Abstract
Adenylation (A) domains found in all nonribosomal peptide synthetase (NRPS) modules are essential catalytic components and function as gatekeepers to select amino acid building blocks during nonribosomal peptide biosynthesis. Leveraging the strict substrate recognition characteristics of these enzymes, we targeted the development of active site-directed proteomic probes for A domains in NRPSs that enable detection, isolation, identification, and enzymatic characterization of A domains in native proteomes. Here, we describe a general strategy for selective chemical labeling of individual A domains in NRPS enzymes using active site-directed proteomic probes coupled to 5′-O-N-(aminoacyl)sulfamoyladenosine (AMS) scaffold with a clickable benzophenone functionality. These probes selectively target individual A domains in natural product producer proteomes by ligand-directed protein labeling. The data demonstrate that these proteomic tools can greatly facilitate the molecular identification, functional characterization, and profiling of virtually any kind of A domains of NRPS enzymes in complex biological systems.
Natural products display biologically interesting activities including a profound number of clinical antimicrobial, anticancer, and immunosuppressive agents, virulence factors, and signaling molecules.1 Many of these small molecules are biosynthesized by large, highly versatile multifunctional enzymes known as polyketide synthases (PKSs), nonribosomal peptide synthetases (NRPSs), and PKS-NRPS hybrids.2 Although significant progress has shed light on the functions of these multifunctional enzymes at the genetic level,3 their understanding at the protein level is much less understood. Analogous to the importance of studying the human proteome encoded by the genome, proteomic investigation of natural product producer microorganisms provides a level of information that cannot be understood solely by genetic approaches.4 In addition, some of these multifunctional enzymes are particularly resistant to routine cloning, expression, and biochemical evaluation as intact recombinant proteins. This is partly because these enzymes are large molecular species with a range between 200 and 1600 kDa, and because of the intractability of the producer organisms to conventional genetic manipulation and heterologous expression.5 Profiling these enzyme family members in the proteomes of diverse organisms could provide highly complementary to genetic approaches by allowing us to not only understand the activity, transcriptional control, and post-translational events, but also facilitate the molecular identification and functional characterization of these enzyme families directly in native proteomic environments.
Several chemistry-based approaches have exploited selective chemical labeling of thioesterase (TE),6 dehydratase (DH),7 and aryl acid adenylating enzymes8 found in any PKS and/or NRPS enzymes in bacterial proteomes. As not all domains are present in each PKS and NRPS enzymes, a more complete set of probes may be critical to identify full modules and realize practical proteomic approaches consisting of activity-based probes to natural product biosynthetic pathways.9,10 The adenylation (A) domain found in all NRPS modules is an essential catalytic component and functions as the gatekeeper of the NRPS assembly line. The A domain selectively incorporates a cognate amino acid into peptide-based natural products from a much larger monomer pool, including all 20 proteinogenic amino acids as well as a number of non-proteinogenic amino acids, aryl acids, fatty acids, and hydroxy acid building blocks.11 Using the strict A-domain active site chemistry, we set out to develop chemical labeling systems for A domains in NRPS enzymes using active site-directed proteomic probes (Figure 1).
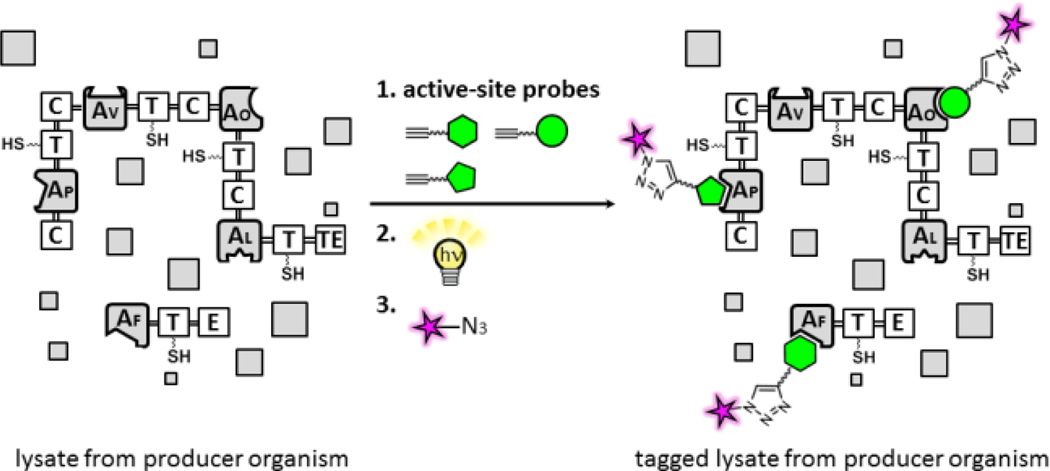
Figure 1.
Methods for proteomic analysis of A domains in NRPS biosynthetic enzymes using active site-directed proteomic probes for A domains. Modules are comprised of thiolation (T), adenylation (A) [Af: l-Phe; Ap: l-Pro; Av: l-Val; Ao: l-Orn specific A domains[, epimerization (E), condensation (C), and thioesterase (TE) domains. In a gel-based analysis, recombinant NRPS enzymes or proteomes treated with individual probes are incubated with a rhodamine (Rh)-azide reporter tag under click chemistry (CC) conditions and separated by SDS-PAGE, and labeled A domains in NRPS enzymes are visualized by in-gel fluorescence imaging.12
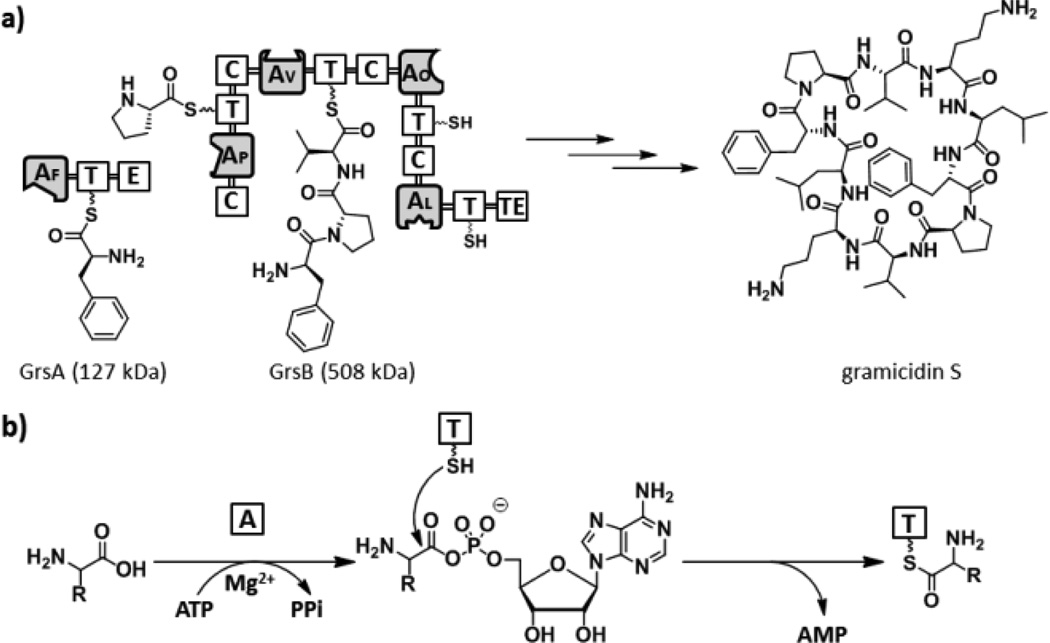
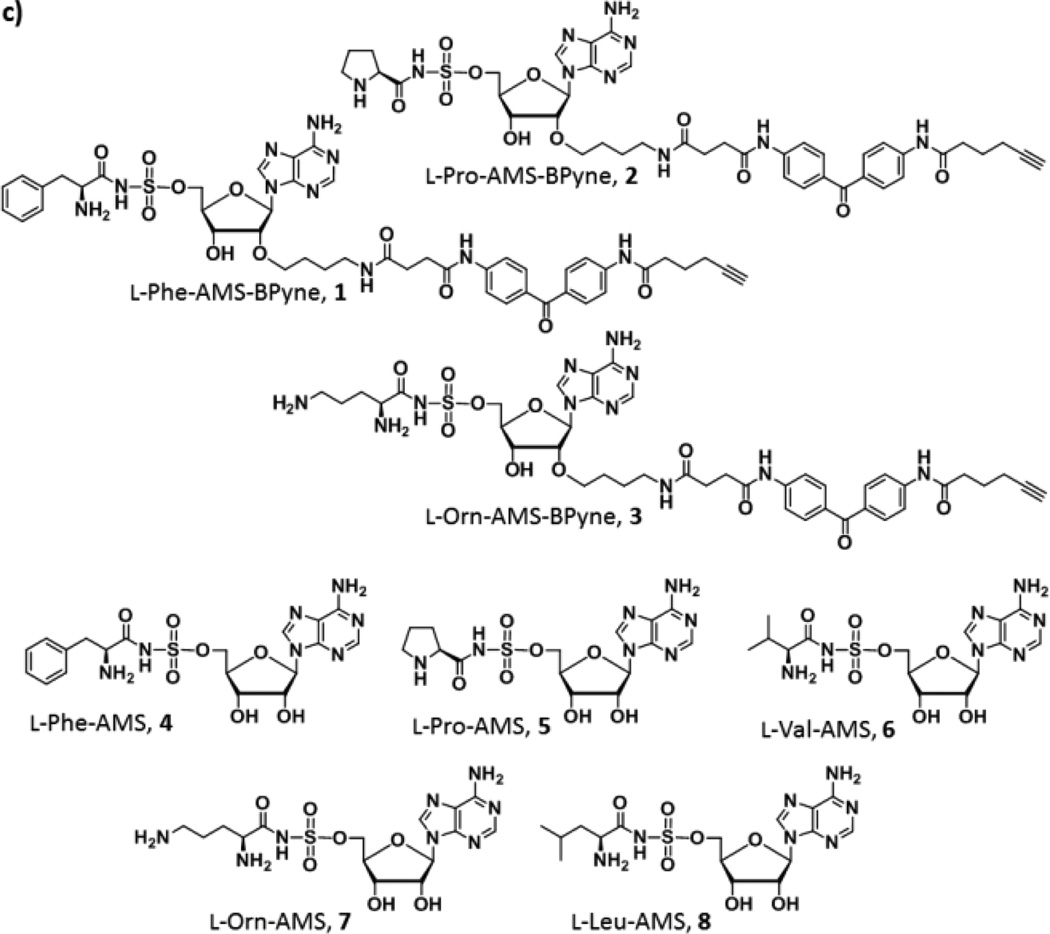
We chose A domains in the gramicidin S biosynthetic enzymes, GrsA and GrsB as the initial targets to demonstrate our strategy (Figure 2a).13 Gramicidin S is a cyclic decapeptide antibiotic with the primary structure cyclo (-d-Phe1-l-Pro2-l-Val3-l-Orn4-l-Leu5-)2, which is biosynthesized by GrsA and GrsB peptide synthetases. GrsA is a single NRPS module containing the domain structure A1 (l-Phe)-T-E and responsible for the incorporation of d-Phe during the gramicidin S biosynthesis. In contrast, GrsB is a large multifunctional megasynthetase with a calculated molecular weight of 508 kDa, and consists of four NRPS modules, C-A2 (l-Pro)-T-C-A3 (l-Val)-T-C-A4 (l-Orn)-T-C-A5 (l-Leu)-T-TE. Individual four A domains (A2–A5) are housed on this single protein and selectively incorporate their cognate amino acids, l-Pro, l-Val, l-Orn, and l-Leu, respectively, in the NRPS assembly line. Briefly, A domains selectively recognize the cognate amino acids and convert them to aminoacyl adenylate intermediates. The adenylated substrates in turn undergo a nucleophilic attack by the terminus thiol group of the 4′-phosphopantetheine arm of a downstream T domain, forming a thioester bound aminoacyl-S-T (Figure 2b). Ligand design exploiting tight binding characteristics for the active sites of A domains was based on the reliable bisubstrate inhibitor scaffold by conjugating amino acids with 5′-O-[N-(aminoacyl)sulfamoyl[adenosine (AMS) wherein the highly reactive acylphosphate linkage has been substituted by a bioisosteric and chemically stable non-hydrolysable acylsulfamate group.14 A complex structure of the GrsA A domain with l-Phe and AMP revealed that chemical modification at the 2′-OH group of the adenosine skeleton would be tolerated.15 Indeed, attachment of a long pegylated biotin to l-Phe-AMS 4 at this position showed no effect of binding for the GrsA A domain.16 We therefore decided to incorporate a clickable benzophenone functionality at this position. We selected l-Phe, l-Pro, and l-Orn as ligands to demonstrate the versatility of this scaffold as labeling reagents for A domains. To this end, active site-directed proteomic probes, l-Phe-AMS-BPyne 1, l-Pro-AMS-BPyne 2, and l-Orn-AMS-BPyne 3 were synthesized by the assembly of the AMS scaffold, a photoreactive benzophenone and a clickable alkyne functionality (Figure 2c, and Schemes S1 and S2). In addition, l-Phe-AMS 4,16 l-Pro-AMS 5, l-Val-AMS 6,17 l-Orn-AMS 7, and l-Leu-AMS 8 were prepared to provide analogs of 1–3 for comparative activity studies (Figure 2c and Scheme S3).
Figure 2.
(a) Biosynthetic pathway of the gramicidine S. GrsA is composed of A1 (l-Phe), T, and E domains. In contrast, GrsB consists of four NRPS modules, C-A2 (l-Pro)-T-C-A3 (l-Val)-T-C-A4 (l-Orn)-T-C-A5 (l-Leu)-T-TE. (b) The adenylation reaction in NRPS. (c) Structures of active site-directed proteomic probes and inhibitors for A domains described in this study.
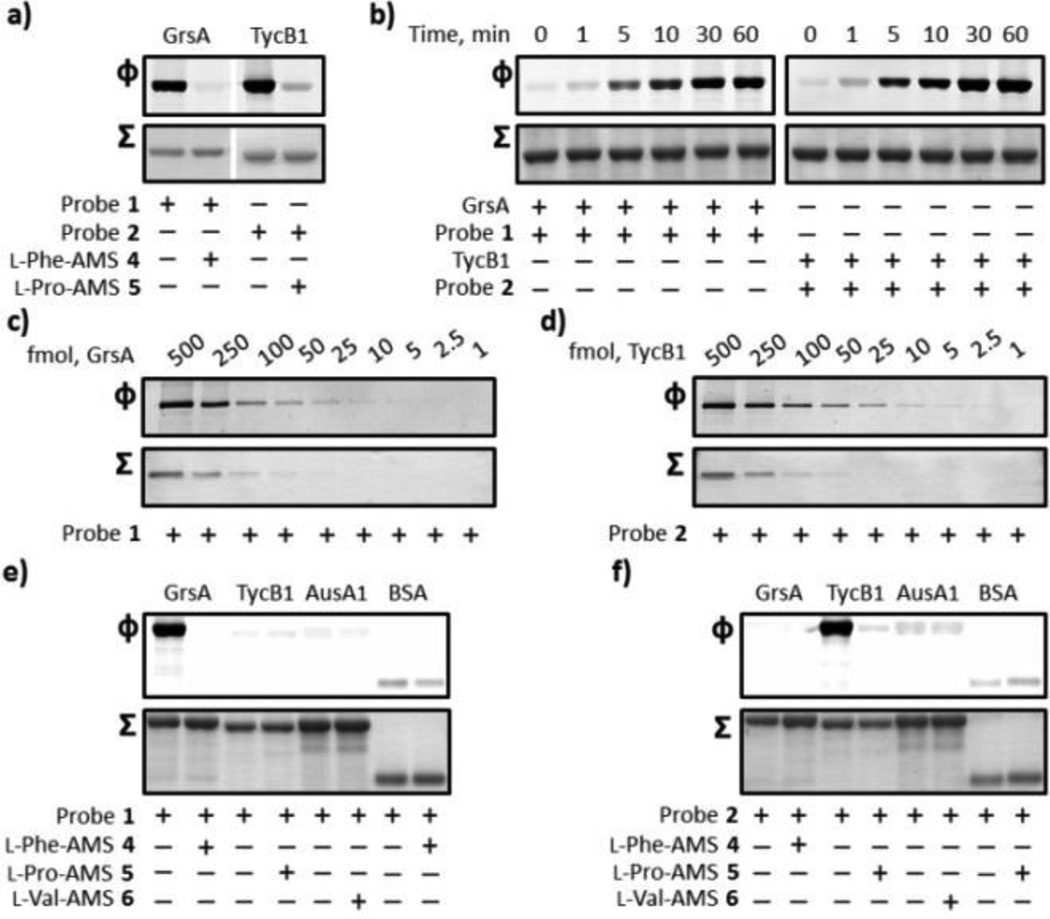
Our biochemical studies began with an examination of the specific inhibitory activities of l-Phe-AMS-BPyne 1, l-Phe-AMS 4, l-Pro-AMS-BPyne 2, and l-Pro-AMS 5 against recombinant holo-GrsA for 1 and 4 and holo-TycB1 for 2 and 4. TycB1 is derived from the tyrocidine biosynthetic enzymes, incorporates l-Pro, and contains the domain structure C-A (l-Pro)-T.18 Apparent Ki values were determined by a coupled hydroxamate-MesG continuous spectrophotometric assay (Figure S1).19 Probes 1 and 2 exhibited tight-binding properties against the A domains of GrsA and TycB1, respectively, with calculated Kiapp values of 1.43 ± 0.19 nM for GrsA and 327 ± 17.0 nM for TycB1 (Figure S2). Inhibitors 4 and 5 gave Kiapp values of 1.20 ± 0.14 nM for GrsA16 and 431 ± 42.0 nM for TycB1, respectively (Figure S2). These experiments demonstrated that the attachment of a bulky photoreactive benzophenone and a clickable alkyne at the 2′-OH group of the adenosine skeleton have no effect on the binding affinity of the A domains present within the NRPS enzymes tested.
Our labeling studies began by examining the ability of probes 1 and 2 to specifically label recombinant proteins, GrsA and TycB1, respectively. Probes 1 (1 µM) and 2 (1µM) were incubated with GrsA (1 µM) and TycB1 (1 µM) in either the absence or presence of excess 4 and 5, respectively, for 10 min at room temperature and pH 8.0. These samples were then photoactivated with ultraviolet light (365 nm) for 30 min at 0 °C, treated with Rh-azide under standard click chemistry (CC) conditions,12 and monitored by SDS-PAGE coupled with in-gel fluorescence scanning. SDS-PAGE analysis demonstrated the clear labeling of GrsA and TycB1. The labeling processes of GrsA with 1 and TycB1 with 2 were completely inhibited by pre-treatment of GrsA with 4 and TycB1 with 5 (Figure 3a). We next performed time course experiments by evaluating the labeling of GrsA and TycB1. Maximum fluorescence band intensity was observed at 30 min for both reactions (Figure 3b). Subsequent characterization allowed us to identify the lower detection limit for each protein labeling. As shown in Figure 3c and 3d, we found that probes 1 and 2 attained highly sensitive detection as low as 10 fmol of GrsA and 5 fmol of TycB1. We finally asked whether probes 1 and 2 could discriminate the cognate A domain activity by ligand-directed protein labeling. AusA1 is the excised didomain (A1-T1) of the aureusimine synthase AusA (A1-T1-C-A2-T2-R) from Staphylococcus aureus.17 Labeling experiments of GrsA, TycB1, AusA1, and BSA with 1 µM 1 resulted in selective labeling of the cognate A domain of GrsA with virtually no labeling of the non-cognate A domains of TycB1 and AusA1 and BSA (Figure 2e). Probe 2 also showed similar selective labeling of the cognate A domain of TycB1 with no detectable labeling of GrsA, AusA1, and BSA (Figure 2f). The A1 domain of AusA incorporates l-Val during the biosynthesis of aureusimines. These results indicate that the strict substrate recognition systems of A domains allow us to selectively label the A domains with substrate specificity identical to an attached amino acid moiety.
Figure 3.
Labeling of recombinant holo-GrsA and holo-TycB1 with probes 1 and 2. (a) Labeling of GrsA and TycB1 and competitive inhibition studies with excess inhibitors 4 and 5. GrsA (1 µM) and TycB1 (1 µM) were individually pre-incubated in either the absence or presence of 100 µM of inhibitors 4 and 5 and treated with 1 µM of the individual probes 1 and 2. (b) Ultraviolet photolysis time course studies of the labeling of GrsA with probe 1 (left) and TycB1 with probe 2 (right). SDS-PAGE analysis denoting the labeling of 1 µM of GrsA and TycB1 with 1 µM probes 1 and 2, respectively. (c, d) Limit of detection of GrsA and TycB1 labeling. GrsA (0.125–62.5 nM) and TycB1 (0.125–62.5 nM) were individually incubated with probes 1 and 2. (e, f) Labeling specificity of probes 1 and 2. GrsA (1 µM), TycB1 (1 µM), AusA1 (1 µM), and BSA (1 µM) were individually treated with 1 µM of probes 1 and 2 in either the absence or presence of 100 µM of the inhibitors 4, 5, and 6. For each panel, Φ depicts the fluorescence observed with λex = 532 nm and λem = 580 nm, and Σ displays the total protein content by staining with Coomassie Blue (a, b, e, and f) or a silver staining method (c and d). Full gels (Figures S3) and experimental procedures are provided in the SI.
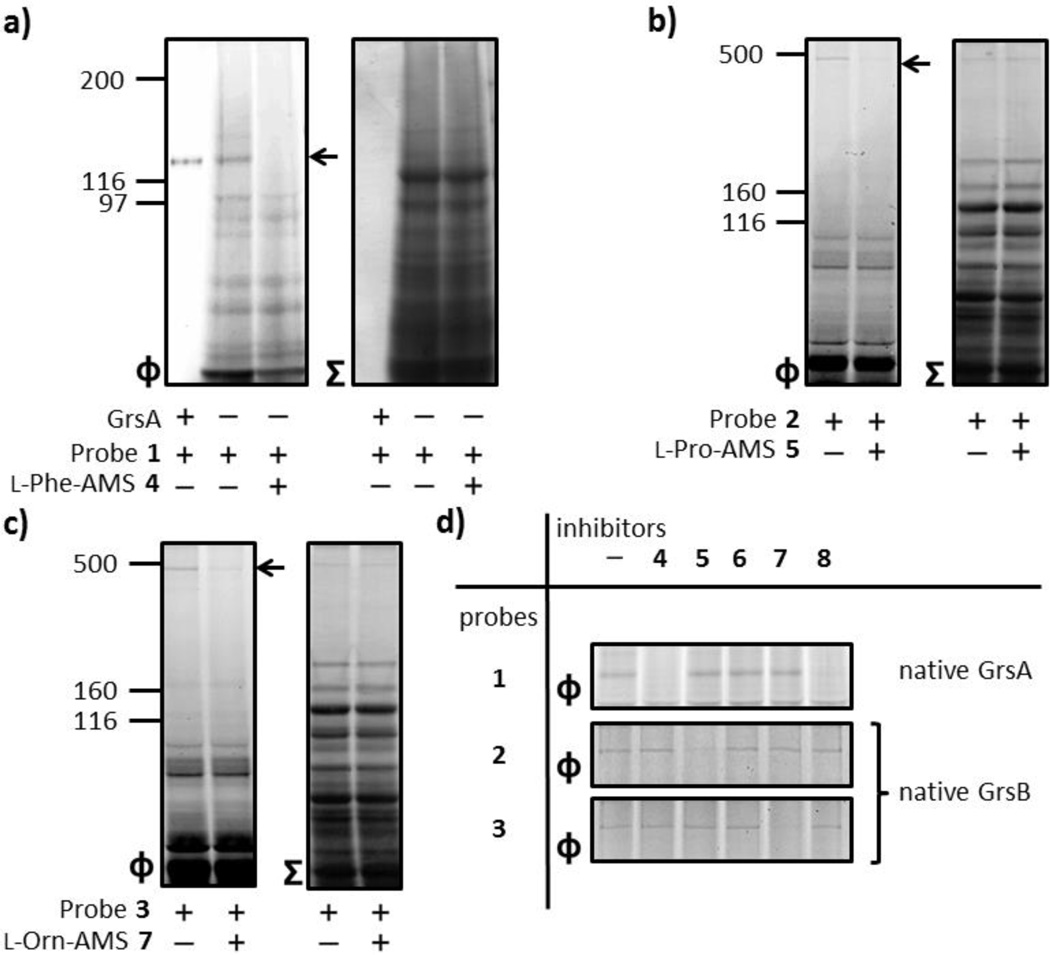
After demonstrating the compatibility of probes 1 and 2 for selective labeling of A domains in recombinant enzyme systems, we sought to demonstrate the applicability of probes 1–3 by applying them to the analysis of natural product producers’ NRPS systems. Two strains of Aneurinibacillus migulanus known as Bacillus brevis, ATCC 9999 and DSM 5759 have been demonstrated as gramicidin S producing strains.20 We first evaluated the availability of probe 1 to detect endogenous GrsA in these producers. The cellular lysate of producing strain ATCC 9999 was adjusted to pH 8.0, incubated with 1 µM of 1 for 10 min at room temperature, irradiated for 30 min at 0 °C and subjected to the click reaction with Rh-azide. In-gel fluorescence analysis showed a distinguishable fluorescence signal at ~120 kDa corresponding to the size of labeled standard recombinant GrsA (Figure 4a).16 In addition, the observed single band was abrogated by pre-treatment with 4 (Figure 4a). Notably, the GrsA was the only specific labeled protein in this proteomic sample, further demonstrating the specificity of this probe. These results validate that probe 1 is a highly specific labeling agent for l-Phe activating A domains that shows no cross-reactivity with other proteins and markedly low levels of background signals within the proteomic environment.
Figure 4.
Proteomic applications of active site-directed proteomic probes 1–3 for A domains. (a) Labeling of endogenous GrsA in an A. migulanus ATCC 9999 cellular lysate by probe 1. The A. migulanus ATCC 9999 lysate (1.5 mg/mL) was treated with 1 µM 1 in either the absence or presence of 100 µM 4. (b) Labeling of endogenous GrsB in the A. migulanus DSM 5759 cellular lysate by probes 2 and 3. The A. migulanus DSM 5759 lysate (1.5 mg/mL) was individually treated with 100 µM 5 and 7 before the addition of individual members of 1 µM probes 2 and 3. (d) Individual labeling of A domains and profiling of A domain functions using a combination of probes 1–3 and inhibitors 4–8. In order to investigate GrsA labeling, the A. migulanus ATCC 9999 lysate (1.5 mg/mL) was preincubated with individual members of inhibitors 4–8 (100 µM) before the addition of 1 µM of probe 1. To evaluate the labeling of GrsB, the A. migulanus DSM 5759 lysate (1.5 mg/mL) was individually treated with 100 µM of inhibitors 4–8 before the addition of individual members of 1 µM probes 2 and 3. For each panel, Φ depicts the fluorescence observed with λex = 532 nm and λem = 580 nm, and Σ displays the total protein content by staining with Coomassie Blue. Full gels (Figure S4) and experimental procedures are provided in the SI.
As an ultimate application of proteomic activity, we evaluated the availability of probes 2 and 3 by applying them to A domains housed on a multifunctional megasynthetase, GrsB in a proteomic context. The DSM 5759 proteome was individually incubated with 1 µM of probes 2 and 3 for 10 min at room temperature, irradiated for 5 min at 0 °C and subsequently treated with Rh-azide under the CC conditions. In-gel fluorescence analysis showed only labeled fluorescence bands at ~500 kDa in the individual probes applied (Figure 4b and 4c). Significantly, this labeling completely disappeared by pre-treatment with the corresponding inhibitors 4 and 5 (Figures 4b and 4c). In particular, the high-molecular-weight (HMW) band was the only species labeled by 2 and 3 in this proteome. Collectively, these labeling experiments led us to identify the labeled protein as GrsB on the knowledge of the molecular weight and the presence of two A domains with substrate specificity for l-Pro and l-Orn on this single protein. A labeled HMW band was visualized with silver staining, excised, analyzed by LC-MS/MS, and found to contain the endogenous GrsB protein with 40% peptide coverage (Figure S5).
With proteomic tools in hand, we finally attempted to demonstrate individual labeling and profiling of substrate preference of A domains in GrsA and GrsB by a combination of probes 1–3 with inhibitors 4–8. In order to investigate GrsA labeling, ATCC 9999 lysates were preincubated with inhibitors 4–8 (100 µM) before the addition of 1 µM of probe 1, and the samples were exposed to ultraviolet light for 5 min and treated with an Rh-azide. To evaluate the labeling of GrsB, DSM 5759 lysates were individually treated with 100 µM of inhibitors 4–8 before the addition of 1 µM probes 2 and 3. As shown in Figure 4d, the labeling of GrsB by probes 2 and 3 disappeared only by the addition of 5 and 7, respectively. These results validated that probes 2 and 3 selectively targeted individual A domains housed on GrsB, A2 (l-Pro) and A4 (l-Orn), respectively. In contrast, the labeling of GrsA by probe 1 was abrogated in the presence of either 4 or 8. This labeling pattern of GrsA correlates well with its substrate preferences, because the A domain of GrsA is known to recognize l-Leu as a miscognate substrate with lower catalytic efficiency than that of l-Phe.21 Concurrently, these experiments emphasized the significant differences with the substrate specificity of individual A domains by comparing the competitive activity-based profiling (ABPP) of the A domains.
In summary, we have demonstrated practical proteomic tools for the study of A domains in NRPS enzymes based on the development of active site-directed proteomic probes for A domains appended to a clickable benzophenone functionality at the 2′-OH of the adenosine skeleton. We have demonstrated the feasibility and general strategy of selective chemical labeling for A domains in NRPSs by applying three A domains in two endogenous NRPS enzymes, GrsA (A1: l-Phe) and GrsB (A2: l-Pro; A4: l-Orn) as well as two recombinant NRPS enzymes, GrsA (A: l-Phe) and TycB1 (A: l-Pro). The chemical labeling strategies of A domains could be used to probe natural product producing bacteria, assign A-domain organization, and characterize A-domain functions by the competitive ABPP platform. By using appropriate amino acids as a ligand, this labeling technique offers easy access to virtually any kind of NRPS A domains in complex biological proteomes. These approaches should be immediately useful for detecting, monitoring and tracking the expression of orphan NRPS and PKS-NRPS hybrid genes in sequenced organisms, optimizing fermentative microorganisms and analyzing pathogenic bacteria for the production of small molecule virulence factors.22 The competitive ABPP approach should be suitable for identifying reversible enzyme inhibitors for blocking the production of NRPS and PKS-NRPS virulence factors.23,24 It may also be possible to use this approach for selective visualization, molecular identification, and functional characterization of NRPS and PKS-NRPS hybrid enzymes associated with the biosynthesis of peptide-based natural products of interest. This is because a large portion of these compounds readily correlate to the amino acid specificity of A domains found on their associated multifunctional enzymes.24 Combinations of these probes should further improve the detection, isolation, and high resolution of low-abundance enzymes in proteomic samples, and we are currently applying cocktails of these probes for studying biosynthetic pathways in a variety of natural product producer organisms.
Supplementary Material
Acknowledgments
This work was supported by the Sasakawa Scientific Research Grant from The Japan Science Society (S.K.), a Grant-in Aid for Young Scientists (B) 26750370 (F.I.), and a research grant from the Japan Society of the Promotion of Science (H.K.). We thank Prof. Mohamed Marahiel (Philipps-Universität Marburg, Germany) for providing the GrsA and TycB1 expression constructs.
Footnotes
ASSOCIATED CONTENT
Supporting Information
Supporting figures; procedures for the syntheses of l-Phe-AMS-BPyne 1, l-Pro-AMS-BPyne 2, l-Orn-AMS-BPyne 3, l-Pro-AMS 5, l-Orn-AMS 7, and l-Leu-AMS 8; complete gel images; full experimental details; and copies of 1H and 13C-NMR spectra. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES
- 1.Walsh CT. Science. 2004;303:1805. doi: 10.1126/science.1094318. [DOI] [PubMed] [Google Scholar]
- 2.Fischbach MA, Walsh CT. Chem. Rev. 2006;106:3468. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- 3.Cane DE, Walsh CT, Khosla C. Science. 1998;282:63. doi: 10.1126/science.282.5386.63. [DOI] [PubMed] [Google Scholar]
- 4.Reyes CP, La Clair JJ, Burkart MD. Chem. Commun. 2010;46:8151. doi: 10.1039/c0cc02876b. [DOI] [PubMed] [Google Scholar]
- 5.(a) Cane DE, Walsh CT. Chem. Biol. 6:R319. doi: 10.1016/s1074-5521(00)80001-0. [DOI] [PubMed] [Google Scholar]; (b) Pfeifer BA, Khosla C. Microbiol. Mol. Biol. Rev. 2001;65:106. doi: 10.1128/MMBR.65.1.106-118.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meier JL, Mercer AC, Burkart MD. J. Am. Chem. Soc. 2008;130:5443. doi: 10.1021/ja711263w. [DOI] [PubMed] [Google Scholar]
- 7.Ishikawa F, Haushalter RW, Burkart MD. J. Am. Chem. Soc. 2012;134:769. doi: 10.1021/ja2082334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duckworth BP, Wilson DJ, Nelson KM, Boshoff HI, Barry CE, III, Aldrich CC. ACS Chem. Biol. 2012;7:1653. doi: 10.1021/cb300112x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meier JL, Niessen S, Hoover HS, Foley TL, Cravatt BF, Burkart MD. ACS Chem. Biol. 2009;4:948. doi: 10.1021/cb9002128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meier JL, Burkart MD. Curr. Opin. Chem. Biol. 2010;15:1. doi: 10.1016/j.cbpa.2010.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.(a) Sieber SA, Marahiel MA. Chem. Rev. 2005;105:715. doi: 10.1021/cr0301191. [DOI] [PubMed] [Google Scholar]; (b) Gulick AM. ACS Chem. Biol. 2009;4:811. doi: 10.1021/cb900156h. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Hur GH, Vickery CR, Burkart MD. Nat. Prod. Rep. 2012;10:1074. doi: 10.1039/c2np20025b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.(a) Speers AE, Adam GC, Cravatt BF. J. Am. Chem. Soc. 2003;125:4686. doi: 10.1021/ja034490h. [DOI] [PubMed] [Google Scholar]; (b) Speers AE, Cravatt BF. Chem. Biol. 2004;11:535. doi: 10.1016/j.chembiol.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 13.(a) Kratzschmar J, Krause M, Marahiel MA. J. Bacteriol. 1989;171:5422. doi: 10.1128/jb.171.10.5422-5429.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Hoyer KM, Mahlert C, Marahiel MA. Chem. Biol. 2007;14:3. doi: 10.1016/j.chembiol.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.(a) Finking R, Neumüller A, Solsbacher J, Konz D, Kretzschmar G, Schweitzer M, Krumm T, Marahiel MA. Chem Bio Chem. 2003;4:903. doi: 10.1002/cbic.200300666. [DOI] [PubMed] [Google Scholar]; (b) Ferreras JA, Ryu J-S, Di Lello F, Tan DS, Quadri LEN. Nat. Chem. Biol. 2005;1:29. doi: 10.1038/nchembio706. [DOI] [PubMed] [Google Scholar]; (c) Qiao C, Gupte A, Boshoff HI, Wilson DJ, Bennett EM, Somu RV, Barry CE, Aldrich CC. J. Med. Chem. 2007;50:6080. doi: 10.1021/jm070905o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Conti E, Stachlhaus T, Marahiel MA, Brick P. EMBO J. 1997;16:4174. doi: 10.1093/emboj/16.14.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishikawa F, Kakeya H. Bioorg. Med. Chem. Lett. 2014;22:865. doi: 10.1016/j.bmcl.2013.12.082. [DOI] [PubMed] [Google Scholar]
- 17.Wilson DJ, Shi C, Teitelbaum AM, Gulick AM, Aldrich CC. Biochemistry. 2013;52:926. doi: 10.1021/bi301330q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mootz HD, Marahiel MA. J. Bacteriol. 1997;179:6843. doi: 10.1128/jb.179.21.6843-6850.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilson DJ, Aldrich CC. Anal. Biochem. 2010;404:56. doi: 10.1016/j.ab.2010.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berditsch M, Afonin S, Ulrich AS. Appl. Environ. Microbiol. 2007;73:6620. doi: 10.1128/AEM.00881-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stachelhaus T, Mootz HD, Marahiel MA. Chem. Biol. 1999;6:493. doi: 10.1016/S1074-5521(99)80082-9. [DOI] [PubMed] [Google Scholar]
- 22.(a) Gross H. Appl. Microbiol. Biotechnol. 2007;75:267. doi: 10.1007/s00253-007-0900-5. [DOI] [PubMed] [Google Scholar]; (b) Cisar JS, Tan DS. Chem. Soc. Rev. 2008;37:1320. doi: 10.1039/b702780j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Evans MJ, Cravatt BF. Chem. Rev. 2006;106:3279. doi: 10.1021/cr050288g. [DOI] [PubMed] [Google Scholar]
- 24.(a) Bergmann S, Schumann J, Scherlach K, Lange C, Brakhage AA, Hertweck C. Nat. Chem. Biol. 2007;3:213. doi: 10.1038/nchembio869. [DOI] [PubMed] [Google Scholar]; (b) Ferreras J, Stirrett KL, Lu X, Ryu J, Soll CE, Tan DS, Quadri LEN. Chem. Biol. 2008;25:51. doi: 10.1016/j.chembiol.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.(a) van Wageningen AMA, Kirkpatrick PN, Williams DH, Harris BR, Kershaw JK, Lennard NJ, Jones M, Jones SJM, Solenberg PJ. Chem. Biol. 1998;5:155. doi: 10.1016/s1074-5521(98)90060-6. [DOI] [PubMed] [Google Scholar]; (b) Schümann J, Hertweck C. J. Am. Chem. Soc. 2007;129:9564. doi: 10.1021/ja072884t. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.