Abstract
Over the past 15 years, laser-based microdissection has improved the precision by which scientists can procure cells of interest from a heterogeneous tissue section. However, for studies that require a large amount of material (e.g., proteomics) or for cells that are scattered and difficult to identify by standard histological stains, an immunostain-based, automated approach becomes essential. In this chapter, we discuss the use of immunohistochemistry (IHC) and immunofluorescence (IF) to guide the microdissection process via manual and software-driven auto-dissection methods. Although technical challenges still exist with these innovative approaches, we present here methods and protocols to successfully perform immuno-based microdissection on commercially available laser dissection systems.
Keywords: Automation, Immunohistochemistry, Microdissection
1. Introduction
Immunohistochemistry (IHC) was developed over 60 years ago for in situ measurements of proteins in histological sections (1). Based on the recognition of target cell antigens by specific primary antibodies, IHC has become a staple for molecular analysis of tissue sections in the research laboratory and clinic. The more recent integration of this commonly used technique with laser microdissection has resulted in a powerful combination that is enabling new studies and is amenable to further automation to improve dissection throughput (2–5).
The first such method developed was called immunoguided laser capture microdissection or immuno-LCM (4) (Fig. 1a). This approach combines immuno-stained tissue with LCM, allowing the investigator to select specific cell types by visually aiming a laser at stained target cells. This procedure is helpful for identifying cell populations that would otherwise not be recognized by standard histological stains, such as hematoxylin and eosin (H&E), and to subsequently dissect a specific subset of cells using a molecular marker. Immuno-LCM can now be performed in an automated fashion using newly developed software packages available for several commercial dissection instruments. These instruments identify target cells by image processing and then automatically dissect the tissue using a motorized stage and a coordinate system. Both manual and automated immuno-LCM can utilize either IHC or immunofluorescence (IF) to guide the dissection process (4–5, 11). As a specific example of an automated immuno-LCM system, the ArcturusXT (Life Technologies) incorporates an image analysis software package called AutoScanXT, which specifically identifies stained areas with minimal supervision by the investigator (Fig. 1b). The use of image analysis software improves the speed of the microdissection process by selecting targets based on a threshold set by the instrument’s operator.
Fig. 1.
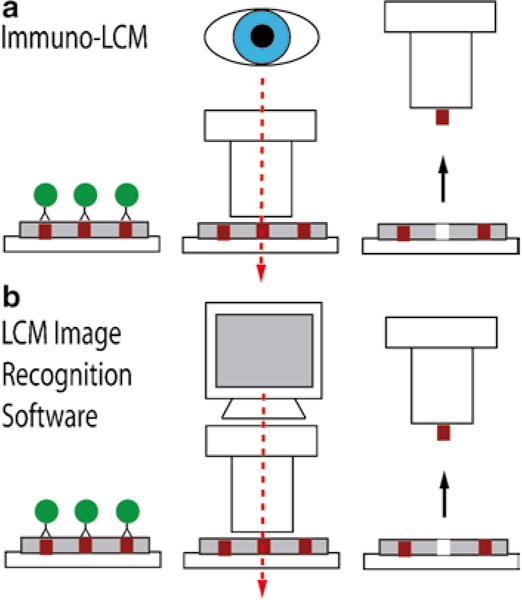
Schematic representation of immunoguided microdissection techniques. Initial immunostaining step represented by the antibody complex attached to a labeling enzyme (circles).
This chapter provides the materials, protocols, and related references that will permit investigators to successfully perform immunoguided microdissection. However, the reader should note that these technologies and associated molecular analysis methods are not static, but are being continually refined and improved. In particular, on-going studies to assess the effects of immunostaining and microdissection on biomolecular recovery and methods to improve extraction techniques are currently areas of active investigation by our group and others (6–11). Moreover, there are often subtle differences in the dissection conditions that are required for each tissue type and target cell. Nonetheless, the protocols described below are amenable to most dissection studies and serve as a good starting template.
2. Materials
2.1. Immunohistochemistry
The following materials are for an IHC protocol that we use to label epithelial cells using monoclonal antibodies against cytokeratins.
IHC horizontal staining tray (Thermo Scientific, Waltham, MA).
Vertical staining tubs and racks (Simport).
Rice steamer (Black & Decker), used for antigen retrieval.
Citrate buffer, pH 6.0 (DAKO Carpinteria, CA) for antigen retrieval, stored at 4°C.
Phosphate-buffered saline (PBS): 150 mM NaCl, 25 mM NaPO4, pH 7.4. Stored at room temperature.
DAKO Envision+ IHC Kit (DAKO) provides the peroxidase blocking solution (0.03% H2O2), the secondary antibody polymer (either mouse or rabbit or dual), and the 3,3′-diaminobenzidine (DAB) substrate solution (see Note 1).
Mouse Anti-Cytokeratin AE1/AE3 diluted 1:50 in Zymed Antibody Diluent (Invitrogen, Carlsbad, CA). Reagents stored at 4°C, but the dilution is made fresh prior to staining and used at room temperature (see Note 2).
DAB Enhancer (Invitrogen) stored at room temperature.
Deionized water (diH2O). Stored at room temperature.
Graded alcohols (70, 95, and 100% ethanol). Stored at room temperature.
Xylenes for drying and dewaxing formalin-fixed, paraffin-embedded (FFPE) tissue. Stored at room temperature.
2.2. Immunofluorescence
Reagents from IHC protocol described above, plus secondary Goat Anti-Mouse antibody with Alexa Fluor® 488 (Invitrogen).
2.3. Immuno‐LCM
CapSur® Macro LCM caps (Life Technologies, Carlsbad, CA).
LCM machine; ArcturusXT, Veritas, or PixCell II (Life Technologies).
2.4. LCM Image Recognition Software
CapSur® Macro LCM caps (Life Technologies).
LCM machine; ArcturusXT (Life Technologies) with optional AutoScanXT software.
3. Methods
Described below are protocols for tissue immunostaining, immuno-LCM, and immuno-LCM using the AutoScanXT software. The initial immunostaining step is essential to successful microdissection using these approaches. A strong staining with minimal background enables precise procurement of the targets of interest.
It is important to note that downstream molecular analysis must be considered when utilizing these immuno-stained microdissection methods. DNA is the most robust biomolecule following IHC; however, yields are typically reduced; yet the overall quality of the DNA is unaffected (6, 11–14). RNA is the most susceptible to the staining process and is often degraded in the initial staining steps (6, 15). If RNA were the downstream molecule of interest, it would be necessary to shorten the staining protocol times significantly and add RNase inhibitors. However, it is highly dependent on the abundance of the RNA of interest and it has been our experience that only small RNA targets are recoverable following standard IHC. Better results for RNA extraction have been obtained with the use of modified protocols for immunofluorescence staining (16, 17). On the contrary, proteins are analyzable via SDS–PAGE and mass spectrometry methods, but mild lysis buffer methods, such as 2D-PAGE, presents a challenge to recover the protein content of the tissue as harsher buffers and treatments are more effective (6, 15). Despite these few caveats, the combination of immunoguided dissection tools with the burgeoning field of molecular biology enables researchers to pursue tissue-based scientific questions that would otherwise not be feasible.
3.1. Immunohistochemistry: Frozen Tissues
A tissue specimen is sectioned utilizing a cryostat machine (Leica) at 8-μm thick sections. The tissue is then placed on positively charged glass slides. Tissue sections can be stored at −80°C for several days prior to microdissection.
To begin IHC, place the tissue slides in a 70% ethanol bath for 2 min at room temperature. Then transfer the slides to a diH2O bath.
Proceed with the endogenous peroxidase block from the DAKO Envision+ kit, for 10–40 min at room temperature (see Note 3).
Wash the slides in PBS bath (3×, 2 min each wash).
Incubate slides with diluted primary antibody for 30 min at room temperature (see Note 4).
Wash the slides in PBS bath (3×, 2 min each wash).
Incubate slides with pre-diluted DAKO Envision+ secondary antibody for 30 min at room temperature.
Wash the slides in PBS bath (3×, 2 min each wash).
Apply DAKO Envision+ DAB solution for 5 min at room temperature.
Wash in diH2O bath (3×, 2 min each wash).
Apply the Zymed DAB Enhancer for 3 min at room temperature.
Wash in diH2O bath (3×, 2 min each wash) (see Note 5).
Dehydrate the sample in graded alcohols (ethanol 70, 95, and 100%) for 2 min each followed by xylenes for 3×, 2 min each.
3.2. Immunofluorescence: Frozen Tissue
Section the tissue specimen using a cryostat (Leica) at 8-μm thickness and place the section on positively charged glass slides. Tissue sections can be stored at −80°C for several days prior to microdissection.
To begin IHC, place the tissue slide in a 70% ethanol bath for 2 min at room temperature. Then transfer the slide to a diH2O bath.
Wash the slides in PBS bath (3×, 2 min each wash).
Incubate slide with diluted primary antibody for 30 min at room temperature (see Note 4).
Wash the slides in PBS bath (3×, 2 min each wash).
Incubate slide with secondary antibody with Alexa Fluor 488 for 30 min at room temperature.
Wash in diH2O bath (3×, 2 min each wash).
Dehydrate the sample in graded alcohols (ethanol 70, 95, and 100%) followed by xylenes for 2 min each.
3.3. Immunohistochemistry: Formalin-Fixed Paraffin-Embedded Tissues
For FFPE tissue, an antigen retrieval step is typically required prior to incubation with antibodies (18). A number of antigen retrieval approaches are possible, including the use of heat induced epitope retrieval (HIER) (19) and the use of enzymes, such as trypsin or pepsin (20–26). HIER will be described here (see Notes 6 and 7).
For HIER, a citrate buffer (300 ml) is pre-heated in a rice steamer for 40 min.
The FFPE tissue is dewaxed in xylenes 3×, 2 min for each incubation. The tissue is then rehydrated through graded alcohols (100, 95, and 70% for 2 min each) to diH2O prior to placement in the hot citrate bath.
Place the rehydrated tissue slides in the hot citrate bath for 25 min.
Remove entire citrate bath containing the slides from the streamer and allow cooling at room temperature for 25 min.
Proceed with the IHC protocol (Subheading 3.1) as stated above beginning with step 3.
3.4. Immuno-LCM
These instructions assume the use of a PixCell II instrument, although they are easily adaptable to other products in the LCM line by Life Technologies.
Following immunohistochemistry (Subheadings 3.1 and 3.3), the dehydrated tissue is placed on the platform of the PixCell II device and the vacuum is applied (see Note 8).
A CapSure® Macro LCM cap (Life Technologies) is placed over the dehydrated tissue via the loading arm.
The IR laser is tested in an open area of the cap to obtain a laser shot “ring”. Adjust the parameters of the laser for optimal lifting. For example, for a 30-μm laser spot size, the ranges of typical parameters are: 20–30 mW power and 5–6 ms duration.
Under microscopic visualization, locate the immuno-stained cells of interest and fire the laser over the desired area.
When the dissection is complete, remove the cap from the slide and proceed with downstream molecular analysis.
3.5. LCM Image Recognition Software
The AutoScanXT Software Module from Life Technologies is an image analysis program that automatically identifies immuno-stained regions of interest as defined by the user (27) (see Note 9). AutoScanXT provides three levels of analysis: pixel, texture, and morphology. In this protocol, we will focus on the pixel analysis tool.
Following immunostaining, the dehydrated tissue is placed on the platform of the ArcturusXT device.
The operator utilizes the ArcturusXT software field of view to select a tissue area of interest and the appropriate magnification. Once selected, a CapSure® Macro LCM cap (Life Technologies) is then placed automatically over the dehydrated tissue.
Select the “AutoScanXT” icon located on the Select Tool Panel of the ArcturusXT operating software.
Select “Acquire Image” and a static image is captured of the area of interest (see Note 10). A new analysis file will need to be created and named by the user.
In the pixel analysis settings, regions of interest (ROI) are selected on the static jpeg image by the user and highlighted by red circles. Likewise, background areas are also selected by the user and highlighted by blue squares on the jpeg image (Fig. 2a) (see Note 11).
The AutoScanXT software then “learns” what the desired ROIs are and highlights the area to be dissected as indicated by empty red circles (Fig. 2b). A list of the identified regions selected by the program is then generated in the “Regions” tab (see Note 12).
The user verifies if this is the desired area of interest to be dissected and if so can “harvest” the area (see Note 13). At this point, the ArcturusXT machine automatically dissects the highlighted area (Fig. 2c, d) (see Note 14).
When the dissection is complete, move the cap to the quality control (QC) station and proceed with downstream molecular analysis.
Fig. 2.
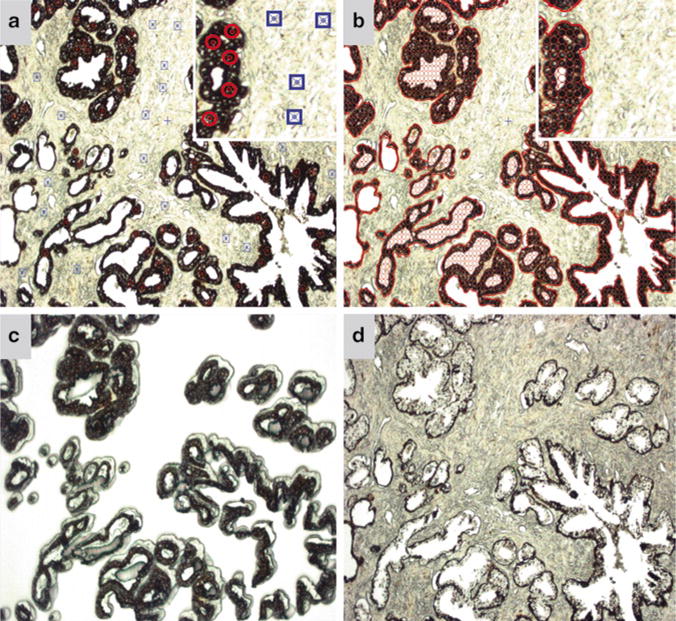
AutoScanXT software guided dissection of a cytokeratin AE1/AE3 IHC-stained prostate tissue. (a) Screen capture of the static jpeg image where ROIs (circles) and background (squares) as determined by the user. Inset highlights the symbols used to select the ROIs and background. The symbols are enhanced in the inset for visualization purposes. (b) Screen capture of the ROIs (circled areas) selected by the AutoScanXT analysis module. (c) Image of the LCM cap showing the dissected epithelial cells. (d) Image of the remaining tissue following AutoScanXT-directed dissection. All images captured at 100× magnification.
Footnotes
Several companies offer similar IHC products.
Cytokeratin AE1/AE3 is presented as an example; however, numerous primary antibodies against particular antigens of interest can be utilized. The antibody dilution needs to be determined empirically by the user for proper staining and can vary from 1:25 to 1:2,000.
The incubation time is dependent on the amount of endogenous peroxidase present in the tissue. Some tissues display more activity than others and thus longer blocking times may be required as determined by the user.
The primary antibody incubation is highly dependent on the relative abundance of the target antigen and the kinetics of the antibody–antigen interaction. A separate protocol that is commonly used is incubation with the primary antibody overnight at 4°C.
No counterstain, such as hematoxylin, is used when performing the immuno-stain procedure for the microdissection methods described, as it will decrease the signal-to-noise between the desired stained areas of the tissue and the undesired background.
Antigen retrieval is highly dependent on the antigen of interest and the amount of fixation. Therefore, a number of strategies and parameters may need to be tested by the user to identify the ideal protocol.
Immunofluorescence is also possible on FFPE tissue, although it is not discussed in detail here. The formalin fixative increases background autofluorescence of the sample, which makes the technique challenging. For a robust protocol of immunofluorescence on FFPE, refer to method described by Robertson et al. (28).
Immunofluorescent-stained slides may also be used for immuno-LCM. This is especially useful for proteomic studies (29). The Life Technologies microdissection device for this approach requires fluorescent capabilities.
It is also worth noting that other companies also offer similar image analysis software, such as the Leica LMD AutoVision Control system.
The AutoScanXT software only works with jpeg images. The program does not recognize tiff files and thus the user must select only jpeg images.
It is important to choose representative ROIs and areas of background in the static jpeg image, as this will help the software decipher between the two in a more comprehensive fashion. A large number of points are not necessary for a successful selection.
The user can either select all of the regions determined to fit the pixel criteria or individual regions of interest via the check-boxes located in the “Regions” tab.
If the highlighted area is incorrect. The user can increase or decrease the stringency accordingly and repeat the process.
For a complete step-by-step instruction of how to use the AutoScanXT, refer to the instruction manual, which can be downloaded at http://www.moldev.com.br/pages/software/autoscanXT.html.
References
- 1.Coons AH, Creech HJ, Jones RN. Immunological properties of an antibody containing a fluorescent group. Proceedings of the Society for Experimental Biology and Medicine. 1941;47:200–202. [Google Scholar]
- 2.Emmert-Buck MR, Bonner RF, Smith PD, Chuaqui RF, Zhuang Z, Goldstein SR, Weiss RA, Liotta LA. Laser capture microdissection. Science. 1996;274:998–1001. doi: 10.1126/science.274.5289.998. [DOI] [PubMed] [Google Scholar]
- 3.Bonner RF, Emmert-Buck M, Cole K, Pohida T, Chuaqui R, Goldstein S, Liotta LA. Laser capture microdissection: molecular analysis of tissue. Science. 1997;278(1481):1483. doi: 10.1126/science.278.5342.1481. [DOI] [PubMed] [Google Scholar]
- 4.Fend F, Emmert-Buck MR, Chuaqui R, Cole K, Lee J, Liotta LA, Raffeld M. Immuno-LCM: laser capture microdissection of immunostained frozen sections for mRNA analysis. Am J Pathol. 1999;154:61–66. doi: 10.1016/S0002-9440(10)65251-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fend F, Kremer M, Quintanilla-Martinez L. Laser capture microdissection: methodical aspects and applications with emphasis on immuno-laser capture microdissection. Pathobiology. 2000;68:209–214. doi: 10.1159/000055925. [DOI] [PubMed] [Google Scholar]
- 6.Tangrea MA, Kreitman MS, Rosenberg AM, Mukherjee S, Eberle FC, Jaffe ES, Wallis BS, Hanson JC, Chuaqui RF, Rodriguez-Canales J, Emmert-Buck MR. Effects of Immunohistochemistry on Biomolecules in Tissue Specimens: Importance for Expression-Based Microdissection Technologies. American Association for Cancer Research Annual Meeting; Washington, D.C.. 2010. [Google Scholar]
- 7.Macdonald JA, Murugesan N, Pachter JS. Validation of immuno-laser capture microdissection coupled with quantitative RT-PCR to probe blood-brain barrier gene expression in situ. J Neurosci Methods. 2008;174:219–226. doi: 10.1016/j.jneumeth.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 8.Liu Y, Wu J, Liu S, Zhuang D, Wang Y, Shou X, Zhu J. Immuno-laser capture microdissection of frozen prolactioma sections to prepare proteomic samples. Colloids Surf B Biointerfaces. 2009;71:187–193. doi: 10.1016/j.colsurfb.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 9.Nakamura N, Ruebel K, Jin L, Qian X, Zhang H, Lloyd RV. Laser capture microdissection for analysis of single cells. Methods Mol Med. 2007;132:11–18. doi: 10.1007/978-1-59745-298-4_2. [DOI] [PubMed] [Google Scholar]
- 10.Fassunke J, Majores M, Ullmann C, Elger CE, Schramm J, Wiestler OD, Becker AJ. In situ-RT and immuno-laser microdissection for mRNA analysis of individual cells isolated from epilepsy-associated glioneuronal tumors. Lab Invest. 2004;84:1520–1525. doi: 10.1038/labinvest.3700165. [DOI] [PubMed] [Google Scholar]
- 11.Eberle FC, Hanson JC, Killian JK, Wei L, Ylaya K, Hewitt SM, Jaffe ES, Emmert-Buck MR, Rodriguez-Canales J. Immunoguided laser assisted microdissection techniques for DNA methylation analysis of archival tissue specimens. J Mol Diagn. 2010;12:394–401. doi: 10.2353/jmoldx.2010.090200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grover AC, Tangrea MA, Woodson KG, Wallis BS, Hanson JC, Chuaqui RF, Gillespie JW, Erickson HS, Bonner RF, Pohida TJ, Emmert-Buck MR, Libutti SK. Tumor-associated endothelial cells display GSTP1 and RARbeta2 promoter methylation in human prostate cancer. J Transl Med. 2006;4:13. doi: 10.1186/1479-5876-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanson JA, Gillespie JW, Grover A, Tangrea MA, Chuaqui RF, Emmert-Buck MR, Tangrea JA, Libutti SK, Linehan WM, Woodson KG. Gene promoter methylation in prostate tumor-associated stromal cells. J Natl Cancer Inst. 2006;98:255–261. doi: 10.1093/jnci/djj051. [DOI] [PubMed] [Google Scholar]
- 14.Rodriguez-Canales J, Hanson JC, Tangrea MA, Erickson HS, Albert PS, Wallis BS, Richardson AM, Pinto PA, Linehan WM, Gillespie JW, Merino MJ, Libutti SK, Woodson KG, Emmert-Buck MR, Chuaqui RF. Identification of a unique epigenetic sub-microenvironment in prostate cancer. J Pathol. 2007;211:410–419. doi: 10.1002/path.2133. [DOI] [PubMed] [Google Scholar]
- 15.Tangrea MA, Mukherjee S, Gao B, Markey SP, Du Q, Armani M, Kreitman MS, Rosenberg AM, Wallis BS, Eberle FC, Duncan FC, Hanson JC, Chuaqui RF, Rodriguez-Canales J, Emmert-Buck MR. Effect of immunohistochemistry on molecular analysis of tissue samples: implications for microdissection technologies. J Histochem Cytochem. 2011;59:591–600. doi: 10.1369/0022155411404704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murakami H, Liotta L, Star RA. IF-LCM: laser capture microdissection of immunofluorescently defined cells for mRNA analysis rapid communication. Kidney Int. 2000;58:1346–1353. doi: 10.1046/j.1523-1755.2000.00295.x. [DOI] [PubMed] [Google Scholar]
- 17.Anthony RM, Urban JF, Jr, Alem F, Hamed HA, Rozo CT, Boucher JL, Van Rooijen N, Gause WC. Memory T(H)2 cells induce alternatively activated macrophages to mediate protection against nematode parasites. Nat Med. 2006;12:955–960. doi: 10.1038/nm1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramos-Vara JA, Beissenherz ME. Optimization of immunohistochemical methods using two different antigen retrieval methods on formalin-fixed paraffin-embedded tissues: experience with 63 markers. J Vet Diagn Invest. 2000;12:307–311. doi: 10.1177/104063870001200402. [DOI] [PubMed] [Google Scholar]
- 19.Shi SR, Key ME, Kalra KL. Antigen retrieval in formalin-fixed, paraffin-embedded tissues: an enhancement method for immunohistochemical staining based on microwave oven heating of tissue sections. J Histochem Cytochem. 1991;39:741–748. doi: 10.1177/39.6.1709656. [DOI] [PubMed] [Google Scholar]
- 20.Battifora H, Kopinski M. The influence of protease digestion and duration of fixation on the immunostaining of keratins. A comparison of formalin and ethanol fixation. J Histochem Cytochem. 1986;34:1095–1100. doi: 10.1177/34.8.2426335. [DOI] [PubMed] [Google Scholar]
- 21.Huang SN. Immunohistochemical demonstration of hepatitis B core and surface antigens in paraffin sections. Lab Invest. 1975;33:88–95. [PubMed] [Google Scholar]
- 22.Huang SN, Minassian H, More JD. Application of immunofluorescent staining on paraffin sections improved by trypsin digestion. Lab Invest. 1976;35:383–390. [PubMed] [Google Scholar]
- 23.Jacobsen M, Clausen PP, Smidth S. The effect of fixation and trypsinization on the immunohistochemical demonstration of intracellular immunoglobulin in paraffin embedded material. Acta Pathol Microbiol Scand A. 1980;88:369–376. doi: 10.1111/j.1699-0463.1980.tb02508.x. [DOI] [PubMed] [Google Scholar]
- 24.Miettinen M. Immunostaining of intermediate filament proteins in paraffin sections. Evaluation of optimal protease treatment to improve the immunoreactivity. Pathol Res Pract. 1989;184:431–436. doi: 10.1016/S0344-0338(89)80039-1. [DOI] [PubMed] [Google Scholar]
- 25.Ordonez NG, Manning JT, Jr, Brooks TE. Effect of trypsinization on the immunostaining of formalin-fixed, paraffin-embedded tissues. Am J Surg Pathol. 1988;12:121–129. doi: 10.1097/00000478-198802000-00005. [DOI] [PubMed] [Google Scholar]
- 26.Pinkus GS, O’Connor EM, Etheridge CL, Corson JM. Optimal immunoreactivity of keratin proteins in formalin-fixed, paraffin-embedded tissue requires preliminary trypsinization. An immunoperoxidase study of various tumours using polyclonal and monoclonal antibodies. J Histochem Cytochem. 1985;33:465–473. doi: 10.1177/33.5.2580883. [DOI] [PubMed] [Google Scholar]
- 27.http://www.moleculardevices.com/pages/software/autoscanXT.html. AutoScanXT Software.
- 28.Robertson D, Savage K, Reis-Filho JS, Isacke CM. Multiple immunofluorescence labelling of formalin-fixed paraffin-embedded (FFPE) tissue. BMC Cell Biol. 2008;9:13. doi: 10.1186/1471-2121-9-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mouledous L, Hunt S, Harcourt R, Harry JL, Williams KL, Gutstein HB. Proteomic analysis of immunostained, laser-capture microdissected brain samples. Electrophoresis. 2003;24:296–302. doi: 10.1002/elps.200390026. [DOI] [PubMed] [Google Scholar]


