Abstract
Facioscapulohumeral muscular dystrophy (FSHD) is an autosomal dominant muscular disorder with a wide clinical variability. Contractions of the D4Z4 macrosatellite repeat on chromosome 4q35 are the molecular basis of the pathophysiology. Recently, in a subset of patients without D4Z4 repeat contractions, variants in the SMCHD1 gene have been identified that lead to hypomethylation of D4Z4 and thus DUX4 transcription, which causes FSHD type 2. In this study, we have screened 55 FSHD1-negative and 40 FSHD1-positive patients from unrelated families for potentially pathogenic variants in SMCHD1 by next-generation sequencing (NGS). We identified variants in SMCHD1 in 11 index patients, including missense, splice site and non-sense mutations. We developed a pyrosequencing assay to determine the methylation status of the D4Z4 repeat array and found significantly lower methylation levels for FSHD2 patients than for healthy controls and FSHD1 patients. Two out of eleven SMCHD1 mutation carriers had moderately contracted D4Z4 alleles thus these patients are suffering from FSHD1 and 2. Comparing the phenotype of patients, all FSHD2 patients were relatively mildly affected while patients with FSHD1+2 were much more severely affected than expected from their D4Z4 copy number. Our findings confirm the role of SMCHD1 mutations in FSHD2 and as a modifier of disease severity. With SMCHD1 variants found in 16.4% of phenotypic FSHD patients without D4Z4 repeat contractions, the incidence of FSHD2 is rather high and hence we suggest including sequencing of SMCHD1, haplotyping and methylation analysis in the workflow of molecular FSHD diagnostics.
INTRODUCTION
Facioscapulohumeral muscular dystrophy (FSHD) is one of the most common autosomal dominant muscular dystrophies with an estimated prevalence of 1:20 000, second only to the myotonic dystrophies.1, 2 Clinically, FSHD is characterized by weakness of facial, shoulder girdle and upper arm muscles, later on affecting also the pelvic girdle and foot extensors. Symptoms typically start in the second decade of life, muscular weakness and atrophy are often asymmetric and slowly progressive. A wide variation in disease severity is observed ranging from asymptomatic carriers to wheelchair dependency.3, 4, 5
The classic form of FSHD, FSHD1 (OMIM #158900), has been associated with a contraction of the polymorphic macrosatellite repeat D4Z4 in the subtelomeric region of chromosome 4q35.6, 7 Normal individuals show 11–150 repeat units of 3.3 kb in size each, arranged head-to-tail, whereas in affected individuals the repeat is truncated to 10 or less units.7, 8 At least one repeat unit is required for developing FSHD.9 For diagnostic purposes of FSHD1, D4Z4 repeat length is determined by Southern blotting. The probe p13E-11 binds to a specific locus proximal to the D4Z4 repeat array.7 Combined restriction enzyme digestion with the enzymes EcoRI, EcoRI/BlnI and XapI allows differentiating the repeat array on chromosome 4q35 from the homologous repeat region on chromosome 10q26.10, 11 Contractions on 10q are not associated with the disease.11, 12
In about 5% of patients fulfilling the clinical criteria of FSHD, no contraction of the D4Z4 repeat array can be found. No clinical signs have been identified yet to distinguish these patients from FSHD1. This second form has been termed as FSHD2 (OMIM #158901).13
Recently, Lemmers et al14, 15, 16 have presented a genetic model for FSHD as a disorder requiring two genetic modifications. According to this model, in both, FSHD1 and FSHD2, a permissive genetic background on chromosome 4q is required to develop the disease. The FSHD permissive allele shows the proximal haplotype A containing the pLAM region with a polyadenylation signal and an SSLP (simple sequence length polymorphism), resulting in the permissive haplotypes 4qA159, 4qA161, 4qA166H and 4qA168. With a frequency of 86% the 4qA161 haplotype is the most common variant among 4qA alleles. Each D4Z4 repeat unit encodes a copy of the retrogene DUX4 (MIM *606009), a germline transcription factor that is normally repressed in somatic cells most likely by a mechanism of heterochromatin condensation.17, 18, 19 Ectopic DUX4 expression in skeletal muscle is highly cytotoxic and causes muscle cell death. In healthy individuals with a ‘full length' repeat array (>10 copies) of D4Z4, the transcription of DUX4 is suppressed by a condensed heterochromatin structure of the surrounding genomic region.20, 21, 22
For FSHD1, the second genetic component required is a contraction of D4Z4 repeats to less than 10 units.10 This is associated with hypomethylation of the D4Z4 repeat array and a relaxation of the chromatin structure allowing for transcription of DUX4.16, 22 On an FSHD permissive haplotype, the most distal copy of DUX4 uses the polyadenylation signal of the adjoining pLAM region whereby this transcript is stabilized.18 Thus, this genomic background is now permissive for the expression of DUX4.16
As the second component in FSHD2, Lemmers et al23 have identified mutations in the gene SMCHD1 (structural maintenance of chromosomes flexible hinge domain containing 1, MIM *614982). Like in FSHD1, an FSHD permissive genetic background is required in addition to develop the disease. Since both genetic components of FSHD2, FSHD permissive haplotype and SMCHD1 are located on different chromosomes, they segregate independently in affected families by a truly codominant digenic inheritance.
The SMCHD1 gene is localized on chromosome 18p11.32, encodes 48 exons and its product is known to be involved in chromosome condensation and cohesion. It has a role in X inactivation and repression of genes through methylation. It was shown that homozygous loss-of-function mutations in SMCHD1 are lethal in mice.24, 25, 26 Heterozygous loss-of-function mutations in SMCHD1 are viable in the mouse model and seem to have an effect on the chromatin structure of the D4Z4 locus resulting in D4Z4 hypomethylation and somatic de-repression of DUX4 transcription in FSHD2 patients.22, 24 In combination with the FSHD-permissive haplotype, haploinsufficiency of SMCHD1 results in expression of DUX4.23
In this study, we analyzed two cohorts of patients with typical clinical FSHD phenotypes for the genetic features of FSHD2. The first group of 55 unrelated patients had clinically ‘classical' FSHD but no contraction in the D4Z4 repeat array (group A). In a second cohort of 40 patients diagnosed as FSHD1 by D4Z4 repeat contractions, we tested for variants in SMCHD1 as potential modifiers of disease severity (group B). This cohort included a subgroup of eight patients with contractions in D4Z4 (2–9 repeats) and particularly severe phenotypes, two with unusual Southern blot results and a group of 30 patients with a borderline number (10 or 11) of D4Z4 repeats showing distinct clinical symptoms. We characterized the genetic background of these patients including the number of repeat units, the haplotype 4qA161 and the methylation status of the D4Z4 repeat array.
On the basis of these results, we present a modified diagnostic approach for FSHD type 1 and 2.
PATIENTS AND METHODS
Patients
To study the role of SMCHD1 in FSHD, we selected 55 unrelated index patients excluded from having FSHD1 by Southern blotting in our routine diagnostic service, but showing a ‘classical' FSHD phenotype as assessed by experienced neurologists according to the diagnostic criteria of Padberg et al3 (group A).
In a second group of 40 FSHD1 patients with D4Z4 repeat contractions, we analyzed 30 patients with a borderline number of repeat units, that is, 10 or 11 repeats, in view of the reduced penetrance of such alleles, 2 patients with an unusual Southern blot result suggesting hybrid 4q/10q alleles and 8 patients showing a more severe phenotype than expected from the number of repeat units detected (2–9 repeats, group B). Available relatives of patients diagnosed as FSHD2 were analyzed to confirm segregation of the identified SMCHD1 variants.
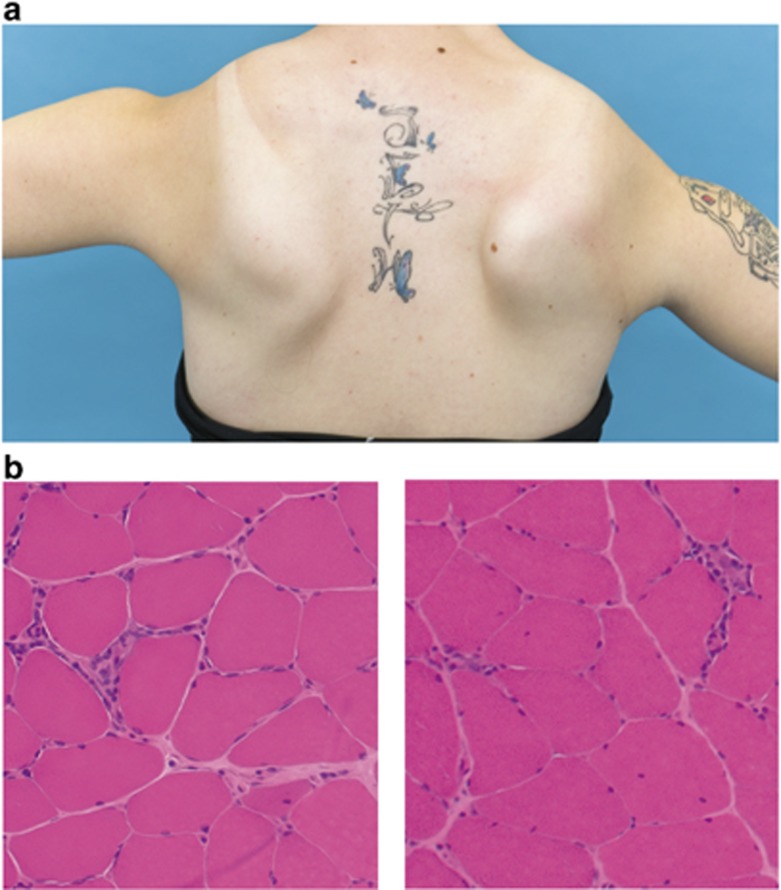
Informed consent for genetic analyses was given by all patients and available family members. In Table 1, the clinical presentation of all index patients tested positive for FSHD2 is summarized. Figure 1a shows the typical muscular weakness of the shoulder girdle in an FSHD2 patient, and in Figure 1b the muscle biopsy of an FSHD2 patient is shown. Genetic analysis was performed on genomic DNA isolated from peripheral-blood lymphocytes with standard methods.
Table 1. Clinical data.
| Distribution of weakness | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient | Gender | FSHD1 diagnostic result (D4Z4 repat units) | CK valuea | Age at onset | Age at data ascertainment | First affected musclesb | Facial muscles | Shoulder girdle | Abdominal muscles | Lower extremity | Asymmetry | Muscle biopsy |
| FSHD2-08 | m | >10 | 2.6 | 43 | 44 | Lower legs | Yes | Yes | Yes | Yes | No | — |
| FSHD2-10 | f | >10 | 5.3 | 20 | 22 | Arms | Yes | Yes | Yes | No | Yes | Signs of myositis |
| FSHD2-13 | f | >10 | 1.5 | 55 | 64 | Lower legs | Yes | Yes | Yes | Yes | Yes | Myositic/myogenic and neurogenic changes |
| FSHD2-28 | f | >10 | 2.2 | 24 | 32 | Shoulder girdle | Yes | Yes | Yes | No | Yes | Myopathic pattern, no inflammation |
| FSHD2-38 | f | >10 | 5.4 | 40 | 47 | Shoulder girdle | Yes | Yes | Yes | Yes | Yes | Dystrophic pattern with inflammatory infiltration |
| FSHD2-82 | f | >10 | 1.1 | 25 | 74 | Shoulder girdle | No | Yes | No | Yes | No | — |
| FSHD2-89 | f | 10 | 1.0 | 11 | 21 | Shoulder girdle | Yes | Yes | No | No | Yes | — |
| FSHD2-92 | m | 10 | 1.9 | 20 | 55 | Lower legs | Yes | Yes | Yes | Yes | Yes | Dystrophic pattern |
| FSHD2-120 | f | >10 | 1.0 | 30 | 40 | Arms | Yes | Yes | Yes | Yes | — | Dystrophic pattern |
| FSHD2-130 | m | >10 | 2.9 | 30 | 41 | Arms | Yes | Yes | Yes | Yes | Yes | Myopathic pattern |
| FSHD2-131 | m | >10 | 2.3 | 60 | 75 | Arms | No | Yes | No | Yes | Yes | Myopathic pattern, desmin accumulation |
Abbreviations: f, female; FSHD, facioscapulohumeral muscular dystrophy; m, male; —, no information available.
CK (creatine kinase) value given as multiples of upper normal.
Subjective estimation of the patient.
Figure 1.
Presentation of a typical FSHD2 patient. (a) Muscular weakness of the shoulder girdle with scapulae alatae in an FSHD2 patient. (b) Muscle histology (M. gastrocnemius, HE stain) of an FSHD2 patient with inflammatory infiltration of non-necrotic muscle fibers, mildly increased endomysial connective tissue and internal nuclei.
4qA161 haplotyping
The subtelomeric 4q haplotype was characterized by the single-nucleotide polymorphism (SNP) assay published by Tsumagari et al.27 The assay uses PCR and a restriction fragment length polymorphism proximal to the D4Z4 repeat locus. This assay distinguishes the most common permissive haplotype 4qA161 from non-4qA161 alleles but cannot detect other permissive alleles or differentiate between one or two 4qA161 alleles nor give the information on which allele the haplotype is located.
Sequencing of the SMCHD1 gene and data analysis
All 48 exons of the SMCHD1 gene were sequenced by next-generation sequencing (NGS) on the 454 GS Junior platform (Roche Diagnostics, Mannheim, Germany).28 For target enrichment, the Access Array System of Fluidigm (South San Francisco, CA, USA) was used that allows parallel amplification of 48 target regions for 48 samples in one single PCR setup.29 For sequencing of SMCHD1, up to 24 patients were processed in parallel to obtain a coverage of >20x for all exons (except for exon 1).
NGS data were evaluated with the software GensearchNGS (PhenoSystems, Lillois Witterzée, Belgium) using the reference sequence GRCh37 (hg19) and SMCHD1 transcript NM_015295.2. Alignment settings: allowed error rate: 8, maximum indel length: 12. Variant filter settings: minimum coverage: 4, frequency:≥20%, nearest exon distance:<21 bp. All sequence variants detected and all amplicons with a coverage of <20x were verified by Sanger sequencing using the Fluidigm primers and standard methods. The interpretation of findings was done by database research using the Alamut software package (Interactive Biosoftware, Rouen, France) including sequence conservation comparison (SIFT30), pathogenicity prediction of cDNA variation (UMD Predictor31) and Grantham score assessment (PolyPhen-232 and MutationTaster33). Splice site prediction was performed using five different prediction tools provided by Alamut: SplicesiteFinder-like (Alamut-specific tool), MaxEntScan,34 NNSPLICE,35 GeneSplicer36 and Human Splicing Finder.37 Variants and phenotypes were submitted to the public gene variant database LOVD (http://www.LOVD.nl/SMCHD1) with DB IDs SMCHD1_00003 and SMCHD1_00053-00061.
D4Z4 methylation analysis
Analysis of the methylation status of the D4Z4 repeat array was done by two different techniques.
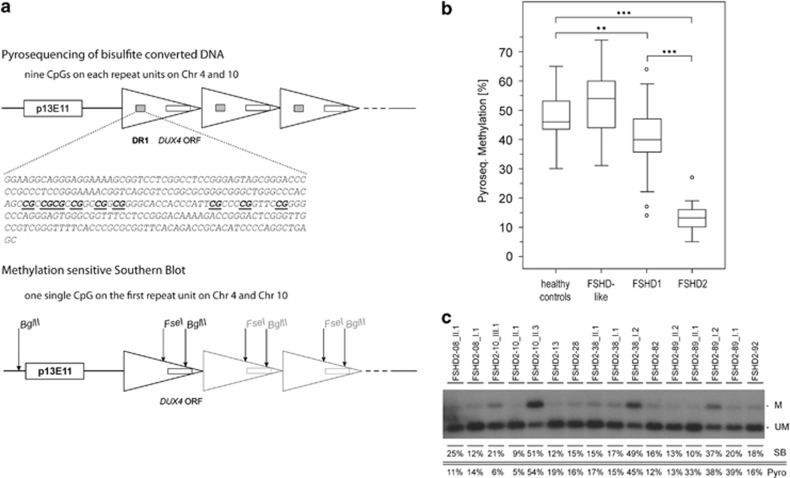
The D4Z4 methylation of the most proximal D4Z4 repeat unit was analyzed by Southern blotting hybridized with probe p13E-11 after digestion with the methylation sensitive restriction enzyme FseI as described previously.23, 38 This assay tests for the methylation status of only one single CpG site and does not distinguish between chromosomes 4q and 10q (Figure 2a). Probe signals were evaluated and quantified using a phosphorimager and ImageQuant software (GE Healthcare, Fairfield, CT, USA).
Figure 2.
Methylation analysis of the D4Z4 locus. (a) Genomic arrangement of the D4Z4 locus. D4Z4 repeat units are depicted as triangles. The binding site of the Southern blot probe p13E11, the region of extreme demethylation (DR1) and the open reading frame (ORF) of DUX4 are depicted as boxes. Restriction enzyme cutting sites are marked with arrows. By the pyrosequencing assay, nine CpGs within the DR1 region on every repeat unit are analyzed (reference sequence shown, analyzed CpGs underlined). The test therefore represents mainly the methylation status of internal repeats. In the methylation-sensitive Southern blot, genomic DNA is digested with restriction enzymes EcoRI (cutting site outside the shown region), BglII and the methylation-sensitive enzyme FseI. Hybridization is performed with the probe p13E11. This test shows the methylation level of the most proximal repeat unit only. Both tests cannot distinguish between chromosomes 4 and 10. (b) Boxplot diagram of methylation levels determined by pyrosequencing. The boxplot is created using the mean methylation level of nine CpGs for each individual sample (sample size: 48 healthy controls, 49 FSHD-like (FSHD phenotype, no contraction in D4Z4, no pathogenic variant in SMCHD1), 37 FSHD1, 13 FSHD2). Outliers are depicted as ‘○'. **P<0.01, ***P<0.001. (c) Methylation-sensitive Southern blot. Methylated (M) and unmethylated (UM) D4Z4 fragments are indicated on the right. For each index patient and available family members methylation data (%) are given as estimated by Southern blotting (SB) and pyrosequencing (Pyro), respectively.
Previously, Hartweck et al reported a domain of extreme demethylation within D4Z4.39, 40 On the basis of these findings, we established a pyrosequencing assay that covers parts of the DR1 domain described by these authors to analyze methylation levels on internal repeat units of D4Z4. DNA was treated with bisulfite for conversion of unmethylated cytosines using the Epi Tect 96 Bisulfite Kit (Qiagen, Hilden, Germany). Primers were designed within the DR1 region using the PyroMark Assay Design 2.0 software (Qiagen). For amplification and pyrosequencing, the following primer set was used: FSHD2-forward: GGGTTGAGGGTTGGGTTTATA, FSHD2-reverse: ACAAAACTCAACCTAAAAATATAC biotinylated, FSHD2-sequencing: GGGTTGGGTTTATAGT (fragment length 172 bp, chromosome position chr4: 190987949–190988113, containing 18 CpGs of which 9 can be analyzed reliably by this assay). Fragments were amplified with Platinum Taq Polymerase (Invitrogen, Life Technologies GmbH, Darmstadt, Germany) using the following PCR conditions: initial denaturation 3 min 95 °C, 35 cycles of 30 s 95 °C, 30 s 59 °C, 20 s 72 °C, final elongation 3 min 72 °C. The used primers do not differentiate between chromosomes 4q and 10q because of the high sequence homology of the loci. Pyrosequencing was performed using the PyroMark Q96 MD Pyrosequencing System (Biotage, Uppsala, Sweden) with the PyroMark Gold Q96 CDT Reagents Kit (Qiagen). Evaluation of the data was done with the Q-CpG software (Biotage). Statistical evaluation was performed with the SPSS Statistics software (IBM, Ehningen, Germany).
RESULTS
4qA161 haplotyping
Of the 95 index patients in this study, 79 individuals were positive for the FSHD-permissive haplotype 4qA161 as determined by the SNP assay of Tsumagari et al27 and were therefore included in the NGS analysis (44 individuals from group A and 35 from group B).
Sequencing of the SMCHD1 gene
After target enrichment and amplicon sequencing, the obtained data were analyzed with the software GenSearch NGS (PhenoSystems). Except for exon 1, all exons were well covered (coverage >20x) with a mean coverage of 73 (min. 0, max. 364). Exon 1 and single failed exons with a coverage of <20x (8% of all exons sequenced) were analyzed by Sanger sequencing.
Among the 79 individuals with an FSHD-permissive haplotype, we identified heterozygous variants in the SMCHD1 gene by NGS in 11 index patients (9 in group A and 2 in group B), all were confirmed by Sanger sequencing and pathogenicity was assessed using the software package Alamut (Interactive Biosoftware). The results are summarized in Table 2. All but one variant were not known in public databases (dbSNP138, http://www.ncbi.nlm.nih.gov/SNP/ and 1000 Genomes Project41) while variant c.3274_3276+2del (c.3276_3276+4del according to HGVS nomenclature) in exon 25 has been reported as pathogenic by Lemmers et al.23 The latter represents a splice site mutation, leading to skipping of exon 25 and a frameshift in the mRNA (p.Val1093Leufs*26) resulting in a premature stop codon. The very same splice site of exon 25 is affected by variant c.3274_3276+1del found in patients FSHD2-13 and FSHD2-120. Formally, it leads to the deletion of lysine1092 and at the same time to skipping of exon 25 during the splicing process. In patient FSHD2-82, variant c.4988T>G replaces the leucine codon 1663 by a stop codon (p.(Leu1663*)). A base exchange in intron 11 leads to the splice mutation c.1957-3C>T in patient FSHD2-130, which causes the predicted loss of the splice donor site and hereupon skipping of exon 11. This splicing event results in disruption of the open reading frame. The remaining six variants result in amino-acid substitutions. Variants c.328G>A (p.(Ala110Thr)), c.1844T>A (p.(Val615Asp)), c.4346G>A (p.(Arg1449Lys)), c.4388A>C (p.(Gln1463Pro)) and c.4454C>T (p.(Pro1485Leu)) are all predicted to be pathogenic by SIFT, PolyPhen2, MutationTaster and UMD Predictor. For variant c.1433G>A (p.(Gly478Glu)), the pathogenicity scores are slightly lower (see Table 2).
Table 2. Variants in SMCHD1.
| Predicted effects by | ||||||
|---|---|---|---|---|---|---|
| Patient | Mutation type | Exon positiona | Chromosome positionb | Transcript positionc | Protein positiond | (a) SIFT,e (b) Mutation Taster,f (c) PolyPhen-2,g (d) UMD Predictor,h (e) Splice effecti |
| FSHD2-08 | Missense | Exon 11 | g.2700627G>A | c.1433G>A | p.Gly478Glu | (a) Deleteriousj (b) Disease causing (c) Possibly damaging (0.618)/benign (0.142) (d) Pathogenic (93) |
| FSHD2-10 | 5‘ Splice site | Exon/intron 25 | g.2732487_2732492del | c.3276_3276+4del | p.Val1093Leufs*26 | Reported as pathogenic |
| FSHD2-13 | 5‘ Splice site | Exon/intron 25 | g.2732488_2732492del | c.3274_3276+1del | p.Lys1092del | (e) Cryptic splicing and skipping of exon 25 |
| FSHD2-28 | Missense | Exon 34 | g.2752550G>A | c.4346G>A | p.Arg1449Lys | (a) Deleteriousj (b) Disease causing (1.0) (c) Probably damaging (0.993/0.967) (d) Pathogenic (100) |
| FSHD2-38 | Missense | Exon 14 | g.2705693T>A | c.1844T>A | p.Val615Asp | (a) Deleteriousj (b) Disease causing (1.0) (c) Probably damaging (0.999/0.996) (d) Pathogenic (100) |
| FSHD2-82 | Non-sense | Exon 40 | g.2771552T>G | c.4988T>G | p.Leu1663* | Reading frame is interrupted by a premature STOP codon |
| FSHD2-89 | Missense | Exon 35 | g.2760691A>C | c.4388A>C | p.Gln1463Pro | (a) Deleteriousj (b) Disease causing (0.994) (c) Probably damaging (0.97/0.93) (d) Pathogenic (90) |
| FSHD2-92 | Missense | Exon 36 | g.2762122C>T | c.4454C>T | p.Pro1485Leu | (a) Deleteriousj (b) Disease causing (1.0) (c) Probably damaging (1.000/0.996) (d) Pathogenic (81) |
| FSHD2-120 | 5‘ Splice site | Exon/intron 25 | g.2732488_2732492del | c.3274_3276+1del | p.Lys1092del | (e) Cryptic splicing and skipping of exon 25 |
| FSHD2-130 | 5‘ Splice site | Intron 11 | g.2700662G>A | c.1463+5G>A | — | (e) Predicted negative effect on splice donor site, probable skipping of exon 11, predicted frameshift |
| FSHD2-131 | Missense | Exon 3 | g.2666964G>A | c.328G>A | p.Ala110Thr | (a) Deleteriousj (b) Disease causing (0.704) (c) Probably damaging (0.998/0.863) (d) Pathogenic (87) |
Exon number is based on GenBank NG_031972.1.
Genomic position is based on hg19.
Transcript position is based on NM_015295.2.
Protein position is based on NP_056110.2.
SIFT score: 0–1, median 0–4.32.
Mutation Taster: P-value 0–1.
PolyPhen-2: score 0–1 for HumDiv/HumVar model.
UMD Predictor: score 0–100.
Splice effect predicted by Alamut splice prediction tools.
SIFT score 0.00, median: 4.32.
D4Z4 methylation assay
For all patients in the study, a methylation analysis was performed to investigate the methylation levels of the D4Z4 repeat array. The methylation-sensitive Southern blot showed methylation levels of ≤25% for all tested patients with potentially pathogenic variants in SMCHD1. For our pyrosequencing assay, patients were divided into four groups according to their diagnostic results: FSHD-like patients negative for FSHD1 and FSHD2, FSHD1 patients, FSHD2 patients and FSHD1+2 patients. In addition, we analyzed 48 healthy controls. The methylation levels in healthy individuals ranged from 30 to 65%, while FSHD2 patients had the lowest levels with 5–27% methylation. FSHD1 patients show methylation levels of 14–64%. The few patients with mutations in both FSHD loci (FSHD1+2) were ranging between 13 and 33% methylation. For FSHD-like patients (FSHD1 and 2 negative) a methylation of 31–74% was found, which is in the range of healthy controls. Methylation levels for both applied techniques are summarized in Figure 2a and b.
Segregation analysis
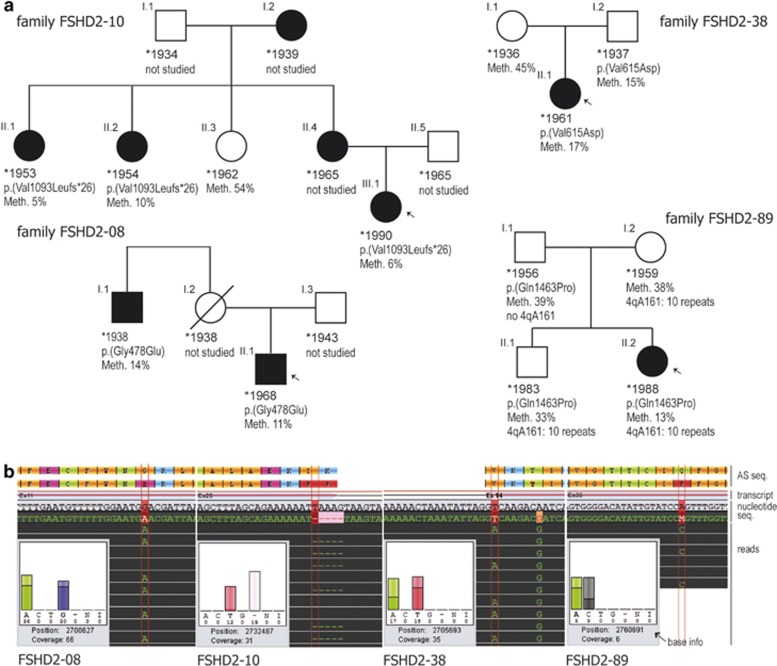
Family members were available from four SMCHD1 variant carriers for studying the segregation within the family. Probands were tested for the familial SMCHD1 variant, the 4qA161 haplotype and the methylation status of the D4Z4 region. The results are given in Figure 3a in the form of pedigrees.
Figure 3.
Genetic characterization of four families with FSHD2. (a) Pedigrees with FSHD2. Affected individuals are depicted in black, index patients are marked with an arrow. For each individual, the following information is given if available: year of birth, variant in SMCHD1 (protein position based on NP_056110.2) and methylation level of D4Z4 in percent as determined by pyrosequencing. For family FSHD2-89, the number of repeat units on a contracted 4qA allele is given. All individuals with no indicated repeat number showed ≥11 repeat units. (b) Sequencing traces of the SMCHD1 variants from families shown above, generated by the software GenSearchNGS. The diagram shows reference and consensus sequence of protein and nucleotide, respectively (indicated as ‘AS seq.', ‘transcript' and ‘nucleotide seq.' on the right), exon/intron structure of the selected transcript (NM_015295.2) as well as base exchanges or deletions found in the displayed reads (marked by vertical red lines). The inset ‘base info' gives data on frequency and balance of the viewed base (‘A C T G' bases, ‘-' deletion, ‘N' base not defined, ‘I' insertion) and the coverage at the genomic position (hg19). In addition to the heterozygous variant, patient FSHD2-38 shows a homozygous SNP (c.1851A>G, rs635132) at g.2705700.
DISCUSSION
In this study, we analyzed two cohorts of unrelated FSHD index cases, 55 of them with normal (>11 copies, group A) and 40 with contracted D4Z4 repeats (<11 copies, group B). We investigated all parameters relevant for FSHD2 as suggested in previous publications: the presence of the most common permissive haplotype 4qA161, mutations in the gene SMCHD1 and the methylation status of D4Z4. Eleven of the ninety-five index patients met all three criteria of FSHD2. Among the analyzed patients of the two cohorts, we identified ten different pathogenic variants in SMCHD1, nine of them were novel.
Six of the detected variants are leading to amino-acid substitutions of which c.328G>A (p.(Ala110Thr)), c.1844T>A (p.(Val615Asp)), c.4346G>A (p.(Arg1449Lys)), c.4388A>C (p.(Gln1463Pro)) and c.4454C>T (p.(Pro1485Leu)) are predicted to be pathogenic by all bioinformatic tools applied. For variant c.1433G>A (p.(Gly478Glu)) in patient FSHD2-08, pathogenicity scores are slightly lower. Therefore, an additional family member was analyzed to investigate the segregation of the variant. The variant was also present in this second affected relative. The low methylation levels of the D4Z4 locus found for both patients support the pathogenicity of this variant (Figure 2a).
Variant c.3276_3276+4del found in patient FSHD2-10 is predicted to induce cryptic splicing, skipping of exon 25 and disruption of the open reading frame in the mRNA (p.(Val1093Leufs*26)). It has been reported before as pathogenic for two unrelated patients.23 The very same splice site of exon 25 is affected by variant c.3274_3276+1del found in patients FSHD2-13 and FSHD-120. From the functional point of view, the effect should be similar to the variant reported by Lemmers et al,23 that is, skipping of exon 25.
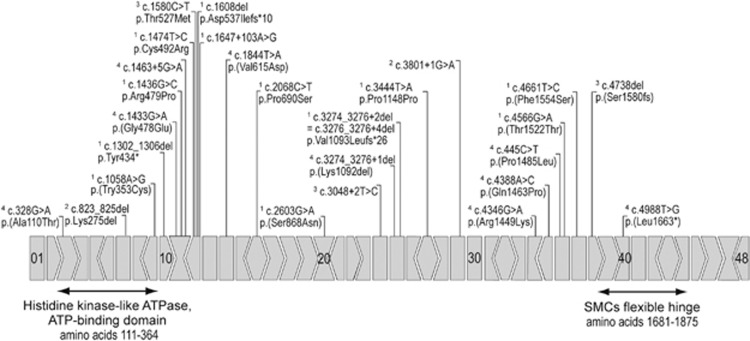
Figure 4 gives an overview of all pathogenic variants published for SMCHD1 to date. Altogether, variants are spread all over the gene. There are two regions with a high density of variants, exon 12 and exon 25. Small deletions in the 3′ region of exon 25 were found in five unrelated families.23 From these findings, a mutational hotspot or a founder effect could be suspected, but nothing is known about the ancestry of the patients. SMCHD1 has two highly conserved domains at the beginning and the end of the amino-acid chain (see Figure 4). Two variants were reported within the highly conserved ATPase domain (amino acids 111–364). Patients were described to be mildly to moderately affected.23, 38 Variant c.328G>A (p.(Ala110Thr)) is located directly proximal to the ATPase domain, the patient (FSHD2-131) shows very mild symptoms with a late onset at the age of 60. Furthermore, we present a patient carrying a non-sense mutation in exon 40. In patient FSHD2-82, the variant c.4988T>G leads to the replacement of the leucine codon 1663 by a stop codon (p.(Leu1663*)), which predicts the loss of the SMC's flexible hinge domain (amino acids 1681–1875). The identified splice variant c.1957-3C>T (FSHD2-130) is predicted to result in skipping of exon 11 and hence frame shifting. Mutations disrupting the open reading frame were already reported before (p.Tyr434*, p.Asp537Ilafs*10, c.1647+103A>G, p.Ser1580fs).23, 42
Figure 4.
Mutations in SMCHD1. All mutations in the gene SMCHD1 known to date are compiled. Exons are indicated with boxes (5′ splice site I +0, >+1, <+2. 3' splice site I +0, >−1, <−2), the SMCHD1 conserved protein domains are marked with arrows. The listed mutations were published by 1Lemmers et al,23 2Mitsuhashi et al,47 3Sacconi et al42 and 4the present study. Mutations are spread all over the gene, not sparing any of the functional domains of the protein.
Comparing the effect of variants with the phenotype, all FSHD2 patients in our first cohort show the typical FSHD distribution of muscle weakness, with a reportedly slow progression of the disease (Table 1). None of the patients had a disease onset in childhood but in the second decade of life or later. Notably, the phenotype of the patients carrying truncation variants is not more severe than in patients with missense mutations. These results suggest that neither mutations in the ATPase domain, nor loss of the SMC's hinge domain result in an aggravation of the phenotype. Little is known about the functional domains of SMCHD1, but if any stable protein is being synthesized apparently this does not impair the phenotype. Hence haploinsufficiency of the remaining intact SMCHD1 allele seems to be the pathomechanism underlying FSHD2.
By screening a number of 40 patients with contractions in D4Z4, we found two unrelated FSHD1 patients (both with 10 D4Z4 repeat units on an FSHD-permissive allele) carrying additional potentially pathogenic variants in SMCHD1. Compared with the disease severity of the other FSHD2 patients, these patients are more severely affected with an age of onset at 11 years in patient FSHD2-89 and a loss of ambulation at the age of 50 years in FSHD2-92. Similarly, Sacconi et al42 have reported three unrelated families with combined FSHD1 plus FSHD2 showing an unusual clinical severity. These observations suggest SMCHD1 as a modifier of disease severity in FSHD1.
The methylation levels of D4Z4 for the FSHD2 patients are summarized in Figure 2. The methylation-sensitive Southern blot shows a methylation level for affected patients of ≤25% in full agreement with previous findings.23 This test analyzes only a single CpG at the first D4Z4 unit on chromosomes 4 and 10 (Figure 2a). Our pyrosequencing assay is focused on a different locus, namely a series of nine CpGs within the previously reported DR1 region, on each D4Z4 unit of chromosomes 4 and 10.40 Thus, the pyrosequencing assay gives the methylation status of internal repeat units, whereas the Southern blot investigates only the first repeat unit. This is likely to be the reason for variances of methylation levels observed between the two methods (Figure 2c). Nevertheless, the pyrosequencing assay shows a significant difference between FSHD2 patients (13.31±5.86%) and controls (47.67±8.05% P-value<0.0001) as well as between FSHD2 and FSHD1 (40.16±11.56% P-value<0.0001). FSHD-like patients (negative for FSHD1 and 2) show a methylation level in the range of the healthy control group (53.12±10.32%), significantly different from FSHD1 and FSHD2. These patients likely suffer from a different muscle disease. The three patients diagnosed as FSHD1+2 are not included in Figure 1b. Their methylation levels are 33% (FSHD2-89 II-1), 13% (FSHD2-89 II-2) and 16% (FSHD2-92). While the results of the pyrosequencing methylation test are comparable to the methylation-sensitive Southern blot, the former is far easier and faster to perform.
Patient FSHD2-38 I.2 (Figure 3a) shows all three genetic findings required for FSHD2, haplotype 4qA161, hypomethylation to a level of 15% and a causal variant in SMCHD1. Upon neurological examination, however, the patient did not display any myopathic signs. His affected daughter showed typical signs of FSHD like facial weakness and scapulae alatae with an age of onset at 40 years (Table 1). This family confirms the incomplete penetrance of FSHD2.23
The relevance of the methylation status for disease manifestation is illustrated by family FSHD2-89 (see Figure 3a). The female index patient (FSHD2-89_II.2) showed first symptoms at the age of 11 with a muscle weakness pattern typical for FSHD. Her older brother had no obvious myopathic signs at the age of 26. Both parents could not be examined neurologically but reportedly have a normal muscle status. The index patient (FSHD2-89_II.2) carries 10 D4Z4 repeats, the FSHD permissive haplotype 4qA161, a pathogenic variant in SMCHD1 and a methylation level of 13%. She is, therefore, affected by FSHD1 and FSHD2. Her healthy brother (II.1) shares the SMCHD1 variant, the 4qA161 haplotype and the 10 D4Z4 repeats but has a methylation level of 33%, well above the upper limit for all other FSHD2 patients (compare Figure 1b, FSHD2). Her father (I.1) carries the SMCHD1 variant but lacks the permissive haplotype and the D4Z4 contraction. His methylation level is in the normal range (39%). Her mother (I.2) has the D4Z4 contraction on the 4qA161 haplotype but no SMCHD1 variant and normal methylation status (38%). Thus, only the index patient fulfills all criteria for FSHD1+FSHD2 while both her brother and father lack one and two factors, respectively, required for manifesting FSHD2. The mother may be a case of reduced penetrance for FSHD1 as is frequently observed especially in individuals with 10 D4Z4 repeat copies.43, 44
Incomplete penetrance was previously reported for FSHD. In Italy, a frequency of 1.2% was observed for the permissive FSHD allele associated with a D4Z4 contraction in the general population.45 For FSHD2, 19% of patients showing all three disease criteria were not affected.23 Another aspect is the known clinical variability even within the families that was already observed by the first describers of the disease, Landouzy and Dejerine in 1886.46 Mutations in SMCHD1 explain only a fraction of patients with typical FSHD phenotype and no D4Z4 contraction. These findings give rise to the assumption for other genetic, epigenetic or environmental disease modifiers in FSHD. In this situation, genetic counselling in FSHD2 pedigrees poses a challenge. Even with a truly co-dominant inheritance of a 4qA permissive haplotype and a SMCHD1 mutation, the recurrence risk will depend on the degree of relation to the index patient, not accounting for non-Mendelian factors.
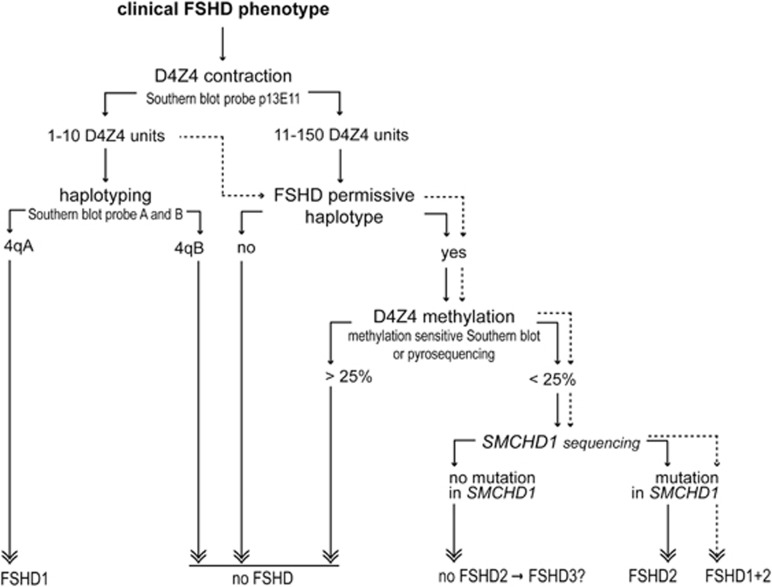
The results of our study confirm mutations in SMCHD1 as causative prerequisite for FSHD2 and a modifier of disease severity in FSHD1.23, 42 With the detection of potentially pathogenic SMCHD1 variants in 16.4% (9 out of 55) of FSHD1-negative cases, FSHD2 contributes significantly to the clinical phenotype. In our second cohort of FSHD1-positive patients, 5% (2 out of 40) of cases showed potentially pathogenic variants in SMCHD1, which therefore appears to be a modifier of disease severity in patients with D4Z4 contractions. Hence we propose to extend the routine FSHD diagnostics by the three tests for FSHD2, that is, haplotype characterization, methylation status of D4Z4 and mutation screening of SMCHD1. Our suggested diagnostic workflow is depicted in Figure 5. For clinical interpretation, all genetic data are needed including D4Z4 repeat size, the presence of an FSHD permissive allele, the methylation level of D4Z4 and mutations in SMCHD1. With the genetic data at hand, it appears worthwhile to re-examine the clinical features of FSHD2 patients with the aim of eventually refining the clinical diagnostic algorithm.
Figure 5.
Diagnostic workflow for FSHD. Suggestion for a diagnostic workflow for patients with a typical FSHD phenotype. Up to 95% of FSHD cases can be explained by contractions in the D4Z4 repeat array, for the remaining cases FSHD2 should be investigated. The dotted lines indicate the suggested diagnostic workflow for FSHD1 patients with a severe phenotype investigating FSHD1 plus FSHD2. Special cases like p13E-11 deletions or hybrid alleles are not considered, but should always be kept in mind.48
Acknowledgments
We are deeply grateful to all patients for their participation in this study. We thank Dr Eberhard Schneider for support in statistical evaluation and Dr Bhupesh Prusty, Isabel Kleinschnitz and Katja Schrooten for technical assistance.
The authors declare no conflict of interest.
References
- Emery AE: Population frequencies of inherited neuromuscular diseases—a world survey. Neuromuscul Disord 1991; 1: 19–29. [DOI] [PubMed] [Google Scholar]
- Mostacciuolo ML, Pastorello E, Vazza G et al: Facioscapulohumeral muscular dystrophy: epidemiological and molecular study in a north-east Italian population sample. Clin Genet 2009; 75: 550–555. [DOI] [PubMed] [Google Scholar]
- Padberg GW, Lunt PW, Koch M, Fardeau M: Diagnostic criteria for facioscapulohumeral muscular dystrophy. Neuromuscul Disorde 1991; 1: 231–234. [DOI] [PubMed] [Google Scholar]
- Pandya S, King WM, Tawil R: Facioscapulohumeral dystrophy. Phys Ther 2008; 88: 105–113. [DOI] [PubMed] [Google Scholar]
- Statland JM, Tawil R: Facioscapulohumeral muscular dystrophy: molecular pathological advances and future directions. Curr Opin Neurol 2011; 24: 423–428. [DOI] [PubMed] [Google Scholar]
- Wijmenga C, Frants RR, Brouwer OF, Moerer P, Weber JL, Padberg GW: Location of facioscapulohumeral muscular dystrophy gene on chromosome 4. Lancet 1990; 336: 651–653. [DOI] [PubMed] [Google Scholar]
- Wijmenga C, Hewitt JE, Sandkuijl LA et al: Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat Genet 1992; 2: 26–30. [DOI] [PubMed] [Google Scholar]
- van Deutekom JC, Wijmenga C, van Tienhoven EA et al: FSHD associated DNA rearrangements are due to deletions of integral copies of a 3.2 kb tandemly repeated unit. Hum Mol Genet 1993; 2: 2037–2042. [DOI] [PubMed] [Google Scholar]
- Tupler R, Berardinelli A, Barbierato L et al: Monosomy of distal 4q does not cause facioscapulohumeral muscular dystrophy. J Med Genet 1996; 33: 366–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyaya M, Maynard J, Rogers MT et al: Improved molecular diagnosis of facioscapulohumeral muscular dystrophy (FSHD): validation of the differential double digestion for FSHD. J Med Genet 1997; 34: 476–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmers RJL, de Kievit P, van Geel M et al: Complete allele information in the diagnosis of facioscapulohumeral muscular dystrophy by triple DNA analysis. Ann Neurol 2001; 50: 816–819. [DOI] [PubMed] [Google Scholar]
- Bakker E, Wijmenga C, Vossen RH et al: The FSHD-linked locus D4F104S1 (p13E-11) on 4q35 has a homologue on 10qter. Muscle Nerve Suppl 1995; 2: S39–S44. [PubMed] [Google Scholar]
- de Greef JC, Lemmers RJ, Camano P et al: Clinical features of facioscapulohumeral muscular dystrophy 2. Neurology 2010; 75: 1548–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmers RJ, de Kievit P, Sandkuijl L et al: Facioscapulohumeral muscular dystrophy is uniquely associated with one of the two variants of the 4q subtelomere. Nat Genet 2002; 32: 235–236. [DOI] [PubMed] [Google Scholar]
- Lemmers RJ, Wohlgemuth M, van der Gaag KJ et al: Specific sequence variations within the 4q35 region are associated with facioscapulohumeral muscular dystrophy. Am J Hum Genet 2007; 81: 884–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmers RJ, van der Vliet PJ, Klooster R et al: A unifying genetic model for facioscapulohumeral muscular dystrophy. Science 2010; 329: 1650–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriels J, Beckers MC, Ding H et al: Nucleotide sequence of the partially deleted D4Z4 locus in a patient with FSHD identifies a putative gene within each 3.3 kb element. Gene 1999; 236: 25–32. [DOI] [PubMed] [Google Scholar]
- Dixit M, Ansseau E, Tassin A et al: DUX4, a candidate gene of facioscapulohumeral muscular dystrophy, encodes a transcriptional activator of PITX1. Proc Natl Acad Sci USA 2007; 104: 18157–18162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider L, Geng LN, Lemmers RJ et al: Facioscapulohumeral dystrophy: incomplete suppression of a retrotransposed gene. PLoS Genet 2010; 6: e1001181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowaljow V, Marcowycz A, Ansseau E et al: The DUX4 gene at the FSHD1A locus encodes a pro-apoptotic protein. Neuromuscul Disord 2007; 17: 611–623. [DOI] [PubMed] [Google Scholar]
- Wallace LM, Liu J, Domire JS et al: RNA interference inhibits DUX4-induced muscle toxicity in vivo: implications for a targeted FSHD therapy. Mol Ther 2012; 20: 1417–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Greef JC, Lemmers RJ, van Engelen BG et al: Common epigenetic changes of D4Z4 in contraction-dependent and contraction-independent FSHD. Hum Mutat 2009; 30: 1449–1459. [DOI] [PubMed] [Google Scholar]
- Lemmers RJ, Tawil R, Petek LM et al: Digenic inheritance of an SMCHD1 mutation and an FSHD-permissive D4Z4 allele causes facioscapulohumeral muscular dystrophy type 2. Nat Genet 2012; 44: 1370–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blewitt ME, Gendrel AV, Pang Z et al: SmcHD1, containing a structural-maintenance-of-chromosomes hinge domain, has a critical role in X inactivation. Nat Genet 2008; 40: 663–669. [DOI] [PubMed] [Google Scholar]
- Gendrel AV, Tang YA, Suzuki M et al: Epigenetic functions of smchd1 repress gene clusters on the inactive X chromosome and on autosomes. Mol Cell Biol 2013; 33: 3150–3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mould AW, Pang Z, Pakusch M et al: Smchd1 regulates a subset of autosomal genes subject to monoallelic expression in addition to being critical for X inactivation. Epigenetics Chromatin 2013; 6: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsumagari K, Chen D, Hackman JR, Bossler AD, Ehrlich M: FSH dystrophy and a subtelomeric 4q haplotype: a new assay and associations with disease. J Med Genet 2010; 47: 745–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE et al: Genome sequencing in microfabricated high-density picolitre reactors. Nature 2005; 437: 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moonsamy PV, Williams T, Bonella P et al: High throughput HLA genotyping using 454 sequencing and the Fluidigm Access Array System for simplified amplicon library preparation. Tissue Antigens 2013; 81: 141–149. [DOI] [PubMed] [Google Scholar]
- Ng PC, Henikoff S: Predicting deleterious amino acid substitutions. Genome Res 2001; 11: 863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederic MY, Lalande M, Boileau C et al: UMD-predictor, a new prediction tool for nucleotide substitution pathogenicity — application to four genes: FBN1, FBN2, TGFBR1, and TGFBR2. Hum Mut 30: 952–959. [DOI] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L et al: A method and server for predicting damaging missense mutations. Nat Methods 2010; 7: 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz JM, Rodelsperger C, Schuelke M, Seelow D: MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods 2010; 7: 575–576. [DOI] [PubMed] [Google Scholar]
- Yeo G, Burge CB: Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol 2004; 11: 377–394. [DOI] [PubMed] [Google Scholar]
- Reese MG, Eeckman FH, Kulp D, Haussler D: Improved splice site detection in Genie. J Comput Biol 1997; 4: 311–323. [DOI] [PubMed] [Google Scholar]
- Pertea M, Lin X, Salzberg SL: GeneSplicer: a new computational method for splice site prediction. Nucleic Acids Res 29: 1185–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmet FO, Hamroun D, Lalande M, Collod-Beroud G, Claustres M, Beroud C: Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res 2009; 37: e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Overveld PG, Lemmers RJ, Sandkuijl LA et al: Hypomethylation of D4Z4 in 4q-linked and non-4q-linked facioscapulohumeral muscular dystrophy. Nat Genet 2003; 35: 315–317. [DOI] [PubMed] [Google Scholar]
- Katargin AN, Pavlova LS, Kisseljov FL, Kisseljova NP: Hypermethylation of genomic 3.3-kb repeats is frequent event in HPV-positive cervical cancer. BMC Med Genet 2009; 2: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartweck LM, Anderson LJ, Lemmers RJ et al: A focal domain of extreme demethylation within D4Z4 in FSHD2. Neurology 2013; 80: 392–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genomes Project ConsortiumGenomes Project Consortium, Abecasis GR Genomes Project Consortium, Auton A et al: An integrated map of genetic variation from 1,092 human genomes. Nature 2012; 491: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacconi S, Lemmers RJ, Balog J et al: The FSHD2 gene SMCHD1 is a modifier of disease severity in families affected by FSHD1. Am J Hum Genet 2013; 93: 744–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunt PW, Jardine PE, Koch MC et al: Correlation between fragment size at D4F104S1 and age at onset or at wheelchair use, with a possible generational effect, accounts for much phenotypic variation in 4q35-facioscapulohumeral muscular dystrophy (FSHD). Hum Mol Genet 1995; 4: 951–958. [DOI] [PubMed] [Google Scholar]
- Zatz M, Marie SK, Cerqueira A, Vainzof M, Pavanello RC, Passos-Bueno MR: The facioscapulohumeral muscular dystrophy (FSHD1) gene affects males more severely and more frequently than females. Am J Med Genet 1998; 77: 155–161. [PubMed] [Google Scholar]
- Scionti I, Fabbri G, Fiorillo C et al: Facioscapulohumeral muscular dystrophy: new insights from compound heterozygotes and implication for prenatal genetic counselling. J Med Genet 2012; 49: 171–178. [DOI] [PubMed] [Google Scholar]
- Cabianca DS, Gabellini D: The cell biology of disease: FSHD: copy number variations on the theme of muscular dystrophy. J Cell Biol 2010; 191: 1049–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuhashi S, Boyden SE, Estrella EA et al: Exome sequencing identifies a novel SMCHD1 mutation in facioscapulohumeral muscular dystrophy 2. Neuromuscul Disord 2013; 23: 975–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmers RJ, O'Shea S, Padberg GW, Lunt PW, van der Maarel SM: Best practice guidelines on genetic diagnostics of Facioscapulohumeral muscular dystrophy: workshop 9th June 2010, LUMC, Leiden, The Netherlands. Neuromusc Disord 2012; 22: 463–470. [DOI] [PubMed] [Google Scholar]