Abstract
Evaluation of new acyl carrier protein hydrolase (AcpH, EC 3.1.4.14) homologs from proteobacteria and cyanobacteria reveals significant variation in substrate selectivity and kinetic parameters for phosphopantetheine hydrolysis from carrier proteins. Evaluation with carrier proteins from both primary and secondary metabolic pathways reveals an overall preference for acyl carrier protein (ACP) substrates from type II fatty acid synthases, as well as variable activity for polyketide synthase ACPs and peptidyl carrier proteins (PCP) from non-ribosomal peptide synthases. We also demonstrate the kinetic parameters of these homologs for AcpP and the 11-mer peptide substrate YbbR. These findings enable the fully reversible labeling of all three classes of natural product synthase carrier proteins as well as full and minimal fusion protein constructs.
Keywords: Protein labeling, peptide labeling, reversible labeling, bio-conjugation, chemoenzymatic synthesis, FRET assay, phosphodiesterase, hydrolase, acyl carrier protein, pantetheine
Introduction
Post-translational protein modification allows engineering of extra utility into biochemical systems for a variety of medically and scientifically useful purposes.1–4 A particularly useful post-translational modification of the carrier proteins (CP) from fatty acid synthase (FAS), polyketide synthase (PKS) and non-ribosomal peptide synthase (NRPS) pathways entails the addition of 4′-phosphopantetheine (PPant) analogs, turning unmodified apo-CP into modified crypto-CP.5–10 The Walsh laboratory identified a minimal peptide (YbbR) that can be modified alone or as a fusion protein.11,12 We have recently shown the ability to selectively remove functional labels from the Escherichia coli type II FAS ACP, AcpP, using the recombinant Pseudomonas aeruginosa acyl carrier protein hydrolase (PaAcpH), which offers the ability to iteratively label and un-label ACPs and ACP fusions with a wide variety of functionalities.13 Here we expand these tools in the evaluation of AcpH homologs from proteobacteria and cyanobacteria that not only display superior kinetics for PPant removal from E. coli AcpP, but also depict the first known specific PPant hydrolase activity against short peptide substrates.
Since the first identification and characterization of E. coli AcpH (EcAcpH),14,15 we have searched for ways to broadly incorporate a specific biocompatible hydrolase activity into the tool set of phosphopantetheine labeling first introduced using the phosphopantetheinyl-transferase (PPTase) Sfp from Bacillus subtilis.5,11,12,16 Despite an otherwise unknown natural role for the AcpH, an enzyme not consistently present in all bacteria, the identification of a more stable AcpH homolog from P. aeruginosa17 and its subsequent characterization using free and fusion-ACPs with phosphopantetheine analogs13 offered the potential of a promiscuous AcpH with which to establish a robust reversible labeling strategy. However, while this PaAcpH has primarily demonstrated promiscuity for a broad range of modified phosphopantetheines appended to the E. coli type II FAS ACP, we were unable to constitute activity on many carrier proteins in our library. These results highlighted the need for a broader AcpH homolog activity analysis.
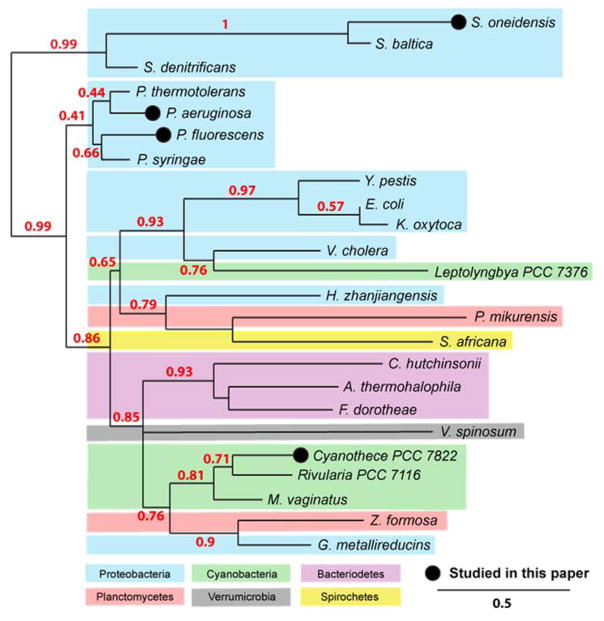
Here we present a thorough substrate evaluation and kinetic analysis of four AcpH homologs identified in proteobacteria and cyanobacteria: P. aeruginosa PAO1 (PaAcpH, NP_253043.1), Cyanothece sp. PCC 7822 (CyAcpH, YP_003888700.1), Shewanella oneidensis MR-1 (SoAcpH, NP_718678.1), and Pseudomoans fluorescens NCIMB 10586 (PfAcpH). We chose these organismic sources to represent a snapshot of currently annotated PaAcpH relatives (Figure 1), comparing proximal phylogenetic relations (P. fluorescens) and more distal relationships (Cyanothece, S. oneidensis) using calculations from protein sequences.18 In addition, we selected AcpH homologs with broad sequence variation to provide additional support of active site residue predictions.19 Finally, to bolster Sfp/AcpH methodology as a site-specific reversible labeling tool, we evaluated function of these homologs with YbbR and S6 peptides discovered for PPant labeling, containing 11 and 12 amino acids respectively.11,12
Figure 1.
Phylogenetic analysis of AcpH sources. AcpH homologs are currently predicted for a diverse set of bacterial phyla, including proteobacteria, cyanobacteria, bacteriodetes, planctomycetes, spirochetes, and verrumicrobia. We analyze the activity for two closely-related AcpH homologs (P. aeruginosa & P. fluorescens) as well as two distantly-related AcpH homologs (Cyanothece PCC 7822 & S. oneidensis). Phylogenetic map calculatings are derived from protein sequences using Phylogeny.fr web utility.18
Results and discussion
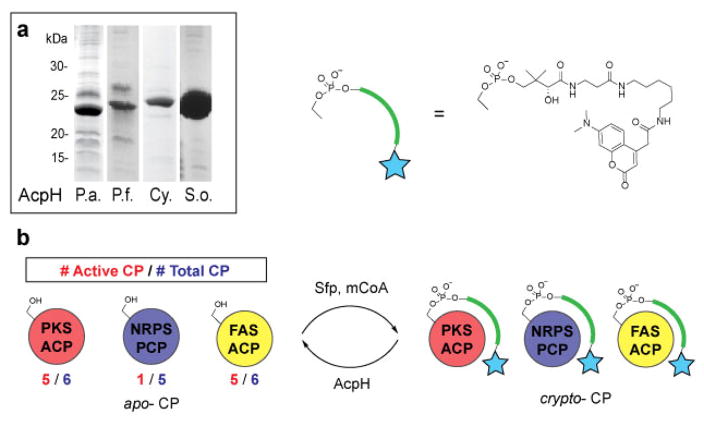
Cloning and expression produced soluble protein for all constructs (Figure 2a & Supplementary Figure 1). The nucleotide sequence for the obtained P. fluorescens NCIMB 10586 AcpH gene is uploaded to NCBI with accession number KF667507, as the source strain’s genome is not sequenced.
Figure 2.
AcpH homolog activity against crypto-CP. a) AcpH homologs were cloned and expressed as soluble proteins. b) Crypto- carrier proteins (CP) were generated by labeling apo-CP with Sfp and modified coenzyme A (mCoA) generated in situ with CoaA, CoaD, CoaE, ATP and coumarin-pantetheine. The most active substrates were from FAS and PKS-type CP.
Our initial goal was to establish the AcpH substrate preference with regards to carrier proteins from FAS, PKS, and NRPS pathways (Supplementary Table 1). We labeled all carrier proteins with a coumarin-pantetheine analog using the established one-pot labeling methodology (Figure 2b)16,20 and quantified AcpH activity as a significant reduction in protein band fluorescence on SDS-PAGE. FAS ACPs were all derived from bacterial protein targets with the exception of the apicoplast ACP from P. falciparum. Evaluated FAS ACPs included E. coli AcpP (EcAcpP, type II), P. aeruginosa AcpP (PaAcpP, type II), S. oneidensis AcpP (SoAcpP, type II), P. falciparum ACP (PfACP, Type II),21 M. tuberculosis AcpM (Type II) and MAS (FAS/PKS hybrid). PaAcpH, PfAcpH, and CyAcpH were active against all type II FAS ACPs (Table 1, Supplementary Figures 2–9). No AcpH activity was observed with the type I M. tuberculosis MAS, which resembles the vertebrate FAS in size and domain organization. SoAcpH showed inactivity with all crypto-ACPs tested, so we implemented holo-SoAcpP as a suitable model of its anticipated natural substrate. This was performed in order to confirm inactivity was not a result of incompatibility with a non-native coumarin-PPant appendage on crypto-ACPs. Interestingly, both crypto-SoAcpP and holo-SoAcpP demonstrated spontaneous, non-enzymatic PPant hydrolysis overnight (Supplementary Figures 6 & 10a), requiring the subsequent holo-SoAcpP analysis be conducted on a shorter timescale (Supplementary Figure 10b).
Table 1.
Qualitative AcpH substrate selectivity for carrier proteins. AcpH homolog activity was determined through overnight reaction with crypto-CP except in the case of holo-SoAcpP. A significant reduction in fluorescence of AcpH-treated CP from controls (Supplementary Figures 2–9, 11–14) or Urea-PAGE gel-shift (Supplementary Figure 10) are labeled as active “+”.
| Carrier Protein | Enzyme | |||||
|---|---|---|---|---|---|---|
| Organism | Name | PaAcpH | PfAcpH | CyAcpH | SoAcpH | |
|
|
||||||
| FAS | E. coli | AcpP | + | + | + | − |
| P. aeruginosa | AcpP | + | + | + | − | |
| S. oneidensis | AcpP | + | + | + | − | |
| P. falciparum | ACP‡ | + | + | + | − | |
| M. tuberculosis | AcpM | + | + | + | − | |
| M. tuberculosis | MAS | − | − | − | − | |
|
|
||||||
| PKS | S. coelicolor | ActACP | − | + | − | − |
| A. parasiticus | PksA | + | + | + | − | |
| G. fujikuroi | Pks4 | − | + | − | − | |
| L. majuscula | JamC | + | + | + | − | |
| L. majuscula | JamF | − | − | − | − | |
| P. agglomerans | AdmA | − | − | − | − | |
|
|
||||||
| NRPS | P. agglomerans | AdmI | − | + | − | − |
| V. cholerae | VibB | − | − | − | − | |
| A. orientalis | CepK | − | − | − | − | |
| P. protogens | PltL | + | − | − | − | |
| P. syringae | SyrB1 | − | − | − | − | |
P. falciparum ACP is derived from the apicoplast FAS-ACP.
PKS-type ACPs were derived from a mixture of bacterial and fungal targets. Evaluated PKS-ACPs included S. coelicolor ActACP,7 A. parasiticus PksA,22 G. fujikuroi Pks4, 23 L. majuscula JamC and JamF, 24,25 and P. agglomerans AdmA.26 Activity in this category was less consistent (Table 1, Supplementary Figures 2–3, 8–9, 11–12), with PfAcpH demonstrating the only activity with crypto- ActACP and Pks4, while all other AcpH except SoAcpH were capable of activity with crypto- PksA and JamC.
NRPS-type peptidyl carrier proteins (PCP) were derived from bacterial protein targets. Evaluated PCPs included P. agglomerans AdmI,26 V. cholerae VibB,27 A. orientalis CepK, P. protogens PltL, P. syringae SyrB1.28 The only detected activities resulted from PfAcpH with AdmI and PaAcpH with PltL (Table 1, Supplementary Figures 4, 8–9, 12–14). It is particularly interesting that an AcpH from P. aeruginosa worked with a PCP from another Pseudomonas species while the P. fluorescens AcpH did not. Without knowing the true overall role of AcpH within each organism, it is difficult to predict how AcpH activity is dictated. However, in analyzing the sequence variation between P. fluorescens from which our PfAcpH was derived, and P. protogens from which PltL was derived, we find that the amino acid sequence identity of PaAcpH to the P. protogens Pf-5 annotated AcpH (not studied) is 70%, while the PfAcpH to P. protogens identity is closer at 82%.
In compiling our results, we noticed that even a modest difference in sequence between PaAcpH and PfAcpH enzymes (62% identity) results in a significant difference in substrate specificity. This indicates that sequence homology alone may not be used to predict substrate compatibility. Despite the promising results demonstrating improved substrate promiscuity of the PfAcpH and CyAcpH homologs compared to the established PaAcpH, the inactivity of SoAcpH raised more questions. We suspected some possibilities for this were that the protein is not purified in its active form (despite high yields and purity), is misannotated as an AcpH while possessing alternate function, or underwent a loss of function mutation in its genetic past that was not detrimental to the survival of S. oneidensis. To further investigate a cause for its inactivity, we evaluated the secondary structure of SoAcpH compared to the known active CyAcpH. Circular dichroism revealed strong alpha helical character (Supplementary Figure 15), indicating a consistent protein fold. Additionally, we aligned the SoAcpH protein sequence to those of PaAcpH, PfAcpH, CyAcpH, and EcAcpH for comparison to the existing EcAcpH analysis based off the phosphodiesterase SPoT.19 This analysis reveals all predicted aspartate active site Mn2+ binding residues in the case of all AcpH except SoAcpH (Supplementary Figure 16), lending additional support to our results demonstrating the S. oneidensis protein is inactive as an AcpH. SoAcpH lacks two out of three AcpH Mn2+-binding aspartates predicted by the Cronan laboratory, whose mutation of a single one of these residues in EcAcpH reduces measurable activity to nearly undetectable levels.19
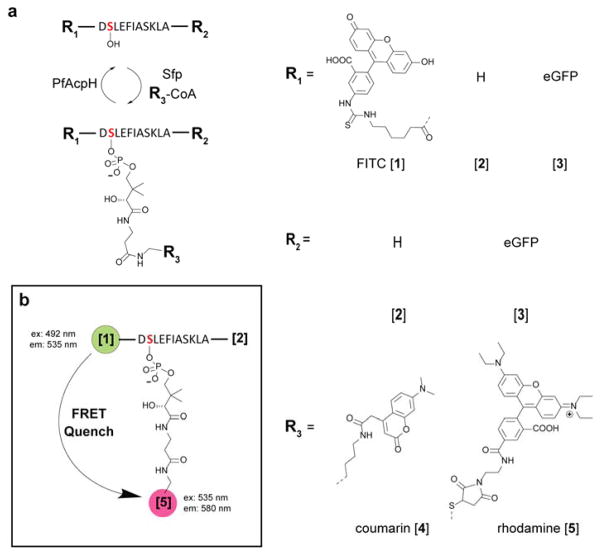
Due to the observations of improved activity of PfAcpH and CyAcpH from the original PaAcpH, our next goal was characterization of activity against phosphopantetheine-labeled YbbR and S6.11,12 We evaluated several variations of YbbR, including free peptide (11AA) (Figure 3a & Supplementary Figure 17a), fluoresceinisothiocyanate YbbR (FITC-YbbR) conjugate (Supplementary Figure 17b),29 and eGFP-YbbR fusions.11 We conjugated coumarin-PPant to YbbR variations and S6 (Supplementary Figure 17c) with one-pot methodology and analyzed with Urea-PAGE using all AcpH homologs, demonstrating qualitative activity for all YbbR constructs with PfAcpH and CyAcpH and no activity for PaAcpH or SoAcpH (Table 2, Supplementary Figures 18–20). Additionally, PfAcpH and PaAcpH demonstrated apparent activity with crypto-S6 peptide while CyAcpH and SoAcpH did not generate results markedly different than negative controls (Supplementary Figure 21). These observations enable a wide variety of bio-conjugation applications using minimal peptides and phosphopantetheine analogs and provides potential advantages for increased substrate variety and experimental flexibility.
Figure 3.
AcpH accommodates YbbR modifications. a) Various YbbR peptide substrate variations were evaluated for AcpH activity qualitatively by SDS-PAGE (Supplementary Figures 8, 18–20), and b) quantitatively using a FRET quench assay (Supplementary Figure 24). PfAcpH was found to accommodate all YbbR appendages, and generate useful kinetic data (Table 3).
Table 2.
AcpH activity with modified YbbR. Various YbbR peptide substrate variations were evaluated for AcpH activity qualitatively (Supplementary Figures 12–14). AcpH from both P. fluorescens and Cyanothece PCC7822 demonstrated detectable activity with qualitative analysis.
| Modification | Enzyme | |||||
|---|---|---|---|---|---|---|
| N-term | C-term | PaAcpH | PfAcpH | CyAcpH | SoAcpH | |
|
| ||||||
| YbbR | N/A | N/A | − | + | + | − |
| FITC | N/A | − | + | + | N/A | |
| GFP | 6xHis | − | + | + | − | |
| 6xHis | GFP | − | + | + | − | |
|
|
||||||
| S6 | N/A | N/A | + | + | − | − |
Given the demonstrated variation in AcpH substrate compatibility for both full carrier proteins, S6 peptide, and variations of the 11-mer YbbR substrate, we sought to further distinguish the new AcpH homologs with kinetic evaluation using the representative E. coli AcpP and YbbR substrates. Kinetic analysis of the AcpH homologs with holo- E. coli AcpP at 37°C resulted in superior kinetic values for CyAcpH compared to PaAcpH and PfAcpH (Table 3, Supplementary Figure 22). Kcat values obtained were 211 min−1 for CyAcpH, 3.7 min−1 for PfAcpH, and 0.6 min−1 for PaAcpH, with kcat/Km values of 3.6 min−1*μM−1 for CyAcpH, 0.06 min−1*μM−1 for PfAcpH, and 0.05 min−1*μM−1 for PaAcpH. Cronan previously determined a kcat of 13.1 min−1 for EcAcpH, and kcat/Km of 2.4 min−1/μM,18 which compares favorably to the more soluble PaAcpH and PfAcpH, but is still significantly lower than CyAcpH. We were at first suspicious of the high apparent turnover for CyAcpH, but these results were further confirmed using EDTA quench to terminate hydrolysis, resulting in enzyme arrest prior to HPLC analysis (Supplementary Figure 23).
Table 3.
AcpH homolog AcpP and YbbR kinetics. AcpH homolog activity was determined at 37°C with holo- E. coli AcpP using HPLC detection (Supplementary Figure 22), as well as crypto- FITC-YbbR in a FRET-quench microwell plate assay (Supplementary Figure 24). Best in-class kinetics for evaluated substrates are CyAcpH for holo-AcpP, and PfAcpH for crypto- FITC-YbbR.
| AcpH | CP | [AcpH], μM | Vmax (min−1) | Km (μM) | kcat (min−1) | kcat/KM (min−1*μM−1) |
|---|---|---|---|---|---|---|
| PaAcpH | holo-ACP (E. coli) | 1 | 0.59 ± 0.02 | 12 ± 2 | 0.59 ± 0.02 | 0.049 |
| PfAcpH | 1 | 3.7 ± 0.1 | 61 ± 7 | 3.7 ± 0.1 | 0.060 | |
| CyAcpH | 0.005 | 1.1 ± 0.1 | 58 ± 9 | 211 ± 11 | 3.6 | |
|
| ||||||
| PaAcpH | crypto-FITC-YbbR (3) | 1 | 0.008 ± 0.016 | 177 ± 744 | 0.008 ± 0.016 | 4.7E-05 |
| PfAcpH | 1 | 0.17 ± 0.01 | 48 ± 5 | 0.17 ± 0.01 | 0.003 | |
| CyAcpH | 1 | 0.004 ± 0.004 | 21 ± 68 | 0.004 ± 0.004 | 2.0E-04 | |
These results indicate the significant finding that turnover of E. coli holo- AcpP by CyAcpH surpasses the kinetic parameters of Sfp for E. coli apo- AcpP (kcat = 5.8 min−1 and kcat/Km = 1 min−1μM−1).30 While the CyAcpH kinetic results are derived from free E. coli AcpP, this result implies a significant advantage for this newly characterized enzyme in designing reversible labeling scenarios with AcpP as a protein handle.
Kinetics of AcpH homologs with the YbbR substrate were determined using a FRET-reporter system with rhodamine WT PPant-labeled FITC-YbbR, as previously utilized to monitor Sfp activity (Figure 3b).29,31 Here FITC-YbbR was conjugated with Rhodamine-CoA31 to generate the crypto- FITC-YbbR substrate (Supplementary Figure 17d). HPLC purification and lyophilization of the crypto- FITC-YbbR supplied the substrate necessary for AcpH kinetic analysis. SoAcpH was not analyzed for kinetics, as it did not display activity in the qualitative YbbR activity analysis. Real-time analysis of AcpH homologs at 37°C in 96-well format provided kinetic data favoring the activity of PfAcpH for crypto- FITC-YbbR. Kcat values obtained were 0.17 min−1 for PfAcpH with kcat/Km of 0.003 min−1μM−1 (Supplementary Figure 24 and Table 3). Compared to the Sfp for YbbR (kcat = 11 min−1, kcat/Km of 0.091 min−1μM−1),11 the PfAcpH demonstrated two orders of magnitude slower reaction rates for label removal.
Following the completion of our substrate panels and the identification of our most promiscuous AcpH from P. fluorescens, we lastly wished to identify any particularly important consensus residues from active protein sequences to guide future substrate prediction. The sequences from 10 amino acids flanking the modified serine were aligned and used to generate a consensus sequence from all carrier proteins active with PfAcpH (Supplementary Figure 25). This procedure generated a core consensus of “DLGXDSLDXVEL” with “X” being a nonconserved amino acid, which displays a strong homology with amino acids residing in type II FAS ACP. This absolute sequence is not required for activity, but our results indicate that most carrier proteins with 5 or fewer matching amino acids are inactive, the exceptions being AdmI and YbbR/S6 peptide substrates. The “DSL” portion appears to be especially important, as only one PKS ACP, JamC, and one NRPS PCP, AdmI, are active with the variation “DSS” and “DSV” respectively. In comparison, none of the inactive NRPS PCPs contain the “DSL” active site sequences or possess more than 6 matching identical amino acids surrounding the active site.
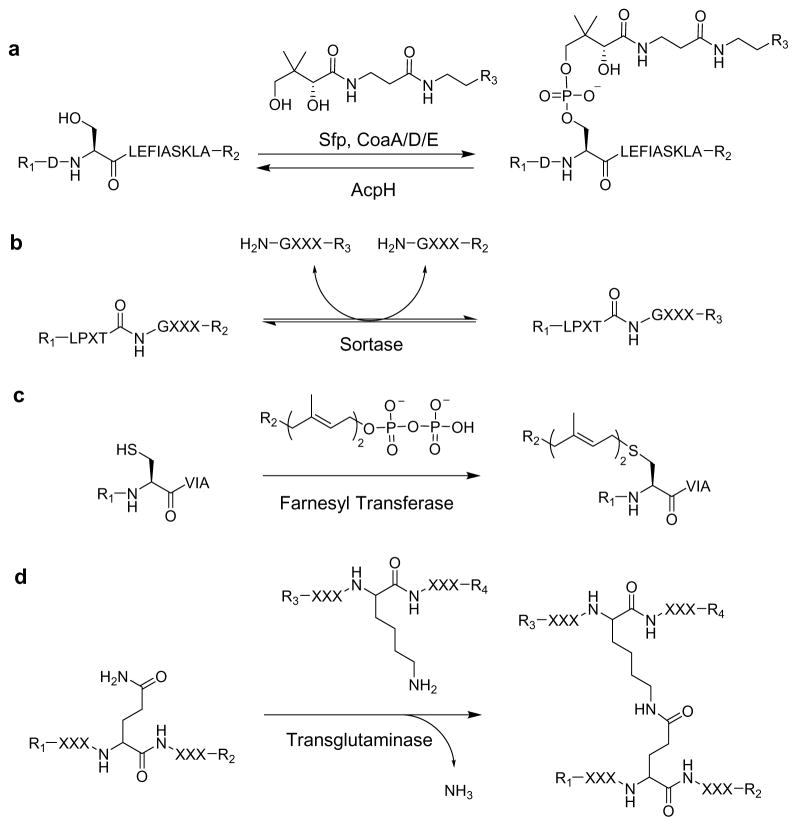
Site-specific PPant labeling may be compared to existing techniques (Scheme 1) allowing peptide labeling, including sortase (5AA),32,33 farnesyl-transferase (4AA),34 and transglutaminase (5AA).35,36 PPant labeling provides various advantages when compared to these other peptide labeling systems, the first of which is label removal now enabled by AcpH. The other reversible labeling system utilizes sortase, which imposes experimental limitations due to its bi-directional conjugation process. Compared to farnesyl-transferase, the Sfp labeling step possesses a similar kcat, but allows flexible YbbR placement at the amino/carboxy-terminus or internal to the target protein.11 In comparison, the farnesyl prosthetic attachment site must be at the protein C-terminus, and removal of the label requires an irreversible carboxypetidase protease cleavage within the target peptide sequence. A negative aspect of farnesyl transferase labeling is that significant modifications of the farnesyl-pyrophosphate substrate reduce kinetic parameters by factors of 10–35 fold.34 Transglutaminase offers perhaps the highest potential kinetics of irreversible peptide conjugation systems with kcat values approximately two orders of magnitude higher than Sfp with YbbR,11,37 but the least site-specificity due to reaction with many possible glutamine-containing sequences.35 Thus, for bioconjugation applications requiring short fusion sequences, easily reversible and site-specific labeling can be implemented with variations of the 11 amino acid YbbR using combined Sfp/PfAcpH methodology.
Scheme 1. Reversible and non-reversible peptide labeling techniques.
Various peptide labeling techniques exist that allow the conjugation of various functional groups (-R) to peptide scaffolds. (a) Reversible phosphopantetheine labeling with Sfp and AcpH allow directional application of probes to various modified YbbR peptide, while (b) sortase reversibly exchanges peptide labels in an equilibrium-driven manner. Other predominant irreversible peptide labeling can be accomplished through (c) addition of farnesyl group with farnesyl transferase, as well as (d) peptide coupling catalyzed by transglutaminase.
The incidental activity of PfAcpH with a peptide discovered originally for Sfp11 implies that there is significant room for AcpH activity improvement, either by modification of the YbbR sequence, or pursuit of a new dual-purpose peptide that possesses desirable kinetics for both Sfp and AcpH activity. Although currently elusive, acquiring a high-resolution crystal structure of an AcpH homolog would open the door to a better understanding of enzyme activity, including substrate interactions and mechanism. These characteristics may only presently be hypothesized or modeled from distantly related phosphodiesterases.19 Identification of residues involved for carrier protein activity through structure studies or mutagenesis could enable rational mutation to alter substrate specificity or improve activity. Alternatively, design and successful implementation of directed evolution for this enzyme class could also provide significant improvement to the AcpH activity profile.
Conclusions
In conclusion, our analysis of three new AcpH gene products and comparison to existing P. aeruginosa AcpH reveals a remarkable array of information regarding substrate compatibility. Despite the apparent inactivity of the annotated hypothetical AcpH from S. oneidensis, the AcpH homologs from P. fluorescens and Cyanothece sp. PCC 7822 present superior alternatives to the existing methods for phosphopantetheine removal, with CyAcpH demonstrating remarkable kinetic values for holo-AcpP hydrolysis, while PfAcpH possesses the best available kinetics for crypto-YbbR hydrolysis, as well as the broadest apparent substrate promiscuity with the evaluated carrier protein panel. These new enzymes demonstrate substantial potential for further substrate truncation and peptide sequence modification, as well as ready implementation with established reversible ACP labeling methods.
Methods
General
Protein concentrations were determined using UV absorbance at 280 nm, with extinction coefficients calculated using ExPASy online tool for each protein.38
Cloning
The P. aeruginosa PAO1 AcpH gene [genID: 881435] identified previously17 was cloned as described previously.13 All primers used for cloning are located in supplemental information (Supplementary Table 2) Cyanothece PCC 7822 AcpH gene [genID: 9739974], and Shewanella oneidensis AcpH gene [genID: 1170805] were cloned from genomic DNA using standard techniques. P. fluorescens NCIMB 10586 AcpH [KF667507] was cloned using homology primers designed from P. fluorescens SBW25 AcpH gene [genID: 7817947]. Cyanothece PCC7822 AcpH PCR product was generated from genomic DNA using forward primer “CyAcpH F1” and reverse primer “CyAcpH R1” with Phusion polymerase. Shewanella oneidensis MR-1 AcpH (SoAcpH) PCR product was generated from genomic DNA using forward primer “SoAcpH F1” and reverse primer “SoAcpH R1” with Phusion polymerase. P. fluorescens NCIMB 10586 AcpH (PfAcpH) PCR product was first generated from genomic DNA using forward primer “PfAcpH F1” and reverse primer “PfAcpH R1” using Phusion polymerase, generating low yields of ~600bp and ~1000bp products. Both ~600bp and ~1000bp products were submitted for sequencing using “PfAcpH F1” and “PfAcpH R1”and the 1000bp product contained a gene coding for a homologous AcpH. Reverse primer “PfAcpH R2” containing the stop codon and “PfAcpH R3” without the stop codon were designed from the derived PCR product sequence and used with forward primer “PfAcpH F1 to generate a new ~600bp band using nested PCR with Pfu polymerase, as Phusion did not generate product with the new primers. All final PCR products and template plasmid pET29b were treated with NdeI and XhoI restriction endonucleases, gel-purified, ligated, transformed into E. coli DH5a, and sequenced for confirmation.
Protein expression and purification
All AcpH expression and purification procedures are previously described.13 E. coli thioesterase TesA39 was expressed and purified in the same manner as AcpH. However, CyAcpH and PfAcpH purification and desalting utilized an adjusted buffer containing 50 mM TrisCl pH 8.0, 250 mM NaCl, and 25% glycerol. The additional glycerol contributed significantly to higher perceived protein recovery and stability. E. coli AcpP, and P. aeruginosa MBP-AcpP were prepared as previously described.13 S. oneidensis AcpP was prepared in the same manner as E. coli AcpP. MtbAcpM, ActACP, JamC, JamF, AdmA, AdmI, SyrB1-AT, PltL, CepK, VibB, and MAS containing cells were grown in LB with appropriate antibiotic at 37°C until they reached an optical density of 0.6. IPTG was added to 1 mM, and the cells were incubated with shaking at 16°C overnight. Plasmodium ACP was grown as previously described.21 All CP constructs were grown in antibiotics appropriate for the contained plasmid. All cells were centrifuged to obtain a pellet, which was re-dissolved in lysis buffer. MtbAcpM expressed as an apo/holo/acyl mixture, and required overnight treatment with Affigel-25 (Bio-Rad, Hercules, CA) conjugates of TesA and PfAcpH prior to labeling. Plasmodium ACP expressed as holo- and was treated with PaAcpH conjugated to Affigel-25 prior to labeling. Spin concentration of all carrier proteins with 3 kDa MWCO centrifugal filters (EMD Millipore, Billerica, Massachusetts) resulted in concentrated protein stocks.
One-pot carrier protein coumarin-labeling strategy
Unless otherwise noted, all coumarin-PPant labeling proceeded as follows: apo- carrier protein or peptide at 50–200 μM were labeled in 50 mM Na-HEPES pH 7.5, 10 mM MgCl2, 8 mM ATP, 0.1 μM MBP-CoaA/D/E, 0.1 μM native Sfp, and 1.1 equivalents of coumarin-pantetheine at 37°C overnight. Crypto- protein samples were re-purified using standard IMAC techniques in pH 8.0 lysis buffer with Ni-NTA resin, and spin-concentrated/buffer exchanged to remove imidazole and concentrate with 0.5 mL 3 kDa MWCO cellulose filters. Crypto- peptide samples were purified via HPLC, lyophilized, and re-dissolved in 50 mM TrisCl pH 8.0 prior to analysis. Peptide products were identified by coupled LC/ESI-MS spectrometry (Supplementary Figures 26–29) and for reaction confirmation by LC using the same method as LC/ESI-MS analysis (Supplementary Figures 30, 31).
General AcpH gel-based activity
Specific procedures for AcpH activity are described previously.13 Briefly, qualitative analysis of crypto- carrier protein or peptide samples proceeded at 37°C overnight reaction with 1 μM AcpH homolog. Following AcpH treatment, an equal volume of 2X SDS-PAGE loading dye was added to crypto-carrier protein samples and heated 5 min at 90°C, and run on 12% or 15% SDS-PAGE. Gels were fixed in 50/40/10% water/methanol/acetic acid for 30 minutes, and washed with water three times before UV imaging. Holo- carrier protein reactions were analyzed with Urea-PAGE as previously described.13 All protein gels were Coomassie stained for evaluating total protein. Crypto- peptides were evaluated on Urea-PAGE, and were imaged immediately after running with no gel fixing.
FITC-YbbR labeling & purification
Preparation of rhodamine-labeled FITC-YbbR proceeded via reaction of 200 μM FITC-YbbR with 200 μM rhodamine WT-CoA synthesized as described previously31 with 1 μM Sfp in 50 mM HEPES pH 7.5 and 10 mM MgCl2 for 2 hours at 37°C. Crypto- peptide samples were purified with HPLC, lyophilized, and re-dissolved in 50 mM TrisCl pH 8.0 at 4 mM prior to analysis.
HPLC AcpH kinetics
Holo- E. coli AcpP was prepared as a serial dilution in 50 mM TrisCl, 250 mM NaCl, 30 mM MgCl2, and 2 mM MnCl2. PaAcpH, and PfAcpH were prepared at 2 μM in similar buffer with 10% glycerol but lacking Mg/Mn. CyAcpH was prepared at 10 nM in the same AcpH buffer. Addition of AcpH into holo-AcpP samples provided final top concentrations of 400, 200, 100, 50, 25, 12.5 μM for PaAcpH/PfAcpH, and 450, 225, 112.5, 56.25, 28.1, 14 μM for CyAcpH. Reactions were transferred to pre-warmed shaker at 37°C and were quenched with 100 mM EDTA pH 7.0 after 10 minutes. Samples were centrifuged and evaluated at 210 nm with HPLC using an acetonitrile gradient to determine apo-AcpP product formation. Michaelis-Menten kinetics were calculated using GraphPad Prism.
FRET AcpH kinetics
Rhodamine-labeled FITC-Ybbr as well as standard 1:1 apo- FITC-YbbR:rhodamine-CoA was subjected to an 11-point serial dilution in 50 mM TrisCl pH 8.0 to achieve final concentrations of 400 – 0.4 μM. PfAcpH, CyAcpH, PaAcpH and buffer blank were prepared to give a final solution concentration of 1 or 0 μM AcpH, 50 mM TrisCl pH 8.0, 15 mM MgCl2, 1 mM MnCl2, and 1 mg/mL BSA. Total reaction volumes were 50 μL and utilized a 96-well Costar 3694 plate (Corning, Lowell, MA). Following mixing, reactions were centrifuged for 2 minutes at 1500 rpm, and incubated at 37°C for 4 hours in a HTS 7000 plus Bioassay Reader (Perkin Elmer, Waltham, MA) in kinetic mode. Comparison of activity from 20–30 minutes to buffer blank and 1:1 apo-FITC-YbbR:rhodamine-CoA standard allowed calculation of product formation and determination of Michaelis-Menten kinetics using GraphPad Prism.
AcpH holo-ACP kinetics sample preparation
E. coli holo-ACP was diluted into 50 mM TrisCl pH 8.0, 250 mM NaCl, 10% glycerol, 30 mM MgCl2 and 2 mM MnCl2 buffer to a concentration of 800 μM. Serial dilution of holo-ACP resulted in a final concentration range of 800-25 μM. AcpH was diluted from lysis buffer into 50 mM TrisCl pH 8.0, 250 mM NaCl, 10% glycerol and added to an equal volume of the holo-ACP serial dilution to initiate the reaction. Reaction tubes were transferred to a pre-warmed rack at 37°C and shaken for the duration of the experiment. Time points were collected at 10 minutes by addition of reaction contents to 100 mM EDTA (pH 8.0). All samples were frozen at −80°C until evaluated by HPLC.
Verification of EDTA quench with PfAcpH and CyAcpH
PfAcpH and CyAcpH were prepared in the same buffer conditions as the HPLC assay format in a 20 μL reaction volume, with the following alterations. Holo- E. coli AcpP was utilized at a final concentration of 150 μM. Each AcpH was prepared in four different manners: with 0 mM Mg2+/Mn2+, 0 mM Mg2+/Mn2+ plus EDTA pre-quench, 15 mM Mg2+/1 mM Mn2+, and 15 mM Mg2+/1 mM Mn2+ plus EDTA pre-quench. EDTA quench involved an equal volume addition of 100 mM EDTA pH 7.5. Samples were incubated 10 minutes at 37°C and then quenched with EDTA if not already pre-quenched. Samples were then incubated overnight at room temperature to simulate the conditions experienced by HPLC samples while awaiting injection. One fourth volume of 5X Native-PAGE loading dye was added to samples prior to running 10 μL of that mixture on 20% Urea-PAGE for analysis.
HPLC detection method
Kinetics samples were mixed briefly with finger flicking, and centrifuged at 13000 rpm for 10 minutes at room temperature prior to transferring contents into HPLC vials. 20 μL of each reaction time point was injected on an Agilent 1100 series HPLC with column (Burdick & Jackson OD5 # 9575, 25cm × 4.6 mm ID) using an acetonitrile/water gradient. Both water and acetonitrile contained 0.05% TFA. Method gradient for each injection: 0–5 min isocratic 10% acetonitrile, 5–30 min gradient 10–100% acetonitrile, 30–35 min isocratic 100% acetonitrile, 35–37 min gradient 100–10% acetonitrile, 37–40 min isocratic 10% acetonitrile. HPLC-grade solvents were used exclusively. Apo- & holo-ACP protein standards were used to validate the retention times identified using 210 nm UV light at approximately 21 and 19 minutes, respectively. Peak integration was performed for all samples, and substrate turnover was calculated from the ratio of apo- to holo- ACP present in the HPLC trace and the known concentration of total ACP in reaction samples. Calculated rates for AcpH versus substrate concentration were obtained through Microsoft Excel data analysis and graphed in Prism GraphPad using the “Michaelis-Menten” function for enzyme kinetics, with a zero data point added for substrate concentration of 0 μM*min−1 and 0 μM substrate for AcpH graphs.
Circular dichroism analysis of select AcpH homologs
CyAcpH and SoAcpH protein preparations in Tris lysis buffer were desalted with PD-10 desalting columns into ice-cold 0.2 μm filtered 50 mM K2HPO4 pH 8.0, and subsequently diluted to 0.2 mg/mL. Samples were kept on ice until transfer to 2 mm width quartz cuvette for analysis at 25°C. Scans were acquired in 0.5 nm increments averaged over 5 seconds from 260 nm to 200 nm. Sample data was subjected to smooth and raw ellipticity was used in conjunction with specific protein residue number and molecular weight to calculate molar ellipticity using established procedures.
Supplementary Material
Acknowledgments
Funding Sources
This research was funded by US National Institutes of Health grants R01GM094924, R01GM095970 and US Air Force Office of Scientific Research grant FA9550-12-1-0414.
We thank W. and L. Gerwick (UCSD) for Cyanothece PCC 7822 genomic DNA and JamC/JamF constructs; M. Rothmann (UCSD) for S. oneidensis AcpP construct; S. Tsai (UC Irvine) for PksA and Pks4 proteins; J. Keasling (UC Berkeley) for AcpM and TesA constructs; T. Foley (UCSD, NIH) for YbbR, FITC-YbbR, and S6 peptides, C. Walsh (Harvard) for eGFP-YbbR fusion, PltL, AdmA/I, and SepB1 constructs; S. Prigge (Johns Hopkins) for PfACP plasmid; L. Tallorin and M. Jaremko (UCSD) for PfACP and CepK/PltL protein; S. Barkho (UCSD) for circular dichroism assistance; Mass Spectrometry Facility (UCSD) for performing LC/ESI-MS analysis.
ABBREVIATIONS
- AcpH
acyl carrier protein hydrolase
- PPant
phosphopantetheine
- PPTase
phosphopantetheinyl transferase
- ACP
acyl carrier protein
- CP
carrier protein
- PKS
polyketide synthesis
- FAS
fatty acid synthesis
- NRPS
non-ribosomal peptide synthesis
- CoA
coenzyme A
- mCoA
modified coenzyme A
- AA
amino acid
- FITC
fluorescein isothiocyanate
- eGFP
enhanced green fluorescent protein
- FRET
Förster resonance energy transfer
- ATP
adenosine triphosphate
- SDS
sodium dodecyl sulfate
- PAGE
polyacrylamide gel electrophoresis
- MBP
maltose binding protein
- HPLC
high-performance liquid chromatography
- UV
ultraviolet
Footnotes
Electronic Supplementary Information (ESI) available: Additional images and tables may be viewed in the supplemental information, including gel images and mass spectrometry data at http://pubs.rsc.org.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
References
- 1.Prescher JA, Bertozzi CR. Nat Chem Biol. 2005;1:13–21. doi: 10.1038/nchembio0605-13. [DOI] [PubMed] [Google Scholar]
- 2.Luchansky SJ, Argade S, Hayes BK, Bertozzi CR. Biochemistry. 2004;43:12358–12366. doi: 10.1021/bi049274f. [DOI] [PubMed] [Google Scholar]
- 3.Batra G, Talha SM, Nemani SK, Dhar N, Swaminathan S, Khanna N. Protein Expres Purif. 2010;74:99–105. doi: 10.1016/j.pep.2010.04.017. [DOI] [PubMed] [Google Scholar]
- 4.Stachler MD, Chen I, Ting AY, Bartlett JS. Mol Ther. 2008;16:1467–1473. doi: 10.1038/mt.2008.129. [DOI] [PubMed] [Google Scholar]
- 5.Quadri LE, Weinreb PH, Lei M, Nakano MM, Zuber P, Walsh CT. Biochemistry. 1998;37:1585–1595. doi: 10.1021/bi9719861. [DOI] [PubMed] [Google Scholar]
- 6.Foley TL, Young BS, Burkart MD. FEBS J. 2009;276:7134–7145. doi: 10.1111/j.1742-4658.2009.07425.x. [DOI] [PubMed] [Google Scholar]
- 7.Haushalter RW, Filipp FV, Ko KS, Yu R, Opella SJ, Burkart MD. ACS Chem Biol. 2011;6:413–418. doi: 10.1021/cb200004k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ishikawa F, Haushalter RW, Burkart MD. J Am Chem Soc. 2012;134:769–772. doi: 10.1021/ja2082334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.La Clair JJ, Foley TL, Schegg TR, Regan CM, Burkart MD. Chem Biol. 2004;11:195–201. doi: 10.1016/j.chembiol.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 10.Meier JL, Haushalter RW, Burkart MD. Bioorg Med Chem Lett. 2010;20:4936–4939. doi: 10.1016/j.bmcl.2010.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yin J, Straight PD, McLoughlin SM, Zhou Z, Lin AJ, Golan DE, Kelleher NL, Kolter R, Walsh CT. P Natl Acad Sci USA. 2005;102:15815–15820. doi: 10.1073/pnas.0507705102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yin J, Lin AJ, Golan DE, Walsh CT. Nat Protoc. 2006;1:280–285. doi: 10.1038/nprot.2006.43. [DOI] [PubMed] [Google Scholar]
- 13.Kosa NM, Haushalter RW, Smith AR, Burkart MD. Nat Meth. 2012;9:981–984. doi: 10.1038/nmeth.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thomas J, Cronan JE. J Biol Chem. 2005;280:34675–34683. doi: 10.1074/jbc.M505736200. [DOI] [PubMed] [Google Scholar]
- 15.Vagelos PR, Larrabee AR. J Biol Chem. 1967;242:1776–1781. [PubMed] [Google Scholar]
- 16.Worthington AS, Burkart MD. Org Biomol Chem. 2006;4:44–46. doi: 10.1039/b512735a. [DOI] [PubMed] [Google Scholar]
- 17.Murugan E, Kong R, Sun H, Rao F, Liang ZX. Protein Expres Purif. 2010;71:132–138. doi: 10.1016/j.pep.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 18.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O. Nucleic Acids Res. 2008;36:W465–469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomas J, Rigden DJ, Cronan JE. Biochemistry. 2007;46:129–136. doi: 10.1021/bi061789e. [DOI] [PubMed] [Google Scholar]
- 20.Clarke KM, Mercer AC, La Clair JJ, Burkart MD. J Am Chem Soc. 2005;127:11234–11235. doi: 10.1021/ja052911k. [DOI] [PubMed] [Google Scholar]
- 21.Prigge ST, He X, Gerena L, Waters NC, Reynolds KA. Biochemistry. 2003;42:1160–1169. doi: 10.1021/bi026847k. [DOI] [PubMed] [Google Scholar]
- 22.Crawford JM, Korman TP, Labonte JW, Vagstad AL, Hill EA, Kamari-Bidkorpeh O, Tsai SC, Townsend CA. Nature. 2009;461:1139–1143. doi: 10.1038/nature08475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Linnemannstons P, Schulte J, del Mar Prado M, Proctor RH, Avalos J, Tudzynski B. Fungal Genet Biol. 2002;37:134–148. doi: 10.1016/s1087-1845(02)00501-7. [DOI] [PubMed] [Google Scholar]
- 24.Dorrestein PC, Blackhall J, Straight PD, Fischbach MA, Garneau-Tsodikova S, Edwards DJ, McLaughlin S, Lin M, Gerwick WH, Kolter R, Walsh CT, Kelleher NL. Biochemistry. 2006;45:1537–1546. doi: 10.1021/bi052333k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edwards DJ, Marquez BL, Nogle LM, McPhail K, Goeger DE, Roberts MA, Gerwick WH. Chem Biol. 2004;11:817–833. doi: 10.1016/j.chembiol.2004.03.030. [DOI] [PubMed] [Google Scholar]
- 26.Fortin PD, Walsh CT, Magarvey NA. Nature. 2007;448:824–827. doi: 10.1038/nature06068. [DOI] [PubMed] [Google Scholar]
- 27.Marshall CG, Burkart MD, Meray RK, Walsh CT. Biochemistry. 2002;41:8429–8437. doi: 10.1021/bi0202575. [DOI] [PubMed] [Google Scholar]
- 28.Matthews ML, Krest CM, Barr EW, Vaillancourt FH, Walsh CT, Green MT, Krebs C, Bollinger JM. Biochemistry. 2009;48:4331–4343. doi: 10.1021/bi900109z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Foley TL, Burkart MD. Anal Biochem. 2009;394:39–47. doi: 10.1016/j.ab.2009.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quadri LE, Weinreb PH, Lei M, Nakano MM, Zuber P, Walsh CT. Biochemistry. 1998;37:1585–1595. doi: 10.1021/bi9719861. [DOI] [PubMed] [Google Scholar]
- 31.Foley TL, Yasgar A, Garcia CJ, Jadhav A, Simeonov A, Burkart MD. Org Biomol Chem. 2010;8:4601–4606. doi: 10.1039/c0ob00322k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williamson DJ, Fascione MA, Webb ME, Turnbull WB. Angew Chem Int Edit. 2012;51:9377–9380. doi: 10.1002/anie.201204538. [DOI] [PubMed] [Google Scholar]
- 33.Hirakawa H, Ishikawa S, Nagamune T. Biotechnol Bioeng. 2012;109:2955–2961. doi: 10.1002/bit.24585. [DOI] [PubMed] [Google Scholar]
- 34.Rashidian M, Song JM, Pricer RE, Distefano MD. J Am Chem Soc. 2012;134:8455–8467. doi: 10.1021/ja211308s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee JH, Song C, Kim DH, Park IH, Lee SG, Lee YS, Kim BG. Biotechnol Bioeng. 2012;110:353–362. doi: 10.1002/bit.24622. [DOI] [PubMed] [Google Scholar]
- 36.Takahara M, Hayashi K, Goto M, Kamiya N. J Biosci Bioeng. 2013 doi: 10.1016/j.jbiosc.2013.05.025. S1389-17231300207-7. [DOI] [PubMed] [Google Scholar]
- 37.Yang MT, Chang CH, Wang JM, Wu TK, Wang YK, Chang CY, Li TT. J Biol Chem. 2011;286:7301–7307. doi: 10.1074/jbc.M110.203315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.ExPASy ProtParam Tool. 2013 http://web.expasy.org/protparam/
- 39.Goh EB, Baidoo EE, Keasling JD, Beller HR. Appl Environ Microb. 2012;78:70–80. doi: 10.1128/AEM.06785-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.