Abstract
6q16 deletions have been described in patients with a Prader–Willi-like (PWS-like) phenotype. Recent studies have shown that certain rare single-minded 1 (SIM1) loss-of-function variants were associated with a high intra-familial risk for obesity with or without features of PWS-like syndrome. Although SIM1 seems to have a key role in the phenotype of patients carrying 6q16 deletions, some data support a contribution of other genes, such as GRIK2, to explain associated behavioural problems. We describe 15 new patients in whom de novo 6q16 deletions were characterised by comparative genomic hybridisation or single-nucleotide polymorphism (SNP) array analysis, including the first patient with fetopathological data. This fetus showed dysmorphic facial features, cerebellar and cerebral migration defects with neuronal heterotopias, and fusion of brain nuclei. The size of the deletion in the 14 living patients ranged from 1.73 to 7.84 Mb, and the fetus had the largest deletion (14 Mb). Genotype–phenotype correlations confirmed the major role for SIM1 haploinsufficiency in obesity and the PWS-like phenotype. Nevertheless, only 8 of 13 patients with SIM1 deletion exhibited obesity, in agreement with incomplete penetrance of SIM1 haploinsufficiency. This study in the largest series reported to date confirms that the PWS-like phenotype is strongly linked to 6q16.2q16.3 deletions and varies considerably in its clinical expression. The possible involvement of other genes in the 6q16.2q16.3-deletion phenotype is discussed.
Introduction
Prader–Willi syndrome (PWS, MIM 176270) is an imprinting disease caused by paternal deletions, maternal uniparental disomy or imprinting anomalies in the 15q11.2q13 region.1 Clinical diagnostic criteria vary with age,2 and consist chiefly of neonatal hypotonia, early-onset obesity, and developmental delay.
A PWS-like phenotype, characterised by hypotonia, obesity, acromicria and variable motor, and cognitive delays,3 has been reported in several conditions, such as maternal uniparental disomy for chromosome 14,4, 5 certain 1p36 deletions,6, 7 2p25 deletions,8 Xq21 duplications,9 Xq23q25 duplications,10 and some cases of fragile X syndrome.11, 12 However, 6q16 deletion is the most common genetic abnormality in patients exhibiting the PWS-like phenotype.
To date >30 patients with 6q deletions, encompassing the q16.2 and/or q16.3 cytogenetic sub-bands, have been reported.3, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36 However, few of them underwent molecular characterisation of their genetic abnormalities, using either chromosomal microarray analysis,3, 13, 15, 16, 17, 18, 22, 23, 25 fluorescence in situ hybridisation (FISH) analysis with bacterial artificial chromosomes (BAC) clones21 or STR analysis.24 Two publications evaluated genotype–phenotype correlations at the 6q16 locus, but included only five and three patients, respectively.13, 17 The first study identified a 4.1-Mb minimal critical region for PWS-like within the 6q16 cytogenetic band.13 Recently, obesity and PWS-like syndrome have been ascribed to loss-of-function variants in the single-minded 1 (SIM1) gene encompassed in 6q16 critical minimal region,37, 38, 39, 40 whereas a role for GRIK2 deletion in behavioural problems has been suggested.13
Here, we describe 15 new patients (including one fetus) with 6q16 deletions, including 6q16.2 and/or 6q16.3 sub-bands, investigated by chromosomal microarray analysis. Genotype–phenotype correlations were assessed. Our results confirm the major role for SIM1 haploinsufficiency in obesity and the PWS-like phenotype.
Subjects and methods
Patients
Seven French centres and one Italian centre recruited one fetus and 14 children or young adults with 6q16 deletions, encompassing the 6q16.2 and/or 6q16.3 sub-bands. Experienced geneticists examined all patients. Informed consent was obtained from all patients and/or parents for a genetic evaluation, an assessment of deletions' parental origin and publication of clinical pictures. For the fetus, the parents provided their written informed consent to an autopsy.
Fetal examination (patient no. 1)
After termination of pregnancy, an autopsy of the fetus (patient no. 1) was performed according to protocols, including radiographs, photographs, and macroscopic and microscopic examination of all organs.41 Biometrics were compared with previously established reference values.42
Cytogenetic studies
The karyotype of the fetus was determined using in situ cultured amniocytes, following conventional procedures. For the other 14 patients, cultured peripheral lymphocytes were used.
Microarray studies were done in all 15 patients. DNA was extracted using standard procedures from cultured amniocytes (patient no. 1) or peripheral blood lymphocytes (patients no. 2–15). Patients no. 1–12 were investigated using the following oligonucleotide arrays: CGX-12 (Roche NimbelGen, Madison, WI, USA) in patient no. 1, Agilent 44 K (Agilent Technologies, Santa Clara, CA, USA) in patients no. 2–8, 10 and 11, Agilent 60 K in patient no. 9, or Agilent 180 K in patient no. 12. Patients no. 13 and 14 were evaluated using HumanHap 300 and HumanCytoSNP-12, respectively (Illumina, San Diego, CA, USA), and patient no. 15 was evaluated using Genome-Wide Human SNP Array 6.0 (Affymetrix, Santa Clara, CA, USA). Results were analysed according to Human Feb. 2009 (GRCh37/hg19) Assembly. All 15 patients have been submitted for registration in the DECIPHER database (https://decipher.sanger.ac.uk/).
FISH was performed using chromosomal preparations according to standard protocols to confirm the 6q deletions characterised by microarray.43 Probes were prepared from bacterial artificial chromosomes BAC using rolling circle amplification followed by nick translation labelling. The absence of parental deletion was checked in 14 cases, the exception being patient no. 4.
Parental origin study
Microsatellites and SNP array analysis were performed in nine patients (patients no. 2, 3, 5, 7, 8, 11, 13, 14, and 15) to investigate the parental origin of the imbalance. We either selected microsatellites at the common deleted region of the UCSC Genome Browser microsatellite or designed simple repeat tracks and primers using the NCBI Primer-BLAST program (D6S1671, D6S475, D6S2079, D6S20CA, D6S15AAT, D6S21TA, and D6S18GT). After PCR, fragment analysis was performed on an ABI 3730 XL DNA sequencing analyser and processed using GeneMapper 3.7 software (Applied Biosystems, Foster City, CA, USA). For the patient no. 15, parental origin study was performed analysing a total of 16 informative SNPs selected from 1008 SNPs located in the deleted region.
Supplementary Table S1 in the Supplementary Information lists the primers used for each microsatellite.
Results
Clinical and fetopathological data
Patient no. 1
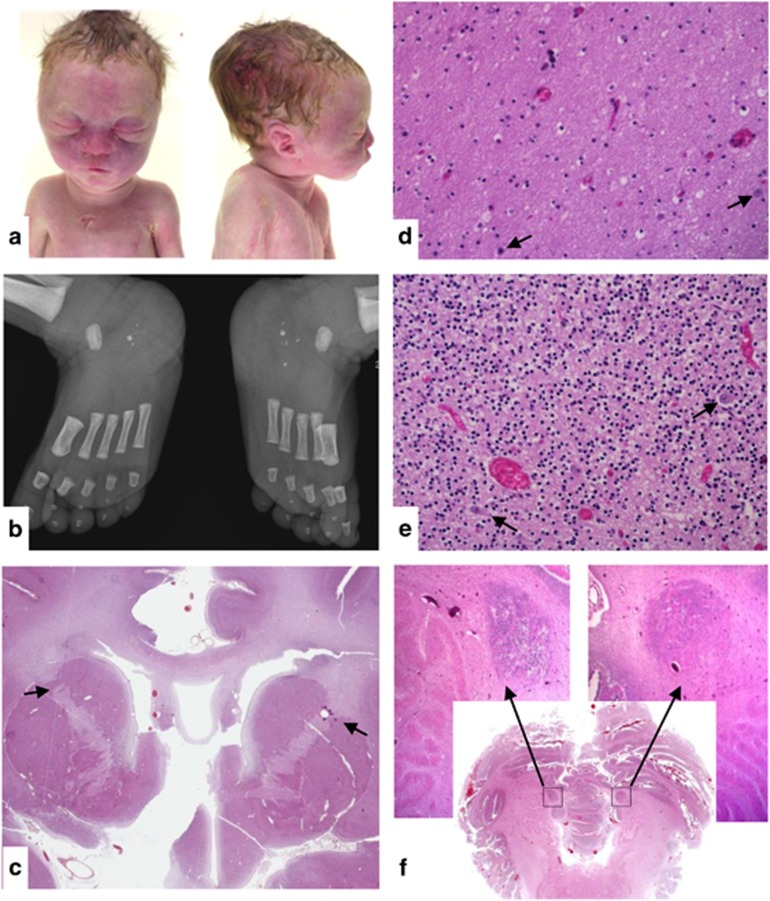
Patient no. 1 was a male fetus at 35 weeks of gestation (WG), who was the product of the first pregnancy of unrelated parents. The mother has unilateral hearing loss and the maternal grandmother has a bilateral hearing loss. A high-risk maternal screening test for Down syndrome prompted karyotype determination on amniotic fluid cells, which showed a 6q14-q16 deletion. Pyelectasis was seen on sonogram at 23 WG. The parents requested termination of pregnancy at 35 WG. Foot length was under the 5th centile and weight was 2140 g (5th centile). The pyelectasis was confirmed. The facial gestalt consisted of a short straight forehead, marked suborbital folds, a broad nasal bridge, prominent philtrum with a thin upper lip, micrognathia, and abnormally overfolded helices with a small horizontal fold along the upper edge (Figure 1a).
Figure 1.
Fetopathological study of patient no. 1. (a) Facial features: short straight forehead, marked suborbital folds, broad nasal bridge, prominent philtrum, thin upper lip, micrognathia, and abnormally hemmed ears with a small horizontal fold along the upper edge of the helix. (b) Radiographs of the feet: bilateral calcaneal fragmentation and hypermineralisation. (c) Sagittal section through the brain: internal capsule dysmorphism with fusion of anterior caudate nucleus and putamen (black arrows). (d) Cerebral white matter containing ectopic neurons (black arrows). (e) Cerebellar grey matter containing ectopic neurons (black arrows). (f) Sagittal section through the cerebellum showing focal neuronal ectopia.
The radiographic skeletal survey showed delayed bone maturation relative to gestational age, absence of ossification of the distal femoral epiphyses, hypoplasia of the sixth cervical vertebral body, sternal dysplasia, bilateral brachymesophalangia of the fifth digits, and bilateral calcaneal fragmentation with increased mineralisation (Figure 1b).
Microscopic examination of the brain evidenced fusion of the anterior caudate nucleus and putamen (Figure 1c), multiple ectopic neurons in the white matter (Figure 1d) and ectopic Purkinje cells in the internal granular layer of the cerebellum (Figure 1e). Two large heterotopias were identified in the white matter of the paravermis (Figure 1f).
Patients no. 2–15
All 14 patients had developmental delay with variable degrees of cognitive deficiency. Table 1 lists the main clinical data and Figure 2 shows photographs of several patients.
Table 1. Clinical description of each of our 15 patients.
| Patient | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gender | M | F | M | M | M | F | M | F | F | M | M | M | M | M | M |
| Age (years) | 35 WG | 17 | 3.5 | 23 | 6 | 10 | 18 | 29 | 23 | 8 | 12 | 10 | 7 | 14 | 8 |
| Perinatal data | |||||||||||||||
| Birth weight (percentile) | 5th | 95th | 50th | 90th | 50th | 25th | 40th | 40th | 50th | 60th | 90th | ||||
| Birth height (percentile) | 50th | >95th | 50th | >95th | 50th | 50th | 40th | 50th | 50th | >75th | |||||
| OFC at birth (percentile) | 20–50th | 80th | 25th | >95th | 25th | 75th | 75th | 90th | 90th | ||||||
| Hypotonia | − | + | − | − | + | + | − | − | − | − | − | ||||
| Feeding difficulties | − | − | + | + | − | − | − | − | − | + | + | − | − | ||
| Clinical features | |||||||||||||||
| Development delay | + | + | + | + | + | + | + | + | + | + | + | + | + | ||
| Learning disabilities | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| Behavioural disorders | + | + | + | + | + | + | + | − | − | + | + | + | |||
| Sleep disorders | − | − | − | + | + | − | − | − | + | + | − | − | |||
| Hyperphagia | + | + | − | + | + | + | − | − | |||||||
| Obesity a | + | + | + | − | + | + | + | + | + | −* | − * | − * | − | + | |
| Craniofacial features | |||||||||||||||
| Round face/full cheeks | + | + | + | + | − | + | + | + | + | + | + | + | |||
| Skull features | − | Macrocephaly | Brachy-, macrocephaly | − | − | Macrocephaly | − | − | − | − | − | Frontal bossing, macrocephaly | Frontal bossing, macrocephaly | Brachycephaly | Macrocephaly |
| Philtrum features | Prominent | Marked | − | − | Marked | − | − | − | − | − | Marked | − | Long | − | Marked |
| Bulbous nose | − | + | + | − | − | − | − | − | − | − | + | − | + | − | + |
| Others | Thin upper lip, marked suborbital folds, broad nasal bridge, micrognathia, abnormally overfolded helices | Horizontal eyebrows | Horizontal eyebrows | Abnormally overfolded helices | Synophris, hirsutism, small mouth | Synophris | Epicanthal folds | Hypertelorism | Triangular face shape | Narrow and horizontal palpebral fissures, bushy eyebrows, broad nasal bridge, protruding and pointed chin, large ears | |||||
| Abnormal extremities | |||||||||||||||
| Hands | Brachymetacarpia | − | − | − | Short and stubby fingers | Stubby fingers | − | Short | Short | Short, bilateral clinodactyly | − | − | − | − | |
| Feet | Short | − | − | − | − | Flat | − | − | − | Short, flat | 2nd and 3rd toes syndactyly | Flat, valgus | − | − | − |
| Genital anomalies | − | − | − | − | − | − | − | − | − | − | − | Unilateral cryptorchidism | − | Unilateral cryptorchidism | − |
| Abnormal brain MRI | − | − | − | − | + (Ventriculo-megaly) | − | − | ||||||||
| Vision anomalies | Severe myopia | − | − | Severe myopia | − | Myopia | Strabismus | − | Astigmatism | Hypermetropia | Nystagmus | − | Strabismus | − | |
| Others | Bilateral pyelectasis, neuronal ectopia, internal capsule dysmorphy | Megacystis | Hypogonadism | Prenatal increased nuchal translucency | |||||||||||
Data on weight and length at the time of examination were used to calculate the BMI and BMI-for-age percentile on the basis of establishing a standard definition for child overweight and obesity worldwide (international survey. Tim Cole et al.57 Results are represented with (+) when values correspond to obesity, (−) for normal BMI or with (−*) when values correspond to overweight.
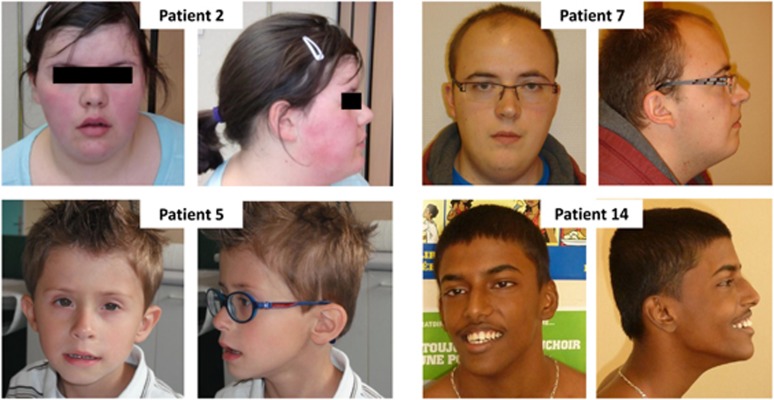
Figure 2.
Photographs of four study patients. Note the round face, full cheeks, bulbous nose, and a prominent philtrum in patient no. 2; horizontal eyebrows and a prominent philtrum in patient no. 5; round face and full cheeks in patient no. 7; and a triangular face shape in patient no. 14.
Cytogenetic and molecular results
Table 2 reports the cytogenetic abnormalities. Microarray analyses showed overlapping 6q deletions, extending from 92 138 719 bp to 108 227 875 bp (hg19). Supplementary Table S2 lists the genes included in the deletions. Except for patients no. 14 and 15, all patients had deletions that included the SIM1 gene. Minimal deletion size across patients ranged from 1.73 to 14 Mb.
Table 2. Cytogenetic characterisation of the 6q16 deletions in our 15 patients.
| Patient | Decipher ID | Description using HGVS recommendations | ISCN description (hg19) | Deletion size (Mb) | Inheritance | Parental origin of the deleted chromosome |
|---|---|---|---|---|---|---|
| 1 | 292285 | chr6.hg19:g.(92,103,929_92,138,719)_(106,099,894_106,140,794)del | 6q16.1q21(92,138,719-106,099,894)x1 | 14 | de novo | Not tested |
| 2 | 275133 | chr6.hg19:g.(98,313,927_98,342,090)_(103,457,328_103,493,161)del | 6q16.1q16.3(98,342,090-103,497,328)x1 | 5.11 | de novo | Paternal |
| 3 | 258874 | chr6.hg19:g.(96,233,216_96,246,431)_(101,346,495_101,352,914)del | 6q16.1q16.3(96,246,431-101,346,495)x1 | 5.1 | de novo | Paternal |
| 4 | 253169 | chr6.hg19:g.(97,588,639_97,985,807)_(102,192,907_102,266,317)del | 6q16.1q16.3(97,985,807-102,192,907)x1 | 4.2 | Not tested | Not tested |
| 5 | 253170 | chr6.hg19:g.(99,143,426_99,284,234)_(102,931,873_103,179,875)del | 6q16.2q16.3(99,284,234-102,931,873)x1 | 3.64 | de novo | Paternal |
| 6 | 253172 | chr6.hg19:g.(99,143,426_99,284,234)_(102,931,873_103,179,875)del | 6q16.2q16.3(99,284,234-102,931,873)x1 | 3.64 | de novo | Not tested |
| 7 | 260579 | chr6.hg19:g.(96,842,941_96,976,463)_(104,454,191_104,668,815)del | 6q16.1q16.3(96,976,463-104,454,191)x1 | 7.47 | de novo | Paternal |
| 8 | 264111 | chr6.hg19 :g.(98,917,989_98,966,909)_(101,858,360_101,869,595)del | 6q16.2q16.3(98,966,909-101,858,360)x1 | 2.89 | de novo | Paternal |
| 9 | 275474 | chr6.hg19:g.(100,260,987_100,382,250)_(102,582,366_102,772,530)del | 6q16.3(100,382,250-102,582,366)x1 | 2.2 | de novo | Not tested |
| 10 | 292291 | chr6.hg19:g.(93,007,836_93,342,048)_(104,454,191_104,668,815)del | 6q16.1q16.3(93,342,048-104,454,191)x1 | 11 | de novo | Not tested |
| 11 | 268590 | chr6.hg19:g.(100,260,987_100,382,309)_(108,227,875_108,278,822)del | 6q16.1(95,078,973-97,278,982)x1, 6q16.2q16.3(98,621,277-103,179,934)x1 | 7.84 | de novo | Maternal |
| 12a | 291928 | chr6.hg19:g.[(94,292,552_95,078,973)_(97,278,982_97,339,291)del | 6q16.1(95,078,973-97,278,982)x1, 6q16.2q16.3(98,621,277-103,179,934)x1 | 2,2 4.55 | de novo | Not tested |
| 13b | 292355 | chr6.hg19:g.(95,947,049_95,977,796)_(101,469,173_101,526,597)del | 6q16.1q16.3(95,977,796-101,469,173)x1 | 5.49 | de novo | Paternal |
| 14 | 292356 | chr6.hg19:g.(98,798,280_98,905,933)_(100,642,867_100,650,387)del | 6q16.2q16.3(98,905,933-100,642,867)x1 | 1.73 | de novo | Maternal |
| 15 | 291784 | chr6.hg19:g.(96,200,773_96,200,844)_(99,629,252_99,629,407)del | 6q16.1q16.2(96,200,844-99,629,252)x1 | 3.42 | de novo | Paternal |
Patient 12 karyotype: 46,XY,t(6;13)(q16.1q21)dn.
Additional CNVs for patient 13: 1q44(245,915,431-246,518,362)x1 mat, 4q31.21q31.2(143,272,775-147,915,323)x3 pat, 16p11.2(29,664,529-30,198,600)x3 pat.
Discussion
Our results obtained in the largest reported series of patients with 6q16 deletion, including 6q16.2 and/or 6q16.3 sub-bands, support a strong association between this chromosomal abnormality and a distinct phenotype reminiscent of PWS.
The 6q16.2q16.3 chromosomal region is not polymorphic: the Database of Genomic Variants (http://dgv.tcag.ca/) contains no large copy number variation in this region in healthy individuals, and all reported 6q16 deletions occurred de novo in symptomatic patients. The region contains no low copy repeats or recurrent breakpoints.
An imprinting effect in 6q16 deletions was hypothesised by Faivre et al14 based on the paternal origin of a de novo 6q16 deletion in a patient with PWS-like. The authors speculated that the phenotype might be ascribable to the haploinsufficiency of paternally expressed genes located in the deleted region. Other observations support this hypothesis.18, 19 In our series, only two of nine deletions in patients, for whom parental-origin data were obtained, were located in the maternal chromosome, which is consistent with the ratio reported previously for interstitial deletions at any site.44 In another study, de novo imbalances not mediated by low copy repeats were significantly more often of paternal than of maternal origin.45 Thus, to date, although there is no strong evidence supporting an imprinting mechanism in the 6q16 region, a parent-of-origin effect cannot be excluded, as none of the three maternally derived deletions, which were currently reported, (patient no. 11, 14, and case 4 from Bonaglia et al report)13 was associated with PWS-like features.
Learning disabilities, behavioural disorders, and obesity are common in 6q16 deletions (Figure 3 and Table 3). Our observations narrow the minimal critical region for PWS-like phenotype (obesity, developmental delay with or without hypotonia and/or short extremities) to a 1-Mb region within the previously reported 4.1-Mb minimal region,13 from nt 100 382 250 bp to nt 101 346 495 bp on Human Feb. 2009 (GRCh37/hg19) Assembly. This region contains the SIM1, MCHR2, and ASCC3 genes. SIM1 encodes a transcription factor that mediates hypothalamic paraventricular nucleus development. In mice, postnatally induced Sim1 deficiency causes hyperphagic obesity, and Sim1 overexpression partially corrects the obesity by normalising food intake.46, 47 Sim1 neuron ablation in adult mice induces hyperphagic obesity.48 In humans, SIM1 disruption due to an apparently balanced translocation caused severe obesity and hyperphagia in a girl.49 Obesity was a feature in several patients with 6q16 deletion and SIM1 deficiency (Figure 3 and Table 3). Loss-of-function variants in SIM1 may cause human obesity with or without PWS-like features.37, 38, 39, 40 However, in our study, SIM1 deletions in patients no. 5, 11, 12, and 13 were not associated with obesity. Thus, although SIM1 may have a critical role in regulating body weight, SIM1 deletion is not sufficient to develop obesity. In patients no. 12 and 13, the impact of other associated chromosomal abnormalities cannot be excluded. In particular, patient no. 13 had an additional 16p11.2 duplication that might have protected against obesity, as this copy number variation is associated with a low body mass index.50 On the contrary, patient no. 15 is obese, despite having a deletion that does not encompass SIM1. Another obese patient with a 6q16 deletion, sparing SIM1, was previously reported, but no gene is known in the overlapping deleted region in these two patients (our patient no. 15 and the patient no. 11 of Rosenfeld and collaborators study).17 A position effect cannot be excluded, although none of the known SIM1 enhancer sequences is deleted in these two patients.51
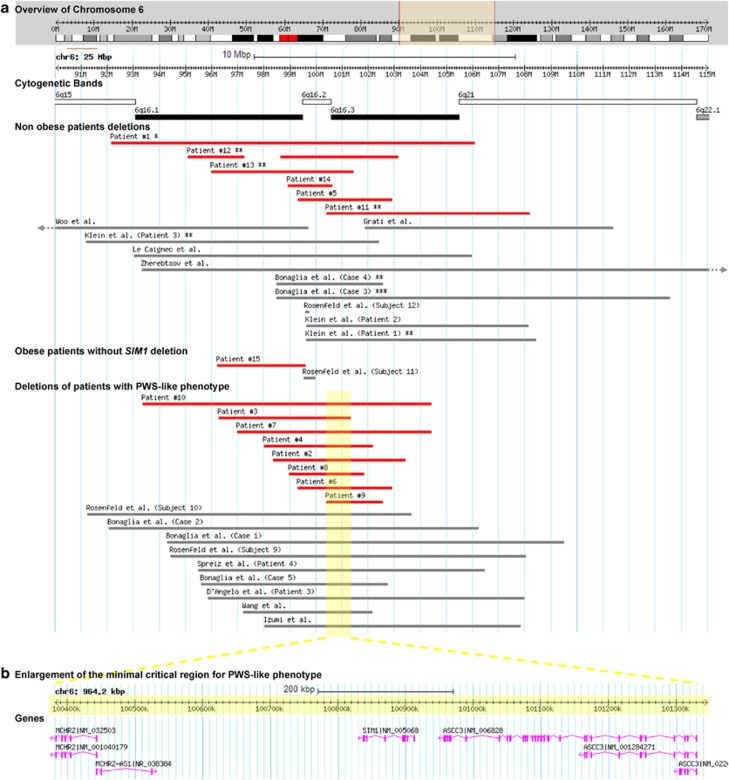
Figure 3.
Schematic alignment of 6q16 deletions obtained using Database of genomic Variants (DGV) Custom Tracks tool (http://dgv.tcag.ca/gb2/gbrowse/dgv2_hg19/). (a) Representation of molecularly defined 6q16 deletions encompassing 6q16.2 and/or 6q16.3 sub-bands, reported here (red bars) or previously (grey bars). Previously reported deletions were characterised by DNA microarray,3, 13, 15, 16, 17, 18, 22, 23, 25 FISH analysis using BAC clones,21 or STR analysis.24 *Fetal case, **overweight, ***only perinatal data were available. (b) Enlargement of the minimal critical region defined by PWS-like patients, excluding patient no. 15 from the present series and Subject 11 from Rosenfeld et al series. The region contains three genes: MCHR2, SIM1, and ASCC3.
Table 3. Summarised description of our patients and published ones.
| Our patients | Previous reportsa | Overall | |
|---|---|---|---|
| Genderb | 11M/4F | 11M/8F | 22M/12F |
| Parental originc | 7P/2M | 7P/1M | 14P/3M |
| Perinatal data | |||
| Hypotonia | 3/11 | 12/20 | 48% |
| Feeding difficulties | 4/13 | 4/20 | 24% |
| Clinical features | |||
| Development delay | 13/13 | 16/17 | 97% |
| Learning disabilities | 14/14 | 15/16 | 97% |
| Behavioural problems | 10/12 | 7/16 | 61% |
| Sleep disorders | 4/12 | 1/19 | 16% |
| Hyperphagia | 5/14 | 6/8 | 50% |
| Obesity | 10/14 | 10/17 | 65% |
| Craniofacial features | |||
| Rounded face/full cheeks | 11/12 | 6/19 | 55% |
| Skull features | 7/15 | 11/19 | 53% |
| Philtrum features | 6/15 | 6/19 | 35% |
| Bulbous nose | 5/15 | 2/19 | 21% |
| Abnormal extremities | 10/14 | 10/17 | 65% |
| Hands | 6/14 | 7/17 | 42% |
| Feet | 4/14 | 5/17 | 29% |
| Genital anomalies | 2/15 | 2/20 | 11% |
| Abnormal brain MRI | 1/7 | 8/10 | 53% |
| Vision anomalies | 8/14 | 7/13 | 56% |
Some patients with SIM1 loss-of-function variants have cognitive impairments and/or behavioural disorders.38, 39, 40 However, none of those described to date had a history of neonatal hypotonia or feeding difficulties early in life.38 Recently, a statistically significant association was demonstrated between the SIM1 SNP rs3734354 (Pro352Thr) and language impairment.52 SIM1 loss-of-function is possibly responsible for neurobehavioural disorders. The penetrance and severity of neurobehavioural disorders in patients with SIM1 loss-of-function variants seem to be lower than of those of obesity. Thus, the very high penetrance of cognitive impairment and behavioural disorders in patients with 6q16 deletions is probably due to haploinsufficiency of other genes in the same region. GRIK2 abnormalities may be associated with autistic-like behaviour in patients with 6q16 deletion.13 In our study, three of eight patients having behavioural disorders (patient no. 3, 13, and 14) were not deleted for GRIK2. Although a position effect on GRIK2 cannot be excluded, an alternative possibility is the involvement of other genes in the deleted region. MCHR2 encodes a melanin-concentrating hormone receptor expressed in the brain, and may contribute to regulate body weight in rodents.53 In humans, a SNP of this gene may exert a moderate effect on food-intake abnormalities.54 Genome-wide association studies identified MCHR2 as a putative risk factor for bipolar affective disorders.55 None of our patients had psychiatric diagnoses, but 80% exhibited MCHR2 haploinsufficiency and displayed behavioural features (emotional instability, fits of anger, aggressiveness, hyperphagia). Patient no. 14, who had severe autistic traits and profound intellectual disability, carried the smallest 6q16 deletion in this patient series, encompassing only the MCHR2 gene within the minimal region described here. The functions of the other deleted genes in our patients would not seem to bear any obvious relationship to their phenotype (Supplementary Table S2).
Patient no. 1 is the second prenatally diagnosed reported case of molecularly characterised 6q16 deletion, but the first one with fetopathological examination.24 Autopsy finding included abnormalities in neuronal migration and grey nuclei. However, he had a large 6q16 deletion (14 Mb) that encompassed several developmental genes, including EPHA7.56 In addition, none of the central nervous system abnormalities observed in this patient has been found by brain-imaging studies in previously reported cases of 6q16 deletion. In one study, various brain malformations were found in 65% of patients with 6q16 deletions.17 Of the seven patients who underwent cerebral magnetic resonance imaging in our study, only one had ventriculomegaly, and none had neuronal migration abnormalities.
To conclude, 6q16 deletion syndrome is a contiguous gene-deletion syndrome, in which SIM1 haploinsufficiency probably explains the incomplete penetrance of the obesity phenotype. Our clinical observations support a role in human neurodevelopment for other genes located in the 6q16 region. Further research on how these genes impact brain development and behaviour, together with the identification of additional individuals carrying 6q16 abnormalities, will improve our understanding of how loss of these genes may contribute to the genesis of neurodevelopmental diseases.
Acknowledgments
We are very thankful to the AChro-Puce network, the French national network of microarray users (http://www.renapa.univ-montp1.fr/), for the publication of our collaboration call. This study makes use of data generated by the DECIPHER Consortium. A full list of centres that contributed to the generation of the data is available from http://decipher.sanger.ac.uk and via email from decipher@sanger.ac.uk. Funding for the DECIPHER project was provided by the Wellcome Trust.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Cassidy SB, Schwartz S, Miller JL, Driscoll DJ: Prader-Willi syndrome. GenetMed 2012; 14: 10–26. [DOI] [PubMed] [Google Scholar]
- Holm VA, Cassidy SB, Butler MG et al: Prader-Willi syndrome: consensus diagnostic criteria. Pediatrics 1993; 91: 398–402. [PMC free article] [PubMed] [Google Scholar]
- D'Angelo CS, Kohl I, Varela MC et al: Obesity with associated developmental delay and/or learning disability in patients exhibiting additional features: report of novel pathogenic copy number variants. Am J Med Genet A 2013; 161A: 479–486. [DOI] [PubMed] [Google Scholar]
- Cox H, Bullman H, Temple IK: Maternal UPD(14) in the patient with a normal karyotype: clinical report and a systematic search for cases in samples sent for testing for Prader-Willi syndrome. Am J Med Genet A 2004; 127A: 21–25. [DOI] [PubMed] [Google Scholar]
- Hosoki K, Kagami M, Tanaka T et al: Maternal uniparental disomy 14 syndrome demonstrates prader-willi syndrome-like phenotype. J Pediatr 2009; 155: 900–903, e901. [DOI] [PubMed] [Google Scholar]
- D'Angelo CS, Da Paz JA, Kim CA et al: Prader-Willi-like phenotype: investigation of 1p36 deletion in 41 patients with delayed psychomotor development, hypotonia, obesity and/or hyperphagia, learning disabilities and behavioral problems. Eur J Med Genet 2006; 49: 451–460. [DOI] [PubMed] [Google Scholar]
- Tsuyusaki Y, Yoshihashi H, Furuya N et al: 1p36 deletion syndrome associated with Prader-Willi-like phenotype. Pediatr Int 2010; 52: 547–550. [DOI] [PubMed] [Google Scholar]
- Doco-Fenzy M, Leroy C, Schneider A et al: Early-onset obesity and paternal 2pter deletion encompassing the ACP1, TMEM18, and MYT1L genes. Eur J Hum Genet 2014; 22: 471–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabbett MT, Peters GB, Carmichael JM, Darmanian AP, Collins FA: Prader-Willi syndrome phenocopy due to duplication of Xq21.1-q21.31, with array CGH of the critical region. Clin Genet 2008; 73: 353–359. [DOI] [PubMed] [Google Scholar]
- Monaghan KG, Van Dyke DL, Feldman GL: Prader-Willi-like syndrome in a patient with an Xq23q25 duplication. Am J Med Genet 1998; 80: 227–231. [DOI] [PubMed] [Google Scholar]
- Schrander-Stumpel C, Gerver WJ, Meyer H, Engelen J, Mulder H, Fryns JP: Prader-Willi-like phenotype in fragile X syndrome. Clin Genet 1994; 45: 175–180. [DOI] [PubMed] [Google Scholar]
- Stalker HJ, Keller KL, Gray BA, Zori RT: Concurrence of fragile X syndrome and 47, XYY in an individual with a Prader-Willi-like phenotype. Am J Med Genet A 2003; 116A: 176–178. [DOI] [PubMed] [Google Scholar]
- Bonaglia MC, Ciccone R, Gimelli G et al: Detailed phenotype-genotype study in five patients with chromosome 6q16 deletion: narrowing the critical region for Prader-Willi-like phenotype. Eur J Hum Genet 2008; 16: 1443–1449. [DOI] [PubMed] [Google Scholar]
- Faivre L, Cormier-Daire V, Lapierre JM et al: Deletion of the SIM1 gene (6q16.2) in a patient with a Prader-Willi-like phenotype. J Med Genet 2002; 39: 594–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein OD, Cotter PD, Moore MW et al: Interstitial deletions of chromosome 6q: genotype-phenotype correlation utilizing array CGH. Clin Genet 2007; 71: 260–266. [DOI] [PubMed] [Google Scholar]
- Le Caignec C, Swillen A, Van Asche E, Fryns JP, Vermeesch JR: Interstitial 6q deletion: clinical and array CGH characterisation of a new patient. Eur J Med Genet 2005; 48: 339–345. [DOI] [PubMed] [Google Scholar]
- Rosenfeld JA, Amrom D, Andermann E et al: Genotype-phenotype correlation in interstitial 6q deletions: a report of 12 new cases. Neurogenetics 2012; 13: 31–47. [DOI] [PubMed] [Google Scholar]
- Spreiz A, Muller D, Zotter S et al: Phenotypic variability of a deletion and duplication 6q16.1 —> q21 due to a paternal balanced ins(7;6)(p15;q16.1q21). Am J Med Genet A 2010; 152A: 2762–2767. [DOI] [PubMed] [Google Scholar]
- Varela MC, Simoes-Sato AY, Kim CA, Bertola DR, De Castro CI, Koiffmann CP: A new case of interstitial 6q16.2 deletion in a patient with Prader-Willi-like phenotype and investigation of SIM1 gene deletion in 87 patients with syndromic obesity. Eur J Med Genet 2006; 49: 298–305. [DOI] [PubMed] [Google Scholar]
- Villa A, Urioste M, Bofarull JM, Martinez-Frias ML: De novo interstitial deletion q16.2q21 on chromosome 6. Am J Med Genet 1995; 55: 379–383. [DOI] [PubMed] [Google Scholar]
- Wang JC, Turner L, Lomax B, Eydoux P: A 5-Mb microdeletion at 6q16.1-q16.3 with SIM gene deletion and obesity. Am J Med Genet A 2008; 146A: 2975–2978. [DOI] [PubMed] [Google Scholar]
- Zherebtsov MM, Klein RT, Aviv H, Toruner GA, Hanna NN, Brooks SS: Further delineation of interstitial chromosome 6 deletion syndrome and review of the literature. Clin Dysmorphol 2007; 16: 135–140. [DOI] [PubMed] [Google Scholar]
- Izumi K, Housam R, Kapadia C et al: Endocrine phenotype of 6q16.1-q21 deletion involving SIM1 and Prader-Willi syndrome-like features. Am J Med Genet A 2013; 161A: 3137–3143. [DOI] [PubMed] [Google Scholar]
- Grati FR, Lalatta F, Turolla L et al: Three cases with de novo 6q imbalance and variable prenatal phenotype. Am J Med Genet A 2005; 136: 254–258. [DOI] [PubMed] [Google Scholar]
- Woo KS, Kim JE, Kim KE et al: A de novo proximal 6q deletion confirmed by array comparative genomic hybridization. Korean J Lab Med 2010; 30: 84–88. [DOI] [PubMed] [Google Scholar]
- Gilhuis HJ, van Ravenswaaij CM, Hamel BJ, Gabreels FJ: Interstitial 6q deletion with a Prader-Willi-like phenotype: a new case and review of the literature. Eur J Paediatr Neurol 2000; 4: 39–43. [DOI] [PubMed] [Google Scholar]
- Hopkin RJ, Schorry E, Bofinger M et al: New insights into the phenotypes of 6q deletions. Am J Med Genet 1997; 70: 377–386. [PubMed] [Google Scholar]
- Evers LJ, Schrander-Stumpel CT, Engelen JJ et al: Deletion of the long arm of chromosome 6: two new patients and literature review. Clin Genet 1996; 50: 138–144. [DOI] [PubMed] [Google Scholar]
- Duran-Gonzalez J, Gutierrez-Angulo M, Garcia-Cruz D, Ayala Mde L, Padilla M, Davalos IP: A de novo interstitial 6q deletion in a boy with a split hand malformation. J Appl Genet 2007; 48: 405–407. [DOI] [PubMed] [Google Scholar]
- Glover G, Lopez I, Gabarron J, Carmona JA: Partial monosomy 6q(q15q21) by de novo interstitial deletion. Clin Genet 1988; 33: 308–310. [DOI] [PubMed] [Google Scholar]
- Pandya A, Braverman N, Pyeritz RE, Ying KL, Kline AD, Falk RE: Interstitial deletion of the long arm of chromosome 6 associated with unusual limb anomalies: report of two new patients and review of the literature. Am J Med Genet 1995; 59: 38–43. [DOI] [PubMed] [Google Scholar]
- Schwartz MF, Kaffe S, Wallace S, Desnick RJ: Interstitial deletion of the long arm of chromosome 6 [del(6) (q16q22)]: case report and review of the literature. Clin Genet 1984; 26: 574–578. [DOI] [PubMed] [Google Scholar]
- Horigome H, Takano T, Hirano T, Kajima T, Ohtani S: Interstitial deletion of the long arm of chromosome 6 associated with absent pulmonary valve. Am J Med Genet 1991; 38: 608–611. [DOI] [PubMed] [Google Scholar]
- Fryns JP, Vandenberghe K, Van den Berghe H: Prune-belly anomaly and large interstitial deletion of the long arm of chromosome 6. Ann Genet 1991; 34: 127. [PubMed] [Google Scholar]
- Nakagome Y, Tanaka T, Hashimoto T, Kuyama M, Maruyama M: Interstitial deletion 6q in a malformed boy. Ann Genet 1980; 23: 49–51. [PubMed] [Google Scholar]
- Wakahama Y, Nakayama M, Fujimura M: Autopsy findings in interstitial deletion 6q. Pediatr Pathol 1991; 11: 97–103. [DOI] [PubMed] [Google Scholar]
- Stutzmann F, Ghoussaini M, Couturier C, Marchand M, Vatin V, Corset L: Loss-of-function mutations in SIM1 cause a specific form of Prader-Willi-like syndrome. Diabetologia 2009; 52: 104. [Google Scholar]
- Ramachandrappa S, Raimondo A, Cali AM et al: Rare variants in single-minded 1 (SIM1) are associated with severe obesity. J clin Invest 2013; 123: 3042–3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnefond A, Raimondo A, Stutzmann F et al: Loss-of-function mutations in SIM1 contribute to obesity and Prader-Willi-like features. J clin Invest 2013; 123: 3037–3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegers D, Beckers S, Hendrickx R et al: Mutation screen of the SIM1 gene in pediatric patients with early-onset obesity. IntJ Obesity (Lond) 2013; 38: 1000–1004. [DOI] [PubMed] [Google Scholar]
- Winter R, Knowles S, Bieber F, Baraitser M: The malformed fetus and stillbirth: a diagnostic approach. New York: Wiley, 1988. [Google Scholar]
- Guihard-Costa AM, Menez F, Delezoide AL: Organ weights in human fetuses after formalin fixation: standards by gestational age and body weight. Pediatr Dev Pathol 2002; 5: 559–578. [DOI] [PubMed] [Google Scholar]
- Lichter P, Cremer T: Chromosome analysis by non-isotopic in situ hybridization; in Rooney DE, Czepulkowski BH (eds):: Human Cytogenetics: a Practical Approach. Oxford: IRL Press, 1992, pp 157–192. [Google Scholar]
- Thomas NS, Durkie M, Van Zyl B et al: Parental and chromosomal origin of unbalanced de novo structural chromosome abnormalities in man. Hum Genet 2006; 119: 444–450. [DOI] [PubMed] [Google Scholar]
- Sibbons C, Morris JK, Crolla JA, Jacobs PA, Thomas NS: De novo deletions and duplications detected by array CGH: a study of parental origin in relation to mechanisms of formation and size of imbalance. Eur J Hum Genet 2012; 20: 155–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolson KP, Gemelli T, Gautron L, Elmquist JK, Zinn AR, Kublaoui BM: Postnatal Sim1 deficiency causes hyperphagic obesity and reduced Mc4r and oxytocin expression. J Neurosci 2010; 30: 3803–3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kublaoui BM, Holder JL Jr., Tolson KP, Gemelli T, Zinn AR: SIM1 overexpression partially rescues agouti yellow and diet-induced obesity by normalizing food intake. Endocrinology 2006; 147: 4542–4549. [DOI] [PubMed] [Google Scholar]
- Xi D, Gandhi N, Lai M, Kublaoui BM: Ablation of Sim1 neurons causes obesity through hyperphagia and reduced energy expenditure. PloS one 2012; 7: e36453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holder JL Jr., Butte NF, Zinn AR: Profound obesity associated with a balanced translocation that disrupts the SIM1 gene. Hum Mol Genet 2000; 9: 101–108. [DOI] [PubMed] [Google Scholar]
- Jacquemont S, Reymond A, Zufferey F et al: Mirror extreme BMI phenotypes associated with gene dosage at the chromosome 16p11.2 locus. Nature 2011; 478: 97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MJ, Oksenberg N, Hoffmann TJ, Vaisse C, Ahituv N: Functional characterization of SIM1-associated enhancers. Hum Mol Gene 2014; 23: 1700–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovey D, Zettergren A, Jonsson L et al: Associations between oxytocin-related genes and autistic-like traits. Soc Neurosci 2014; 9: 378–386. [DOI] [PubMed] [Google Scholar]
- Ludwig DS, Tritos NA, Mastaitis JW et al: Melanin-concentrating hormone overexpression in transgenic mice leads to obesity and insulin resistance. J Clin Invest 2001; 107: 379–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoussaini M, Vatin V, Lecoeur C et al: Genetic study of the melanin-concentrating hormone receptor 2 in childhood and adulthood severe obesity. J Clin Endocrinol Metab 2007; 92: 4403–4409. [DOI] [PubMed] [Google Scholar]
- Abou Jamra R, Schulze TG, Becker T et al: A systematic association mapping on chromosome 6q in bipolar affective disorder—evidence for the melanin-concentrating-hormone-receptor-2 gene as a risk factor for bipolar affective disorder. Am J Med Genet B Neuropsychiatr Genet 2010; 153B: 878–884. [DOI] [PubMed] [Google Scholar]
- Traylor RN, Fan Z, Hudson B et al: Microdeletion of 6q16.1 encompassing EPHA7 in a child with mild neurological abnormalities and dysmorphic features: case report. Mol Cytogenet 2009; 2: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole TJ, Bellizzi MC, Flegal KM, Dietz WH: Establishing a standard definition for child overweight and obesity worldwide: international survey. BMJ 2000; 320: 1240–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.