Abstract
This protocol is used for the isolation and analysis of protein complexes using the tandem affinity purification (TAP) tag system. The protocol describes the purification of a protein fused to a TAP tag comprised of two protein A domains and the calmodulin binding peptide separated by a TEV cleavage site. This is a powerful technique for rapid purification of protein complexes and the analysis of their stoichiometric composition, posttranslational modifications, structure, and functional activities.
1. THEORY
This is a two-step purification protocol that allows for the identification of specific protein complexes with a low background of contaminating proteins (Rigaut et al., 1999; Puig et al., 2001). The strategy has been used successfully for purification and identification of complexes from Saccharomyces cerevisiae, Schizosaccharomyces pombe, and mammalian cells (Tanny et al., 2004; Verdel et al., 2004; Chen et al., 2001). The TAP-tagged protein is purified, first, by binding of its protein A tag to an IgG resin. After cleavage with TEV (tobacco etch virus) protease, the remaining calmodulin-binding peptide (CBP) fusion protein is bound to a calmodulin resin. The target protein is then eluted by the addition of a calcium chelator, resulting in purified material that is free of peptide or IgG, which is ideal for identifying interacting partners using tandem mass spectrometry approaches. However, since the TAP tag is quite large, not all proteins are functional when fused with this tag. It is critical to test functionality prior purification. In addition, weak or transient interactions may not be detected using this two-step purification method and therefore, a one-step purification approach may also need to be tried. Other combinations of tags have also been successfully tried (Bürckstümmer et al., 2006; Gloeckner et al., 2007).
TAP purifications have emerged as ideal methods for the isolation and characterization endogenous proteins or complexes that are not particularly abundant. The two purification steps provide sufficient purity for the biochemical and proteomic analysis of most complexes under study in our laboratory. This protocol can easily be scaled up or down depending on protein expression level and desired protein yield.
2. EQUIPMENT
Refrigerated centrifuge
Bead beater (BioSpec Bead Beater, model 1107900)
0.5-mm glass beads (BioSpec)
Refrigerated microcentrifuge
SDS-PAGE gel rig
Platform rotator/mixer
Poly-prep columns (BioRad)
Poly-prep column caps and tip closures (BioRad)
Square stand (ring stand)
Stand clamps
50-ml polypropylene conical tubes
15-ml polypropylene conical tubes
Pipet-aid
Pipettes
Micropipettors
Pipettor tips
1.7-ml polypropylene microcentrifuge tubes
3. MATERIALS
Tris base
Sodium chloride (NaCl)
Calcium chloride (CaCl2)
EDTA
EGTA
Magnesium acetate (Mg(OAc)2)
Sodium phosphate dibasic (Na2HPO4)
Sodium phosphate monobasic monohydrate (NaH2PO4·H2O)
Sodium fluoride (NaF)
Na2VO4
Imidazole
NP-40
Trichloroacetic acid
2-Mercaptoethanol (BME)
Glycerol
Potassium acetate (KOAc)
1,3-Dithiothreitol (DTT)
Leupeptin
Benzamidine
Phenylmethylsulfonyl fluoride (PMSF)
EDTA-free complete protease inhibitor tablet (Roche)
IgG-Sepharose (GE Healthcare)
Calmodulin-Sepharose (GE Healthcare)
TEV protease, recombinant (Invitrogen)
Ethanol
Acetone
Anti-TAP antibody (Thermo Fisher)
Bromophenol Blue
Sodium dodecyl sulfate (SDS)
3.1. Solutions & buffers
| Step 1 Lysis Buffer | |||
|---|---|---|---|
| Component | Final concentration | Stock | Amount |
| Na2PO4 | 6 mM | – | 0.85 g |
| NaH2PO4·H2O | 4 mM | – | 0.55 g |
| NP-40 | 1% | 100% | 10 ml |
| NaCl | 150 mM | – | 8.8 g |
| EDTA | 2 mM | 0.5 M | 4 ml |
| EGTA | 1 mM | 0.5 M | 2 ml |
| NaF | 50 mM | – | 2.1 g |
| Leupeptin | 4 μg ml−1 | – | 4 mg |
| Na2VO4 | 0.1 mM | 0.5 M | 0.2 ml |
| Glycerol | 5% | 100% | 50 ml |
Add water to 1 l
Filter-sterilize, wrap in foil and store at 4 °C until use
Aliquot 50 ml immediately prior to use and add 1 complete protease inhibitor cocktail tablet (Roche) – or – pepstatin, bestatin, and aprotinin to 1 μg ml−1;
130 μl of 0.5 M benzamidine, freshly prepared in 100% ethanol
500 μl of 100 mM PMSF, freshly prepared in 100% ethanol
| Step 2 Wash Buffer | |||
|---|---|---|---|
| Component | Final concentration | Stock | Amount |
| Tris–HCl, pH 8.0 | 10 mM | 1 M | 1 ml |
| NaCl | 150 mM | 5 M | 3 ml |
| NP-40 | 0.1% | 10% | 1 ml |
Add water to 100 ml
| Step 3 TEV Cleavage (TEV-C) Buffer | |||
|---|---|---|---|
| Component | Final concentration | Stock | Amount |
| Tris–HCl, pH 8.0 | 10 mM | 1 M | 0.5 ml |
| KOAc | 150 mM | 5 M | 1.5 ml |
| NP-40 | 0.1% | 10% | 0.5 ml |
| EDTA | 0.5 mM | 0.5 M | 50 μl |
| DTT* | 1 mM | 1 M | 50 μl |
Add DTT immediately before use
Add water to 50 ml
| Step 4 Calmodulin Binding (CAM-B) Buffer | |||
|---|---|---|---|
| Component | Final concentration | Stock | Amount |
| Tris–HCl, pH 8.0 | 10 Mm | 1 M | 1 ml |
| NaCl | 150 mM | 5 M | 3 ml |
| Mg(OAc)2 | 1 mM | 1 M | 0.1 ml |
| Imidazole | 1 mM | 1 M | 0.2 ml |
| CaCl2 | 2 mM | 1 M | 0.2 ml |
| BME* | 14.3 M | 14.3 M | 69.7 μl |
Add BME immediately before use
Add water to 100 ml
| Calmodulin Elution (CAM-E) Buffer | |||
|---|---|---|---|
| Component | Final concentration | Stock | Amount |
| Tris–HCl, pH 8.0 | 10 mM | 1 M | 100 μl |
| NaCl | 150 mM | 5 M | 300 μl |
| Mg(OAc)2 | 1 mM | 1 M | 10 μl |
| Imidazole | 1 mM | 1 M | 10 μl |
| EGTA | 10 mM | 0.5 M | 200 μl |
| BME* | 10 mM | 14.3 M | 6.9 μl |
Add BME immediately before use
Add water to 10 ml
| Step 5 2×Sample Buffer | |||
|---|---|---|---|
| Component | Final concentration | Stock | Amount |
| Tris–HCl, pH 6.8 | 100 mM | 1 M | 100 μl |
| SDS | 4% | 25% | 160 μl |
| Glycerol | 20% | 50% | 400 μl |
| DTT | 200 mM | 1 M | 200 μl |
| PMSF | 2 mM | 100 mM | 20 μl |
| Bromophenol blue | 0.2% | 2 mg | |
Add water to 1 ml
4. PROTOCOL
4.1. Preparation
Grow 3–6 l of yeast to OD600 = 1–2. Pellet the cells by centrifugation and flash-freeze the cell pellet in liquid nitrogen. The pellets are usually about 5–10 g. Store all pellets at −80 °C.
For each purification, include an untagged strain isogenic to the strain expressing the TAP-tagged protein. During proteomics analysis, the untagged strain will be used as a comparison for nonspecific background binding.
4.2. Duration
| Preparation | About 1 day |
| Protocol | About 6–10 h |
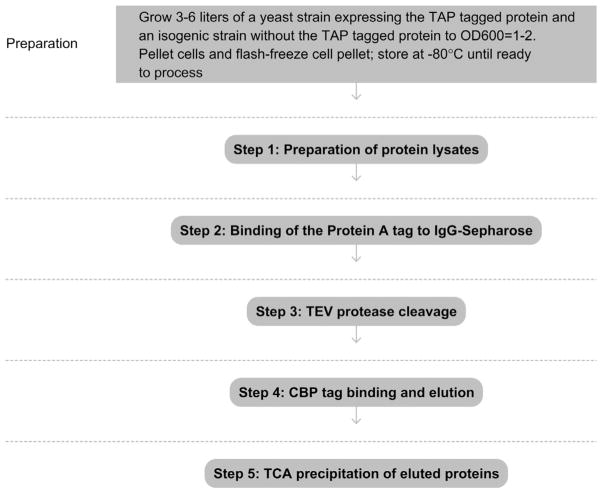
See Fig. 1 for the flowchart of the complete protocol.
Figure 1.
Flowchart of the complete protocol, including preparation.
5. STEP 1 PREPARATION OF PROTEIN LYSATES
5.1. Overview
Pelleted, frozen yeast cells will be lysed by bead beating and the resulting lysates will be cleared by centrifugation to remove cell debris before beginning the purification. Other lysis methods, which generate less heat than bead beating, may be more suitable for some protein purifications and should be considered. These include lysis in a coffee grinder in the presence of dry ice (Schultz et al., 1997), lysis under liquid nitrogen using a mortar and pestle, or lysis in a liquid nitrogen-cooled steel chamber (e.g., Retsch Cryomill).
5.2. Duration
1–1.5 h
-
1.1
Add frozen yeast pellets to the medium BioSpec bead beating chamber (89 ml) filled about half way with 0.5-mm glass beads (so that the beads just cover the screw that holds the rotor in place). Then add lysis buffer with freshly added protease inhibitors until the chamber is completely filled.
-
1.2
Fill the outer chamber with ice water. Pulse the bead beater 7–10 s up to 8 times with 1 min between each pulse.
-
1.3
Using a pipet-aid and pipette, transfer the lysate to a 50-ml conical tube, leaving behind the glass beads. Wash the beads with 5 ml of lysis buffer containing protease inhibitors and add the wash to the same 50-ml tube.
-
1.4
Spin in a refrigerated centrifuge (such as a Sorvall RC5, with SH-3000 rotor) for 10 min at 1500×g.
-
1.5
Transfer the supernatant to a clean conical tube, 15 or 50 ml, depending on the lysate volume. Remove 50 μl of each supernatant (supernatant fraction, S1), to be analyzed later (see Step 6.1).
5.3. Tip
Filling the bead beating chamber completely prevents trapping air bubbles, which promote protein denaturation during lysis.
5.4. Tip
The larger 250-ml lysis chamber can also be used for pellets ranging in size from about 10 to 100 g of cells. The larger chamber appears to generate less heat during lysis; however, with smaller pellets, this results in a less concentrated supernatant.
5.5. Tip
To check for efficient lysis, a small aliquot can be taken from the bead beater and visualized under a microscope. >50% lysis is good.
See Fig. 2 for the flowchart of Step 1.
Figure 2.
Flowchart of Step 1.
6. STEP 2 BINDING OF THE PROTEIN A TAG TO IgG-SEPHAROSE
6.1. Overview
This is the first binding step of the tandem affinity tag purification. Each supernatant will be incubated with IgG-Sepharose beads to capture the target protein. After this binding reaction, the beads will be washed before proceeding with the purification. All steps are carried out in the cold room (at 4 °C).
6.2. Duration
2.5–3 h
-
2.1
For every 50 ml of lysate, remove 400 μl of 50% slurry of IgG-Sephaorse beads (200 μl of packed beads). Wash beads 3 times in lysis buffer (without protease inhibitors added), spinning the beads at 500×g between washes.
-
2.2
Add the washed beads to the conical tubes containing each supernatant and put all tubes on a rotating or nutating platform for 2 h at 4 °C.
-
2.3
Pour the supernatant and beads from the conical tube into a large BioRad poly-prep column. Allow the beads to settle by gravity as the unbound supernatant flows through. Save 50 μl of the flow-through (unbound fraction, UB1), to be analyzed later (see Step 6.1).
-
2.4
Wash the beads 3 times with 10 ml of wash buffer.
6.3. Tip
Using a vacuum aspirator during bead washing can lead to accidental bead loss. Therefore, use a pipetor instead.
6.4. Tip
When working with Sepharose or agarose beads, always cut the tip of your pipette using a clean razor blade prior to pipetting the beads. Otherwise the beads will clog the tip during pipetting leading to the uptake of more buffer and fewer beads.
6.5. Tip
Be gentle with the IgG-Sepharose beads. Never spin the beads at >500×g, and always resuspend the beads by gently pipetting, never vortexing.
6.6. Tip
Do not add more than the recommended amount of beads for a given purification. The beads have a high binding capacity, and increasing bead volume does not lead to more target protein binding, but will increase background binding.
See Fig. 3 for the flowchart of Step 2.
Figure 3.
Flowchart of Step 2.
7. STEP 3 TEV PROTEASE CLEAVAGE
7.1. Overview
The TAP tag is composed of two protein A domains and a CBP domain separated by a TEV protease cleavage site. During this step, the target protein, now bound to IgG beads, is eluted by cleavage using the TEV protease before proceeding to the second binding step.
7.2. Duration
2 h
-
3.1
Add 10 ml of TEV-C buffer to the column of washed beads from Step 2.4 and allow to flow through the column.
-
3.2
Close the bottom of the column with a tip closure and move it to room temperature.
-
3.3
Add 1 ml of TEV-C buffer containing the recommended amount of recombinant TEV protease (300–500 U ml−1 for Invitrogen TEV pro-tease, or as described by manufacturer). Incubate for 1–1.5 h at room temperature. Every 20 min, gently resuspend the beads by tapping the tube or by pipetting up and down.
-
3.4
Align the column with a fresh poly-prep column (with the bottom capped). Carefully remove the stopper and allow the elutate to drain into the new column.
-
3.5
Wash out the old column with 0.5 ml of TEV cleavage buffer, and drain into the new column containing the eluate.
-
3.6
Save a small amount of the IgG-Sepharose beads (IgG beads fraction, B1), to analyze later (see Step 6.1).
7.3. Tip
Alternatively, the TEV cleavage can be done overnight at 4 °C on an end-over-end rotator.
See Fig. 4 for the flowchart of Step 3.
Figure 4.
Flowchart of Step 3.
8. STEP 4 CBP TAG BINDING AND ELUTION
8.1. Overview
This is the second binding step of the tandem affinity tag purification. The target protein, now cleaved by TEV protease, has the CBP tag remaining and will be incubated with Calmodulin-Sepharose beads. After this binding reaction, the beads will be washed and the protein will be eluted and will be ready for analysis. All steps are performed at 4 °C.
8.2. Duration
2 h
-
4.1
To the poly-prep column containing the TEV eluate (from Step 3.5), add 3 μl of 1 M CaCl2 for each ml of the TEV elution (e.g., 4.5 μl 1 M CaCl2 to 1.5 ml of eluate, adjust as necessary for other volumes).
-
4.2
For each purification, remove 300 μl of 50% slurry of Calmodulin-Sepharose beads (150 μl of packed beads). Wash the beads 3 times in CAM-B buffer, spinning the beads at 500×g between washes.
-
4.3
Add 3 ml of CAM-B buffer and the washed beads to the poly-prep column. Cap and seal the top of the column, and incubate on a rotating platform at 4 °C for 1 h.
-
4.4
After binding, allow the column to drain, but keep analiquot of the flow-through (CAM unbound fraction, UB2), to analyze later (see Step 6.1).
-
4.5
Wash the beads in the column 3 times 10 ml with CAM-B buffer. Keep an aliquot of the beads to analyze later (CAM beads, B2).
-
4.6
Elute the protein into 1.7-ml microcentrifuge tubes. Obtain three fractions by gently pipetting 550 μl (E1), 450 μl (E2), and 450 μl (E3) of CAM-E buffer to the top of the column bed and allowing it to drain into the collection tubes.
8.3. Tip
To ensure effective washes, for each wash, allow for the entire 10 ml to flow through the column before adding the next wash.
8.4. Tip
Keep a close eye on your column during the washes. Waiting too long between washes can cause the beads to dry out.
8.5. Tip
To get the last drop of each elution fraction, carefully touch the tip of the column to the side of the collection tube while pressing the top of the column with your gloved thumb.
8.6. Tip
Step 4.6 suggests collecting three fractions, but this can be adjusted to obtain a more concentrated peak fraction if necessary.
See Fig. 5 for the flowchart of Step 4.
Figure 5.
Flowchart of Step 4.
9. STEP 5 TCA PRECIPITATION OF ELUTED PROTEINS
9.1. Overview
The purified proteins often need to be concentrated by TCA precipitation prior to gel electrophoresis followed by Coomassie Blue staining or silver staining or analysis by mass spectrometry.
9.2. Duration
1–3 h
-
5.1
Split each eluate fraction equally into two microcentrifuge tubes. Adjust the volume with 100% TCA to a final concentration of 20% TCA for each eluate. Vortex well and keep on ice for 20 min.
-
5.2
Spin in a microcentrifuge at maximum speed (~15 000×g) at 4 °C for 20 min.
-
5.3
Carefully remove the supernatant with a pipettor and discard. Add 1 ml of 10% TCA, cap the tube, invert once to wash the sides of the tube, and spin for 10 min at maximum speed at 4 °C. Carefully remove the supernatant with a pipettor and discard.
-
5.4
Add 1 ml cold acetone (stored at −20 °C), cap the tube, invert, and spin at maximum speed at 4 °C for 10 min. Carefully remove the supernatant with a pipettor and discard. Briefly spin again, and remove the residual liquid with a pipettor. Allow the pellet to air-dry at room temperature for 5–10 min.
-
5.5
To one of the TCA pellets, add 15–20 μl of 1×SDS sample buffer. Leave the tubes at room temperature for at least 30 min, vortexing vigorously every 5 min or so, heat at 65 °C for 10 min, and resolve the proteins by SDS-PAGE (see One-dimensional SDS-Polyacrylamide Gel Electrophoresis (1D SDS-PAGE)). Stain the gel with either colloidal Coomassie Blue or silver to visualize purified proteins (see Coomassie Blue Staining or Silver Staining of SDS-polyacrylamide Gel).
-
5.6
The second TCA pellet can be used for analysis by mass spectrometry, which can also be performed on the whole mixture of proteins or excised bands from silver or Coomassie stained gels (Haas et al., 2006; Shevchenko et al., 1996).
9.3. Tip
Many times there will be no visible pellet after the TCA precipitation, so be careful to gently remove washes with a pipettor, not a vacuum aspirator.
9.4. Tip
Large TCA pellets may take longer than 30 min to dissolve in sample buffer. It may be necessary to leave the tubes at room temperature overnight prior to loading the gel to ensure that the pellets have completely dissolved.
See Fig. 6 for the flowchart of Step 5.
Figure 6.
Flowchart of Step 5.
10. STEP 6 TROUBLESHOOTING
10.1. Overview
In the event that the purification fails to yield the desired protein, the following steps can be taken to troubleshoot.
-
6.1
The fractions that were collected throughout the procedure can help diagnose the step that is problematic. For instance, some proteins can stick to the IgG beads even after TEV cleavage. Performing a Western blot on the collected fractions would indicate that the protein remained on the beads. Take fractions, S1, UB1, B1, UB2, B2, and an aliquot of each eluate and add 2×sample buffer to a final concentration of 1×. See Chapter Western Blotting using Chemiluminescent Substrates for a detailed protocol for Western blotting. After resolving the fractions by SDS-PAGE, probe the membrane with an anti-TAP antibody (Thermo Fisher, binds the CBP portion of the TAP tag).
-
6.2
Proteins with very low expression levels may result in a low yield, in which case you can scale up the procedure to increase yield. For some proteins, we have had to use 100 g of yeast cells instead of the standard 5–10 g described above (Hong et al., 2005; Motamedi et al., 2004).
-
6.3
For proteins that are prone to degradation, alternative lysis methods that prevent heat generation during cell breakage are more desirable. These include lysis in a coffee grinder with dry ice (Schultz et al., 1997), grinding by mortar and pestle under liquid nitrogen, or using a cooled steel mill device (e.g., Retsch Cryomill).
Footnotes
Referenced Protocols in Methods Navigator
One-dimensional SDS-Polyacrylamide Gel Electrophoresis (1D SDS-PAGE).
Coomassie Blue Staining.
Silver Staining of SDS-polyacrylamide Gel.
Western Blotting using Chemiluminescent Substrates.
References
- Bürckstümmer T, Bennett KL, Preradovic A, et al. An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nature Methods. 2006;3(12):1013–1019. doi: 10.1038/nmeth968. [DOI] [PubMed] [Google Scholar]
- Chen CY, Gherzi R, Ong SE, et al. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107(4):451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- Gloeckner CJ, Boldt K, Schumacher A, Roepman R, Ueffing M. A novel tandem affinity purification strategy for the efficient isolation and characterisation of native protein complexes. Proteomics. 2007;7(23):4228–4234. doi: 10.1002/pmic.200700038. [DOI] [PubMed] [Google Scholar]
- Haas W, Faherty BK, Gerber SA, et al. Optimization and use of peptide mass measurement accuracy in shotgun proteomics. Molecular & Cellular Proteomics. 2006;5(7):1326–1337. doi: 10.1074/mcp.M500339-MCP200. [DOI] [PubMed] [Google Scholar]
- Hong EE, Villén J, Gerace EL, Gygi SP, Moazed D. A cullin E3 ubiquitin ligase complex associates with Rik1 and the Clr4 histone H3-K9 methyltransferase and is required for RNAi-mediated heterochromatin formation. RNA Biology. 2005;2(3):106–111. doi: 10.4161/rna.2.3.2131. [DOI] [PubMed] [Google Scholar]
- Motamedi MR, Verdel A, Colmenares SU, Gerber SA, Gygi SP, Moazed D. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell. 2004;119(6):789–802. doi: 10.1016/j.cell.2004.11.034. [DOI] [PubMed] [Google Scholar]
- Puig O, Caspary F, Rigaut G, et al. The tandem affinity purification (TAP) method: A general procedure of protein complex purification. Methods. 2001;24(3):218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Séraphin B. A generic protein purification method for protein complex characterization and prote-ome exploration. Nature Biotechnology. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- Schultz MC, Hockman DJ, Harkness TAA, Garinther WI, Althiem BA. Chromatin assembly in a yeast whole-cell extract. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(17):9034–9039. doi: 10.1073/pnas.94.17.9034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Analytical Chemistry. 1996;68(5):850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Tanny JC, Kirkpatrick DS, Gerber SA, Gygi SP, Moazed D. Budding yeast silencing complexes and regulation of Sir2 activity by protein–protein interactions. Molecular and Cellular Biology. 2004;24(16):6931–6946. doi: 10.1128/MCB.24.16.6931-6946.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, et al. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303(5658):672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]