Abstract
Recent genome-wide association studies implicated that the nicotinic acetylcholine receptors (nAChRs) are common susceptible genes of two contextual diseases: chronic obstructive pulmonary disease (COPD) and lung cancer. We aimed to test whether the copy number variations (CNVs) in nAChRs have hereditary contributions to development of the two diseases. In two, two-stage, case–control studies of southern and eastern Chinese, a common CNV-3956 that duplicates the cholinergic receptor, nicotinic, α7 (CHRNA7) gene was genotyped in a total of 7880 subjects and its biological phenotype was assessed. The ≥4-copy of CNV-3956 increased COPD risk (≥4-copy vs 2/3-copy: OR=1.44, 95% CI=1.23–1.68) and caused poor lung function, and it similarly augmented risk (OR=1.49, 95% CI=1.29–1.73) and worsened prognosis (hazard ratio (HR)=1.25, 95% CI=1.07–1.45) of lung cancer. The ≥4-copy was estimated to account for 1.56% of COPD heritability and 1.87% of lung cancer heritability, respectively. Phenotypic analysis further showed that the ≥4-copy of CNV-3956 improved CHRNA7 expression in vivo and increased the carriers' smoking amount. The CNV-3956 of CHRNA7 contributed to increased risks and poor prognoses of both COPD and lung cancer, and this may be a genetic biomarker of the two diseases.
Introduction
Chronic obstructive pulmonary disease (COPD) and lung cancer are two major striking lung diseases that are contextual.1, 2 It has been reported that 40–70% of lung cancer patients complicate with COPD,3 whereas COPD patients have an extremely high incidence of lung cancer, up to 16.7 cases per 1000 person-years.4 In addition, a meta-analysis revealed that COPD patients suffer a 2.76-fold risk of lung cancer compared with the common population.5 COPD is now assumed to be a potentially intermediate phenotype ahead of lung malignant transformation, on account of the mediation effect that COPD has on lung cancer development as earlier reported.6 Early screening of COPD is reported to be very important during lung cancer surveillance.7
Epidemiological evidences indicated that COPD and lung cancer have similar pathogenesis with regard to shared etiological factors, including environmental exposures and genetic susceptibility.8, 9, 10 The most predominant commonality of COPD and lung cancer in hereditary is the overlap of susceptible loci residing on the nicotinic acetylcholine receptors (nAChRs). Several single-nucleotide polymorphisms (SNPs) in the gene cluster CHRNA3/CHRNB4/CHRNA5 were reported to be associated with the risk of both diseases.11, 12, 13 These variants are also associated with prognosis of lung cancer.11, 14, 15 However, these SNPs can only explain a small proportion of disease heritability as shown by the relatively small increments in risk they have, reflecting ‘Missing heritability' exists. Copy number variations (CNVs) are suggested to account for a large proportion of this ‘Missing heritability'. As a prevalent genetic aberration covering >1 kb duplication or deletion, CNVs encompass genes leading to dosage imbalances and aberrant expression of genes,16 and thus influence development of human diseases. The CNVs located in these nAChRs genes may affect their function. We therefore hypothesized that the CNVs in nAChRs may alter the carriers' susceptibility and prognosis to COPD and lung cancer.
To the above object, we screened all common CNVs (altered copy number frequency, ACNF >5%) in the 11 neuronal nAChRs genes and genotyped the common CNVs in two, two-stage, case–control studies of southern and eastern Chinese with a total of 1791 COPD patients and 1940 normal lung function controls, as well as 2072 lung cancer cases and 2077 cancer-free controls. A functional assay was further performed to assess the biological effect of the CNVs.
Materials and methods
Study subjects
Two, two-stage, case–control studies were conducted respectively for COPD and lung cancer in southern and eastern Han Chinese. The southern population was used as a discovery set, whereas the eastern population was the validation set. In brief, 1025 COPD patients and 1061 normal lung function controls from Guangzhou city, as well as 766 COPD patients and 879 normal controls from Suzhou city, were enrolled as described in previous studies.11, 17 In addition, 1056 lung cancer cases and 1056 cancer-free controls from Guangzhou city, as well as 1016 lung cancer cases and 1021 cancer-free controls from Suzhou city, were used as described previously.18, 19 All controls were age (±5 years) and sex frequency matched with the cases. Having signed a written informed consent, each subject provided data regarding the demographic characters and potential risk factors and donated one-time 5 ml blood sample. Among these subjects, there were 427 individuals having at least 4-year follow-up of lung function between 2002 and 2010 with annual spirometric detection,20 and 570 lung cancer patients from the southern Chinese and 569 cases from the eastern Chinese having complete survival follow-up data.19 Detailed information about subjects' recruitment and definitions of variables are presented in Supplementary Methods. These studies were approved by the institutional review boards of Guangzhou Medical University and Soochow University.
CNV selection and genotyping
Based on the Database of Genomic Variants (DGV: http://projects.tcag.ca/variation/ last access: 1 January 2013), we screened all CNVs in the 11 neuronal nAChRs genes and found that two CNVs, chr15.hg19:g.31766300_32986579dup (ie, CNV-3956 in DGV database) and chr15.hg19:g.32446792_32899507del (ie, CNV-32018 in DGV database) of CHRNA7 (OMIM: 118511), are common in Chinese (Supplementary Table S1). Because the CNV-3956 absolutely encompasses the CNV-32018 (Supplementary Figure S1) and there is no available probe specially for the CNV-32018, we just genotyped the CNV-3956 in all 7880 subjects by the Taqman copy number assay with a special probe for CNV-3956 (ie, cat. no. Hs00059157) according to the protocol of Applied BioSystems, Carlsbad, CA, USA (Supplementary Figure S2A). We also randomly selected 200 samples to genotype the CNV-3956 by the Accucopy assay to validate the genotyping results of the Taqman assay (Supplementary Figure S2B).The results were 95.0% concordant between the two assays. Detailed protocol for the Taqman assay and Accucopy assay are presented in Supplementary Methods. In addition, we submitted the genotype information and frequency of the CNV-3956 to the LOVD 3.0/shared database linked to the cholinergic receptor, nicotinic, α7 (CHRNA7) gene by creating three individuals (ie, nos. 00017868, 00017869 and #00017870) representing lung cancer patients, COPD patients and healthy controls, respectively (http://databases.lovd.nl/shared/genes/CHRNA7).
CHRNA7 expression detection
The CNV-3956 covers the whole CHRNA7 gene and thus may influence the CHRNA7 expression. We detected CHRNA7 expression in 37 lung cancer tissues and their adjacent normal tissues that were collected during surgical excision in the Second and Tumor Hospitals of Guangzhou Medical University (Guangzhou, China) and immediately placed in liquid nitrogen as described previously.18 The mRNA level of CHRNA7 was relatively quantitated to β-actin by the SYBR-Green real-time PCR with a detection kit from Promega company (Madison, WI, USA). The protocol for real-time PCR was routine as described previously.18, 19
Statistical analysis
Differences in the distribution of demographic characteristics and selected factors between cases and controls were analyzed by the χ2 test. The associations between CNV-3956 and two diseases risk were calculated by logistical regression analysis with age, sex, pack-years smoked, biomass fueling and sets as covariables. Homogeneity test was performed with the Breslow–Day test. Interaction between risk factors and variant genotypes was assessed by multiplicative interaction.21 The Kaplan–Meier method, log-rank test and Cox model with adjustment for age, sex, pack-years smoked, biomass fueling, sets, histological types and clinical stages were used to evaluate the effect of the CNV-3956 on lung cancer survival. The REML model was used to assess the heritability explained by the genetic variant.22 A mediation model was used to test the indirect effect that the CNV had on lung cancer via COPD. Detailed statistical protocols for heritability, mediation and interaction test are presented in Supplementary Methods. Differences in CHRNA7 expression and lung function between the CNV-3956 genotypes were analyzed by one-way ANOVA test and Student's t-test. The study power was calculated using the PS Software.23 All tests were two sided and evaluated using the SAS software (version 9.3; SAS Institute, Cary, NC, USA). P<0.05 was considered statistically significant.
Results
As shown in Table 1, in the discovery set, the copy number of CNV-3956 was significantly associated with increased risks of COPD and lung cancer in a copy number-dependent manner (P=4.41 × 10−5 for COPD; P=0.001 for lung cancer). Compared with individuals carrying 2-copy, those carrying ≥4-copy had an increased risk for both COPD and lung cancer, but 3-copy did not. When data were dichotomized, we found that compared with 2/3-copy (ie, 2- or 3-copy genotype), individuals carrying 4- or more copy genotype (ie, ≥4-copy) had 1.44-fold increase in risk for developing COPD (OR=1.44, 95% CI=1.17–1.78; P=6.00 × 10−4) and 1.47-fold increase for lung cancer (OR=1.47, 95% CI=1.20–1.80; P=1.86 × 10−4). Consistent with the above findings, in the validation set, the ≥4-copy also had a significantly increased risk of COPD (OR=1.44, 95% CI=1.14–1.81; P=0.002) and lung cancer (OR=1.52, 95% CI=1.23–1.87; P=8.91 × 10−5) in comparison with the 2/3-copy. Because the above associations between the two populations were homogeneous (P=0.642 for COPD and P=0.958 for lung cancer), we merged the two sets (ie, discovery set and validation set) and found that the ≥4-copy exerted a 1.44-fold increased risk of COPD (OR=1.44, 95% CI=1.23–1.68; P=4.98 × 10−6), whereas it conferred a 1.49-fold increased risk of lung cancer (OR=1.49, 95% CI=1.29–1.73; P=5.54 × 10−8). Furthermore, the heritability test showed that ≥4-copy of CNV-3956 accounts for 1.56% of COPD heritability and 1.87% of lung cancer heritability, respectively. The distribution of demographic characteristics and risk factors of COPD and lung cancer among studied subjects are presented in Supplementary Tables S2 and S3, respectively.
Table 1. Genotype frequency of CHRNA7 CNV-3956 in cases and controls and its associations with COPD and lung cancer risks.
| Southern Chinese (discovery set) | Eastern Chinese (validation set) | Pooled set | |||||||
|---|---|---|---|---|---|---|---|---|---|
| CNV-3956 genotypesa | Cases, n (%) | Controls, n (%) | Adjusted OR (95% CI)b | Cases, n (%) | Controls, n (%) | Adjusted OR (95% CI)b | Cases, n (%) | Controls, n (%) | Adjusted OR (95% CI)b |
| COPD studies | 1025 | 1061 | 766 | 879 | 1791 | 1940 | |||
| 2-Copy | 589 (58.4) | 707 (67.5) | 1.00 (ref.) | 443 (58.6) | 571 (66.2) | 1.00 (ref.) | 1032 (58.5) | 1278 (66.9) | 1.00 (ref.) |
| 3-Copy | 147 (14.6) | 129 (12.3) | 1.38 (1.06–1.80) | 102 (13.5) | 103 (12.0) | 1.28 (0.94–1.74) | 249 (14.1) | 232 (12.2) | 1.34 (1.10–1.64) |
| ≥4-Copy | 273 (27.0) | 212 (20.2) | 1.53 (1.23–1.89) | 211 (27.9) | 188 (21.8) | 1.50 (1.18–1.90) | 484 (27.4) | 400 (20.9) | 1.51 (1.29–1.77) |
| Trend test P-value | 4.41 × 10−5 | 0.001 | 1.00 × 10−7 | ||||||
| 2/3-Copy | 736 (72.9) | 836 (79.8) | 1.00 (ref.) | 545 (72.1) | 674 (78.2) | 1.00 (ref.) | 1281 (72.6) | 1510 (79.1) | 1.00 (ref.) |
| ≥4-Copy | 273 (27.1) | 212 (20.2) | 1.44 (1.17–1.78) | 211 (27.9) | 188 (21.8) | 1.44 (1.14–1.81) | 484 (27.4) | 400 (20.9) | 1.44 (1.23–1.68) |
| Lung cancer studies | 1056 | 1056 | 1016 | 1021 | 2072 | 2077 | |||
| 2-Copy | 591 (57.1) | 666 (64.0) | 1.00 (ref.) | 570 (58.2) | 645 (63.7) | 1.00 (ref.) | 1161 (57.6) | 1311 (63.9) | 1.00 (ref.) |
| 3-Copy | 144 (13.9) | 151 (14.5) | 1.11 (0.86–1.43) | 127 (13.0) | 150 (14.8) | 0.98 (0.75–1.28) | 271 (13.5) | 301 (14.6) | 1.03 (0.86–1.24) |
| ≥4-Copy | 301 (29.0) | 224 (21.5) | 1.50 (1.22–1.85) | 282 (28.8) | 217 (21.4) | 1.51 (1.22–1.87) | 583 (28.9) | 441 (21.5) | 1.50 (1.30–1.74) |
| Trend test P-value | 0.001 | 4.00 × 10−4 | 2.97 × 10−7 | ||||||
| 2/3-Copy | 735 (71.0) | 817 (78.5) | 1.00 (ref.) | 697 (71.2) | 795 (78.6) | 1.00 (ref.) | 1432 (71.1) | 1612 (78.5) | 1.00 (ref.) |
| ≥4-Copy | 301 (29.0) | 224 (21.5) | 1.47 (1.20–1.80) | 282 (28.8) | 217 (21.4) | 1.52 (1.23–1.87) | 583 (28.9) | 441 (21.5) | 1.49 (1.29–1.73) |
The success rate of genotyping was 98.2%.
Adjusted in a logistic regression model with age, sex, pack-years smoked, biomass fueling and sets as covariables. Bold entries indicate a statistical significance.
In stratification analysis as shown in Supplementary Figure S3, the ≥4-copy of CNV-3956 exerted an intuitively higher OR in individuals with pre-existing COPD (OR=2.01) than those without (OR=1.42). Moreover, the ≥4-copy significantly interacted with smoking on increasing risks of both diseases (P=0.006 for COPD and P=0.003 for lung cancer). Taking the nonsmokers carrying the 2/3-copy genotype as reference (OR=1.0), smokers carrying the ≥4-copy genotype exerted the highest OR on risk of COPD (OR=3.18; 95% CI=2.47–4.10) and lung cancer (OR=2.60; 95% CI=2.08–3.26) in comparison with smokers with the 2/3-copy genotype (COPD: OR=1.91; 95% CI=1.59–2.29; lung cancer: OR=1.46; 95% CI=1.24–1.72) and nonsmokers with the ≥4-copy genotype (COPD: OR=1.32; 95% CI=1.07–1.62; lung cancer: OR=1.26; 95% CI=1.03–1.55).
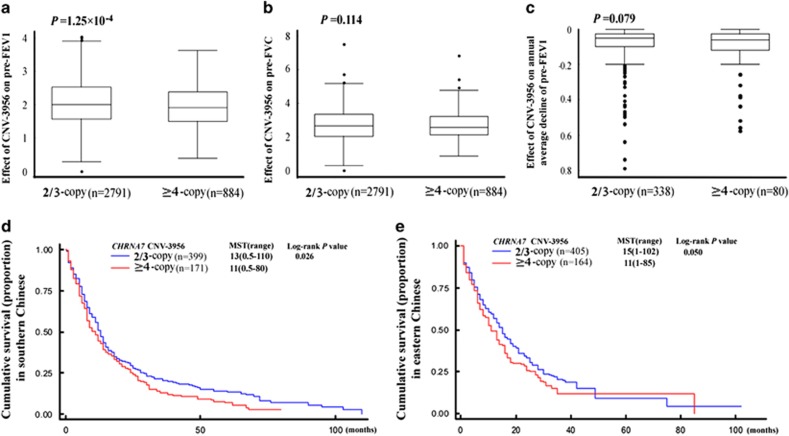
We next examined the effect of CNV-3956 on prognosis of the two diseases. As shown in Figure 1a–c, individuals carrying the ≥4-copy had a significantly lower pre-bronchodilator forced expiratory volume in 1 s (pre-FEV1) than those carrying the 2/3-copy in the total population of COPD study (≥4-copy vs 2/3-copy: mean±SD: 1.94±0.66 vs 2.04±0.70; P=1.25 × 10−4), and the former displayed more annual average decline of pre-FEV1 than the latter in the 427 subjects who had at least 4-year follow-up of lung function (−0.113±0.144 vs −0.087±0.108), but the difference was not significant (P=0.079). However, no significant difference of pre-bronchodilator forced vital capacity (pre-FVC) was observed (P=0.114). Besides, for the total southern and eastern lung cancer patients, the median follow-up periods were 12 months and 15 months, whereas the follow-up periods ranged from 0.5 to 110 months and from 1 to 102 months, respectively. As shown in Figure 1d and e and Table 2, the median survival time (MST) of lung cancer patients from the southern Chinese was significantly decreased, being lowered by 2 months in the ≥4-copy carriers (MST: 13 months) than in the 2/3-copy carriers (MST: 11 months; log-rank test P=0.026). The ≥4-copy genotype also conferred a higher death rate in comparison with the 2/3-copy (hazard ratio (HR)=1.24; 95% CI=1.03–1.50; P=0.025). Results from the eastern Chinese confirmed the above finding, the ≥4-copy exerted a shorter MST and a higher mortality compared with the 2/3-copy (11 vs 15 months, log-rank test P=0.050; HR=1.29; 95% CI=1.01–1.64, P=0.044). We then merged the two populations and found that the ≥4-copy caused a 3-month decrease in MST than the 2/3-copy (14 vs 11; log-rank test P=0.003) and 25% increase in mortality (HR=1.25; 95% CI=1.07–1.45; P=0.004). In addition, stratification analysis of survival revealed that the above effect was lost in some subgroups such as smoking avoiders. However, no significant interaction was observed between the ≥4-copy and selected variables on lung cancer survival (Supplementary Figure S4).
Figure 1.
Effect of the CHRNA7 CNV-3956 on lung function of COPD and survival of lung cancer. (a) The effect of CNV-3956 on pre-FEV1; (b) the effect of CNV-3956 on pre-FVC; (c) the effect of CNV-3956 on annual average decline of pre-FEV1; (d) Kaplan–Meier survival curve for individuals with lung cancer by the CNV-3956 genotypes in the southern Chinese; and (e) Kaplan–Meier survival curve for individuals with lung cancer by the CNV-3956 genotypes in the eastern Chinese. Columns indicate mean and bars indicate SD; Student's t-test was used to test the differences in lung function between the CNV-3956 genotypes, and the log-rank test was used to assess the effect of CNV-3956 on lung cancer survival. As shown, individuals carrying ≥4-copy had a significantly lower pre-FEV1 but not pre-FVC as well as annual average decline of pre-FEV1 than those carrying 2/3-copy in the total population of COPD study. In addition, the median survival time (MST) of lung cancer patients was significantly decreased in the ≥4-copy carriers than in the 2/3-copy carriers.
Table 2. Effect of CHRNA7 CNV-3956 on lung cancer survival in lung cancer patients.
| CNV-3956 genotypes | n (%) | Death | MST (months) | Log-rank P-value | HR (95% CI)a | Cox mode P-valuea |
|---|---|---|---|---|---|---|
| Southern Chinese | 570 | 515 | 12 | |||
| 2-Copy | 330 (57.9) | 291 | 13 | 0.082 | 1.00 (ref.) | |
| 3-Copy | 69 (12.1) | 62 | 13 | 1.06 (0.80–1.39) | 0.694 | |
| ≥4-Copy | 171 (30.0) | 162 | 11 | 1.25 (1.03–1.52) | 0.023 | |
| Trend test P-value | 0.026 | |||||
| 2/3-Copy | 399 (70.0) | 353 | 13 | 0.026 | 1.00 (ref.) | |
| ≥4-Copy | 171 (30.0) | 162 | 11 | 1.24 (1.03–1.50) | 0.025 | |
| Eastern Chinese | 569 | 408 | 15 | |||
| 2-Copy | 336 (59.1) | 230 | 15 | 0.141 | 1.00 (ref.) | |
| 3-Copy | 69(12.1) | 48 | 15 | 0.90 (0.62–1.29) | 0.551 | |
| ≥4-Copy | 164 (28.8) | 130 | 11 | 1.26 (0.98–1.62) | 0.076 | |
| Trend test P-value | 0.101 | |||||
| 2/3-Copy | 405 (71.2) | 278 | 15 | 0.050 | 1.00 (ref.) | |
| ≥4-Copy | 164 (28.8) | 130 | 11 | 1.29 (1.01–1.64) | 0.044 | |
| Merged population | 1139 | 923 | 12 | |||
| 2-Copy | 666 (58.5) | 521 | 14 | 0.011 | 1.00 (ref.) | |
| 3-Copy | 138 (12.1) | 110 | 13 | 1.02 (0.82–1.27) | 0.881 | |
| ≥4-Copy | 335 (29.4) | 292 | 11 | 1.25 (1.07–1.46) | 0.004 | |
| Trend test P-value | 0.006 | |||||
| 2/3-Copy | 804 (70.6) | 631 | 14 | 0.003 | 1.00 (ref.) | |
| ≥4-Copy | 335 (29.4) | 292 | 11 | 1.25 (1.07–1.45) | 0.004 |
Abbreviation: MST, median survival time. Bold entries indicate a statistical significance.
Cox regression analysis was adjusted for age, sex, pack-years smoked, biomass fueling, sets, histological types and clinical stages.
We further examined the association between the CNV-3956 and smoking behavior in the total 7880 subjects (Table 3). The individuals carrying the ≥4-copy were more likely to become heavy smokers (pack-years smoked ≥43) than those carrying the 2/3-copy (P=3.00 × 10−4). The mean pack-years smoked was also significantly higher in the former than the latter (≥4-copy vs 3-copy vs 2-copy: 35.1±25.9 vs 31.4±25.1 vs 31.6±23.2; P=0.001). However, the CNV seemed to have no effect on smoking status (i.e., never, former or current).
Table 3. Distribution of genotypes of the CNV-3956 by smoking status in Chinese populations.
| CNV-3956 genotypes | |||
|---|---|---|---|
| 2-Copy, n % | 3-Copy, n % | ≥4-Copy, n % | |
| Smoking status | |||
| Never (n=4050) | 2517 (52.6) | 539 (51.2) | 994 (52.1) |
| Former (n=1463) | 887 (18.6) | 220 (20.9) | 356 (18.7) |
| Current (n=2230) | 1378 (28.8) | 294 (27.9) | 558 (29.2) |
| Group test P-valuea | 0.499 | ||
| Pack-years smokedb | |||
| ≤ 15 (n=1016) | 657 (29.0) | 156 (30.4) | 203 (22.2) |
| 15–43 (n=1749) | 1052 (46.5) | 248 (48.2) | 449 (49.1) |
| ≥43 (n=928) | 556 (24.5) | 110 (21.4) | 262 (28.7) |
| Group test P-valuea | 3.00 × 10−4 | ||
The two-sided χ2 test between difference groups.
Categorized by using the 25th and 75th percentile pack-years smoked as the cutoff points.
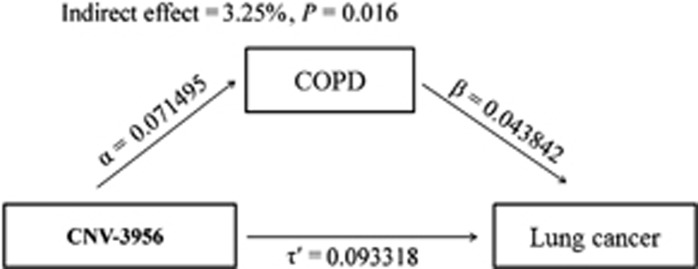
Because the CNV-3956 was significantly associated with pack-years smoked, COPD and lung cancer, and pack-years smoked and COPD are well-established risk factors of lung cancer, we analyzed whether pack-years smoked or COPD acted as a mediator on the CNV-caused development of lung cancer using the mediation test. The results showed that 3.25% of the effect caused by CNV-3956 went through COPD (Sobel test: P=0.016; Figure 2). However, no significant mediation effect was observed for pack-years smoked (data not shown). Moreover, we also did not find significant mediation effect of pack-years smoked on association between the CNV and COPD risk (data not shown).
Figure 2.
Path model for mediating effect of COPD on association between the CNV-3956 and lung cancer risk. α Was the comparable regression coefficient for association between the CNV-3956 and COPD; β was the comparable regression coefficient for association between COPD and lung cancer; τ′ was the comparable regression coefficient for association between the CNV-3956 and lung cancer; αβ was the indirect effect of COPD on lung cancer, and τ′ was the direct effect of CNV-3956 on lung cancer. The total effect of CNV-3956 on lung cancer τ=τ′+αβ and COPD explained αβ/τ × 100% effect of factor on lung cancer risk. Here, the mediation effect of COPD on association between the CNV-3956 and lung cancer risk was αβ/τ=0.071495 × 0.043842/(0.071495 × 0.043842+0.093318)=3.25%.
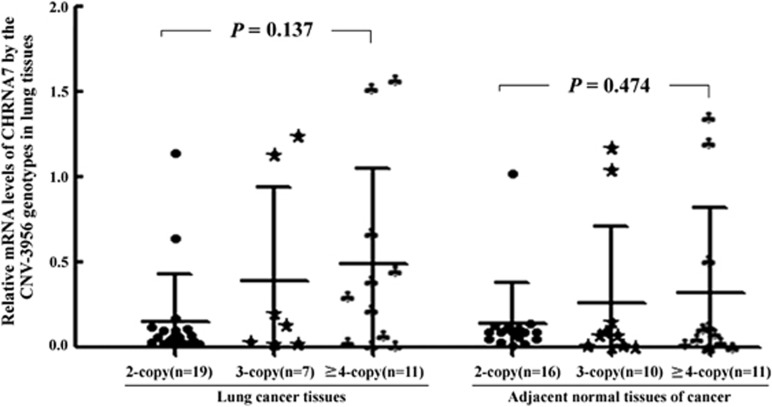
The CNV-3956 may alter the CHRNA7 expression underlying a dosage effect. As shown in Figure 3, although a significant difference in CHRNA7 mRNA levels between the three genotypes of CNV-3956 in cancer tissues was not observed (ANOVA test: P=0.137), the ≥4-copy was associated with a significantly higher CHRNA7 expression than the 2-copy (Student's t-test: P=0.037). However, the above effect was not observed in the normal tissues, reflecting that there are some unrecognized mechanisms modulating the genetic effect of CNV-3956, such as gene–environment interaction.
Figure 3.
Correlation between the CHRNA7 CNV-3956 genotypes and CHRNA7 mRNA level in lung cancer tissues and their adjacent normal lung tissues. The mRNA level of CHRNA7 was relatively quantitated to β-actin. The horizontal lines refer to mean and SD of two sides. Columns indicate mean from three independent experiments and bars indicate SD; the ANOVA test was used to analyze the difference in relative mRNA levels of CHRNA7 between the three genotypes of CNV-3956. Although a significant difference in CHRNA7 mRNA levels between the three genotypes of CNV-3956 in cancer tissues was not observed (P=0.137), ≥4-copy had a significantly higher CHRNA7 expression than 2-copy (Student's t-test: P=0.037).
Discussion
In the current two, two-stage, case–control studies of southern and eastern Chinese, we found that the ≥4-copy of CNV-3956 of CHRNA7 conferred to an increased risk of COPD and lung cancer and an enhancive decline in lung function as well as poor survival of lung cancer, which may be due to an underlying mechanism that ≥4-copy of CNV-3956 improved CHRNA7 expression and exerted an increase in smoking amount. The CNV could explain ∼1.56% of COPD heritability and 1.87% of lung cancer heritability. Furthermore, the effect of CNV-3956 on lung cancer risk was mediated by COPD, 3.25% of which was exerted indirectly through COPD. To the best of our knowledge, we first reported the associations between the CNVs in the nAChR genes and development of lung diseases.
CHRNA7 is one of the most predominant nAChRs expressed in both central nervous system and pulmonary tissue,24, 25, 26 and is responsible for mediating the effects of nicotine addict and toxicological effect caused by nicotine metabolites, such as nicotine-derived nitrosamine ketone (NNK).27, 28, 29 CHRNA7 may play a role in the pathogenesis of COPD and lung cancer because of its effect on nicotine addiction. Evidence that active CHRNA7 can induce lung fibroblast and regulate lung inflammation also supports it may involve the development of both diseases.30, 31 In the current study, we found that the ≥4-copy of CNV-3956 that duplicates the CHRNA7 gene conferred increased risks and poor prognoses of lung cancer and COPD, underlying a possible biological mechanism by increasing CHRNA7 expression and inducing carriers to consume more cigarettes because individuals with higher CHRNA7 expressions would be more nicotine sensitive. A challenge on this mechanism is that the loss genotype of CNV-32018, which we did not test due to technical limitation, would possibly induce exon deletion of CHRNA7 and nonfunctional CHRNA7, and thus the ≥4-copy of CNV-3956 may cause a upregulation of nonfunctional CHRNA7. To rule out such a possibility, we queried the Ensemble database and found no CHRNA7 transcript with a loss of the last exons (http://asia.ensembl.org/Homo_sapiens/Gene/Summary?db=core;g=ENSG00000175344;r=15:32322691-32464722). It means that there may be some unknown mechanisms on limiting the effect of CNV-32018. Anyway, on account of the fact that smoking contributes to the development of COPD and lung cancer,32 it is conceivable from our finding that CNV-3956 was associated with increased risks and poor prognoses of both diseases. Moreover, the adverse effect could be modulated by smoking because the CNV had a significant interaction with smoking on increasing the risk of both diseases. Expression of CHRNA7 is nicotine sensitive,26 and therefore the adverse effect of CNV-3956 might be smoking dependent.
Remarkably, we found that COPD had a significant mediation effect on the CNV-3956–lung cancer association and explained ∼3.25% of the total risk effect that CNV had on lung cancer, reflecting that COPD might be an intermediate phenotype in the process of CNV-induced development of lung cancer. The ≥4-copy carriers of CNV-3956 are more likely to develop COPD, and in turn lead to a high risk of lung cancer.
In addition, the association between CNV-3956 and survival of lung cancer is noteworthy, although previous studies have reported SNPs in other nAChRs genes with effects on lung cancer survival.11, 14, 15 Compared with other nAChRs, CHRNA7 is the major responser of tobacco stimulation that induces tumor cell resistance to cell apoptosis through Akt- dependent proliferation and NF-κB-dependent survival pathways.33 Inhibition of CHRNA7 can reduce the tumorigenicity of lung cancer cells.34 It is well accepted now that CHRNA7 is a valuable molecular target for therapy of lung cancer and its antagonists (eg, D-tubocurarine or a-CbT) might provide ‘efficacious adjuvant therapy' for lung cancer.25 Therefore, on the fact of that the CNV-3956 caused a different CHRNA7 expression, it should be in consideration for future individual's treatment.
Human CHRNA7 gene is located on the chromosome 15q14. By now, several studies have demonstrated that genetic variants in CHRNA7 were associated with various human diseases and traits, such as the associations between CNVs in CHRNA7 and various neurological disorders.35, 36, 37, 38 In addition, one study conducted in African and European Americans showed that one SNP of CHRNA7 was associated with nicotine dependence,27 suggesting that the SNPs of CHRNA7 might show association with smoking-related diseases. Yet, till now, no published studies including genome-wide association studies (GWASs) have reported a significant association between CHRNA7 SNPs and the risk of smoking-related diseases, and this may be because of the stringent criterion of statistical significance (P<10−5) of GWASs and a lack of attention to SNPs of other nAChR genes. Moreover, the CNVs that lie in the CHRNA7 region might cause inadequate cover in this region of GWAS platforms. Here, we showed that CNV-3956 of CHRNA7exerted an adverse effect on the development of COPD and lung cancer, and explained ∼1.56% of COPD heritability and 1.87% of lung cancer heritability.
This study has several strengths. Based on two populations with a relatively large sample size, we have achieved consistent results for both COPD and lung cancer studies in the two studied groups. The study power was strong (99.9% for COPD to detect an OR of 1.44 for ≥4-copy and 99.8% for lung cancer to detect an OR of 1.49 for ≥4-copy compared with other genotypes). The genotype–phenotype correlation also supported our findings. However, as these were hospital-based case–control studies, some limitations like selection bias or information bias are unavoidable.
Taken together, our study first reported significant associations between CNV-3956 in the CHRNA7 gene and risks as well as prognoses of COPD and lung cancer, possibly underlying a biological mechanism that CNV-3956 affected CHRNA7 expression and smoking. The effect of CNV-3956 on lung cancer risk was mediated by COPD. All these results suggest that CNV-3956 in CHRNA7 may be a common genetic biomarker to predict risks and prognoses of COPD and lung cancer. Validations with larger population-based studies in different ethnic groups are warranted.
Acknowledgments
We thank Dr Zhanhong Xie, Ms Wanmin Zeng and Ling Liu for their assistance in recruiting the subjects. This study was supported by the National Natural Scientific Foundation of China Grants 30671813, 30872178, 81072366, 81273149 and 81473040 (to J Lu), and partly by 81001278 and 81171895 (to Y Zhou) and 81402753 (to L Yang), the Guangdong Provincial Scientific Research Grants 8251018201000005 (to J Lu), Guangdong Provincial High Level Experts Grants 2010-79, Changjiang Scholars and Innovative Research Team in University Grant IRT0961 and Guangdong Natural Science Foundation Team Grant 10351012003000000 (to J Lu).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Houghton AM: Mechanistic links between COPD and lung cancer. Nat Rev Cancer 2013; 13: 233–245. [DOI] [PubMed] [Google Scholar]
- El-Zein RA, Young RP, Hopkins RJ, Etzel CJ: Genetic predisposition to chronic obstructive pulmonary disease and/or lung cancer: important considerations when evaluating risk. Cancer Prev Res (Phila) 2012; 5: 522–527. [DOI] [PubMed] [Google Scholar]
- Young RP, Hopkins RJ, Christmas T, Black PN, Metcalf P, Gamble GD: COPD prevalence is increased in lung cancer, independent of age, sex and smoking history. Eur Respir J 2009; 34: 380–386. [DOI] [PubMed] [Google Scholar]
- de Torres JP, Marin JM, Casanova C et al: Lung cancer in patients with chronic obstructive pulmonary disease— incidence and predicting factors. Am J Respir Crit Care Med 2011; 184: 913–919. [DOI] [PubMed] [Google Scholar]
- Wang H, Yang L, Zou L et al: Association between chronic obstructive pulmonary disease and lung cancer: a case-control study in Southern Chinese and a meta-analysis. PLoS One 2012; 7: e46144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Spitz MR, Amos CI, Wilkinson AV, Wu X, Shete S: Mediating effects of smoking and chronic obstructive pulmonary disease on the relation between the CHRNA5-A3 genetic locus and lung cancer risk. Cancer 2010; 116: 3458–3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine Y, Katsura H, Koh E, Hiroshima K, Fujisawa T: Early detection of COPD is important for lung cancer surveillance. Eur Respir J 2012; 39: 1230–1240. [DOI] [PubMed] [Google Scholar]
- Schwartz AG, Ruckdeschel JC: Familial lung cancer: genetic susceptibility and relationship to chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2006; 173: 16–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RP, Hopkins RJ: How the genetics of lung cancer may overlap with COPD. Respirology 2011; 16: 1047–1055. [DOI] [PubMed] [Google Scholar]
- Yang L, Yang X, Ji W et al: Effects of a functional variant c.353T>C in Snai1 on risk of two contextual diseases: COPD and lung cancer. Am J Respir Crit Care Med 2014; 189: 139–148. [DOI] [PubMed] [Google Scholar]
- Yang L, Qiu F, Lu X et al: Functional polymorphisms of CHRNA3 predict risks of chronic obstructive pulmonary disease and lung cancer in Chinese. PLoS One 2012; 7: e46071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorgeirsson TE, Gudbjartsson DF, Surakka I et al: Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat Genet 2010; 42: 448–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone NL, Culverhouse RC, Schwantes-An TH et al: Multiple independent loci at chromosome 15q25.1 affect smoking quantity: a meta-analysis and comparison with lung cancer and COPD. PLoS Genet 2010; 6: pii e1001053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin G, Bae EY, Yang E et al: A functional polymorphism on chromosome 15q25 associated with survival of early stage non-small-cell lung cancer. J Thorac Oncol 2012; 7: 808–814. [DOI] [PubMed] [Google Scholar]
- Niu X, Chen Z, Shen S et al: Association of the CHRNA3 locus with lung cancer risk and prognosis in Chinese Han population. J Thorac Oncol 2010; 5: 658–666. [DOI] [PubMed] [Google Scholar]
- Zhou J, Lemos B, Dopman EB, Hartl DL: Copy-number variation: the balance between gene dosage and expression in Drosophila melanogaster. Genome Biol Evol 2011; 3: 1014–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Yang L, Liu Y et al: Functional polymorphisms in NFkappaB1/IkappaBalpha predict risks of chronic obstructive pulmonary disease and lung cancer in Chinese. Hum Genet 2013; 132: 451–460. [DOI] [PubMed] [Google Scholar]
- Yang L, Liu B, Huang B et al: A functional copy number variation in the WWOX gene is associated with lung cancer risk in Chinese. Hum Mol Genet 2013; 22: 1886–1894. [DOI] [PubMed] [Google Scholar]
- Liu B, Yang L, Huang B et al: A functional copy-number variation in MAPKAPK2 predicts risk and prognosis of lung cancer. Am J Hum Genet 2012; 91: 384–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Hu G, Wang D et al: Community based integrated intervention for prevention and management of chronic obstructive pulmonary disease (COPD) in Guangdong, China: cluster randomised controlled trial. BMJ 2010; 341: c6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Liu B, Yang L et al: Functional genetic variants of c-Jun and their interaction with smoking and drinking increase the susceptibility to lung cancer in southern and eastern Chinese. Int J Cancer 2012; 131: E744–E758. [DOI] [PubMed] [Google Scholar]
- Vattikuti S, Guo J, Chow CC: Heritability and genetic correlations explained by common SNPs for metabolic syndrome traits. PLoS Genet 2012; 8: e1002637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont WD, Plummer WD Jr.: Power and sample size calculations for studies involving linear regression. Control Clin Trials 1998; 19: 589–601. [DOI] [PubMed] [Google Scholar]
- Albuquerque EX, Pereira EF, Alkondon M, Rogers SW: Mammalian nicotinic acetylcholine receptors: from structure to function. Physiol Rev 2009; 89: 73–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paleari L, Cesario A, Fini M, Russo P: alpha7-Nicotinic receptor antagonists at the beginning of a clinical era for NSCLC and Mesothelioma? Drug Discov Today 2009; 14: 822–836. [DOI] [PubMed] [Google Scholar]
- Lam DC, Girard L, Ramirez R et al: Expression of nicotinic acetylcholine receptor subunit genes in non-small-cell lung cancer reveals differences between smokers and nonsmokers. Cancer Res 2007; 67: 4638–4647. [DOI] [PubMed] [Google Scholar]
- Saccone NL, Schwantes-An TH, Wang JC et al: Multiple cholinergic nicotinic receptor genes affect nicotine dependence risk in African and European Americans. Genes Brain Behav 2010; 9: 741–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Improgo MR, Tapper AR, Gardner PD: Nicotinic acetylcholine receptor-mediated mechanisms in lung cancer. Biochem Pharmacol 2011; 82: 1015–1021. [DOI] [PubMed] [Google Scholar]
- Schuller HM: Is cancer triggered by altered signalling of nicotinic acetylcholine receptors? Nat Rev Cancer 2009; 9: 195–205. [DOI] [PubMed] [Google Scholar]
- Roman J, Ritzenthaler JD, Gil-Acosta A, Rivera HN, Roser-Page S: Nicotine and fibronectin expression in lung fibroblasts: implications for tobacco-related lung tissue remodeling. FASEB J 2004; 18: 1436–1438. [DOI] [PubMed] [Google Scholar]
- Wang H, Yu M, Ochani M et al: Nicotinic acetylcholine receptor alpha7 subunit is an essential regulator of inflammation. Nature 2003; 421: 384–388. [DOI] [PubMed] [Google Scholar]
- Thun MJ, Carter BD, Feskanich D et al: 50-year trends in smoking-related mortality in the United States. N Engl J Med 2013; 368: 351–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurutani J, Castillo SS, Brognard J et al: Tobacco components stimulate Akt-dependent proliferation and NFkappaB-dependent survival in lung cancer cells. Carcinogenesis 2005; 26: 1182–1195. [DOI] [PubMed] [Google Scholar]
- Paleari L, Sessa F, Catassi A et al: Inhibition of non-neuronal alpha7-nicotinic receptor reduces tumorigenicity in A549 NSCLC xenografts. Int J Cancer 2009; 125: 199–211. [DOI] [PubMed] [Google Scholar]
- Williams NM, Franke B, Mick E et al: Genome-wide analysis of copy number variants in attention deficit hyperactivity disorder: the role of rare variants and duplications at 15q13.3. Am J Psychiatry 2012; 169: 195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szafranski P, Schaaf CP, Person RE et al: Structures and molecular mechanisms for common 15q13.3 microduplications involving CHRNA7: benign or pathological? Hum Mutat 2010; 31: 840–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzen EL, Need AC, Hayden KM et al: Genome-wide scan of copy number variation in late-onset Alzheimer's disease. J Alzheimers Dis 2010; 19: 69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DT, Shen Y, Weiss LA et al: Microdeletion/duplication at 15q13.2q13.3 among individuals with features of autism and other neuropsychiatric disorders. J Med Genet 2009; 46: 242–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.