Abstract
Liver disease is a major global health concern. Liver cirrhosis is one of the leading causes of death in the world and currently the only therapeutic option for end-stage liver disease (e.g., acute liver failure, cirrhosis, chronic hepatitis, cholestatic diseases, metabolic diseases, and malignant neoplasms) is orthotropic liver transplantation. Transplantation of hepatocytes has been proposed and used as an alternative to whole organ transplant to stabilize and prolong the lives of patients in some clinical cases. Although these experimental therapies have demonstrated promising and beneficial results, their routine use remains a challenge due to the shortage of donor livers available for cell isolation, variable quality of those tissues, the potential need for lifelong immunosuppression in the transplant recipient, and high costs. Therefore, new therapeutic strategies and more reliable clinical treatments are urgently needed. Recent and continuous technological advances in the development of stem cells suggest they may be beneficial in this respect. In this review, we summarize the history of stem cell and induced pluripotent stem cell (iPSC) technology in the context of hepatic differentiation and discuss the potential applications the technology may offer for human liver disease modeling and treatment. This includes developing safer drugs and cell-based therapies to improve the outcomes of patients with currently incurable health illnesses. We also review promising advances in other disease areas to highlight how the stem cell technology could be applied to liver diseases in the future.
1.0 INTRODUCTION: STEM CELLS
The concept of one cell type that can give rise to all cells of the human body is very exciting to the medical and scientific world. This pluripotent stem cell could theoretically be used to produce unlimited amounts of terminally differentiated cells that may be used to correct a disease phenotype or stabilize a patient until an organ for transplant becomes available. Stem cells could also be used for in vitro drug discovery and screening. Furthermore, starting from biological material from patients with genetic diseases, cells can be differentiated to the cell type affected by the disease and used to model and understand disease-related mechanisms. Disease-specific cells can also be genetically corrected and used for cell transplantation to correct the phenotype. All of the above applications have been reported.
Classically, pluripotent stem cells are derived from embryonic sources and are called embryonic stem cells (ESCs). However, pluripotency can also be achieved in somatic cells by forced expression of specific transcription factors, nuclear transfer to an unfertilized oocyte, or fusion to an ESC. Stem cells have also been isolated from various adult tissues, without the need for reprogramming, and are called adult stem cells. These stem cells are considered multipotent, rather than pluripotent, because they demonstrate limited differentiation capabilities in culture. Adult stem cells and their potential use to study and treat liver diseases are beyond the scope of this report.
2.0 GENERAL STEM CELL INFORMATION – FROM ESCS TO IPSCS
ESCs are derived from the inner cell mass of the blastocyst. They have normal karyotypes, express high levels of telomerase activity, and maintain the developmental potential to differentiate to all 3 embryonic germ layers. Almost twenty years ago, Thompson and colleagues reported the first derivation and characterization of human ESCs (Thomson et al., 1998).
The ethical dilemma behind the formation of the various human ESC lines remains a challenging issue for their use in regenerative medicine and in vitro drug discovery. The creation of each ESC line involves developing a fertilized egg to the pre-implantation blastocyst stage and isolating the cells from the inner cell mass, which will eventually become the ESC line. Some believe this process to be destruction of a human life, therefore making this a topic for debate. However, if an alternative pluripotent cell source were to become available that is similar to the proliferative and developmental potential of ESCs, there would no longer be an ethical issue. The discovery and creation of induced pluripotent stem cells (iPSCs) circumvents this ethical issue.
Mammalian embryogenesis begins with the fertilization of the female oocyte by a male sperm to form a single cell that contains the proper 2N content of genetic material. Successive cell divisions lead to the formation of the pre-implantation blastocyst that contains the outer trophectoderm and inner cell mass. Directed differentiation of the cells within the blastocyst to their appropriate cell type is coordinated by highly specific, spatially and temporally controlled, molecular signaling events. These observations indicate that development potential is controlled by manipulation of global DNA gene expression within the cell rather than direct genetic code alterations.
When researchers successfully performed somatic cell nuclear transfer in sheep they experimentally proved that development potential is controlled by manipulation of global DNA gene expression within the mammalian cell (Wilmut et al., 1997). Briefly, Wilmut and colleagues transferred a somatic cell nucleus to an enucleated unfertilized egg and implanted the egg within a host uterus, which led to the birth of a live lamb. This experiment provided evidence that differentiation of a cell toward a somatic state is not accomplished by irreversible genetic alterations. A cell becomes specified not by changing its DNA sequence, but by controlling the expression of certain genes through specific controls. Cell differentiation and pluripotency was no longer seen as unidirectional. Somatic cell nuclear transfer has yet to be successfully accomplished with human cells.
Since the successful somatic cell nuclear transfer experiments, researchers have determined ways to manipulate a cell’s developmental potential back to an embryonic-like state where, theoretically, developmental potential is at its highest plasticity. One approach to generate pluripotent stem cells from somatic cells was discovered when mouse embryonic or adult fibroblasts were reprogrammed to an ESC-like state by forced transcription of Oct3/4, Sox2, c-Myc, and Klf4 by lentiviral induction (Takahashi and Yamanaka, 2006). These cells were similar to ESCs in morphology, growth properties, expression of ESC marker genes, teratoma formation, and injection into a blastocyst yielded cells that contributed to multiple tissues during mouse development, including the germ line. Further characteristics of these ESC-like cells include DNA demethylation of key pluripotency genes and regulators, endogenous expression of pluripotency genes, and down-regulation of viral genes used to induce pluripotency. These cells were called iPSCs. A year later, generation of iPSCs was confirmed to be possible in adult human dermal fibroblasts by the same method (Takahashi et al., 2007) and by forced expression of a new set of 4 factors: Oct4, Sox2, Nanog, and Lin28 (Yu et al., 2007).
It is important to know the molecular mechanisms associated with the reprogramming process. This information will enable researchers to determine more efficient and reproducible ways to generate iPSCs. It will also help in assessing if iPSCs are safe to use in a clinical setting. This is based on what pathways are being manipulated and if this can potentially lead to complications if the cells are transplanted into a patient. Important pathways and events involved in the reprogramming process include: p53 and its various downstream targets (Choi et al., 2011; Mali et al., 2008; Zhao et al., 2008), innate immune response pathways and regulators (Angel and Yanik, 2010), pathways downstream of hypoxia inducible factors (Yoshida et al., 2009), MEK-ERK and TGFβ pathways (Lin et al., 2009), retinoic acid signaling (Wang et al., 2011), AID-dependent DNA demethylation (Bhutani et al., 2010), downstream targets of the orphan nuclear receptor Esrrb (Feng et al., 2009), and up-regulation of epithelial cell adhesion molecule complex proteins (Huang et al., 2011). Furthermore, it has been determined that reprogramming factor stoichiometry influences the epigenetic state of resulting iPSCs (Carey et al., 2011). Different epigenetic states of iPSCs influence the resulting pluripotency and developmental potential of generated iPSCs and can affect the tumorigenicity of cells when transplanted in vivo. Finally, high resolution time-lapsed imaging showed that successfully reprogrammed cells undergo a rapid shift in their proliferative rate that coincides with a reduction in cell size (Smith et al., 2010). This observation suggests that part of the reprogramming process follows defined rather than stochastic steps.
Since generating iPSCs involves forced expression of transcription factors it is not surprising that there are variations among the developmental and growth potential of different cell lines. Through the use of live cell imaging, observations were made that lead to the conclusion that true reprogrammed colonies express: TRA1-60, DNMT3B, and REX1 and are able to silence viral DNA (Chan et al., 2009). It was also determined that partially reprogrammed intermediates are alkaline phosphatase positive as well as express SSEA-4, GDF4, hTERT, and NANOG (Chan et al., 2009). This information is important because iPSCs that have the greatest developmental potential need to be identified early in the generation process so resources are not spent expanding and investigating the use of cell lines with limited differentiation capabilities.
True pluripotency is determined by the ability of stem cells to differentiate to all 3 germ layers. Therefore in order to claim iPSCs are pluripotent, investigators needed to describe in vitro and in vivo methods to differentiate iPSCs to all cell types of the 3 germ layers. Generation of a chimeric embryo that contains cells from the host and the donor in all tissues is the gold-standard method to establish pluripotency. However, teratoma formation is the “gold standard” in vivo method researchers use to determine pluripotency in cells derived from human sources because creating a chimeric embryo is not possible. In this assay, if an injected iPSC line forms tumors consisting of all cell types of the 3 germ layers then the cell line is fully reprogrammed and pluripotent. As for in vitro methods, investigators used prior studies performed on ESCs as a platform for differentiation. Researchers began to determine chemically defined culture conditions that model the signaling events known to occur in tissue specification during embryogenesis. Reports have been generated where iPSCs were differentiated to ventral midbrain dopaminergic neurons (Cooper et al., 2010), neurospheres (Nori et al., 2011), hematopoietic and endothelial cells (Choi et al., 2009), cardiomyocytes (Tanaka et al., 2009; Yokoo et al., 2009; Zhang et al., 2009), and insulin secreting islet-like clusters (Tateishi et al., 2008). These reports of in vitro differentiation of iPSCs represent lineages from all 3 germ layers. In each instance iPSCs were derived from fibroblasts and differentiation to desired cell type was achieved, however the phenotype observed in culture is frequently not 100% comparable to the level of a primary cell. Thus in vitro differentiation needs to be optimized.
Figure 1 summarizes some of the potential uses and applications of human stem cell technology in bioresearch and medicine.
Figure 14.13.1.
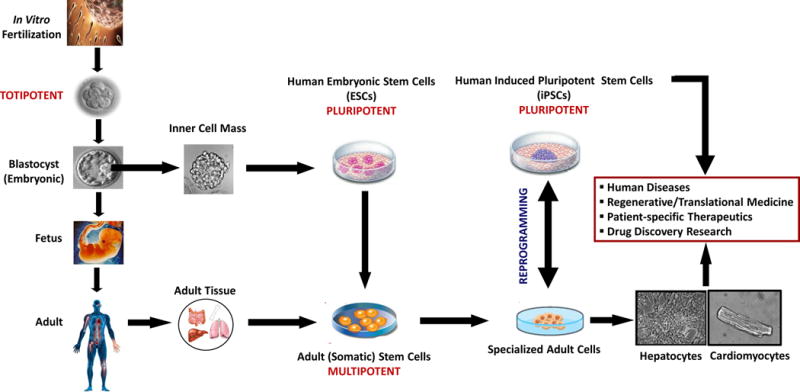
Potential use and application of human stem cell technology in bioresearch and medicine. When a sperm fertilizes an egg, a single cell is created; in a few hours, this fertilized egg divides into identical cells, capable of forming an entire organism (totipotent cells). Approximately four days after fertilization, these totipotent cells begin to form the blastocyst, which is a hollow sphere of cells with an inner cell mass. The inner cell mass of the blastocyst can be isolated and cultured to form colonies of cells known as human embryonic stem cells (hESCs). These pluripotent stem cells can form cell types of all three germ layers of the human body (ectoderm, endoderm, and mesoderm) and may undergo further specialization to give rise to more distinctive cells known as multipotent stem cells. Multipotent stem cells are capable of generating some adult cell types but not all cell types of the organism. Adult cells can be reprogrammed in vitro to cells that have cell surface markers, gene expression, and other characteristics of pluripotent stem cells. These adult cells are reprogrammed by introducing a set of key transcription factors into the cells. These cells are known as human induced pluripotent stem cells (hiPSCs) and they are able to form cell types of all three germ layers. Human stem cell technology has the potential to provide unprecedented opportunities to study, treat, and prevent a vast array of human diseases by establishing physiologically relevant cell-based models for biomedical research, regenerative medicine, and personalized drug discovery and testing. Modified from Davila et al., 2009.
3.0 IPSC DERIVATION – VARIOUS METHODS AND CELL SOURCES
In order to make iPSC technology clinically relevant, it is preferred to induce pluripotency without viral DNA integration into the target cell genome, which can create unwanted mutations due to random integration of DNA with uncontrolled copy numbers. Mouse fibroblasts and hepatocytes were shown to be successfully reprogrammed by using non integrating adenoviruses to deliver the reprogramming factors demonstrating that reprogramming to pluripotency could be achieved through transient expression of viral genes (Stadtfeld et al., 2008). Another report recently reported that temperature-sensitive Sendai viral vectors can be used, resulting in transgene-free iPSCs (Ban et al., 2011). Vector integration-free human iPSCs have been derived by using nonviral techniques. These techniques include using episomes derived from the Epstein-Barr virus (Yu et al., 2009) and transient transfection of plasmid DNA encoding reprogramming factors (Si-Tayeb et al., 2010b). Furthermore, it has been demonstrated that a novel nonviral minicircle vector, free of bacterial DNA, can be used to induce pluripotency (Jia et al., 2010). These reports demonstrated that somatic cell reprogramming does not require transgene genomic integration or continued expression of exogenous DNA. Other non-integrating methods used to generate iPSCs include using mRNA transfection (Warren et al., 2010; Yakubov et al., 2010) and direct delivery of reprogramming proteins (Kim et al., 2009). Investigators have also reported being able to use a bacteriophage ΦC31 integrase to reprogram cells with only a single integration site with locations favoring intron regions, thus producing cells with undisturbed endogenous gene function (Ye et al., 2010). It has also been shown that poly-β-amino esters can be used to transfect plasmids. Although this technique still resulted in iPSCs that contain transgenes, it did not require the use of a virus to deliver the genes (Montserrat et al., 2010). Researchers were able to induce pluripotency without the use of Oct4 under entirely feeder-free conditions, although still using integrating lentiviral vectors (Montserrat et al., 2011). Finally, a report demonstrated that reprogramming can be achieved using Dox-inducible lentiviral vectors (Hockemeyer et al., 2008).
The use of the oncogenes, such as c-Myc and Klf4, to induce pluripotency is also a problem when trying to make iPSCs more clinically relevant. Studies have shown that reprogramming could be successful without using c-Myc and Klf4. Human fetal gut mesentery-derived cells and human amnion using Oct4, Sox2, and Nanog (Li et al., 2010; Zhao et al., 2010) and primary human fibroblasts using only Oct4 and Sox2 (Huangfu et al., 2008) were shown to be successfully reprogrammed to pluripotent cells. It is important to point out that although iPSCs can be derived without forced expression of Klf4 or c-Myc, the efficiency of reprogramming is drastically reduced. Table 1 summarizes the first reports of the different ways to induce pluripotency and if the method results in transgene-free cells.
Table 1. Methods to Generate iPSCs.
This table summarizes the first reports of different ways to induce pluripotency, species of cells that were used, and if that method leads to transgene-free cells. This table is not an exhaustive list and is meant to be representative.
| Reference | Species | Method | Transgene-free |
|---|---|---|---|
| (Takahashi and Yamanaka, 2006) | Mouse | Lentivirus | No |
| (Takahashi et al., 2007), (Yu et al., 2007) | Human | Lentivirus | No |
| (Stadtfeld et al., 2008) | Mouse | Adenovirus | Yes |
| (Ban et al., 2011) | Human | Sendai Virus | Yes |
| (Yu et al., 2009) | Human | Episomal | Yes |
| (Si-Tayeb et al., 2010b) | Human | Plasmid DNA | Yes |
| (Jia et al., 2010) | Human | Novel ‘minicircle’ DNA | Yes |
| (Warren et al., 2010), (Yakubov et al., 2010) | Human | mRNA transfection | Yes |
| (Kim et al., 2009) | Human | Protein transfection | Yes |
| (Montserrat et al., 2010) | Human | Poly-β-amino esters to transfect plasmids | No |
| (Ye et al., 2010) | Mouse and Human | Bacteriophage ΦC31 integrase | No |
| (Hockemeyer et al., 2008) | Human | Drug inducible lentivirus | No |
| (Li et al., 2010) and (Zhao et al., 2010) | Human | Lentivirus delivering only Oct4, Sox2 and Nanog | No |
| (Huangfu et al., 2008) | Human | Retrovirus delivering only Oct4 and Sox2 along with valproic acid treatment | No |
| (Montserrat et al., 2011) | Pig | Lentivirus delivering only Sox2, Klf4 and c-Myc in feeder-free conditions | No |
In order to make iPSC technology relevant for both in vitro diagnostics and cell transplantation for regenerative medicine purposes, iPSCs need to be efficiently and reproducibly generated, at low-cost and in a non-labor intensive manner.
It is known that reprogramming is enhanced by p53 deficiency and that p21, a downstream target of p53, contributes to p53-dependent repression of iPSC formation. This led researchers to postulate that p53 targets may help to hold back cells from becoming reprogrammed. The particular miRNA, miR-34, is a downstream target of p53 and induced during iPSC development. It has been reported that miR-34 deficiency, in mice, significantly increases reprogramming efficiency, kinetics, and iPSC generation (Choi et al., 2011). Furthermore, unlike p53 deficiency which enhances reprogramming at the expense of iPSC differentiation potential, miR-34 deficiency increases iPSC formation without compromising self renewal or differentiation. Therefore miR-34 inhibitors can be used to increase reprogramming efficiencies and decrease the time needed to establish iPSC lines. This also increases the amount of cell lines that can be selected for to determine the best line to differentiate to the desired cell type.
Transfection of somatic cells with protein-encoding RNA of reprogramming genes is an attractive method to produce clinically relevant iPSCs free of genomic modification. However this method has low efficiency and is labor intensive. It has been determined that multiple rounds of RNA transfection trigger an innate immune response that leads to growth arrest and eventual cell death. Therefore researchers hypothesized that if they desensitize cells to exogenous RNA with various siRNA cocktails targeting the innate immune response they can enable multiple rounds of RNA transfection without causing growth arrest and cell death. Specifically, siRNA has been used to knockdown interferon-β, Eif2ak2 and Stat2 to rescue transfected cells from the innate immune response triggers by frequent RNA transfection (Angel and Yanik, 2010).
Reprogramming efficiency can be enhanced through forcing the transcription of all six transcription factors, rather than just using Oct4, Sox2, Klf4, and c-Myc (also known as Yamanaka factors) or Oct4, Sox2, Nanog, and Lin28 (also known as Thomson factors) (Liao et al., 2008). This was shown using lentiviruses, however it is likely that this concept would be applicable to all types of reprogramming methods. It was known that TGFβ and MEK-ERK pathway inhibitors promote mesenchymal to epithelial transition, which is an important step in the reprogramming process. As expected, using chemical compounds that inhibit the TGFβ and MEK-ERK pathways have increased reprogramming efficiency of human fibroblasts and decrease the time to cell line establishment (Lin et al., 2009).
Other methods to enhance the reprogramming process include: adding SV40 large T antigen (which inactivates p53) to either set of 4 reprogramming genes enhances colony formation up to 70-fold higher with formation 1–2 weeks earlier (Mali et al., 2008); co-expressing RAR-γ and Lrh-1 with the Yamanaka factors (Wang et al., 2011); reprogramming cells under hypoxic conditions (Yoshida et al., 2009); using UTF1 forced transcription with p53 siRNA along with the Yamanaka factors (Zhao et al., 2008); and enrichment of the cell population for progenitor cells (Kleger et al., 2012). Table 2 summarizes some of the different strategies to increase reprogramming efficiencies and iPSC colony formation.
Table 2. Summary of the Different Strategies to Increase Reprogramming Efficiencies and iPSC Colony Formation.
This table summarizes the various ways researchers have increased reprogramming efficiencies and iPSC colony formation. Table includes the specific reference associated with the method and the species of cells used. Method described in table is what is included in addition to the usual forced transcription of the reprogramming genes. This table is not an exhaustive list and is meant to be representative.
| Reference | Species | Method |
|---|---|---|
| (Choi et al., 2011) | Mouse | miR-34 deficiency |
| (Angel and Yanik, 2010) | Human | Innate immune suppression |
| (Liao et al., 2008) | Human | Combination of six transcription factors |
| (Yoshida et al., 2009) | Mouse and Human | Hypoxia |
| (Lin et al., 2009) | Human | TGFβ and MEK-ERK pathway inhibition |
| (Mali et al., 2008) | Human | Addition of SV40 large T antigen |
| (Wang et al., 2011) | Mouse and Human | Co-expression of RAR-γ and Lrh-1 |
| (Zhao et al., 2008) | Human | Addition of UTF1 forced transcription and p53 siRNA |
| (Kleger et al., 2012) | Mouse | Enrichment of the cell population for progenitor cells |
Researchers have confirmed, so far, iPSC induction in the following cell types: fetal lung fibroblasts, neonatal foreskin fibroblasts, mesenchymal stem cells and dermal fibroblasts taken from healthy patients (Park et al., 2008b), adult mouse liver and stomach cells (Aoi et al., 2008), primary human adult hepatocytes (Liu et al., 2010), fetal gut mesentery-derived cells (Li et al., 2010), human and mouse extra-embryonic cells (Nagata et al., 2009), human mesenchymal cells from the umbilicial cord matrix and amniotic membrane (Cai et al., 2010), human amnion (Zhao et al., 2010), juvenile human primary keratinocytes (Aasen et al., 2008), mouse hematopoietic and myogenic cells (Polo et al., 2010), mouse fetal hepatocytes (Kleger et al., 2012; Lee et al., 2012), human fetal hepatocytes (Hansel et al., 2013), and human lymphoblastoid cells (Barrett et al., 2014). Table 3 summarizes some of the first successful reports in reprogramming different somatic cell types.
Table 3. Generation of iPSCs from Various Somatic Cell Sources.
This table summarizes some of the first reports of iPSCs generated from various somatic cell sources and species the iPSCs were derived from. This table is not an exhaustive list and is meant to be representative.
| Reference | Species | Cell Type |
|---|---|---|
| (Liu et al., 2010) | Human | Adult Hepatocytes |
| (Aoi et al., 2008) | Mouse | Adult mouse liver and stomach cells |
| (Park et al., 2008b) | Human | Fetal lung fibroblasts, neonatal skin fibroblasts, mesenchymal stem cells and dermal fibroblasts |
| (Li et al., 2010) | Human | Fetal gut mesentery-derived cells |
| (Nagata et al., 2009) | Human and Mouse | Extra-embryonic cells |
| (Cai et al., 2010) | Human | Mesenchymal cells from the umbilical cord matrix and amniotic membrane |
| (Zhao et al., 2010) | Human | Amnion |
| (Aasen et al., 2008) | Human | Keratinocytes |
| (Polo et al., 2010) | Mouse | Hematopoietic and Myogenic cells |
| (Kleger et al., 2012) (Lee et al., 2012) | Mouse | Fetal Hepatocytes |
| (Hansel et al., 2013) | Human | Fetal Hepatocytes |
| (Barrett et al., 2014) | Human | Lymphoblastoid cells |
4.0 DIFFERENTIATION TO HEPATOCYTES
Protocols for differentiation to hepatocyte-like cells were first established with ESCs. When iPSC technology was published, ESC protocols were applied to iPSCs. The stem cell field is constantly optimizing the most efficient and reproducible protocols to generate stem-cell derived somatic cells that most closely resemble primary cells isolated from human tissues. Nevertheless, to date, no consensus protocol has been established and no protocol has generated cells that are functionally equivalent to primary hepatocytes.
Researchers began determining ways to differentiate ESCs in vitro to desired cell types by exploiting observations made by developmental biologists during the study of normal embryonic development. Chemically defined culture conditions were determined by mimicking the same signaling events that occur during natural development. Therefore the first approach to generating functional hepatocytes from ESCs was to prime the cells to become endoderm, the embryonic derivative from which hepatocytes are developed. It has been shown that ESC differentiation toward hepatic endoderm requires activin A and Wnt3a signaling (Hay et al., 2008a) as well as FGF signaling (Morrison et al., 2008). Furthermore, researchers have determined that activin A will only specify definitive endoderm from ESCs when phosphatidylinositol 3-kinase (PI3K) signaling is suppressed, thus establishing the PI3K inhibitor LY294002 as an important small molecule to use during endoderm differentiation (McLean et al., 2007). PI3K signaling is usually activated by insulin/IGF exposure. It has been reported that inhibition of glycogen synthase kinase 3 (GSK-3) is also important in forming definite endoderm from ESCs (Bone et al., 2011). Furthermore this group showed prolonged treatment with their novel GSK inhibitor generated a population of cells that displayed hepatoblast characteristics such as expression of α-fetoprotein and HNF4α, with further treatment leading to the generation of hepatocyte-like cells capable of producing albumin. Small molecules have also been demonstrated to direct ESCs to definitive endoderm at a higher efficiency than that achieved by activin A (Borowiak et al., 2009). The two most efficient compounds were benzoic acid- and oxoheptanoic acid-based. Researchers have been able to establish endoderm progenitors through constitutive expression of SOX17 via a Cre-inducible expression vector (Seguin et al., 2008). These cell lines represent a constant source of endoderm that can be differentiated on demand to endoderm derivatives that could be used for cell transplant or in vitro analysis, such as hepatocytes. To increase the efficiency of differentiation protocols, highly defined culture conditions and extracellular matrices (ECMs) were applied in later experiments. These approaches facilitated the definitive endoderm formation from human pluripotent stem cells and resulted in better hepatocyte-like cells. The mentioned protocols were effective in a vast variety of hESC and hiPSC lines but specific cell line optimization is necessary (Cai et al., 2008; DeLaForest et al., 2011; Si-Tayeb et al., 2010a). Some of the important observations and advancements for in vitro endoderm formation are summarized in Table 4.
Table 4. Formation of Endoderm from ESCs and iPSCs.
This table summarizes some of the important observations, advancements and protocols for in vitro formation of endoderm from ESCs. This table is not an exhaustive list and is meant to be representative.
| Reference | Species | Importance for in vitro Endoderm Formation |
|---|---|---|
| (Hay et al., 2008a) | Human | Activin A and Wnt3a signaling |
| (Morrison et al., 2008) | Mouse | FGF signaling |
| (McLean et al., 2007) | Human | PI3K inhibition |
| (Bone et al., 2011) | Human | GSK-3 inhibition |
| (Borowiak et al., 2009) | Mouse and Human | Novel small molecules to induce endoderm |
| (Seguin et al., 2008) | Human | Endoderm cell lines established by constitutive SOX17 expression |
| (D’Amour et al., 2005) | Human | Activin A signaling and low serum |
| (Si-Tayeb et al., 2010a) | Human | 5 days RPMI media containing B27 supplements and Activin A |
| (DeLaForest et al., 2011) | Human | PI3-kinase inhibitor LY294002 and B-27 supplement without insulin |
| (Cai et al., 2008) | Human | Activin A, BMP4, FGF2 signaling. NS21 as a substitute for B27 |
| (Teo et al., 2012) | Human | Activin and BMP4 Synergism |
| (Tahamtani et al., 2014) | Human | Stauprimide (Spd) for priming cells/Activin A |
The next step to generating stem cell-derived hepatocytes from ESCs involves the formation of hepatic progenitors that can be matured to cells functionally similar to primary hepatocytes. Many protocols have been established using this stepwise approach with chemically defined media mimicking the molecular signaling events during normal embryonic development of the liver (Agarwal et al., 2008; Basma et al., 2009; Bukong et al., 2012; Cai et al., 2007; Duan et al., 2010; Hay et al., 2008b; Medine et al., 2011; Sharma et al., 2009; Touboul et al., 2010; Tuleuova et al., 2010). All of these protocols have varying degrees of success in terms of generating ESC-derived hepatocytes that have liver-specific gene expression, protein expression, protein secretion, CYP450 metabolism and induction and glycogen storage comparable to primary human hepatocytes. Mature hepatic function has yet to be accomplished through in vitro differentiation of ESCs.
Small molecules and xeno-free ECM were utilized – instead of the conventional growth factors and animal-derived ECMs – to develop a reliable source of functional human hepatocytes with mature phenotypes for cell-based therapies and research (Farzaneh et al., 2014; Shan et al., 2013). If the technology was moved towards clinical development the use of small molecules in xeno-free conditions would lessen medical concerns associated with transplantation of the cells into humans.
Finally, a recent publication highlights how to derive functional hepatocytes from ESCs in a highly detailed and step-by-step procedure (Szkolnicka et al., 2014). Table 5 summarizes the various protocols that have been published regarding differentiation of ESCs to hepatocytes.
Table 5. Various Reports of ESC Differentiation to Hepatocytes.
This table summarizes the various reports on the generation of ESC-derived hepatocytes from different species’ cells. Table includes the reference, species of cell used and a brief summary of the growth factors, cytokines and culture techniques used within each reference’s differentiation protocol. The forward slashes in the “summary of protocol” column denote a change in culture conditions consisting of a basal medium or growth factor change. This table is not an exhaustive list and is meant to be representative.
| Reference | Species | Summary of Protocol |
|---|---|---|
| (Agarwal et al., 2008) | Human | ActA/FGF4, HGF/BSA, FGF4, HGF/FGF4, HGF, OSM, DEX |
| (Basma et al., 2009) | Human | EB formation/ActA, FGF2/HGF, DMSO/DEX |
| (Bukong et al., 2012) | Human | ActA, FGF2/FGF4, BMP2, LY294002/FGF10, RA, FGF4, HGF, EGF, SB431542/FGF10, RA, FGF4, HGF, EGF, SB431542, OSM, DEX |
| (Cai et al., 2007) | Human | ActA/FGF4, BMP2/HGF/OSM, DEX |
| (Duan et al., 2010) | Human | ActA/FGF4, HGF, BMP2, BMP4/FGF4, HGF, BMP2, BMP4, DMSO/FGF4, HGF, OSM, DEX, DMSO |
| (Hay et al., 2008b) | Human | ActA, NaBut/ActA, NaBut/DMSO/HGF, OSM |
| (Medine et al., 2011) | Human | ActA, Wnt3a/DMSO/HGF, OSM |
| (Sharma et al., 2009) | Human | DMSO/NaBut/SNAP |
| (Touboul et al., 2010) | Human | ActA, FGF2/LY294002, ActA, BMP4, FGF2/FGF10/FGF10, RA, SB431542/FGF4, HGF, EGF |
| (Tuleuova et al., 2010) | Mouse | Use of various growth factor arrays and micropatterned co-cultures to induce hepatic differentiation |
| (Farzaneh et al., 2014) | Human | Serum- and xenobiotic-free, simple, and cost-effective ECM, “RoGel,” developed |
| (Szkolnicka et al., 2014) | Human | Wnt3a, ActA/KO-serum, DMSO/HGF, OSM |
| (Sengupta et al., 2014) | Human | Albumin fraction V, ActA/FGF4, BMP2/HGF, OSM, DEX |
Many investigators are interested in using iPSCs for drug discovery and regenerative medicine applications to treat liver disease. Therefore, researchers are interested in determining the best in vitro protocol for the derivation of high quality iPSC-derived hepatocytes that is efficient and reproducible. There have been many reports claiming differentiation of iPSCs to hepatocytes using chemically defined culture conditions that include using various growth factors and cytokines, plating techniques and transduction of key liver-specific transcription factors (Asgari et al., 2013; Chen et al., 2012; Ghodsizadeh et al., 2010; Rashid et al., 2010; Sancho-Bru et al., 2011; Si-Tayeb et al., 2010a; Song et al., 2009; Takayama et al., 2012; Zhang et al., 2011). All of these reports use different gene expression characteristics and hepatocyte-specific function tests to characterize their iPSC-derived hepatocytes. The take home message from all of these reports is that in most function tests, these stem-cell derived hepatocytes were never on the same level as compared to a primary hepatocyte. In fact, iPSC-derived hepatocytes showed only 0.3–10% activity when compared to primary hepatocytes in certain hepatocyte-specific function tests (Song et al., 2009). A protocol to produce iPSC-derived hepatocytes that can be used for drug discovery or regenerative medicine applications has yet to be developed.
Although the potential of developing and improving differentiation protocols is promising, there are still major challenges to translate this science to clinical practice and biomedical applications. The ability to mass produce functional stem cell derived hepatocytes using a reliable and reproducible differentiation protocol remains a formidable barrier to many industrial and biomedical applications of stem cell technology. The early experiments only addressed the efficiency of differentiation in small-scale adherent cell culture conditions, which resulted in poor enzyme inducibility and limited metabolic function. Integrated expansion and differentiation platforms to produce hepatocytes in suspension and 3D culture were introduced to address this issue (Vosough et al., 2013). The combination of cell aggregation (3D culture) and cAMP signaling enhanced the maturation of hepatocyte-like cells (HLCs) derived from human pluripotent stem cells (hPSCs). These cells showed similar expression profiles and metabolic activity to primary human hepatocytes (Ogawa et al., 2013). 3D matrices were used to enable further maturation and to improve the function of the HLCs. These protocols resulted in polarized structures of HLCs that are crucial in high throughput assays and extended the metabolic activity of cells up to 75 days (Gieseck et al., 2014).
The hPSC-derived HLCs in 3D suspension culture demonstrated better inducibility and enhanced enzyme activity while treated with specific inducers. A significant increase in CYP1A2 and CYP3A4 was reported and end metabolites of omeprazole and RIF were produced respectively (Sengupta et al., 2014).
Table 6 summarizes the various reports of iPSC differentiation to hepatocytes in various species’ cells with a brief summary of their novel differentiation protocol.
Table 6. Various Reports of iPSC Differentiation to Hepatocytes.
This table summarizes the various reports within the literature on the generation of iPSC-derived hepatocytes from different species’ cells. Table includes the reference, species of cell used and a brief summary of the growth factors, cytokines and transduction techniques used within each reference’s differentiation protocol. The forward slashes in the “summary of protocol” column denote a change in culture conditions consisting of a basal medium or growth factor change. This table is not an exhaustive list and is meant to be representative.
| Reference | Species | Summary of Protocol |
|---|---|---|
| (Asgari et al., 2013) | Human | ActA/FGF4, HGF/OSM, DEX |
| (Chen et al., 2012) | Human | ActA, Wnt3a, HGF/DMSO/OSM, DEX, ITS |
| (Sancho-Bru et al., 2011) | Mouse | ActA, Wnt3a/BMP4, FGF2/FGF1, FGF4, FGF8/HGF, Follistatin |
| (Si-Tayeb et al., 2010a) | Human | ActA/BMP4, FGF2/HGF/OSM |
| (Song et al., 2009) | Human | ActA/BMP2, FGF4/HGF, KGF/OSM, DEX/OSM, DEX, N2, B27 |
| (Takayama et al., 2012) | Human | ActA, FGF2, transduction w. HNF3β/BMP4, FGF4, transduction w. HNF3β and HNF1α/HGF, FGF1, FGF4, FGF10, transduction w. HNF3β and HNF1α/HGF, OSM, DEX |
| (Rashid et al., 2010) | Human | ActA, FGF2, BMP4, LY294002/ActA/HGF, OSM |
| (Ghodsizadeh et al., 2010) | Human | EB Formation/ActA/DMSO HGF/DEX |
| (Zhang et al., 2011) | Human | ActA/FGF4, BMP2/HGF, KGF/OSM, DEX/OSM, DEX, N2, B27 |
| (Vosough et al., 2013) | Human | 3D aggregates and integrated differentiation/ActA, Rapa/FGF4, HGF/OSM, DEX |
| (Ogawa et al., 2013) | Human | Combination of 3D aggregation and cAMP signaling/ActA, Wnt3a, BMP4/B27, FGF10, FGF2, BMP4/HGF, DEX, OSM |
| (Gieseck et al., 2014) | Human | ActA, FGF2, BMP4, LY-294002, CHIR99021/Hepatozyme-SFM, HGF, OSM |
| (Shan et al., 2013) | Human | Using small molecules to increase maturation of iPSC-derived hepatocytes |
5.0 EPIGENETIC MEMORY, DONOR DIFFERENCES, AND CELL SOURCE INFLUENCING IPSC DIFFERENTIATION POTENTIALS
Many iPSC lines show an ability to differentiate to one lineage more efficiently and reproducibly than others. This experimental observation prompted investigations to determine why there are differences in developmental potential among cell lines. An initial concept was that there might be subtle genetic differences between the iPSC lines created, even if the cell lines were derived from the same initial cell source.
It has been determined that background mutations in parental cells account for most of the genetic heterogeneity of iPSCs (Young et al., 2012). This was initially determined by sequencing the genome of ten mouse iPSC clones derived from 3 independent experiments and comparing them to the parental cell genomes. Hundreds of single nucleotide variants in every clone were detected, with an average of 11 in coding regions. In two of the experiments all variants were unique for each clone and did not cluster in pathways, but in the third all four clones contained 157 shared genetic variants, which could also be detected in rare cells within the parental cell pool. In short, these researchers concluded that most of the genetic variation in iPSC lines is not caused by reprogramming, but is rather a consequence of cloning individual cells within the parental pool and “capturing” that single reprogrammed cells unique genetic mutational history. These genetically unique iPSC lines could be the reason why some lines will differentiate to one lineage more efficiently and reproducibly than others. This is especially true if a line has a unique mutation in a coding region of a protein essential for specification to a certain lineage. For example lines that have mutations in the HNF4α gene could potentially have difficulty differentiating to hepatocytes, since HNF4α is an important transcription factor that controls expression of many key hepatocyte-specific genes.
Another group has determined that aberrant silencing of the Dlk1-Dio3 gene cluster on chromosome 12qF1 in mouse iPSCs leads to poor developmental potential of the clones when comparing genetically identical iPSCs (Stadtfeld et al., 2010). This is consistent with the developmental role that this gene cluster is known to be involved in. More specifically, it was shown that when this gene cluster is silenced the iPSC clones contributed poorly to chimeras and failed to support the development of entirely iPSC-derived animals. In comparison, iPSCs that had normal expression of this locus had the ability to form high-grade chimeras and generated viable all-iPSC mice. Furthermore, treatment of a clone that had silencing of the locus with a histone deacetylase inhibitor, such as valproic acid, reactivated the locus and rescued the lines developmental capabilities.
Evidence has been generated that show mouse iPSCs contain epigenetic memory of the donor cell from which they were made from (Kim et al., 2010) and cell type of origin influences in vitro differentiation potentials (Polo et al., 2010). It was shown that iPSC lines have DNA methylation signatures that are characteristic of their somatic tissue of origin, which in turn favors the differentiation of that cell line to lineages related to the original cell source. Therefore, according to this data, if hepatocytes were the target cell, iPSCs generated from hepatocytes would represent the best cell source to generate stem cell-derived hepatocytes that most closely resemble their primary cell counterparts. However it was shown that continuous passing of iPSC lines largely attenuated the differences in differentiation capabilities, suggesting that early passage iPSCs have superior re-differentiation capacity.
Investigations into donor-dependent variations in hepatic differentiation of iPSCs have been conducted (Kajiwara et al., 2012). In these investigations 28 iPSC lines originating from various somatic cells were analyzed. The iPSCs were derived from various methods including retroviruses, Sendai viruses, or episomal plasmids. The study concluded that the origins, not the derivation methods, may be a major determinant of variation in hepatic differentiation. Specifically, it was determined that clones derived from peripheral blood cells consistently showed efficient differentiation, whereas many clones derived from adult dermal fibroblasts showed poor differentiation. When lines from peripheral blood and dermal fibroblasts from the same individual were compared, they found that variations in hepatic differentiation were attributable to donor differences, rather than to the cell type of origin of the iPSC line.
A second group has experimental evidence, like the previously discussed report, that regardless of cell origin the different human iPSC lines all showed the same ability to differentiate into hepatic cells in vitro (Liu et al., 2011). They differentiated iPSCs derived from hepatocytes, fibroblasts and keratinocytes, all of which showed the same ability to differentiate into hepatic cells.
Recently, investigators have determined that there is an increased reprogramming capacity of mouse liver progenitor cells, compared with differentiated liver cells (Kleger et al., 2012). Specifically, adult and fetal liver cells from mice were enriched for their progenitor cells and it was determined that enriching for these cells dramatically increased reprogramming efficiencies. Enriching fetal liver cells for progenitor cells resulted in a 275-fold higher reprogramming efficiency compared with unsorted fetal cells. It was determined that this reprogramming efficiency increase was associated with endogenous expression of reprogramming factors and members of the BAF-complex, which mediate epigenetic changes during reprogramming.
Other investigators demonstrated that there is a contribution of hepatic lineage stage-specific donor memory to the differentiation potential of mouse iPSCs (Lee et al., 2012). Specifically, iPSCs derived from fetal liver cells retain superior hepatic re-differentiation compared to iPSCs derived from adult liver cells. However once again, continuous passaging negated the differences in differentiation capabilities.
Taken together, and if translated to humans, human fetal hepatocytes should have a superior reprogramming capacity and re-differentiation potential compared to adult hepatocytes. Therefore fetal hepatocyte-derived iPSCs may represent the most efficient and cost effective source to generate iPSC-derived hepatocytes that most closely resemble primary human hepatocytes in terms of liver-specific gene expression and function. Theoretically, these differentiated cells can be used for drug discovery, molecular biology disease research, and regenerative medicine for liver-based diseases.
Recently, we reported the generation of 40 iPSC lines from primary human hepatocytes under feeder-free conditions (Hansel et al., 2013). Of these, 37 iPSC lines were generated from fetal hepatocytes, 2 iPSC lines from normal adult hepatocytes and 1 iPSC line from adult hepatocytes of a patient diagnosed with Crigler-Najjar syndrome type-1 (CN-1). Human fetal liver cells need to be reprogrammed because they have limited self renewal, proliferation, and differentiation capabilities. Taken together, human fetal liver cells are not an unlimited cell source to be used for transplant and research purposes. Select iPSC lines were confirmed reprogrammed and expressed markers of pluripotency by gene expression, flow cytometry, immunocytochemistry, and teratoma formation. Fetal hepatocytes were reprogrammed at a frequency over 50-fold higher than adult hepatocytes. Adult hepatocytes were only reprogrammed with 6 factors, while fetal hepatocytes could be reprogrammed with 3 (OCT4, SOX2, NANOG) or 4 factors (OCT4, SOX2, NANOG, LIN28 or OCT4, SOX2, KLF4, C-MYC). The increased reprogramming efficiency of fetal cells was not due to increased transduction efficiency or vector toxicity. These studies confirm that iPSCs can be generated from adult and fetal hepatocytes including those with genetic diseases. Fetal hepatocytes reprogram much more efficiently than adult, although both could serve as useful sources of iPSC-derived hepatocytes for basic research or transplantation. Further experimentation needs to be performed on these adult and fetal hepatocyte-derived iPSCs to determine if the fetal-derived cells retain superior hepatic differentiation as compared to their adult-derived counterparts.
In short, it has been shown that both donor differences as well as the cell type of origin of the iPSC contribute to the developmental potential and differentiation capacities of clonally expanded iPSCs. This means both factors must be taken into account when determining the best iPSC to be differentiated to hepatocytes.
6.0 IN VITRO AND IN VIVO LIVER DISEASE MODELING WITH IPSCS – AND GENETIC CORRECTION
Now that iPSC technology has been described, investigators are generating disease-specific iPSCs to study disease both in vitro and in vivo. These cells will allow investigators to have an unlimited supply of disease-specific cells if the iPSCs can be differentiated to the cell type affected by the disease. These differentiated cells can be used for disease modeling, drug discovery, and autologous cell replacement therapies after genetic correction.
Genetic correction was originally made possible through the usage of genome modifying zinc-finger nucleases (ZFNs) (Carbery et al., 2010). They generate site-specific double strand breaks, leading to insertions or deletions via DNA repair by the non-homologous end joining pathway. ZFNs have corrected genetic defects in iPSCs from disease-specific cells, specifically in those generated from patients with alpha-1 antitrypsin (A1AT) deficiency (Yusa et al., 2011) and sickle cell anemia (Sebastiano et al., 2011).
Accurate genetic manipulations by directed nucleases are a promising approach to correct genetic abnormalities in patient-specific iPSCs as well. The RNA-guided Cas9 nuclease from the clustered regularly interspaced short palindromic repeats (CRISPR) bacterial adaptive immune system can be used to mediate genome editing in iPSCs by specifying a 20 nucleotide targeting sequence within its guide RNA. This system, more commonly referred to as the CRISPR-Cas9 system, leverages its genome editing capabilities by harnessing the non-homologous end joining (NHEJ) or homology-directed repair (HDR) found in all mammalian cells (Ran et al., 2013). This powerful “genome editing” technology has been used by researchers to replace, disrupt, or insert into an organism’s genome. The CRISPR-Cas9 system successfully corrected point mutations in A1AT deficiency disease-specific iPSCs (Smith et al., 2014). Also, by this advanced method, the Fah mutation in murine hepatocytes of the tyrosinemia mouse model was corrected. The modified cells repopulated the murine liver and increased survival rate and body weight in mice (Yin et al., 2014). In other studies, the PCSK9 gene was mutated by CRISPR-Cas9 system in mouse liver. The loss-of-function in this gene successfully changed the lipid profile of animals, such that the plasma cholesterol level decreased 35–40% and LDL receptors increased (Ding et al., 2014). An adenovirus-based CRISPR-Cas9 system for in vivo gene editing precisely knocked-out the CEBP-alpha gene, which is an important transcription factor for metabolic genes in the liver (Cheng et al., 2014). Also, liver cancer models were established when tumor suppressor genes Pten and p53 were simultaneously targeted in wild-type mice by this system. The results were very similar to when liver tumors were induced by Cre–LoxP-mediated deletion of those genes (Xue et al., 2014).
iPSCs have been generated from patients with many liver-based genetic inborn errors in metabolism. These diseases include: A1AT deficiency (Rashid et al., 2010; Yusa et al., 2011), familial hypercholesterolemia (FH) (Rashid et al., 2010), glycogen storage disease (GSD) (Ghodsizadeh et al., 2010; Rashid et al., 2010), CN-1 (Ghodsizadeh et al., 2010; Hansel et al., 2013; Rashid et al., 2010), tyrosinemia (Ghodsizadeh et al., 2010; Rashid et al., 2010), progressive familial hereditary cholestasis (Ghodsizadeh et al., 2010), and Wilson’s disease (Zhang et al., 2011).
Disease-specific iPSCs also have utility outside of liver disease research and the following diseases have had iPSCs generated: adenosine deaminase deficiency-related severe combined immunodeficiency (Park et al., 2008a), Shwachman-Bodian-Diamond syndrome (Park et al., 2008a), Gaucher disease type III (Park et al., 2008a), muscular dystrophy (Kazuki et al., 2010; Park et al., 2008a), Parkinson disease (Park et al., 2008a; Soldner et al., 2009), Huntington disease (Park et al., 2008a), juvenile-onset type 1 diabetes mellitus (Park et al., 2008a), down syndrome/trisomy 21 (Park et al., 2008a), carrier state of Lesch-Nyhan syndrome (Park et al., 2008a), spinal muscular atrophy (Ebert et al., 2009), familial dysautonomia (Lee et al., 2009), lysosomal storage disorders (Meng et al., 2010), Timothy syndrome (Pasca et al., 2011), Fanconi anemia (Raya et al., 2009), sickle cell anemia (Sebastiano et al., 2011), rheumatoid arthritis (Lee et al., 2014), and osteoarthritis (Lee et al., 2014). iPSCs have also been generated from skin fibroblasts of an 82 year old woman diagnosed with amyotrophic lateral sclerosis and subsequently differentiated back into motor neurons (Dimos et al., 2008). This report demonstrates that iPSC technology can even be used to generate cells from patients who are in the latter stages of life. Recently, disease-specific phenotypes were demonstrated in dopamine neurons derived from human iPSCs generated from patients with genetic and sporadic Parkinson’s disease (Sanchez-Danes et al., 2012).
In short, disease-specific iPSCs represent an invaluable opportunity to recapitulate disease pathologies in vitro, thereby enabling disease investigation, drug development, and genetic correction for autologous cell replacement therapy. Table 7 summarizes some of the various disease-specific iPSCs that have been generated.
Table 7. Summary of Various Reports of Disease-specific iPSC Generation.
This table is a summary of the various diseases that have had iPSCs generated from patient-specific cells. This table is not an exhaustive list and is meant to be representative.
| Reference | Species | Disease |
|---|---|---|
| Liver Disease-specific Applications | ||
| (Hansel et al., 2013) | Human | CN-1 |
| (Rashid et al., 2010) | Human | A1AT, FH, GSD, CN-1, tyrosinemia |
| (Ghodsizadeh et al., 2010) | Human | Tyrosinemia, GSD, progressive familial hereditary cholestasis, CN-1 |
| (Yusa et al., 2011) | Human | A1AT |
| (Zhang et al., 2011) | Human | Wilson’s disease |
| Other Disease Applications | ||
| (Park et al., 2008a) | Human | adenosine deaminase deficiency-related severe combined immunodeficiency, Shwachman-Bodian-Diamond syndrome, Gaucher disease type III, muscular dystrophy, Parkinson disease, Huntington disease, juvenile-onset type 1 diabetes mellitus, down syndrome/trisomy 21, carrier state of Lesch-Nyhan syndrome |
| (Kazuki et al., 2010) | Mouse and Human | muscular dystrophy |
| (Soldner et al., 2009) | Human | Parkinson’s disease (transgene-free) |
| (Ebert et al., 2009) | Human | spinal muscular atrophy |
| (Lee et al., 2009) | Human | familial dysautonomia |
| (Meng et al., 2010) | Mouse | lysosomal storage disorders |
| (Pasca et al., 2011) | Human | Timothy syndrome |
| (Raya et al., 2009) | Human | Fanconi anemia |
| (Sebastiano et al., 2011) | Human | sickle cell anemia |
| (Dimos et al., 2008) | Human | Amyotrophic lateral sclerosis |
| (Sanchez-Danes et al., 2012) | Human | Genetic and sporadic Parkinson’s disease |
| (Lee et al., 2014) | Human | Rheumatoid arthritis and osteoarthritis |
In vitro disease models currently rely on tumor cells lines and transformed derivatives of native tissue, which may not be the best approaches to model authentic disease states. If disease-specific iPSCs can be generated and differentiated back to the cell type affected by the disease, then these cells would represent a better in vitro disease model than what is currently available.
In terms of liver-specific diseases, iPSCs have been shown to be a powerful tool to model liver-based inborn errors in metabolism in vitro. The following diseases have been modeled in vitro using iPSCs: CN-1, tyrosinemia, A1AT, FH, GSD, progressive familial hereditary cholestasis, and Wilson’s disease (Ghodsizadeh et al., 2010; Rashid et al., 2010; Yusa et al., 2011; Zhang et al., 2011). The resulting iPSCs from A1AT, FH and GSD were found to recapitulate key pathological features of the diseases affecting the patients from with they were derived, such as aggregation of misfolded A1AT in the endoplasmic reticulum, deficient LDL receptor-mediated cholesterol uptake and glycogen accumulation (Rashid et al., 2010). Careful controls for such studies are needed, such that iPSC-derived hepatocytes derived from “normal” patients have to demonstrate normal protein folding, cholesterol uptake, glycogen accumulation, or any other function being investigated. Therefore a failure of the iPSCs to differentiate properly to hepatocytes with mature functions could also provide inaccurate evidence of disease state simply due to the immaturity of the cells being investigated.
The iPSCs derived from a patient with A1AT were corrected for the point mutation in the A1AT gene that was responsible for the deficiency (Yusa et al., 2011). This genetic correction restored the structure and function of A1AT in subsequently derived liver cells in vitro. The Wilson’s disease-specific iPSCs were shown to be differentiated into hepatocyte-like cells that displayed abnormal cytoplasmic localization of mutated ATP7B and defective copper transport, as would be expected in Wilson’s disease primary hepatocytes (Zhang et al., 2011). More importantly, gene correction using a lentiviral vector that expresses the corrected ATP7B gene or treatment with the chaperone drug curcumin could reverse the functional defect within the hepatocyte in vitro. These observations demonstrate the power of disease-specific iPSCs to model and treat liver diseases with gene therapy and potential autologous cell replacement. These cells can also be used for new drug discovery by treatment of disease-specific iPSC-derived hepatocytes with new small molecules to reverse pathologies.
Many in vivo disease models utilize transgenic or pharmaceutical/chemical based approaches that do not control for off target effects and some diseases do not have a relevant model. A Fah(−/−)Rag2(−/−)Il2rg(−/−) mouse has been developed that allows for the robust expansion of human hepatocytes within the host liver (Azuma et al., 2007; Strom et al., 2010). This specialized knockout mouse can be repopulated up to 90% with human hepatocytes. Primary cells have a limited life span in culture and metabolic disease hepatocytes are not readily available. Gramignoli and colleagues have recently reported the high-level repopulation (>80%) of mouse liver with human hepatocytes from a variety of liver diseases, including biliary atresia, Crigler-Najjar, Ornithine Transcarbamylase (OTC) deficiency, thus creating “humanized” versions of these diseases that can be studied in greater detail in the laboratory (Gramignoli et al., 2013). Since the cells from a founder mouse can be isolated and transplanted into subsequent generations, the cells of interest can be maintained longer than the life-span of the original mice, and can be continually used to create additional mice with the same genetic/metabolic defect. Not all laboratories have such access to diseases human liver tissue, thus, if iPSCs were generated from patients with genetic metabolic diseases and differentiated back to a hepatocyte-like state, these cells could serve as an unlimited source of cells for research. If these genetically defective hepatocytes highly repopulate the mouse liver it is likely that the animal would recreate the phenotype similar to what is seen in the human patient. These mice that contain “humanized” livers from either primary or iPSC-derived hepatocytes will be excellent models for metabolic disorders because the basis for disease in the animal is human cell-derived while the host is an easily manipulated animal. These mice could provide the microenvironment milieu to support the tissue’s physiological function within the context of the whole organism, enabling greater understanding of disease pathogenesis and providing a platform for preclinical testing of drug candidates. Figure 2 outlines the potential surrogate sources of human hepatocytes from iPSCs for the study and treatment of liver diseases (Cantz et al., 2014; Strom et al., 2010).
Figure 14.13.2.
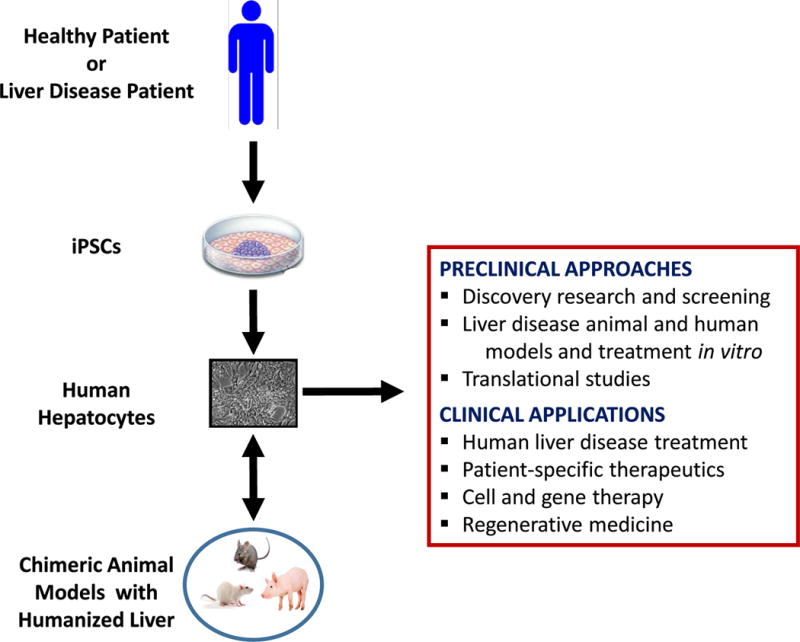
Potential surrogate sources of human hepatocytes from iPSCs for the study and treatment of liver diseases. iPSC-derived hepatocytes from healthy or liver disease patient can be transplanted into chimeric animal models and repopulate the host liver with human cells. Humanized liver animal models provides a biomedical tool not only for drug screening, metabolism and toxicity but also for studying liver pathogenesis of viral etiology, liver regenerations, liver cancer, and in the near future, for cell and gene therapy purposes. Although the potential therapeutic applications of these cells can be foreseen, more preclinical and clinical data (safety, efficacy, mechanisms of action involved in their therapeutic effects) are needed before large scale clinical studies are conducted.
Many diseases have been modeled using iPSCs outside of a liver context. For example, a recent study generated disease-specific iPSCs from patients with familial dysautonomia and differentiated the cells back to neural crest precursor cells that maintain disease phenotype (Lee et al., 2009). These neural crest precursor cells were then treated with a candidate drug and shown to respond to the drug as expected, demonstrating iPSC technology’s utility in drug development. There was a report of a complete genetic correction of iPSCs from a muscular dystrophy patient using human artificial chromosome technology to deliver a correct copy of the dystrophin gene (Kazuki et al., 2010). These corrected iPSCs were differentiated to muscle-like tissues that expressed dystrophin. iPSCs have been generated from a patient with ALS and were successfully directed to differentiate into motor neurons, the cell type destroyed in ALS (Dimos et al., 2008). Patients suffering from sickle cell anemia have had iPSCs generated from their cells and then had their genetic mutation corrected for using zinc finger nuclease technology (Sebastiano et al., 2011). Fanconi anemia disease-specific iPSCs have been generated (Raya et al., 2009). These disease-specific iPSCs were shown to be corrected for their defect and could give rise to hematopoietic progenitors of the myeloid and erythroid lineages that are phenotypically normal. Patients afflicted with Timothy syndrome have had iPSCs generated from their somatic cells (Pasca et al., 2011). Neurons derived from these cells have given new insights into the cellular phenotypes associated with Timothy syndrome that would not have been possible with primary cells from these patients. This observation demonstrates the true power of disease modeling with iPSC technology and how it can provide new targets to treat underlying disease pathologies. Other reports using iPSCs to model disease like the above examples include disease-specific cells from patients with: lysosomal storage disorders (Meng et al., 2010), spinal muscular atrophy (Ebert et al., 2009) and Parkinson’s disease (Soldner et al., 2009).
Combining gene editing with disease-specific iPSC technology provides opportunities never before available to identify the contribution of specific genetic changes to disease, as well as opportunities to identify the effects of small molecules and drugs on these processes (Merkle and Eggan, 2013).
7.0 PATIENT-SPECIFIC IPSCS – ARE THEY CLINICALLY RELEVANT?
The promise of iPSC technology is that it will allow for patient-specific cell therapy. Theoretically, patient-specific cells can be generated, corrected for their defect, and then transplanted back to the patient, without the need for immunosuppression. It was recently reported that undifferentiated, syngeneic mouse iPSCs are immunogenic and elicit strong immune responses when transplanted (Zhao et al., 2011). However, more recent data suggest that while undifferentiated iPSCs may elicit an immune response, when these cells are induced to differentiate into embryoid bodies or more mature cell types, there was a lack of immune recognition and rejection of the cells (Guha et al., 2013). Later studies clearly determined that endothelial cells generated from mouse iPSCs survived long-term when transplanted and induced a tolerance the authors described as “similar to self-tolerance” associated with an increased IL-10 expression (de Almeida et al., 2014). As with previous studies, the undifferentiated iPSCs elicited a strong immunological response including lymphocytic infiltration, and elevated interferon-gamma, granzyme-B, and perforin levels. Thus, while undifferentiated autologous iPSCs are readily detected and rejected by the immune system, more differentiated cell types may be well tolerated by recipients.
Another concern with the use of iPSCs and ESCs for cellular therapy is the tumorigenic potential of undifferentiated stem cells. It is likely that in any experiment where iPSC are differentiated to a specific cell type, there could remain a small proportion of undifferentiated cells. Even small numbers of undifferentiated cells within a differentiated cell population could lead to teratoma formation. Therefore methods have been developed to remove tumor-forming cells from differentiated cultures. A monoclonal antibody against SSEA-5, and separation based on SSEA-5 expression through FACS greatly reduced teratoma-formation with poorly differentiated ESC cultures (Tang et al., 2011). To ensure complete removal of potential cancer causing cells, they identified other pluripotency surface makers such as CD9, CD30, CD50, CD90 and CD200. Moreover, when subjecting ESCs to the SSEA-5 antibody and antibodies to 2 of the above mentioned other markers, they completely removed teratoma-formation potential from the poorly differentiated ESC cultures. Thus demonstrating experimentally it is possible to deplete undifferentiated teratoma-forming cells from cells that have been differentiated from ESCs. Additional studies showed that pluripotent stem cells rely heavily on oleate as a lipid source. Inhibition of stearoyl-coA desaturase inhibits the biosynthesis of oleate and selectively kills pluripotent cells and can be used as a method to remove residual tumor-forming pluripotent stem cells from a population of more differentiated cells (Ben-David and Benvenisty, 2014). These investigators also demonstrated that Claudin-6, one of family of the tight junction proteins, is highly expressed in pluripotent stem cells. Clostridium perfringens enterotoxin, which is known to specifically bind to many of the claudins, was employed to selectively and specifically kill pluripotent stem cells in a mixed population of differentiated and undifferentiated cells. However, with the claudin-directed toxins, one must take into account the gene expression profile in the desired differentiated population, as this toxin will also kill cells of endodermal and epithelial pathways because of their continued claudin expression. Both ablative therapies were shown to reduce or eliminate the tumorigenicity of an otherwise mixed population of cells (Ben-David and Benvenisty, 2014).
8.0 CHALLENGES OF IPSC APPLICATIONS AND THE PATH MOVING FORWARD
While there have been significant clinical advances in the field of stem cell therapy, challenging and controversial debates about standards, their potential applications and commercialization, still exist. The lack of a comprehensive database, nomenclature, or minimum information standards has led to confusion and ineffectiveness in collecting and sharing information across the scientific community (Lowenthal et al., 2012; Luong et al., 2011; Martell et al., 2010). Furthermore, the lack of ethical, cultural, regulatory and legal harmonization has had profound effects towards global applications and commercialization (Andrews et al., 2014; Arcidiacono et al., 2012; Isasi and Knoppers, 2011). Stem cell products, such as iPSCs, portray unique regulatory challenges and face substantial translational barriers under existing regulatory frameworks (Asakura, 2014; Seifinejad et al., 2010). For example, some methods to generate iPSCs require viral integration and further genetic modification for their potential use as cell therapy (e.g., autologous and allogenic; see Figure 3). These facts underscore another layer of complexity, making the implementation of prospective strategies to foster unified cross-jurisdictional collaborations difficult (Grieshammer and Shepard, 2014; Peterson and Loring, 2014; Seifinejad et al., 2010; Thompson and Manilay, 2011). Regulatory frameworks for cell therapy are in different states of development worldwide and each country has their own regulatory policies and practices. The stem cell community, scientists, and stakeholders (e.g., governmental delegates, industry, lawyers, and representatives from funding bodies) have acknowledged the necessity of establishing short- and long term- strategies to address these gaps and concerns. Several concerted plans have been proposed that could set the stage for improving collaboration toward stem cell research goals. These integrated plans have been focused on: 1.) implementing “minimum standards” for stem cell protocols and practices; 2.) creating a centralized database repository with computational approaches; 3.) establishing global stem cell registry information; and 4.) establishing a hub for harmonizing the technical, ethical, legal, and regulatory frameworks for cell therapy products worldwide (Andrews et al., 2014; Arcidiacono et al., 2012; Lowenthal et al., 2012; Luong et al., 2011; Turner et al., 2013). However, these proposals are still under discussion and no consensus has been reached yet due to the complexity of the issues involved and the lack of organization and coordination. Perhaps appropriated plans, led by proper groups, should initially follow the strategies that have been successful in addressing similar complex initiatives, such as in the bone marrow donor, tuberculosis, and stem cell bank programs (2014; Ekins et al., 2010; Turner et al., 2013). The creation of a centralized computer database for relevant scientific information and the unification of legal, ethical, and safety standards is extremely important. This may facilitate and accelerate the path forward, as well as promote trust in the stem cell community and foster successful worldwide collaborations among investigators.
Figure 14.13.3.
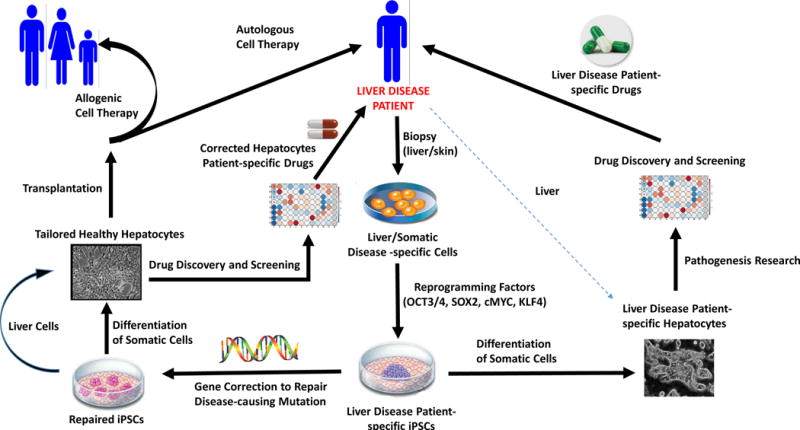
Potential applications of iPSC technology for the study and treatment of liver diseases. iPSC technology is a remarkable tool for researchers and clinicians to leverage to model liver disease mechanisms and develop suitable strategies for therapy. Briefly, the process starts by performing a biopsy (e.g., liver, skin, etc.) on a disease patient. Cells isolated from the tissue are reprogrammed to establish patient- and disease-specific iPSC lines by forcing expression of specific transcription factors (e.g., OCT3/4, SOX2, cMYC, KLF4, NANOG, LIN28). Genetic mutations can then be corrected by gene targeting approaches (e.g., ZFN, RNA-guided nucleases, etc.) and corrected cells can be differentiated in vitro into viable and functional patient-specific hepatocytes. These corrected hepatocytes can be transplanted back to the disease patient (autologous cell therapy) or to a different patient (allogenic cell therapy). Furthermore, noncorrected disease-specific cells can be differentiated to hepatocytes that recapitulate key disease phenotypes and used for drug development and disease-related research. Specifically, iPSC-derived disease-specific hepatocytes can be used to establish disease models, study pathogenesis of the disease, or to select appropriate and optimal patient-specific drug therapy.
9.0 OVERALL CONCLUSIONS
iPSC technology provides unprecedented opportunities to study, prevent, and treat a vast array of human diseases, including those of the liver. These cells’ ability of self-renewal and differentiation allow for the generation of an unlimited amount of hepatocyte-like cells for transplant and research. For example, these differentiated cells have the potential to be used for cell therapy to repair, replace, and regenerate functional liver tissue to cure, stabilize, or prolong a patient’s life until an organ for orthotopic liver transplant becomes available. Recent studies have demonstrated that iPSC-derived hepatocytes are useful for in vitro investigation of genetic liver disorders (Choi et al., 2013; Dianat et al., 2013; Hussain et al., 2011; Li et al., 2014; Rashid et al., 2010; Yusa et al., 2011). Disease-specific iPSC lines of inherited metabolic disorders (e.g., A1AT, FH, GSD and Wilson’s disease) have been generated and their specific disease was successfully modeled (Hussain et al., 2011; Irudayam et al., 2014; Yusa et al., 2011; Zhang et al., 2011). In addition, the genetic and molecular mechanisms of other widely known liver disorders, such as nonalcoholic and alcoholic liver diseases, viral hepatitis, tyrosinemia type-I, centrilobular necrosis, and cirrhosis, are currently being investigated (Choi et al., 2013; Egashira et al., 2013; Eggenschwiler et al., 2013; Grieshammer and Shepard, 2014; Inoue et al., 2014; Merkle and Eggan, 2013; Moriya et al., 2008; Schwartz et al., 2012; Soldner and Jaenisch, 2012). Investigators utilizing a pharmaceutical approach to therapy can use these iPSC-derived hepatocytes for in vitro or in vivo disease modeling and personalized medicine. iPSCs from various genetic backgrounds are valuable resources for evaluating drug-drug interactions, metabolism, and toxicity of drugs and environmental agents (Inoue and Yamanaka, 2011; Kiskinis and Eggan, 2010; Sengupta et al., 2014). Furthermore, iPSCs hold great promise for reducing drug attrition rates during drug discovery and development, which is one of the biggest bottlenecks that pharmaceutical and biotech companies have had for decades. These cells represent a valuable source of materials for identifying and developing newer and safer drugs, as they can be used to validate new relevant targets and generate suitable cell-based human models to improve existing predictive toxicity assays (Baxter et al., 2010; Choi et al., 2013; Davila et al., 2009; Engle and Puppala, 2013; Kolaja, 2014; Medine et al., 2013; Rowntree and McNeish, 2010; Sengupta et al., 2014).
The continuous and rapid technological advances observed in the stem cell field have been possible by coupling iPSC technology with new approaches such as “omics”-related research, nuclear reprogramming, gene-editing technology, RNAi, tissue engineering, medical devices, high-throughput screens (HTS), and humanized chimeric animal models. However, there are still several obstacles and limitations that need to be resolved before the technology is routinely used for clinical therapy. For example, information about iPSC cell lines and protocols of reprogramming and differentiation into hepatocyte-like cells should be optimized and standardized to increase efficiency and eliminate potential tumor formation and immunogenicity. In addition, discussions for improving collaboration on iPSC clinical applications and commercialization worldwide must continue. Although it is clear that additional work is needed to overcome these barriers, the continuous and rapid technological advances observed in the past few years make the iPSC field a primary area for investment, with significant potential impact in biomedical research and translational medicine.
Figure 3 summarizes the potential applications of iPSC technology for the study and treatment of liver diseases.
Acknowledgments
Funding
Marc C. Hansel was funded by National Institutes of Health grant T32EB001026 (CATER Predoctoral Fellowship). Stephen C. Strom was funded by Pfizer, Inc.; the National Institutes of Health grant NIDDKRC1DK086135; and the Torsten och Ragnar Sodeberg Stiftelse, Swedish Research Council (Vetenskapsradet). Roberto Gramignoli was funded by the National PKU Alliance.
References
- Aasen T, Raya A, Barrero MJ, Garreta E, Consiglio A, Gonzalez F, Vassena R, Bilic J, Pekarik V, Tiscornia G, Edel M, Boue S, Izpisua Belmonte JC. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat Biotechnol. 2008;26:1276–1284. doi: 10.1038/nbt.1503. [DOI] [PubMed] [Google Scholar]
- Agarwal S, Holton KL, Lanza R. Efficient differentiation of functional hepatocytes from human embryonic stem cells. Stem Cells. 2008;26:1117–1127. doi: 10.1634/stemcells.2007-1102. [DOI] [PubMed] [Google Scholar]
- Andrews PW, Cavanagro J, Deans R, Feigel E, Horowitz E, Keating A, Rao M, Turner M, Wilmut I, Yamanaka S. Harmonizing standards for producing clinical-grade therapies from pluripotent stem cells. Nat Biotechnol. 2014;32:724–726. doi: 10.1038/nbt.2973. [DOI] [PubMed] [Google Scholar]
- Angel M, Yanik MF. Innate immune suppression enables frequent transfection with RNA encoding reprogramming proteins. PLoS One. 2010;5:e11756. doi: 10.1371/journal.pone.0011756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Anonymous. Yamanaka announces plan to establish global iPS cell bank. The Asahi Shimbun. 2014 [This reference does not have a pre-defined format and therefore uses the “Generic” format] [Google Scholar]
- Aoi T, Yae K, Nakagawa M, Ichisaka T, Okita K, Takahashi K, Chiba T, Yamanaka S. Generation of pluripotent stem cells from adult mouse liver and stomach cells. Science. 2008;321:699–702. doi: 10.1126/science.1154884. [DOI] [PubMed] [Google Scholar]
- Arcidiacono JA, Blair JW, Benton KA. US Food and Drug Administration international collaborations for cellular therapy product regulation. Stem Cell Res Ther. 2012;3:38. doi: 10.1186/scrt129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura A. Grand challenges in the field of stem cell research. Front Cell Dev Biol. 2014;2 doi: 10.3389/fcell.2014.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asgari S, Moslem M, Bagheri-Lankarani K, Pournasr B, Miryounesi M, Baharvand H. Differentiation and transplantation of human induced pluripotent stem cell-derived hepatocyte-like cells. Stem Cell Rev. 2013;9:493–504. doi: 10.1007/s12015-011-9330-y. [DOI] [PubMed] [Google Scholar]
- Azuma H, Paulk N, Ranade A, Dorrell C, Al-Dhalimy M, Ellis E, Strom S, Kay MA, Finegold M, Grompe M. Robust expansion of human hepatocytes in Fah−/−/Rag2−/−/Il2rg−/− mice. Nat Biotechnol. 2007;25:903–910. doi: 10.1038/nbt1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban H, Nishishita N, Fusaki N, Tabata T, Saeki K, Shikamura M, Takada N, Inoue M, Hasegawa M, Kawamata S, Nishikawa S. Efficient generation of transgene-free human induced pluripotent stem cells (iPSCs) by temperature-sensitive Sendai virus vectors. Proc Natl Acad Sci U S A. 2011;108:14234–14239. doi: 10.1073/pnas.1103509108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett R, Ornelas L, Yeager N, Mandefro B, Sahabian A, Lenaeus L, Targan SR, Svendsen CN, Sareen D. Reliable generation of induced pluripotent stem cells from human lymphoblastoid cell lines. Stem Cells Transl Med. 2014;3:1429–1434. doi: 10.5966/sctm.2014-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basma H, Soto-Gutierrez A, Yannam GR, Liu L, Ito R, Yamamoto T, Ellis E, Carson SD, Sato S, Chen Y, Muirhead D, Navarro-Alvarez N, Wong RJ, Roy-Chowdhury J, Platt JL, Mercer DF, Miller JD, Strom SC, Kobayashi N, Fox IJ. Differentiation and transplantation of human embryonic stem cell-derived hepatocytes. Gastroenterology. 2009;136:990–999 e994. doi: 10.1053/j.gastro.2008.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter MA, Rowe C, Alder J, Harrison S, Hanley KP, Park BK, Kitteringham NR, Goldring CE, Hanley NA. Generating hepatic cell lineages from pluripotent stem cells for drug toxicity screening. Stem Cell Res. 2010;5:4–22. doi: 10.1016/j.scr.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-David U, Benvenisty N. Chemical ablation of tumor-initiating human pluripotent stem cells. Nat Protoc. 2014;9:729–740. doi: 10.1038/nprot.2014.050. [DOI] [PubMed] [Google Scholar]
- Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010;463:1042–1047. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bone HK, Nelson AS, Goldring CE, Tosh D, Welham MJ. A novel chemically directed route for the generation of definitive endoderm from human embryonic stem cells based on inhibition of GSK-3. J Cell Sci. 2011;124:1992–2000. doi: 10.1242/jcs.081679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowiak M, Maehr R, Chen S, Chen AE, Tang W, Fox JL, Schreiber SL, Melton DA. Small molecules efficiently direct endodermal differentiation of mouse and human embryonic stem cells. Cell Stem Cell. 2009;4:348–358. doi: 10.1016/j.stem.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukong TN, Lo T, Szabo G, Dolganiuc A. Novel developmental biology-based protocol of embryonic stem cell differentiation to morphologically sound and functional yet immature hepatocytes. Liver Int. 2012;32:732–741. doi: 10.1111/j.1478-3231.2011.02743.x. [DOI] [PubMed] [Google Scholar]
- Cai J, DeLaForest A, Fisher J, Urick A, Wagner T, Twaroski K, Cayo M, Nagaoka M, Duncan SA. Protocol for directed differentiation of human pluripotent stem cells toward a hepatocyte fate. 2008 [PubMed] [Google Scholar]
- Cai J, Li W, Su H, Qin D, Yang J, Zhu F, Xu J, He W, Guo X, Labuda K, Peterbauer A, Wolbank S, Zhong M, Li Z, Wu W, So KF, Redl H, Zeng L, Esteban MA, Pei D. Generation of human induced pluripotent stem cells from umbilical cord matrix and amniotic membrane mesenchymal cells. J Biol Chem. 2010;285:11227–11234. doi: 10.1074/jbc.M109.086389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Zhao Y, Liu Y, Ye F, Song Z, Qin H, Meng S, Chen Y, Zhou R, Song X, Guo Y, Ding M, Deng H. Directed differentiation of human embryonic stem cells into functional hepatic cells. Hepatology. 2007;45:1229–1239. doi: 10.1002/hep.21582. [DOI] [PubMed] [Google Scholar]
- Cantz T, Sharma AD, Ott M. Concise review: cell therapies for hereditary metabolic liver diseases-concepts, clinical results, and future developments. Stem Cells. 2014;33:1055–1062. doi: 10.1002/stem.1920. [DOI] [PubMed] [Google Scholar]
- Carbery ID, Ji D, Harrington A, Brown V, Weinstein EJ, Liaw L, Cui X. Targeted genome modification in mice using zinc-finger nucleases. Genetics. 2010;186:451–459. doi: 10.1534/genetics.110.117002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey BW, Markoulaki S, Hanna JH, Faddah DA, Buganim Y, Kim J, Ganz K, Steine EJ, Cassady JP, Creyghton MP, Welstead GG, Gao Q, Jaenisch R. Reprogramming factor stoichiometry influences the epigenetic state and biological properties of induced pluripotent stem cells. Cell Stem Cell. 2011;9:588–598. doi: 10.1016/j.stem.2011.11.003. [DOI] [PubMed] [Google Scholar]
- Chan EM, Ratanasirintrawoot S, Park IH, Manos PD, Loh YH, Huo H, Miller JD, Hartung O, Rho J, Ince TA, Daley GQ, Schlaeger TM. Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nat Biotechnol. 2009;27:1033–1037. doi: 10.1038/nbt.1580. [DOI] [PubMed] [Google Scholar]
- Chen YF, Tseng CY, Wang HW, Kuo HC, Yang VW, Lee OK. Rapid generation of mature hepatocyte-like cells from human induced pluripotent stem cells by an efficient three-step protocol. Hepatology. 2012;55:1193–1203. doi: 10.1002/hep.24790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng R, Peng J, Yan Y, Cao P, Wang J, Qiu C, Tang L, Liu D, Jin J, Huang X, He F, Zhang P. Efficient gene editing in adult mouse livers via adenoviral delivery of CRISPR/Cas9. FEBS Lett. 2014;588:3954–3958. doi: 10.1016/j.febslet.2014.09.008. [DOI] [PubMed] [Google Scholar]
- Choi KD, Yu J, Smuga-Otto K, Salvagiotto G, Rehrauer W, Vodyanik M, Thomson J, Slukvin I. Hematopoietic and Endothelial Differentiation of Human Induced Pluripotent Stem Cells. Stem Cells. 2009;27:559–567. doi: 10.1634/stemcells.2008-0922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SM, Kim Y, Shim JS, Park JT, Wang RH, Leach SD, Liu JO, Deng C, Ye Z, Jang YY. Efficient drug screening and gene correction for treating liver disease using patient-specific stem cells. Hepatology. 2013;57:2458–2468. doi: 10.1002/hep.26237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YJ, Lin CP, Ho JJ, He X, Okada N, Bu P, Zhong Y, Kim SY, Bennett MJ, Chen C, Ozturk A, Hicks GG, Hannon GJ, He L. miR-34 miRNAs provide a barrier for somatic cell reprogramming. Nat Cell Biol. 2011;13:1353–1360. doi: 10.1038/ncb2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper O, Hargus G, Deleidi M, Blak A, Osborn T, Marlow E, Lee K, Levy A, Perez-Torres E, Yow A, Isacson O. Differentiation of human ES and Parkinson’s disease iPS cells into ventral midbrain dopaminergic neurons requires a high activity form of SHH, FGF8a and specific regionalization by retinoic acid. Mol Cell Neurosci. 2010;45:258–266. doi: 10.1016/j.mcn.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Amour KA, Agulnick AD, Eliazer S, Kelly OG, Kroon E, Baetge EE. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat Biotechnol. 2005;23:1534–1541. doi: 10.1038/nbt1163. [DOI] [PubMed] [Google Scholar]
- Davila JC, Donaldson DB, Engle SJ, Pryor HI, Vacanti JP. Stem Cell Technology for Embryotoxicity and Hepatotoxicity Evaluation. In: Ekins S, Xu J, editors. Drug Efficacy, Safety, and Biologics Discovery: Emerging Technologies and Tools. John Wiley & Sons; New Jersey: 2009. pp. 175–213. [Google Scholar]
- de Almeida PE, Meyer EH, Kooreman NG, Diecke S, Dey D, Sanchez-Freire V, Hu S, Ebert A, Odegaard J, Mordwinkin NM, Brouwer TP, Lo D, Montoro DT, Longaker MT, Negrin RS, Wu JC. Transplanted terminally differentiated induced pluripotent stem cells are accepted by immune mechanisms similar to self-tolerance. Nat Commun. 2014;5:3903. doi: 10.1038/ncomms4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLaForest A, Nagaoka M, Si-Tayeb K, Noto FK, Konopka G, Battle MA, Duncan SA. HNF4A is essential for specification of hepatic progenitors from human pluripotent stem cells. Development. 2011;138:4143–4153. doi: 10.1242/dev.062547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dianat N, Steichen C, Vallier L, Weber A, Dubart-Kupperschmitt A. Human pluripotent stem cells for modelling human liver diseases and cell therapy. Curr Gene Ther. 2013;13:120–132. doi: 10.2174/1566523211313020006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimos JT, Rodolfa KT, Niakan KK, Weisenthal LM, Mitsumoto H, Chung W, Croft GF, Saphier G, Leibel R, Goland R, Wichterle H, Henderson CE, Eggan K. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321:1218–1221. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- Ding Q, Strong A, Patel KM, Ng SL, Gosis BS, Regan SN, Cowan CA, Rader DJ, Musunuru K. Permanent alteration of PCSK9 with in vivo CRISPR-Cas9 genome editing. Circ Res. 2014;115:488–492. doi: 10.1161/CIRCRESAHA.115.304351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Y, Ma X, Zou W, Wang C, Bahbahan IS, Ahuja TP, Tolstikov V, Zern MA. Differentiation and characterization of metabolically functioning hepatocytes from human embryonic stem cells. Stem Cells. 2010;28:674–686. doi: 10.1002/stem.315. [DOI] [PubMed] [Google Scholar]
- Ebert AD, Yu J, Rose FF, Jr, Mattis VB, Lorson CL, Thomson JA, Svendsen CN. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature. 2009;457:277–280. doi: 10.1038/nature07677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egashira T, Yuasa S, Fukuda K. Novel insights into disease modeling using induced pluripotent stem cells. Biol Pharm Bull. 2013;36:182–188. doi: 10.1248/bpb.b12-00960. [DOI] [PubMed] [Google Scholar]
- Eggenschwiler R, Loya K, Wu G, Sharma AD, Sgodda M, Zychlinski D, Herr C, Steinemann D, Teckman J, Bals R, Ott M, Schambach A, Scholer HR, Cantz T. Sustained knockdown of a disease-causing gene in patient-specific induced pluripotent stem cells using lentiviral vector-based gene therapy. Stem Cells Transl Med. 2013;2:641–654. doi: 10.5966/sctm.2013-0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekins S, Bradford J, Dole K, Spektor A, Gregory K, Blondeau D, Hohman M, Bunin BA. A collaborative database and computational models for tuberculosis drug discovery. Mol Biosyst. 2010;6:840–851. doi: 10.1039/b917766c. [DOI] [PubMed] [Google Scholar]
- Engle SJ, Puppala D. Integrating human pluripotent stem cells into drug development. Cell Stem Cell. 2013;12:669–677. doi: 10.1016/j.stem.2013.05.011. [DOI] [PubMed] [Google Scholar]
- Farzaneh Z, Pakzad M, Vosough M, Pournasr B, Baharvand H. Differentiation of human embryonic stem cells to hepatocyte-like cells on a new developed xeno-free extracellular matrix. Histochem Cell Biol. 2014;142:217–226. doi: 10.1007/s00418-014-1183-4. [DOI] [PubMed] [Google Scholar]
- Feng B, Jiang J, Kraus P, Ng JH, Heng JC, Chan YS, Yaw LP, Zhang W, Loh YH, Han J, Vega VB, Cacheux-Rataboul V, Lim B, Lufkin T, Ng HH. Reprogramming of fibroblasts into induced pluripotent stem cells with orphan nuclear receptor Esrrb. Nat Cell Biol. 2009;11:197–203. doi: 10.1038/ncb1827. [DOI] [PubMed] [Google Scholar]
- Ghodsizadeh A, Taei A, Totonchi M, Seifinejad A, Gourabi H, Pournasr B, Aghdami N, Malekzadeh R, Almadani N, Salekdeh GH, Baharvand H. Generation of liver disease-specific induced pluripotent stem cells along with efficient differentiation to functional hepatocyte-like cells. Stem Cell Rev. 2010;6:622–632. doi: 10.1007/s12015-010-9189-3. [DOI] [PubMed] [Google Scholar]
- Gieseck RL, 3rd, Hannan NR, Bort R, Hanley NA, Drake RA, Cameron GW, Wynn TA, Vallier L. Maturation of induced pluripotent stem cell derived hepatocytes by 3D-culture. PLoS One. 2014;9:e86372. doi: 10.1371/journal.pone.0086372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramignoli R, Tahan V, Dorko K, Skvorak KJ, Hansel MC, Zhao W, Venkataramanan R, Ellis EC, Jorns C, Ericzon BG, Rosenborg S, Kuiper R, Soltys KA, Mazariegos GV, Fox IJ, Wilson EM, Grompe M, Strom SC. New potential cell source for hepatocyte transplantation: discarded livers from metabolic disease liver transplants. Stem Cell Res. 2013;11:563–573. doi: 10.1016/j.scr.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieshammer U, Shepard KA. Proceedings: consideration of genetics in the design of induced pluripotent stem cell-based models of complex disease. Stem Cells Transl Med. 2014;3:1253–1258. doi: 10.5966/sctm.2014-0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha P, Morgan JW, Mostoslavsky G, Rodrigues NP, Boyd AS. Lack of immune response to differentiated cells derived from syngeneic induced pluripotent stem cells. Cell Stem Cell. 2013;12:407–412. doi: 10.1016/j.stem.2013.01.006. [DOI] [PubMed] [Google Scholar]
- Hansel MC, Gramignoli R, Blake W, Davila J, Skvorak K, Dorko K, Tahan V, Lee BR, Tafaleng E, Guzman-Lepe J, Soto-Gutierrez A, Fox IJ, Strom SC. Increased reprogramming of human fetal hepatocytes compared with adult hepatocytes in feeder-free conditions. Cell Transplant. 2013;23:27–38. doi: 10.3727/096368912X662453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay DC, Fletcher J, Payne C, Terrace JD, Gallagher RC, Snoeys J, Black JR, Wojtacha D, Samuel K, Hannoun Z, Pryde A, Filippi C, Currie IS, Forbes SJ, Ross JA, Newsome PN, Iredale JP. Highly efficient differentiation of hESCs to functional hepatic endoderm requires ActivinA and Wnt3a signaling. Proc Natl Acad Sci U S A. 2008a;105:12301–12306. doi: 10.1073/pnas.0806522105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay DC, Zhao D, Fletcher J, Hewitt ZA, McLean D, Urruticoechea-Uriguen A, Black JR, Elcombe C, Ross JA, Wolf R, Cui W. Efficient differentiation of hepatocytes from human embryonic stem cells exhibiting markers recapitulating liver development in vivo. Stem Cells. 2008b;26:894–902. doi: 10.1634/stemcells.2007-0718. [DOI] [PubMed] [Google Scholar]
- Hockemeyer D, Soldner F, Cook EG, Gao Q, Mitalipova M, Jaenisch R. A drug-inducible system for direct reprogramming of human somatic cells to pluripotency. Cell Stem Cell. 2008;3:346–353. doi: 10.1016/j.stem.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HP, Chen PH, Yu CY, Chuang CY, Stone L, Hsiao WC, Li CL, Tsai SC, Chen KY, Chen HF, Ho HN, Kuo HC. Epithelial cell adhesion molecule (EpCAM) complex proteins promote transcription factor-mediated pluripotency reprogramming. J Biol Chem. 2011;286:33520–33532. doi: 10.1074/jbc.M111.256164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Osafune K, Maehr R, Guo W, Eijkelenboom A, Chen S, Muhlestein W, Melton DA. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotechnol. 2008;26:1269–1275. doi: 10.1038/nbt.1502. [DOI] [PubMed] [Google Scholar]
- Hussain K, Challis B, Rocha N, Payne F, Minic M, Thompson A, Daly A, Scott C, Harris J, Smillie BJ, Savage DB, Ramaswami U, De Lonlay P, O’Rahilly S, Barroso I, Semple RK. An activating mutation of AKT2 and human hypoglycemia. Science. 2011;334:474. doi: 10.1126/science.1210878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue H, Nagata N, Kurokawa H, Yamanaka S. iPS cells: a game changer for future medicine. EMBO J. 2014;33:409–417. doi: 10.1002/embj.201387098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue H, Yamanaka S. The use of induced pluripotent stem cells in drug development. Clin Pharmacol Ther. 2011;89:655–661. doi: 10.1038/clpt.2011.38. [DOI] [PubMed] [Google Scholar]
- Irudayam JI, Contreras D, Sivasubramaniam S, Arumugaswami V. Modeling Liver Diseases Using Induced Pluripotent Stem Cell (iPSC)-Derived Hepatocytes. J Stem Cell Res Ther. 2014;4 [Google Scholar]
- Isasi R, Knoppers BM. From banking to international governance: fostering innovation in stem cell research. Stem Cells Int. 2011;2011:498132. doi: 10.4061/2011/498132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia F, Wilson KD, Sun N, Gupta DM, Huang M, Li Z, Panetta NJ, Chen ZY, Robbins RC, Kay MA, Longaker MT, Wu JC. A nonviral minicircle vector for deriving human iPS cells. Nat Methods. 2010;7:197–199. doi: 10.1038/nmeth.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajiwara M, Aoi T, Okita K, Takahashi R, Inoue H, Takayama N, Endo H, Eto K, Toguchida J, Uemoto S, Yamanaka S. Donor-dependent variations in hepatic differentiation from human-induced pluripotent stem cells. Proc Natl Acad Sci U S A. 2012;109:12538–12543. doi: 10.1073/pnas.1209979109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazuki Y, Hiratsuka M, Takiguchi M, Osaki M, Kajitani N, Hoshiya H, Hiramatsu K, Yoshino T, Kazuki K, Ishihara C, Takehara S, Higaki K, Nakagawa M, Takahashi K, Yamanaka S, Oshimura M. Complete genetic correction of ips cells from Duchenne muscular dystrophy. Mol Ther. 2010;18:386–393. doi: 10.1038/mt.2009.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Kim CH, Moon JI, Chung YG, Chang MY, Han BS, Ko S, Yang E, Cha KY, Lanza R, Kim KS. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell. 2009;4:472–476. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LI, Yabuuchi A, Takeuchi A, Cunniff KC, Hongguang H, McKinney-Freeman S, Naveiras O, Yoon TJ, Irizarry RA, Jung N, Seita J, Hanna J, Murakami P, Jaenisch R, Weissleder R, Orkin SH, Weissman IL, Feinberg AP, Daley GQ. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–292. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiskinis E, Eggan K. Progress toward the clinical application of patient-specific pluripotent stem cells. J Clin Invest. 2010;120:51–59. doi: 10.1172/JCI40553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleger A, Mahaddalkar PU, Katz SF, Lechel A, Joo JY, Loya K, Lin Q, Hartmann D, Liebau S, Kraus JM, Cantz T, Kestler HA, Zaehres H, Scholer H, Rudolph KL. Increased Reprogramming Capacity of Mouse Liver Progenitor Cells, Compared With Differentiated Liver Cells, Requires the BAF Complex. Gastroenterology. 2012 doi: 10.1053/j.gastro.2012.01.004. [DOI] [PubMed] [Google Scholar]
- Kolaja K. Stem cells and stem cell-derived tissues and their use in safety assessment. J Biol Chem. 2014;289:4555–4561. doi: 10.1074/jbc.R113.481028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G, Papapetrou EP, Kim H, Chambers SM, Tomishima MJ, Fasano CA, Ganat YM, Menon J, Shimizu F, Viale A, Tabar V, Sadelain M, Studer L. Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature. 2009;461:402–406. doi: 10.1038/nature08320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Kim Y, Yi H, Diecke S, Kim J, Jung H, Rim YA, Jung SM, Kim M, Kim YG, Park SH, Kim HY, Ju JH. Generation of disease-specific induced pluripotent stem cells from patients with rheumatoid arthritis and osteoarthritis. Arthritis Res Ther. 2014;16:R41. doi: 10.1186/ar4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SB, Seo D, Choi D, Park KY, Holczbauer A, Marquardt JU, Conner EA, Factor VM, Thorgeirsson SS. Contribution of Hepatic Lineage Stage-Specific Donor Memory to the Differential Potential of Induced Mouse Pluripotent Stem Cells (IPSC) Stem Cells. 2012 doi: 10.1002/stem.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Suzuki K, Kim NY, Liu GH, Izpisua Belmonte JC. A cut above the rest: targeted genome editing technologies in human pluripotent stem cells. J Biol Chem. 2014;289:4594–4599. doi: 10.1074/jbc.R113.488247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhao H, Lan F, Lee A, Chen L, Lin C, Yao Y, Li L. Generation of human-induced pluripotent stem cells from gut mesentery-derived cells by ectopic expression of OCT4/SOX2/NANOG. Cell Reprogram. 2010;12:237–247. doi: 10.1089/cell.2009.0103. [DOI] [PubMed] [Google Scholar]
- Liao J, Wu Z, Wang Y, Cheng L, Cui C, Gao Y, Chen T, Rao L, Chen S, Jia N, Dai H, Xin S, Kang J, Pei G, Xiao L. Enhanced efficiency of generating induced pluripotent stem (iPS) cells from human somatic cells by a combination of six transcription factors. Cell Res. 2008;18:600–603. doi: 10.1038/cr.2008.51. [DOI] [PubMed] [Google Scholar]
- Lin T, Ambasudhan R, Yuan X, Li W, Hilcove S, Abujarour R, Lin X, Hahm HS, Hao E, Hayek A, Ding S. A chemical platform for improved induction of human iPSCs. Nat Methods. 2009;6:805–808. doi: 10.1038/nmeth.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Kim Y, Sharkis S, Marchionni L, Jang YY. In vivo liver regeneration potential of human induced pluripotent stem cells from diverse origins. Sci Transl Med. 2011;3:82ra39. doi: 10.1126/scitranslmed.3002376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Ye Z, Kim Y, Sharkis S, Jang YY. Generation of endoderm-derived human induced pluripotent stem cells from primary hepatocytes. Hepatology. 2010;51:1810–1819. doi: 10.1002/hep.23626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowenthal J, Lipnick S, Rao M, Hull SC. Specimen collection for induced pluripotent stem cell research: harmonizing the approach to informed consent. Stem Cells Transl Med. 2012;1:409–421. doi: 10.5966/sctm.2012-0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luong MX, Auerbach J, Crook JM, Daheron L, Hei D, Lomax G, Loring JF, Ludwig T, Schlaeger TM, Smith KP, Stacey G, Xu RH, Zeng F. A call for standardized naming and reporting of human ESC and iPSC lines. Cell Stem Cell. 2011;8:357–359. doi: 10.1016/j.stem.2011.03.002. [DOI] [PubMed] [Google Scholar]
- Mali P, Ye Z, Hommond HH, Yu X, Lin J, Chen G, Zou J, Cheng L. Improved efficiency and pace of generating induced pluripotent stem cells from human adult and fetal fibroblasts. Stem Cells. 2008;26:1998–2005. doi: 10.1634/stemcells.2008-0346. [DOI] [PubMed] [Google Scholar]
- Martell K, Trounson A, Baum E. Stem cell therapies in clinical trials: workshop on best practices and the need for harmonization. Cell Stem Cell. 2010;7:451–454. doi: 10.1016/j.stem.2010.09.004. [DOI] [PubMed] [Google Scholar]
- McLean AB, D’Amour KA, Jones KL, Krishnamoorthy M, Kulik MJ, Reynolds DM, Sheppard AM, Liu H, Xu Y, Baetge EE, Dalton S. Activin a efficiently specifies definitive endoderm from human embryonic stem cells only when phosphatidylinositol 3-kinase signaling is suppressed. Stem Cells. 2007;25:29–38. doi: 10.1634/stemcells.2006-0219. [DOI] [PubMed] [Google Scholar]
- Medine CN, Lucendo-Villarin B, Storck C, Wang F, Szkolnicka D, Khan F, Pernagallo S, Black JR, Marriage HM, Ross JA, Bradley M, Iredale JP, Flint O, Hay DC. Developing high-fidelity hepatotoxicity models from pluripotent stem cells. Stem Cells Transl Med. 2013;2:505–509. doi: 10.5966/sctm.2012-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medine CN, Lucendo-Villarin B, Zhou W, West CC, Hay DC. Robust generation of hepatocyte-like cells from human embryonic stem cell populations. J Vis Exp. 2011:e2969. doi: 10.3791/2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng XL, Shen JS, Kawagoe S, Ohashi T, Brady RO, Eto Y. Induced pluripotent stem cells derived from mouse models of lysosomal storage disorders. Proc Natl Acad Sci U S A. 2010;107:7886–7891. doi: 10.1073/pnas.1002758107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkle FT, Eggan K. Modeling human disease with pluripotent stem cells: from genome association to function. Cell Stem Cell. 2013;12:656–668. doi: 10.1016/j.stem.2013.05.016. [DOI] [PubMed] [Google Scholar]
- Montserrat N, de Onate L, Garreta E, Gonzalez F, Adamo A, Eguizabal C, Hafner S, Vassena R, Izpisua Belmonte JC. Generation of feeder-free pig induced pluripotent stem cells without Pou5f1. Cell Transplant. 2011;21:815–825. doi: 10.3727/096368911X601019. [DOI] [PubMed] [Google Scholar]
- Montserrat N, Garreta E, Gonzalez F, Gutierrez J, Eguizabal C, Ramos V, Borros S, Izpisua Belmonte JC. Simple generation of human induced pluripotent stem cells using poly-beta-amino esters as the non-viral gene delivery system. J Biol Chem. 2010;286:12417–12428. doi: 10.1074/jbc.M110.168013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya K, Yoshikawa M, Ouji Y, Saito K, Nishiofuku M, Matsuda R, Ishizaka S, Fukui H. Embryonic stem cells reduce liver fibrosis in CCl4-treated mice. Int J Exp Pathol. 2008;89:401–409. doi: 10.1111/j.1365-2613.2008.00607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison GM, Oikonomopoulou I, Migueles RP, Soneji S, Livigni A, Enver T, Brickman JM. Anterior definitive endoderm from ESCs reveals a role for FGF signaling. Cell Stem Cell. 2008;3:402–415. doi: 10.1016/j.stem.2008.07.021. [DOI] [PubMed] [Google Scholar]
- Nagata S, Toyoda M, Yamaguchi S, Hirano K, Makino H, Nishino K, Miyagawa Y, Okita H, Kiyokawa N, Nakagawa M, Yamanaka S, Akutsu H, Umezawa A, Tada T. Efficient reprogramming of human and mouse primary extra-embryonic cells to pluripotent stem cells. Genes Cells. 2009;14:1395–1404. doi: 10.1111/j.1365-2443.2009.01356.x. [DOI] [PubMed] [Google Scholar]
- Nori S, Okada Y, Yasuda A, Tsuji O, Takahashi Y, Kobayashi Y, Fujiyoshi K, Koike M, Uchiyama Y, Ikeda E, Toyama Y, Yamanaka S, Nakamura M, Okano H. Grafted human-induced pluripotent stem-cell-derived neurospheres promote motor functional recovery after spinal cord injury in mice. Proc Natl Acad Sci U S A. 2011;108:16825–16830. doi: 10.1073/pnas.1108077108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa S, Surapisitchat J, Virtanen C, Ogawa M, Niapour M, Sugamori KS, Wang S, Tamblyn L, Guillemette C, Hoffmann E, Zhao B, Strom S, Laposa RR, Tyndale RF, Grant DM, Keller G. Three-dimensional culture and cAMP signaling promote the maturation of human pluripotent stem cell-derived hepatocytes. Development. 2013;140:3285–3296. doi: 10.1242/dev.090266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IH, Arora N, Huo H, Maherali N, Ahfeldt T, Shimamura A, Lensch MW, Cowan C, Hochedlinger K, Daley GQ. Disease-specific induced pluripotent stem cells. Cell. 2008a;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008b;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- Pasca SP, Portmann T, Voineagu I, Yazawa M, Shcheglovitov A, Pasca AM, Cord B, Palmer TD, Chikahisa S, Nishino S, Bernstein JA, Hallmayer J, Geschwind DH, Dolmetsch RE. Using iPSC-derived neurons to uncover cellular phenotypes associated with Timothy syndrome. Nat Med. 2011;17:1657–1662. doi: 10.1038/nm.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson SE, Loring JF. Genomic instability in pluripotent stem cells: implications for clinical applications. J Biol Chem. 2014;289:4578–4584. doi: 10.1074/jbc.R113.516419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo JM, Liu S, Figueroa ME, Kulalert W, Eminli S, Tan KY, Apostolou E, Stadtfeld M, Li Y, Shioda T, Natesan S, Wagers AJ, Melnick A, Evans T, Hochedlinger K. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat Biotechnol. 2010;28:848–855. doi: 10.1038/nbt.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid ST, Corbineau S, Hannan N, Marciniak SJ, Miranda E, Alexander G, Huang-Doran I, Griffin J, Ahrlund-Richter L, Skepper J, Semple R, Weber A, Lomas DA, Vallier L. Modeling inherited metabolic disorders of the liver using human induced pluripotent stem cells. J Clin Invest. 2010;120:3127–3136. doi: 10.1172/JCI43122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raya A, Rodriguez-Piza I, Guenechea G, Vassena R, Navarro S, Barrero MJ, Consiglio A, Castella M, Rio P, Sleep E, Gonzalez F, Tiscornia G, Garreta E, Aasen T, Veiga A, Verma IM, Surralles J, Bueren J, Izpisua Belmonte JC. Disease-corrected haematopoietic progenitors from Fanconi anaemia induced pluripotent stem cells. Nature. 2009;460:53–59. doi: 10.1038/nature08129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowntree RK, McNeish JD. Induced pluripotent stem cells: opportunities as research and development tools in 21st century drug discovery. Regen Med. 2010;5:557–568. doi: 10.2217/rme.10.36. [DOI] [PubMed] [Google Scholar]
- Sanchez-Danes A, Richaud-Patin Y, Carballo-Carbajal I, Jimenez-Delgado S, Caig C, Mora S, Di Guglielmo C, Ezquerra M, Patel B, Giralt A, Canals JM, Memo M, Alberch J, Lopez-Barneo J, Vila M, Cuervo AM, Tolosa E, Consiglio A, Raya A. Disease-specific phenotypes in dopamine neurons from human iPS-based models of genetic and sporadic Parkinson’s disease. EMBO Mol Med. 2012;4:380–395. doi: 10.1002/emmm.201200215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancho-Bru P, Roelandt P, Narain N, Pauwelyn K, Notelaers T, Shimizu T, Ott M, Verfaillie C. Directed differentiation of murine-induced pluripotent stem cells to functional hepatocyte-like cells. J Hepatol. 2011;54:98–107. doi: 10.1016/j.jhep.2010.06.014. [DOI] [PubMed] [Google Scholar]
- Schwartz RE, Trehan K, Andrus L, Sheahan TP, Ploss A, Duncan SA, Rice CM, Bhatia SN. Modeling hepatitis C virus infection using human induced pluripotent stem cells. Proc Natl Acad Sci U S A. 2012;109:2544–2548. doi: 10.1073/pnas.1121400109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastiano V, Maeder ML, Angstman JF, Haddad B, Khayter C, Yeo DT, Goodwin MJ, Hawkins JS, Ramirez CL, Batista LF, Artandi SE, Wernig M, Joung JK. In situ genetic correction of the sickle cell anemia mutation in human induced pluripotent stem cells using engineered zinc finger nucleases. Stem Cells. 2011;29:1717–1726. doi: 10.1002/stem.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seguin CA, Draper JS, Nagy A, Rossant J. Establishment of endoderm progenitors by SOX transcription factor expression in human embryonic stem cells. Cell Stem Cell. 2008;3:182–195. doi: 10.1016/j.stem.2008.06.018. [DOI] [PubMed] [Google Scholar]
- Seifinejad A, Tabebordbar M, Baharvand H, Boyer LA, Salekdeh GH. Progress and promise towards safe induced pluripotent stem cells for therapy. Stem Cell Rev. 2010;6:297–306. doi: 10.1007/s12015-010-9121-x. [DOI] [PubMed] [Google Scholar]
- Sengupta S, Johnson BP, Swanson SA, Stewart R, Bradfield CA, Thomson JA. Aggregate culture of human embryonic stem cell-derived hepatocytes in suspension are an improved in vitro model for drug metabolism and toxicity testing. Toxicol Sci. 2014;140:236–245. doi: 10.1093/toxsci/kfu069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan J, Schwartz RE, Ross NT, Logan DJ, Thomas D, Duncan SA, North TE, Goessling W, Carpenter AE, Bhatia SN. Identification of small molecules for human hepatocyte expansion and iPS differentiation. Nat Chem Biol. 2013;9:514–520. doi: 10.1038/nchembio.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma NS, Wallenstein EJ, Novik E, Maguire T, Schloss R, Yarmush ML. Enrichment of hepatocyte-like cells with upregulated metabolic and differentiated function derived from embryonic stem cells using S-NitrosoAcetylPenicillamine. Tissue Eng Part C Methods. 2009;15:297–306. doi: 10.1089/ten.tec.2008.0303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si-Tayeb K, Noto FK, Nagaoka M, Li J, Battle MA, Duris C, North PE, Dalton S, Duncan SA. Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology. 2010a;51:297–305. doi: 10.1002/hep.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si-Tayeb K, Noto FK, Sepac A, Sedlic F, Bosnjak ZJ, Lough JW, Duncan SA. Generation of human induced pluripotent stem cells by simple transient transfection of plasmid DNA encoding reprogramming factors. BMC Dev Biol. 2010b;10:81. doi: 10.1186/1471-213X-10-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C, Abalde-Atristain L, He C, Brodsky BR, Braunstein EM, Chaudhari P, Jang YY, Cheng L, Ye Z. Efficient and Allele-Specific Genome Editing of Disease Loci in Human iPSCs. Mol Ther. 2014;23:570–577. doi: 10.1038/mt.2014.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Nachman I, Regev A, Meissner A. Dynamic single-cell imaging of direct reprogramming reveals an early specifying event. Nat Biotechnol. 2010;28:521–526. doi: 10.1038/nbt.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldner F, Hockemeyer D, Beard C, Gao Q, Bell GW, Cook EG, Hargus G, Blak A, Cooper O, Mitalipova M, Isacson O, Jaenisch R. Parkinson’s disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136:964–977. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldner F, Jaenisch R. Medicine. iPSC disease modeling. Science. 2012;338:1155–1156. doi: 10.1126/science.1227682. [DOI] [PubMed] [Google Scholar]
- Song Z, Cai J, Liu Y, Zhao D, Yong J, Duo S, Song X, Guo Y, Zhao Y, Qin H, Yin X, Wu C, Che J, Lu S, Ding M, Deng H. Efficient generation of hepatocyte-like cells from human induced pluripotent stem cells. Cell Res. 2009;19:1233–1242. doi: 10.1038/cr.2009.107. [DOI] [PubMed] [Google Scholar]
- Stadtfeld M, Apostolou E, Akutsu H, Fukuda A, Follett P, Natesan S, Kono T, Shioda T, Hochedlinger K. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010;465:175–181. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Nagaya M, Utikal J, Weir G, Hochedlinger K. Induced pluripotent stem cells generated without viral integration. Science. 2008;322:945–949. doi: 10.1126/science.1162494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom SC, Davila J, Grompe M. Chimeric mice with humanized liver: tools for the study of drug metabolism, excretion, and toxicity. Methods Mol Biol. 2010;640:491–509. doi: 10.1007/978-1-60761-688-7_27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szkolnicka D, Farnworth SL, Lucendo-Villarin B, Hay DC. Deriving functional hepatocytes from pluripotent stem cells. Curr Protoc Stem Cell Biol. 2014;30:1G 5 1–1G 5 12. doi: 10.1002/9780470151808.sc01g05s30. [DOI] [PubMed] [Google Scholar]
- Tahamtani Y, Azarnia M, Farrokhi A, Moradmand A, Mirshahvaladi S, Aghdami N, Baharvand H. Stauprimide Priming of Human Embryonic Stem Cells toward Definitive Endoderm. Cell J. 2014;16:63–72. [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takayama K, Inamura M, Kawabata K, Sugawara M, Kikuchi K, Higuchi M, Nagamoto Y, Watanabe H, Tashiro K, Sakurai F, Hayakawa T, Furue MK, Mizuguchi H. Generation of metabolically functioning hepatocytes from human pluripotent stem cells by FOXA2 and HNF1alpha transduction. J Hepatol. 2012;57:628–636. doi: 10.1016/j.jhep.2012.04.038. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Tohyama S, Murata M, Nomura F, Kaneko T, Chen H, Hattori F, Egashira T, Seki T, Ohno Y, Koshimizu U, Yuasa S, Ogawa S, Yamanaka S, Yasuda K, Fukuda K. In vitro pharmacologic testing using human induced pluripotent stem cell-derived cardiomyocytes. Biochem Biophys Res Commun. 2009;385:497–502. doi: 10.1016/j.bbrc.2009.05.073. [DOI] [PubMed] [Google Scholar]
- Tang C, Lee AS, Volkmer JP, Sahoo D, Nag D, Mosley AR, Inlay MA, Ardehali R, Chavez SL, Pera RR, Behr B, Wu JC, Weissman IL, Drukker M. An antibody against SSEA-5 glycan on human pluripotent stem cells enables removal of teratoma-forming cells. Nat Biotechnol. 2011;29:829–834. doi: 10.1038/nbt.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateishi K, He J, Taranova O, Liang G, D’Alessio AC, Zhang Y. Generation of insulin-secreting islet-like clusters from human skin fibroblasts. J Biol Chem. 2008;283:31601–31607. doi: 10.1074/jbc.M806597200. [DOI] [PubMed] [Google Scholar]
- Teo AK, Ali Y, Wong KY, Chipperfield H, Sadasivam A, Poobalan Y, Tan EK, Wang ST, Abraham S, Tsuneyoshi N, Stanton LW, Dunn NR. Activin and BMP4 synergistically promote formation of definitive endoderm in human embryonic stem cells. Stem Cells. 2012;30:631–642. doi: 10.1002/stem.1022. [DOI] [PubMed] [Google Scholar]
- Thompson HL, Manilay JO. Embryonic stem cell-derived hematopoietic stem cells: challenges in development, differentiation, and immunogenicity. Curr Top Med Chem. 2011;11:1621–1637. doi: 10.2174/156802611796117702. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Touboul T, Hannan NR, Corbineau S, Martinez A, Martinet C, Branchereau S, Mainot S, Strick-Marchand H, Pedersen R, Di Santo J, Weber A, Vallier L. Generation of functional hepatocytes from human embryonic stem cells under chemically defined conditions that recapitulate liver development. Hepatology. 2010;51:1754–1765. doi: 10.1002/hep.23506. [DOI] [PubMed] [Google Scholar]
- Tuleuova N, Lee JY, Lee J, Ramanculov E, Zern MA, Revzin A. Using growth factor arrays and micropatterned co-cultures to induce hepatic differentiation of embryonic stem cells. Biomaterials. 2010;31:9221–9231. doi: 10.1016/j.biomaterials.2010.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner M, Leslie S, Martin NG, Peschanski M, Rao M, Taylor CJ, Trounson A, Turner D, Yamanaka S, Wilmut I. Toward the development of a global induced pluripotent stem cell library. Cell Stem Cell. 2013;13:382–384. doi: 10.1016/j.stem.2013.08.003. [DOI] [PubMed] [Google Scholar]
- Vosough M, Omidinia E, Kadivar M, Shokrgozar MA, Pournasr B, Aghdami N, Baharvand H. Generation of functional hepatocyte-like cells from human pluripotent stem cells in a scalable suspension culture. Stem Cells Dev. 2013;22:2693–2705. doi: 10.1089/scd.2013.0088. [DOI] [PubMed] [Google Scholar]
- Wang W, Yang J, Liu H, Lu D, Chen X, Zenonos Z, Campos LS, Rad R, Guo G, Zhang S, Bradley A, Liu P. Rapid and efficient reprogramming of somatic cells to induced pluripotent stem cells by retinoic acid receptor gamma and liver receptor homolog 1. Proc Natl Acad Sci U S A. 2011;108:18283–18288. doi: 10.1073/pnas.1100893108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, Meissner A, Daley GQ, Brack AS, Collins JJ, Cowan C, Schlaeger TM, Rossi DJ. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KH. Viable offspring derived from fetal and adult mammalian cells. Nature. 1997;385:810–813. doi: 10.1038/385810a0. [DOI] [PubMed] [Google Scholar]
- Xue W, Chen S, Yin H, Tammela T, Papagiannakopoulos T, Joshi NS, Cai W, Yang G, Bronson R, Crowley DG, Zhang F, Anderson DG, Sharp PA, Jacks T. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature. 2014;514:380–384. doi: 10.1038/nature13589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakubov E, Rechavi G, Rozenblatt S, Givol D. Reprogramming of human fibroblasts to pluripotent stem cells using mRNA of four transcription factors. Biochem Biophys Res Commun. 2010;394:189–193. doi: 10.1016/j.bbrc.2010.02.150. [DOI] [PubMed] [Google Scholar]
- Ye L, Chang JC, Lin C, Qi Z, Yu J, Kan YW. Generation of induced pluripotent stem cells using site-specific integration with phage integrase. Proc Natl Acad Sci U S A. 2010;107:19467–19472. doi: 10.1073/pnas.1012677107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Xue W, Chen S, Bogorad RL, Benedetti E, Grompe M, Koteliansky V, Sharp PA, Jacks T, Anderson DG. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol. 2014;32:551–553. doi: 10.1038/nbt.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoo N, Baba S, Kaichi S, Niwa A, Mima T, Doi H, Yamanaka S, Nakahata T, Heike T. The effects of cardioactive drugs on cardiomyocytes derived from human induced pluripotent stem cells. Biochem Biophys Res Commun. 2009;387:482–488. doi: 10.1016/j.bbrc.2009.07.052. [DOI] [PubMed] [Google Scholar]
- Yoshida Y, Takahashi K, Okita K, Ichisaka T, Yamanaka S. Hypoxia enhances the generation of induced pluripotent stem cells. Cell Stem Cell. 2009;5:237–241. doi: 10.1016/j.stem.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Young MA, Larson DE, Sun CW, George DR, Ding L, Miller CA, Lin L, Pawlik KM, Chen K, Fan X, Schmidt H, Kalicki-Veizer J, Cook LL, Swift GW, Demeter RT, Wendl MC, Sands MS, Mardis ER, Wilson RK, Townes TM, Ley TJ. Background mutations in parental cells account for most of the genetic heterogeneity of induced pluripotent stem cells. Cell Stem Cell. 2012;10:570–582. doi: 10.1016/j.stem.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, Slukvin II, Thomson JA. Human Induced Pluripotent Stem Cells Free of Vector and Transgene Sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Yusa K, Rashid ST, Strick-Marchand H, Varela I, Liu PQ, Paschon DE, Miranda E, Ordonez A, Hannan NR, Rouhani FJ, Darche S, Alexander G, Marciniak SJ, Fusaki N, Hasegawa M, Holmes MC, Di Santo JP, Lomas DA, Bradley A, Vallier L. Targeted gene correction of alpha1-antitrypsin deficiency in induced pluripotent stem cells. Nature. 2011;478:391–394. doi: 10.1038/nature10424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Wilson GF, Soerens AG, Koonce CH, Yu J, Palecek SP, Thomson JA, Kamp TJ. Functional cardiomyocytes derived from human induced pluripotent stem cells. Circ Res. 2009;104:e30–41. doi: 10.1161/CIRCRESAHA.108.192237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Chen S, Li W, Guo X, Zhao P, Xu J, Chen Y, Pan Q, Liu X, Zychlinski D, Lu H, Tortorella MD, Schambach A, Wang Y, Pei D, Esteban MA. Rescue of ATP7B function in hepatocyte-like cells from Wilson’s disease induced pluripotent stem cells using gene therapy or the chaperone drug curcumin. Hum Mol Genet. 2011;20:3176–3187. doi: 10.1093/hmg/ddr223. [DOI] [PubMed] [Google Scholar]
- Zhao HX, Li Y, Jin HF, Xie L, Liu C, Jiang F, Luo YN, Yin GW, Wang J, Li LS, Yao YQ, Wang XH. Rapid and efficient reprogramming of human amnion-derived cells into pluripotency by three factors OCT4/SOX2/NANOG. Differentiation. 2010;80:123–129. doi: 10.1016/j.diff.2010.03.002. [DOI] [PubMed] [Google Scholar]
- Zhao T, Zhang ZN, Rong Z, Xu Y. Immunogenicity of induced pluripotent stem cells. Nature. 2011;474:212–215. doi: 10.1038/nature10135. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Yin X, Qin H, Zhu F, Liu H, Yang W, Zhang Q, Xiang C, Hou P, Song Z, Liu Y, Yong J, Zhang P, Cai J, Liu M, Li H, Li Y, Qu X, Cui K, Zhang W, Xiang T, Wu Y, Liu C, Yu C, Yuan K, Lou J, Ding M, Deng H. Two supporting factors greatly improve the efficiency of human iPSC generation. Cell Stem Cell. 2008;3:475–479. doi: 10.1016/j.stem.2008.10.002. [DOI] [PubMed] [Google Scholar]


