Summary
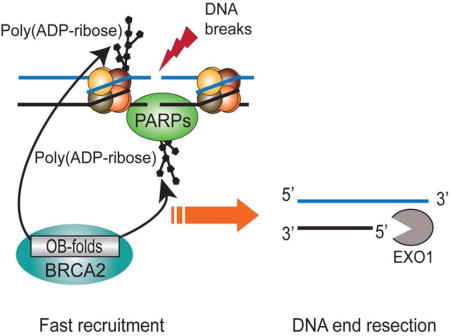
Breast Cancer Susceptibility Gene 2 (BRCA2) plays a key role in DNA damage repair for maintaining genomic stability. Previous studies have shown that BRCA2 contains three tandem oligonucleotide/oligosaccharide binding folds (OB-folds) that are involved in DNA-binding during DNA double-strand break repair. However, the molecular mechanism of BRCA2 in DNA damage repair remains elusive. Unexpectedly, we found that the OB-folds of BRCA2 recognize poly(ADP-ribose) (PAR) and mediate the fast recruitment of BRCA2 to DNA lesions, which is suppressed by PARP inhibitor treatment. Cancer-associated mutations in the OB-folds of BRCA2 disrupt the interaction with PAR and abolish the fast relocation of BRCA2 to DNA lesions. The quickly recruited BRCA2 is important for the early recruitment of exonuclease 1(EXO1) and is involved in DNA end resection, the first step of homologous recombination (HR). Thus, these findings uncover a molecular mechanism by which BRCA2 participates in DNA damage repair.
eTOC Blurb
Zhang et al. show that BRCA2 OB-folds act as a poly(ADP-ribose)-binding module and mediate the fast recruitment of BRCA2 to DNA lesions. The early recruitment of BRCA2 is important for EXO1-dependent DNA end resection, the initial step of homologous recombination (HR) repair.
Introduction
Human cells encounter numerous environmental and internal hazards daily. Fortunately, with DNA damage repair systems, cells faithfully repair DNA lesions for maintaining genomic stability. Loss of DNA damage repair pathways induces accumulation of DNA damage lesions, and ultimately leads to genomic instability and tumorigenesis. Thus, DNA damage repair plays a key role in tumor suppression, and many DNA damage repair proteins are important tumor suppressors. One typical example is Breast Cancer Susceptibility Gene 2 (BRCA2).
Carriers of germline mutations of BRCA2 are predisposed to early onset breast and ovarian cancers as well as Fanconi Anemia (Howlett et al., 2002; Nathanson et al., 2001; Tavtigian et al., 1996; Wooster et al., 1995; Wooster and Weber, 2003). Mutations of BRCA2 are also associated with other types of cancer such as prostate and pancreatic cancers (Breast Cancer Linkage, 1999). Accumulated evidence suggests that BRCA2 plays a key role during DNA damage repair (Scully and Livingston, 2000; Venkitaraman, 2002). Cells with BRCA2 mutations are hypersensitive to cross-linking agents and fail to repair DNA double-stranded breaks (DSBs) via homologous recombination (HR) pathway(Chen et al., 1999; Moynahan et al., 2001; Patel et al., 1998), an error-free repair mechanism for DSBs. BRCA2 has been revealed to facilitate the relocation of RAD51, a key HR recombinase, to DNA lesions and promote the formation of RAD51 nucleoprotein filament, which is required for a strand invasion step during HR repair(Davies et al., 2001; Jensen et al., 2010; Liu et al., 2010; Powell et al., 2002; Thorslund et al., 2010; Yang et al., 2005a; Yuan et al., 1999).
The human BRCA2 gene encodes a 3418 amino acids nuclear protein that shares little sequence homology with any other protein in human. BRCA2 comprises an N-terminal region, a central region with eight BRC repeats, three tandem oligonucleotide/oligosaccharide-binding folds (OB-folds) and a C-terminal motif. The N-terminal region of BRCA2 is known to associate with PALB2(Oliver et al., 2009; Xia et al., 2006), whereas the eight BRC repeats and the C-terminal motif are important for the interaction with RAD51(Carreira et al., 2009; Chen et al., 1998; Mizuta et al., 1997; Pellegrini et al., 2002; Sharan et al., 1997; Wong et al., 1997). The primary sequence of the three tandem OB-folds of BRCA2 is highly conserved during evolution. They associate with DSS1, a small scaffold protein that maintains the correct folding of the tandem OB-folds of BRCA2 (Marston et al., 1999; Yang et al., 2002). Moreover, the OB-folds of BRCA2 are critical for the DNA repair function of BRCA2 as cancer-associated mutations have been identified in this region (Guidugli et al., 2014; Karchin et al., 2008).
Previous study has shown that the OB-folds of BRCA2 are capable of binding to single-stranded DNA (ssDNA) overhangs generated at DSB ends (Yang et al., 2002). Here, we found that the OB-folds of BRCA2 are important for targeting BRCA2 to DNA lesions within 30 seconds. In such short time, DSB ends are unlikely to be processed into ssDNA overhangs to host BRCA2. Interestingly, poly(ADP-ribose) (PAR), a type of oligosaccharide sharing basic component with ssDNA, is synthesized by poly(ADP-ribose) polymerases (PARPs) immediately following DNA damage. Accumulated evidence shows that poly(ADP-ribosyl)ation plays an important role in DNA damage response (Gibson and Kraus, 2012; Hassa and Hottiger, 2008; Schreiber et al., 2006). Upon DNA damage induction, PARP1, the founding member of PARPs, rapidly detects DNA single-strand breaks (SSBs) or double-strand breaks (DSBs), and is activated for PAR synthesis at DNA lesions within a few seconds in response to DNA damage(Kim et al., 2005; Luo and Kraus, 2012). Moreover, our previous study suggests that the solo OB-fold motif is able to recognize iso-ADP-ribose, the linkage between two ADP-ribose unites (Zhang et al., 2014). Here, our results suggested that the tandem OB-folds of BRCA2 recognize DNA damage-induced PAR and target BRCA2 to DNA lesions. BRCA2 and PARylation regulate the recruitment of EXO1, which is one of the mechanisms to regulate DNA end resection and HR repair.
Results
The OB-folds mediate the fast recruitment of BRCA2 to DNA lesions
BRCA2 has been shown to relocate to DNA lesions in response to DSBs. However, the kinetics of the recruitment of BRCA2 to DNA lesions is unclear. Here, we added a GFP tag at the N-terminus of BRCA2 and expressed GFP-BRCA2 in mouse embryonic fibroblasts (MEFs). Cells were treated with laser microirradiation, and the localization of GFP-BRCA2 was monitored with live cell imaging. Interestingly, we found that BRCA2 was recruited to DNA lesions within 30 seconds, which is much faster than we expected (Figures 1A and S1A). BRCA2 was also retained at DNA lesions for prolonged time (Figure 1A). To validate the results, we used anti-BRCA2 antibody to examine the recruitment of endogenous BRCA2 to DNA lesions in U2OS cells, and obtained similar kinetics (Figures 1B and S1B). Collectively, these results suggest that BRCA2 quickly relocates to DNA lesions in response to DNA damage.
Figure 1. The OB-folds mediate the fast recruitment of BRCA2 to DNA lesions.
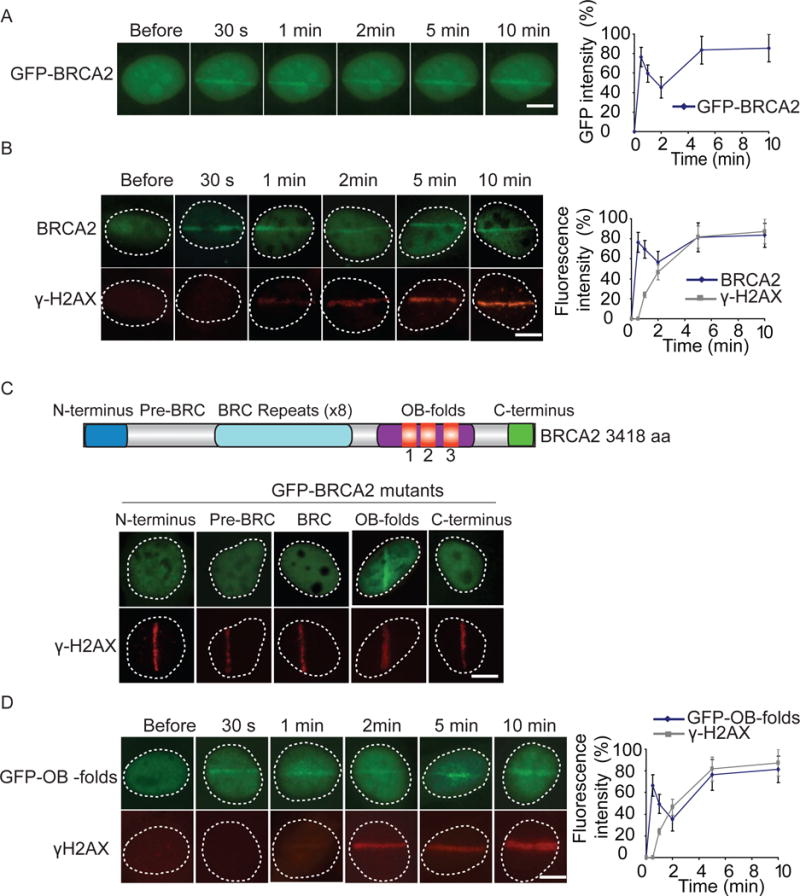
(A) The relocation kinetics of GFP-BRCA2 to DNA damage sites. GFP-tagged BRCA2 was expressed in MEFs, and the relocation kinetics was monitored in a time course following laser microirradiation. For quantitative and comparative imaging, signal intensities at the laser line were converted into a numerical value using Axiovision software (version 4.5). Normalized fluorescent curves from 20 cells were averaged. The error bars represent the standard deviation. Scale bar = 10 μm. Signal intensities were plotted using Excel. (B) The relocation kinetics of BRCA2 to DNA damage sites. U2OS cells were examined with laser microirradiation and stained with anti-BRCA2 and anti-γH2AX at indicated time points. Scale bar = 10 μm. (C) Only the OB-folds of BRCA2 are able to relocate to the sites of DNA damage. Individual domain of BRCA2 was tagged with GFP and expressed in U2OS cells. Cells were treated with laser microirradiation and were immunostained with anti-γH2AX. Scale bar = 10 μm. (D) The relocation kinetics of GFP-OB-folds to DNA damage sites. GFP-tagged BRCA2 OB-folds was expressed in MEFs, and the relocation kinetics was monitored in a time course following laser microirradiation. The cells were co-stained with anti-γH2AX. Scale bar = 10 μm.
Since BRCA2 contains several domains to interact with its binding partners, we tagged each individual domain with GFP, and found that only the OB-folds of BRCA2 were able to relocate to the sites of DNA damage (Figure 1C). We also measured the kinetics of the recruitment. Similar to the full length of BRCA2, the OB-folds were quickly recruited to DNA lesions within 30 seconds and stayed for prolonged time (Figure 1D). Collectively, these results suggest that the OB-folds mediate the recruitment of BRCA2 to DNA lesions.
Fast recruitment of BRCA2 to DNA lesions is mediated by PAR
Previous report shows that the OB-folds of BRCA2 bind to ssDNA (Yang et al., 2002). However, the fast recruitment of BRCA2 indicates that other binding partner similar to ssDNA might be recognized by the OB-folds of BRCA2. In response to DNA damage, massive PAR is synthesized by PARPs in a few seconds (Kim et al., 2005; Luo and Kraus, 2012). Similar to oligo-nucleotide, PAR also contains phosphate groups, ribose and adenine. The substrates of DNA damage-induced PARylation is PARP1, the enzyme itself, as well as histones at or adjacent to DNA lesions (Kim et al., 2005; Luo and Kraus, 2012). Interestingly, PAR is a type of oligosaccharide that can be recognized by OB-fold motifs (Zhang et al., 2014). Thus, we examined whether DNA damage-induced PARylation mediates the recruitment of the OB-folds of BRCA2. We treated fibroblasts with olaparib, a potent PARP1 inhibitor to suppress PAR synthesis, and found that the recruitment of the OB-folds of BRCA2 is significantly delayed (Figures 2A and S2A). Similarly, the recruitment of endogenous BRCA2 to DNA lesions was also significantly delayed in the presence of olaparib treatment (Figures 2B and S2B). Moreover, to validate the results, we used an inducible I-SceI system generating a solo DNA double strand break to examine the early recruitment of BRCA2 (Li and Yu, 2013). Consistently, in the absence of PAR, the recruitment of BRCA2 was remarkably delayed (Figure S2C). Taken together, these results suggest that the fast recruitment of BRCA2 is regulated by DNA damage-induced PARylation. Since most DNA damage-induced PARylation is mediated by PARP1 and PARP2 (Gibson and Kraus, 2012; Luo and Kraus, 2012), we knocked down PARP1 and PARP2 respectively (Figure 2C). In the absence of PARP1 but not PARP2, the early recruitment of BRCA2 was significantly suppressed (Figure 2C), suggesting that PARP1-mediated PARylation facilitates the early recruitment of BRCA2. We confirmed the results in PARP1-deficient mouse embryonic fibroblasts (Parp1−/− MEFs) (Figure 2C). Compared with that in wild type MEFs, the OB-folds of BRCA2 failed to be quickly recruited to DNA lesions in Parp1−/− MEFs. Collectively, these results demonstrate that PAR mediates the fast recruitment of BRCA2 to DNA lesions.
Figure 2. Fast recruitment of BRCA2 to DNA lesions is mediated by PAR.
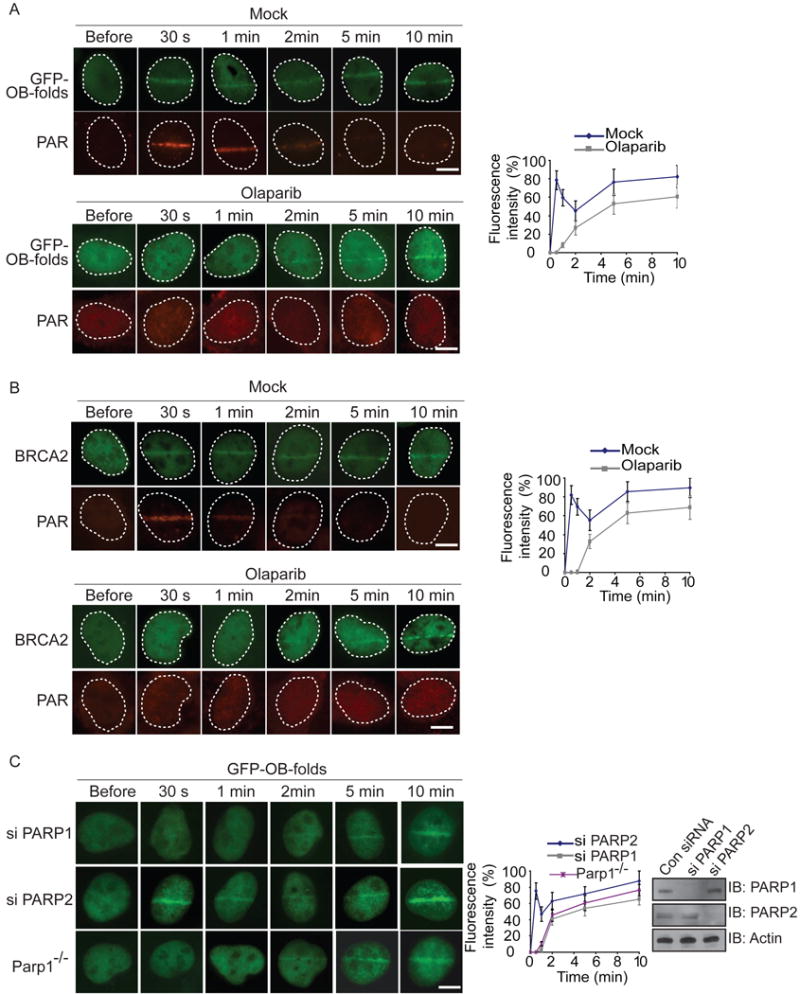
(A) The effect of PARP inhibitor treatment on the recruitment of GFP-OB-folds to DNA damage sites. GFP-tagged BRCA2 OB-folds (GFP-OB-folds) was expressed in U2OS cells, and the relocation kinetics was monitored in a time course following laser microirradiation. The cells were co-stained with anti-PAR. Scale bar = 10 μm. (B) PARP inhibitor treatment suppresses the recruitment of BRCA2 to DNA damage sites. U2OS cells were examined with laser microirradiation and stained with anti-BRCA2 and anti-PAR at indicated time points. Scale bar = 10 μm. (C) The effect of depletion of PARP1 or PARP2 on the recruitment of GFP-OB-folds to DNA damage sites. GFP-OB-folds was expressed in PARP1 deficient MEFs (Parp1−/−) or U2OS cells depletion of PARP1 or PARP2 with siRNA. The relocation of GFP-OB-folds was monitored in a time course following laser microirradiation. Depletion of endogenous PARP1 and PARP2 by siRNA was examined by western blotting. Scale bar = 10 μm.
OB-folds domain of BRCA2 binds to PAR in vitro
Since PAR shares the basic components including ribose sugar, phosphate and adenosine with oligonucleotide, we ask if the OB-folds of BRCA2 interact with PAR. We first generated and purified PAR from in vitro PARylation assay. It has been shown that DSS1 is a binding partner of the OB-folds and facilitates their folding of the OB-folds. Thus, we co-purified the recombinant OB-folds/DDS1 complex and incubated with PAR. We found that the OB-folds of BRCA2 directly bound to PAR in an in vitro pull-down assay (Figure 3A). Previous studies have shown that the integrity of the tertiary structure of the OB-folds is required for the interaction with ssDNA(Yang et al., 2002). Similarly, deleting part of the OB-folds or DSS1 alone abolished the interaction, indicating that the folding of the OB-folds is important for the interaction with PAR. In a reciprocal pull-down experiment, PAR was also found to interact with the OB-folds of BRCA2 (Figure 3B).
Figure 3. OB-folds domain of BRCA2 binds to PAR in vitro.
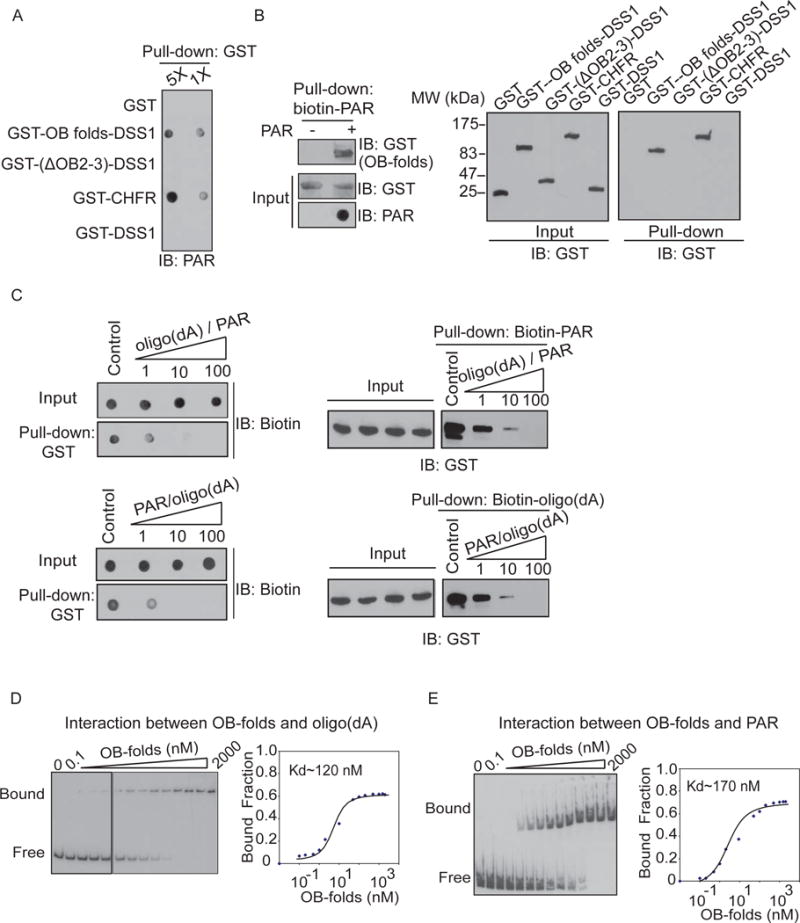
(A) The recombinant GST-fusion proteins were incubated with PAR. Protein-associated PAR was examined by glutathione agarose beads pull down and dot blotting with anti-PAR antibody. Recombinant GST and GST-CHFR were used as the negative and positive controls respectively. (B) The OB-folds domain of BRCA2 interacts with biotin-PAR. Left panel: the recombinant GST-OB-folds was incubated with or without biotin-PAR. The interaction was examined by streptavidin beads pull-down assay and Western blotting with anti-GST antibody. Right panel: The OB2 and OB3 of BRCA2 are essential for the interaction with PAR. GST fusion proteins were incubated with biotin-PAR. The interaction was examined by streptavidin beads pull-down assay and Western blotting with anti-GST antibody. GST and GST-CHFR were used as the negative and positive controls respectively. (C) PAR competes out oligo(dA) (50 mer) to interact with the OB-folds domain of BRCA2. Biotinylated oligo(dA) or PAR was incubated with GST-OB-folds. Pull down assays were performed with GST beads (left) or streptavidin beads (right). 1, 10 and 100 indicate the molar ration between oligo(dA) and PAR in each sample. (D–E) Analysis of the affinity of the BRCA2 OB-folds to oligo(dA) (D) or PAR (E) by EMSA. EMSA reactions were carried out in the presence of 0.5 nM radiolabeled oligo(dA) or PAR and varying amounts of BRCA2 OB-folds as indicated. To calculate dissociation constant (Kd), the percentage of radiolabeled DNA-protein complexes was quantified and plotted against the quantity of protein. The data are the average of two independent saturation binding experiments.
It has been shown that the OB-folds of BRCA2 recognize ssDNA without sequence specificity (Yang et al., 2002). To compare the binding affinity, we performed in vitro competition assays using oligo(dA) that might be similar to PAR. In these assays, 50 mer oligo(dA) has been used to compete with 30 ~ 50-mer PAR with increased molar ratio from 1:1 to 100:1. PAR and oligo(dA) could compete with each other to bind the OB-folds of BRCA2 (Figure 3C). Moreover, with electrophoretic mobility shift assays (EMSA), we quantitatively measured the affinities of these interactions. The dissociation constant of both interaction is between 100–200 nM (Figure 3D, E). Taken together, these results suggest that the affinity between the OB-folds and PAR is very similar to that between the OB-folds and ssDNA.
The OB-folds of BRCA2 associate with PAR in response to DNA damage in vivo
Next, we ask if BRCA2 interacts with PAR in vivo. Using immunoprecipitation (IP) with anti-BRCA2 antibody and dot blotting with anti-PAR antibody, we found that BRCA2 associated with PAR in vivo (Figure 4A). Moreover, since DNA damage induces PAR synthesis at the sites of DNA damage (Kim et al., 2005; Luo and Kraus, 2012), the interaction between PAR and BRCA2 was significantly increased following IR treatment (Figure 4A). This interaction was further confirmed by reciprocal IP (Figure 4B). To exclude the possibility that the interaction between BRCA2 and PAR is mediated by DNA, we treated samples with DNase I to digest DNA. Similar results were obtained (Figure S3). In addition, to exclude the possibility that BRCA2 itself was PARylated, we treated the precipitates with 1 % SDS that abolished the non-covalent interactions. In the presence of SDS, BRCA2 was dissociated with PAR (Figure 4C), suggesting that BRCA2 itself is not PARylated in response to DNA damage. We also used olaparib to suppress PAR synthesis. When PAR synthesis was suppressed by olaparib treatment, BRCA2 no longer interacted with PAR following DNA damage (Figure 4D). Taken together, these results suggest that BRCA2 recognizes PAR in vivo, especially when PAR is massively synthesized following DNA damage.
Figure 4. The OB-folds of BRCA2 associate with PAR in response to DNA damage in vivo.
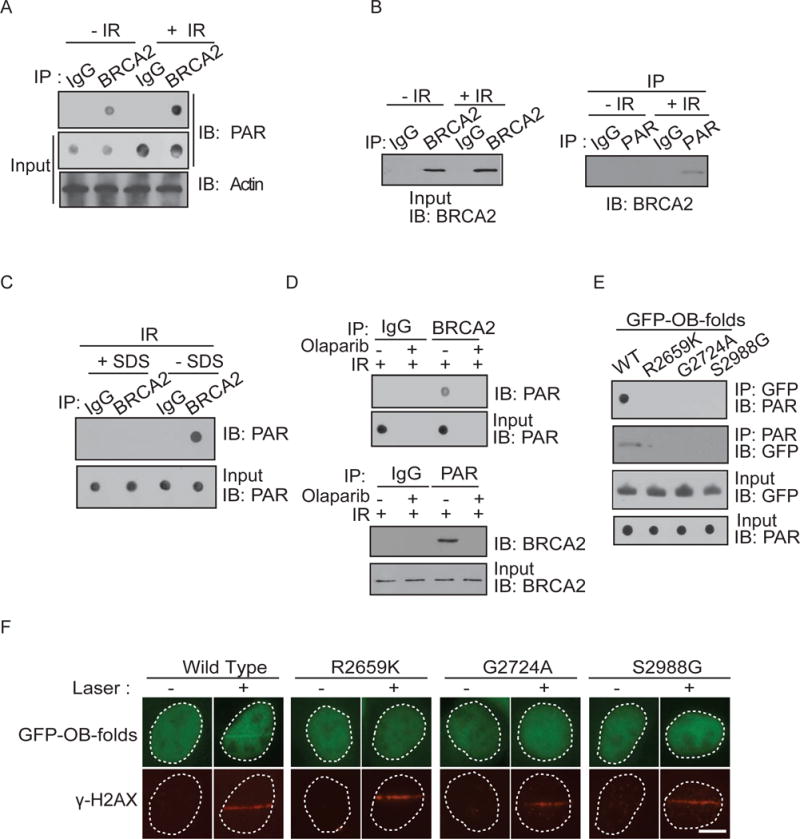
(A and B) The in vivo interaction between BRCA2 and PAR was examined by co-IP (A) and reciprocal co-IP (B). U2OS cells were treated with 0 or 10 Gy of IR. 5 minutes after IR, cells were lysed and analyzed with indicated antibodies. Input or IPed samples were analyzed by dot blotting (A) or Western blotting (B) with the indicated antibodies. (C) BRCA2 itself is not PARylated in response to IR. Same as (A) except that the precipitation of co-IP was treated with or without 1 % SDS. (D) The in vivo interaction between BRCA2 and PAR was examined by co-IP and reciprocal co-IP in the presence or absence of olaparib (100 nM) and IR treatment (10 Gy) with the indicated antibodies. Irrelevant IgG was included as the IP control. (E) The cancer-associated missense mutations surrounding or inside of the OB-folds abolish the interaction with PAR. GFP-tagged OB-folds and the indicated mutants were expressed in 293T cells. Cell lysates were analyzed by the indicated antibodies. The expression levels of exogenous OB-folds proteins were examined by Western blotting with anti-GFP antibody. (F) The OB-folds with cancer-associated mutations fail to relocate to the sites of DNA damage. GFP-tagged BRCA2 OB-folds mutant was expressed in MEFs, and the relocation was monitored at 1 minute following laser microirradiation. The cells were co-stained with anti-γH2AX. Scale bar = 10 μm.
Cancer-associated BRCA2 mutations have been identified surrounding and inside of the OB-folds of BRCA2 (Guidugli et al., 2014; Karchin et al., 2008). Next, we ask if these mutations regulate the interaction between the OB-folds of BRCA2 and PAR. We analyzed three breast cancer-associated mutations: R2659K, G2724A and S2988G. Among these pathogenic mutations, R2659 locates in the helical domain just prior to the OB1; G2724 is inside of the OB1; and S2988 is in the OB2 of BRCA2. Interestingly, all three mutations abolished the interaction between the OB-folds of BRCA2 and PAR (Figure 4E). Moreover, all three mutations abolished the recruitment of the BRCA2 OB-folds to DNA lesions (Figure 4F).
BRCA2 and PARylation regulates the recruitment of EXO1 and DNA end resection
Since PAR mediates early recruitment of BRCA2 to DNA lesions, we explored the biological function of the PAR-dependent recruitment of BRCA2 to DNA lesions. BRCA2 is known for loading RAD51 to 3′ ssDNA overhangs at DSB ends, which is an essential step for RAD51-mediated strand invasion (Carreira et al., 2009; Davies et al., 2001; Esashi et al., 2007; Jensen et al., 2010; Liu et al., 2010; Pellegrini et al., 2002; Powell et al., 2002; Thorslund et al., 2010; Yang et al., 2005a). However, 3′ ssDNA overhangs are unlikely to be processed within 30 seconds. Moreover, the kinetics of the recruitment of RAD51 was much slower than that of BRCA2 (Figure S4A), suggesting that the early recruitment of BRCA2 to DNA lesions is important for another biological process. The first step of HR is DNA end resection. A group of endo- and exo-nucleases including MRE11, CtIP and EXO1 function together to process DNA ends into 3′ ssDNA overhangs(Huertas, 2010; Longhese et al., 2010; Mimitou and Symington, 2011; Raynard et al., 2008; Symington and Gautier, 2011). We ask if early recruited BRCA2 plays a role in DNA end resection by examining if BRCA2 facilitates the loading of these nucleases to DNA ends. Among these nucleases, MRE11 and EXO1 reach DNA lesions within 30 seconds (Figures 5 and S4B). However, the recruitment of CtIP is much slower compared to that of MRE11 or EXO1 (Figure S4C), suggesting that CtIP is unlikely to be regulated by early recruited BRCA2. In U2OS cells lacking BRCA2, MRE11 was still recruited to the sites of DNA damage within 30 seconds (Figure S4B). However, the recruitment of EXO1 was significantly delayed in the absence of BRCA2 (Figures 5 and S4D). Moreover, we treated cells with olaparib to suppress early recruitment of BRCA2 to DNA lesions. Lacking early recruited BRCA2, the early recruitment of EXO1 was also impaired (Figures 5 and S4E–F). Taken together, our results indicate that BRCA2 facilitates the early recruitment of EXO1.
Figure 5. BRCA2 and PARylation regulate the recruitment of EXO1.
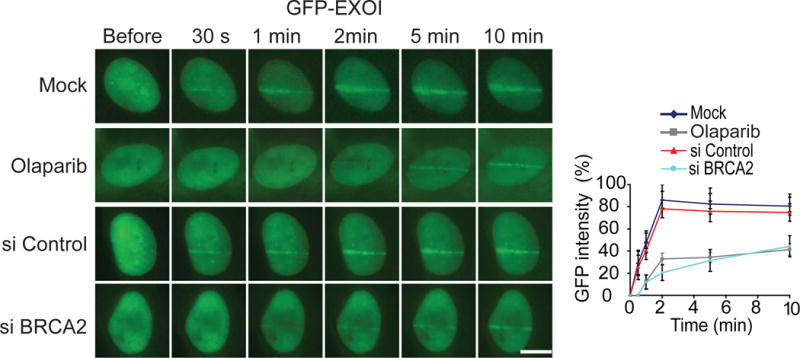
Depletion of BRCA2 or suppression of PAR synthesis abolishes the early recruitment of EXO1 to DNA lesions. U2OS cells were treated with siBRCA2 or olaparib, the relocation kinetics of GFP-EXO1 to the sites of DNA damage was examined. Scale bar = 10 μm. GFP fluorescence at the laser line was converted into a numerical value (relative fluorescence intensity) using Axiovision software (version 4.5). Normalized fluorescent curves from 20 cells from three independent experiments were averaged. Signal intensities were plotted using Excel. Error bars represent the SD.
Since EXO1 mediates DNA end resection, we ask if BRCA2 is also important for DNA end resection. We performed a DNA damage end resection assay by examining the BrdU exposure at DNA lesions in S phase (Figures 6A and S5A–B). Usually, BrdU is incorporated into genomic DNA and buried in the DNA double-strand helix, thus could not be detected by anti-BrdU Ab. However, once DNA lesions occur and induce DNA end resection, BrdU at 3′ ssDNA overhangs is exposed and is able to be detected by anti-BrdU following laser microirradiation in an immunostaining assay (Figure 6A). We found that BrdU started to be exposed from 10 minutes following DNA damage (Figure 6B, upper panel). The exposure of BrdU at processed DNA ends was regulated by EXO1 as the BrdU exposure at DNA lesions was suppressed in cells lacking EXO1, which is consistent with previous observations that EXO1 is required for DNA end resection (Mimitou and Symington, 2011). Interestingly, in BRCA2-deficient cells, the BrdU exposure at DNA lesions was significantly delayed (Figures 6C and S5A). Similar results were observed in cells with knockdown of BRCA2 (Figures 6B, S5B). When BRCA2-deficient cells were reconstituted with wild type BRCA2, the kinetics of BrdU exposure at DNA ends were restored (Figure 6C). However, when BRCA2-deficient cells were reconstituted with the R2659K mutant that are defective for PAR binding, the BrdU exposure at DNA lesions (Figure 6A), and the recruitment of EXO1 were also significantly delayed (Figure S4D,). Moreover, when cells were treated with olaparib to suppress PARylation, the BrdU exposure was also remarkably delayed (Figure 6D), suggesting that the early recruited BRCA2 by PAR might be important for EXO1-mediated DNA end resection.
Figure 6. BRCA2 and PARylation are important for DNA end resection.
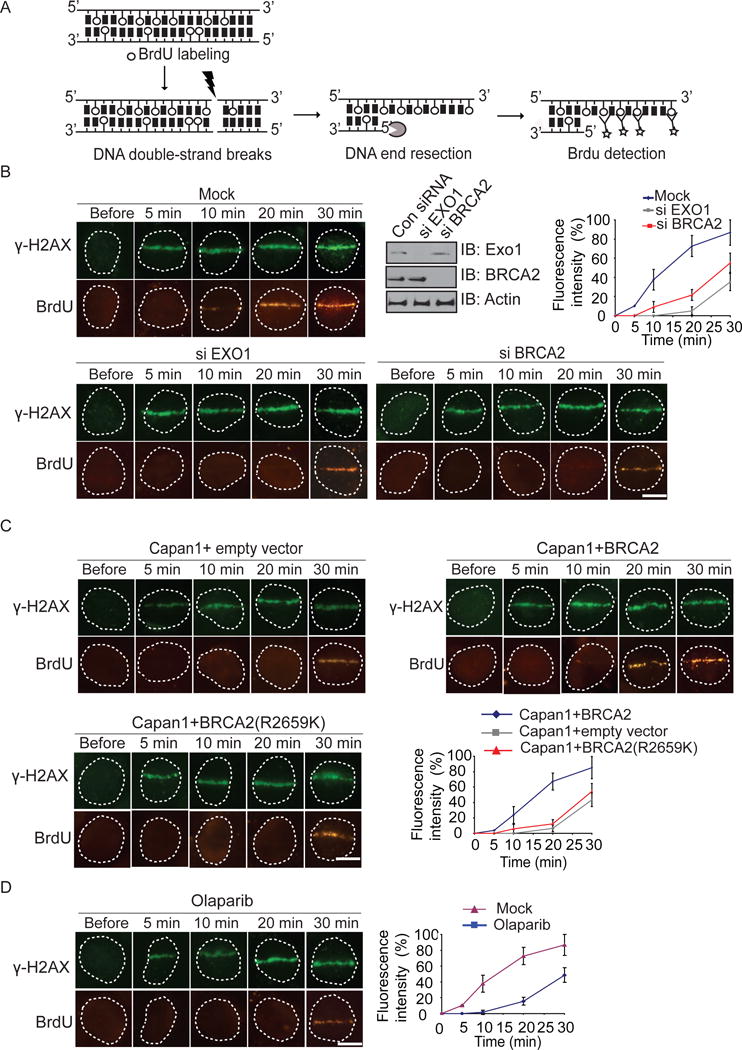
(A) Schematic diagram of DNA damage end resection assay. (B) Depletion of EXO1 or BRCA2 suppresses DNA end resection. U2OS cells treated with siEXO1 or siBRCA2 were examined with laser microirradiation and stained with anti-BrdU and anti-γH2AX at indicated time points. Scale bar = 10 μm. Depletion of endogenous EXO1 and BRCA2 by siRNA was examined by western blotting. (C) BRCA2 is important for EXO1-mediated DNA end resection. Capan1 cells and Capan1 reconstituted with wild type BRCA2 cells were examined with laser microirradiation and stained with anti-BrdU and anti-γH2AX at indicated time points. Scale bar = 10 μm. (D) U2OS cells were treated with olaparib. Scale bar = 10 μm. The end resection was analyzed as same as in (B).
Discussion
In this study, we found that the tandem OB-folds of BRCA2 is a PAR-binding module. DNA damage-induced PARylation mediates the fast recruitment of BRCA2 to DNA lesions, which is important for the recruitment of EXO1 and EXO1-dependent DNA end resection. With both in vitro and in vivo analyses, we found that the OB-folds recognize DNA damage-induced PAR at DNA lesions. Since PAR is an oligo-nucleotide like polymer sharing the basic components with oligo-nucleotide, it is likely that the OB-folds use the same binding sites to recognize both ssDNA and PAR. Our study shows that the affinity between the OB-folds and PAR is similar to that between the OB-folds and ssDNA. Previous study demonstrates that three tandem OB-folds are organized to form a single domain and interacts with ssDNA at multiple sites (Yang et al., 2002). Here, we also show that several cancer-associated mutations near or inside of the OB-folds abolish the interaction between PAR and the OB-folds, suggesting that tertiary structure of the BRCA2 OB-folds plays a key role for the interaction. Future structural analysis will reveal the details of the interaction between PAR and the OB-folds.
The function of the interaction between PAR and the OB-folds is to quickly relocate BRCA2 to DNA lesions. It has been reported that PARPs can use up to 90 % cellular NAD+ to synthesize massive amount of PAR at DNA lesions (Berger et al., 1983; Yu et al., 2002). Thus, ample amount of PAR is sufficient to recruit BRCA2 to the sites of DNA damage. Moreover, PARylation occurs within a few seconds following DNA damage (Kim et al., 2005; Luo and Kraus, 2012), which is consistent with the kinetics of the early recruitment of BRCA2. Importantly, direct suppression of PAR synthesis by PARP inhibitor remarkably delays the recruitment of BRCA2. And we have used two different systems to validate the results. Thus, these lines of evidence suggest that DNA damage-induced PARylation mediates the fast wave recruitment of BRCA2 to the sites of DNA damage. PARylation is also a short-lived posttranslational modification during DNA damage response. Within a few minutes, DNA damage-induced PAR is hydrolyzed by PARG and other PAR degradases(D’Amours et al., 1999; Kim et al., 2005; Luo and Kraus, 2012). Thus, DNA damage-induced PARylation mediates early and fast recruitment of BRCA2. Along with PAR degradation, DNA end resection is started and ssDNA overhangs are generated by resection enzymes. Since the OB-fold of BRCA2 also recognizes ssDNA, the 3′ ssDNA overhangs are important for the retention of BRCA2 at DNA lesions. Consistently, we found two waves of BRCA2 at DNA lesions (Figure 1A and B). The first wave recruitment of BRCA2 is clearly mediated by PAR. With PAR degradation, BRCA2 is released and recognizes the ssDNA overhangs. This two-wave retention at DNA lesions also occurs in other DNA damage repair machineries, such as BRCA1 (Li and Yu, 2013), which indicates the potentially different biological functions during this two-wave retention because these repair machineries recognize different partners in each stage.
Previously studies have demonstrated that the second wave of BRCA2 retention is to load RAD51 to DNA lesions and facilitates RAD51/ssDNA filament invasion into sister chromatids(Carreira et al., 2009; Davies et al., 2001; Esashi et al., 2007; Jensen et al., 2010; Liu et al., 2010; Pellegrini et al., 2002; Powell et al., 2002; Thorslund et al., 2010; Yang et al., 2005a). Our study also verified that the recruitment kinetics of RAD51 is consistent with the timing of the second wave of BRCA2 at DNA lesions. Here, we have dissected the biological function of the first wave of BRCA2 that is mediated by DNA damage-induced PAR. We found that BRCA2 is important for DNA end resection by facilitating EXO1, a key resection enzyme, to DNA lesions. However, we did not detect the direct interaction between BRCA2 and EXOI, suggesting that BRCA2 does not directly recruit EXOI to the site of DNA damage. It is possible that BRCA2 sets up the local environment for the recruitment of EXO1. The analog situation is the RNF8-mediated protein ubiquitination. In the previous studies, we and others have shown that RNF8 mediates the recruitment of BRCA1 (Huen et al., 2007; Kolas et al., 2007; Mailand et al., 2007; Wu et al., 2009). However, RNF8 does not directly interact with BRCA1 for the recruitment. Instead, RNF8 mediates histone ubiquitination that is recognized by the BRCA1-associated partner RAP80. The interaction between ubiquitin and the ubiquitin-binding domain of RAP80 mediates the recruitment of BRCA1. Similarly, BRCA2 is quickly recruited by PAR and is likely to modify local chromatin at the sites of DNA damage for the recruitment of EXO1. Recent studies indicate that EXO1 has several binding partners such as PCNA, MSH2 and MSH3 etc. (Dherin et al., 2009; Liberti et al., 2011; Schmutte et al., 2001; Tishkoff et al., 1997; Tran et al., 2001). Especially, it has been shown that PCNA mediates the recruitment of EXO1(Chen et al., 2013). Thus, it is likely that BRCA2 may regulate PCNA for the recruitment of EXO1.
Moreover, it has been shown that the MRN complex is involved in the recruitment of EXO1 in budding yeast (Nicolette et al., 2010). However, we did not observe that the early recruitment of EXO1 is regulated by the MRN complex in U2OS cells. The recruitment mechanism of EXO1 in budding yeast could be very different from mammals. Especially, yeast does not have protein poly-ADP-ribosylation and PARPs. And the PAR-binding module in NBS1 is degenerated in yeast. The kinetics of the recruitment of DNA damage response factors in yeast remains elusive. It is possible that the early and fast recruitment mechanism mediated by poly-ADP-ribosylation occurs during the evolution, which becomes more important for mammals to keep the genomic stability.
Besides RAD51, PAR and ssDNA, BRCA2 also interacts with PALB2 via an N-terminal region (Oliver et al., 2009; Xia et al., 2006). It has been shown that PALB2 facilitates the recruitment of BRCA2 to DNA lesions as well (Xia et al., 2007; Xia et al., 2006). However, the N-terminus of BRCA2 per se could not be recruited to the sites of DNA damage (Figure 1C), suggesting that only the interaction between PALB2 and BRCA2 is insufficient to mediate the recruitment of BRCA2. Alternatively, PALB2 may interact with BRCA2 through multiple contacts. But like BRCA2 and RAD51, PALB2 is also an ssDNA-binding protein, and facilitates the loading RAD51 onto the ssDNA overhangs (Dray et al., 2010). PALB2 even directly interacts with RAD51 (Park et al., 2014). Thus, PALB2 is likely to play a key role for the second wave of BRCA2 at DNA lesions. PALB2, BRCA2 and RAD51 may form a complex that is important for RAD51/ssDNA filament formation.
Taken together, our study demonstrates a molecular mechanism of BRCA2 in DNA damage repair, which is helpful for understanding the BRCA2 mutation-induced tumorigenesis.
Experimental procedures
GFP-BRCA2 and GFP-ExoI plasmids were a gift from laboratory of Drs. Bing Xia and Zhongsheng You, respectively. CtIP, MRE11 and N-terminus (a.a. 1–300), Pre-BRC (a.a. 1–933), BRC (a.a. 934–2467), OB-folds (a.a. 2477–3196) or C-terminus (a.a. 3163–3418) of BRCA2 were clone into pEGFP-C1 vector. The PAR-binding mutants and siRNA resistant form of BRCA2 OB-folds (a.a. 2477–3196) were generated using the QuikChange site-directed mutagenesis kit (Stratagene).
The siRNA sequences targeting PARP1, PARP2, BRCA2 and Exo1 are 5′-CAAAGUAUCCCAAGAAGUUdTdT-3′, 5′-GGAGAAGGAUGGUGAGAAAdTdT-3′, 5′-GAAGAACAAUAUCCUACUAdTdT-3′ and 5′-CAAGCCUAUUCUCGUAUUUdTdT-3′ respectively. The siRNAs were transfected into cells using oligofectamine (Invitrogen) according to manufacturer’s instructions.
Anti β-actin, anti-biotin, anti-EXO1 and anti-GFP antibodies were purchased from Sigma. Anti-PARP1, anti-PARP2 and anti-BRCA2antibodies were purchased from Millipore. Anti-PAR antibody was purchased from Trevigen. Anti- Rad51, phospho-H2AX (γ-H2AX) were generated as described previously (Ward et al., 2003; Yamane et al, 2002; Yu et al., 2003).
Baculoviruses and recombinant protein purification from insect cells
For baculovirus, DNA fragment containing the BRCA2 OB-folds (a.a. 2477–3196) and full-length DSS1, were subcloned into pFastBac vector with or without an GST tag, and baculoviruses were generated according to manufacturer’s instructions. After cells were infected with combinations of baculoviruses for 48 h, the SF9 cells were harvested, washed with PBS and lysed with 10 ml ice-cold NETN-100 (0.5 % NP-40, 2 mM EDTA, 100 mM Tris-HCl pH 7.5, 100 mM NaCl) buffer. The soluble fraction was incubated with 0.5 ml glutathione-Sepharose beads (for GST tag). The protein/complexes were affinity purified according to manufacturer’s instructions.
Synthesis, purification and fractionation of PAR
His tagged human poly(ADP-ribose) polymerase1 (PARP1) was expressed in bacteria and purified by Ni-NTA affinity resin. PAR was synthesized and purified as described previously except for the following modifications(Fahrer et al., 2007). PAR was synthesized in a 20 ml incubation mixture containing 100 mM Tris-HCl pH7.8, 10 mM MgCl2, 1 mM NAD+, 10 mM DTT, 60 μg calf thymus histone, 50 μg octameric oligonucleotide GGAATTCC and 2 mg PARP1. To generate biotinyl-PAR, 10 μM biotinyl-NAD+ (Trevigen) was included in the reaction. The mixture was incubated at 30 °C for 60 minutes and stopped by addition of 20 ml ice-cold 20% TCA. Oligo DNA was removed by DNase I and proteins were digested by proteinase K. Purified PAR was fractionated according to chain length by anion exchange HPLC protocol as described previously(Fahrer et al., 2007).
Laser micro-irradiation and imaging of cells
U2OS cells and MEFs with or without transfection of indicated plasmids were plated on glass-bottomed culture dishes (Mat Tek Corporation). Laser micro-irradiation was performed using an IX 71 microscope (Olympus) coupled with the MicoPoint Laser Illumination and Ablation System (Photonic Instruments, Inc.). A 337.1 nm laser diode (3.4 mW) transmits through a specific Dye Cell and then yields 365 nm wavelength laser beam that is focused through ×60 UPlanSApo/1.35 oil objective to yield a spot size of 0.5–1 μm. Cells were exposed to the laser beam for about3.5 ns. The pulse energy is 170 μJ at 10 Hz. Images were taken by the same microscope with CellSens software (Olympus).
For quantitative and comparative imaging, signal intensities at the laser line were converted into a numerical value using Axiovision software (version 4.5). To compensate for nonspecific fluorescent bleaching during the repeated image acquisition, in every image, we first measured the average fluorescent intensity (at the laser line) as a function of time, and then divided it by the average fluorescent intensity measured elsewhere in the cell (background) as a function of time. To get normalized GFP-tagged proteins accumulation curve for each cell, the fluorescent intensity (RF) at the laser line was calculated by the following formula: RF(t) = [(I−Ipre IR)/(Imax−Ipre IR)], where Ipre IR is the fluorescent intensity of the laser line region before irradiation, and Imax represents the maximum fluorescent intensity at the laser line. Normalized fluorescent curves from 20 cells were averaged. The error bars represent the standard deviation. Scale bar = 10 μm. Signal intensities were plotted using Excel.
Electrophoretic mobility shift assay (EMSA)
The affinity between the OB-folds of BRCA2 and PAR or oligo(dA) was measured by electrophoretic mobility shift assay (EMSA) essentially as described(Hellman and Fried, 2007; Yang et al., 2005b). Briefly, the BRCA2 OB-folds recombinant protein was incubated with 32P-labeled DNA or PAR probes (20,000 cpm, 0.5 nM) in a 30 μl reaction containing buffer A (50 mM Tris–HCl, pH 8.0, 50 mM NaCl, 1 mM DTT, 0.05% NP-40,100 ng poly (dI-dC) and 6% glycerol) for 30 min at 4°C. The reaction mixtures were electrophoresed at 4°C on 5% non-denaturing polyacrylamide gel in a buffer containing 7 mM Tris–HCl (pH 7.4), 3 mM boric acid and 1 mM EDTA. Gels were dried and autoradiographed. The apparent dissociation constant (Kd) was estimated as the protein concentration at which half of the radiolabeled DNA probe was shifted in an EMSA essentially as described(Yang et al., 2005b).
Dot blotting
Recombinant proteins (10 pmol) were conjugated to Glutathione beads and incubated with PAR (100 pmol, calculated as the ADP-ribose unit) for 2 hours at 4°C. Beads were washed with NETN-100 buffer for four times. GST-fusion proteins were eluted from beads by glutathione and spotted onto a nitrocellulose membrane. The membrane was blocked with TBST buffer (0.15 M NaCl, 0.01 M Tris-HCl pH 7.4, 0.1 % Tween 20) supplemented with 5 % milk, extensively washed with TBST. After drying in the air, the membrane was examined by anti-PAR antibody.
GST fusion protein expression and pull-down assay
GST fusion proteins were expressed in Escherichia coli and purified using standard procedures. Purified GST fusion proteins (1 pmol) were incubated with biotin-labeled PAR (5 pmol) and streptavidin beads for 2 hours at 4 °C. After washing with NETN-100 buffer four times, samples were boiled in SDS-sample buffer and elutes were analyzed by Western blotting with anti-GST antibody.
Cell Culture, Cell lysis, Immunoprecipitation and Western blotting
Human cancer cell lines were maintained in RPMI 1640 medium with 10 % fetal calf serum and cultivated at 37 °C in 5 % CO2 (v/v). For ionizing radiation, cells were irradiated by using JL Shepherd 137Cs radiation source at indicated doses. Cells were lysed with NETN-100 buffer containing 10 mM NaF and 50 mM β-glycerophosphate. Immunoprecipitation and western blotting were performed following standard protocol.
Flow Cytometry
Cells were trypsinized and washed with PBS twice, and centrifuged (800 rpm) at 4°C. Cells were fixed in 500 μL ice-cold 70% ethanol overnight at 4°C. Fixed cells were centrifuged at 500 rpm at 4°C for 10 minutes and suspended in 500 μL PBS containing 50 μL of 50 μg/mL propidium iodide (Sigma) and 10 μL of 5 μg/mL RNase for 45 to 60 minutes at 4°C. Flow cytometric analysis was done within 24 hours of propidium iodide staining on a Beckman-Coulter Epics XL (Fullerton, CA). EXPO32 software was used for optimum analysis of cell cycle (G1, G2, and S phases)
Poly(ADP-ribose) (PAR) binding assays
Approximately 2 μM (5X) or 0.4 μM (1X) of each recombinant protein were incubated with 10 μM PAR, 30 μl glutathione agarose in the buffer containing 10 mM NaH2PO4 (pH 7.5), 100 mM NaCl. After incubation for 1 hour at room temperature, the beads were extensively washed with PBS, and bound proteins were released by adding 30 μl sample buffer (150 mM Tris-HCl, 10 % SDS, 10 mM EDTA) followed by heating at 80 °C for 10 minutes. 2 μl aliquots of samples were dot-blotted onto nitrocellulose membranes. After incubation for 1 hour at 60 °C, membranes were subjected to dot blotting analysis with anti-PAR antibodies.
Immunofluorescence staining
Cells grown on coverslips were fixed with 3 % paraformaldehyde for 20 minutes and permeabilized with 0.5 % Triton X-100 in PBS for 5 minutes at room temperature. Samples were blocked with 5 % goat serum and then incubated with primary antibody for 60 minutes. Samples were washed three times and incubated with secondary antibody for 30 minutes. The coverslips were mounted onto glass slides and visualized by a fluorescence microscope.
DNA damage end resection assay
Cells were grown on 15 mm glass bottom cell culture dish such that they are rapidly dividing (usually 50 % confluent). Incubate cells in 10 μM BrdU for 24 hours. Cells were exposed to the laser beam for about 3.5 ns. The pulse energy is 170 μJ at 10 Hz. Cells were immediately pretreated with ice-cold NETN-300 buffer (0.5 % NP-40, 2 mM EDTA, 100 mM Tris-HCl pH 7.5, 300 mM NaCl) and fixed with 3 % paraformaldehyde for 30 minutes. Immunostaining was performed as described before(Zhang et al., 2009).
Inducible DSB System
A DSB system was established before(Li and Yu, 2013). Briefly, U2OS cells stably expressing RFP-I-SceI-GR were used in this system. The synthetic glucocorticoid ligand TA (Sigma) was added (0.1 mM) to induce the translocation of RFP-I-SceI-GR from cytoplasm into nucleus, which generated a solo DSB. The kinetics of the recruitment of DNA damage repair factors were examined. Images were taken using the same microscope of laser microirradiation with the CellSens software (Olympus).
Supplementary Material
Highlights.
-
-
OB-folds of BRCA2 recognize DNA damage-induced poly(ADP-ribosyl)ation
-
-
PARylation and OB-folds of BRCA2 mediate the fast recruitment of BRCA2 to DNA lesions
-
-
The early recruitment of BRCA2 is important for EXO1-dependent DNA end resection
Acknowledgments
This work was supported in part by grants from National Institutes of Health (CA132755, CA130899 and CA187209 to X.Y.), National Natural Science Foundation of China (81572775) and the Program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institutions of Higher Learning. X.Y. is a recipient of Era of Hope Scholar Award from the Department of Defense and Research Scholar Award from Leukemia and Lymphoma Society.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
X.Y. designed the research. F.Z., J.S., C.B., and X.Y. performed the research and analyzed the data. F.Z. and X.Y. wrote the paper.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Berger NA, Sims JL, Catino DM, Berger SJ. Poly(ADP-ribose) polymerase mediates the suicide response to massive DNA damage: studies in normal and DNA-repair defective cells. Princess Takamatsu Symp. 1983;13:219–226. [PubMed] [Google Scholar]
- Breast Cancer Linkage, C. Cancer risks in BRCA2 mutation carriers. Journal of the National Cancer Institute. 1999;91:1310–1316. doi: 10.1093/jnci/91.15.1310. [DOI] [PubMed] [Google Scholar]
- Carreira A, Hilario J, Amitani I, Baskin RJ, Shivji MK, Venkitaraman AR, Kowalczykowski SC. The BRC repeats of BRCA2 modulate the DNA-binding selectivity of RAD51. Cell. 2009;136:1032–1043. doi: 10.1016/j.cell.2009.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CF, Chen PL, Zhong Q, Sharp ZD, Lee WH. Expression of BRC repeats in breast cancer cells disrupts the BRCA2-Rad51 complex and leads to radiation hypersensitivity and loss of G(2)/M checkpoint control. J Biol Chem. 1999;274:32931–32935. doi: 10.1074/jbc.274.46.32931. [DOI] [PubMed] [Google Scholar]
- Chen PL, Chen CF, Chen Y, Xiao J, Sharp ZD, Lee WH. The BRC repeats in BRCA2 are critical for RAD51 binding and resistance to methyl methanesulfonate treatment. Proc Natl Acad Sci U S A. 1998;95:5287–5292. doi: 10.1073/pnas.95.9.5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Paudyal SC, Chin RI, You Z. PCNA promotes processive DNA end resection by Exo1. Nucleic Acids Res. 2013;41:9325–9338. doi: 10.1093/nar/gkt672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Amours D, Desnoyers S, D’Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J. 1999;342(Pt 2):249–268. [PMC free article] [PubMed] [Google Scholar]
- Davies AA, Masson JY, McIlwraith MJ, Stasiak AZ, Stasiak A, Venkitaraman AR, West SC. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol Cell. 2001;7:273–282. doi: 10.1016/s1097-2765(01)00175-7. [DOI] [PubMed] [Google Scholar]
- Dherin C, Gueneau E, Francin M, Nunez M, Miron S, Liberti SE, Rasmussen LJ, Zinn-Justin S, Gilquin B, Charbonnier JB, et al. Characterization of a highly conserved binding site of Mlh1 required for exonuclease I-dependent mismatch repair. Mol Cell Biol. 2009;29:907–918. doi: 10.1128/MCB.00945-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dray E, Etchin J, Wiese C, Saro D, Williams GJ, Hammel M, Yu X, Galkin VE, Liu D, Tsai MS, et al. Enhancement of RAD51 recombinase activity by the tumor suppressor PALB2. Nat Struct Mol Biol. 2010;17:1255–1259. doi: 10.1038/nsmb.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esashi F, Galkin VE, Yu X, Egelman EH, West SC. Stabilization of RAD51 nucleoprotein filaments by the C-terminal region of BRCA2. Nat Struct Mol Biol. 2007;14:468–474. doi: 10.1038/nsmb1245. [DOI] [PubMed] [Google Scholar]
- Fahrer J, Kranaster R, Altmeyer M, Marx A, Burkle A. Quantitative analysis of the binding affinity of poly(ADP-ribose) to specific binding proteins as a function of chain length. Nucleic Acids Res. 2007;35:e143. doi: 10.1093/nar/gkm944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson BA, Kraus WL. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol. 2012;13:411–424. doi: 10.1038/nrm3376. [DOI] [PubMed] [Google Scholar]
- Guidugli L, Carreira A, Caputo SM, Ehlen A, Galli A, Monteiro AN, Neuhausen SL, Hansen TV, Couch FJ, Vreeswijk MP, et al. Functional assays for analysis of variants of uncertain significance in BRCA2. Hum Mutat. 2014;35:151–164. doi: 10.1002/humu.22478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassa PO, Hottiger MO. The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front Biosci. 2008;13:3046–3082. doi: 10.2741/2909. [DOI] [PubMed] [Google Scholar]
- Hellman LM, Fried MG. Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions. Nat Protoc. 2007;2:1849–1861. doi: 10.1038/nprot.2007.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlett NG, Taniguchi T, Olson S, Cox B, Waisfisz Q, De Die-Smulders C, Persky N, Grompe M, Joenje H, Pals G, et al. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002;297:606–609. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P. DNA resection in eukaryotes: deciding how to fix the break. Nat Struct Mol Biol. 2010;17:11–16. doi: 10.1038/nsmb.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen RB, Carreira A, Kowalczykowski SC. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 2010;467:678–683. doi: 10.1038/nature09399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karchin R, Agarwal M, Sali A, Couch F, Beattie MS. Classifying Variants of Undetermined Significance in BRCA2 with protein likelihood ratios. Cancer Inform. 2008;6:203–216. doi: 10.4137/cin.s618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MY, Zhang T, Kraus WL. Poly(ADP-ribosyl)ation by PARP-1: ‘PARlaying’ NAD+ into a nuclear signal. Genes Dev. 2005;19:1951–1967. doi: 10.1101/gad.1331805. [DOI] [PubMed] [Google Scholar]
- Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, Panier S, Mendez M, Wildenhain J, Thomson TM, et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 2007;318:1637–1640. doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Yu X. Function of BRCA1 in the DNA damage response is mediated by ADP-ribosylation. Cancer Cell. 2013;23:693–704. doi: 10.1016/j.ccr.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberti SE, Andersen SD, Wang J, May A, Miron S, Perderiset M, Keijzers G, Nielsen FC, Charbonnier JB, Bohr VA, et al. Bi-directional routing of DNA mismatch repair protein human exonuclease 1 to replication foci and DNA double strand breaks. DNA Repair (Amst) 2011;10:73–86. doi: 10.1016/j.dnarep.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Doty T, Gibson B, Heyer WD. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat Struct Mol Biol. 2010;17:1260–1262. doi: 10.1038/nsmb.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longhese MP, Bonetti D, Manfrini N, Clerici M. Mechanisms and regulation of DNA end resection. The EMBO journal. 2010;29:2864–2874. doi: 10.1038/emboj.2010.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Kraus WL. On PAR with PARP: cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes Dev. 2012;26:417–432. doi: 10.1101/gad.183509.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- Marston NJ, Richards WJ, Hughes D, Bertwistle D, Marshall CJ, Ashworth A. Interaction between the product of the breast cancer susceptibility gene BRCA2 and DSS1, a protein functionally conserved from yeast to mammals. Mol Cell Biol. 1999;19:4633–4642. doi: 10.1128/mcb.19.7.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS. DNA end resection–unraveling the tail. DNA repair. 2011;10:344–348. doi: 10.1016/j.dnarep.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuta R, LaSalle JM, Cheng HL, Shinohara A, Ogawa H, Copeland N, Jenkins NA, Lalande M, Alt FW. RAB22 and RAB163/mouse BRCA2: proteins that specifically interact with the RAD51 protein. Proc Natl Acad Sci U S A. 1997;94:6927–6932. doi: 10.1073/pnas.94.13.6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynahan ME, Pierce AJ, Jasin M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol Cell. 2001;7:263–272. doi: 10.1016/s1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- Nathanson KL, Wooster R, Weber BL. Breast cancer genetics: what we know and what we need. Nat Med. 2001;7:552–556. doi: 10.1038/87876. [DOI] [PubMed] [Google Scholar]
- Nicolette ML, Lee K, Guo Z, Rani M, Chow JM, Lee SE, Paull TT. Mre11-Rad50-Xrs2 and Sae2 promote 5′ strand resection of DNA double-strand breaks. Nat Struct Mol Biol. 2010;17:1478–1485. doi: 10.1038/nsmb.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver AW, Swift S, Lord CJ, Ashworth A, Pearl LH. Structural basis for recruitment of BRCA2 by PALB2. EMBO Rep. 2009;10:990–996. doi: 10.1038/embor.2009.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JY, Singh TR, Nassar N, Zhang F, Freund M, Hanenberg H, Meetei AR, Andreassen PR. Breast cancer-associated missense mutants of the PALB2 WD40 domain, which directly binds RAD51C, RAD51 and BRCA2, disrupt DNA repair. Oncogene. 2014;33:4803–4812. doi: 10.1038/onc.2013.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel KJ, Yu VP, Lee H, Corcoran A, Thistlethwaite FC, Evans MJ, Colledge WH, Friedman LS, Ponder BA, Venkitaraman AR. Involvement of Brca2 in DNA repair. Mol Cell. 1998;1:347–357. doi: 10.1016/s1097-2765(00)80035-0. [DOI] [PubMed] [Google Scholar]
- Pellegrini L, Yu DS, Lo T, Anand S, Lee M, Blundell TL, Venkitaraman AR. Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature. 2002;420:287–293. doi: 10.1038/nature01230. [DOI] [PubMed] [Google Scholar]
- Powell SN, Willers H, Xia F. BRCA2 keeps Rad51 in line. High-fidelity homologous recombination prevents breast and ovarian cancer? Mol Cell. 2002;10:1262–1263. doi: 10.1016/s1097-2765(02)00789-x. [DOI] [PubMed] [Google Scholar]
- Raynard S, Niu H, Sung P. DNA double-strand break processing: the beginning of the end. Genes Dev. 2008;22:2903–2907. doi: 10.1101/gad.1742408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutte C, Sadoff MM, Shim KS, Acharya S, Fishel R. The interaction of DNA mismatch repair proteins with human exonuclease I. J Biol Chem. 2001;276:33011–33018. doi: 10.1074/jbc.M102670200. [DOI] [PubMed] [Google Scholar]
- Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- Scully R, Livingston DM. In search of the tumour-suppressor functions of BRCA1 and BRCA2. Nature. 2000;408:429–432. doi: 10.1038/35044000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharan SK, Morimatsu M, Albrecht U, Lim DS, Regel E, Dinh C, Sands A, Eichele G, Hasty P, Bradley A. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature. 1997;386:804–810. doi: 10.1038/386804a0. [DOI] [PubMed] [Google Scholar]
- Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annu Rev Genet. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- Tavtigian SV, Simard J, Rommens J, Couch F, Shattuck-Eidens D, Neuhausen S, Merajver S, Thorlacius S, Offit K, Stoppa-Lyonnet D, et al. The complete BRCA2 gene and mutations in chromosome 13q-linked kindreds. Nat Genet. 1996;12:333–337. doi: 10.1038/ng0396-333. [DOI] [PubMed] [Google Scholar]
- Thorslund T, McIlwraith MJ, Compton SA, Lekomtsev S, Petronczki M, Griffith JD, West SC. The breast cancer tumor suppressor BRCA2 promotes the specific targeting of RAD51 to single-stranded DNA. Nat Struct Mol Biol. 2010;17:1263–1265. doi: 10.1038/nsmb.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff DX, Boerger AL, Bertrand P, Filosi N, Gaida GM, Kane MF, Kolodner RD. Identification and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an exonuclease that interacts with MSH2. Proc Natl Acad Sci U S A. 1997;94:7487–7492. doi: 10.1073/pnas.94.14.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran PT, Simon JA, Liskay RM. Interactions of Exo1p with components of MutLalpha in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2001;98:9760–9765. doi: 10.1073/pnas.161175998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkitaraman AR. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell. 2002;108:171–182. doi: 10.1016/s0092-8674(02)00615-3. [DOI] [PubMed] [Google Scholar]
- Ward IM, Minn K, van Deursen J, Chen J. p53 Binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Mol Cell Biol. 2003;23:2556–2563. doi: 10.1128/MCB.23.7.2556-2563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong AK, Pero R, Ormonde PA, Tavtigian SV, Bartel PL. RAD51 interacts with the evolutionarily conserved BRC motifs in the human breast cancer susceptibility gene brca2. J Biol Chem. 1997;272:31941–31944. doi: 10.1074/jbc.272.51.31941. [DOI] [PubMed] [Google Scholar]
- Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J, Collins N, Gregory S, Gumbs C, Micklem G. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378:789–792. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- Wooster R, Weber BL. Breast and ovarian cancer. N Engl J Med. 2003;348:2339–2347. doi: 10.1056/NEJMra012284. [DOI] [PubMed] [Google Scholar]
- Wu J, Huen MS, Lu LY, Ye L, Dou Y, Ljungman M, Chen J, Yu X. Histone ubiquitination associates with BRCA1-dependent DNA damage response. Mol Cell Biol. 2009;29:849–860. doi: 10.1128/MCB.01302-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia B, Dorsman JC, Ameziane N, de Vries Y, Rooimans MA, Sheng Q, Pals G, Errami A, Gluckman E, Llera J, et al. Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat Genet. 2007;39:159–161. doi: 10.1038/ng1942. [DOI] [PubMed] [Google Scholar]
- Xia B, Sheng Q, Nakanishi K, Ohashi A, Wu J, Christ N, Liu X, Jasin M, Couch FJ, Livingston DM. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Molecular cell. 2006;22:719–729. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- Yamane K, Wu X, Chen J. A DNA damage-regulated BRCT-containing protein, TopBP1, is required for cell survival. Mol Cell Biol. 2002;22:555–566. doi: 10.1128/MCB.22.2.555-566.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Jeffrey PD, Miller J, Kinnucan E, Sun Y, Thoma NH, Zheng N, Chen PL, Lee WH, Pavletich NP. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science. 2002;297:1837–1848. doi: 10.1126/science.297.5588.1837. [DOI] [PubMed] [Google Scholar]
- Yang H, Li Q, Fan J, Holloman WK, Pavletich NP. The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA-ssDNA junction. Nature. 2005a;433:653–657. doi: 10.1038/nature03234. [DOI] [PubMed] [Google Scholar]
- Yang Y, Sass LE, Du C, Hsieh P, Erie DA. Determination of protein-DNA binding constants and specificities from statistical analyses of single molecules: MutS-DNA interactions. Nucleic Acids Res. 2005b;33:4322–4334. doi: 10.1093/nar/gki708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu SW, Wang H, Poitras MF, Coombs C, Bowers WJ, Federoff HJ, Poirier GG, Dawson TM, Dawson VL. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science. 2002;297:259–263. doi: 10.1126/science.1072221. [DOI] [PubMed] [Google Scholar]
- Yu X, Chini CC, He M, Mer G, Chen J. The BRCT domain is a phosphoprotein binding domain. Science. 2003;302:639–642. doi: 10.1126/science.1088753. [DOI] [PubMed] [Google Scholar]
- Yuan SS, Lee SY, Chen G, Song M, Tomlinson GE, Lee EY. BRCA2 is required for ionizing radiation-induced assembly of Rad51 complex in vivo. Cancer Res. 1999;59:3547–3551. [PubMed] [Google Scholar]
- Zhang F, Chen Y, Li M, Yu X. The oligonucleotide/oligosaccharide-binding fold motif is a poly(ADP-ribose)-binding domain that mediates DNA damage response. Proc Natl Acad Sci U S A. 2014;111:7278–7283. doi: 10.1073/pnas.1318367111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Ma J, Wu J, Ye L, Cai H, Xia B, Yu X. PALB2 links BRCA1 and BRCA2 in the DNA-damage response. Curr Biol. 2009;19:524–529. doi: 10.1016/j.cub.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


