Abstract
Species' functional roles in key ecosystem processes such as predation, pollination or seed dispersal are determined by the resource use of consumer species. An interaction between resource and consumer species usually requires trait matching (e.g. a congruence in the morphologies of interaction partners). Species' morphology should therefore determine species' functional roles in ecological processes mediated by mutualistic or antagonistic interactions. We tested this assumption for Neotropical plant–bird mutualisms. We used a new analytical framework that assesses a species's functional role based on the analysis of the traits of its interaction partners in a multidimensional trait space. We employed this framework to test (i) whether there is correspondence between the morphology of bird species and their functional roles and (ii) whether morphologically specialized birds fulfil specialized functional roles. We found that morphological differences between bird species reflected their functional differences: (i) bird species with different morphologies foraged on distinct sets of plant species and (ii) morphologically distinct bird species fulfilled specialized functional roles. These findings encourage further assessments of species' functional roles through the analysis of their interaction partners, and the proposed analytical framework facilitates a wide range of novel analyses for network and community ecology.
Keywords: ecological networks, fleshy-fruited plants, frugivorous birds, functional diversity, mutualistic interactions, traits
1. Introduction
Many ecological processes, such as predation, pollination or seed dispersal, involve species interactions. These interactions are usually governed by the trait matching between interaction partners in food webs and other types of ecological networks [1–6]: species preferentially interact with the species whose trait combinations match best because it allows them to exploit resources most efficiently [7,8]. By contrast, incongruence in traits might render an interaction impossible (forbidden link hypothesis) [9]. On the level of species assemblages, the trait matching between individual interacting species should translate into a covariation in corresponding traits in the interacting species groups [5], either because the groups have coevolved or because species from one group can only occur where they find suitable interaction partners with matching morphology (ecological fitting) [10].
Species morphology is often used as a surrogate for species' functional roles in ecological assemblages. For instance, analyses of functional diversity [11–14] assume that differences in species' resource use are reflected in species morphology [14,15]. However, the link between the morphology of species and their functional roles in ecological assemblages is still not fully understood [14,16]. In predator–prey relationships or mutually beneficial interactions, the functional role of a species is defined by its interaction with other species in the sense of an Eltonian niche [17,18]; species that interact with different sets of species fulfil different functional roles in a species assemblage (e.g. they forage on small or large prey items or pollinate different kinds of flowers). Hence, in these types of interactions, it should be possible to characterize a species's role by the traits of its interaction partners (e.g. by the morphology of its prey or resource species). Ecosystem processes that are mediated by species interactions are therefore suitable systems to investigate whether species' morphologies reflect their functional roles, because one can compare the traits of species with the traits of their interaction partners.
Morphologically specialized species are supposed to fulfil specialized functions in species assemblages [19,20], and there is indication that the loss of morphologically specialized species may cause the loss of distinct functional roles in species assemblages [21–24]. The specialization of species, however, is usually only assessed by the degree to which a species's morphology differs from those of other species in the species assemblage (species originality) or from that of the most similar species in the assemblage (species uniqueness), but not by the originality and uniqueness of its functional role. Although the inference of functional specialization from morphological specialization would be very valuable for the identification of key species in an assemblage [25], the relationship between morphological and functional specialization has never been investigated.
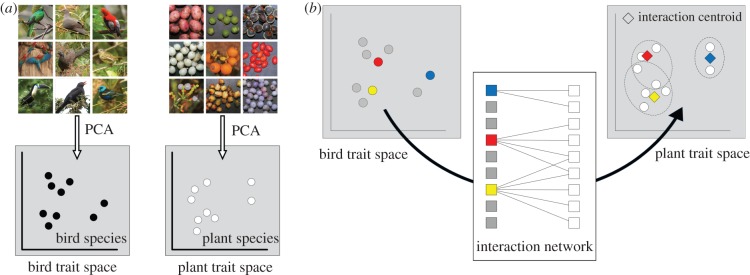
Here, we present a new analytical framework for assessing species' functional roles that combines methods from functional diversity research and network analysis (figures 1 and 2). The framework characterizes the functional role of a species by analysing the traits of species' interaction partners in a multidimensional trait space (figure 1). The framework can be applied to all types of mutualistic and antagonistic interactions, and we tested it by investigating the functional roles in two diversified interacting species groups, Neotropical frugivorous birds and fleshy-fruited plants, using an extensive dataset of plant–frugivore interactions from the tropical Andes [5]. These two groups are well suited for this analysis, because the functional roles of birds and plants are mutually beneficial [26] and involve several co-adaptations in bird and plant traits (e.g. between beak size and fruit size, body mass and fruit availability, and wing shape and foraging layer) [5,24,27–32]. We tested the relationships between (i) the morphology of bird species and their functional roles in plant–frugivore networks (figure 2a,b), and between (ii) the morphological and functional specialization of bird species (figure 2c,d). We hypothesized that (i) the morphology of bird species reflect their functional roles (i.e. which plant species they consume), and that (ii) morphologically specialized bird species fulfil specialized functional roles in the species assemblage (i.e. they forage on plant species that few other species consume).
Figure 1.
A new analytical framework for assessing species' functional roles by analysing the traits of species' interaction partners. (a) Bird and plant species are projected into their respective multidimensional trait spaces according to their morphological differences. (b) Using data on plant–bird interactions, bird species (exemplified by the red, yellow and blue circles in bird trait space) are projected into the plant trait space by calculating their interaction centroids (i.e. the mean position of the plant species that each bird species consumes, symbolized by the red, yellow and blue diamonds in the plant trait space). The position of the bird interaction centroids in plant trait space can then be analysed in the same way as the position of bird species in bird trait space. All photos © Matthias Dehling, CC license does not apply.
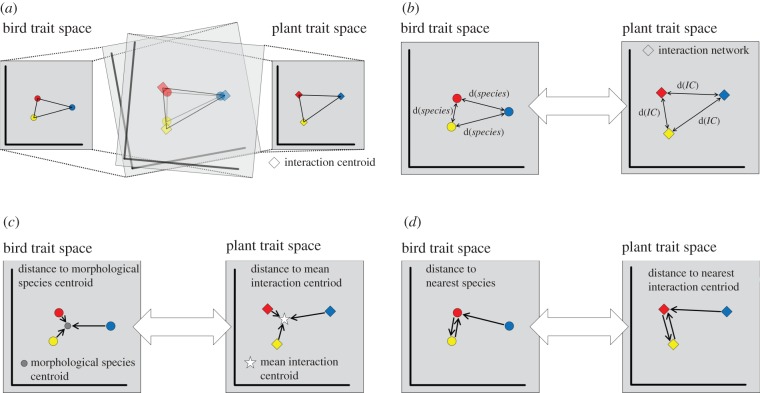
Figure 2.
Comparison between morphology and functional roles (a,b) and between morphological and functional specialization (c,d) of bird species. (a) The relative position of bird species in bird trait space (the red, yellow and blue circles) and the relative position of their interaction centroids in plant trait space (the red, yellow and blue diamonds) are compared with Procrustes rotation which superimposes two ordinations until the best fit is reached. (b) The distances between bird species in bird trait space, d(species), and the distances between their interaction centroids, d(IC), in plant trait space are compared with Mantel tests. (c) The morphological originality of a bird species (the distance from a species to the mean position of all species in bird trait space) is compared with the functional originality (the distance from a species's interaction centroid to the mean interaction centroid in plant trait space). (d) Likewise, the morphological uniqueness (the distance of a species to the next similar species in bird trait space) is compared with the functional uniqueness (the distance from a species's interaction centroid to that of the functionally most similar species in plant trait space).
2. Material and methods
(a). Plant–bird interaction networks
We sampled weighted plant–bird interaction networks at two sites in the Manú Biosphere Reserve in the Andes of southeast Peru (‘Manú’ hereafter): Wayqecha (13.2° S, 71.6° W, 3000 m, upper montane rainforest) and San Pedro (13.1° S, 71.6° W, 1500 m, lower montane rainforest). At each site, we installed plots of 100 × 30 m (six plots in Wayqecha, eight plots in San Pedro) and sampled networks four times approximately every three months between December 2009 and September 2010. In each round, we observed each transect for 30 h on five consecutive days and recorded which bird species fed on which plant species (total observation time 720 h in Wayqecha and 960 h in San Pedro; further details are provided in [5]). The Wayqecha network consisted of 1344 interaction events (a bird visiting a plant and consuming fruits) between 26 bird and 51 plant species, and the San Pedro network consisted of 4988 interaction events between 61 bird and 53 plant species. The networks were robustly sampled (see [5] for an assessment of sampling completeness).
(b). Projection of species into their respective functional trait spaces
We calculated the morphological differences between all bird species from the interaction networks based on traits that are relevant for the foraging of frugivorous birds: beak length and width relating to fruit choice preferences [28,29], pointedness of the wing relating to the preferred foraging layer of a bird species [5,28,30], and body mass relating to energy requirements [31,32]. We calculated the morphological differences between plant species based on plant traits that are related to the presentation of fruits and correspond to the foraging preferences of birds: fruit size and diameter, crop mass and plant height [5,24,28–32]. Bird traits were measured on museum specimens or compiled from literature; plant traits were sampled in the field (see [5,33] for details). From these traits, we calculated the morphological differences between bird species and between plant species as pairwise Mahalanobis distances. According to these distances, we projected bird and plant species into their respective four-dimensional morphological trait spaces using principal coordinates analysis (PCA) [13] (figure 1a).
(c). Projection of species into the trait space of their interaction partners
We assessed the functional roles of species by analysing the traits of the species with which they interact. For that, we projected bird species into the functional trait space of the plant species: for every bird species, we calculated its weighted interaction centroid (the mean position of the plant species it consumed; figure 1b) in plant trait space. The calculation of the interaction centroids was weighted by the observed interaction frequencies between pairs of species. This ensures that frequently observed interactions receive more weight than rarely observed links. The relationships between the interaction centroids of bird species in the plant trait space can then be analysed concordant to an analysis of the relationships between the positions of bird species in bird trait space (figure 1b). Different to indirect ordination approaches [34] or the matching of specific, individual trait pairs [5,6], this allows a direct association between the trait combinations of interacting species groups. Moreover, it facilitates a direct analysis of species' functional differences that is based on the comparison of species' foraging preferences, and that is independent of species' own morphology.
(d). The relationship between species' morphologies and their functional roles
To assess the relationship between species' morphologies and their functional roles (hypothesis i), we used two complementary approaches. First, we compared the relative positions of the bird species in bird trait space with the relative positions of their interaction centroids in plant trait space with Procrustes rotation [35] (figure 2a). Procrustes rotation analyses the similarity of the relative arrangement of points in two ordinations by superimposing the two ordinations until the best fit is reached and then quantifying the distances between the corresponding points. Procrustes rotation requires that the ordinations have the same number of dimensions; hence, in cases where bird and plant trait spaces are built from differing numbers of traits, the number of PCA axes has to be adjusted. Because our trait spaces were built from the same number of traits, this was not necessary. To test if Procrustes results were expected from chance, we randomized the interactions between species in the networks (1000 iterations per network) once with the algorithm of Patefield [36], which generates random tables with fixed margin totals, and once with the approach of Vázquez et al. [37], which keeps the connectance between species in the randomized matrix identical to that in the original matrix. For each iteration and null model algorithm, we repeated the Procrustes rotations. As an additional test, we compared the morphological and functional differences between species by relating the pairwise distances between bird species in bird trait space with the pairwise distances between their interaction centroids in plant trait space with Mantel tests [38] (figure 2b). The distance between the interaction centroids as a measure for functional dissimilarity is analogous to the measure for functional beta-diversity proposed by Dehling et al. [33].
(e). Relationship between morphological and functional specialization
To test if morphologically extreme species fulfilled specialized functional roles in the network (hypothesis ii), we compared the morphological specialization of species with their functional specialization (figure 2c,d). We calculated the degree of morphological specialization of bird and plant species as the degree to which their morphology differed from those of other species in the assemblage, using two measures of specialization: morphological originality (the distance of a point in trait space to the centroid of the trait space; figure 2c) and morphological uniqueness (the distance of a point in trait space to the nearest neighbouring point in the trait space; figure 2d) [21,39]. Accordingly, we calculated the functional specialization of bird species as the degree to which each species's interaction centroid differed from those of the other species in the assemblage as functional originality (the distance of an interaction centroid to the mean interaction centroid across all other species; figure 2c) and functional uniqueness (the distance of an interaction centroid to the nearest interaction centroid; figure 2d). To test if morphological specialization reflected functional specialization, we used Pearson's correlation tests to compare species' morphological and functional originality (both square-root-transformed) and morphological and functional uniqueness (both square-root-transformed). Non-parametric (Spearman rank) correlation tests resulted in qualitatively identical results. For all analyses, we used R v. 2.15 [40].
3. Results and discussion
(a). Relationship between morphology and functional role
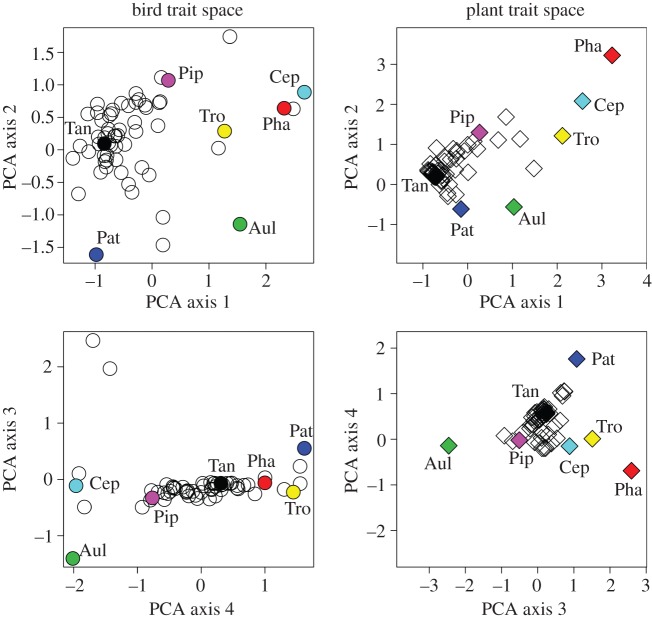
Consistent with our first hypothesis, the relative position of bird species in bird trait space correlated significantly with the relative position of their interaction centroids in plant trait space (site 1, Wayqecha: Procrustes SS = 0.66, Procrustes r = 0.59, p = 0.003; site 2, San Pedro: Procrustes SS = 0.63, Procrustes r = 0.61, p < 0.001; figure 3). Separate analyses for the seasonal networks yielded qualitatively identical results. The correlations from the observed interaction networks were significantly stronger than the correlations based on randomized interactions (p < 0.001 for both null models). The Mantel test showed that pairwise morphological distances between bird species in bird trait space correlated significantly with the pairwise functional distances between their interaction centroids in the plant trait space (Wayqecha: Mantel r = 0.51, p = 0.004; San Pedro: r = 0.62, p < 0.001). Hence, bird species with different morphologies interacted with different sets of plant species from different parts of the plant trait space. Accordingly, the degree to which bird species differed morphologically corresponded to the degree to which their consumed plant species differed. Our findings show that bird species prefer morphologically corresponding plant species which indicates a trend towards reciprocal morphological specialization in seed-dispersal systems. In contrast, classic network analysis highlighted the prevalence of asymmetric specialization in plant–animal mutualisms (i.e. species with many interaction partners tend to interact with species that have few interaction partners, and vice versa) [37,41]. Accounting for the morphological matching between interaction partners appears to reverse this asymmetry in specialization and instead demonstrates a high degree of reciprocal dependence between morphologically co-adapted plant and animal species. This suggests that network stability may depend both on the asymmetry in the number of interaction partners [42], and on the presence of morphologically corresponding interaction partners.
Figure 3.
Visualization of the positions of bird species in bird trait space and their interaction centroids in plant trait space exemplified for the species assemblage in San Pedro (1500 m). PCA axes 1 and 2 are shown in the first row, axes 3 and 4 in the second. Birds in bird trait space are symbolized as circles, interaction centroids of birds in plant trait space are symbolized as diamonds. The match between the positions of bird species in bird trait space and their interaction centroids in plant trait space is exemplified for a selection of bird species that lie at the edges of the bird trait space (coloured circles and diamonds). Note that the match between the positions is stronger in the four-dimensional trait space than in the two-dimensional visualizations. Bird species: Aul, Aulacorhynchus derbianus; Cep, Cephalopterus ornatus; Pat, Patagioenas plumbea; Pha, Pharomachrus auriceps; Pip, Pipreola intermedia; Tan, Tangara xanthocephala; Tro, Trogon personatus. (Online version in colour.)
The strong match between the relative positions of species and their interaction centroids showed that there was a strong non-random matching in the morphologies of interacting bird and plant species. This corroborates that trait matching in species from different trophic levels strongly influences whether or not species interact [1,3,5,6]. In addition, our results show that morphological matching on the scale of individual interactions leads to an overall congruency in the trait spaces of interacting species groups. Species with certain morphologies (i.e. combinations of species traits) from one trophic level interact with species of corresponding morphologies from the other trophic level, leading to a compartmentalization of the interaction networks (modularity) [43,44], and also of the trait spaces of the interacting species groups. The congruency in the trait spaces also corroborates the finding that trait matching in individual interactions leads to covariation in the functional diversity of interacting species groups on large spatial scales [5].
The correspondence between the morphological and functional differences between species shows that morphological differences correspond to differences in resource use. This is in contrast to previous studies that found weak relationships when comparing morphological differences between species with the species turnover of their interaction partners (see Albouy et al. [16] and references therein). However, these studies considered only the morphologies of the consumer species, but not those of their interaction partners, whereas in this study, we considered the functional traits of consumer species and the corresponding functional traits of their interaction partners [5]. Hence, we show that the foraging preferences of consumer species can be described either by combinations of species traits that describe species' adaptations to their resource use, or by combinations of resource traits that describe the characteristics of a species's interaction partners. The degree to which consumer and resource traits reflect the functional roles of species depends on the selection of traits and their relevance for the investigated ecological process [5]. Traits that are irrelevant will obscure the functional differences between species. It is therefore important to include only those traits in the analyses that are meaningful for the ecological process that is studied. We therefore recommend using quantitative tests (e.g. the fourth-corner analysis for species interactions in ecological networks [5,45]) to identify relevant functional traits that determine the interaction.
(b). Relationship between morphological and functional specialization
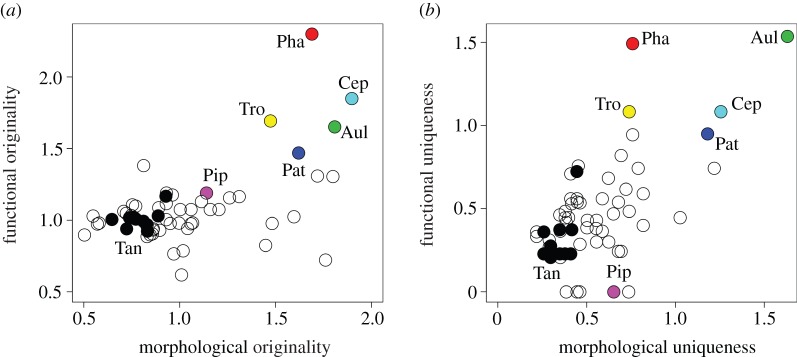
Consistent with our second hypothesis, morphological specialization corresponded to functional specialization. The morphological originality of bird species correlated significantly with the originality of their interaction centroids (Wayqecha: r = 0.55, p = 0.003; San Pedro: r = 0.54, p < 0.001; figure 4a), and the morphological uniqueness of bird species correlated significantly with the uniqueness of their interaction centroids (Wayqecha: r = 0.56, p = 0.003; San Pedro: r = 0.62, p < 0.001; figure 4b). Hence, bird species with extreme morphologies fulfilled specialized functional roles in the species assemblages. For instance, in San Pedro, Pharomachrus auriceps (Trogonidae), Trogon personatus (Trogonidae), Cephalopterus ornatus (Cotingidae) and Aulacorhynchus derbianus (Ramphastidae) were both very original and unique in their functional roles (figure 4). This corroborates that the degree of morphological specialization can be used to identify functionally specialized species in species assemblages, yielding important information for the identification of key species in ecosystems.
Figure 4.
Visualization of the relationship between (a) morphological and functional originality, and (b) morphological and functional uniqueness of bird species, exemplified for the San Pedro assemblage. Functional originality is the distance from a species's interaction centroid to the mean interaction centroid, functional uniqueness the distance from a species's interaction centroid to that of the functionally most similar species in plant trait space. Measures of originality and uniqueness were square-root-transformed. Bird species: Aul, Aulacorhynchus derbianus; Cep, Cephalopterus ornatus; Pat, Patagioenas plumbea; Pha, Pharomachrus auriceps; Pip, Pipreola intermedia; Tan, Tangara xanthocephala; Tro, Trogon personatus. Tanager species (Tangara spp.) are shown by black circles. (Online version in colour.)
The analysis of functional specialization can be used to assess which functional roles might be affected if a certain species or group of species disappears from a species assemblage (e.g. owing to climatic or land-use changes). The trait compatibility between resource and consumer species in the newly composed species assemblages could be used to assess whether certain functional roles are going to be lost [46–48]. Based on the functional uniqueness of a species, one can assess whether lost functional roles are likely to be compensated by other species in the assemblage. For instance, in the case of our study system, tanager (Tangara) species were functionally not very unique, sharing their role with many other species (figure 4b), and the local extinction of a single tanager species may therefore have relatively little impact on the associated plant species. By contrast, toucan (Aulacorhynchus) and quetzal (Pharomachrus) species were very unique in their functional roles and their extinction would probably trigger a functional disruption for their associated plant species in the tropical Andes (e.g. a marked reduction in seed dispersal). Hence, our framework could be used to assess the secondary extinction risks for species' interaction partners, and allows testing which species from other trophic levels are likely to be affected if a particular species is lost from a site. By accounting for the morphological similarity among species, our approach may be used to predict likelihoods of secondary species extinctions, and it would go beyond predictions of classic network analysis, which cannot account for the flexibility of species in switching to other, morphologically similar interaction partners.
(c). Analyses of functional diversity across taxa
Another important application of our framework could be the comparison of functional roles of unrelated taxa in mutualistic or antagonistic ecological networks. Similar functional roles in species assemblages can be fulfilled by species from distantly related taxa whose morphologies cannot be directly compared (e.g. mammals and birds in seed-dispersal networks or several vertebrate and invertebrate taxa in pollination networks) [49–51]. Analyses of functional diversity calculated from morphological traits have therefore been restricted to analyses within a single taxon [5,16,33,39,52]. If species from different taxa fulfil similar, converging functional roles in different regions, then this might result in incomplete and misleading patterns of functional diversity. The projection of species into the trait space of their interaction partners, as proposed in this study, allows for a direct comparison of the functional roles of unrelated taxa with differing morphologies, and hence a quantification of the functional diversity of species assemblages across taxa and regions. This represents an important conceptual advance that will facilitate the analysis of the factors underlying the evolution of functional roles in species assemblages.
Acknowledgements
We thank Robert Junker and one anonymous referee for valuable comments. Fieldwork in Manú was logistically supported by PerúVerde, and the Amazon Conservation Association, and conducted under the permits 041-2010-AG-DGFFS-DGEFFS, 008-2011-AG-DGFFS-DGEFFS, 01-C/C-2010-SERNANP-JPNM and 01-2011-SERNANP-PNM-JEF.
Data accessibility
All underlying data are deposited in the BiK-F Data & Metadata Repository (http://www.dx.doi.org/10.12761/SGN.2016.01.021).
Authors' contributions
D.M.D. and M.S. conceived general ideas. D.M.D. developed methodology, collected data and performed the analyses. All authors discussed the results. D.M.D. wrote the manuscript, with contributions from all authors. All authors contributed to subsequent revisions and approved the final version of the manuscript.
Competing interests
We have no competing interests.
Funding
D.M.D., K.B.-G. and M.S. received support from the research funding programme ‘LOEWE—Landes-Offensive zur Entwicklung Wissenschaftlich-ökonomischer Exzellenz’ of Hesse's Ministry of Higher Education, Research, and the Arts. Fieldwork in Peru was also supported by a grant from the German Academic Exchange Service (DAAD) to D.M.D. P.J. had support from a Severo Ochoa Excellence Award from the Spanish MINECO (SEV-2012-0262).
References
- 1.Stang M, Klinkhamer PG, Waser NM, Stang I, van der Meijden E.. 2009. Size-specific interaction patterns and size matching in a plant–pollinator interaction web. Ann. Bot. 103, 1459–1469. ( 10.1093/aob/mcp027) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stouffer DB, Rezende EL, Amaral LAN. 2011. The role of body mass in diet contiguity and food-web structure. J. Anim. Ecol. 80, 632–639. ( 10.1111/j.1365-2656.2011.01812.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eklöf A, et al. 2013. The dimensionality of ecological networks. Ecol. Lett. 16, 577–583. ( 10.1111/ele.12081) [DOI] [PubMed] [Google Scholar]
- 4.Junker RR, Blüthgen N, Brehm T, Binkenstein J, Paulus J, Schaefer HM, Stang M. 2013. Specialisation on traits as basis for the niche-breadth of flower visitors and as structuring mechanism of ecological networks. Funct. Ecol. 27, 329–341. ( 10.1111/1365-2435.12005) [DOI] [Google Scholar]
- 5.Dehling DM, Töpfer T, Schaefer HM, Jordano P, Böhning-Gaese K, Schleuning M.. 2014. Functional relationships beyond species richness patterns: trait matching in plant–bird mutualisms across scales. Glob. Ecol. Biogeogr. 23, 1085–1093. ( 10.1111/geb.12193) [DOI] [Google Scholar]
- 6.Maglianesi MA, Blüthgen N, Böhning-Gaese K, Schleuning M.. 2014. Morphological traits determine specialization and resource use in plant–hummingbird networks in the Neotropics. Ecology 95, 3325–3334. ( 10.1890/13-2261.1) [DOI] [Google Scholar]
- 7.Fleming TH. 1979. Do tropical frugivores compete for food? Am. Zool. 19, 1157–1172. ( 10.1093/icb/19.4.1157) [DOI] [Google Scholar]
- 8.Fenster CB, Armbruster WS, Wilson P, Dudash MR, Thomson JD. 2004. Pollination syndromes and floral specialization. Annu. Rev. Ecol. Syst. 35, 375–403. ( 10.1146/annurev.ecolsys.34.011802.132347) [DOI] [Google Scholar]
- 9.Jordano P, Bascompte J, Olesen JM. 2003. Invariant properties in coevolutionary networks of plant-animal interactions. Ecol. Lett. 6, 69–81. ( 10.1046/j.1461-0248.2003.00403.x) [DOI] [Google Scholar]
- 10.Janzen DH. 1985. On ecological fitting. Oikos 45, 308–310. ( 10.2307/3565565) [DOI] [Google Scholar]
- 11.Tilman D. 2001. Functional diversity. In Encyclopedia of biodiversity (ed. Levin SA.), pp. 109–120. San Diego, CA: Academic Press. [Google Scholar]
- 12.Petchey OL, Gaston KJ. 2002. Functional diversity (FD), species richness and community composition. Ecol. Lett. 5, 402–411. ( 10.1046/j.1461-0248.2002.00339.x) [DOI] [Google Scholar]
- 13.Villéger S, Mason NW, Mouillot D.. 2008. New multidimensional functional diversity indices for a multifaceted framework in functional ecology. Ecology 89, 2290–2301. ( 10.1890/07-1206.1) [DOI] [PubMed] [Google Scholar]
- 14.Ricklefs RE. 2012. Species richness and morphological diversity of passerine birds. Proc. Natl Acad. Sci. USA 109, 14 482–14 487. ( 10.1073/pnas.1212079109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kraft NJB, Godoy O, Levine JM. 2015. Plant functional traits and the multidimensional nature of species coexistence. Proc. Natl Acad. Sci. USA 112, 797–802. ( 10.1073/pnas.1413650112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Albouy C, Guilhaumon F, Villéger S, Mouchet M, Mercier L, Culioli JM, Tomasini JA, Le Loc'h F, Mouillot D.. 2011. Predicting trophic guild and diet overlap from functional traits: statistics, opportunities and limitations for marine ecology. Mar. Ecol. Prog. Ser. 436, 17–28. ( 10.3354/meps09240) [DOI] [Google Scholar]
- 17.Elton C. 1927. Animal ecology. London, UK: Sidgwick and Jackson. [Google Scholar]
- 18.Chase JM, Leibold MA. 2003. Ecological niches: linking classical and contemporary approaches. Chicago, IL: The University of Chicago Press. [Google Scholar]
- 19.Pavoine S, Ollier S, Dufour AB. 2005. Is the originality of a species measurable?. Ecol. Lett. 8, 579–586. ( 10.1111/j.1461-0248.2005.00752.x) [DOI] [Google Scholar]
- 20.Mouillot D, Culioli JM, Pelletier D, Tomasini JA. 2008. Do we protect biological originality in protected areas? A new index and an application to the Bonifacio Strait Natural Reserve. Biol. Conserv. 141, 1569–1580. ( 10.1016/j.biocon.2008.04.002) [DOI] [Google Scholar]
- 21.Bellwood DR, Wainwright PC, Fulton CJ, Hoey AS. 2006. Functional versatility supports coral reef biodiversity. Proc. R. Soc. B 273, 101–107. ( 10.1098/rspb.2005.3276) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clavel J, Julliard R, Devictor V. 2011. Worldwide decline of specialist species: toward a global functional homogenization? Front. Ecol. Environ. 9, 222–228. ( 10.1890/080216) [DOI] [Google Scholar]
- 23.Estes JA, et al. 2011. Trophic downgrading of planet Earth. Science 333, 301–306. ( 10.1126/science.1205106) [DOI] [PubMed] [Google Scholar]
- 24.Galetti M, et al. 2013. Functional extinction of birds drives rapid evolutionary changes in seed size. Science 340, 1086–1090. ( 10.1126/science.1233774) [DOI] [PubMed] [Google Scholar]
- 25.Tylianakis JM, Laliberté E, Nielsen A, Bascompte J.. 2010. Conservation of species interaction networks. Biol. Conserv. 143, 2270–2279. ( 10.1016/j.biocon.2009.12.004) [DOI] [Google Scholar]
- 26.Howe HF, Smallwood J.. 1982. Ecology of seed dispersal. Annu. Rev. Ecol. Syst. 13, 201–228. ( 10.1146/annurev.es.13.110182.001221) [DOI] [Google Scholar]
- 27.Ricklefs RE. 1977. A discriminant function analysis of assemblages of fruit-eating birds in Central America. Condor 79, 228–231. ( 10.2307/1367166) [DOI] [Google Scholar]
- 28.Moermond TC, Denslow JS. 1985. Neotropical avian frugivores: patterns of behavior, morphology, and nutrition, with consequences for fruit selection. Ornithol. Monogr. 36, 865–897. ( 10.2307/40168322) [DOI] [Google Scholar]
- 29.Wheelwright NT. 1985. Fruit size, gape width and the diets of fruit-eating birds. Ecology 66, 808–818. ( 10.2307/1940542) [DOI] [Google Scholar]
- 30.Gill FG. 2007. Ornithology. New York, NY: Freeman. [Google Scholar]
- 31.Blendinger PG, Villegas M.. 2011. Crop size is more important than neighborhood fruit availability for fruit removal of Eugenia uniflora (Myrtaceae) by bird seed dispersers. Plant Ecol. 212, 889–899. ( 10.1007/s11258-010-9873-z) [DOI] [Google Scholar]
- 32.Corlett RT, Primack RB. 2011. Tropical rain forests: an ecological and biogeographical comparison. Chichester, UK: Wiley-Blackwell. [Google Scholar]
- 33.Dehling DM, Fritz SA, Töpfer T, Päckert M, Estler P, Böhning-Gaese K, Schleuning M.. 2014. Functional and phylogenetic diversity and assemblage structure of frugivorous birds along an elevational gradient in the tropical Andes. Ecography 37, 1047–1055. [Google Scholar]
- 34.Flörchinger M, Braun J, Böhning-Gaese K, Schaefer HM. 2010. Fruit size, crop mass, and plant height explain differential fruit choice of primates and birds. Oecologia 164, 151–161. ( 10.1007/s00442-010-1655-8) [DOI] [PubMed] [Google Scholar]
- 35.Peres-Neto PR, Jackson DA. 2001. How well do multivariate data sets match? The advantages of a Procrustean superimposition approach over the Mantel test. Oecologia 129, 169–178. ( 10.1007/s004420100720) [DOI] [PubMed] [Google Scholar]
- 36.Patefield WM. 1981. Algorithm AS 159: an efficient method of generating random R×C tables with given row and column totals. J. R. Stat. Soc. C Appl. Stat. 30, 91–97. ( 10.2307/2346669) [DOI] [Google Scholar]
- 37.Vázquez DP, Melián CJ, Williams NM, Blüthgen N, Krasnov BR, Poulin R.. 2007. Species abundance and asymmetric interaction strength in ecological networks. Oikos 116, 1120–1127. ( 10.1111/j.0030-1299.2007.15828.x) [DOI] [Google Scholar]
- 38.Mantel N. 1967. The detection of disease clustering and a generalized regression approach. Cancer Res. 27, 209–220. [PubMed] [Google Scholar]
- 39.Buisson L, Grenouillet G, Villéger S, Canal J, Laffaille P.. 2013. Toward a loss of functional diversity in stream fish assemblages under climate change. Glob. Change Biol. 19, 387–400. ( 10.1111/gcb.12056) [DOI] [PubMed] [Google Scholar]
- 40.R Core Team. 2012. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; See http://www.R-project.org/. [Google Scholar]
- 41.Bascompte J, Jordano P.. 2007. Plant–animal mutualistic networks: the architecture of biodiversity. Annu. Rev. Ecol. Evol. Syst. 38, 567–593. ( 10.1146/annurev.ecolsys.38.091206.095818) [DOI] [Google Scholar]
- 42.Bascompte J, Jordano P, Olesen JM. 2006. Asymmetric coevolutionary networks facilitate biodiversity maintenance. Science 312, 431–433. ( 10.1126/science.1123412) [DOI] [PubMed] [Google Scholar]
- 43.Thébault E. 2013. Identifying compartments in presence–absence matrices and bipartite networks: insights into modularity measures. J. Biogeogr. 40, 759–768. ( 10.1111/jbi.12015) [DOI] [Google Scholar]
- 44.Schleuning M, et al. 2014. Ecological, historical and evolutionary determinants of modularity in weighted seed-dispersal networks. Ecol. Lett. 17, 454–463. ( 10.1111/ele.12245) [DOI] [PubMed] [Google Scholar]
- 45.Spitz J, Ridoux V, Brind'Amour A.. 2014. Let's go beyond taxonomy in diet description: testing a trait-based approach to prey–predator relationships. J. Anim. Ecol. 83, 1137–1148. ( 10.1111/1365-2656.12218) [DOI] [PubMed] [Google Scholar]
- 46.Zhang F, Hui C, Terblanche JS. 2011. An interaction switch predicts the nested architecture of mutualistic networks. Ecol. Lett. 14, 797–803. ( 10.1111/j.1461-0248.2011.01647.x) [DOI] [PubMed] [Google Scholar]
- 47.Ramos-Jiliberto R, Valdovinos FS, Moisset de Espanés P, Flores JD. 2012. Topological plasticity increases robustness of mutualistic networks. J. Anim. Ecol. 81, 896–904. ( 10.1111/j.1365-2656.2012.01960.x) [DOI] [PubMed] [Google Scholar]
- 48.Pellissier L, Rohr RP, Ndiribe C, Pradervand JN, Salamin N, Guisan A, Wisz M.. 2013. Combining food web and species distribution models for improved community projections. Ecol. Evol. 3, 4572–4583. ( 10.1002/ece3.843) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Olesen JM, Bascompte J, Dupont YL, Jordano P.. 2007. The modularity of pollination networks. Proc. Natl Acad. Sci. USA 104, 19 891–19 896. ( 10.1073/pnas.0706375104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dalsgaard B, Martín González AM, Olesen JM, Timmermann A, Andersen LH, Ollerton J.. 2008. Pollination networks and functional specialization: a test using Lesser Antillean plant–hummingbird assemblages. Oikos 17, 789–793. ( 10.1111/j.0030-1299.2008.16537.x) [DOI] [Google Scholar]
- 51.Mello MAR, Marquitti FMD, Guimaraes PR Jr, Kalko EKV, Jordano P, de Aguiar MAM. 2011. The modularity of seed dispersal: differences in structure and robustness between bat– and bird–fruit networks. Oecologia 167, 131–140. ( 10.1007/s00442-011-1984-2) [DOI] [PubMed] [Google Scholar]
- 52.Calba S, Maris V, Devictor V.. 2014. Measuring and explaining large-scale distribution of functional and phylogenetic diversity in birds: separating ecological drivers from methodological choices. Glob. Ecol. Biogeogr. 23, 669–678. ( 10.1111/geb.12148) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All underlying data are deposited in the BiK-F Data & Metadata Repository (http://www.dx.doi.org/10.12761/SGN.2016.01.021).