre+ showed SFB overgrowth compared to co-housed cre− littermate control mice (Figure S5E). These data demonstrated that intestinal epithelial cell specific IL-17RA and IL-17RC signaling regulates SFB colonization.
Figure 3. Enteric IL-17R signaling regulates SFB colonization.
(A) Fecal contents of multiple (co-housed or co-housed then separated) Il17rafl/fl x villin cre mice litters were analyzed for SFB colonization by qPCR. Individual litters SFB abundance data of 6 weeks (co-housed, left panel) and 10 weeks age (separated for 4 weeks after 6 week of co-housing, right panel) mice were shown.
(B) Terminal ileum of Il17rafl/fl x villin cre+ (n=4) and co-housed littermate cre− (n=5) control mice were analyzed for SFB colonization by qPCR.
(C) Terminal ileum of Il17rafl/fl x villin cre+ and littermate cre− mice were analyzed for SFB abundance by SEM microscopy. SEM images were acquired at 2K (upper panel) and 0.5K (lower panel) magnifications.
(D and E) Lymphocytes were isolated from lamina propria of Il17rafl/fl x villin cre+ (n=3) and littermate cre− (n=4) control mice. CD4 T cells (gated on CD3+CD4+) producing IL-17A and IL-22 were analyzed by flow cytometer. Percentages of IL-17 or both IL-17 and IL-22-producing CD4+ T cells from multiple mice were shown in figure E.
(F) Terminal ileum of Il17rafl/fl x villin cre+ (n=8) and littermate control cre− (n=7) mice were analyzed for the expression of Il17a and Il22 by qPCR data.
SEM images are representative of n=4 mice per group. Figure 3F were generated from 2 independent experiments. Figure 3B is representative of 3 independent experiments. Each symbol indicates experiments from an individual animal. Data presented as mean ± SEM on relevant graphs. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.005 (Mann-Whitney test, Two-tailed). See also Figure S5.
Commensal dysbiosis in enteric Il17ra−/− mice
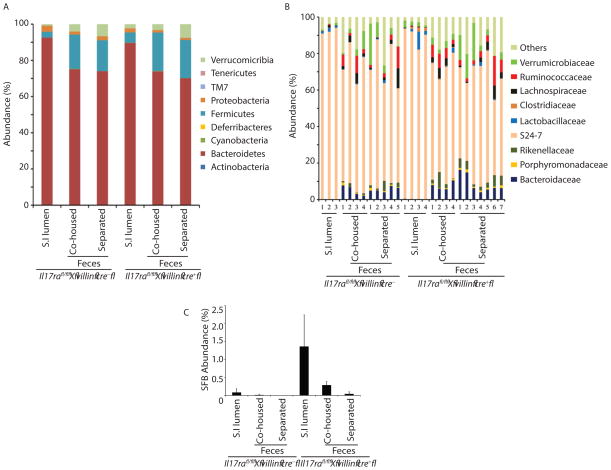
The significant role of epithelial IL-17R signaling in SFB colonization led us to hypothesize a similar role for IL-17 in the colonization of other commensal bacteria. High throughput 16S rRNA microbial community analysis on stool and terminal ileum luminal contents of co-housed (6 week) or separated (separated for 4 weeks after co-housing) mice were utilized to study the overall influence of IL-17R signaling on the microbial community. Analysis demonstrated the overall microbial community remained largely unchanged in epithelial-specific Il17ra deficient mice, suggesting a more subtle and specific microbial control mechanism for epithelial IL-17R signaling on the commensal microbial community (Figure 4A and 4B). In addition, 16S rRNA microbial sequencing results further confirmed our qPCR data, revealing SFB overgrowth in Il17rafl/fl x villin cre+ mice (Figure 4C). A detailed analysis of operational taxonomic units (OTUs) suggested differential abundance of Candidatus arthromitus (SFB), S24-7 and the Clostridiales family (Table 1 top and bottom panel). These data demonstrated that intestinal epithelial cell specific IL-17RA signaling had a minor role in regulating major commensal microbial community.
Figure 4. Commensal dysbiosis in enteric Il17ra−/− mice.
(A and B) Fecal (n=4–7) and terminal ileum luminal contents (S.I lumen, n=3–4) of Il17rafl/fl x villin cre+ and littermate control cre− mice were analyzed for commensal diversity at A) phyla, and B) selected family level by 16S microbial sequencing.
(C) Fecal (n=4–7) and terminal ileum luminal contents (S.I lumen, n=3–4) of Il17rafl/fl x villin cre+ and littermate control cre− mice were analyzed for SFB abundance by 16S microbial sequencing. See also Table 1.
Intestinal IL-17R signaling regulates Nox1, Pigr and α-defensin expression in vivo
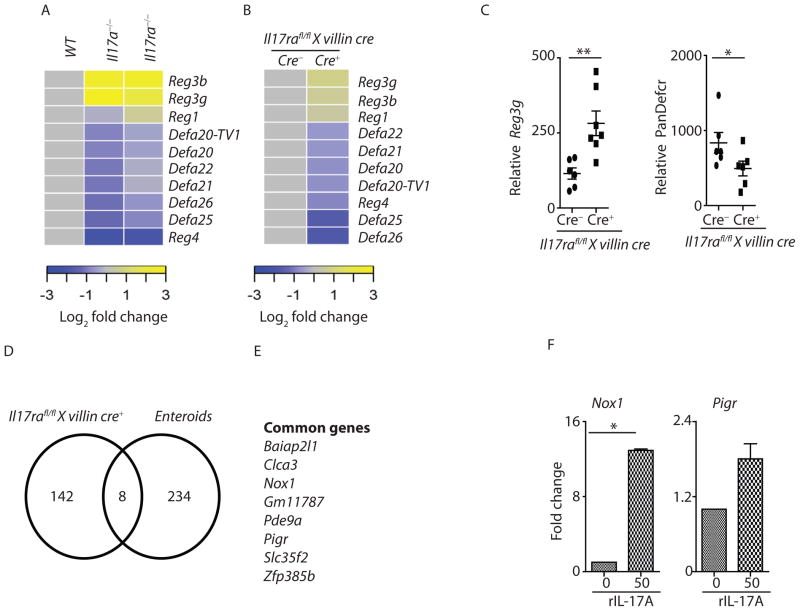
Next, we determined the possible mechanisms of IL-17-dependent regulation of SFB. Enteric anti-microbial peptides, including regenerating-islet derived (Reg)3 family members (Reg3α,β,γ) and α-defensins play important roles in regulating the commensal microbiota (Salzman et al., 2010; Vaishnava et al., 2011). RNA sequencing data from terminal ileum of SFB-colonized Il17a−/− and Il17ra−/− mice revealed an elevated expression of Reg3b and Reg3g consistent with expanded numbers of IL-22+ cells in these mice, but a substantial reduction of several α-defensins genes (Figure 5A). Furthermore, RNA sequencing data from distal small intestine of SFB-colonized Il17rafl/fl x villin cre+ and co-housed littermate control cre− mice confirmed a role for enteric IL-17R signaling in the regulation of α-defensins in vivo (Figure 5B). qPCR data further confirmed increased Reg3g and reduced global α-defensin (pan-defensin) transcripts in the terminal ileum of Il17rafl/fl x villin cre+ mice (Figure 5C). In line with these observations, Rorc−/− mice, which lack Th17 cells and ILC3s, also showed reduced α-defensin expression in the terminal ileum (data not shown). To further confirm a role of IL-17 in regulating defensin expression in vivo, we injected adenovirus expressing IL-17A into Rorc−/− mice. Administration of recombinant adenovirus expressing mouse IL-17A led to increased Il17a transcript as well as an increase in the expression of several α-defensins genes in the terminal ileum of Rorc−/− mice (Figure S6A). Furthermore, we cultured primary intestinal enteroids from C57BL/6 mice and stimulated with recombinant IL-17A for 24 hours. To determine the genes that are acutely regulated by IL-17A signaling, we compared RNAseq data from IL-17A stimulated enteroids (significantly upregulated) versus Il17rafl/fl x villin cre+ mice terminal ileum data (significantly downregulated) (Figure 5D). We found 8 common genes directly regulated by IL-17 both in vitro and in vivo (Figure 5D and 5E). IL-17-dependent genes include Nox1, an apical NADPH oxidase, Polymeric immunoglobulin receptor (Pigr) and Clca3 chloride channel. qPCR was performed to confirm differences in Nox1 and Pigr expression (Figure 5F). These genes (Pigr, Clca3 and Nox1) were not induced in IL-17A stimulated enteroids generated from Il17rafl/fl x villin cre+ (Figure S6B and S6C and data not shown). Collectively, these data suggest that intestinal IL-17R signaling regulates α-defensins, Nox1 and Pigr expression.
Figure 5. Enteric IL-17R signaling regulates expression of host defense genes.
(A and B) Terminal ileum of WT (n=3), Il17a−/− (n=3) and Il17ra−/− (n=3) mice as well as B) Il17rafl/fl x villin cre+ (n=4) and littermate control cre− (n=4) mice were analyzed for the expression of antimicrobial peptides by RNA sequencing. Defa20-TV1 is predicted gene similar to defensin-related cryptdin 20, transcript variant 1 (LOC100041890), mRNA.
(C) Terminal ileum of Il17rafl/fl x villin cre+ (n=6) and littermate control cre− (n=6) mice were analyzed for the expression of Reg3g and global α-defensin (PanDefcr) by qPCR.
(D) Enteroids from C57BL/6 mice were cultured, and stimulated with recombinant IL-17A. Control and IL-17A stimulated enteroids were subjected to RNA sequencing analysis. Enteroids RNA sequencing results were compared to Il17rafl/fl x villin cre mouse terminal ileum RNA sequencing data. Venn diagram was created to compare significantly upregulated genes in IL-17A stimulated enteroids (in vitro) (n=3–4) to significantly downregulated genes in the terminal ileum (in vivo) of Il17rafl/fl x villin cre+ mice.
(E) List of genes in IL-17A stimulated enteroids shared with terminal ileum of Il17rafl/fl x villin cre+ mice.
(F) Enteroids qPCR data confirmed a role for IL-17A in Nox1 and Pigr expression (n=3).
Each symbol indicates results from single mouse. Data presented as mean ± SEM on relevant graphs. *P ≤ 0.05; **P ≤ 0.01, (Mann-Whitney test, Two tailed). See also Figure S6.
IL-17RA-dependent α-defensin, Nox1 and Pigr regulate SFB colonization
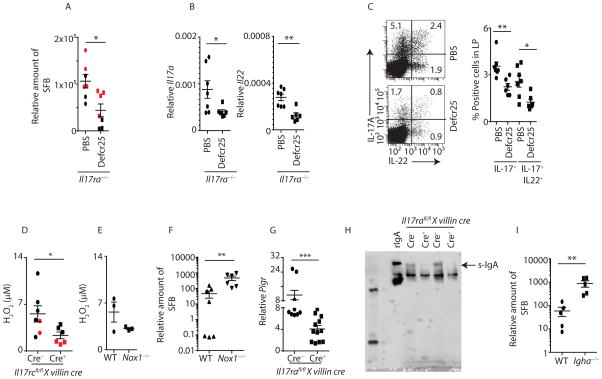
As the expression of specific α-defensins were reduced in Il17a−/−, Il17ra−/− and Il17rafl/fl x villin cre+ mice in vivo (Figure 5A and 5B) and transgenic overexpression of human α-defensins in mouse gut tissue effectively clear SFB (Salzman et al., 2010), we investigated the potential role of exogenous α-defensins in regulating SFB overgrowth in Il17ra−/− mice. To examine this, we synthesized the active form of defensin-related cryptdin-25 (defcr25) peptide and tested its ability to control SFB colonization. We administered defcr25 in the intestinal lumen as demonstrated in supplemental figure S6D. Indeed, Il17ra−/− treated with local defcr25 peptide administration showed reduced SFB colonization in terminal ileum as well as reduced Th17 responses in the lamina propria of Il17ra−/− mice (Figure 6A to 6C). We observed a 2–4 fold reduction in SFB following α-defensin treatment, suggesting additional pathways in regulating SFB colonization. We next determined the role of α-defensins in commensal species diversity. High throughput 16S rRNA microbial community analysis in the feces suggested limited changes in Bacteroidetes (increased), Firmicutes (decreased) and other commensal bacteria composition following local delivery of defcr25 in Il17ra−/− mice (Figure S6E and S6F).
Figure 6. α-defensin, Pigr and Nox1 regulate SFB colonization.
(A) SFB colonized Il17ra−/− mice were treated with PBS or defcr25 peptide. After 12 days, terminal ileum of defcr25 (n=6) or vehicle (n=7) treated Il17ra−/− mice were analyzed for SFB colonization by qPCR.
(B) Terminal ileum of defcr25 (n=6) or vehicle (n=7) treated Il17ra−/− mice were analyzed for the expression of Il17a and Il22 by qPCR.
(C) Lamina propria lymphocytes were isolated on day 12 post defcr25 (n=6) or vehicle (n=6) treated Il17ra−/− mice. CD4 T cells (gated on CD3+CD4+) producing IL-17A and IL-22 were analyzed by flow cytometer (left panel). Percentages of IL-17 or both IL-17 and IL-22 producing CD4+ T cells from multiple mice shown in right panel.
(D) The terminal ileum luminal lavage were collected, and analyzed immediately. Amplex red assay was performed to detect luminal H2O2 concentration in the terminal ileum of Il17rcfl/fl x villin cre+ (n=6) mice and littermate cre− control (n=7) mice.
(E) Amplex red assay was performed to detect luminal H2O2 concentrations in the terminal ileum of WT (n=3) and Nox1−/− (n=3) mice.
(F) Fecal contents of co-housed WT (n=7) and Nox1−/− (n=6) mice at 6 weeks age were analyzed for SFB colonization by qPCR.
(G) Terminal ileum of Il17rafl/fl x villin cre+ (n=12) as well as littermate control cre− (n=9) mice were analyzed for the expression of Pigr by qPCR.
(H) Fecal contents of Il17rafl/fl x villin cre+ as well as littermate cre− control mice were immunoblotted for dimeric IgA.
(I) Fecal contents of age matched WT (n=5) and Igha−/− (n=5) mice were analyzed for SFB colonization by qPCR.
Each symbol indicates results from a single mouse. Figure 6A, 6B, 6C, 6D and 6G were generated from two independent experiments. Individual mouse data of experiment one (black symbol) and experiment two (red symbol) in figure 6A and 6E were separated. Data presented as mean ± SEM on relevant graphs. *P ≤ 0.05; **P ≤ 0.01, (Mann-Whitney test, Two-tailed). See also Figure S6.
As our RNAseq data revealed a role of IL-17 in regulating Nox1 expression, we next determined H2O2 concentrations in the terminal ileum of Il17rcfl/fl x villin cre+ mice using an Amplex red assay. These data showed a significant reduction in H2O2 concentrations in the terminal ileum of Il17rcfl/fl x villin cre+ mice as compared to littermate cre− control mice (Figure 6D). We observed a similar reduction in Nox1−/− mice as a positive control (Figure 6E). Next, we determined the role of Nox1 in regulating SFB colonization. We acquired SFB free WT and Nox1−/− mice from The Jackson Laboratory and have maintained them at our animal facility after SFB inoculation. To determine the role of Nox1 in SFB regulation, WT and Nox1−/− mice were co-housed for 2 weeks immediately after weaning. We measured SFB colonization one and two weeks after the co-housing period. WT and Nox1−/− mice exhibited similar colonization of SFB within one week of the co-housing period (data not shown). However, after two weeks, Nox1−/− mice showed a higher degree of SFB colonization in the feces and terminal ileum as compared to co-housed WT mice (Figure 6F and S6G). As the pIgR-IgA axis has been shown to regulate commensal bacteria (Suzuki et al., 2004), we also observed reduced Pigr expression in the intestine of SFB colonized Il17rafl/fl x villin cre+ mice compared to littermate control cre− mice (Figure 6G). Furthermore, western blot analysis of fecal dimeric secretory IgA was substantially reduced in Il17rafl/fl x villin cre+ mice, consistent with enteric IL-17R signaling being a key regulator of Pigr expression and transportation of secretory IgA in vivo. (Figure 6H). Furthermore, we observed high SFB colonization in the fecal contents of Igha−/− (IgA−/−) mice compared to WT mice, demonstrating a role of IgA in regulating SFB (Figure 6I). Collectively, our data indicate IL-17R-dependent intestinal signaling controls SFB-overgrowth by regulating expression of α-defensins, Nox1 and Pigr.
Intestinal IL-17R signaling regulates autoimmune inflammation
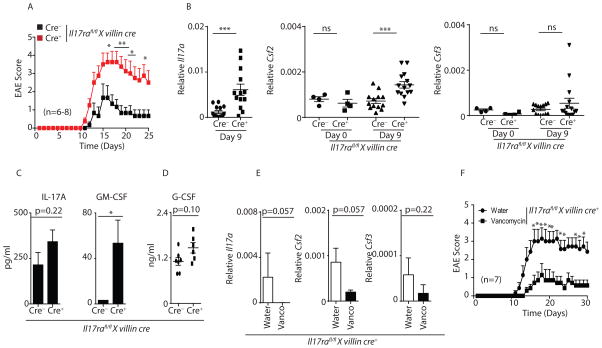
We next determined the functional consequences of disrupted enteric IL-17R signaling, SFB dysbiosis and autoimmunity using experimental autoimmune encephalomyelitis (EAE), a mouse model of MS. As control of Th17 cells occurs in the intestine (Esplugues et al., 2011; Gaboriau-Routhiau et al., 2009; Ivanov et al., 2009) and mice lacking IL-17A and IL-17F signaling (Il17rc−/−) or Th17 differentiation (Batf−/−, Il23p19−/−) have milder EAE disease (Hu et al., 2010; McGeachy et al., 2009; Schraml et al., 2009), we determined if the persistent SFB overgrowth and expanded Th17 cells in gut specific Il17rafl/fl x villin cre+ mice were more susceptible to EAE. Indeed, EAE onset, peak severity, as well incidence scores were higher in Il17rafl/fl x villin cre+ mice compared to littermate control cre− mice (Figure 7A and S7A). To determine the mechanisms underlying the increased EAE severity in Il17rafl/fl x villin cre+ mice, we first analyzed the IL-17 precursor frequency in the spleen on day 9 post EAE when disease scores were 0 in both groups. We observed a small trend towards higher myelin oligodendrocyte glycoprotein (MOG)-specific IL-17A cells in the spleen of Il17rafl/fl x villin cre+ mice as compared to littermate control cre− mice (Figure S7B), however this was not statistically significant. Also, we did not detect SFB-specific T cells in the spleen and spinal cord of MOG immunized mice (data not shown). Next, we examined IL-17 and other encephalitogenic factors such as the cytokines GM-CSF and G-CSF, which have been implicated in EAE severity (Codarri et al., 2011; Rumble et al., 2015). We analyzed Il17a, Csf2 (encodes GM-CSF) and Csf3 (encodes G-CSF) expression in the terminal ileum, inguinal lymph node (ILN), spleen and spinal cord on day 9 post MOG immunization when disease scores were 0 in both groups. Compared to unimmunized (day 0) mice, Il17a expression in the terminal ileum remains upregulated in Il17rafl/fl x villin cre+ mice on day 9 post MOG-immunization as compared to littermate cre− control mice (Figure 3C – 3E and 7B). However, there were no differences in Il17a expression in the spleen, ILN, or spinal cord in Il17rafl/fl x villin cre+ compared to littermate cre− mice on day 9 post immunization (Figure S7C). Csf2 expression was also unchanged in unimmunized Il17rafl/fl x villin cre+ and littermate cre− mice, suggesting that the degree of SFB colonization alone does not regulate its expression (Figure 7B). However, we found that Csf2 but not Csf3 expression was significantly upregulated in the terminal ileum (but not in the ILN, spleen or spinal cord) in response to MOG immunization in Il17rafl/fl x villin cre+ mice compared to littermate cre− control mice (Figure 7B and S7C). Additionally, serum GM-CSF concentrations were also significantly elevated in Il17rafl/fl x villin cre+ mice (Figure 7C). We further demonstrated that isolated lamina propria lymphocytes from day 9 MOG immunized Il17rafl/fl x villin cre+ mice produce substantially higher GM-CSF in response to MOG re-stimulation as compared to cells derived from cre− control mice (Figure S7D). This suggests that development of encephalitogenic GM-CSF producing cells may occur in the lamina propria. There was a trend towards higher serum IL-17A and G-CSF concentrations in Il17rafl/fl x villin cre+ mice, however these differences were not statistically significant (Figure 7C and 7D). To further confirm the role of the gut microbiota on GM-CSF and disease severity, we administered vancomycin in the drinking water of Il17rafl/fl x villin cre+ mice for two weeks prior to EAE induction. Vancomycin treated Il17rafl/fl x villin cre+ mice had substantially reduced Csf2 and Il17a expression in the gut on day 9 post immunization (Figure 7E) and significantly milder EAE scores compared to control mice (Figure 7F). Collectively, our data indicate intestinal IL-17R controls SFB dysbiosis, which is associated with enhanced local and systemic GM-CSF after MOG immunization.
Figure 7. Il17rafl/fl x villin cre+ mice demonstrate enhanced EAE disease severity.
(A) Data shown are EAE onset, peak and severity score in Il17rafl/fl x villin cre+ (n=8) mice and littermate control cre− (n=6) mice.
(B) Terminal ileum of Il17rafl/fl x villin cre+ (n=12) and littermate control cre− (n=13) mice were isolated on day 9 post MOG immunization as well as from unimmunized mice (n=4), and analyzed for the expression of Il17a, Csf2 and Csf3 by qPCR.
(C) Il17rafl/fl x villin cre+ (n=7) and littermate cre− (n=7) control mice serum were isolated on day 9 post MOG immunization. IL-17A and GM-CSF concentrations were determined in the serum by Luminex assay.
(D) Data shown are G-CSF concentration in the serum of Il17rafl/fl x villin cre+ (n=7) and littermate cre− control mice (n=7) on day 9 post EAE.
(E) Terminal ileum of control water (n=7) or vancomycin (n=7) treated MOG-immunized Il17rafl/fl x villin cre+ mice (on day 9 post MOG immunization) were analyzed for the expression of Il17a, Csf2 and Csf3 by qPCR.
(F) Data shown are EAE onset, peak and severity score in control water (n=7) or vancomycin (n=7) treated MOG-immunized Il17rafl/fl x villin cre+ mice.
Figures 7A, 7C, 7D and 7F were generated from 2 independent experiments. Figure 7B was generated from 3 independent experiments. Data presented as mean ± SEM on relevant figures. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.005 (Mann-Whitney test, Two-tailed). See also Figure S7.
Discussion
Our results indicate the existence of a reciprocal relationship between SFB, Th17 lineage commitment, and subsequent IL-17R signaling controls SFB and at the same time constrains the size of the Th17 pool in the lamina propria. Our data showed that abrogation of enteric IL-17RA signaling pathway led to commensal dysbiosis, increased serum GM-CSF concentration and enhanced predisposition to neuroinflammation.
Several studies have found that Il17ra−/− mice displayed exacerbated Th17 responses in vivo (Nagata et al., 2008; Ye et al., 2001a). It has been shown that T cells lacking the IL-17RC receptor were unstable and displayed exacerbated Th17 responses (Wang et al., 2013). However, this study did not investigate the possible role of microbiota in driving hyper Th17 responses. Our data indicate that exacerbated Th17 responses in Il17ra−/− may be influenced by environmental factors, such as colonization with SFB. Furthermore, Il17rafl/fl x Cd4 cre+ mice did not show expanded Th17 responses as compared to littermate control cre− mice both in vivo and in vitro, suggesting T cell instability may not be responsible for exacerbated IL-17 responses observed in Il17ra−/− and Il17rafl/fl x villin cre+ mice. Of note, Il17rafl/fl x Cd4 cre mice were SFB free.
Global Il17ra−/−, Il17rafl/fl x EIIa cre+ and enteric IL-17RA knockouts mice have SFB overgrowth and expanded Th17 responses in the lamina propria, suggesting a reciprocal relationship between SFB and induced IL-17 responses. As both IL-17A and IL-17F use IL-17RA and IL-17RC receptor chains, we expect an important role for both cytokines in SFB regulation, although local concentrations of these individual cytokines have a dominant effect. It has been reported that IL-17F responses are 10–30 fold weaker than IL-17A; however the IL-17A-IL-17F heterodimer can signal (McAllister et al., 2005; Wright et al., 2007).
A previous study by Shih et al suggested that SFB colonization is regulated by IL-22 but not IL-17 signaling (Shih et al., 2014) using Il17f−/− and Il17rc−/− mice. However, it is unclear what the SFB exposure was in these colonies as there was no positive control reported with these animals. Moreover, much of the subsequent data in that paper are in an Il23r−/− background, which would also compromise IL-17 responses. Furthermore, Shih et al. demonstrated that the adoptive transfer of WT CD4+ cells controlled SFB nearly 100% whereas Il22−/− CD4+ cells still controlled SFB to 50% in Il23r−/−Rag2−/− mice, strongly implicating IL-22 independent mechanisms of SFB control. Our data indicate a requirement of IL-17R in controlling SFB using three independent genetic lines—Il17rafl/fl x EIIa cre+, Il17rafl/fl x villin cre+ and Il17rcfl/fl x villin cre+ mice as well as an anti-IL17RA antibody treatment approach. Moreover, our data shows Il17ra−/− and Il17rafl/fl x villin cre+ mice have high SFB colonization despite increased endogenous IL-22 and Reg3γ. This led us to postulate that both IL-22 and IL-17 are regulating SFB colonization by an interdependent or two independent mechanisms.
It is possible that both IL-17 and IL-22 regulate SFB colonization by two independent pathways. In support of this, we have observed very high SFB colonization in Rorc−/− mice deficient in both IL-17 and IL-22 cytokines as compared to individual Il17r−/− or Il22−/− mice. Detailed OTUs analysis from the terminal ileum of our floxed mice suggested a critical role of intestinal IL-17R signaling in regulating many other commensal bacteria. However, it is possible that SFB overgrowth or relatively high Il22 expression in Il17rafl/fl x villin cre+ mice accounted for these changes. Indeed, IL-22 can sequester essential metal ions from microbes and allow colonization of pathogenic bacteria (Behnsen et al., 2014).
Commensal dysbiosis and associated immune responses in Il17rafl/fl x villin cre+ mice were positively correlated with reduced expression of α-defensins, Pigr and Nox1. In addition, adenovirus mediated of expression of IL-17A in Th17 deficient Rorc−/− mice increased α-defensin expression. Interestingly, acute in vitro stimulation of mouse enteroids with IL-17A did not lead to significant changes in α-defensin expression. α-defensin synthesis and release are regulated by multiple factors including developmental and microbial signals (Ayabe et al., 2000; Durand et al., 2012). It is likely that enteric IL-17 either synergizes with other factors to regulate α-defensin expression or the developmental program takes longer than the 24 hours incubation period we used in vitro. Our data suggest a role for IL-17-dependent α-defensins in regulating SFB colonization. Our data are further supported by observations where overexpression of the human α-defensin, HBD-5, in the GI tract reduced colonization of SFB (Salzman et al., 2010). Using an unbiased RNAseq approach, we identified for the first time a unique role of IL-17A in regulating Nox1 expression and SFB colonization. Consistent with published literature, our data also suggest that IL-17 regulates Pigr expression and luminal IgA responses (Cao et al., 2012; Moon et al., 2014). Importantly, Igha−/− mice have SFB overgrowth, suggesting IL-17R signaling activates multiple pathways to regulate SFB overgrowth and maintain gut homeostasis.
Our data further demonstrate that IL-17R-dependent gut immunity plays a critical role in regulating autoimmune inflammation. As SFB modulates Th17 immune responses, we expected to see an increase in IL-17 precursor frequency in the lymph nodes of Il17rafl/fl x villin cre+ mice. Although we observed a trend towards higher MOG-specific IL-17A precursor frequency in the spleen of Il17rafl/fl x villin cre+ mice, these data did not reach statistical significance, suggesting other factors are contributing to EAE severity. It has been shown that Csf2−/− mice are highly resistant to EAE development (Codarri et al., 2011; El-Behi et al., 2011). Terminal ileum Csf2 expression and serum GM-CSF concentrations were significantly higher in Il17rafl/fl x villin cre+ mice after MOG immunization, suggesting IL-17R-dependent regulation of encephalitogenic GM-CSF may have a dominant role in inducing more severe EAE in these animals. It has been shown that IL-1β and/or IL-23 can drive Csf2 expression in Rorγt+ T cells (Codarri et al., 2011; El-Behi et al., 2011). It is possible that expanded Th17 (Rorγt+ T cells) cells in Il17rafl/fl x villin cre+ mice may able to acutely respond to MOG immunization and provide source of encephalitogenic GM-CSF. In support of this notion, we have found that isolated lamina propria lymphocytes from day 9 post MOG immunized Il17rafl/fl x villin cre+ mice produce significantly higher GM-CSF in response to MOG peptide re-stimulation as compared to cre− control mice. Vancomycin treatment reduces the Csf2 expression as well as disease severity, suggesting a possible role of gut-derived GM-CSF in EAE pathogenesis. Our data supports the concept that IL-17 is beneficial in controlling dysbiosis in the gut, but may be harmful if dysregulated or elicited against auto-antigens. Overall, our study highlights the importance of intestinal IL-17R signaling in the host-microbiome interaction and its impact on intestinal and peripheral autoimmune inflammation. These findings may have a tremendous impact on our understanding of intestinal and autoimmune disorder pathogenesis and may provide a therapeutic target for these diseases.
Experimental Procedures
Mice
WT, Rorc−/−, Il17a−/−, Il17ra−/− and Thy1.1 IL-17F reporter in C57BL/6 background were housed in pathogen-free conditions at Children’s Hospital of Pittsburgh. Il17rafl/fl and Il17rcfl/fl mice were generated at Ozgene. Stat3fl/fl x Cd4 cre, ll17rafl/fl x Cd4 cre and ll17rafl/fl x villin cre and ll17rcfl/fl x villin cre on C57BL/6 background were generated and maintained at Children’s Hospital of Pittsburgh. All of the animal studies were conducted with the approval of the University of Pittsburgh Institutional Animal Care and Use Committee. Additional mice strains (Stat3fl/fl, Nox1−/−,Tcrb−/− and Igha−/−) details and generation of conditional knockout mice procedures are provided in the Supplemental Experimental Procedures.
Reagents
SFB-tetramer (I-Ab/3340-A6 tetramer) as described before (Yang et al., 2014) was obtained from Dr. Marc Jenkins, Department of Microbiology, Center for Immunology, University of Minnesota.
Animal treatment
For in vivo defensin administration, 6 weeks old mice were gavaged with PBS or 10 μg/mouse of active defcr25 peptide on days 0, 2, 4, 6, 8, 10. For in vivo neutralization of IL-17RA, mice were intraperitoneally injected with anti-IL-17RA (Amgen) or isotype control (500 μg/mouse) on day 0, 3 and 6.
16S rRNA microbial community analysis
Fecal and terminal ileum luminal DNA were isolated using QiAMP stool DNA extraction kit. Microbial community analysis utilized PCR amplification of the V4 region of 16S rRNA followed by sequencing on an Illumina MiSeq as previously described (Caporaso et al., 2012). See Supplemental Experimental Procedures for details on methods.
qPCR and RNA sequencing
Total RNA from terminal ileum of 6 weeks old mice and IL-17A (0 or 50 ng/ml) stimulated enteroids or sorted CD4+ T cell were isolated using Trizol RNA extraction methods as per manufacturer’s instructions (Life Technologies), and reverse transcribed into cDNA using Biorad IScript kit. Bio-rad SsoAdvanced supermix were used for qPCR. For RNA sequencing, purified RNA were used as starting material for deep sequencing using the in-house Illumina TrueSeq RNA Sample Preparation v2 Guide. See Supplemental Experimental Procedures for details on primers and methods.
Microbiota transplantation and co-housing
SFB-colonization into SFB-free mice strains was achieved by gavaging SFB-containing fecal suspension from Rorc−/− or Taconic C57BL/6 mice. See Supplemental Experimental Procedures for details on co-housing, SFB screening and primers sequences.
EAE and cytokine measurement
Mice were immunized at both sites on the hind flank with 100 μg MOG peptide in 200 μl CFA containing 100 μg M. tuberculosis strain H37Ra as described previously (McGeachy et al., 2009). ELISPOT was performed to detect MOG-specific IL-17A-producing CD4 T cells from the spleen on day 9 after immunization. Serum from these mice was analyzed for IL-17A, GM-CSF and G-CSF concentrations by Luminex or ELISA. See Supplemental Experimental Procedures for details on methods.
Enteroid cultures
Approximately 5 cm of mouse proximal jejunal tissue was used for enteroid culture. Enteroids were stimulated with rIL-17A (0 or 50 ng/mL) for 24 hours followed by RNA extraction for qPCR and RNA-seq. See Supplemental Experimental Procedures for details on methods.
Ligand clearance
Recombinant mouse IL-17A (1 μg/mouse) was injected intravenously into WT or Il17ra−/− mice. Serum IL-17A concentrations were measured using BioLegend IL-17A ELISA kit and plotted as the percentage clearance at different time points.
Th17 differentiation, lymphocytes isolation, activation and staining
For in vitro differentiation, naïve CD4 (CD4+CD62L+) cells from WT, Il17ra−/−, or Thy1.1 reporter (IL-17F) mice were polarized under Th17 differentiating conditions. See Supplemental Experimental Procedures for details on Th17 differentiation methods. For in vivo analysis, lamina propria lymphocytes were isolated and stained with anti-CD3, anti-CD4, anti-IL-17A and anti-IL-22 e-biosciences antibodies. Stained lymphocytes were analyzed using a Becton Dickinson (BD) LSR II flow cytometer and data were analyzed with the BD Diva software.
Scanning Electron Microscopy (SEM)
1 cm portions of 6 week old mouse distal small intestine were cut open and fixed in 2.5% glutaraldehyde and 2% paraformaldehyde in PBS (pH 7.2) for 4 hours and processed for standard SEM at Center of Biologic Imaging, University of Pittsburgh. See Supplemental Experimental Procedures for details on methods.
Amplex red assay and western blot
50 μl of intestinal lavage were subjected to Amplex red assay according to manufacturer’s instructions (Life Technologies). Fecal suspensions containing 20μg protein were used for IgA immunoblot under non-reducing SDS-page. See Supplemental Experimental Procedures for details on methods.
Statistical analyses
GraphPad Prism software was used to analyze experimental groups. To compare differences between two groups, we used the unpaired Mann-Whitney test. For multiple group comparisons, we used ANOVA with Bonferroni’s post-hoc test. Percentage incidence graphs were analyzed using Log-rank test. Data were reported as significant if P ≤ 0.05.
Supplementary Material
Highlights.
SFB-induced Th17 cell expansion in the gut controls the degree of SFB colonization
IL-17R-dependent regulation of α-defensin, Nox1 and Pigr controls SFB
Intestinal IL-17R signaling regulates dysbiosis and autoimmune inflammation.
Acknowledgments
We thank Ozgene for the generation of our floxed mice. We thank Dr. Donna Beer Stolz and Jonathan Franks for SEM imaging. We thank Dr. Mark Jenkins for providing SFB-tetramer. We thank Dr. Dan Littman for providing control and SFB-tetramer and also for critically reading the manuscript. We thank Drs. Timothy Sanders and Anuradha Ray for critically reading manuscript. We thank Dr. Mandy McGeachy for advice on EAE experiments. We thank Dr. Avijit Ray, Blood Center of Wisconsin for advice on EAE experiments. P.K. and L.M. are supported from Children’s Hospital of Pittsburgh Research Advisory Committee Grant from Children’s Hospital of Pittsburgh of the UPMC Health System. P.C is supported by NIH grant T32GM008208. T.E. is supported by NIH grant F30AI114146. M.G. is supported by K08DK101608. The authors would like to acknowledge support from the following PHS grants: P50HL084932, 5R01HL061271, and R37HL079142 to J.K.K.
Footnotes
Author Contributions:
P.K. and J.K.K designed the experiments and wrote the manuscript. P.K., P.C, and J.K.K. performed data analyses. M.G. performed IgA western blot experiments. P.C performed qPCR analysis of Il17ra−/−. W.E. performed Th17 polarization experiment. P.K., T.E., A.V. and K.B. performed 16S microbial sequencing data collection and analysis. W.H performed RNA sequencing experiments and data analyses. D.W.M provided critical experimental samples. A.S.J and A.A.S performed enteroids culture and stimulation. L.M and P.K. performed EAE experiments. R.C.M assisted in designing of defcr25 peptides.
Accession Number
The raw RNA sequencing data has been deposited into the sequencing read archive under SRA accession number SRP069071. 16S microbial sequencing data was deposited under SRA accession 4677462.3 to 4677494.3 on MG-RAST.
Supplemental information includes seven figures, one table and Supplemental Experimental Procedures.
Conflict of interest
R.C.M holds stock in Peptilogics, Inc. and serves on the scientific advisory board for the company. Although a potential financial conflict of interest was identified based on the author’s relationship with Peptilogics, the research findings included in this publication may not necessarily be related to the interests of Peptilogics. J.K.K is cofounder of MiniVax, Inc. No work in this paper was supported by MiniVax, Inc.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ayabe T, Satchell DP, Wilson CL, Parks WC, Selsted ME, Ouellette AJ. Secretion of microbicidal [alpha]-defensins by intestinal Paneth cells in response to bacteria. Nat Immunol. 2000;1:113–118. doi: 10.1038/77783. [DOI] [PubMed] [Google Scholar]
- Behnsen J, Jellbauer S, Wong Christina P, Edwards Robert A, George Michael D, Ouyang W, Raffatellu M. The Cytokine IL-22 Promotes Pathogen Colonization by Suppressing Related Commensal Bacteria. Immunity. 2014;40:262–273. doi: 10.1016/j.immuni.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch TCG, Augustin R, Anton-Erxleben F, Fraune S, Hemmrich G, Zill H, Rosenstiel P, Jacobs G, Schreiber S, Leippe M, et al. Uncovering the evolutionary history of innate immunity: The simple metazoan Hydra uses epithelial cells for host defence. Developmental & Comparative Immunology. 2009;33:559–569. doi: 10.1016/j.dci.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Cao AT, Yao S, Gong B, Elson CO, Cong Y. Th17 Cells Upregulate Polymeric Ig Receptor and Intestinal IgA and Contribute to Intestinal Homeostasis. The Journal of Immunology. 2012;189:4666–4673. doi: 10.4049/jimmunol.1200955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, McAleer Jeremy P, Lin Y, Paterson David L, Zheng M, Alcorn John F, Weaver Casey T, Kolls Jay K. Th17 Cells Mediate Clade-Specific, Serotype-Independent Mucosal Immunity. Immunity. 2011;35:997–1009. doi: 10.1016/j.immuni.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codarri L, Gyulveszi G, Tosevski V, Hesske L, Fontana A, Magnenat L, Suter T, Becher B. ROR[gamma]t drives production of the cytokine GM-CSF in helper T cells, which is essential for the effector phase of autoimmune neuroinflammation. Nat Immunol. 2011;12:560–567. doi: 10.1038/ni.2027. [DOI] [PubMed] [Google Scholar]
- Dardalhon V, Korn T, Kuchroo VK, Anderson AC. Role of Th1 and Th17 cells in organ-specific autoimmunity. Journal of Autoimmunity. 2008;31:252–256. doi: 10.1016/j.jaut.2008.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand A, Donahue B, Peignon G, Letourneur F, Cagnard N, Slomianny C, Perret C, Shroyer NF, Romagnolo B. Functional intestinal stem cells after Paneth cell ablation induced by the loss of transcription factor Math1 (Atoh1) Proceedings of the National Academy of Sciences. 2012;109:8965–8970. doi: 10.1073/pnas.1201652109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Behi M, Ciric B, Dai H, Yan Y, Cullimore M, Safavi F, Zhang GX, Dittel BN, Rostami A. The encephalitogenicity of TH17 cells is dependent on IL-1- and IL-23-induced production of the cytokine GM-CSF. Nat Immunol. 2011;12:568–575. doi: 10.1038/ni.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esplugues E, Huber S, Gagliani N, Hauser AE, Town T, Wan YY, O/’Connor W, Rongvaux A, Van Rooijen N, Haberman AM, et al. Control of TH17 cells occurs in the small intestine. Nature. 2011;475:514–518. doi: 10.1038/nature10228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzenburg S, Fraune S, Künzel S, Baines JF, Domazet-Lošo T, Bosch TCG. MyD88-deficient Hydra reveal an ancient function of TLR signaling in sensing bacterial colonizers. Proceedings of the National Academy of Sciences. 2012;109:19374–19379. doi: 10.1073/pnas.1213110109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaboriau-Routhiau V, Rakotobe S, Lécuyer E, Mulder I, Lan A, Bridonneau C, Rochet V, Pisi A, De Paepe M, Brandi G, et al. The Key Role of Segmented Filamentous Bacteria in the Coordinated Maturation of Gut Helper T Cell Responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- Hu Y, Ota N, Peng I, Refino CJ, Danilenko DM, Caplazi P, Ouyang W. IL-17RC Is Required for IL-17A– and IL-17F–Dependent Signaling and the Pathogenesis of Experimental Autoimmune Encephalomyelitis. The Journal of Immunology. 2010;184:4307–4316. doi: 10.4049/jimmunol.0903614. [DOI] [PubMed] [Google Scholar]
- Hueber W, Sands BE, Lewitzky S, Vandemeulebroecke M, Reinisch W, Higgins PDR, Wehkamp J, Feagan BG, Yao MD, Karczewski M, et al. Secukinumab, a human anti-IL-17A monoclonal antibody, for moderate to severe Crohn’s disease: unexpected results of a randomised, double-blind placebo-controlled trial. Gut. 2012;61:1693–1700. doi: 10.1136/gutjnl-2011-301668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. Induction of Intestinal Th17 Cells by Segmented Filamentous Bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Frutos RdL, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, Finlay BB, Littman DR. Specific Microbiota Direct the Differentiation of IL-17-Producing T-Helper Cells in the Mucosa of the Small Intestine. Cell Host & Microbe. 2008;4:337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock C, Hermans G, Pedotti R, Brendolan A, Schadt E, Garren H, Langer-Gould A, Strober S, Cannella B, Allard J, et al. Gene-microarray analysis of multiple sclerosis lesions yields new targets validated in autoimmune encephalomyelitis. Nat Med. 2002;8:500–508. doi: 10.1038/nm0502-500. [DOI] [PubMed] [Google Scholar]
- Malley R, Srivastava A, Lipsitch M, Thompson CM, Watkins C, Tzianabos A, Anderson PW. Antibody-Independent, Interleukin-17A-Mediated, Cross-Serotype Immunity to Pneumococci in Mice Immunized Intranasally with the Cell Wall Polysaccharide. Infection and Immunity. 2006;74:2187–2195. doi: 10.1128/IAI.74.4.2187-2195.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister F, Henry A, Kreindler JL, Dubin PJ, Ulrich L, Steele C, Finder JD, Pilewski JM, Carreno BM, Goldman SJ, et al. Role of IL-17A, IL-17F, and the IL-17 Receptor in Regulating Growth-Related Oncogene-α and Granulocyte Colony-Stimulating Factor in Bronchial Epithelium: Implications for Airway Inflammation in Cystic Fibrosis. The Journal of Immunology. 2005;175:404–412. doi: 10.4049/jimmunol.175.1.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeachy MJ, Chen Y, Tato CM, Laurence A, Joyce-Shaikh B, Blumenschein WM, McClanahan TK, O’Shea JJ, Cua DJ. The interleukin 23 receptor is essential for the terminal differentiation of interleukin 17-producing effector T helper cells in vivo. Nat Immunol. 2009;10:314–324. doi: 10.1038/ni.1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon C, VanDussen KL, Miyoshi H, Stappenbeck TS. Development of a primary mouse intestinal epithelial cell monolayer culture system to evaluate factors that modulate IgA transcytosis. Mucosal Immunol. 2014;7:818–828. doi: 10.1038/mi.2013.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata T, Mckinley L, Peschon JJ, Alcorn JF, Aujla SJ, Kolls JK. Requirement of IL-17RA in Con A Induced Hepatitis and Negative Regulation of IL-17 Production in Mouse T Cells. The Journal of Immunology. 2008;181:7473–7479. doi: 10.4049/jimmunol.181.11.7473. [DOI] [PubMed] [Google Scholar]
- Roberts S, Gueguen Y, de Lorgeril J, Goetz F. Rapid accumulation of an interleukin 17 homolog transcript in Crassostrea gigas hemocytes following bacterial exposure. Developmental & Comparative Immunology. 2008;32:1099–1104. doi: 10.1016/j.dci.2008.02.006. [DOI] [PubMed] [Google Scholar]
- Rumble JM, Huber AK, Krishnamoorthy G, Srinivasan A, Giles DA, Zhang X, Wang L, Segal BM. Neutrophil-related factors as biomarkers in EAE and MS. The Journal of Experimental Medicine. 2015;212:23–35. doi: 10.1084/jem.20141015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman NH, Hung K, Haribhai D, Chu H, Karlsson-Sjoberg J, Amir E, Teggatz P, Barman M, Hayward M, Eastwood D, et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol. 2010;11:76–82. doi: 10.1038/ni.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schraml BU, Hildner K, Ise W, Lee WL, Smith WAE, Solomon B, Sahota G, Sim J, Mukasa R, Cemerski S, et al. The AP-1 transcription factor Batf controls TH17 differentiation. Nature. 2009;460:405–409. doi: 10.1038/nature08114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih VFS, Cox J, Kljavin NM, Dengler HS, Reichelt M, Kumar P, Rangell L, Kolls JK, Diehl L, Ouyang W, Ghilardi N. Homeostatic IL-23 receptor signaling limits Th17 response through IL-22–mediated containment of commensal microbiota. Proceedings of the National Academy of Sciences. 2014;111:13942–13947. doi: 10.1073/pnas.1323852111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E, Stark MA, Zarbock A, Burcin TL, Bruce AC, Vaswani D, Foley P, Ley K. IL-17A Inhibits the Expansion of IL-17A-Producing T Cells in Mice through “Short-Loop” Inhibition via IL-17 Receptor. The Journal of Immunology. 2008;181:1357–1364. doi: 10.4049/jimmunol.181.2.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Meek B, Doi Y, Muramatsu M, Chiba T, Honjo T, Fagarasan S. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:1981–1986. doi: 10.1073/pnas.0307317101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, Ley R, Wakeland EK, Hooper LV. The Antibacterial Lectin RegIIIγ Promotes the Spatial Segregation of Microbiota and Host in the Intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Wu L, Bulek K, Martin BN, Zepp JA, Kang Z, Liu C, Herjan T, Misra S, Carman JA, et al. The psoriasis-associated D10N variant of the adaptor Act1 with impaired regulation by the molecular chaperone hsp90. Nat Immunol. 2013;14:72–81. doi: 10.1038/ni.2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright JF, Guo Y, Quazi A, Luxenberg DP, Bennett F, Ross JF, Qiu Y, Whitters MJ, Tomkinson KN, Dunussi-Joannopoulos K, et al. Identification of an Interleukin 17F/17A Heterodimer in Activated Human CD4+ T Cells. Journal of Biological Chemistry. 2007;282:13447–13455. doi: 10.1074/jbc.M700499200. [DOI] [PubMed] [Google Scholar]
- Wüthrich M, Gern B, Hung CY, Ersland K, Rocco N, Pick-Jacobs J, Galles K, Filutowicz H, Warner T, Evans M, et al. Vaccine-induced protection against 3 systemic mycoses endemic to North America requires Th17 cells in mice. The Journal of Clinical Investigation. 2011;121:554–568. doi: 10.1172/JCI43984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Torchinsky MB, Gobert M, Xiong H, Xu M, Linehan JL, Alonzo F, Ng C, Chen A, Lin X, et al. Focused specificity of intestinal TH17 cells towards commensal bacterial antigens. Nature. 2014;510:152–156. doi: 10.1038/nature13279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye P, Garvey PB, Zhang P, Nelson S, Bagby G, Summer WR, Schwarzenberger P, Shellito JE, Kolls JK. Interleukin-17 and Lung Host Defense againstKlebsiella pneumoniae Infection. American Journal of Respiratory Cell and Molecular Biology. 2001a;25:335–340. doi: 10.1165/ajrcmb.25.3.4424. [DOI] [PubMed] [Google Scholar]
- Ye P, Rodriguez FH, Kanaly S, Stocking KL, Schurr J, Schwarzenberger P, Oliver P, Huang W, Zhang P, Zhang J, et al. Requirement of Interleukin 17 Receptor Signaling for Lung Cxc Chemokine and Granulocyte Colony-Stimulating Factor Expression, Neutrophil Recruitment, and Host Defense. The Journal of Experimental Medicine. 2001b;194:519–528. doi: 10.1084/jem.194.4.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.