Abstract
Small RNAs (miRNA, siRNA, and piRNA) regulate gene expression through targeted destruction or translational repression of specific messenger RNA in a fundamental biological process called RNA interference (RNAi). The Argonaute proteins, which derive from a highly conserved family of genes found in almost all eukaryotes, are critical mediators of this process. Four AGO genes are present in humans, three of which (AGO 1, 3, and 4) reside in a cluster on chromosome 1p35p34. The effects of germline AGO variants or dosage alterations in humans are not known, however, prior studies have implicated dysregulation of the RNAi mechanism in the pathogenesis of several neurodevelopmental disorders. We describe five patients with hypotonia, poor feeding, and developmental delay who were found to have microdeletions of chromosomal region 1p34.3 encompassing the AGO1 and AGO3 genes. We postulate that haploinsufficiency of AGO1 and AGO3 leading to impaired RNAi may be responsible for the neurocognitive deficits present in these patients. However, additional studies with rigorous phenotypic characterization of larger cohorts of affected individuals and systematic investigation of the underlying molecular defects will be necessary to confirm this.
Introduction
RNA interference (RNAi), also called posttranscriptional gene silencing (PTGS), is a process in which small noncoding RNA molecules regulate gene expression by interfering with translation of corresponding messenger RNA (mRNA).1 MicroRNA (miRNA) are the primary RNAi effectors, and it is estimated that miRNAs regulate the translation of at least one-third of proteins encoded by the human genome.2 MiRNA undergo a complex biosynthetic process starting in the nucleus where the Drosha-Pasha microprocessor complex effects the first cleavage, transforming primary transcripts, or pri-miRNA, into pre-miRNA.3 Pre-miRNA is then exported to the cytoplasm where Dicer performs the second cleavage, generating mature miRNA.3 The miRNA is then integrated into an RNA-induced silencing complex (RISC), which guides recognition of a complementary sequence of mRNA. Argonaute proteins (AGO1-4), which constitute the catalytic component of the RISC, cleave the mRNA, effectively blocking translation.3 AGO proteins are also directly involved in miRNA biogenesis4 and thus fulfill several essential roles in the RNA interference pathway.
Animal models carrying variants in DGCR8 (which encodes Pasha), DICER1, AGO1, and AGO2 display severe neurological impairment and defective development and function of the central nervous system.5, 6, 7, 8 In humans, the neuropsychiatric features seen in patients with 22q11.2 microdeletion syndrome are thought to result from haploinsufficiency of the DGCR8 gene and associated perturbations in miRNA biogenesis.9
We present five patients with different but overlapping deletions involving the 1p34.3 locus, ranging in size from 1.1 to 3.1 Mb. The shared deleted region is approximately 289 kb and encompasses the AGO1 and AGO3 genes. The effect of AGO dosage alterations in humans has not been previously reported. We hypothesize that loss of AGO1 and AGO3 in these patients accounts for the shared neurocognitive phenotype, which includes hypotonia, poor feeding, and developmental delay.
Materials and methods
Contact between physicians caring for the patients described was facilitated by the Decipher Database (https://www.decipher.sanger.ac.uk). Variants for each patient have been submitted to this web resource and the corresponding Decipher ID specified at the end of each clinical report.
Clinical reports
Proband 1 was the first child born to non-consanguineous parents of Norwegian and German heritage. She was born at term by spontaneous vaginal delivery weighing 3000 g (10th–25th percentile) and measuring 49.5 cm in length (50th percentile) following an unremarkable pregnancy. Speech delays were noted between 18 and 24 months of age. At her initial genetics visit at age 2 years and 10 months, she was communicating primarily with poorly articulated one word utterances, gestures, and tonal inflections.
At her most recent evaluation at 3 years 9 months, her weight was 13.5 kg (16th percentile), height 95.9 cm (25th percentile), and occipitofrontal circumference (OFC) 48.1 cm (10th–25th percentile). She had mildly impaired balance but was able to walk and jump without difficulty. She was using six-word phrases and approximately 50% of her speech was understandable. She was able to name body parts and count to 15. Receptive language was intact and age-appropriate. Pertinent exam findings included: high nasal bridge, upturned nasal tip, mildly down-slanting palpebral fissures, high-arched palate, elongated-appearing digits, tented upper lip, and retrognathia. She had mild hypotonia with pronated feet.
Diagnostic workup included fragile X testing which was normal, and array CGH (Roche Nimblegen, CGX-6, 135K, Madison, WI, USA) which showed a 2.7 Mb deletion at 1p34.3 (chr1.hg18:g.(36,103,080-36,130,907)_(38,861,099-38,893,034)del) (NCBI 36/hg18), encompassing 27 OMIM genes. The proband's parents have declined carrier testing to date (Decipher ID: 291841).
Proband 2 was the first child of non-consanguineous parents of French origin. She was born at term following an unremarkable pregnancy. Her birth weight was 2810 g (10th percentile), length 48 cm (25th percentile), and OFC 34.5 cm (25th–50th percentile). She had neonatal hypotonia and feeding difficulties. Development was subsequently delayed. She walked independently at 21 months, was able to string two words together at 2 years, and was speaking in short sentences at 3 years of age.
At her most recent evaluation at 10 years 6 months, her weight was 25 kg (3rd percentile), height was 137 cm (25th percentile), and OFC was 53 cm (25th percentile). She displayed ongoing learning delays and was just starting to learn how to read and write. She had velopharyngeal insufficiency that affected her speech. On exam, she had bilateral epicanthal folds, a triangular upper lip, fullness of the periorbital region, and long thin fingers.
Array CGH (Agilent Catalog, 44K) showed a 2.2 Mb deletion at 1p34.3 (chr1.hg18:g.(35,878,982-35,927,274)_(38,364,135-38,603,709)del) (NCBI 36/hg18), including 31 OMIM genes. Parental testing confirmed this deletion to be de novo (Decipher ID: 004679).
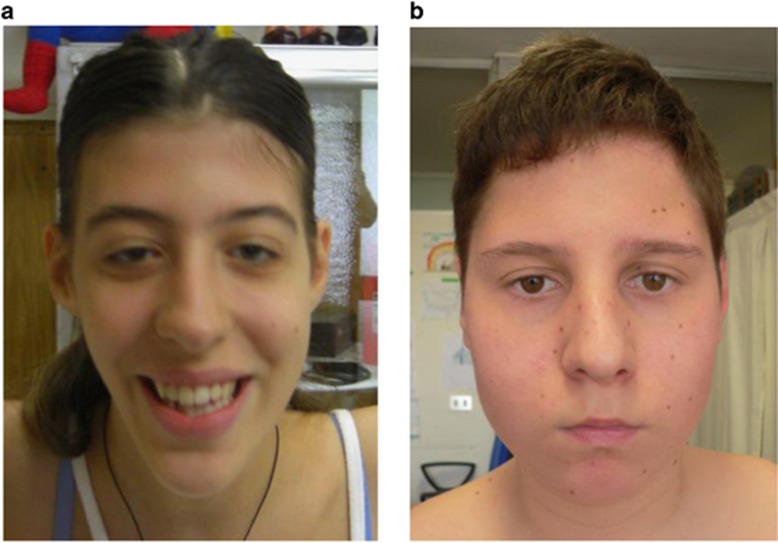
Proband 3 was born to healthy, non-consanguineous parents of Italian origin after an unremarkable pregnancy at 42 weeks gestational age. Birth weight was 3000 g (10th–25th percentile). She had neonatal breastfeeding difficulties and severe bilateral hip dislocation requiring surgical management. She did not walk independently until 3 years of age and also had speech delays. At age 17, she scored less than 55 on formal IQ testing consistent with moderate intellectual disability. At her most recent evaluation at 18 years of age, she weighed 46 kg (5th percentile), was 166 cm tall (50th percentile), and had an OFC of 50 cm (<1st percentile; −3 to 4 SD). She had a friendly disposition, poor vocabulary, and a limited attention span. Her voice had a nasal quality due to velopharyngeal insufficiency. Facial exam revealed bilateral ptosis, bitemporal narrowing, down-slanting palpebral fissures, small ears, retrognathia, a high-arched palate, maxillary hypoplasia, and a long mandible (Figure 1a). She also had moderate joint laxity, hypotonia, and flat feet.
Figure 1.
Facial phenotypes of proband 3 (a) and proband 5 (b).
Diagnostic workup included hematologic and biochemical testing, urinalysis, ECG, and echocardiography, all of which were normal. Brain MRI showed mild cortical atrophy in the occipital and parietal lobes. Array CGH (Cytochip, BluGnome, 180 K with 75–100 kb resolution) showed a 1.1 Mb deletion at 1p34.3 (chr1.hg18: g.(35,691,738_35,705,605)_(36,825,269_36,848,510)del) (NCBI 36/hg18) encompassing 18 OMIM genes. The result was confirmed by FISH and was found to be de novo (Decipher ID: 256601).
Proband 4 was the second child born to non-consanguineous healthy parents of German origin. He was born at 41 weeks gestational age after a pregnancy complicated by intrauterine growth restriction (IUGR) diagnosed at 23 weeks. His birth weight was 2660 g (<1st percentile, −3.29 SD), length 47 cm (<1st percentile, −2.66 SD), and OFC 34 cm (10th percentile). Following delivery, he required treatment for neonatal sepsis, showed marked muscular hypotonia with failure to thrive, and required nasogastric tube feeding for several weeks. At 7 months of age, his OFC dropped below the 3rd percentile. At 11 months of age he was 69 cm long (−3.1 SD) and had an OFC of 44 cm (−3 SD). He had a friendly disposition with prolonged eye contact and excessive babbling. He was able to turn independently from front to back and from back to front. Physical exam showed a prominent forehead, flat nasal bridge, deep-set eyes with short palpebral fissures, a small chin, and hypospadias. At his most recent follow-up at age 17 months, his muscular hypotonia had improved, however, he was not yet able to sit unsupported.
Oligo/SNP Array analysis (Affymetrix, CytoScan HD, Santa Clara, CA, USA) showed a 3.1 Mb deletion in chromosomal region 1p34.3 (chr1.hg18:g.(35,543,882_35,544,184)_(38,659,938_38,667,647)del) (GRCH36/hg18) encompassing 33 OMIM genes. Metaphase FISH analysis of the patient and his parents confirmed the deletion was a de novo occurrence (Decipher ID: 291728).
Proband 5 was the first child born to healthy, non-consanguineous parents of Romanian descent. He was delivered at 38 weeks gestational age for IUGR by cesarean section, which was complicated by perinatal asphyxia. Birth weight was 2400 g (<10th percentile), birth length was 49 cm (50th percentile), and he was reportedly turricephalic. At delivery, he had poor suction but could breastfeed.
Developmentally, he started babbling at 6 months, was speaking single words at 18 months and short phrases by 2½ years. He walked independently at 16 months and was toilet-trained by 2½ years. At his initial genetics evaluation at 8 years 5 months, his height was 130 cm (50th–75th percentile), weight 32.5 kg (90th–97th percentile), and OFC 50 cm (≥2nd percentile). He had a flat occiput, straight eyebrows, small mouth, bifid uvula, highly arched palate, dorsal left convex kyphoscoliosis, bilateral pes planus, mild hypotonia, and mildly lax joints. High resolution banding karyotype, molecular analysis of the FRAXA locus, echocardiography, abdominal ultrasonography, and EEG were normal. Brain MRI showed a large cavum septum pellucidum. He had moderate cognitive impairment (IQ: 50) with some degree of hyperactivity and impulsivity.
At his most recent evaluation at 13 years of age (Figure 1b), the patient's behavioral phenotype had improved. On physical examination, he was 162 cm tall (90th–97th percentile), weighed 60 kg (>97th percentile), and had an OFC of 52.5 cm (2nd–10th percentile). A leg length discrepancy of approximately 1 cm was noted.
Oligo/SNP array analysis (Affymetrix, GeneChip 6.0) showed a 1.2 Mb deletion involving 1p34.3 (chr1.hg18:g.(35,212,692_35,219,831)_(36,415,737_36,420,093)del) (GRCH36/hg18) and encompassing 18 OMIM genes. Parental testing confirmed de novo occurrence of the deletion (Decipher ID: 271479).
Results
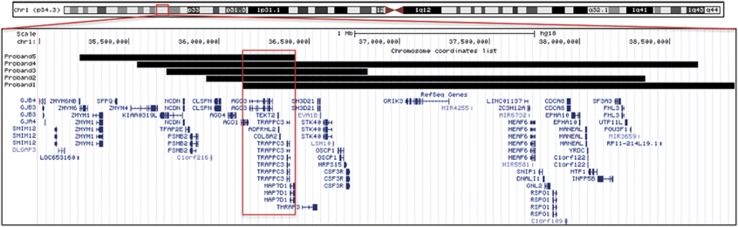
The clinical features of all five patients are summarized in Table 1. The position and extent of each deletion are shown in Figure 2. The minimal critical region is a segment of approximately 289 Kb encompassing the following OMIM genes: AGO1, AGO3, TEKT2, ADPRHL2, COL8A2, TRAPPC3, and THRAP3. Notably, deletion of AGO4 is also present in four of the five patients (probands 2, 3, 4, and 5).
Table 1. Summary of the clinical and cytogenetic findings of the 1p34.3 cohort (n=5).
| Proband 1 | Proband 2 | Proband 3 | Proband 4 | Proband 5 | |
|---|---|---|---|---|---|
| Array platform (hg18) | Roche Nimblegen CGX-6 135K | Agilent Catalog 44K Array | BluGnome 180K Cytochip | Affymetrix CytoScan HD | Affymetrix GeneChip 6.0 |
| Deletion | 1p34.3 | 1p34.3 | 1p34.3 | 1p34.3 | 1p34.3 |
| Coordinates (hg18) | 36 130 907–38 861 099 | 35 927 274–38 364 135 | 35 705 605–36 825 269 | 35 544 184–38 659 938 | 35 219 831–36 415 737 |
| Size | ~2.7 Mb | ~2.2 Mb | ~1.1 Mb | ~3.1 Mb | ~1.2 Mb |
| Inheritance | Unknown | De novo | De novo | De novo | De novo |
| Age at last evaluation | 3 years 9 months | 10 years 6 months | 18 years | 17 months | 13 years |
| Development | Motor and speech delay | Motor and speech delay. Special education | Motor and speech delay. Moderate intellectual disability | Motor and speech delay | Speech delay. Moderate intellectual disability |
| Height (percentile) | 24th | 25th | 50th | <1st (−3.2 SD) | 90th–97th |
| Weight (percentile) | 15th | 2nd | 5th | 1st (−2.4 SD) | >97th |
| OFC (percentile) | 3rd–10th | 25th | <<3rd (−3 to 4 SD) | <1st (−3.3 SD) | 2nd–10th |
| Dysmorphic features | High nasal bridge, upturned nasal tip, mildly down-slanting palpebral fissures, high-arched palate with dental crowding, retrognathia, long-appearing fingers, dysplastic toenails, pronated feet | Bilateral epicanthal folds, triangular upper lip, fullness of periorbital region, long thin fingers | Long hypotonic face, bitemporal narrowing, down-slanting palpebral fissures, ptosis, small ears, retrognathia, high-arched palate, maxillary hypoplasia, long mandible, flat feet | Prominent forehead, deep-set eyes, short palpebral fissures, small chin | Flat occiput, straight eyebrows, small mouth, bifid uvula, highly arched palate, dorsal left convex kyphoscoliosis, bilateral pes planus |
| Poor feeding | + | + | + | + | ± |
| Hypotonia | + | + | + | + | + |
| Velopharyngeal insufficiency | + | + | + | − | − |
| Other medical problems | Astigmatism | Buccofacial dyspraxia | Severe bilateral hip dislocation, joint laxity, nasal voice | Hypospadias | Joint laxity |
| Pregnancy and delivery | Uncomplicated | Uncomplicated | Uncomplicated | IUGR at 23 weeks, neonatal sepsis | IUGR, perinatal asphyxia |
| Age at delivery | 38 weeks | 37 weeks | 42 weeks | 41 weeks | 38 weeks |
| Family history | Male cousin with possible pervasive developmental delay | Not available | Healthy parents | Healthy parents | Healthy parents |
Figure 2.
Approximate alignment of the deletions present in the five probands (UCSC Genome Browser, hg18) showing the region of overlap and relevant genes.
Discussion
We describe five patients with similar clinical features (summarized in Table 1) who were found on chromosomal microarray to harbor deletions of the 1p34.3 locus, ranging in size from 1.1 to 3.1 Mb and with a shared (critical) region of approximately 289 Kb (Figure 2). These five affected individuals share several characteristics including early feeding difficulties, hypotonia, motor and speech delay, and variable dysmorphic features.
Individuals with sizeable deletions involving this chromosomal location have not been well described in the literature or documented in studies of phenotypically normal individuals per the Database of Genomic Variants (http://www.dgv.tcag.ca/dgv/app/home). A single report describes two siblings with learning disabilities and attention-deficit hyperactivity disorder, both of whom had a 1p34.1p34.3 deletion in addition to a paracentric inversion of chromosome 1 [inv(1)(p22.3p34.1)]. The authors postulate that the siblings' phenotype is accounted for by traumatic postnatal experience (parental abuse and neglect) and suggest that the 1p34.1p34.3 deletion may be of little clinical consequence.10 Our report suggests that deletions of 1p34.3 may be associated with a clear phenotype.
The overlapping region of deletion was examined to identify candidate genes that could explain the phenotype shared by our patients. OMIM genes encompassed by this 289 Kb region include AGO1, AGO3, TEKT2, ADPRHL2, COL8A2, TRAPPC3, and THRAP3. Of these genes, only COL8A2 has been linked to a phenotype in humans. Heterozygous point variants in COL8A2 have been identified in individuals with early onset Fuchs endothelial corneal dystrophy (FECD),11, 12, 13 a relatively common condition affecting approximately 5% of the US population in which bilateral progressive loss of corneal endothelium occurs.12 Mouse models of COL8A2 missense variants recapitulate the disease phenotype.14 None of the patients described carry a diagnosis of corneal dystrophy, however, all five are younger than the typical age of onset for FECD, in which early-onset disease typically manifests in the third decade of life.11, 12, 13
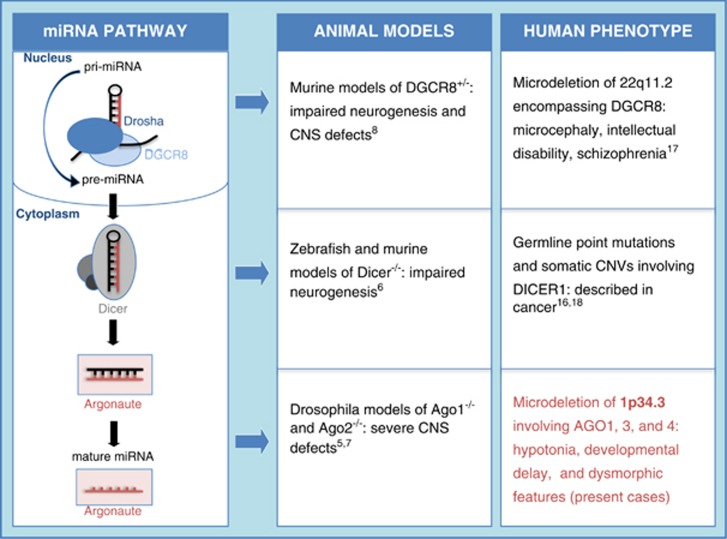
Three mammalian AGO genes (AGO 1, 3, and 4) reside in a cluster of closely related family members on chromosome 1p35p34. Two of the three genes, AGO1 and AGO3, fall within the deleted region shared by our patients and are thus of particular interest. The importance of the AGO proteins in RNAi has been well established.3,4 Interestingly, principal effectors of RNAi – miRNA and siRNA – have been shown to have an essential role in the development and function of the central nervous system (CNS).6,8,15 In addition, several authors have demonstrated relevant developmental brain defects in animal models deficient for AGO proteins5,7 including a reduction in total number of neurons and glia.5 On the basis of these findings, we speculate that haploinsufficiency of AGO genes at this locus has contributed to the neurocognitive deficits observed in our patients. Intriguingly, brain MRI studies performed on probands 3 and 5 showed occipitoparietal cortical brain atrophy and a large cavum of the septum pellucidum, respectively, suggesting that there may be an association between AGO haploinsufficiency and morphologic brain defects in humans as well (Figure 3). Finally, although only AGO1 and AGO3 are present in the shared deleted region, probands 2, 3, 4, and 5 have deletions encompassing all three AGO gene loci. The more severe phenotypes seen in probands 2–5 relative to proband 1 highlight the possibility of an additive effect of AGO gene-family haploinsufficiency.
Figure 3.
MiRNA biogenesis pathway and consequences of dysfunction observed in animal models and in humans (adapted from Yang and Lai19).
In summary, we describe five patients with deletions involving 1p34.3 and hypothesize that the features shared by these patients including early feeding problems, developmental delay, and hypotonia are the direct consequence of haploinsufficiency at this locus. We suspect deletion of the AGO genes, in particular, has led to the neurocognitive features present in these patients; however, we cannot exclude the possibility that other genes in the deleted region may be causative of or contribute to the phenotype. As such, additional studies of larger numbers of affected individuals in conjunction with functional assays will be necessary to more clearly define the developmental consequences of AGO gene haploinsufficiency.
Acknowledgments
We thank the patients and their families for their kind cooperation. This study makes use of data generated by the DECIPHER Consortium. A full list of centers who contributed to the generation of the data is available from http://decipher.sanger.ac.uk and via email from decipher@sanger.ac.uk. Funding for the project was provided by the Wellcome Trust. Electronic database information is available from Online Mendelian Inheritance in Man (http://www.omim.org); Decipher–Wellcome Trust Sanger Institute (https://www.decipher.sanger.ac.uk). This case report was supported in part by a grant from the Italian Ministry of Health (Ricerca Corrente 2013). MJT is funded by the University of Washington Medical Genetics Postdoctoral Fellowship (5T32 GM007454).
The authors declare no conflict of interest.
References
- Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev 2006; 20: 515–524. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005; 120: 15–20. [DOI] [PubMed] [Google Scholar]
- Meister G. Argonaute proteins: functional insights and emerging roles. Nat Rev Genet 2013; 14: 447–459. [DOI] [PubMed] [Google Scholar]
- Yang JS, Lai EC. Dicer-independent, Ago2-mediated microRNA biogenesis in vertebrates. Cell Cycle 2010; 9: 4455–4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka Y, Takeichi M, Uemura T. Developmental roles and molecular characterization of a Drosophila homologue of Arabidopsis Argonaute1, the founder of a novel gene superfamily. Genes Cells 2001; 6: 313–325. [DOI] [PubMed] [Google Scholar]
- Giraldez AJ, Cinalli RM, Glasner ME et al. MicroRNAs regulate brain morphogenesis in zebrafish. Science 2005; 308: 833–838. [DOI] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science 2004; 305: 1437–1441. [DOI] [PubMed] [Google Scholar]
- Stark KL, Xu B, Bagchi A et al. Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nat Genet 2008; 40: 751–760. [DOI] [PubMed] [Google Scholar]
- Forstner AJ, Degenhardt F, Schratt G, Nothen MM. MicroRNAs as the cause of schizophrenia in 22q11.2 deletion carriers, and possible implications for idiopathic disease: a mini-review. Front Mol Neurosci 2013; 6: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez JE, Tuck-Muller CM, Gasparrini W, Li S, Wertelecki W. 1p microdeletion in sibs with minimal phenotypic manifestations. Am J Med Genet 1999; 82: 107–109. [PubMed] [Google Scholar]
- Biswas S, Munier FL, Yardley J et al. Missense mutations in COL8A2, the gene encoding the alpha2 chain of type VIII collagen, cause two forms of corneal endothelial dystrophy. Hum Mol Genet 2001; 10: 2415–2423. [DOI] [PubMed] [Google Scholar]
- Gottsch JD, Sundin OH, Liu SH et al. Inheritance of a novel COL8A2 mutation defines a distinct early-onset subtype of Fuchs corneal dystrophy. Invest Ophthalmol Vis Sci 2005; 46: 1934–1939. [DOI] [PubMed] [Google Scholar]
- Mok JW, Kim HS, Joo CK. Q455V mutation in COL8A2 is associated with Fuchs corneal dystrophy in Korean patients. Eye 2009; 23: 895–903. [DOI] [PubMed] [Google Scholar]
- Meng H, Matthaei M, Ramanan N et al. L450W and Q455K Col8a2 knock-in mouse models of Fuchs endothelial corneal dystrophy show distinct phenotypes and evidence for altered autophagy. Invest Ophthalmol Vis Sci 2013; 54: 1887–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt G. MicroRNAs at the synapse. Nat Rev Neurosci 2009; 10: 842–849. [DOI] [PubMed] [Google Scholar]
- Rio Frio T, Bahubeshi A, Kanellopoulou C et al. DICER1 mutations in familial multinodular goiter with and without ovarian Sertoli-Leydig cell tumors. JAMA 2011; 305: 68–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorstman JA, Staal WG, van Daalen E, van Engeland H, Hochstenbach PF, Franke L. Identification of novel autism candidate regions through analysis of reported cytogenetic abnormalities associated with autism. Mol Psychiatry 2006; 1: 18–28. [DOI] [PubMed] [Google Scholar]
- Zhang F, Gu W, Hurles ME, Lupski JR. Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet 2009; 10: 451–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JS, Lai EC. Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Mol Cell 2011; 43: 892–903. [DOI] [PMC free article] [PubMed] [Google Scholar]