Abstract
Hydrogen sulfide (H2S) elicits pleiotropic physiological effects ranging from modulation of cardiovascular to CNS functions. A dominant method for transmission of sulfide-based signals is via posttranslational modification of reactive cysteine thiols to persulfides. However, the source of the persulfide donor and whether its relationship to H2S is as a product or precursor is controversial. The transsulfuration pathway enzymes can synthesize cysteine persulfide (Cys−SSH) from cystine and H2S from cysteine and/or homocysteine. Recently, Cys−SSH was proposed as the primary product of the transsulfuration pathway with H2S representing a decomposition product of Cys−SSH. Our detailed kinetic analyses demonstrate a robust capacity for Cys−SSH production by the human transsulfuration pathway enzymes, cystathionine beta-synthase and γ-cystathionase (CSE) and for homocysteine persulfide synthesis from homocystine by CSE only. However, in the reducing cytoplasmic milieu where the concentration of reduced thiols is significantly higher than of disulfides, substrate level regulation favors the synthesis of H2S over persulfides. Mathematical modeling at physiologically relevant hepatic substrate concentrations predicts that H2S rather than Cys−SSH is the primary product of the transsulfuration enzymes with CSE being the dominant producer. The half-life of the metastable Cys−SSH product is short and decomposition leads to a mixture of polysulfides (Cys−S−(S)n−S−Cys). These in vitro data, together with the intrinsic reactivity of Cys−SSH for cysteinyl versus sulfur transfer, are consistent with the absence of an observable increase in protein persulfidation in cells in response to exogenous cystine and evidence for the formation of polysulfides under these conditions.
Graphical abstract
INTRODUCTION
Hydrogen sulfide (H2S)-dependent signaling elicits varied effects in the cardiovascular system including smooth muscle relaxation, cytoprotection during reperfusion injury, inhibition of platelet aggregation and inhibition of vascular smooth muscle cell proliferation.1 In the nervous system, H2S is a neuromodulator inducing long-term potentiation when applied with a weak tetanic stimulus.2 H2S also protects against glutamate-induced cytotoxicity3,4 and is a modulator of inflammation.5,6 While the mechanism of sulfide-based signaling is poorly understood, persulfidation at reactive cysteine residues in proteins is accepted as a likely route for signal transmission.7 The low intrinsic reactivity of H2S toward oxidized thiols like disulfides8,9 has, however, raised questions regarding the direct involvement of H2S in signaling and led to the consideration of alternative sulfur donors in protein persulfidation reactions10–13 In this context, polysulfide (R−S−(S)n−S−R), a catenated sulfur chemotype that is more reactive than H2S, has been suggested to be a biologically relevant signaling species14,15 although its origin in mammalian cells has received only scant attention.16,17 The biological effects of H2S have been ascribed to contaminating polysulfides present in commercial NaHS samples.18 A second reactive chemotype invoked in sulfide-based signaling is the metastable persulfide (R−SSH) or hydrodisulfide species.19 Persulfides are intermediates in various enzymatic reactions involved in sulfur metabolism.20–22 Persulfides are also synthesized by the transsulfuration pathway enzymes,15,23 which are also a source of H2S.24–26 Although low molecular weight (LMW) persulfides can react with nucleophiles and electrophiles,27 cysteine and glutathione persulfides (Cys−SSH and GSSH) were reported to exist at high steady-state concentrations in cells.15 Since H2S can be liberated as a side product during decomposition of Cys−SSH and GSSH, it raises even further questions regarding the identity of the primary reactive sulfur species generated by the transsulfuration pathway enzymes at physiologically relevant substrate concentrations, i.e., whether it is H2S versus LMW persulfides.
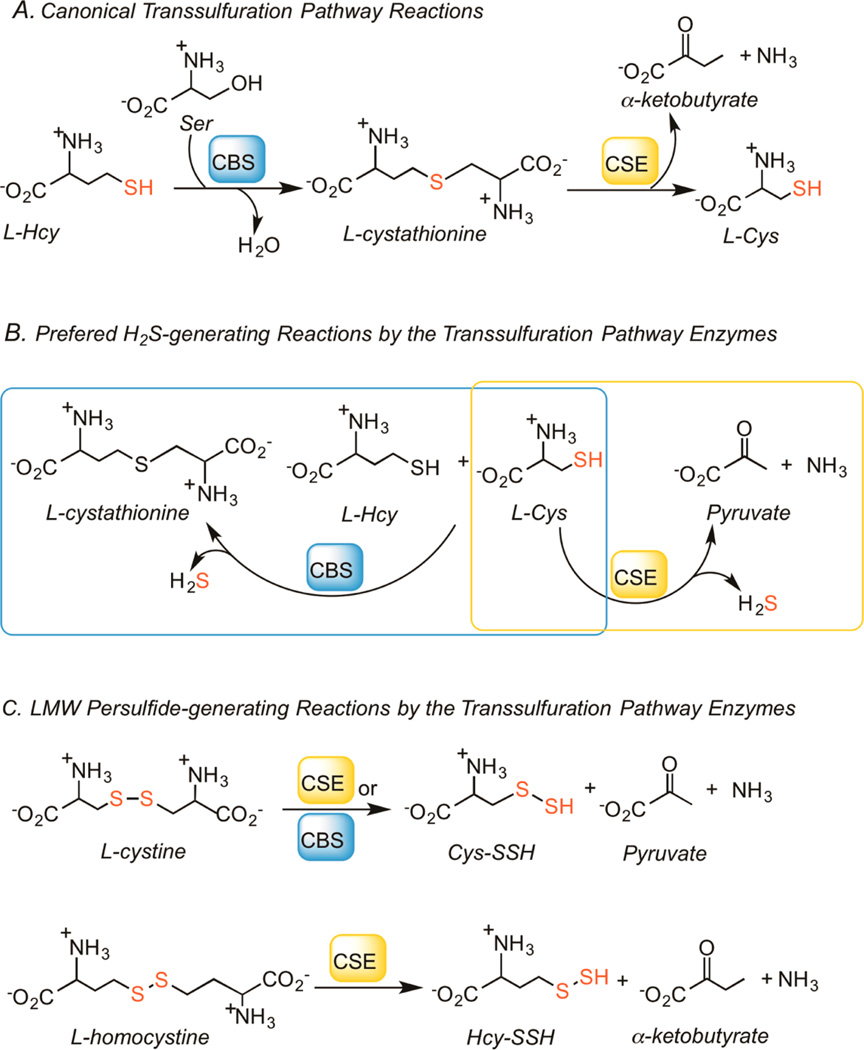
The canonical transsulfuration pathway converts homocysteine to cysteine via the successive actions of cystathionine β-synthase (CBS) and γ-cystathionase (CSE) (Figure 1A). However, the transsulfuration pathway enzymes exhibit a high level of substrate ambiguity and reaction promiscuity. Thus, CBS and CSE are also involved in H2S biogenesis from cysteine and homocysteine (Figure 1B).24,28,29 The third enzyme that produces H2S is mercaptopyruvate sulfurtransferase, which forms an enzyme-bound Cys−SSH intermediate and releases H2S in the presence of reductants or transfers the sulfane sulfur moiety to proteins such as thioredoxin.21 Rat CSE generates Cys−SSH from cystine23 and more recently, rat CBS was shown to catalyze the same reaction (Figure 1C).15 Formation of homocysteine persulfide (HCy−SSH) from homocystine by CSE or CBS has not been previously reported.
Figure 1.
Reactions catalyzed by the transsulfuration pathway enzymes. (A) The canonical reactions catalyzed by CBS and CSE. (B) The preferred H2S-generating reactions via condensation of cysteine and homocysteine (by CBS) and α,β-elimination of cysteine (by CSE). (C) Formation of Cys−SSH from cystine by CBS or CSE and Hcy−SSH from homocystine by CSE.
Another LMW persulfide, GSSH, was postulated to form via persulfide exchange reactions in solution (e.g., between Cys−SSH and GSH).15 GSSH is also produced enzymatically during H2S oxidation by the mitochondrial sulfide-quinone oxidoreductase.20,30 In contrast to the production of H2S during Cys−SSH decomposition, the direct formation of H2S by CBS and CSE is well characterized.24,28,31,32 In the absence of kinetic data on the reaction rates for Cys−SSH and potentially, Hcy−SSH production by the transsulfuration pathway enzymes, it has not been possible to assess their relative contributions to H2S versus to LMW persulfide formation. This information is critically important for assessing whether the transsulfuration pathway is a substantial source of LMW persulfides, which could play a role in signaling.
In addition to the intrinsic catalytic efficiencies of CBS and CSE for H2S versus LMW persulfide production, the ratio of these products is modulated by the concentrations of the relevant substrates in the cell, i.e., via substrate level regulation. In this context, it is important to remember that the cellular milieu is reducing and the concentrations of cystine, homocystine and glutathione disulfide are significantly lower than that of the corresponding thiols, cysteine, homocysteine and glutathione, respectively.15,33 While the absolute concentrations of these thiol and disulfide metabolites vary, the cytoplasmic ratio of the oxidized to reduced species is tightly regulated in a cell- and tissue-specific manner. The intracellular imbalance in favor of thiols is predicted a priori to favor H2S production (from cysteine and homocysteine) versus Cys−SSH and Hcy−SSH formation (from cystine and homocystine, respectively). In this study, we have determined the kinetics of Cys−SSH and Hcy−SSH formation by the human transsulfuration pathway enzymes, simulated their relative contributions to H2S versus persulfide generation at physiologically relevant substrate concentrations, and determined an upper limit for the half-life of Cys−SSH in buffer at pH 7.4. The decay of Cys−SSH is accompanied by the formation of polysulfides (Cys−S−(S)n−S−Cys, n ≥ 1). Our study demonstrates that steady-state persulfide levels are indistinguishable between normal and cystinotic human fibroblasts from patients with pathologically high lysosomal cystine content. Persulfide levels increase in response to exogenous supplementation of cells with cystine but their polysulfide decomposition products rather than enhanced protein persulfidation appear to result under these conditions.
EXPERIMENTAL SECTION
Materials
l-Cystine, l-homocystine, l-serine, cystathionine, iodoacetamide, pyridoxal phosphate, potassium cyanide and potassium thiocyanate were purchased from Sigma-Aldrich. All other chemicals were purchased from Fisher Scientific. Cystinotic fetal lung fibroblasts (GM00090, donor age: 24 weeks) and normal fetal lung fibroblasts (GM01379, donor age: 12 weeks) were purchased from the Coriell Institute Biorepository (Camden, New Jersey).
Expression and Purification of Human CSE and CBS-Recombinant
Human CSE (polymorphic variant S403) and wild-type human CBS were expressed and purified as described previously.34,35
Persulfide Formation Assay
Cys−SSH and Hcy−SSH formation was determined using cold cyanolysis as described previously.36 Briefly, the reaction mixture contained 100 mM HEPES buffer, pH 7.4, 50 µM pyridoxal 5′-phosphate, l-cystine (0–1.5 mM for CSE and 0–10 mM for CBS) and 20 µg of CSE or CBS, in a final volume of 1 mL. Due to the limited solubility of cystine, a stock solution was prepared in 0.1 M NaOH and the pH of the enzymatic reaction mixture was checked following addition of cystine to ensure that it was unaffected. For reactions with CBS, 100 µM AdoMet was also added and the reaction mixtures were incubated at 37 °C for 20 min in parallel with controls lacking enzyme. The reactions were terminated by addition of 40 µL each of 2 M ammonium hydroxide and 1.25 M potassium cyanide to 900 µL of the reaction mixture and incubation was continued at 25 °C for 30 min. Then, 20 µL of formaldehyde (38% v/v) and 200 µL of Goldstein’s reagent36 were added and mixed. The mixture was centrifuged for 5 min at 10 000g and the absorbance of the supernatant was recorded at 460 nm. A calibration curve was prepared using sodium thiocyanate of known concentrations.
Due to the low solubility of homocystine, formation of Hcy−SSH by CSE was monitored in 100 mM Tris buffer, pH 8.5. A stock solution of l-homocystine was prepared in 0.2 M NaOH. The reaction mixture (1 mL final volume) contained 100 mM Tris buffer, pH 8.5, l-homocystine (0–20 mM) and 20 µg CSE incubated at 37 °C for 20 min. Formation Hcy−SSH was detected as described above for Cys−SSH.
Cell Culture and Persulfide Detection
Cells were cultured in MEM supplemented with 15% FBS, 1% penicillin (5000 U/mL)/streptomycin (5000 µg/mL) and 1 mM l-glutamine and incubated in 5% CO2/20% O2 atmosphere. For visualization of persulfide levels by fluorescence microscopy, cells were plated in ibidi dishes (ibidi, Martinsried, Germany) until they reached confluency, fixed using methanol and permeabilized with acetone and then labeled for persulfide detection using the CN-biotin tag-switch assay method, as previously described.37,38 Briefly, fixed cells were first incubated with 10 mM 2-(methylsulfonyl)-1,3-benzothiazole overnight, washed 5 times with PBS and then incubated for 1 h at 37 °C with 2 mM CN-biotin, prepared as described.37,38 DyLight 488 Streptavidin (Pierce, ThermoFisher Scientific) was used to visualize biotinylated proteins. Images of 1024 × 1024 pixels were obtained using a LSM 780 confocal laser scanning system (Carl Zeiss MicroImaging) equipped with an argon laser (458, 488, 514 nm), a diode laser (405 nm), a DPSS-laser (561 nm, LASOS Lasertechnik, Jena, Germany), mounted on an inverted Axio Observer Z1. The filter settings of the confocal scanner were 488 nm excitation for Alexa 488 ((MBS 488/561/633), filter 493–543 nm) and 405 nm excitation for 40,6-diamidino-2-phenyl-indole hydrochloride (MBS-405). A ×63 oil objective lens (numerical aperture 1.4) was used. Sequential scanning and appropriate pinhole settings minimized spectral bleed-through. Experiments were performed in triplicates.
Persulfidation of proteins was also assessed in whole cell lysates (n = 4). Cells were lysed in HEN buffer (50 mM HEPES buffer, pH 7.4, 0.1 mM EDTA, 1% NP-40) containing 1.5% SDS, 1% protease inhibitors (Sigma-Aldrich) and 10 mM 2-methylsulfonyl-1,3-benzothiazole (Santa Cruz Biotechnology, TX). Protein persulfides were labeled using the tag-switch assay method as previously described.37,38. Proteins were separated by SDS-PAGE on a 10% gel, transferred to a nitrocellulose membrane and biotinylation was visualized using DyLight 488 Streptavidin (Pierce, ThermoFisher Scientific) in the ChemiDoc MP System (BioRad). Membranes were also blotted for glyceraldehyde 3-phosphate dehydrogenase, which served as an equal loading control.
To assess the response of persulfide levels to exogenous cystine, normal fetal lung fibroblasts (GM01379), untreated or treated with 200 µM cystine for 1 h, were incubated with 10 µM SSP4 (Dojindo Molecular Technologies, MD) for 15 min. Fluorescence microscopy was carried out using a Carl Zeiss Axiovert 40 CLF inverted microscope, equipped with green fluorescent filter, ×100 oil objective lens and AxioCam ICm1. Experiments were performed in triplicates. Images were postprocessed and semiquantification of fluorescence intensity measured using ImageJ. Protein persulfidation was also assessed in cells ± cystine supplementation using the CN-biotin tag switch reagent as described above.37,38.
Inhibition of Persulfide Formation
The inhibitory effect of serine on Cys−SSH formation by CBS was evaluated under Vmax conditions. Briefly, the reaction contained 100 mM HEPES buffer, pH 7.4, 50 µM pyridoxal 5′-phosphate, 4 mM l-cystine, 100 µM AdoMet, l-serine (0–15 mM) and 20 µg CBS, in a final volume of 1 mL. The reaction mixture was incubated at 37 °C for 20 min. Control reactions lacking CBS were run in parallel. Formation of Cys−SSH was detected by cold cyanolysis as described above. The inhibition constant for serine was calculated using the Michaelis Menten equation describing competitive inhibition kinetics.
In this equation, Ki is the inhibition constant, KM is the Michaelis constant, I is the inhibitor concentration, Vmax is the maximal reaction rate, v is the reaction rate at a given concentration of substrate and inhibitor, and S is the substrate concentration.
LC–MS Analysis of Persulfide Formation
To generate persulfide samples for MS analysis, the reactions were performed in 100 mM ammonium bicarbonate, pH 7.8, containing: (i) 1 mM l-cystine or 4 mM l-homocystine and 50 µg CSE, or (ii) 4 mM l-cystine and 50 µg CBS in a final volume of 1 mL. The mixtures were incubated at 37 °C for 20 min. The reactions were terminated by the addition of 10 mM iodoacetamide and incubated at room temperature for 4 h to block the thiol and persulfide groups. Reactions were run in parallel for estimation of sulfane sulfur formation using the cold cyanolysis procedure described above. The LMW reactants were separated from protein by centrifugation using 10 kDa Amicon filters. The filtrate was frozen at −80 °C until use for MS analysis. Samples were thawed, loaded into vials in an autosampler maintained at 4 °C. Compounds were separated using a HILIC column (BEH XBridge, 4.6 × 50 mm and 3.5 µM particle size from Waters, Milford MA). The column was eluted at a flow rate of 0.5 mL min−1 with a linear gradient ranging from 95% mobile phase B (HPLC-grade acetonitrile) to 95% mobile phase A (20 mM each ammonium acetate and ammonium hydroxide in LC-grade water pH ~ 9.45) over 15 min, followed by 95% A for 5 min and finally, 95% B for 5 min for reequilibration. The effluent of the column was connected to a 4000QTrap Mass Spectrometer (Sciex, Framingham, MA) via the Turbo V electrospray ion source, operating in scans from 50 to 550 m/z over 1 s in positive mode. Other conditions were: declustering potential = 80 V, entrance potential = 10 V, IS = 5500 V, Curtain Gas = 20; GS1 = GS2 = 50 L/min (N2) and ESI temperature = 450 °C with nitrogen as the ion source and collision gas. Peaks for the expected [M + H]+ precursors were identified by extraction of the total ion chromatograms with a mass window of 1.0 a.m.u. To confirm the assigned peaks for cystine, homocystine, Cys−SS−CAM and Hcy-SS−CAM and the reduced amino acids, MS/MS spectra were performed for the mentioned precursors with 1.0 s scans at the range of 50 to 550 mass units. Other conditions were the same as the Q1+ scans except for a collision energy of 30 V, collision cell entrance potential of 10 V and collision cell exit potential of 15 V. Methanol was injected as a blank between the sample runs to reduce sample carryover and water was used as the strong solvent rinse after each injection.
Persulfide Decay Kinetics
The stability of Cys−SSH was determined as follows. Cys−SSH was initially generated in a reaction mixture containing 50 mM Tris, pH 7.4 containing 1 mM l-cystine and 0.7 µM CSE (calculated based on the molecular mass of the monomer) in a final volume of 3 mL, which was incubated at 37 °C for 15 min. Then, the reaction mixture was centrifuged for 5 min at 4 °C using Amicon ultracentrifugal filters with a 10-kDa cutoff to separate the protein from the mixture of substrate and product. The filtrates were then pooled and incubated at 37 °C. The concentration of Cys−SSH was determined by removing 150 µL aliquots at the desired time intervals, mixing with 3 µL of 50 mM Ellman’s reagent (5,5′-dithiobis(2-nitrobenzoic acid) and monitoring the reaction at 412 nm. Persulfides, thiols and sulfide but not disulfides react rapidly with Ellman’s reagent.39
Kinetic Simulation of Reactions Catalyzed by CBS
The three H2S-producing reactions catalyzed by CBS were included in the simulations (Figure S4A, [2–4]).28 The equations describing the velocities for these reactions are presented in Table S1 (eqs 1–3) were developed based on published kinetic data28 and include terms describing the inhibition of CBS by substrates. The rate of Cys−SSH formation by CBS is described by eq 4 and includes terms to describe competitive inhibition by serine and cysteine.
Kinetic Simulation of Reactions Catalyzed by CSE
The H2S producing reactions catalyzed by CSE were included in the simulations (Figure S4B, [2–6]).24 The equations describing the velocities for these reactions are presented in Table S2 (eqs 5–9 were developed based on published kinetic data24 and include terms describing inhibition of the enzyme by substrates). The rates of Cys−SSH and Hcy−SSH formation by CSE are described by eqs 10 and 11, respectively and include terms for competitive inhibition by cysteine, cystathionine, homocysteine, and homocystine (eq 10) or cysteine, cystine, cystathionine, and homocysteine (eq 11).
RESULTS
Kinetics of Persulfide Formation and Characterization of Reaction Products
CSE catalyzes the formation of Cys−SSH and Hcy−SSH from cystine and homocystine, respectively while CBS utilizes only cystine as substrate (Table 1, Figure S1). Under Vmax conditions at pH 7.4 and at 37 °C, the catalytic efficiency for Cys−SSH formation by CSE was ~20-fold greater than by CBS. Due to the low solubility of homocystine at neutral pH, the kinetics of Hcy−SSH formation were characterized at pH 8.5, where the specific activity of CSE is ~5-fold higher than at pH 7.4. By extrapolation, the catalytic efficiency of CSE for Cys−SSH formation at physiological pH is estimated to be ~40-fold higher than for Hcy−SSH formation assuming that the KM for homocystine is unaffected by the change in pH.
Table 1.
Summary of Kinetic Parameters for CBS and CSE-Catalyzed Formation of Persulfidesa
| enzyme (substrate) |
KM (mM) |
Vmax (µmol mg−1 h−1) |
kcat (s−1) |
kcat/KM (M−1 s−1) |
|---|---|---|---|---|
| CBS | ||||
| cystine | 1.3 ± 0.1 | 6.1 ± 0.1 | 0.11 | 85 |
| CSE | ||||
| cystine | 0.12 ± 0.02 | 17 ± 1 | 0.21 | 1.75 × 103 |
| homocystineb | 6.8 ± 1.5 | 120 ± 12 | 1.5 | 221 |
The kinetic parameters were determined in 100 mM HEPES, pH 7.4 with cystine and 100 mM Tris, pH 8.5 with homocystine as described under Experimental Section.
For comparison, the specific activity for Hcy−SSH formation from 1 mM homocystine in 100 mM HEPES, pH 7.4 was 2.1 ± 0.15 versus 11.2 ± 0.5 µmol mg−1 h−1 in 100 mM Tris, pH 8.5 at 37 °C.
Formation of Cys−SSH and Hcy−SSH in the CBS and CSE-catalyzed reactions and the decomposition products of the LMW persulfides were monitored by mass spectrometry (MS). Control reactions lacking protein showed the presence of [Cystine + H+] with m/z = 241, [Cystine + Na+] with m/z = 262.8 and [Cystine +2Na+] with m/z = 284.8 (Figure S2A). Upon reaction with either CSE or CBS followed by derivatization with iodoacetamide, cystine was converted to Cys−SSH as evidenced by the peak with m/z = 211, corresponding to the protonated form of carboxyamidomethylated (CAM) derivative, Cys−SS-CAM (Figure S2B, C). The chromatograms also show an m/z = 121.7 coeluting (at 7.4 min) with Cys−SS-CAM and attributable to protonated cysteine formed via fragmentation of Cys−SS-CAM at the ion source (cysteine itself elutes at 8.76 min). Similarly, the LC-ESI-MS analysis of the reaction mixture containing homocystine showed the expected peak with an m/z = 268.9 and [Homocystine + Na+] with m/z = 290.7 (Figure S3A). In the presence of CSE, Hcy-SS-CAM was seen (m/z = 225.1) (Figure S3B). Since clear separation of homocystine and HCy-SS-CAM was obtained in the HILIC column employed (9 and 7.5 min respectively), it is clear that both protonated compounds appear to exhibit common in-source fragments (m/z =134 and 88). These two fragments are the homologues of those observed at m/z = 120 and m/z = 74 for in-source fragmentation of cystine and Cy-SS-Cam. Also peaks at m/z = 242 and m/z = 247 were observed, which are attributed to the NH4+ (coming from the ammonium acetate in the mobile phase) and Na+ adducts of HCy-SS-CAM.
The LC-ESI-MS/MS data were used to confirm the MS assignments using the Human Metabolome Database (http://www.hmdb.ca/) as a reference for peak assignment. Fragmentation of cystine yielded species with m/z = 119.9 and m/z = 74 corresponding to 2-ammonio-1-acetylene methyl disulfide (H3N+—C≡C—S—S—CH3) and 2-ammonio-1-thia acetylene (HSC≡C—NH3+), respectively (Figure S4A). Similarly, the MS/MS spectrum of homocystine showed peaks at m/z = 133.8 and m/z = 88 generated by fragmentation of the disulfide (Figure S4B) analogous to the fragments observed for cystine but increased by a methylene group. The MS/MS analysis of the CSE reaction with cystine showed 5 major fragment peaks (Figure S4C), two corresponding to the protonated 2-thiaacetamide ion (m/z = 90) and its corresponding deamidation secondary product (OC−CH2−S, m/z = 74). Also observed were the peaks for protonated cysteine (m/z = 122) and the deamidation product of cysteine (m/z = 105) originating from in-source fragmentation of the disulfide precursor ion (m/z = 211). The peak at m/z = 120 is attributable to either the loss of formic acid, CO and NH3 or of the thio-acetamido group from the Cys−SS-CAM precursor ion, which is also seen in the MS/MS spectrum of cystine (Figure S2A). MS/MS analysis of the homocystine-containing reaction mixture showed the expected fragments (m/z = 133.9 and 88) analogous to those seen with Cys−SSH, but with the additional methylene group (Figure S4D). These results confirm CSE-catalyzed formation of Cys−SSH and Hcy−SSH from cystine and homocystine, respectively.
The major side products of enzyme-catalyzed Cys−SSH from cystine were the trisulfide, Cys−S−S−S−Cys (m/z = 273.2) and the pentasulfide Cys−S−S−S−S−S−Cys (m/z = 337) (Figure S5A,B). The peak corresponding to Cys−S−S− S−S−Cys (m/z = 305) was also observed, although at a lower abundance than the other two species. The major side products of CSE-catalyzed Hcy−SSH formation from homocystine were the tri- and tetra-sulfides, Hcy−S−S−S−Hcy (m/z = 301.0) and Hcy−S−S−S−S−Hcy (m/z = 333.0) (Figure S5C,D). As discussed later, these polysulfide (R−(S)n−R) species are formed during decomposition of the initially formed persulfide products.
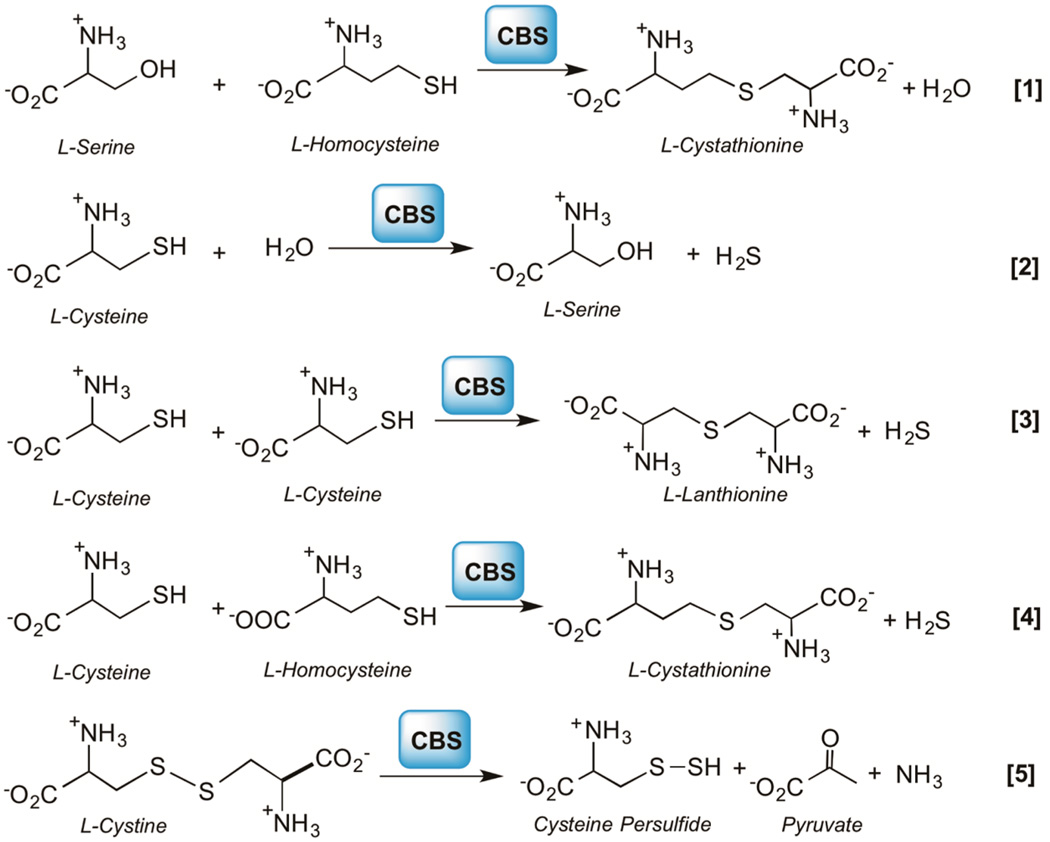
Simulation of Cys−SSH Formation Kinetics by CBS and CSE
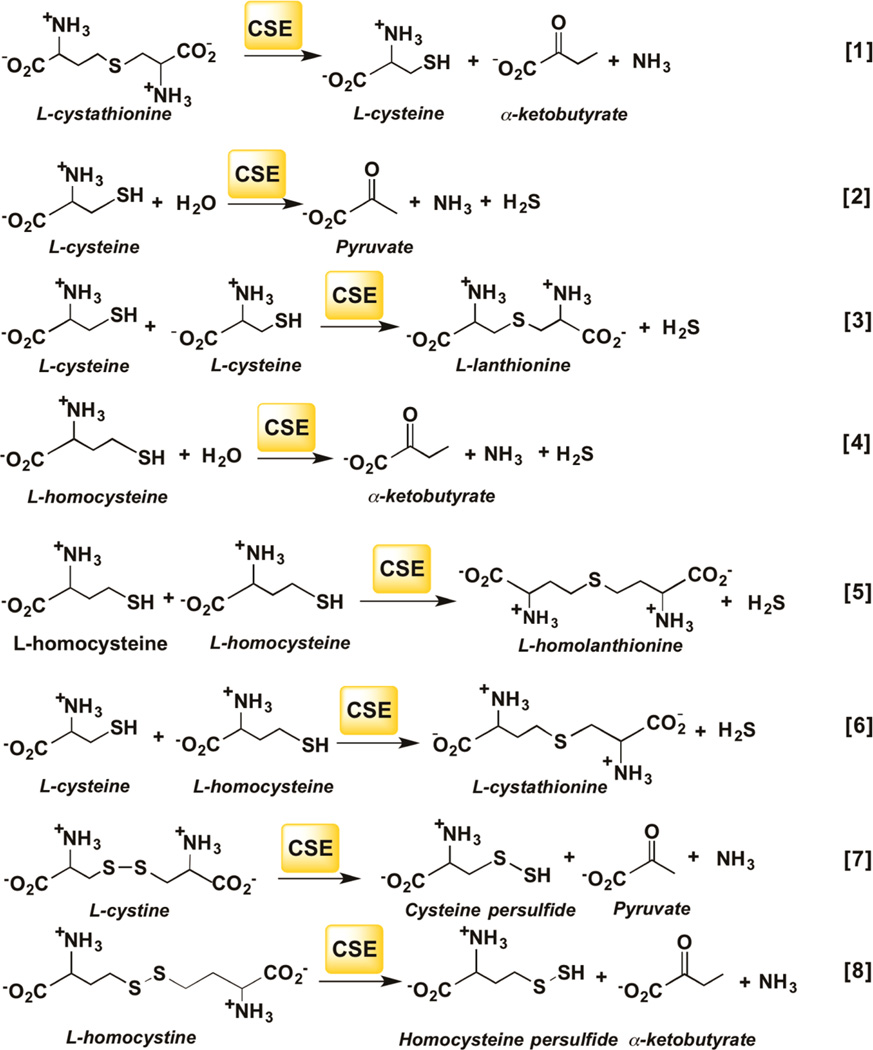
CBS and CSE each catalyzes multiple H2S generating reactions that have been characterized previously.24,28 These reactions have to be taken into account in simulations of the net contribution of each enzyme to H2S versus LMW persulfide formation. The canonical reaction catalyzed by CBS in the transsulfuration pathway involves the β-replacement of serine by homocysteine to give cystathionine (Figure 2, [1]). When cysteine substitutes for serine in the β-replacement reactions [2–4], H2S is eliminated. CBS also catalyzes the α,β-elimination of cystine forming Cys−SSH [5]. The kinetic scheme corresponding to reactions [1–5] is shown in Figure S6A and the KM1 and KM2 values correspond to Michaelis constants for the substrates occupying the first and second amino acid binding sites in CBS as defined.28 The canonical reaction catalyzed by CSE involves the α–γ elimination of cystathionine to give cysteine, α-ketobutyrate and ammonia (Figure 3, [1]). CSE catalyzes a greater variety of H2S producing reactions than CBS (Figure 3, [2–6]). Furthermore, in addition to catalyzing the α,β-elimination of cystine to form Cys−SSH [7], CSE also forms Hcy−SSH from homocystine [8]. The kinetic scheme corresponding to reactions [1–8] is shown in Figure S6B. The KM1 and KM2 values correspond to Michaelis Menten constants for the substrates occupying the first and second amino acid binding sites in CSE.24
Figure 2.
Reactions catalyzed by CBS. CBS catalyzes the condensation of serine and homocysteine to give cystathionine and water [1] or reactions [2–4] that utilize cysteine to produce H2S. Reaction [5] leads to Cys−SSH synthesis from cystine.
Figure 3.
Reactions catalyzed by CSE. CSE catalyzes the α,γ elimination of cystathionine [1]. Reactions [2–6] lead to the production of H2S. Reactions [7] and [8] lead to Cys−SSH and Hcy−SSH synthesis from cystine and homocystine, respectively.
An additional factor that needs to be taken into consideration for simulations is the potential inhibition of LMW persulfide formation by serine and cystathionine, which are substrates for the canonical reactions catalyzed by CBS and CSE, respectively. For this, the inhibition constant for serine for Cys−SSH formation by CBS was determined (KiSer = 0.07 ± 0.03 mM) while the Ki value for cystathionine used in the simulations was the KM for cystathionine (0.28 ± 0.03 mM) reported previously.24 The reaction rates were simulated for H2S, Cys−SSH and Hcy-SSH formation at physiologically relevant liver concentrations of metabolites (Table 2) using eqs 1–11 (Tables S1 and S2) and the kinetic parameters listed in Table S3.
Table 2.
Physiological Concentrations of Metabolites in Murine Liver Used in the Simulations
| metabolite | concentration (µM) | reference |
|---|---|---|
| cysteinea | 100 | 15,33,53,61 |
| cystine | 0.2 | 15 |
| homocysteine | 1.25 | 61 |
| homocystineb | 0.06 | – |
| cystathionine | 8.0 | 61 |
| serine | 560 | 62 |
The study reporting hepatic cystine concentrations of 0.20 ± 0.01 µM reported a cysteine concentration of 68 ± 17 µM.15 Other studies have reported hepatic cysteine concentrations of 129 ± 12 µM,53 115 ± 52 µM33 and 75 ± 3 µM61 leading to the average value of 100 µM used here.
The concentration of homocystine in murine liver has not been reported to our knowledge. We arbitrarily set the concentration of homocystine to be 5% of the homocysteine concentration.
Our simulations confirm that at physiologically relevant substrate concentrations, the dominant mechanism for CBS-catalyzed H2S production is via condensation of cysteine and homocysteine (Figure 2, reaction [4]).28 This reaction accounts for 89% of the total H2S producing activity of CBS (Table 3). In comparison to the sum of the activities for the H2S producing reactions (3.0 µmol mg−1 h−1), Cys−SSH production (0.0001 µmol mg−1 h−1) is negligible. For CSE, the dominant H2S-producing reaction at physiologically relevant substrate concentrations is the α,β-elimination of cysteine (Figure 3, reaction [2]) as reported previously24 and accounts for 98.5% of the total H2S produced by CSE. The specific activities for H2S (1.97 µmol mg−1 h−1) and Cys−SSH (0.026 µmol mg−1 h−1) production by CSE represent 98.7% and 1.3% of the total CSE activity. The specific activity for Hcy−SSH production by CSE (0.0002 µmol mg−1 h−1) is negligible (Table 3).
Table 3.
Simulated Rates of H2S and LMW Persulfide Production at Physiologically Relevant Substrate Concentrationsa
| enzyme/ reaction |
activity (µmol mg−1 h−1) |
enzyme/ reaction |
activity (µmol mg−1 h−1) |
|---|---|---|---|
| CBS | CSE | ||
| H2S from 2 | 0.31 | H2S from 2 | 1.94 |
| H2S from 3 | 0.032 | H2S from 3 | 0.000002 |
| H2S from 4 | 2.66 | H2S from 4 | 0.031 |
| Total H2S (2 + 3 + 4) |
3.0 | H2S from 5 | 0.0001 |
| Cys−SSH from 5 |
0.0001 | H2S from 6 | 0.000005 |
| Total H2S (2 + 3+4 + 5+6) |
1.97 | ||
| Cys−SSH from 7 |
0.026 | ||
| Hcy−SSH from 8 |
0.0002 |
The concentrations of the metabolites used in the simulations are given in Table 2.
Relative Contributions of CBS and CSE to H2S versus Cys−SSH Formation
To compare the relative output of H2S and LMW persulfides by CBS versus CSE in vivo, differences in their protein concentrations in tissues must be taken into account. These numbers are available for murine liver where the amount of CBS is 62-fold lower than CSE.26 Hence, despite the 1.5-fold higher specific activity of CBS than CSE for total H2S production (Table 3), adjustment for differences in their concentration in liver, yields a specific activity for H2S production by CBS that is <3% that of CSE. The rate of Cys−SSH formation by CBS is negligible. Hence, our combined experimental and simulation data indicate that the transsulfuration enzymes are a poor source of LMW persulfides at the values for cellular concentrations of metabolites shown in Table 2.
Half-Life of Cys−SSH in Buffer at Physiological pH
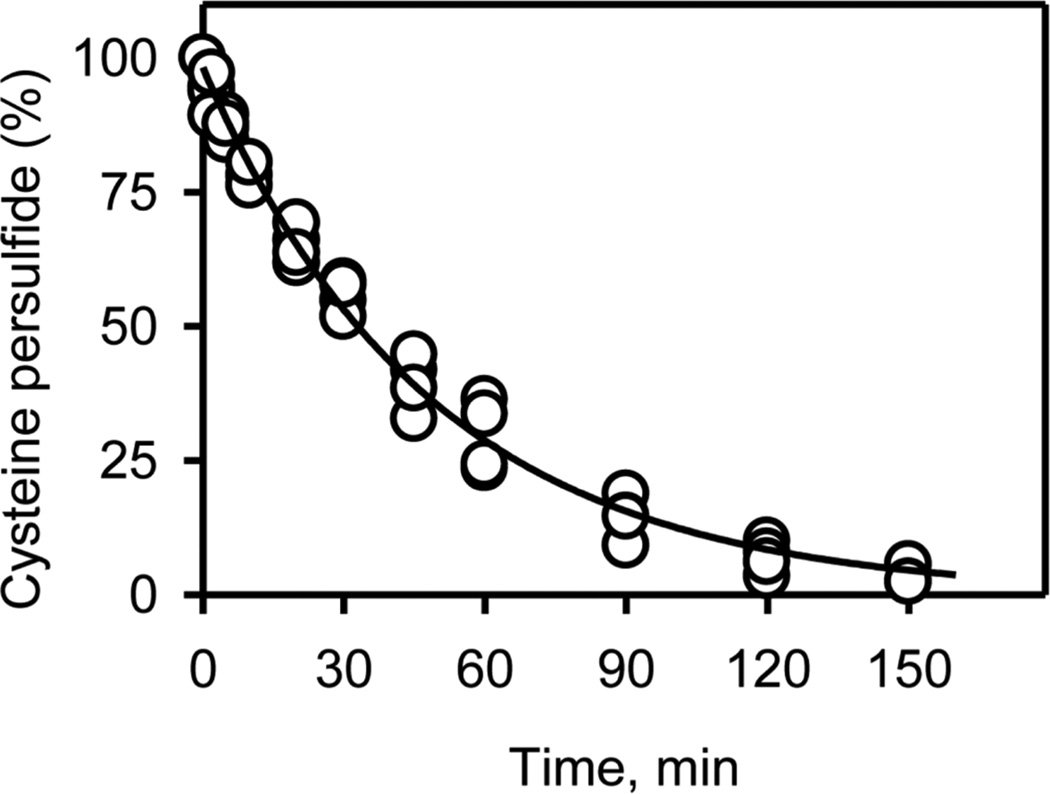
Persulfides are metastable and considerably more reactive than the corresponding thiols.8 Hence a quantitative analysis of their stability at physiological pH is of considerable interest. The half-life of Cys−SSH generated in situ by CSE was determined using Ellman’s reagent to monitor the presence of persulfide.39 The average of three independent experiments yielded a t½ of 35 ± 3.5 min at pH 7.4 and 37 °C (Figure 4). In our experimental set up, unreacted cystine, which is electrophilic, was present together with the product, Cys−SSH. Hence, as discussed later, the decay kinetics represents the sum of the decay of the initial persulfide and intermediate thiol and sulfide species, all of which react with Ellman’s reagent. The half-life of Cys−SSH in vivo is likely to be considerably lower since cells contain high (millimolar) concentrations of LMW and protein thiols40 in addition to other nucleophiles and electrophiles, which can react with Cys−SSH.
Figure 4.
Stability of Cys−SSH in buffer at physiological pH. The kinetics of decay of Cys−SSH formed in the CSE reaction was monitored using Ellman’s reagent as described under Methods. The data are the aggregate of three independent experiments and the line represents a single exponential fit giving a t½ for decay of 35 ± 3.5 min.
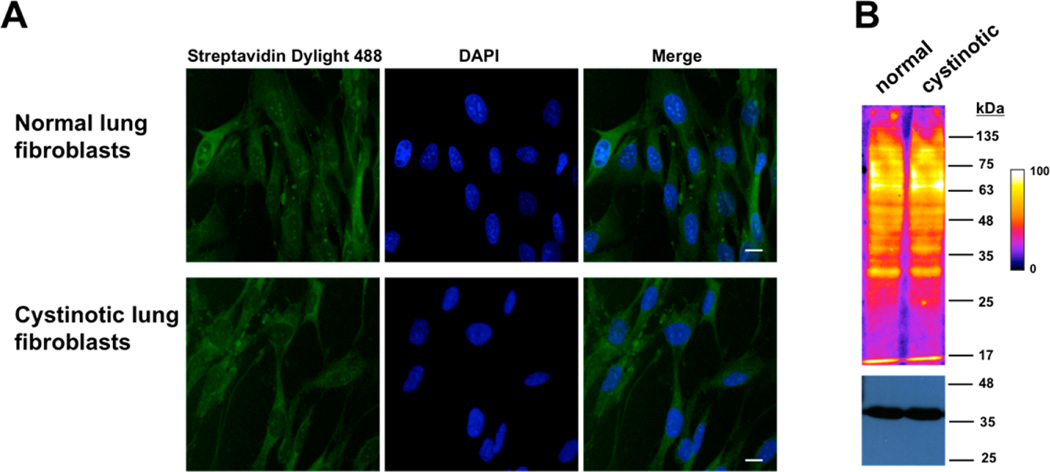
Persulfide Levels in Cystinotic Cells
We used two methods for the in situ detection of persulfides in normal (GM01379) versus cystinotic (GM00090) fetal lung fibroblast cells: the fluorescence-based CN-biotin tag-switch method and the fluorescent sulfane sulfur probe 4, SSP4.37,38,41 Human fibroblasts express both CBS and CSE as evident from protein or activity analyses.42–44 We confirmed the presence of CBS and CSE in both GM00090 and GM01379 by Western blot analysis (not shown). Cystinotic cells accumulate high levels of cystine in the lysosomal compartment due to an underlying defect in a lysosomal cystine transporter.45 The total cystine concentration in the cystinotic GM00090 cell line is ~15-fold higher than in the normal GM01379 cell line44 although it is not known whether cytoplasmic cystine levels are also affected. In contrast, the total cysteine concentrations in these cell lines are comparable.44 Analysis of the fluorescence signal using the CN-biotin tag switch method (n = 30 cells, experiments performed in triplicates) revealed no significant difference between the two different cell types (1 ± 0.2 in control versus 0.9 ± 0.1 in cystinotic fibroblasts) (Figure 5A).
Figure 5.
Visualization of persulfides in normal and cystinotic fibroblasts using the CN-biotin tag switch method. (A) Representative photomicrographs of normal and cystinotic human lung fibroblasts labeled with CN biotin tag-switch assay. The nuclei were stained with DAPI. (B) Intracellular persulfide levels in normal and cystinotic human lung fibroblast cell lysates, labeled using the CN-biotin tag switch reagent. Proteins were visualized using Streptavidin Dylight 488. Artificial color intensity was used and the gradient scale is shown on the right. GAPDH was used as a loading control (bottom). Scale bar: 20 µm.
As a complementary approach, persulfidation levels were also monitored in whole cell lysates. For this, cell lysates were labeled with CN-biotin, the biotinylated proteins separated by SDS PAGE, transferred to a nitrocellulose membrane and visualized using fluorescently labeled streptavidin. As in the whole cell imaging experiments, no significant difference was observed between the two cell lines (1.00 ± 0.03 in GM01379 versus 0.91 ± 0.12 in GM00090, n = 4), following normalization to GAPDH protein levels (Figure 5B).
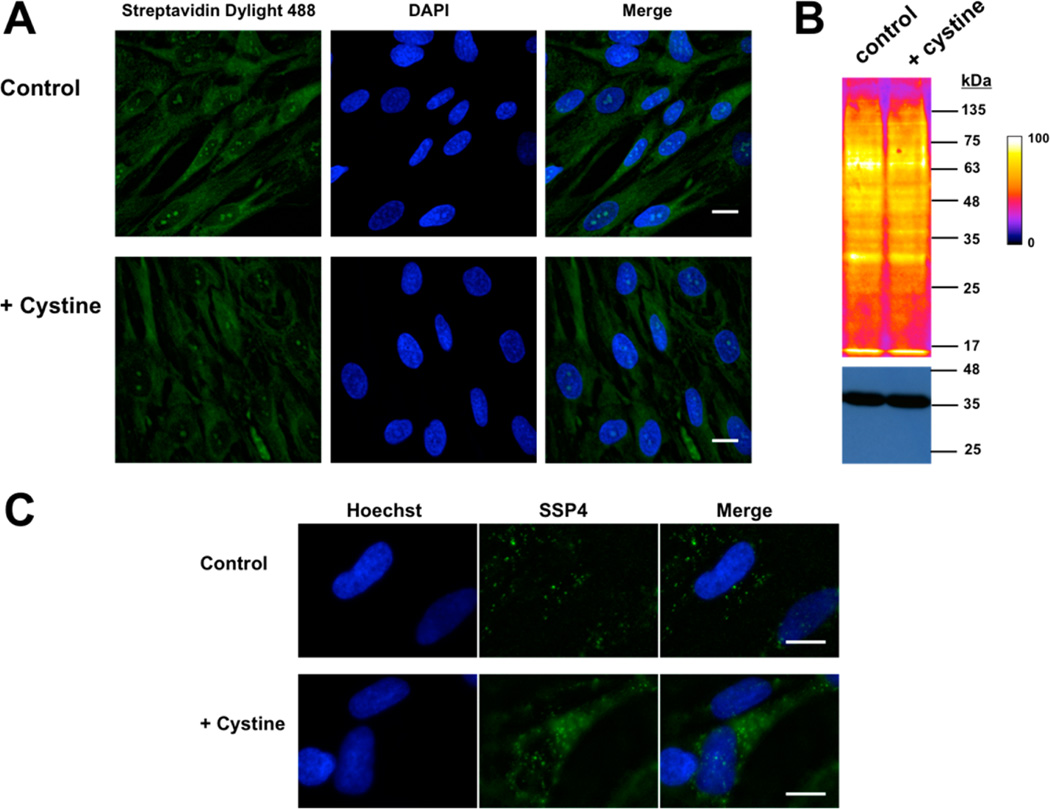
Protein Persulfide Levels are not Increased by Cystine Supplementation
Supplementation of normal fibroblast cells with exogenous cystine (200 µM) for 1 h did not show any difference in persulfide levels as visualized using the CN-biotin tag-switch method in whole cells (Figure 6A) or in cell lysates (Figure 6B). However, an ~2-fold increase in cellular polysulfides (as discussed below) was observed using the SSP4 probe in cystine-treated (11 ± 3, n = 12) versus untreated (5 ± 2, n = 12) cells (Figure 6C).
Figure 6.
Visualization of persulfides and polysulfides in normal fibroblasts in response to exogenous cystine treatment. (A) Photomicrographs of human lung fibroblasts cultured ±200 µM cystine for 1 h and labeled by the CN biotin tag-switch method. Nuclei were stained with DAPI. (B) Western blot analysis of proteins from (A) shows a similar labeling intensity in cells grown ± cystine supplementation. GAPDH was used as a loading control (bottom). (C) Representative photomicrographs show a clear increase in fluorescence in cells treated with cystine when probed with the SSP4 reagent. Nuclei were stained with Hoechst. Scale bar: 20 µm.
DISCUSSION
Despite the growing literature on protein persufidation7 and the cellular and physiological effects of sulfide and polysulfide treatment,46 the identity and sources of reactive sulfur species important for signaling are not known. CBS and CSE, which are believed to be the major sources of H2S29 and of Cys−SSH,15 also generate cysteine from homocysteine via the canonical transsulfuration pathway (Figure 1). In a new twist, it was recently suggested that Cys−SSH is the primary reactive sulfur species produced by CBS and CSE and that H2S is formed via Cys−SSH degradation.15 However, the rates of formation of Cys−SSH versus H2S at physiologically relevant substrate concentrations and the stability of Cys−SSH in the cellular milieu, were not assessed. In this study, we have examined the hypothesis that substrate level regulation controls the competing flux of sulfur-containing amino acids through the transsulfuration pathway using combined kinetic, computational and cellular approaches to evaluate the relative rates of H2S versus Cys−SSH biogenesis at physiologically relevant substrate concentrations. We also demonstrate for the first time that CSE converts homocystine to Hcy−SSH and report the simulated rates of Hcy−SSH formation under normal and homocystinuric conditions.
While the production of Cys−SSH by CSE has been known for some time,47 a significant in vitro rate for Cys−SSH formation by CBS was reported only recently.15 In contrast to the high concentration of cystine (0.5–1.25 mM) used to study Cys−SSH production by purified rat CBS and CSE, tissue cystine concentrations were reported to be 0.05 µM in lung and 0.2 µM in liver while being undetectable in heart and brain.15 Cysteine concentrations were reported to be 2–3 orders of magnitude higher in the same tissues. The cystine/cysteine ratio is very different in the extra- versus intracellular compartments.48 While cystine is the dominant form of the amino acid in plasma, it is present at low concentrations inside cells. Cystine is transported via the xc- cystine/glutamate antiporter49 and once inside, is reduced to cysteine by thioredoxin or thioredoxin related proteins.50
Under Vmax conditions, both CBS and CSE exhibit robust rates of Cys−SSH formation (Table 1). Furthermore, the Vmax for Hcy−SSH and Cys−SSH formation by CSE are comparable at physiological pH (obtained by extrapolation of the kinetic data for Hcy−SSH formation measured at pH 8.5 to pH 7.4). However, the low solubility of homocystine at pH 7.4 combined with the high KM value (6.8 mM), suggests that the catalytic efficiency of Hcy−SSH production is of minor importance under normal cellular conditions where homocystine concentrations are very low. Unlike CSE, human CBS does not generate Hcy−SSH from homocystine. This result is consistent with the inability of homocysteine to form a Schiff base in CBS, which is needed to initiate the elimination reaction.28
In murine liver, where the most complete data set needed to model the kinetics of the transsulfuration pathway, is available,51 the specific activity of H2S production in the presence of 100 µM cysteine is ~484 µmol h−1 kg−1 tissue.52 Since CSE is the major source of cysteine-derived H2S in liver,26 we can estimate by extrapolation from the data in Table 3 that Cys−SSH production at 0.2 µM intracellular cystine is 6.5 µmol h−1 kg−1 tissue or a mere ~1.3% of H2S production.
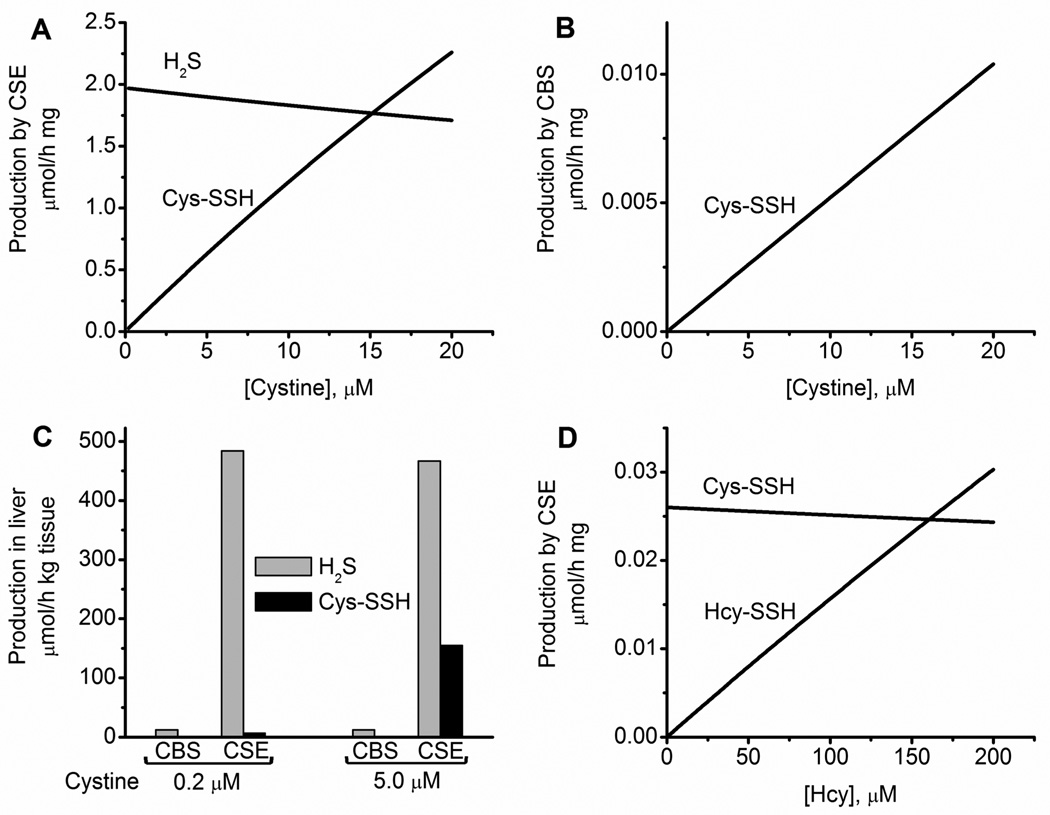
Next, we simulated the dependence of Cys−SSH formation on cystine concentration. As expected, the specific activity of Cys−SSH production increases linearly with cystine concentration (Figure 7A). At cystine concentrations >15 µM, the specific activity for Cys−SSH production is predicted to exceed that of H2S production due to the low KM for cystine (0.12 mM) and the high kcat/KM(cystine) value (1750 M−1 s−1) (Table 1) compared to the corresponding values for H2S production (KM,(Cys) = 1.7 mM and kcat/KM(Cys) = 277 M−1 s−1) by CSE.24 The specific activity for Cys−SSH production by CBS remains negligible at all cystine concentrations (Figure 7B) compared to the specific activity for H2S production (Table S4).
Figure 7.
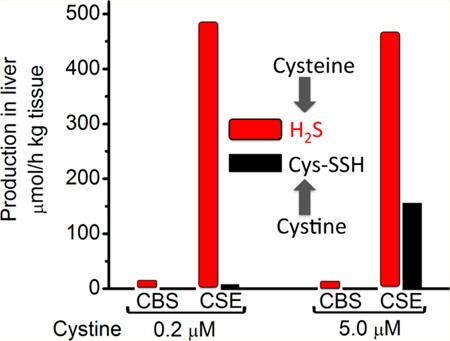
Simulation of H2S and LMW persulfide production rates by CBS and CSE in murine liver. Dependence of the specific rates of total H2S and Cys−SSH production by CSE (A) and of Cys−SSH production by CBS (B) on cystine concentration. A and B, show data from simulations at varying concentrations of cystine and at physiological concentrations of other metabolites (reported in Table 2). The simulations in A predict a modest decline in H2S production rate with increasing cystine concentration due to competition between cysteine and cystine for the CSE active site. (C). Simulated results comparing the rates of H2S and −SSH production by CBS and CSE in murine liver at two concentrations of cystine (0.2 and 5.0 µM) and physiologically relevant concentrations of other metabolites (reported in Table 2). The simulations took into account the difference in CBS and CSE protein levels and the experimentally determined rate of H2S production in murine liver.26,52 The rate of Cys−SSH production by CBS is too low to be visible. (D). Dependence of the specific rates of Cys−SSH and Hcy-SSH production by CSE on homocysteine [Hcy]. The concentration of homocystine was 5% that of homocysteine. The concentration of the other metabolites used for the simulations are reported in Table 2.
In our assessment, the low concentration of cystine (0.2 µM) reported for murine liver15 and used in our initial simulations, could be an underestimation and in fact, cystine concentration is definitely higher in some tissues, e.g., kidney.53 We therefore reran the simulations assuming that cystine represents 5% of the cysteine concentration in liver (like glutathione disulfide represents ~5% of the murine liver glutathione pool). At this concentration of cystine (i.e., 5.0 µM), the specific activity of Cys−SSH production by CSE is 0.63 µmol mg−1 h−1 versus 1.90 µmol mg−1 h− for H2S production (Table S4). The hepatic activity of Cys−SSH production by CBS compared to CSE, which takes into account the differences in tissue levels of CBS and CSE, is negligible at both 0.2 µM and 5.0 µM cystine concentration (Figure 7C). In contrast, the hepatic activity of Cys−SSH production by CSE increases from 1.3% to 33% of H2S synthesis as cystine concentration is increased from 0.2 µM to 5.0 µM (Figure 7C).
Next, we examined the extent to which elevated cellular homocystine concentration can modulate Hcy−SSH formation. Hereditary homocystinuria is a clinical condition that leads to catastrophically elevated homocysteine levels.54 To our knowledge, the concentration of intracellular homocystine has not been reported. Hence, for our simulations, we assumed that homocystine represents 5% of the homocysteine pool (Table 2). The specific activity of Hcy−SSH production from homocystine increases linearly with increasing homocysteine concentration and becomes equal to that of Cys−SSH formation by CSE (from 0.2 µM intracellular cystine) at ~150 µM homocysteine (Figure 7D), suggesting that Hcy−SSH synthesis could be a factor in homocystinuria where homocysteine levels can exceed 200 µM.55
On the basis of our experimental and simulated results, we conclude that H2S rather than Cys−SSH is the major product of the transsulfuration pathway in the reducing intracellular milieu (Figure 7C). However, Cys−SSH production might become more significant under some conditions. First, if the import of cystine from the extracellular medium is coupled by spatial proximity of the transporter to CSE, then the local concentration of available cystine would be higher than its bulk cytoplasmic concentration and would accelerate Cys−SSH formation. However, CSE is normally present diffusely in the cytoplasm and there is no evidence that it localizes near the membrane.56 Second, cystine accumulation as in the pathological condition, cystinuria,57 could enhance Cys−SSH synthesis. Due to its limited solubility, cystine tends to crystallize forming cystine stones, a clinical hallmark of cystinuria,57 which might limit cystine availability in this disease state. Third, acute or chronic oxidative stress conditions leading to increased cytoplasmic cystine levels would stimulate Cys−SSH formation and this possibility warrants further examination.
The absence of substantial cystine-derived protein persulfidation was confirmed by bioimaging studies in cystinotic and normal fibroblasts grown with or without cystine supplementation (Figures 5 and 6). A priori, two factors argue against the utility of LMW persulfides as sulfur donors in protein persulfidation reactions. First, persulfides are highly reactive, which raises questions about how target selectivity would be achieved in a cellular milieu where high concentrations of thiols and other nucleophiles prevail.10 Second, the reaction between Cys−SSH and a thiolate anion is expected to lead to the transfer of the cysteinyl moiety (i.e., protein cysteinylation, eq 1) since the sulfide anion (pKa = 7) is a better leaving group than the cysteine thiolate anion (pKa = 8.3).8 By analogy, in our experiments, the decay of Cys−SSH was accompanied by the formation of polysulfides (Cys−S−(S)n−S−Cys), resulting from transfer of the cysteinyl moiety and elimination of sulfide (eq 2).
| (1) |
| (2) |
| (3) |
Tri- and tetra-sulfides were also reported to form during decomposition of an organic persulfide in aqueous buffer at pH 7.439. In contrast, mixtures of hydropolysulfides (Cys−S− (S)n−H and GS−(S)n−H) were reported to form during the decomposition of Cys−SSH in the presence of glutathione at pH 7.5,15 which is not expected to be favored based on the chemical principles as discussed above (the pKa of glutathione is 8.9 58). A different reactivity, i.e., disproportionation, was observed for an organic persulfide in dichloromethane leading to formation of the organic thiol and elemental sulfur (S8)59.
Cell imaging studies showed an increase in fluorescence intensity in cells treated with exogenous cystine when the SSP4 reagent was used (Figure 6C) but not when the CN-biotin reagent was employed (Figure 6A). Given the increasing utilization of these reagents in bioimaging studies, it is important to understand the reactive sulfur species that each reagent reports on and thereby, to understand the basis of the apparently discrepant results obtained with them. SSP4 like the earlier SSP1/2 reagents purportedly detects persulfides41 but based on chemical principles, is more likely to detect thiosulfoxide tautomers of polysulfides (eq 4) and hydropolysulfides (e.g Cys—S(═S)—SH). The oxidation state of both sulfur atoms in LMW persulfides (e.g., Cys−SSH) is −1. The oxidation state of the bridging sulfurs that are between the terminal sulfurs in hydropolysulfides (e.g., Cys−S−(S)n−SH) and in polysulfides (e.g., Cys−S−(S)n−S−Cys) is 0. On the basis of pKa arguments, the reaction of Cys−SSH with the nucleophile in the SSP probes will result in addition to the cysteine sulfur and displacement of the sulfide anion (eq 5), in analogy to the reaction shown above in eq 1.
| (4) |
| (5) |
The addition of the SSP reagent to the outer sulfur of Cys−SSH could, however, occur in native proteins where the microenvironment results in a perturbation of the cysteine pKa to a value below that of H2S. Reactive cysteines on proteins, which are susceptible to oxidative modifications, do in fact have lower pKa values.60 In addition, enzymes like mercaptopyruvate sulfurtransferase and rhodanese, which stabilize Cys−SSH intermediates in their active sites, use stereoelectronic control to transfer the outer sulfur to thiophilic acceptors.20,21 Hence, SSP4 can detect hydropolysulfides, polysulfides and protein bound persulfides at cysteines with low pKa values but is unlikely to detect LMW persulfides in solution.
Persulfide detection with the CN-biotin reagent involves nucleophilic addition of the reagent to the inner sulfur atom of a benzothiazole-blocked persulfide to generate a CN-biotintagged protein that is subsequently visualized with streptavidin.37 The thio-benzothiazole derived from the reaction of a persulfide with the 2-methylsulfonyl-1,3-benzothiazole blocking group, is a better leaving group than the cysteine thiolate and leads to preferential tagging of the protein (eq 6)37. Hence unlike the SSP reagents, the CN-biotin reagent should be effective in labeling LMW as well as protein persulfides.
| (6) |
Since the CN-biotin reagent did not reveal changes in fluorescence intensity between cells cultured in the presence or absence of cystine supplementation (Figure 6A,B), we conclude that this treatment did not lead to observable changes in protein persulfide levels. On the other hand, an ~2-fold increase in fluorescence intensity in cystine-treated cells was observed using the SSP4 reagent (Figure 6C), which can be attributed to its reaction with the thiosulfoxide tautomer of LMW polysulfides formed by decomposition of Cys−SSH. The synthesis of Cys−SSH could have been transiently increased by the enhanced import of cystine in supplemented cells prior to re-equilibration of the cystine/cysteine ratio via the action of thioredoxin-like proteins.
CONCLUSIONS
Our kinetic results provide a comparative analysis of the inherent capacity for Cys−SSH synthesis by human CBS and CSE under in vitro conditions while our mathematical simulations reveal that H2S rather than Cys−SSH is the major product of the transsulfuration pathway enzymes at physiologically relevant substrate concentrations. Our study also reveals for the first time, the capacity of CSE but not CBS to form Hcy−SSH from homocystine, although the rate of production of this metabolite is predicted to be negligible under normal conditions but to increase in homocystinuria. The steady-state concentration of cellular polysulfides was increased in response to bolus administration of cystine but did not lead to observable changes in protein persulfide levels. Our simulations predict that under conditions of oxidative stress or other conditions leading to increased intracellular cystine concentrations, Cys−SSH synthesis by CSE would rise leading to polysulfide formation. Based on the chemical reactivity of LMW persulfides, they are unlikely to be direct sulfur donors in protein persulfidation reactions.
Supplementary Material
Acknowledgments
This work was supported by a grant from the National Institutes of Health (HL58984 to RB) and the American Heart Association (14POST18760003 to PKY).
Footnotes
ASSOCIATED CONTENT
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.5b10494.
Tables S1–S4 and Figures S1–S6. (PDF)
The authors declare no competing financial interest.
REFERENCES
- 1.Lavu M, Bhushan S, Lefer D. J. Clin. Sci. 2011;120:219. doi: 10.1042/CS20100462. [DOI] [PubMed] [Google Scholar]
- 2.Abe K, Kimura H. J. Neurosci. 1996;16:1066. doi: 10.1523/JNEUROSCI.16-03-01066.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kimura Y, Goto Y, Kimura H. Antioxid. Redox Signaling. 2010;12:1. doi: 10.1089/ars.2008.2282. [DOI] [PubMed] [Google Scholar]
- 4.Kimura Y, Kimura H. FASEB J. 2004;18:1165. doi: 10.1096/fj.04-1815fje. [DOI] [PubMed] [Google Scholar]
- 5.Zanardo RC, Brancaleone V, Distrutti E, Fiorucci S, Cirino G, Wallace JL. FASEB J. 2006;20:2118. doi: 10.1096/fj.06-6270fje. [DOI] [PubMed] [Google Scholar]
- 6.Li L, Bhatia M, Zhu YZ, Zhu YC, Ramnath RD, Wang ZJ, Anuar FB, Whiteman M, Salto-Tellez M, Moore PK. FASEB J. 2005;19:1196. doi: 10.1096/fj.04-3583fje. [DOI] [PubMed] [Google Scholar]
- 7.Mustafa AK, Gadalla MM, Sen N, Kim S, Mu W, Gazi SK, Barrow RK, Yang G, Wang R, Snyder SH. Sci. Signaling. 2009;2:ra72. doi: 10.1126/scisignal.2000464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cuevasanta E, Lange M, Bonanata J, Coitino EL, Ferrer-Sueta G, Filipovic MR, Alvarez B. J. Biol. Chem. 2015;290:26866. doi: 10.1074/jbc.M115.672816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carballal S, Trujillo M, Cuevasanta E, Bartesaghi S, Moller MN, Folkes LK, Garcia-Bereguiain MA, Gutierrez-Merino C, Wardman P, Denicola A, Radi R, Alvarez B. Free Radical Biol. Med. 2011;50:196. doi: 10.1016/j.freeradbiomed.2010.10.705. [DOI] [PubMed] [Google Scholar]
- 10.Mishanina TV, Libiad M, Banerjee R. Nat. Chem. Biol. 2015;11:457. doi: 10.1038/nchembio.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kabil O, Banerjee R. J. Biol. Chem. 2010;285:21903. doi: 10.1074/jbc.R110.128363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kabil O, Banerjee R. Antioxid. Redox Signaling. 2014;20:770. doi: 10.1089/ars.2013.5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kabil O, Motl N, Banerjee R. Biochim. Biophys. Acta, Proteins Proteomics. 2014;1844:1355. doi: 10.1016/j.bbapap.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kimura Y, Mikami Y, Osumi K, Tsugane M, Oka J, Kimura H. FASEB J. 2013;27:2451. doi: 10.1096/fj.12-226415. [DOI] [PubMed] [Google Scholar]
- 15.Ida T, Sawa T, Ihara H, Tsuchiya Y, Watanabe Y, Kumagai Y, Suematsu M, Motohashi H, Fujii S, Matsunaga T, Yamamoto M, Ono K, Devarie-Baez NO, Xian M, Fukuto JM, Akaike T. Proc. Natl. Acad. Sci. U. S. A. 2014;111:7606. doi: 10.1073/pnas.1321232111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vitvitsky V, Yadav PK, Kurthen A, Banerjee R. J. Biol. Chem. 2015;290:8310. doi: 10.1074/jbc.M115.639831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kimura Y, Toyofuku Y, Koike S, Shibuya N, Nagahara N, Lefer D, Ogasawara Y, Kimura H. Sci. Rep. 2015;5:14774. doi: 10.1038/srep14774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greiner R, Palinkas Z, Basell K, Becher D, Antelmann H, Nagy P, Dick TP. Antioxid. Redox Signaling. 2013;19:1749. doi: 10.1089/ars.2012.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Toohey JI. Biochem. J. 1989;264:625. doi: 10.1042/bj2640625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Libiad M, Yadav PK, Vitvitsky V, Martinov M, Banerjee R. J. Biol. Chem. 2014;289:30901. doi: 10.1074/jbc.M114.602664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yadav PK, Yamada K, Chiku T, Koutmos M, Banerjee R. J. Biol. Chem. 2013;288:20002. doi: 10.1074/jbc.M113.466177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mueller EG. Nat. Chem. Biol. 2006;2:185. doi: 10.1038/nchembio779. [DOI] [PubMed] [Google Scholar]
- 23.Cavallini D, Mondovi B, De Marco C, Sciosciasantoro A. Arch. Biochem. Biophys. 1962;96:456. doi: 10.1016/0003-9861(62)90436-8. [DOI] [PubMed] [Google Scholar]
- 24.Chiku T, Padovani D, Zhu W, Singh S, Vitvitsky V, Banerjee R. J. Biol. Chem. 2009;284:11601. doi: 10.1074/jbc.M808026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roman-Morales E, Pietri R, Ramos-Santana B, Vinogradov SN, Lewis-Ballester A, Lopez-Garriga J. Biochem. Biophys. Res. Commun. 2010;400:489. doi: 10.1016/j.bbrc.2010.08.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kabil O, Vitvitsky V, Xie P, Banerjee R. Antioxid. Redox Signaling. 2011;15:363. doi: 10.1089/ars.2010.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan J, Carroll KS. ACS Chem. Biol. 2013;8:1110. doi: 10.1021/cb4001052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh S, Padovani D, Leslie RA, Chiku T, Banerjee RJ. Biol. Chem. 2009;284:22457. doi: 10.1074/jbc.M109.010868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singh S, Banerjee R. Biochim. Biophys. Acta, Proteins Proteomics. 2011;1814:1518. doi: 10.1016/j.bbapap.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hildebrandt TM, Grieshaber MK. FEBS J. 2008;275:3352. doi: 10.1111/j.1742-4658.2008.06482.x. [DOI] [PubMed] [Google Scholar]
- 31.Stipanuk MH, Beck PW. Biochem. J. 1982;206:267. doi: 10.1042/bj2060267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen X, Jhee KH, Kruger WD. J. Biol. Chem. 2004;279:52082. doi: 10.1074/jbc.C400481200. [DOI] [PubMed] [Google Scholar]
- 33.Vitvitsky V, Dayal S, Stabler S, Zhou Y, Wang H, Lentz SR, Banerjee R. Am. J. Physiol.: Regul., Integr. Comp. Physiol. 2004;287:R39. doi: 10.1152/ajpregu.00036.2004. [DOI] [PubMed] [Google Scholar]
- 34.Zhu W, Lin A, Banerjee R. Biochemistry. 2008;47:6226. doi: 10.1021/bi800351a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taoka S, Ohja S, Shan X, Kruger WD, Banerjee R. J. Biol. Chem. 1998;273:25179. doi: 10.1074/jbc.273.39.25179. [DOI] [PubMed] [Google Scholar]
- 36.Wood JL. Methods Enzymol. 1987;143:25. doi: 10.1016/0076-6879(87)43009-7. [DOI] [PubMed] [Google Scholar]
- 37.Zhang D, Macinkovic I, Devarie-Baez NO, Pan J, Park CM, Carroll KS, Filipovic MR, Xian M. Angew. Chem., Int. Ed. 2014;53:575. doi: 10.1002/anie.201305876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park CM, Macinkovic I, Filipovic MR, Xian M. Methods Enzymol. 2015;555:39. doi: 10.1016/bs.mie.2014.11.033. [DOI] [PubMed] [Google Scholar]
- 39.Artaud I, Galardon E. ChemBioChem. 2014;15:2361. doi: 10.1002/cbic.201402312. [DOI] [PubMed] [Google Scholar]
- 40.Hansen RE, Roth D, Winther JR. Proc. Natl. Acad. Sci. U. S. A. 2009;106:422. doi: 10.1073/pnas.0812149106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen W, Liu C, Peng B, Zhao Y, Pacheco A, Xian M. Chem. Sci. 2013;4:2892. doi: 10.1039/C3SC50754H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bittles AH, Carson NA. J. Med. Genet. 1974;11:121. doi: 10.1136/jmg.11.2.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prudova A, Bauman Z, Braun A, Vitvitsky V, Lu SC, Banerjee R. Proc. Natl. Acad. Sci. U. S. A. 2006;103:6489. doi: 10.1073/pnas.0509531103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vitvitsky V, Witcher M, Banerjee R, Thoene J. Mol. Genet. Metab. 2010;99:384. doi: 10.1016/j.ymgme.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Town M, Jean G, Cherqui S, Attard M, Forestier L, Whitmore SA, Callen DF, Gribouval O, Broyer M, Bates GP, van’t Hoff W, Antignac C. Nat. Genet. 1998;18:319. doi: 10.1038/ng0498-319. [DOI] [PubMed] [Google Scholar]
- 46.Kimura H. Antioxid. Redox Signaling. 2015;22:347. doi: 10.1089/ars.2014.6082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamanishi T, Tuboi S. J. Biochem. 1981;89:1913. doi: 10.1093/oxfordjournals.jbchem.a133393. [DOI] [PubMed] [Google Scholar]
- 48.Jones DP, Go YM, Anderson CL, Ziegler TR, Kinkade JM, Jr, Kirlin WG. FASEB J. 2004;18:1246. doi: 10.1096/fj.03-0971fje. [DOI] [PubMed] [Google Scholar]
- 49.Bannai S. J. Biol. Chem. 1986;261:2256. [PubMed] [Google Scholar]
- 50.Pader I, Sengupta R, Cebula M, Xu J, Lundberg JO, Holmgren A, Johansson K, Arner ES. Proc. Natl. Acad. Sci. U. S. A. 2014;111:6964. doi: 10.1073/pnas.1317320111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prudova A, Martinov MV, Vitvitsky V, Ataullakhanov F, Banerjee R. Biochim. Biophys. Acta, Mol. Basis Dis. 2005;1741:331. doi: 10.1016/j.bbadis.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 52.Vitvitsky V, Kabil O, Banerjee R. Antioxid. Redox Signaling. 2012;17:22. doi: 10.1089/ars.2011.4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vitvitsky V, Martinov M, Ataullakhanov F, Miller RA, Banerjee R. Mech. Ageing Dev. 2013;134:321. doi: 10.1016/j.mad.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mudd SH, Finkelstein JD, Irreverre F, Laster L. Science. 1964;143:1443. doi: 10.1126/science.143.3613.1443. [DOI] [PubMed] [Google Scholar]
- 55.Refsum H, Ueland PM, Nygard O, Vollset SE. Annu. Rev. Med. 1998;49:31. doi: 10.1146/annurev.med.49.1.31. [DOI] [PubMed] [Google Scholar]
- 56.Fu M, Zhang W, Wu L, Yang G, Li H, Wang R. Proc. Natl. Acad. Sci. U. S. A. 2012;109:2943. doi: 10.1073/pnas.1115634109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thomas K, Wong K, Withington J, Bultitude M, Doherty A. Nat. Rev. Urol. 2014;11:270. doi: 10.1038/nrurol.2014.51. [DOI] [PubMed] [Google Scholar]
- 58.Portillo-Ledesma S, Sardi F, Manta B, Tourn MV, Clippe A, Knoops B, Alvarez B, Coitino EL, Ferrer-Sueta G. Biochemistry. 2014;53:6113. doi: 10.1021/bi500389m. [DOI] [PubMed] [Google Scholar]
- 59.Bailey TS, Zakharov LN, Pluth MD. J. Am. Chem. Soc. 2014;136:10573. doi: 10.1021/ja505371z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weerapana E, Wang C, Simon GM, Richter F, Khare S, Dillon MB, Bachovchin DA, Mowen K, Baker D, Cravatt BF. Nature. 2010;468:790. doi: 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roman HB, Hirschberger LL, Krijt J, Valli A, Kozich V, Stipanuk MH. Antioxid. Redox Signaling. 2013;19:1321. doi: 10.1089/ars.2012.5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Finkelstein JD, Martin JJ. J. Biol. Chem. 1986;261:1582. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.