Abstract
One of the most endangered vertebrates, the Devils Hole pupfish Cyprinodon diabolis, survives in a nearly impossible environment: a narrow subterranean fissure in the hottest desert on earth, Death Valley. This species became a conservation icon after a landmark 1976 US Supreme Court case affirming federal groundwater rights to its unique habitat. However, one outstanding question about this species remains unresolved: how long has diabolis persisted in this hellish environment? We used next-generation sequencing of over 13 000 loci to infer the demographic history of pupfishes in Death Valley. Instead of relicts isolated 2–3 Myr ago throughout repeated flooding of the entire region by inland seas as currently believed, we present evidence for frequent gene flow among Death Valley pupfish species and divergence after the most recent flooding 13 kyr ago. We estimate that Devils Hole was colonized by pupfish between 105 and 830 years ago, followed by genetic assimilation of pelvic fin loss and recent gene flow into neighbouring spring systems. Our results provide a new perspective on an iconic endangered species using the latest population genomic methods and support an emerging consensus that timescales for speciation are overestimated in many groups of rapidly evolving species.
Keywords: demographic inference, population genomics, speciation, conservation genetics, genetic accommodation, introgression
1. Introduction
Simple environments, isolated habitats with minimal biotic interactions, serve as natural laboratories for examining the extremes of ecological and evolutionary processes [1,2]. From adaptive radiation on remote depauperate islands [3–5] to laboratory microcosms [6–8], simple ecosystems may be isolated from complex community-level processes and often provide unique opportunities for testing theory. For example, what are the simplest environments in which speciation can occur without physical barriers [9–12]? How does effective population size affect the risk of extinction or the potential for adaptation [13–16]? Small inbred populations in simple natural environments may also provide a window into the future of managed endangered populations [17–19].
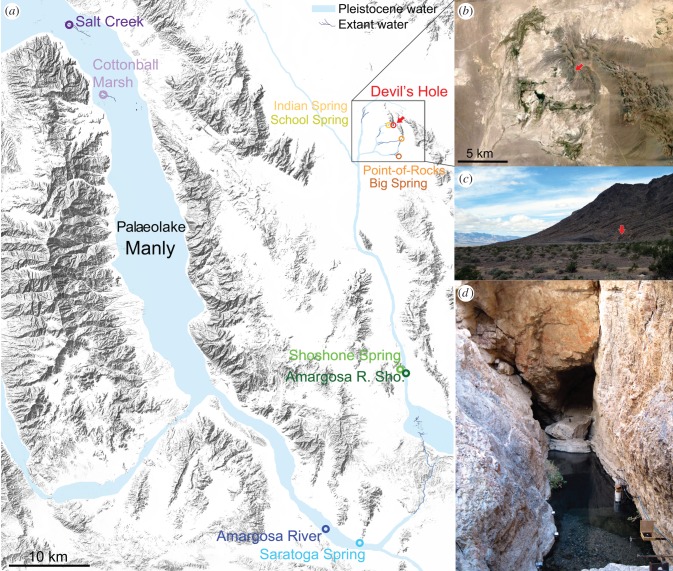
Devils Hole is the smallest natural range of any vertebrate species, comprising a 3.5 × 22 m opening into a flooded fault cavern greater than 130 m deep (figure 1d; [21,22]). The census population size of Cyprinodon diabolis within Devils Hole has varied from only 35–548 individuals since semi-annual monitoring began in 1972 [23], reaching its record low in 2013 and second lowest point (38) in 2006–2007. In addition to limited spawning habitat (a single 2.6 × 6.1 m shelf [22]: figure 1d), the population is limited by the productivity of Devils Hole, which receives no direct sunlight for two months each year and produces 200 times less algal biomass than neighbouring springs [24]. The population size typically decreases in response to reduced winter food availability and rebounds in the spring. Year-round water temperatures in Devils Hole remain near 32°C [21]; however, broad thermal tolerances in Cyprinodon from −1.9°C to 45.5°C [25–27], such as Cyprinodon pachycephalus which completes its entire life cycle at 44°C [28], suggest that diabolis should tolerate higher water temperatures (contra [29]).
Figure 1.
(a) Sample sites within Death Valley National Park, California and Ash Meadows National Wildlife Refuge, Nevada overlaid on the surrounding topography (Google Maps), extant desert springs (dark blue lines) and approximate Pleistocene waterways (light blue shading) present in the region 10–35 kyr ago [20]. Colours of each desert spring correspond to population colours in figure 2. (b) Aerial view of Ash Meadows (Google Earth) with the position of Devils Hole indicated in red. (c,d) Close-up views of Devils Hole in November 2013. The white pipe (for daily supplemental feeding which began in 2007) marks the edge of the spawning shelf. (Online version in colour.)
The improbability of a vertebrate species surviving at such low population sizes in a suboptimal habitat, even for a short time, would seem to defy basic principles of conservation biology. For example, the probability of diabolis extinction within 50 years exceeds 80% based on historical census population sizes [23]. Initial time-calibrated mitochondrial phylogenies suggested that Death Valley pupfishes were relicts isolated for 2–3 Myr [30–32] and estimated the age of the diabolis mitochondrial lineage to be 500 000 years [30,31]. However, early studies did not detect any unique mitochondrial haplotypes in diabolis relative to other Death Valley pupfishes [33], consistent with more recent colonization. Geological evidence strongly indicates that Devils Hole opened to the surface only 60 000 years ago based on the abrupt cessation of mammillary calcite deposition [34], which places an upper bound on the isolation of diabolis within this extreme habitat. Previous studies attributed overestimation of species divergence times to incomplete lineage sorting [30,31]; however, this cannot account for such a large discrepancy [35,36]. Instead, this conflict appears to be caused by 11 Myr priors placed on the root age of Cyprinodon + Megupsilon + Cualac [30,31] based on the assumption that pupfish populations were isolated by the formation of ancient land barriers [32]. Thus, the discrepancy between geological and mtDNA evidence for the age of diabolis is not due to any underlying conflict in the data (e.g. time-calibration is extremely sensitive to priors [37]), but rather the assumption that Cyprinodon cannot disperse over land. Devils Hole was never connected to neighbouring springs by surface flow, thus the current consensus is that diabolis colonized approximately 10–20 000 years ago when water levels were higher [22,31].
A recent study estimated the age of diabolis from the coalescence of 12 microsatellite loci using approximate Bayesian computation without assumptions about overland dispersal and placed the age between 90 and 14 400 years (95% credible intervals in table 1: [38]). However, additional uncertainty in microsatellite mutation rates (10−3–10−6) was not accounted for by use of a point estimate from mammals (10−3), nor was the known failure of a molecular clock for microsatellite mutation rates [39]. Thus, despite substantial attention, diabolis age estimates still range from 90 to 20 000 years [38,40]. Furthermore, additional outstanding questions about the survival of this species within its habitat remain: was Devils Hole colonized only once? Did phenotypic plasticity facilitate adaptation to this harsh environment [41]? How did so many small Death Valley pupfish populations survive inbreeding depression if they were isolated for millions of years [42]?
Table 1.
Genetic diversity (π), private alleles, total nucleotide sites examined and percentage of polymorphic sites in Death Valley pupfishes and additional pupfish outgroups for comparison (EW = extinct in the wild based on IUCN designation or unpublished observations). (The number of individuals sequenced in each population is indicated (electronic supplementary material, appendix S1).)
| species/subspecies | location | genetic diversity | private alleles | total sites | %polymorphic |
|---|---|---|---|---|---|
| Cyprinodon diabolis (n = 4) | Devils Hole | 0.0009 | 1019 | 1 017 310 | 0.0938 |
| C. nevadensis mionectes (n = 8) | Point-of-Rocks | 0.0023 | 1572 | 1 229 973 | 0.2348 |
| C. nevadensis mionectes (n = 1) | Big Spring | 0.0006 | 272 | 403 729 | 0.0577 |
| C. nevadensis pectoralis (n = 7) | Indian Spring | 0.0001 | 21 | 141 445 | 0.0099 |
| C. nevadensis amargosae (n = 7) | Amargosa River | 0.002 | 1008 | 1 261 354 | 0.1954 |
| C. nevadensis shoshone (n = 5) | Shoshone Spring | 0.0013 | 539 | 874 517 | 0.1337 |
| C. nevadensis nevadensis (n = 7) | Saratoga Spring | 0.0021 | 1360 | 1 314 878 | 0.2135 |
| C. salinus salinus (n = 2) | Salt Creek | 0.0009 | 1244 | 1 184 303 | 0.0888 |
| C. salinus milleri (n = 1) | Cottonball Marsh | 0.0004 | 264 | 367 191 | 0.0400 |
| all Death Valley species | Death Valley | 0.0036 | 10 224 | 1 140 524 | 0.7117 |
| C. artifrons (n = 1) | coastal Cancun, Mexico | 0.0034 | 3212 | 1 185 394 | 0.3354 |
| C. variegatus (n = 1) | coastal San Salvador, Bahamas | 0.0039 | 9900 | 1 311 769 | 0.3862 |
| C. alvarezi (n = 1) | captive colony—EW | 0.0011 | 2211 | 965 072 | 0.1149 |
| C. maya (n = 1) | captive colony—EW | 0.0027 | 657 | 964 452 | 0.2722 |
| C. veronicae (n = 1) | captive colony—EW | 0.0015 | 5160 | 1241 190 | 0.1491 |
We examined population structure and gene flow among Death Valley populations, estimated the pupfish mutation rate from a new time-calibrated phylogeny containing all major Cyprinodon lineages, and then used this rate to calibrate a demographic model for the divergence between two closely related pupfish populations in Death Valley and approximate an age for diabolis. We sampled all extant Death Valley and Ash Meadows species and subspecies as well as outgroup species from across the Cyprinodon clade (electronic supplementary material, appendix S1). We conducted all analyses at the population level (i.e. each desert spring) and for clarity we refer to these populations by only their subspecies designation and desert spring name. In some cases, the same subspecies was sampled from multiple desert springs and we treated these as different populations. We used double-digest restriction-site-associated DNA sequencing (ddRADseq [43]) to genotype over 13 000 loci in 56 individuals. This provided 1.3 million sites for demographic inference, 100 000 times more data than previous studies. Our analyses reshape our understanding of population isolation and speciation in this highly endangered group of extremophile fishes and suggest a surprisingly rapid timescale for speciation, genetic assimilation and the evolution of intrinsic reproductive incompatibilities in this group.
2. Material and Methods
(a). Sample collection and bioinformatics
Cyprinodon diabolis is one of the most endangered fish on earth and thus collecting tissue from live animals was impossible. Highly degraded samples putrefied by the 32°C water were obtained from dead specimens collected by National Park staff. Other Death Valley samples used archived specimens from a previous study [44]. DNA was extracted with Qiagen Blood & Tissue kits and ddRADseq libraries [43] were prepared as described previously [45]. One hundred and ninety-eight million Illumina HiSeq 2000 single-end 100 bp reads were aligned to the Cyprinodon variegatus reference genome (v. 1.0) and called using the Stacks pipeline [46]. Loci genotyped in at least six of the 56 high-coverage individuals and sequenced to a minimum depth of eight reads were used for analyses. Overall mean coverage depth per locus per individual for aligned reads was 138×. Further details are provided in the electronic supplementary material, Supplemental Methods.
(b). Population genetic structure and analyses of gene flow
Population genetic structure among populations was evaluated by principal components analysis of genetic variance and Bayesian unsupervised clustering using Structure [47]. Gene flow among populations was estimated using four complementary methods: formal tests of introgression using ABBA/BABA ratios (D-statistics) [48], ancestry proportions across independent Structure runs, non-zero migration parameters in our dadi model, and by using Treemix [49] to estimate secondary gene flow among branches in a maximum-likelihood tree based on the residual covariance in allele frequencies among populations. Although we sampled small numbers of individuals per population, this will not necessarily bias our estimates of genetic differentiation owing to the large number of genetic markers sampled per individual [50].
(c). Phylogenetic analyses and time-calibration
We used BEAST (v. 1.8.1; [51]) for phylogenetic inference from 16 567 concatenated 100 bp loci based on a separate Stacks pipeline sampling the most divergent lineages across Cyprinodon. Estimation of divergence times and mutation rates is highly sensitive to the choice of priors [37] and requires internal calibrations [36,52,53], thus we chose the only well-defined internal calibration event known for Cyprinodon (options discussed in [54]): the 8000 ± 200 year age of Laguna Chichancanab [55,56], an endorheic basin which contains an endemic species flock of Cyprinodon pupfishes (figure 2d; [57]). It is highly unlikely that the Chichancanab species flock diverged before the basin formed based on microsatellite evidence [58,59] and because these species cannot coexist with fish predators found in all neighbouring surface waters (the flock is now extinct due to invasive fishes [54,60]). We conservatively placed a lower bound on the stem age of this clade using a normal prior for the divergence between Cyprinodon artifrons (the most closely related species) and the Chichancanab lineage with a mean of 8000 years and standard deviation of 100 years. This age and associated error (95% confidence interval (CI): ± 200 years) were estimated in geological studies of this lake using multiple core samples and multiple lines of evidence, including stable isotope data and shifts from terrestrial to aquatic invertebrate communities [55,56]. Our analysis is still highly dependent on this choice of prior and would support much older ages for diabolis if we used ancient land barriers or other mutation rates for calibration as in previous studies of this group [30,32]. For example, calibration with the human mutation rate would unrealistically place the age of the Laguna Chichancanab species flock at 4.9 Myr (see the electronic supplementary material, Supplemental Methods).
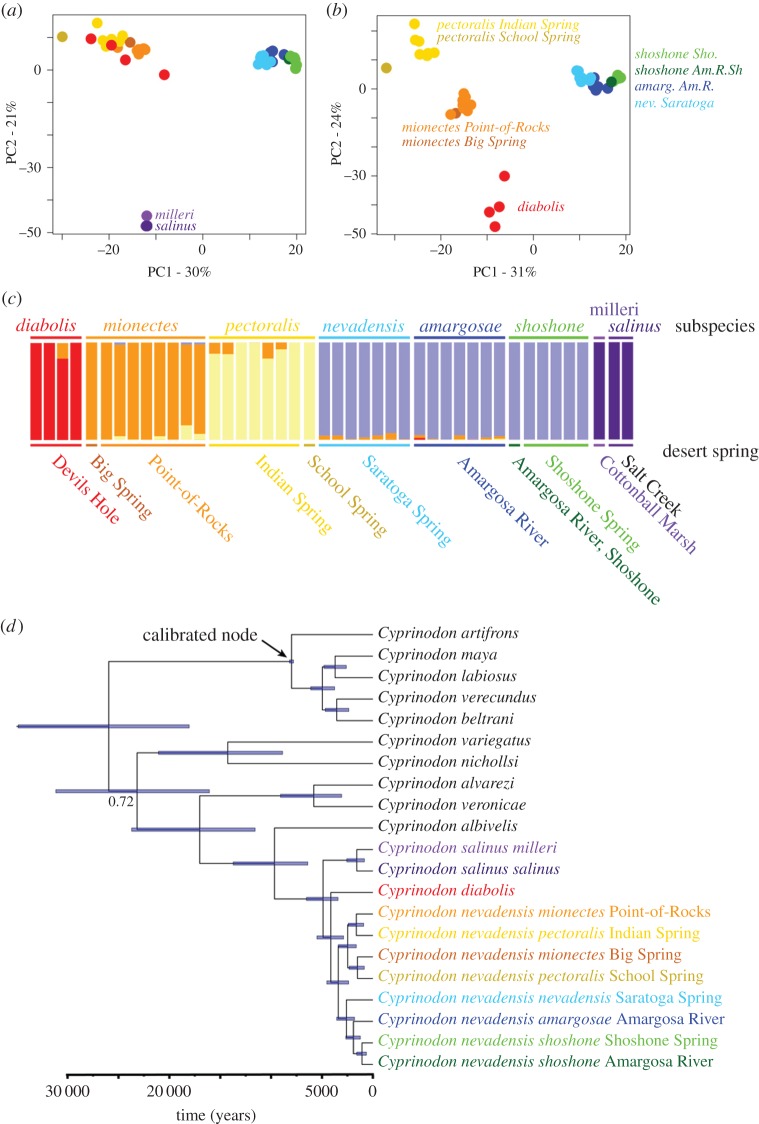
Figure 2.
Genetic structure (a–c) and time-calibrated phylogenetic tree (d) for Death Valley pupfish populations. (a) First two principal components of genetic variance for 4679 single nucleotide polymorphisms (SNPs) showing three main clusters of Death Valley populations. (b) Excluding the distant salinus populations reveals four to five distinct genetic clusters. (c) Bayesian clustering analyses using Structure indicating the proportion of shared ancestry among individuals with k = 5 groups for 4679 SNPs, supported by a reduced rate of change of the log-likelihood at this value (electronic supplementary material, table S5). Species and subspecies designations are indicated on the top row and desert spring systems are indicated on the bottom row, corresponding to sample sites in figure 1a. (d) Time-calibrated maximum clade credibility tree for the Death Valley populations plus all major lineages across Cyprinodon. A single haplotype was sampled from one high-coverage individual per population. All nodes were supported with 100% posterior probability except the one annotated node (0.72). Blue bars indicate 95% credible intervals for the estimated age of each node. (Online version in colour.)
(d). Estimation of the mutation rate in pupfishes
We estimated a median global substitution rate of 5.37 × 10−7 mutations site−1 yr−1 using our original time-calibrated phylogeny for Cyprinodontidae (electronic supplementary material, table S2). This is much greater than the human genome-wide mutation rate estimated from pedigrees (0.5 × 10−9 mutations site−1 yr−1: [61]). Differences between human generation time (20–30 years) and pupfish generation time (less than 4 months in the wild) only partially account for this difference. It is unknown whether this reflects a bias in our RADseq data or a real biological difference in pupfishes, such as increased metabolism in extreme thermal or hypersaline environments [62]. Alternative phylogenetic models, more stringent filtering, and taxon subsets did not substantially change our estimate of the mutation rate (electronic supplementary material, table S2). This estimate was also robust to completely rerunning the pipeline from raw reads trimmed to 53 bp to remove later positions with decreased read qualities and estimation of a new time-calibrated phylogeny (electronic supplementary material, figure S4 and table S2).
(e). Demographic modelling
Our time-calibrated phylogeny probably overestimates the age of young taxa such as diabolis owing to incomplete lineage sorting of only a single concatenated haplotype [35]. Therefore, we used demographic modelling in dadi [63] to estimate divergence time from allele frequencies. Rather than using an approximate simulation of the likelihood (e.g. [38]), dadi directly calculates the likelihood of the joint-site frequency spectrum under various demographic scenarios using a diffusion approximation [63].
Owing to the extreme degradation of diabolis samples, we were not able to obtain a good estimate of diabolis allele frequencies. Instead, we bounded the age of diabolis from the divergence time between the two main population clusters in Ash Meadows: mionectes and amargosae/shoshone/nevadensis (figure 2a–c) and the older divergence of the salinus/milleri outgroup. Our phylogenetic analyses place the diabolis split in between these two branches (figure 2) or within the mionectes group in the trimmed dataset (electronic supplementary material, figure S4). In both cases, the diabolis divergence time is very close to the mionectes and amargosae/shoshone/nevadensis divergence time. We then approximated the diabolis age by correcting for the relative difference in branch lengths. Nonetheless, this approximation is a weakness of our study and should be interpreted with caution.
3. Results
(a). Population structure and genetic diversity
We found evidence of genetic structure among five major groups in Death Valley: diabolis, salinus/milleri, mionectes, pectoralis and amargosae/shoshone/nevadensis (figure 2a–c; electronic supplementary material, figure S5). This is consistent with the expectation that genetic drift in small populations will accelerate lineage sorting and accumulation of genetic structure [64]. Indeed, Death Valley populations exhibited up to 40-fold lower genetic diversity than coastal Cyprinodon populations (table 1). The lowest diversity observed in pectoralis was equal to the lowest genetic diversity ever documented in a wild population ([24]: Lynx lynx) and 10-fold lower than a captive pupfish population extinct in the wild for over 15 years (table 1: Cyprinodon alvarezi [65]). Despite low diversity within each spring, if we pool all Death Valley populations into one large interconnected population, the genetic diversity represented in the entire valley is similar to high diversity coastal populations (table 1).
(b). Time-calibrated phylogeny
Our phylogeny places the stem divergence of Death Valley pupfishes at approximately 10 kyr ago (95% CI: 5–20 kyr ago; figure 2d; electronic supplementary material, figure S4). This interval is centered directly on the most recent flooding of the Death Valley basin from 10 to 35 kyr ago observed in core samples [20]. Death Valley pupfishes shared a common ancestor with Northeastern Mexican species approximately 17 kyr ago (95% CI: 9–34 kyr ago) and Atlantic coastal populations 25 kyr ago (95% CI: 12–41 kyr ago; figure 2d). These ages are consistent with increased population mixing expected from the formation of large pluvial lakes throughout North America (e.g. Palaeolake Manly: figure 1a) during the most recent glacial period 12–25 kyr ago [66]. It is not apparent how low-lying desert populations could have remained isolated within large inland seas because nearly all pupfish species readily interbreed on secondary contact and produce viable offspring (see Discussion).
(c). Demographic analysis of divergence time
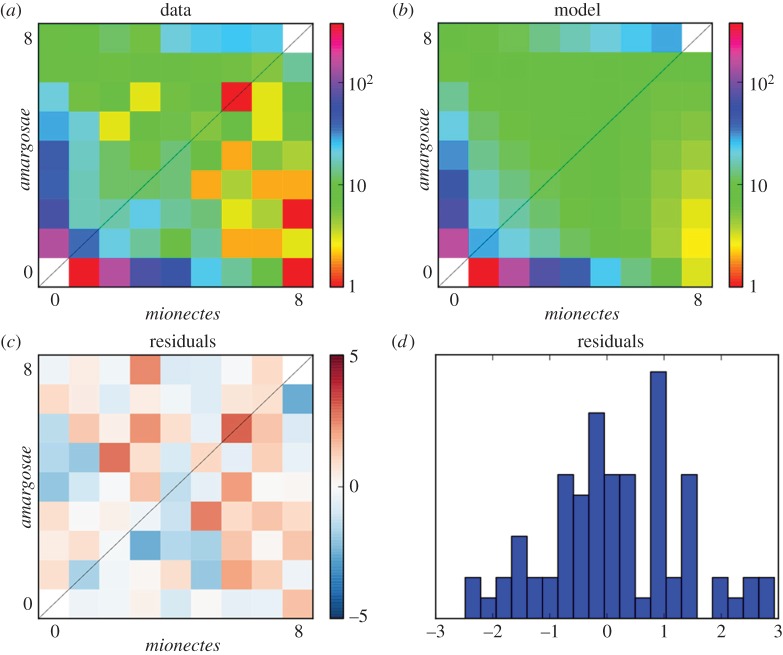
Our maximum-likelihood estimate of the divergence between amargosae/shoshone/nevadensis and mionectes was 210 years (95% CI: 60–363; figure 3; electronic supplementary material, table S2). Encouragingly, this interval contains the Great Flood of 1862, the largest recorded rainfall for California and Nevada [67], which probably flooded Ash Meadows and mixed mionectes with amargosae/shoshone/nevadensis populations downstream (figure 1a). We next compared the median divergence time of diabolis relative to the median divergence between mionectes and amargosae/shoshone/nevadensis in our phylogeny (figure 2d) to rescale an approximate age for diabolis based on our demographic model and our mutation rate estimate for pupfishes (electronic supplementary material, table S2: 5.37 × 10−7 site−1 yr−1). We estimate the age of diabolis to be 255 years (electronic supplementary material, table S2: 95% CI: 105–408), suggesting that the current population colonized only 159 years before the first historical record of Devils Hole pupfish in 1893 [21]. Alternatively, using the slowest and fastest mutation rates across the range of models and datasets we explored, diabolis ranged in age from 830 years to as young as 105 years (younger than the first historical record), respectively (electronic supplementary material, table S2: max and min 95% confidence boundaries).
Figure 3.
Demographic model of amargosae/shoshone/nevadensis and mionectes divergence time inferred from allele frequency data using dadi. (a) Unfolded joint site frequency spectrum collapsed to eight chromosomes, based on 578 557 sites. (b) Predicted site frequency spectrum under our demographic model. (c) Residuals from the demographic model across the site frequency spectrum. (d) Residuals were approximately normally distributed. (Online version in colour.)
(d). Recent gene flow
Unexpectedly, we also found evidence of recent gene flow between diabolis and both amargosae and pectoralis supported by formal tests for introgression (electronic supplementary material, table S4: amargosae D-statistic = 0.17, p = 0.0007; pectoralis D-statistic = 0.15, p = 0.002). Cyprinodon diabolis introgression was also consistent with Structure analyses (figure 2c) and Treemix (electronic supplementary material, figure S1). One amargosae individual contained a median of 1.6% diabolis ancestry overall (figure 2c); however, 95% credible intervals of diabolis ancestry proportions for this individual contained zero in seven out of eight independent Structure runs. Introgression from diabolis into amargosae was also supported by the residual covariance in allele frequencies between these populations estimated using Treemix (electronic supplementary material, figure S1).
The small amount of diabolis ancestry within amargosae (figure 2c), lack of significant introgression with other Death Valley and Ash Meadows populations besides pectoralis (electronic supplementary material, table S4), and directionality of gene flow inferred by Treemix (electronic supplementary material, figure S1) is consistent with gene flow from Devils Hole into Amargosa River sometime after the divergence of amargosae/shoshone/nevadensis and mionectes 210 years ago. Biologists transferred 75 diabolis to a spring in southern Ash Meadows in 1946 [68] which might explain the pectoralis signal, but does not appear to explain the amargosae signal since no introgression was observed in the intervening shoshone population.
Overall, genetic covariance among Death Valley populations indicates additional introgression from salinus and pectoralis into nevadensis (electronic supplementary material, figure S1) and suggests overland dispersal on 100-year timescales. Similarly, our demographic model supported significant low levels of migration between amargosae/shoshone/nevadensis and mionectes after divergence (electronic supplementary material, table S3). We also confirmed a previous study [69] which found that a refuge population of diabolis within a concrete pond hybridized with introduced mionectes (electronic supplementary material, figure S2: 85.3% mionectes ancestry), despite its isolation from neighbouring spring systems.
4. Discussion
Our population genomic analyses paint a dramatically different picture of survival in Death Valley pupfishes than previous assumptions. Rather than vicariance-driven speciation of isolated relicts surviving millions of years throughout flooding of the entire region lasting thousands of years (figure 1a), we found evidence for frequent overland dispersal and mixing of Death Valley pupfish populations on a scale of hundreds–thousands of years. Although overland dispersal across kilometres of desert by an aquatic organism is difficult to fathom, this view is much more consistent with established theories of population persistence and known hydrological connectivity over historical and geological timescales. For example, our time-calibrated phylogeny reasonably indicates that Death Valley pupfishes were isolated from other desert basins after the drying of the most recent pluvial lake in the region 10 kyr ago [20]. Our demographic analysis of the most recent mixing between two desert spring populations is coincident with the largest recorded rainfall in the area in 1862 [67] which would have flooded the dry riverbed connecting these two habitats (figure 1a). We explored a range of phylogenetic models which suggest that diabolis was isolated after salinus/milleri and only slightly earlier than mionectes and amargosae/shoshone/nevadensis, and bound its age between 105 and 830 years (min/max 95% confidence boundaries in the electronic supplementary material, table S2). This very recent origin is consistent with early reports of no unique mitochondrial haplotypes in diabolis relative to other Death Valley pupfishes in the region [33]. However, unbiased sampling of genomic diversity by genomic resequencing and divergence time estimation from the distribution of haplotype block lengths [70,71] are needed to conclusively estimate divergence time. Finally, expectations of rapidly increasing genetic load at the low population sizes frequently observed in desert springs [42], particularly Devils Hole, suggest that frequent dispersal among these populations may be necessary for their long-term persistence. For example, genetic diversity in Death Valley pupfishes was up to 40-fold lower than coastal populations and included the lowest genetic diversity ever observed in a wild population (table 1: Cyprinodon nevadensis pectoralis was equivalent to L. lynx [24]). Nonetheless, our results suggest that the stem lineage of pupfishes in Death Valley survived at least 5000 years before the appearance of extant populations (figure 2d), possibly by maintaining a stable metapopulation despite frequent local extirpations. This perspective may inform conservation strategies for these fishes.
The young age of diabolis is surprising given its large number of unique traits: reduced aggression, sexual dimorphism and size; larger eyes; darker coloration; and the absence of pelvic fins [21,22]. For example, although Death Valley pupfishes do not show exceptional rates of morphological diversification as in the San Salvador and Chichancanab sympatric radiations (which diversified up to 130 times faster than background rates for certain jaw traits [54]), morphological diversification of the Death Valley clade containing diabolis was five times faster than background rates in other pupfishes [54], comparable to rates observed for similar traits in coral reef fishes [72] and Malawi cichlids [73] over longer timescales. Some diabolis-like traits were observed in amargosae when reared in the laboratory at 32°C on a restricted diet to simulate conditions in Devils Hole, including pelvic fin loss in up to 86% of fish [41]. However, laboratory-rearing experiments indicate that pelvic fin loss in diabolis has a genetic basis (electronic supplementary material, figure S3). Combined, these observations suggest that colonists of Devils Hole initially lost pelvic fin expression as a plastic response to higher temperatures and reduced resources, only later evolving a broader norm of reaction for pelvic fin loss at lower temperatures. This process, known as genetic assimilation, has rarely been demonstrated in nature [74,75]. Pelvic fin loss may be adaptive in a benthic habitat, as suggested for stickleback [37]. However, the haphazard pelvic fin loss observed in several other lineages of Cyprinodontiform fishes with no known environmental commonalities (including Empetrichthyids, Megupsilon, Orestias, two Rivulus, one Simpsonichthys and one Aphanius species) is consistent with genetic drift.
Our study adds to an emerging consensus that species ages may frequently be overestimated by time-calibrated mtDNA phylogenies [53,70,76]. There appear to be three main reasons. First, similar demographic modelling studies have consistently found that species divergence times were overestimated in mtDNA phylogenies by millions of years, e.g. in flycatchers [76], due to a systematic bias when sampling a single haplotype per species. Similarly, molecular estimates for the age of polar bears spanned 600 kyr–5 Myr ago using conventional demographic modelling approaches, but were revised to 400 kyr ago, consistent with the fossil evidence, by modelling with dadi [70]. Second, many studies calibrate with distantly related outgroup fossils or ancient vicariance events, such as the breakup of Gondwanaland, assuming no subsequent dispersal was possible [30,32,77]. For example, the age of the modern bowhead whale was revised from 1.1 Myr ago to 150 kyr ago (1 order of magnitude, similar to this study) when calibrating with radiocarbon dates for ancient DNA samples rather than outgroup fossils [36]. Third, substitution rates appear to be at least an order of magnitude slower in older lineages in both coding and non-coding regions, possibly owing to purifying selection or saturation of mutational hotspots, resulting in the overestimation of species ages when these rates are used to calibrate more recently diverged taxa [78,79]. Estimates based on only the single mitochondrial haplotype also contain substantially more uncertainty than multi-locus nDNA estimates.
Overestimation of species divergence times challenges existing assumptions about the timescale for speciation. For example, current models assume that the accumulation of postzygotic intrinsic isolation takes millions of years due to substantial work in many groups [80,81], including pupfishes [82,83]. Nearly all studies of rapid speciation in fishes report no intrinsic incompatibilities in hybrid crosses [84–87], most notably in East African cichlid clades diversifying over millions of years [88]. These assumptions have inspired recent models of diversification, such as ephemeral speciation [89] and the million-year-waiting-time for macroevolutionary bursts [90]. However, these long timescales conflict with observations of reproductive incompatibilities segregating in natural populations [91] which predict much shorter times for the evolution of intrinsic isolation [92,93]. Indeed, our results suggest that intrinsic genetic incompatibilities documented in two different pupfish species [94,95] evolved within less than 30 000 years. Similarly, recent studies indicate that non-lethal intrinsic genetic incompabitilities abound after secondary contact in both swordtail fishes [96] and hominids [93], suggesting that traditional progeny screens are insufficient for detecting subtle incompatibilities. Overall, our study points to the evolution of intrinsic incompatibilities, phenotypic change, and genetic assimilation on extremely rapid timescales, challenging existing models of speciation [89,90].
Our results also provide context for the management of highly inbred wild and captive populations [17,97] and the recent decline of diabolis. High egg survival rates in captivity ([98]: 55%), rebounding population sizes [23], and lower genetic diversity in neighbouring stable populations (table 1) all suggest that the decline is not owing to an exceptional genetic load causing mutational meltdown as previously speculated [69]. However, theory predicts that extinction due to increasing genetic load is highly likely for this small population over timescales of 100 years or less [13,42]. Combined with the 60 000 year age of Devils Hole, this suggests that pupfish extinction and gene flow may have happened many times before in this unique habitat and that diabolis may not be the first pupfish population to have survived there. Protecting the connectivity of this region will be essential for this cycle of rebirth to continue.
Supplementary Material
Acknowledgements
We thank Anthony Echelle for supplying tissue samples from Indian Spring and the refuge diabolis population; Kevin Wilson, Bailey Gaines, and Jeffrey Goldstein for providing access to preserved diabolis specimens; Patrik Nosil, Anthony Echelle, and two anonymous reviewers for insightful comments on the manuscript. We thank the National Park Service and the US Fish and Wildlife Service for permission to conduct this research.
Data accessibility
All datasets (SNP matrices) used for this study are deposited in Dryad: http://dx.doi.org/10.5061/dryad.s30t4.
Authors' contributions
C.H.M. designed the study, prepared samples for sequencing, conducted phylogenetic and population genetic analyses, and wrote the manuscript. J.E.C. ran demographic analyses with dadi and calculated D-statistics. L.H.S. provided logistical support, funding, samples, and motivation for the study. B.J.T. collected most of the samples. All authors commented on the manuscript and gave final approval for publication.
Competing interests
We have no competing interests.
Funding
This study was funded by a research stipend from the Miller Institute for Basic Research in the Sciences to C.H.M. and a small informal expense fund from the National Park Service.
Disclaimer
The findings and conclusions in this article are those of the author(s) and do not necessarily represent the views of the US Fish and Wildlife Service.
References
- 1.Tobler M, Schlupp I, Heubel KU, Riesch R, de León FJG, Giere O, Plath M. 2006. Life on the edge: hydrogen sulfide and the fish communities of a Mexican cave and surrounding waters. Extremophiles 10, 577–585. ( 10.1007/s00792-006-0531-2) [DOI] [PubMed] [Google Scholar]
- 2.Schliewen UK, Tautz D, Pääbo S. 1994. Sympatric speciation suggested by monophyly of crater lake cichlids. Nature 6472, 629–632. ( 10.1038/368629a0) [DOI] [PubMed] [Google Scholar]
- 3.Wagner CE, Harmon LJ, Seehausen O. 2012. Ecological opportunity and sexual selection together predict adaptive radiation. Nature 487, 366–369. ( 10.1038/nature11144) [DOI] [PubMed] [Google Scholar]
- 4.Gavrilets S, Losos JB. 2009. Adaptive radiation: contrasting theory with data. Science 323, 732–737. ( 10.1126/science.1157966) [DOI] [PubMed] [Google Scholar]
- 5.Losos J, Mahler D. 2010. Adaptive radiation: the interaction of ecological opportunity, adaptation, and speciation. In Evolution since Darwin: the first 150 years, pp. 381–420. Sunderland, MA: Sinauer Associates; (cited 12 Jan 2015). [Google Scholar]
- 6.Lenski RE, Travisano M. 1994. Dynamics of adaptation and diversification: a 10,000-generation experiment with bacterial populations. Proc. Natl Acad. Sci. USA 91, 6808–6814. ( 10.1073/pnas.91.15.6808) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rainey P, Travisano M. 1998. Adaptive radiation in a heterogeneous environment. Nature 394, 69–72. ( 10.1038/27900) [DOI] [PubMed] [Google Scholar]
- 8.Bono LM, Gensel CL, Pfennig DW, Burch CL. 2013. Competition and the origins of novelty: experimental evolution of niche-width expansion in a virus. Biol. Lett. 9, 20120616 ( 10.1098/rsbl.2012.0616) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blount ZD, Borland CZ, Lenski RE. 2008. Historical contingency and the evolution of a key innovation in an experimental population of Escherichia coli. Proc. Natl Acad. Sci. USA 105, 7899–7906. ( 10.1073/pnas.0803151105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin C. 2012. Weak disruptive selection and incomplete pheonytpic divergence in two classic examples of sympatric speciation: Cameroon crater lake cichlids. Am. Nat. 180, E90–E109. ( 10.1086/667586) [DOI] [PubMed] [Google Scholar]
- 11.Martin CH. 2013. Strong assortative mating by diet, color, size, and morphology but limited progress toward sympatric speciation in a classic example: Cameroon crater lake cichlids. Evolution 67, 2114–2123. ( 10.1111/evo.12090) [DOI] [PubMed] [Google Scholar]
- 12.Papadopulos AST, et al. 2014. Evaluation of genetic isolation within an island flora reveals unusually widespread local adaptation and supports sympatric speciation. Phil. Trans. R. Soc. B 369, 20130342 ( 10.1098/rstb.2013.0342) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lynch M, Conery J, Burger R. 1995. Mutation accumulation and the extinction of small populations. Am. Nat. 146, 489–518. ( 10.1086/285812) [DOI] [Google Scholar]
- 14.Lanfear R, Kokko H, Eyre-Walker A. 2014. Population size and the rate of evolution. Trends Ecol. Evol. 29, 33–41. ( 10.1016/j.tree.2013.09.009) [DOI] [PubMed] [Google Scholar]
- 15.Bijlsma R, Bundgaard J, Boerema AC. 2000. Does inbreeding affect the extinction risk of small populations? predictions from Drosophila. J. Evol. Biol. 13, 502–514. ( 10.1046/j.1420-9101.2000.00177.x) [DOI] [Google Scholar]
- 16.Willi Y, Van Buskirk J, Hoffmann AA. 2006. Limits to the adaptive potential of small populations. Annu. Rev. Ecol. Evol. Syst. 37, 433–458. ( 10.1146/annurev.ecolsys.37.091305.110145) [DOI] [Google Scholar]
- 17.Kennedy ES, Grueber CE, Duncan RP, Jamieson IG. 2014. Severe inbreeding depression and no evidence of purging in an extremely inbred wild species: the Chatham Island black robin. Evolution 68, 987–995. ( 10.1111/evo.12315) [DOI] [PubMed] [Google Scholar]
- 18.Eldridge MDB, King JM, Loupis AK, Spencer PBS, Taylor AC, Pope LC, Hall GP. 1999. Unprecedented low levels of genetic variation and inbreeding depression in an island population of the black-footed rock-wallabi. Conserv. Biol. 13, 531–541. ( 10.1046/j.1523-1739.1999.98115.x) [DOI] [Google Scholar]
- 19.O'Grady JJ, Brook BW, Reed DH, Ballou JD, Tonkyn DW, Frankham R. 2006. Realistic levels of inbreeding depression strongly affect extinction risk in wild populations. Biol. Conserv. 133, 42–51. ( 10.1016/j.biocon.2006.05.016) [DOI] [Google Scholar]
- 20.Lowenstein T, Li J, Brown C, Roberts S. 1999. 200 ky paleoclimate record from Death Valley salt core. Geology 27, 3–6. () [DOI] [Google Scholar]
- 21.Deacon JE, Taylor FR, Pedretti JW, Pedretti W. 1995. Egg viability and ecology of Devils Hole pupfish: insights from captive propagation. Southwest. Nat. 40, 216–223. [Google Scholar]
- 22.Riggs A, Deacon J. 2002. Connectivity in desert aquatic ecosystems: the Devils Hole story. In Proc. Spring-fed Wetl. Important Sci. Cult. Resour. Intermt. Reg., 7–9 May 2002, Las Vegas, Nevada (eds DW Sada, SE Sharpe). DHS Publication no. 41210. [Google Scholar]
- 23.Beissinger SR. 2014. Digging the pupfish out of its hole: risk analyses to guide harvest of Devils Hole pupfish for captive breeding. PeerJ 2, e549 ( 10.7717/peerj.549) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilson KP, Blinn DW. 2007. Food web structure, energetics, and importance of allochthonous carbon in a desert cavernous limnocrene: Devils Hole, Nevada. West. North Am. Nat. 67, 185–198. ( 10.3398/1527-0904(2007)67%5B185:FWSEAI%5D2.0.CO;2) [DOI] [Google Scholar]
- 25.Heath AG, Turner BJ, Davis WP. 1993. Temperature preferences and tolerances of three fish species inhabiting hyperthermal ponds on mangrove islands. Hydrobiologia 259, 47–55. ( 10.1007/BF00005964) [DOI] [Google Scholar]
- 26.Bennett W, Beitinger T. 1997. Temperature tolerance of the sheepshead minnow, Cyprinodon variegatus. Copeia 1997, 77–87. ( 10.2307/1447842) [DOI] [Google Scholar]
- 27.Carson EW, De la Maza-Benignos M, de Lourdes Lozano-Vilano M, Vela-Valladares L, Banda-Villanueva I, Turner TF. 2013. Conservation genetic assessment of the critically endangered Julimes pupfish, Cyprinodon julimes. Conserv. Genet. 15, 483–488. ( 10.1007/s10592-013-0548-x) [DOI] [Google Scholar]
- 28.Minckley W, Minckley C. 1986. Cyprinodon pachycephalus, a new species of pupfish (Cyprinodontidae) from the Chihuahuan desert of northern Mexico. Copeia 1986, 184–192. ( 10.2307/1444904) [DOI] [Google Scholar]
- 29.Hausner M, Wilson K, Gaines D, Suarez F, Scoppettone G, Tyler S. 2014. Life in a fishbowl: prospects for the endangered Devils Hole pupfish (Cyprinodon diabolis) in a changing climate. Water Resour. Res. 50, 7020–7034. ( 10.1002/2014WR015511) [DOI] [Google Scholar]
- 30.Echelle AA, Carson EW, Echelle AF, Van Den Bussche RA, Dowling TE, Meyer A. 2005. Historical biogeography of the New-World pupfish genus Cyprinodon (Teleostei: Cyprinodontidae). Copeia 2005, 320–339. ( 10.1643/CG-03-093R3) [DOI] [Google Scholar]
- 31.Echelle A. 2008. The western North American pupfish clade (Cyprinodontidae: Cyprinodon): mitochondrial DNA divergence and drainage history. Geol. Soc. Am. Spec. Pap. 439, 27–38. ( 10.1130/2008.2439(02)) [DOI] [Google Scholar]
- 32.Hrbek T, Meyer A. 2003. Closing of the Tethys Sea and the phylogeny of Eurasian killifishes (Cyprinodontiformes: Cyprinodontidae). J. Evol. Biol. 16, 17–36. ( 10.1046/j.1420-9101.2003.00475.x) [DOI] [PubMed] [Google Scholar]
- 33.Echelle A, Dowling T. 1992. Mitochondrial DNA variation and evolution of the Death Valley pupfishes (Cyprinodon, Cyprinodontidae). Evolution (N. Y). 46, 193–206. ( 10.2307/2409814) [DOI] [PubMed] [Google Scholar]
- 34.Winograd IJ, Coplen TB, Landwehr JM, Riggs AC, Ludwig KR, Szabo BJ, Kolesar PT, Revesz KM. 1992. Continuous 500,000-year climate record from vein calcite in Devils Hole, Nevada. Science 258, 255–260. ( 10.1126/science.258.5080.255) [DOI] [PubMed] [Google Scholar]
- 35.Heled J, Drummond AJ. 2010. Bayesian inference of species trees from multilocus data. Mol. Biol. Evol. 27, 570–580. ( 10.1093/molbev/msp274) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ho SYW, Saarma U, Barnett R, Haile J, Shapiro B. 2008. The effect of inappropriate calibration: three case studies in molecular ecology. PLoS ONE 3, e1615 ( 10.1371/journal.pone.0001615) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Warnock RCM, Parham JF, Joyce WG, Lyson TR, Donoghue PCJ. 2015. Calibration uncertainty in molecular dating analyses: there is no substitute for the prior evaluation of time priors. Proc. R. Soc. B 282, 20141013 ( 10.1098/rspb.2014.1013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reed J, Stockwell C. 2014. Evaluating an icon of population persistence: the Devil's Hole pupfish. Proc. R. Soc. B 281, 1648–1658. ( 10.1098/rspb.2014.1648) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ellegren H. 2004. Microsatellites: simple sequences with complex evolution. Nat. Rev. Genet. 5, 435–445. ( 10.1038/nrg1348) [DOI] [PubMed] [Google Scholar]
- 40.Smith G, Dowling T, Gobalet K, Lugaski T, Shiozawa D, Evans P. 2002. Biogeography and rates of evolution of Great Basin fishes. In Great Basin aquatic systems history: Smithsonian contributions to the earth sciences (eds Hershler R, Madsen D, Curry D), pp. 175–234. New York, NY: Food and Agriculture Organization of the United Nations. [Google Scholar]
- 41.Lema S, Nevitt G. 2006. Testing an ecophysiological mechanism of morphological plasticity in pupfish and its relevance to conservation efforts for endangered Devils Hole pupfish. J. Exp. Biol. 209, 3499–3509. ( 10.1242/jeb.02417) [DOI] [PubMed] [Google Scholar]
- 42.Lynch M, Conery J, Burger R. 1995. Mutational meltdowns in sexual populations. Evolution 49, 1067–1080. ( 10.2307/2410432) [DOI] [PubMed] [Google Scholar]
- 43.Peterson BK, Weber JN, Kay EH, Fisher HS, Hoekstra HE. 2012. Double digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS ONE 7, e37135 ( 10.1371/journal.pone.0037135) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duvernell DD, Turner BJ. 1998. Variation and divergence of death valley pupfish populations at retrotransposon-defined loci. Mol. Biol. Evol. 16, 363–371. ( 10.1093/oxfordjournals.molbev.a026117) [DOI] [Google Scholar]
- 45.Martin CH, Cutler JS, Friel JP, Dening T, Coop G, Wainwright PC. 2015. Complex histories of repeated colonization and hybridization cast doubt on the clearest examples of sympatric speciation in the wild. Evolution (N. Y). 69, 1406–1422. [DOI] [PubMed] [Google Scholar]
- 46.Catchen J, Hohenlohe PA, Bassham S, Amores A, Cresko WA. 2013. Stacks: an analysis tool set for population genomics. Mol. Ecol. 22, 3124–3140. ( 10.1111/mec.12354) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155, 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heliconius T, Consortium G. 2012. Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature 487, 94–98. ( 10.1038/nature11041) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pickrell JK, Pritchard JK. 2012. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 8, e1002967 ( 10.1371/journal.pgen.1002967) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Willing E-M, Dreyer C, van Oosterhout C. 2012. Estimates of genetic differentiation measured by F(ST) do not necessarily require large sample sizes when using many SNP markers. PLoS ONE 7, e42649 ( 10.1371/journal.pone.0042649) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 8, 1–8. ( 10.1186/1471-2148-7-214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ho SYW, Phillips MJ. 2009. Accounting for calibration uncertainty in phylogenetic estimation of evolutionary divergence times. Syst. Biol. 58, 367–380. ( 10.1093/sysbio/syp035) [DOI] [PubMed] [Google Scholar]
- 53.Ho SYW, Phillips MJ, Cooper A, Drummond AJ. 2005. Time dependency of molecular rate estimates and systematic overestimation of recent divergence times. Mol. Biol. Evol. 22, 1561–1568. ( 10.1093/molbev/msi145) [DOI] [PubMed] [Google Scholar]
- 54.Martin CH, Wainwright PC. 2011. Trophic novelty is linked to exceptional rates of morphological diversification in two adaptive radiations of Cyprinodon pupfish. Evolution 65, 2197–2212. ( 10.1111/j.1558-5646.2011.01294.x) [DOI] [PubMed] [Google Scholar]
- 55.Covich A, Stuiver M. 1974. Changes in the oxygen 18 as a measure of long-term fluctuations in tropical lake levels and molluscan populations. Limnol. Oceanogr. 19, 682–691. ( 10.4319/lo.1974.19.4.0682) [DOI] [Google Scholar]
- 56.Hodell D, Curtis J, Brenner M. 1995. Possible role of climate in the collapse of Classic Maya civilization. Nature 375, 391–394. ( 10.1038/375391a0) [DOI] [Google Scholar]
- 57.Humphries J, Miller RR. 1981. A remarkable species flock of pupfishes, genus Cyprinodon, from Yucatan, Mexico. Copeia 1981, 52–64. ( 10.2307/1444040) [DOI] [Google Scholar]
- 58.Strecker U. 2006. Characterization and cross-species amplification of microsatellite loci in a Cyprinodon species flock. Mol. Phylogenet. Evol. 39, 843–846. ( 10.1016/j.ympev.2006.01.008) [DOI] [PubMed] [Google Scholar]
- 59.Strecker U, Meyer CG, Sturmbauer C, Wilkens H. 1996. Genetic divergence and speciation in an extremely young species flock in Mexico formed by the genus Cyprinodon (Cyprinodontidae, Teleostei). Mol. Phylogenet. Evol. 6, 143–149. ( 10.1006/mpev.1996.0066) [DOI] [PubMed] [Google Scholar]
- 60.Strecker U. 2006. The impact of invasive fish on an endemic Cyprinodon species flock (Teleostei) from Laguna Chichancanab, Yucatan, Mexico. Ecol. Freshw. Fish 15, 408–418. ( 10.1111/j.1600-0633.2006.00159.x) [DOI] [Google Scholar]
- 61.Scally A, Durbin R. 2012. Revising the human mutation rate: implications for understanding human evolution. Nat. Rev. Genet. 13, 824 ( 10.1038/nrg3353) [DOI] [PubMed] [Google Scholar]
- 62.Brix KV, Grosell M. 2012. Comparative characterization of Na+ transport in Cyprinodon variegatus variegatus and Cyprinodon variegatus hubbsi: a model species complex for studying teleost invasion of freshwater. J. Exp. Biol. 215, 1199–1209. ( 10.1242/jeb.067496) [DOI] [PubMed] [Google Scholar]
- 63.Gutenkunst R, Hernandez R. 2009. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 5, e1000695 ( 10.1371/journal.pgen.1000695) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fisher R. 1930. Genetical theory of natural selection. Oxford, UK: Clarendon Press. [Google Scholar]
- 65.Contreras-Balderas S, Almada-Villela P.1996. Cyprinodon alvarezi. The IUCN Red List of Threatened Species. Gland, Switzerland: International Union for Conservation of Nature.
- 66.Easterbrook D. 1999. Surface processes and landforms. Upper Saddle River, NJ: Prentice-Hall, Inc. [Google Scholar]
- 67.Brewer W. 1930. Up and down California in 1860–1864. New Haven, CT: Yale University Press. [Google Scholar]
- 68.Rivers IR. 1950. The Dryopoidea known or expected to occur in the Nevada Area (Coleoptera). Wasmann J. Biol. 8, 97–112. [Google Scholar]
- 69.Martin AP, Echelle AA, Zegers G, Baker S, Keeler-Foster CL. 2011. Dramatic shifts in the gene pool of a managed population of an endangered species may be exacerbated by high genetic load. Conserv. Genet. 13, 349–358. ( 10.1007/s10592-011-0289-7) [DOI] [Google Scholar]
- 70.Liu S. et al. . 2014. Population genomics reveal recent speciation and rapid evolutionary adaptation in polar bears. Cell 157, 785–794. ( 10.1016/j.cell.2014.03.054) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Harris K, Nielsen R. 2013. Inferring demographic history from a spectrum of shared haplotype lengths. PLoS Genet. 9, e1003521 ( 10.1371/journal.pgen.1003521) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Price SA, Holzman R, Near TJ, Wainwright PC. 2011. Coral reefs promote the evolution of morphological diversity and ecological novelty in labrid fishes. Ecol. Lett. 14, 462–469. ( 10.1111/j.1461-0248.2011.01607.x) [DOI] [PubMed] [Google Scholar]
- 73.Hulsey CD, Mims MC, Parnell NF, Streelman JT. 2010. Comparative rates of lower jaw diversification in cichlid adaptive radiations. J. Evol. Biol. 23, 1456–1467. ( 10.1111/j.1420-9101.2010.02004.x) [DOI] [PubMed] [Google Scholar]
- 74.Pfennig DW, Wund MA, Snell-Rood EC, Cruickshank T, Schlichting CD, Moczek AP. 2010. Phenotypic plasticity's impacts on diversification and speciation. Trends Ecol. Evol. 25, 459–467. ( 10.1016/j.tree.2010.05.006) [DOI] [PubMed] [Google Scholar]
- 75.Braendle C, Flatt T. 2006. A role for genetic accommodation in evolution? BioEssays 28, 868–873. ( 10.1002/bies.20456) [DOI] [PubMed] [Google Scholar]
- 76.Nadachowska-Brzyska K, Burri R, Olason PI, Kawakami T, Smeds L, Ellegren H. 2013. Demographic divergence history of pied flycatcher and collared flycatcher inferred from whole-genome re-sequencing data. PLoS Genet. 9, e1003942 ( 10.1371/journal.pgen.1003942) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Friedman M, Keck BP, Dornburg A, Eytan RI, Martin CH, Hulsey CD, Wainwright PC, Near TJ. 2013. Molecular and fossil evidence place the origin of cichlid fishes long after Gondwanan rifting. Proc. R. Soc. B 280, 20131733 ( 10.1098/rspb.2013.1733) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Woodhams M. 2006. Can deleterious mutations explain the time dependency of molecular rate estimates? Mol. Biol. Evol. 23, 2271–2273. ( 10.1093/molbev/msl107) [DOI] [PubMed] [Google Scholar]
- 79.Ho SYW, Lanfear R, Bromham L, Phillips MJ, Soubrier J, Rodrigo AG, Cooper A. 2011. Time-dependent rates of molecular evolution. Mol. Ecol. 20, 3087–3101. ( 10.1111/j.1365-294X.2011.05178.x) [DOI] [PubMed] [Google Scholar]
- 80.Bolnick DI, Turelli M, López-Fernández H, Wainwright PC, Near TJ. 2008. Accelerated mitochondrial evolution and ‘Darwin's corollary’: asymmetric viability of reciprocal F1 hybrids in Centrarchid fishes. Genetics 178, 1037–1048. ( 10.1534/genetics.107.081364) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Coyne JA, Orr HA. 2004. Speciation. Sunderland, MA: Sinauer Associates. [Google Scholar]
- 82.Turner B, Liu R. 1977. Extensive interspecific genetic compatibility in the New World killifish genus Cyprinodon. Copeia 1977, 259–269. ( 10.2307/1443907) [DOI] [Google Scholar]
- 83.Cokendolpher JC. 2012. Hybridization experiments with the genus Cyprinodon (Teleostei: Cyprinodontidae), vol. 1980, pp. 173–176. American Society of Ichthyologists and Herpetologists (http://www.jstor.org/s).
- 84.Martin CH, Wainwright PC. 2013. Multiple fitness peaks on the adaptive landscape drive adaptive radiation in the wild. Science 339, 208–211. ( 10.1126/science.1227710) [DOI] [PubMed] [Google Scholar]
- 85.Arnegard ME. et al. . 2014. Genetics of ecological divergence during speciation. Nature 511, 307–311. ( 10.1038/nature13301) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schluter D. 2000. Ecology of adaptive radiation. Oxford, UK: Oxford University Press. [Google Scholar]
- 87.Seehausen O, Wagner CE. 2013. Speciation in freshwater fishes. Annu. Rev. Ecol. Evol. Syst. 45, 621–651. ( 10.1146/annurev-ecolsys-120213-091818) [DOI] [Google Scholar]
- 88.Stelkens RB, Young KA, Seehausen O. 2010. The accumulation of reproductive incompatibilities in African cichlid fish. Evolution 64, 617–633. ( 10.1111/j.1558-5646.2009.00849.x) [DOI] [PubMed] [Google Scholar]
- 89.Rosenblum EB, Sarver BAJ, Brown JW, Des Roches S, Hardwick KM, Hether TD, Eastman JM, Pennell MW, Harmon LJ. 2012. Goldilocks meets Santa Rosalia: an ephemeral speciation model explains patterns of diversification across time scales. Evol. Biol. 39, 255–261. ( 10.1007/s11692-012-9171-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Uyeda JC, Hansen TF, Arnold SJ, Pienaar J. 2011. The million-year wait for macroevolutionary bursts. ( 10.5061/dryad.7d580) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cutter AD. 2012. The polymorphic prelude to Bateson-Dobzhansky-Muller incompatibilities. Trends Ecol. Evol. 27, 209–218. ( 10.1016/j.tree.2011.11.004) [DOI] [PubMed] [Google Scholar]
- 92.Wright KM, Lloyd D, Lowry DB, Macnair MR, Willis JH. 2013. Indirect evolution of hybrid lethality due to linkage with selected locus in Mimulus guttatus. PLoS Biol. 11, e1001497 ( 10.1371/journal.pbio.1001497) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sankararaman S, Mallick S, Dannemann M, Prüfer K, Kelso J, Pääbo S, Patterson N, Reich D. 2014. The genomic landscape of Neanderthal ancestry in present-day humans. Nature 507, 354–357. ( 10.1038/nature12961) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tech C. 2006. Postzygotic incompatibilities between the pupfishes, Cyprinodon elegans and Cyprinodon variegatus: hybrid male sterility and sex ratio bias. J. Evol. Biol. 19, 1830–1837. ( 10.1111/j.1420-9101.2006.01173.x) [DOI] [PubMed] [Google Scholar]
- 95.Haas R. 1979. Intergeneric hybridization in a sympatric pair of Mexican cyprinodontid fishes. Copeia 1979, 149–152. ( 10.2307/1443742) [DOI] [Google Scholar]
- 96.Schumer M, Cui R, Powell DL, Dresner R, Rosenthal GG, Andolfatto P. 2014. High-resolution mapping reveals hundreds of genetic incompatibilities in hybridizing fish species. Elife 3, 1–21. ( 10.7554/eLife.02535) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hedrick P, Kalinowski S. 2000. Inbreeding depression in conservation biology. Annu. Rev. Ecol. Syst. 31, 139–162. ( 10.1146/annurev.ecolsys.31.1.139) [DOI] [Google Scholar]
- 98.Feuerbacher O, Weissenfluh D, Chaudoin A, Barlics R, Simons L, Linares-Casenave J. 2014. Ash Meadows fish conservation facility: establishing a Devils Hole pupfish population. Proc. Desert Fishes Counc. Ann. Meet., 21 November, San Jose de Cabo, Baja, California Sur, Mexico. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets (SNP matrices) used for this study are deposited in Dryad: http://dx.doi.org/10.5061/dryad.s30t4.